Abstract
Leukocyte integrin-dependent downregulation of the transcription factor FOXP1 is required for monocyte differentiation and macrophage functions, but the precise gene regulatory mechanism is unknown. Here, we identify multi-promoter structure (P1, P2, and P3) of the human FOXP1 gene. Clustering of the β2-leukocyte integrin Mac-1 downregulated transcription from these promoters. We extend our prior observation that IL-1 receptor-associated kinase 1 (IRAK1) is physically associated with Mac-1 and provide evidence that IRAK1 is a potent suppressor of human FOXP1 promoter. IRAK1 reduced phosphorylation of histone deacetylase 4 (HDAC4) via inhibiting phosphorylation of calcium/calmodulin dependent protein kinase II delta (CaMKIIδ), thereby promoting recruitment of HDAC4 to P1 chromatin. A novel human FOXP1 intronic transcript 1 (FOXP1-IT1) long non-coding RNA (lncRNA), whose gene is embedded within that of FOXP1, has been cloned and found to bind directly to HDAC4 and regulate FOXP1 in cis manner. Overexpression of FOXP1-IT1 counteracted Mac-1 clustering-dependent downregulation of FOXP1, reduced IRAK1 downregulation of HDAC4 phosphorylation, and attenuated differentiation of THP-1 monocytic cells. In contrast, Mac-1 clustering inhibited FOXP1-IT1 expression with reduced binding to HDAC4 as well as phosphorylation of CaMKIIδ to activate the IRAK1 signaling pathway. Importantly, both IRAK1 and HDAC4 inhibitors significantly reduced integrin clustering-triggered downregulation of FOXP1 expression in purified human blood monocytes. Identification of this Mac-1/IRAK-1/FOXP1-IT1/HDAC4 signaling network featuring crosstalk between lncRNA and epigenetic factor for the regulation of FOXP1 expression provides new targets for anti-inflammatory therapeutics.
Keywords: Integrin, FOXP1, promoter, IRAK1, HDAC4, lncRNA
1. Introduction
Lineage determination and maturation of hematopoietic cells are orchestrated by transcription factors, chemokines, and cytokines (1-3). Identification of these transcription factors and the mechanism(s) by which they activate their target genes are important for understanding the development of monocytes and macrophages. Based on experimental evidence obtained from knockdown and gain-in-function strategies, several transcription factors, including runt-related transcription factor 1 (RUNX1), PU.1, CCAAT/enhancer-binding proteins (C/EBPs), early growth response-1 (EGR-1), MAF bZIP transcription factor B (MAFB), interferon regulatory factor-8/interferon consensus sequence-binding protein (IRF-8/ICSBP), and Kruppel-like factor-4 (KLF-4), have been implicated in monocyte differentiation (4, 5). The transcriptional regulation of the c-Fms gene, which encodes for the macrophage colony-stimulating factor receptor (M-CSFR), is a focal point of interest because it is required for the differentiation, proliferation, and survival of monocytes (6, 7). However, the precise external signals that control differentiation of peripheral blood monocytes to tissue macrophages are incompletely defined.
We were intrigued by the possibility that cell adhesion molecules participating in the firm arrest and transmigration of blood-borne monocytes across endothelial and extracellular matrix barriers could provide these “outside-in” signals. Monocytes express the β2-integrin Mac-1 (αMβ2, CD11b/CD18), a heterodimeric, transmembrane cell adhesion receptor. Mac-1 binds a broad repertoire of ligands, including counter-receptors, extracellular matrix proteins, plasma proteins, and microbial ligands (8, 9). Ligand engagement by Mac-1 induces receptor clustering that triggers “outside-in” signaling pathways that regulate gene expression, monocyte differentiation and monocyte/macrophage function (10, 11).
Our previous studies have focused on identifying the molecular mechanisms for “outside-in” signaling by Mac-1. Clustering of Mac-1 promotes activation of NFκB through a cascade involving the physical association of Mac-1 with IRAK1 and downstream signaling via TNF receptor associated factor 6 (TRAF6) and TGF-β-activated kinase 1 (TAK1)(12). Follow on studies indicated that Mac-1 engagement and clustering downregulated the expression of the transcription factor FOXP1, which operates as a repressor of c-Fms (13). Importantly, deficiency of Mac-1 was associated with altered regulation of FOXP1 and monocyte maturation in vivo. We directly tested whether FOXP1 plays a critical role in monocyte differentiation and macrophage function in vivo by generating transgenic mice that overexpress human FOXP1 in monocyte/macrophage lineage cells using the CD68 promoter (macFOXP1tg) (14). Macrophage activity, osteoclastogenesis, and bone resorption were found globally impaired in macFOXP1tg. In vivo bacterial challenge showed that macFOXP1tg mice exhibited reduced macrophage accumulation, bacterial clearance, and survival. Taken together, these observations define important physiological roles for FOXP1 in monocyte differentiation and macrophage function.
FOXP1 also has broad functional roles in lung, cardiac, and lymphocyte development, as well as in cancer (15). Thus, investigating the mechanism by which Mac-1 regulates FOXP1 gene expression would be highly informative in advancing our understanding of how transmembrane receptors direct signals to transcriptional networks and downstream target genes. The precise signaling pathways linking Mac-1 clustering and FOXP1 expression are unknown, thereby proving the rationale for detailed characterization of the FOXP1 promoter region. Our current work identifies human FOXP1 as a multi-promoter gene regulated by Mac-1 through a complex signaling network involving IRAK-1, HDAC4, CaMKIIδ and a novel cloned FOXP1-IT1 long non-coding (lncRNA), whose gene is embedded within FOXP1 itself.
2. Materials and methods
2.1. Materials and biological reagents (See Supplementary Materials)
2.2. Antibodies
Hybridoma capable of producing β2 integrin-activating KIM185 antibody was purchased from American Type Culture Collection (ATCC, Manassas, VA, Cat.# CRL-2839). Antibody from the hybridoma cell culture medium was purified by Pierce chromatography cartridges protein A/G (Thermo Scientific). Kim185 was also produced by ProMab Biotechnologies (Richmond, CA). Anti-FOXP1 and αM-blocking antibody LPM19c were generated as previously described (13). Anti-HDAC4 (phospho Ser632)(Cat.# ab39408, lot# GR121980-2), ChIP grade anti-Histone H3 (acetyl K27)(Cat.# ab4729), anti-CaMKII delta (Cat.# ab181052), and anti-CaMKII (phosphor T286)(Cat.# ab32678) were purchased from Abcam Inc (Cambridge, MA). CaMKII (pan) (Cat.# 4436) and Phosphor-CaMKII (Thr286) were from Cell Signaling Technology (Danvers, MA). ChIP applicable anti-HDAC4 antibody, Cat.# 40969, was from Active Motif (Carlsbad, CA). Anti-β-actin (Cat.# A1978), anti-rabbit (Cat.# A0545), and anti-mouse (Cat.# A9044) IgG-peroxidase conjugated antibodies were from Sigma. Anti-HDAC4 mAb (Cat.# sc-46672), Anti-Integrin αM (2LPM19c)(Cat.# sc-20050), Anti-IRAK1 (Cat.# sc-5288), anti-lamin B (Cat.# sc-6216), and anti-goat IgG-peroxidase conjugated (Cat.# sc-2384) antibodies were purchased from Santa Cruz Biotechnology (Dallas, TX). eFluo450 conjugated IgG control and anti-CD11b antibody were from eBioscience/Thermal Fisher Scientific.
2.3. Reporter constructs and subcloning (See Supplementary Materials)
2.4. Mammalian expression plasmid constructs (See Supplementary Materials)
2.5. DNA and RNA purification (See Supplementary Materials)
2.6. Cell culture and transfection
Human THP-1 and HEK293, and mouse RAW 264.7 cell lines were obtained from ATCC and cultured in conditions recommended by ATCC. HEK293 and RAW264.7 cells were transfected by Lipofectamine 2000 (Invitrogen/Thermo Fisher Scientific) and were performed in Falcon 12 well tissue culture plates (Corning Life Sciences, Tewksbury, MA) with triplicate transfection wells for each sample reaction mix. Generally, 5ng or 100ng/well of internal control pCMV-β-gal (Stratagene/Agilent Technologies, Santa Clara, CA) DNA was combined with 10ng or 30ng/well pNL1.1 [Nluc] reporter construct DNA in the absence or presence of 1μg or 4μg/well of respective gene expression plasmid DNA for HEK293 and RAW264.7 cells respectively. pGL3 series reporter construct DNA utilized 0.5-1.0 μg/well in transfection and any specified protocols are noted in Figure Legends. THP-1 cells were transfected by either the DEAE/Dextran (GE Healthcare Life Sciences, Marlborough, MA) method (35) as previously described (12) for plasmid DNA, or by Amaxa Cell Line Nucleofector® Kit V (Lonza, Allendale, NJ) with Nucleofection device for siRNA and plasmid DNA. For siRNA-mediated knockdown of the αM-subunit of Mac-1,THP-1 cells were co-transfected by pNL1.1 [Nluc]-P1tata, pCMV-β-gal and ΙμM either siGENOME Human ITGAM siRNA SMARTpool or siGENOME Non-Targeting siRNA control #5 by nucleofection with AMAXA Kit V. After 3 days, cytokine-treated the above cells as described in Materials and Method 2.9. were subjected to either protein lysates harvest for immunoblot (E) or to Mac-1 clustering/fibrinogen adhesion assay for 3 hours. Both clustered and non-clustered control cells were harvested for nanoluc assays (F).
2.7. Reporter assays (See Supplementary Materials)
2.8. Oligo primers
Primers for RLM-RACE, gene expressions, ChIP, and RIP were designed by Primer 3 software (36) and were chemically synthesized by Invitrogen/Thermo Fisher Scientific. Primer sequences are listed in the Supplementary Table. Specificity of all PCR primers listed in this table was verified by sequencing either the PCR/qPCR or cloning products that the primers pairs/primers amplified. Internal sequencing primers were also verified by sequencing.
2.9. Mac-1 clustering adhesion assays
Mac-1 clustering for cell adhesion to fibrinogen, a ligand of Mac-1, was achieved as previously described (12) with slight modification. Briefly, petri dishes were coated with fibrinogen (100μg/ml in PBS) at 37°C overnight and blocked by 0.1% Polyvinylpyrrolidone (PVP, Sigma) 37°C for 1 hour or just PVP coated for control samples. Cytokine-treated (1ng/mL TGF-β1 and 50 nmol/L l,25-(OH)2 vitamin D3) native or transfected THP-1 cells were washed and suspended in serum-free RPMI1640 medium at a cell concentration of 5×l05/ml, and then were induced for Mac-1 clustering and adhesive to fibrinogen-coated dishes with the β2-stimulating KIM185 (5-10μg/mL) at 37°C for 2, 4, 8 hours or other time as experiments specified. Anti-CD11b blocking mAh LPM19c (5-10μg/mL) treated non-clustered cells were applied to PVP coated dishes. Human non-clustered monocytes were incubated in Teflon plates. Mac-1 clustered adherent cells on fibrinogen dishes and non-clustered floating cells on LPM19c/PVP dishes or Teflon plates were harvested and washed three times with 1×DPBS for subsequent analysis.
2.10. Reverse transcription (RT), polymerase chain reaction (PCR) and quantitative PCR (qPCR) (See Supplementary Materials)
2.11. RLM-RACE and colony enrichment analysis
Cloning of 5’end transcripts of the human FOXP1 gene was carried out using RNA Ligase Mediated Rapid Amplification of cDNA Ends (RLM-RACE) kit (Ambion/Thermo Fisher Scientific) according to the manufacturer’s protocol. 10μg of purified RNA from proliferating THP-1 cells was used as starting material in combination with FOXP1 gene specific primers (Supplementary Table, primers no. j, R3, R4 and R5) designed based on our previously reported human FOXP1/12CC4 (GenBank accession AF250920) sequence. The RLM-RACE kit enriched transcript cDNA was then cloned into the pGEM-T Easy vector (Promega) for sequencing.
2.12. Candidate promoter cloning and construction
Primers were designed based on transcript cluster results from RLM-RACE. BAC colonies RP11-154H23, 148D13 and 69L8 of human genomic DNA from BACPAC Resources/CHORI (Oakland, CA) served as templates of cloning. The 1.45kb P1-1.4, 1.1kb P2 and 2kb P3 were cloned by PCR with primer pair no. 15/n, no. 17/o and no. 19/q (Supplementary Table) respectively by Elongase enzyme mix with 2μg BAC colony RP11-154H23 DNA template. Purified PCR products were then ligated with the pGL3-Basic-T/A vector for subcloning. Orientation of the inserted promoter was verified by sequencing. pGL3-Basic-P3 truncation mutants were generated by employing ExoIII/ Mung Bean Deletion kit (Stratagene/Agilent Technologies) and performed according to the manufacture’s protocol. The 3.9kb full length P1, 1.75kb TATA box-containing P1tata, and the 2.14kb P1cpgi were PCR amplified with primer pair no. 15/b, 15/a and 16/b (Supplementary Table) respectively with LongAmp Taq DNA polymerase and BAC colony 148D13 DNA template, followed by subcloning into the pNL1.1 [Nluc] vector. Authenticity of all constructs was confirmed by sequencing with RV primer 3, primer no.22, 28, aa and z (Supplementary Table).
2.13. Cloning of FOXP1-IT1 long non-coding RNA
Primers were designed based on a NCBI computationally predicted sequence of GenBank Accession XR_109994.1 GI:310119232. mRNA purified from 10 μg total RNA with RiboMinus™ Human/Mouse Transcriptome Isolation Kit (Ambion/Thermo Fisher Scientific) or 2μg total RNA of proliferating THP-1 cells was subjected to DNase I treatment and reverse transcribed in 25μl volume reaction as described above. This was followed by PCR amplification with LongAmp Taq DNA polymerase with primer pair no. 29/ab (Supplementary Table) in 50μl total volume using the following conditions: 94°C 8 min, 35 cycles of 94°C 30 s, 60°C 30 s, 65°C 5 min followed by one cycle of 65°C 10 min. The 4.0kb and 3.5kb PCR products were then cloned into pcDNA3.1/myc-his (−) A vector. Both PCR products and extracted plasmid DNA from positive E.coli colonies were fully sequenced by T7 Universal, BGH rev primers and internal primers no. 30, 31, 32, 33, 34, 35, 36, 37, ad, ae, af, ag, ah, aj, and r (Supplementary Table). Expression of cloned FOXP1-IT1 lncRNA in transfected cells was confirmed by RT-qPCR with splicing-specific primer pair no. 30/ac or 31/ad (Supplementary Table).
2.14. RNA immunoprecipitation (RIP)
Nuclear lysates of 2-4×107 proliferating or Mac-1 non-clustered and clustered THP-1 cells were used in each experiment as protocol described previously (37). Input control RNA was extracted by TRIzol Reagent following the manufacture’s protocol. Percent enrichment was assessed by amplification of enriched FOXP1-IT1 normalized to either total FOXP1-IT1 (Figure 8.A) or β-actin (Figure 8.C) input control.
2.15. ChIP
ChIP was carried out using ChIP-IT High Sensitivity kit (Active Motif) following the manufacture’s recommendation. Specifically, for THP-1 cells lysates, chromatin was sheared by sonication with 2 cycles of 30 sec on and 30 sec off x 6 min in ice-water bath using a Bioruptor ultrasonicator device (Diagenode, Denville, NJ). qPCR results were normalized by input 28S (Supplementary Table, primer pair no. 24/v) and relative enrichment fold was assessed by normalization of enriched target amplicon vs. those of IgG controls. Amplifications by Human Negative Control Primer Set 1 (Active Motif) were served as negative control.
2.16. Protein phosphorylation profiling of Mac-1 clustered cells (See Supplementary Materials)
2.17. Establishment of G418-resistant THP-1 cell lines transfected by FOXP1-IT1
THP-1 cells were transfected by DEAE-Dextran method with pcDNA3.1 vector or pcDNA3.1/FOXP1-IT1 and selected by 150-800 μg/ml G418 for at least one month. Overexpression of FOXP1-IT1 in survived colonies was identified by RT-qPCR with primers no.30/ac (splicing-specific primers), 37/aj and 43/ap for neomycin gene (Supplementary Table). Selected cell lines were maintained in 100μg/ml G418 for culture except during experiments.
2.18. Human blood monocytes
Human whole blood samples were collected and supplied by the Hematopoietic Biorepository and Cellular Therapy Shared Resource of the Case Comprehensive Cancer Center, Case Western Reserve University. Highly purified human monocytes were isolated by RossetteSep Human Monocyte Enrichment Cocktail with Histoque-1077 density gradient medium as manufacturer’s protocol. Similar to Mac-1 clustering of THP-1 cells, purified human blood monocytes were suspended in serum-free RPMI1640 medium, and then were induced for Mac-1 clustering and adhesive to fibrinogen-coated petri dishes with the β2-stimulating KIM185 mAb (10μg/mL), or with anti-CD11b blocking mAb LPM19c (10μg/mL) on Teflon plates. All plated cells were incubated for 4 hours at 37°C in the presence of either vehicle control, IRAK1 inhibitor (10μM), or Trichostatin A (TSA) (500nM). Mac-1 clustered adherent cells on fibrinogen dishes and non-clustered floating cells on Teflon plates were harvested and washed three times with 1xDPBS for subsequent RNA extraction, RT-qPCR analysis.
2.19. Immunoblotting (See Supplementary Materials)
2.20. Flow Cytometry (See Supplementary Materials)
2.21. Sequencing and data analysis (See Supplementary Materials)
2.22. Statistics
Average values and standard deviations of multiple results were calculated (as mean ± SD), and P values of results comparison were performed with two-tailed Student’s T test, as well as adjusted P values from Sidak’s, Tukey’s or Dunnett’s multiple comparisons test. Interaction of factors was analyzed by two-way ANOVA.
3. Results
3.1. Initial characterization of the FOXP1 promoter: Analysis of 5’end transcripts of human FOXP1 mRNA
Having successfully cloned the human FOXP1 cDNA 12CC4 (GenBank Accession No. AF250920) (13), we turned to THP-1 cells for characterization of the FOXP1 promoter. cDNA colonies from full-length capped mRNA of THP-1 were amplified with specific human FOXP1 gene primers (Supplementary Table, primers no. j, R3, R4 and R5) by the RNA Ligase Mediated Rapid Amplification of cDNA Ends (RLM-RACE) method, subcloned, and sequenced for analysis.
Comparing sequences of 36 transcripts against NCBI genomic DNA assembly covering human FOXP1 gene (nucleotide 70954714..71583989, complement, NC_000003.12, Assembly GRCh38.p12) showed that these transcripts can be categorized into three unique and distinct RACE clusters 1, 2 and 3. They start from upstream regions 383 kb, 379 kb, and 345 kb, respectively, from the AUG start codon of the FOXP1 gene ORF (Figure 1.A, upper panel). Importantly, all 36 transcripts share a downstream common exon (CE), the exon 2 in 12CC4/FOXP1 cDNA (nucleotides 134-267, GenBank Acc. AF250920) which we have previously reported (13) (Figure 1.A). This key feature indicates these colonies truly represent the 5’end transcripts/exons species of human FOXP1 gene.
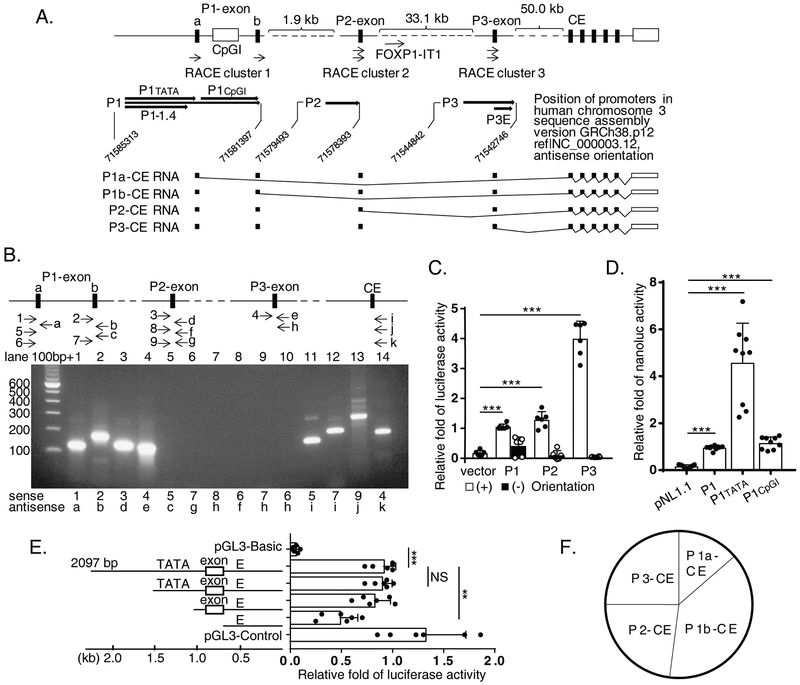
Figure 1. Multiple alternative promoter structure of human FOXP1 gene.
(A) Diagram of the FOXP1 promoter structure (upper panel). Three Rapid Amplification of cDNA Ends (RACE) clusters 1, 2 and 3 as well as P1, P2 and P3, the three candidates of alternative promoter regions in 5’ close proximity to each cluster, were marked. Lower panel represents RNA species transcribed from different promoter. CE is defined as “common exon”. (B) Transcription from human FOXP1 gene promoters. Arrowheads represent position and direction of primers amplifying transcripts (upper panel). PCR products of transcripts designated by upper panel are shown on agarose gel (lower panel). (C) Transcriptional activity of multiple promoters from the human FOXP1 gene in transfected THP-1 cells. (D) Dominant transcriptional activity of P1 in the TATA box area. (E) Transcriptional activity from P3 retained in an enhancer (E). (F) Use of promoters in proliferating THP-1 cells demonstrated based on transcripts copy numbers calibrated by standard curve of quantitative PCR (qPCR) amplified amplicons shown in B, lane 11, 12, 13 and 14. Equal molar concentrations of constructs were used for each experiment in C, D and E. Results of representative experiments performed at least 2 - 4 times in B depending on amplicon; n = 3 separate transfection experiments for each C, D and E. Data represent the mean ± SD. **, P<0.01; ***, P<0.001, two tailed T-test; and adjusted P values from Sidak’s multiple comparisons test for E.
To determine whether these 5’end portions were transcribed from distinct promoters or by alternative splicing of pre-mRNA generated from the same promoter, exon-exon PCR was performed using primers of each 5’end-specific exon and the common exon (CE) in different combinations (Figure 1.B, upper panel). We found that, while primer pairs within certain cloned 5’ exon, i.e. P1-exon a, P1-exon b, P2-exon or P3-exon produced amplified products as expected (Figure 1.B, lower panel; lanes 1, 2, 3, 4; Supplementary Table, sense/antisense primer pairs no. 1/a, 2/b, 3/d, and 4/e), only primers pairing 5’ specific exon(s) with downstream common exons were capable of producing amplified DNA products (lanes 11, 12, 13, and 14; Supplementary Table, primer pairs no. 5/i, 7/i, 9/j and 4/k). Primers pairing between P1-exon a, P1-exon b, P2-exon or P3-exon were unable to generate product (lanes 5, 6, 7, 8, 9 and 10; Supplementary Table, primer pairs no. 5/c, 7/g, 8/h, 6/f, 7/h and 6/h). Taken together, these results indicate that 3 RACE cluster exons could be transcribed independently from three putative promoter regions termed P1, P2 and P3 which are localized proximal to each transcript cluster as shown in Figure 1.A, upper panel. Searching the National Institutes of Health ENCyclopedia Of DNA Elements (ENCODE) project database (16) [http://genome.ucsc.edu/. Feb. 2009 (GRCh37/hg19)] provided evidence that both P1 and P3 regions possess strong Histone 3 lysine 27 acetylation (H3K27ac) activity (see Supplementary Figure 1), a mark of active chromatin, thus supporting the hypothesis that these regions are capable of directing active transcription. We note here the P2 region showed weaker H3K27ac activity compared to the P1 and P3 regions.
3.2. Identification of multiple promoters of the human FOXP1 gene
Based on the above results, genomic PCR primers flanking the P1, P2 and P3 regions were designed. Candidate promoter fragments including a 1,424 bp of P1-1.4, 1,101 bp P2, and 2,097 bp P3 (Figure 1.A), were amplified from bacterial artificial chromosome (BAC) colony RP11-154H23 with primer pairs 15/n, 17/o and 19/q (Supplementary Table), sequencing verified, and subcloned into pGL3-Basic luciferase reporter. THP-1 cells transfected with pGL3-Basic-P1-1.4, -P2, or -P3 reporter constructs in the forward (+), but not reverse (−), orientation expressed robust transcriptional activity (Figure 1.C). As such, these findings identify three active human FOXP1 gene promoter candidates.
The result that PCR failed to amplify a presumably continuous transcript between P1-exon a and b (Figure 1.B, lane 5) prompted further investigation into the versatility of P1. A long P1 region (P1, Figure 1.A) covering 3,916 bp upstream area of P1-exon b was then cloned from BAC colony 148D13 with primer pair 15/b (Supplementary Table). Within this region, sequence analysis by TRANSFAC showed that a 1.75 kb region upstream of P1-exon a (termed as P1tata) contains a −49 putative TATA box (TATTTAT), a typical CCGCGCC TFIIB Recognition Elements (BRE), and an array of 3 BRE-like CGCCGCC from −109, and a −651 Sp1 transcription factor binding site (17). The 2.14 kb region between P1-exon a and b (termed as P1cpgi) contains five Sp1 binding sites embedded in four large CpG islands expanding 368 bp, 707 bp, 218 bp, and 441 bp respectively (analyzed by EMBOSS CpG Plot, Observed/Expected ratio > 0.60, Percent C+G > 50.00, Length > 200. Graph data not shown). P1cpgi does not contain a TATA box, but rather starts from a typical C(+1)TACTCC Initiator element (INR) with two −28 and −52 BRE and a +28 Downstream Core Promoter Element (DPE) AGTCG upstream of P1-exon b (17). Existence of these elements are consistent with independent P1-exon a and b transcription initiation (Figure 1.B, lanes 11 and 12), suggesting that while P1tata directed P1-exon a transcription, either P1cpgi or whole 3.9 kb P1 may be used alternatively for transcription of P1-exon b.
Based on the above analysis, all three 1.75 kb P1tata, 2.14 kb P1cpgi and the longest 3.9 kb P1 versions were subcloned into sensitive nanoluciferase reporter vector pNL1.1[Nluc], called pNL1.1[Nluc]-P1tata, pNL1.1[Nluc]-P1cpgi, and pNL1.1[Nluc]-P1, respectively (Figure 1.A, upper panel). Transfection of these constructs into THP-1 cells demonstrated that all three possessed promoter activities (Figure 1.D). However, P1tata promoter activity was greater than the longest 3.9 kb P1 version, suggesting a potential regulatory effect of P1cpgi.
TRANSFAC analysis also showed that the P2 promoter region contains TATA-like boxes 223-bp upstream of P2-exon. The P3 promoter region directs transcription of P3-exon, the first exon of 12CC4/FOXP1 we identified previously (GenBank Accession No. AF250920). As such, a −423 bp TATA box, TATATATA, upstream of P3-exon may play a role in 12CC4 transcription.
The promoter activity of the 2.0 kb P3 was further investigated by generating a series of truncated mutants. Unexpectedly, strong promoter activity was retained despite deletion of the putative TATA box. A 635-bp 3’ fragment downstream of P3-exon/exon 1 of 12CC4 contributed 53% of the P3 promoter activity (48.44 ± 17.53% vs. 91.41± 11.81%, 635 bp truncate labelled as “E”, vs. 2,097 bp full length; adjusted P value = 0.0024, Sidak’s multiple comparison test; Figure 1.E). Within this region, interestingly, 312 bp (nt 71543381..71543070, NC_000003.12, complement) are highly conserved in genomes of multiple species including chimpanzee, dog, rabbit, and guinea pig (all 99%); monkey, sheep, cow, and pig (all 98%); rat (96%); mouse (95%); at least 14 kinds of birds (93-95%); and zebra fish (75%). Such evolutionary conservation suggests an essential role of this region as a strong enhancer (Figure 1.E) in regulation of FOXP1 gene expression.
The multi-promoter structure of human FOXP1 gene and unique features of each candidate promoter, P1, P2 and P3, provide versatile platforms for gene regulation capable of responding to a variety of cellular conditions and physiological signals. In THP-1 monocytic cells, transcription starting from P1tata (P1a-CE), P1 (P1b-CE), P2 (P2-CE) and P3 (P3-CE) (Figure 1.A, lower panel) accounted for 14%, 38%, 23%, and 25% respectively of overall gene expression (Figure 1.F), indicating that P1 is likely responsible for more than half of FOXP1 transcriptional activity based on a direct method quantifying RNA levels by calculation of real cDNA copy number of each promoter transcribed exon normalized by standard curve of each amplicon. Therefore, our efforts investigating the mechanisms of FOXP1 gene regulation were largely focused on the P1 promoter.
3.3. Mac-1 clustering and PMA-induced macrophage differentiation downregulate FOXP1 promoter transcriptional activity
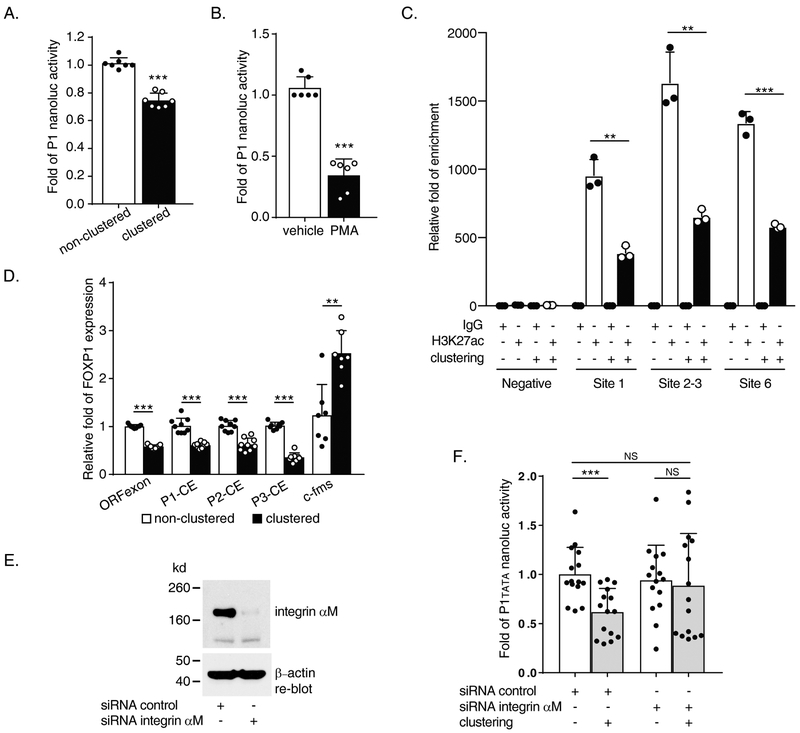
We previously reported that RNA and protein levels of human FOXP1 are downregulated by Mac-1 clustering in THP-1 cells, and human blood monocyte and phorbol myristate acetate (PMA)-induced HL60 cell macrophage differentiation (13). To explore the mechanism of FOXP1 downregulation, we transfected THP-1 cells with pNL1.1[Nluc]-P1 and then examined P1 transcriptional activity after Mac-1 clustering or PMA stimulation. P1 promoter activities were significantly inhibited after Mac-1 clustering (% inhibition = 26.8 ± 3.8%, P<0.001; Figure 2.A) and PMA-induced cellular differentiation (% inhibition = 71.4± 11.6%, P<0.001; Figure 2.B). Moreover, acetylated histone H3 lysine 27 (H3K27ac) ChIP revealed that clustering of Mac-1 reduced P1 transcriptional activity of areas spanning Sp1 binding sites 1, 2-3, and 6 in P1 by 59.0 ± 8.3, 59.5 ± 6.6, 56.6 ± 2.3 % with P = 0.0014, 0.002 and <0.001 respectively (Figure 2.C). Mac-1 clustering also significantly downregulated P2 and P3 promoter activity (data not shown). Transcription was further assessed post-Mac-1 clustering from the respective P1, P2, or P3 exons to the downstream common exon (CE), as depicted in Figure 1.A and 1.B, which was marked by significant increase of c-fms/MCSFR (to 2.53± 0.47 folds, P = 0.001; Figure 2.D). Mac-1 clustering reduced transcription of ORF exon, P1-CE, P2-CE and P3-CE by 42.5 ± 4.3%, 40.5 ± 14.9%, 38.9 ± 16.1%, and 66.2 ± 11.8% respectively (all P<0.001, Figure 2.D). Finally, to confirm that “outside-in” signaling by Mac-1 (αMβ2) is required for downregulation of P1 promoter activity, we first treated THP-1 cells with αM-subunit siRNA and verified appropriate knockdown of αM-subunit protein expression (Figure 2.E). Clustering of Mac-1 significantly downregulated P1tata promoter activity in control siRNA-treated cells (1.0 ± 0.28 vs. 0.62 ± 0.24 folds, non-clustered vs. clustered; P <0.001); in contrast, clustering-induced downregulation of P1TATA promoter activity was abrogated in αM-subunit siRNA-treated cells (0.94 ± 0.36 vs. 0.89 ± 0.53-fold, P = 0.75 non-clustered vs. clustered αM-subunit siRNA; 1.0 ± 0.28 vs. 0.89 ± 0.53-fold, P = 0.47 non-clustered siRNA control vs. clustered αM-subunit siRNA)(Figure 2.F). Taken together, these results indicate that the three candidate promoters are qualified to be responsible for Mac-1 clustering-induced downregulation of FOXP1 gene expression starting from each promoter area.
Figure 2. Mac-1 clustering and PMA induction downregulate both promoter activity and transcription of human FOXP1 gene.
(A) Reporter data from 2 hours Mac-1 clustered pNL1.1 [Nluc]P1 transfected THP-1 cells. (B) Reporter data from 5 nM, 20 hours PMA treated pNL1.1 [Nluc]-P1 transfected THP-1 cells. (C) Acetylated histone H3 lysine 27 (H3K27ac) detection in P1 area via ChIP assay following 2 hours of Mac-1-clustering of THP-1 using primer pairs no. 21/s, 22/t and 23/u (Supplementary Table) and negative control primer set 1 for P1 area sequences. (D) Expression analysis by Reverse Transcription-quantitative PCR (RT-qPCR) following 4 hours Mac-1 clustering of THP-1 cells. Primer pairs used include no. 12/l, 13/j, 10/p, 4/k and 14/m (Supplementary Table) for FOXP1 open reading frame (ORFexon), P1(P1-exon b)-common exon (CE), P2(P2-exon)-CE, P3(P3-exon)-CE and c-fms control, respectively. (E and F) siRNA-mediated knockdown of the αM-subunit of Mac-1 (E) abrogated Mac-1 clustering-induced downregulation of P1tata promoter activity (F). Data shown are results of n = 3 for A; n = 4 for B, separate transfection / clustering or treatment / nanoluc assay experiments; and n = 3 separate clustering / RNA purification / RT-qPCR experiments for D; n = 3 separate co-transfection / Mac-1 clustering / nanoluc assay experiments each of which performed 3-6 separate nucleofection reactions for F. Data represent the mean ± SD. *, P<0.05; **, P<0.01; ***, P<0.001; two tailed T-test. PMA treated vs. non-treated (B); clustered vs. non-clustered (A, C, D & F).
3.4. IRAK1 significantly inhibits the P1tata promoter activity in a pathway that diverges from canonical NFκB signaling
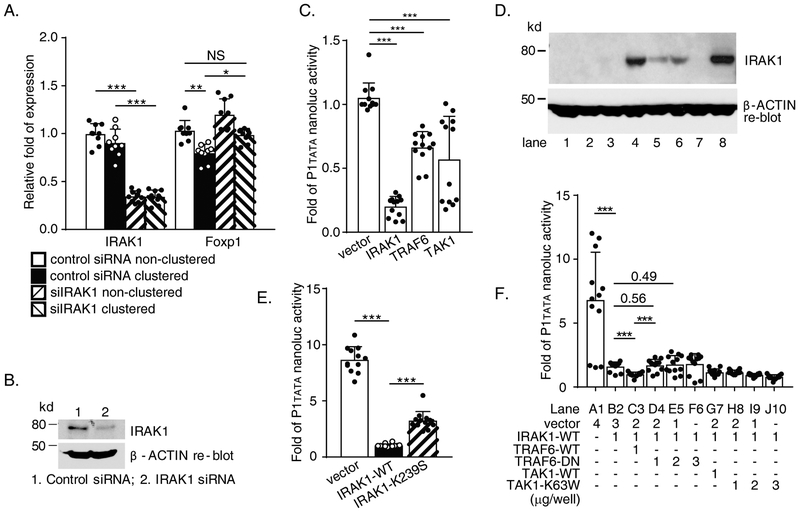
We showed previously that Mac-1 recruits a Toll/IL-1 receptor family-like cascade to modulate NFκB activity (12). IRAK1, a kinase immediately upstream of TRAF6, co-immunoprecipitated with Mac-1. Given that NFκB is capable of both activating and repressing gene transcription (18), we hypothesized that Mac-1-dependent NFκB signaling might be responsible for downregulation of FOXP1 gene expression. To investigate this possibility, we knocked down IRAK1 expression with siRNA in THP-1 cells followed by Mac-1 clustering. As shown in Figure 3.A, IRAK1 siRNA efficiently reduced endogenous IRAK1 mRNA to 34.5 ± 5.8% and 34.0 ± 7.0% in both non-clustered and clustered cells respectively (control siRNA vs. IRAK1 siRNA treatment, P<0.001), and IRAK1 protein level (Figure 3.B). Mac-1 clustering inhibited FOXP1 mRNA significantly to remaining 79.6 ± 9.2% in control siRNA group (non-clustered vs. clustered, adjusted P = 0.0017, Tukey’s multiple comparisons). FOXP1 expression was elevated to 98.4 ± 7.1% in clustered cells of the IRAK1 siRNA treated comparing to non-clustered in control siRNA group. Importantly, this elevated level was significantly different compared to clustered cells in control siRNA group (98.4 ± 7.1% vs. 79.6 ± 9.2 %, adjusted P = 0.012), but did not show statistical significance compared to non-clustered cells of control siRNA treatment (98.4 ± 7.1% vs. 103.0 ± 10.8 %, adjusted P = 0.85). These results suggest an important role for IRAK1 in Mac-1 signaling. Co-transfection experiments with key Toll/IL-1 receptor family signaling intermediates, including wild type IRAK1, TRAF6, and TAK1, and pNL1.1[Nluc]-P1tata were performed in RAW264.7 mouse macrophages. Overexpression of IRAK1 (shown as Figure 3.D) compared to vector alone significantly inhibited P1tata activity (% inhibition = 85.1 ± 10.4, P<0.001, Figure 3.C). Transcriptional inhibition was also observed with overexpression of TRAF6 (% inhibition = 38.9 ± 17.0, P<0.001) and TAK1 (% inhibition = 48.3 ± 35.5, P<0.001; Figure 3.C). To determine whether the kinase activity of IRAK1 is required for inhibition of transcriptional activity, co-transfection with a kinase-inactive mutant of IRAK1, termed IRAK1-K239S (19), was also performed. Strong inhibition of P1tata activity by overexpression of wild type IRAK1 compared to vector alone (% inhibition = 87.9 ± 2.1, P<0.001, vector vs. IRAK1-WT) was blunted by overexpression of IRAK1-K239S (% inhibition = 62.2 ± 9.8; P<0.001, % inhibition comparison, IRAK1-WT vs. IRAK1-K239S, Figure 3.E), implying at least part of the IRAK1 kinase activity is required in this process.
Figure 3. Wild type IL-1 receptor-associated kinase 1 (IRAK1) suppresses human FOXP1 gene promoter activity and expression.
(A) THP-1 cells were transfected by 100nM either IRAK1 or control siRNA by nucleofection with AMAXA Kit V. After 3 days, cells were subject to Mac-1 clustering/fibrinogen adhesion assay for 4 hours. Both clustered and non-clustered control cells were harvested for RNA purification/Reverse Transcription-quantitative PCR (RT-qPCR). (B) Representative immunoblots showing downregulation of endogenous IRAK1 protein level by IRAK1 siRNA in THP-1 cells. (C) Reporter data from RAW264.7 cells co-transfected with various expression genes and pNL1.1[Nluc]-P1tata. (D) Protein expression of transfected IRAK1-WT or IRAK1-K239S in RAW 264.7 cells. Lane 1, 2, 3, vector; Lane 4, 5, 6, IRAK1-WT; lane 8, IRAK1-K239S. (E) Reporter data from RAW264.7 cells co-transfected with either wild type or kinase-deficient (K239S) IRAK1 with pNL1.1 [Nluc]-P1tata. (F) Reporter data from RAW264.7 cells co-transfected by pNL1.1[Nluc]-P1tata, wild type IRAK1 and wild type or dominant negative TNF receptor associated factor 6 (TRAF6) or TGF-β-activated kinase 1 (TAK1) in different dosage. Combinations of constructs co-transfected are listed in the lower panel. Results are n = 4 separate siRNA nucleofection/clustering/RNA purification/RT-qPCR experiments for A. Results are n = 4 separate co-transfections/nanoluc assays experiments for C, E and F respectively; Data represent the mean ± SD. Two tailed T-test; and adjusted P values from Tukey’s multiple comparisons test for A. *, P<0.05; **, P<0.01; ***, P<0.001.
To further explore the nature of NFκB signaling intermediates in FOXP1 gene regulation, additional co-transfection assays using multiple combinations of pNL1.1[Nluc]-P1tata with wild-type and dominant negative IRAK1, TRAF6 and TAK1 constructs were then performed. As demonstrated in Figure 3.F, IRAK1-mediated inhibition of transcriptional activity (IRAK1 alone in lane B2, remaining activity of P1tata = 23.5 ± 5.4 %; P<0.001, B2 vs. A1 vector control) was further augmented by co-transfection with wild-type TRAF6 (IRAK1+TRAF6-WT in lane C3, remaining activity of P1tata = 14.7 ± 2.6 %; P<0.001, B2 vs. C3), but not by dominant negative TRAF6 (TRAF6-DN) (IRAK1+TRAF6-DN in lanes D4 and E5 compared to C3. P<0.001, D4 vs. C3; P = 0.56, lanes D4 vs. B2; P=0.49, lanes E5 vs. B2). These data suggest that IRAK1 and TRAF6 operate in a coordinate manner to inhibit P1tata promoter activity. In contrast, overexpression of a dominant negative TAK1, TAK1-K63W, which we have shown is capable of blocking Mac-1-dependent NFκB activity (12), failed to abrogate or significantly reduce IRAK1-mediated inhibition of P1tata activity (Figure 3.F, lanes H8, I9, J10 compared to lane B2). Of note, TAK1 is a downstream signaling molecule of the IRAK1/TRAF6 in NFκB pathway. Taken together, these findings indicate that inhibition of FOXP1 transcriptional activity requires a novel signaling pathway involving IRAK1 and TRAF6 that diverges from canonical NFκB signaling at TAKI.
3.5. FOXP1-IT1, a lncRNA whose gene is embedded within the FOXP1 gene, counteracts Mac-1 clustering dependent downregulation of FOXP1
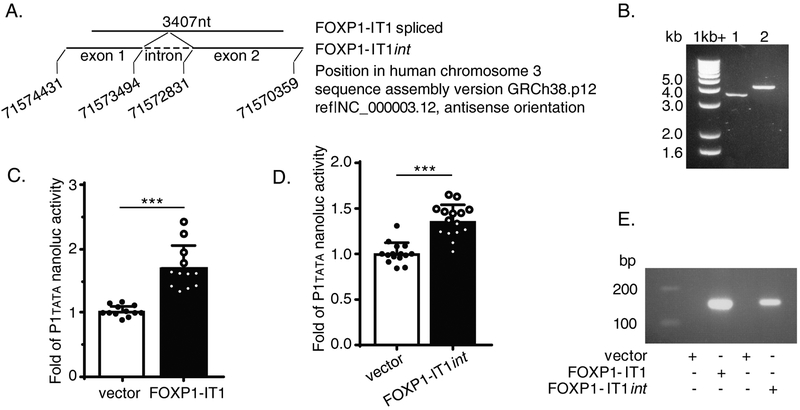
Inspection of the P2 promoter region revealed a computationally predicted long non-coding FOXP1 intronic transcript 1 (FOXP1-IT1, NCBI Accession: XR_109994.1 GI:310119232), whose genomic template is embedded within the FOXP1 gene itself (nucleotides 71574457..71570274 of Homo sapiens chromosome 3, GRCh38.p12 Primary Assembly, NC_000003.12) and is 3,936 bp downstream of P2. Since lncRNAs are capable of regulating transcription (20), we performed a series of experiments to clone this previously unreported lncRNA from human THP-1 cells and investigate whether FOXP1-IT1 regulates FOXP1 expression in Mac-1 signaling. Northern blots, using either random primer/oligo-dT-purified THP-1 mRNA or RiboMinus kit-purified whole THP-1 mRNA hybridized to radioactively labeled FOXP1-IT1 probes, did not detect this lncRNA (data not shown). This result is not unexpected given that FOXP1-IT1 is likely a rare lncRNA species. However, utilizing purified RNA from either DNase I treated random primer/oligo-dT or RiboMinus Transcriptome Isolation methods, we were able to clone both 4.0kb and 3.4kb versions of FOXP1-IT1 using the same pair of PCR primers (Supplementary Table, primers no. 29/ab) designed on the basis of the above NCBI computationally predicted FOXP1-IT1 template (Figure 4.A). Sequencing of the two putative lncRNA PCR products confirmed that the shorter version is a mature 3.4 kb lncRNA in which the 663 bp intron has been spliced (called FOXP1-IT1, lane 1 of Figure 4.B) and the 4.0kb version is one with an intron (called FOXP1-IT1int, lane 2 of Figure 4.B). These results support the co-existence of both spliced and non-spliced versions of FOXP1-IT1 in the same cells. Both versions were subcloned into expression vectors and transfected into RAW264.7 mouse macrophages. Results demonstrated that both versions were capable of activating the transcriptional activity of P1tata 1.70 ± 0.34 and 1.36 ± 0.18-fold, respectively (both P<0.001 compared to vector alone, Figure 4.C and D). We observed that the intron within FOXP1-IT1int can be spliced in transfected RAW264.7, as detected by splicing-specific primer pair crossing two exon boundary of the lncRNA (Supplementary Table, primer pair 30/ac, Figure 4.E).
Figure 4. FOXP1-IT1, a newly cloned novel human long non-coding RNA (lncRNA) enhances human FOXP1 gene promoter activity.
(A) Schematic Diagram of gene position of FOXP1-IT1 lncRNA embedded within gene of FOXP1 in chromosome 3. (B) Cloned FOXP1-IT1 versions run on 1% agarose gel. Lane 1, FOXP1-IT1, intron spliced; lane 2, FOXP1-IT1int, with intron. (C) Reporter data of co-transfection with FOXP1-IT1 and pNL1.1[Nluc]-P1tata constructs in RAW247.6 cells. (D) Reporter data of co-transfection with FOXP1-IT1int and pNL1.1 [Nluc]-P1tata constructs in RAW247.6 cells. (E) Expression of transfected FOXP1-IT1 and FOXP1-IT1int in RAW264.7 cells as qualitative PCR amplified by splicing-specific primer pair no. 30/ac (Supplementary Table) which crosses the two exons boundary of the lncRNA run on 2% agarose gel. Results are n = 4 separate transfections/nanoluc assays experiments for each of C and D; Data represent the mean ± SD. ***, P<0.001; two tailed T-test.
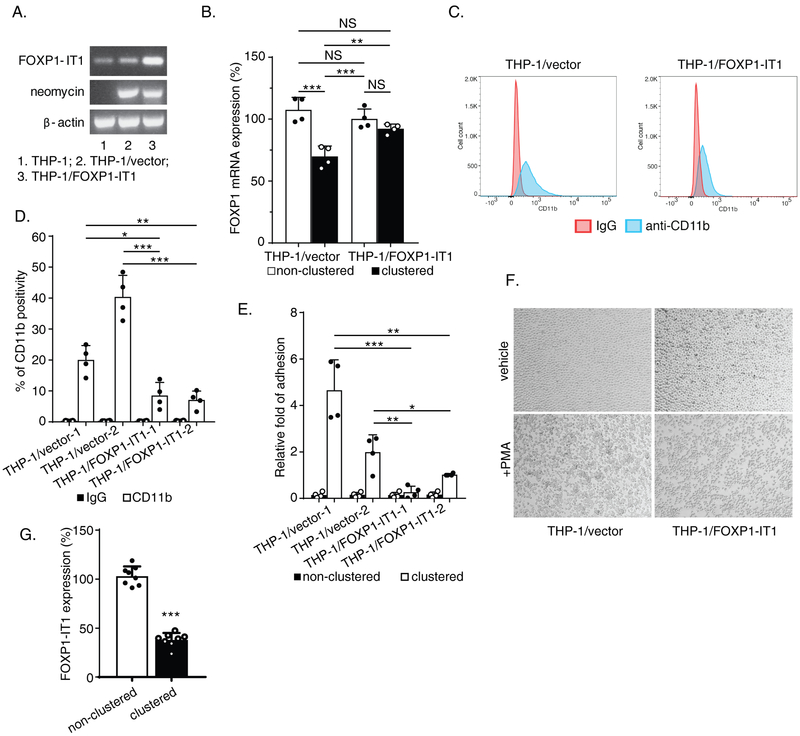
To further investigate potential functions of FOXP1-IT1 in Mac-1-expressing THP-1 cells, we established permanent G418 resistant THP-1 cell lines after transfection with either vector control or FOXP1-IT1 (Figure 5.A). As expected, Mac-1 clustering significantly inhibited FOXP1 mRNA expression in THP-1/vector cells (% inhibition=37.7 ± 10.0 compared to non-clustered control, adjusted P = 0.0001, Tukey’s multiple comparisons) (Figure 5.B). Importantly, however, inhibition of FOXP1 expression was abrogated after clustering of THP-1 cells overexpressing FOXP-1-IT1 (adjusted P = 0.51). Indeed, FOXP1 mRNA expression levels after Mac-1 clustering in these two cell lines were significantly different (clustered THP-1/vector vs. clustered THP-1/FOXP1-IT1, adjusted P = 0.0083). These results are consistent with a significant counteractive effect of FOXP1-IT1 lncRNA against Mac-1 signaling-induced downregulation of FOXP1 (significant interaction, Mac-1 clustering and FOXP1-IT1, P = 0.0027, 2 way ANOVA).
Figure 5. Counteractive effects of FOXP1-IT1 long non-coding RNA (lncRNA) against Mac-1 clustering-induced downregulation of FOXP1 expression, membrane presentation of CD11b/Mac-1 and macrophage differentiation.
(A) Overexpression of transfected FOXP1-IT1 in established G418-resistant permanent cell lines THP-1/ FOXP1-IT1 identified by PCR with splicing-specific primer pair no. 30/ac (Supplementary Table)(for FOXP1-IT1), 43/ap (for neomycin) and 26/x (for β-actin). β-actin calibrated FOXP1-IT1 PCR products at cycle No. 38 were saved for 2% agarose gel electrophoresis. (B) THP-1/vector control and THP-1/FOXP1-IT1 cells were treated by 2 hours Mac-1 clustering. FOXP1 mRNA expression were measured by Reverse Transcription-quantitative PCR (RT-qPCR) with primer pair 12/l. (C) Representative Flow cytometry image performed by staining cells of both THP-1/vector and THP-1/FOXP1-IT1 lines with eFluor-450 conjugated anti-CD11b antibody. (D) Summarized FACS results from 2 THP-1/vector and 2 THP-1/FOXP1-IT1 cell lines. (E) Summarized Mac-1 clustering/cell adhesion efficiency results after 1 hour adhesion to fibrinogen-coated plates from 2 THP-1/vector and 2 THP-1/FOXP1-IT1 cell lines. (F) Representative images of morphological change of early - stage differentiated cells induced by PMA (5nM, 5.5 hours). (G) Endogenous FOXP1-IT1 level downregulated in 4 hours Mac-1-clustered THP-1 cells detected by RT-qPCR with primer pair no.30/ac. Results are n = 4 for B, n = 3 for G separate Mac-1 clustering/RNA purification/RT-qPCR experiments; n = 4 separate FACS experiments for D; n = 4 separate Mac-1 clustering/cell adhesion assays for E; n = 3 separate PMA induction and morphology comparison experiments for F. Data represent the mean ± SD. *, P<0.05; **, P<0.01; ***, P<0.001; two tailed T-test; and adjusted P values from Tukey’s multiple comparisons test, two-way ANOVA for B.
Downregulation of FOXP1 is required in vitro and in vivo for monocyte differentiation and macrophage pro-inflammatory functions—namely, overexpression of FOXP1 impairs monocyte differentiation and attenuates pro-inflammatory functions, including Mac-1 expression, ROS production, and phagocytosis (14). Since FOXP1-IT1 enhanced P1 transcriptional activity, we hypothesized that overexpression of FOXP1-IT1 lncRNA would phenocopy overexpression of FOXP1 itself. Indeed, expression of the αM-subunit of Mac-1 (CD11b), as detected by FACS (Figure 5.C), decreased significantly in two THP-1/FOXP1-IT1 cell lines (8.5 ± 4.2% and 7.1 ± 2.9% CD11b-positive cells) compared to two THP-1/vector cell lines (20.0 ± 4.6% and 40.4 ± 6.9% CD11b-positive cells; quantified in Figure 5.D). In Mac-1 clustering assays with the same cell lines, cell adhesion to the Mac-1 ligand fibrinogen was also reduced in two THP-1/FOXP1-IT1 lines (relative fold of adhesion=0.27 ± 0.25 and 1.02 ± 0.04) compared to two THP-1/vector controls (relative fold of adhesion=4.66 ± 1.3 and 1.99 ± 0.74). Differences of Mac-1-dependent cell adhesion by these two types of cell lines were significant as shown in Figure 5.E. Thus, FOXP1-IT1 is positioned to modulate Mac-1 signaling by both reducing CD11b expression and ligand engagement by Mac-1, which is required for adhesion-induced clustering. Subsequent studies of effect of FOXP1-IT1 on PMA-induced monocyte-macrophage differentiation found that FOXP-IT1 also influenced cell morphology after PMA treatment. In response to PMA stimulation, THP-1/vector cells spread and formed homotypic aggregates; in contrast, THP-1/FOXP1-IT1 cells show reduced spreading and homotypic aggregate formation (Figure 5.F). These results are consistent with FACS and adhesion data above, and imply that overexpression of FOXP1-IT1 lncRNA functions to regulate not only FOXP1 expression level during integrin clustering, but also PMA-induced THP-1 cell monocytic differentiation.
Interestingly, Mac-1 signaling appears to regulate FOXP1-IT1 expression itself. FOXP-1-IT1 lncRNA levels were reduced by 65.4 ± 9.6% (P<0.001, Figure 5.G) 4 hours after Mac-1 clustering, supporting a mechanism in which “outside-in” signaling by Mac-1 downregulates FOXP1, in part, by inhibiting expression of FOXP1-IT1 lncRNA, an activator of FOXP1 expression.
3.6. HDAC4 inhibits FOXP1 expression
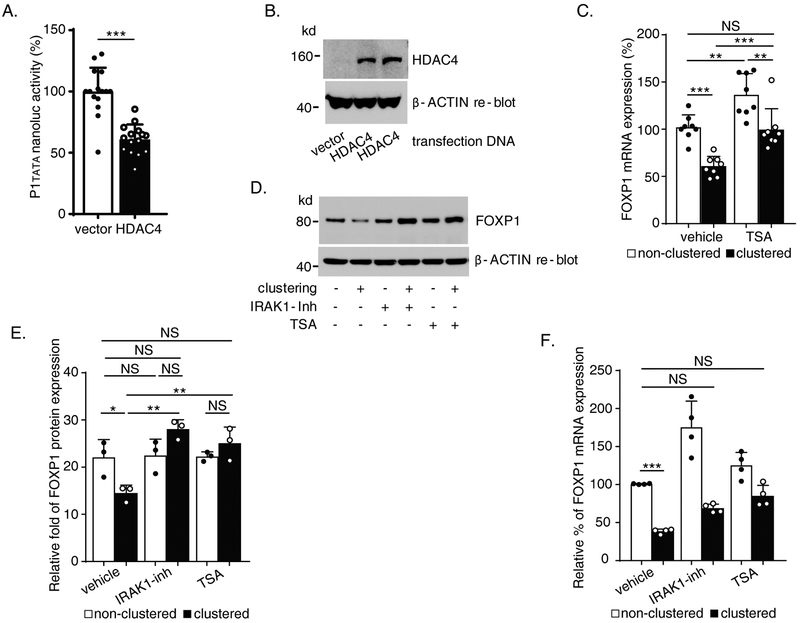
Given that H3K27ac is a known enhancer marker of active gene transcription (21), and that Mac-1 clustering is associated with reduced H3K27ac in the P1 CpGI region (Figure 2.C), we set out to determine the specific histone deacetylase (HDAC) involved in the acetylation/deacetylation of P1 and whether it is associated with IRAK1-dependent inhibition of FOXP1. Indeed, results of our previous studies that employed co-transfection of P3 reporter with six different HDAC (HDAC1-6) expression constructs showed that HDAC4 and HDAC5 inhibited P3 promoter activity (data not shown). We confirmed this observation by co-transfection of P1tata with HDAC4 construct in RAW264.7 cells and showed that HDAC4 inhibited P1tata nanoluc activity significantly (% inhibition = 39.2 ± 20.1%, P<0.001; Figure 6.A and B). To verify a role for HDACs in Mac-1-dependent downregulation of FOXP1, THP-1 cells were pre-treated with the Class II HDAC inhibitor Trichostatin A (TSA) (500 nM, 48 hours) followed by 4 hours Mac-1 clustering in the presence of the inhibitor. As shown in Figure 6.C, TSA treatment attenuated Mac-1 clustering-dependent inhibition of FOXP1 mRNA expression (% inhibition 40.34 ± 12.96% in vehicle-treated clustered cells vs. 26.95 ± 16.48% in TSA-treated clustered cells; vehicle-treated clustered vs. TSA-treated clustered, adjusted P < 0.001, Tukey’s multiple comparisons). Interestingly, TSA pretreatment also increased FOXP1 mRNA expression significantly in non-clustered cells (relative expression, vehicle treated 102.0 ± 12.4% vs. 136.5 ± 21.0% in TSA-treated cells, adjusted P = 0.003). Importantly, no significant difference was observed in FOXP1 mRNA level in TSA-treated clustered cells (99.7 ± 20.7%) compared to vehicle-treated non-clustered cells (102.0 ± 12.4%) (Figure 6.C, adjusted P = 0.994).
Figure 6. Histone deacetylase 4 (HDAC4) inhibits human FOXP1 gene promoter activity and expression.
(A) Reporter data from RAW264.7 cells co-transfected by HDAC4 and pNL1.1 [Nluc]-P1tata. (B) Protein expression of transfected HDAC4 in RAW264.7 cells shown by immunoblot. (C) FOXP1 mRNA expression detected by Reverse Transcription/quantitative PCR (RT-qPCR) with primer pair no. 12/l (Supplementary Table) in proliferating THP-1 treated with Trichostatin A followed by 4 hours Mac-1 clustering. (D) Representative immunoblot demonstrating reduction of Mac-1 clustering-induced downregulation of FOXP1 protein level by IL-1 receptor-associated kinase 1 (IRAK1) inhibitor and Trichostatin A. (E) Summarized results of separate immunoblot experiments represented by D as graph showing quantification of FOXP1 protein bands on blots images analyzed by ImageJ normalized to β-ACTIN controls. (F) FOXP1 mRNA expression detected by RT-qPCR (primer pair no. 12/l, Supplementary Table) in purified human blood monocytes 4 hours after Mac-1 clustering in the presence of either IRAK1 inhibitor or Trichostatin A. Results are n = 5 separate transfections/nanoluc assays experiments for A; n = 3 separate Trichostatin A treatment/Mac-1 clustering/RNA purification/RT -qPCR experiments for C; n = 3 separate inhibitors treatment/Mac-1 clustering/protein immunoblots experiments for E; n = 4 separate inhibitor treatment/Mac-1 clustering/RNA purification/RT -qPCR experiments for F. Data represent the mean ± SD. *, P<0.05; **, P < 0.01; ***, P<0.001. Two tailed T-test; and adjusted P values from Tukey’s or Dunnett’s multiple comparisons test for C, E and F, and two-way ANOVA for E and F (separate analysis of IRAK1 inhibitor vs. vehicle or TSA vs. vehicle).
Since HDAC4 interacts directly with chromatin to suppress gene expression, we hypothesized that the upstream events of Mac-1 signaling mediated by IRAK1 may ultimately target HDAC4, which modifies FOXP1 gene chromatin. To test this hypothesis, THP-1 cells were treated with IRAK1 inhibitor (10μM) or TSA (500nM) followed by Mac-1 clustering and immunoblotting. Represented by Figure 6.D, clustering of Mac-1 downregulated FOXP1 protein level and this downregulation was completely abrogated by treatment with either IRAK1 inhibitor or TSA. Quantified results of separate experiments confirmed the consistency of significant effects of both the inhibitors during Mac-1 clustering in FOXP1 protein translation level (Figure 6.E) (significant interaction, IRAK inhibitor vs. clustering or TSA vs. clustering, P = 0.004 and 0.011 respectively, 2-way ANOVA), which also implies potential linkage of IRAK1 and HDAC4 functions.
The role of IRAK1 and HDAC4 in regulating FOXP1 expression was also explored in authentic human blood monocytes. Human monocytes purified from whole blood were subjected to Mac-1 clustering in the presence of either IRAK1 inhibitor (10μM) or TSA (500nM) (Figure 6.F). Mac-1 clustering in vehicle-treated monocytes significantly downregulated FOXP1 mRNA expression from 100.8 ± 0.6% in non-clustered monocytes to 38.5 ± 3.1% in Mac-1 clustered monocytes (P<0.001, vehicle-treated non-clustered control vs. the vehicle-treated Mac-1 clustered). IRAK1 inhibitor treatment increased FOXP1 mRNA expression in both non-clustered (175.4 ± 34.4%) and Mac-1 clustered (68.9 ± 5.6%; 2-way ANOVA for IRAK1 inhibitor effect P=0.03) cells. Importantly, IRAK1 inhibitor partial rescue of FOXP1 expression in Mac-1 clustered cells from 38.5 ± 3.1% to 68.9 ± 5.6% showed no statistically significant difference in expression compared to vehicle-treated non-clustered control (100.8 ± 0.61%, adjusted P = 0.06 by Dunnett’s multiple comparisons). Treatment with the HDAC inhibitor TSA also increased FOXP1 mRNA expression in both non-clustered (125.4 ± 17.0%) and Mac-1 clustered (85.1 ± 14.0%) monocytes. Inhibition in vehicle-treated clustered monocytes (% inhibition = 61.8 ± 6.1) was partially abrogated by TSA treatment (% inhibition = 32.1 ± 13.5%; adjusted P < 0.001 by Tukey’s multiple comparisons, vehicle-treated clustered vs. the TSA-treated clustered). TSA rescue of FOXP1 expression in Mac-1 clustered monocytes from 38.5± 3.1% to 85.1 ± 14.0% showed no statistically significant difference in expression compared to vehicle-treated non-clustered control monocytes (100.8 ± 0.61%; adjusted P = 0.16 by Dunnett’s multiple comparisons). Taken together, these results indicate that the IRAK1/HDAC4 regulation of FOXP1 is evident not only in monocyte/macrophage cell lines, but also in authentic human blood monocytes.
3.7. IRAK1 inhibits HDAC4 phosphorylation to facilitate HDAC4 recruitment to chromatin by downregulating CaMKII phosphorylation
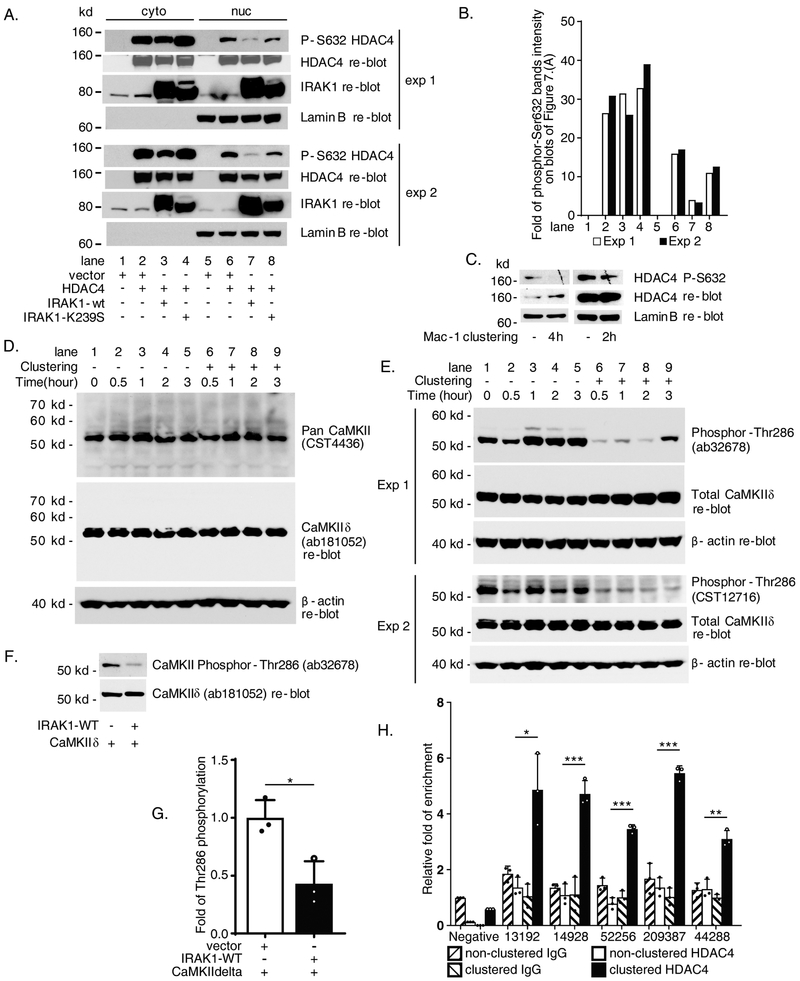
Nuclear/cytoplasmic shuttling of HDAC4 has been well investigated in cardiomyocytes and other types of cells (22). Dephosphorylation of HDAC4 promotes nuclear retention and target gene suppression; in contrast, phosphorylation of HDAC4 is associated with nuclear export to the cytoplasm as well as loss of chromatin-remodeling effects (23). To examine the potential effect of IRAK1 on HDAC4 phosphorylation status, HEK293 cells were co-transfected with HDAC4 and either wild-type IRAK1 or the kinase-deficient IRAK1-K239S mutant. Interestingly, wild-type, but not kinase-deficient mutant IRAK1, significantly reduced nuclear HDAC4-Ser632 phosphorylation (74.9% and 80.4 % reductions in experiment 1 and experiment 2, respectively, lane 7 in Figure 7.A blot images and quantification in Figure 7.B). These results are suggestive of a new mechanism by which IRAK1-WT promotes nuclear localization of HDAC4. Importantly, clustering of Mac-1 also reduced phosphorylation of HDAC4 based on immunoblotting nuclear extracts from Mac-1-clustered THP-1 cells with the same phosphorylation-specific (Ser632) antibody 2 and 4 hours post-Mac-1 clustering (Figure 7.C).
Figure 7. IL-1 receptor-associated kinase 1 (IRAK1) pathway via Calcium/calmodulin-dependent protein kinase II delta (CaMKIIδ) to downregulate histone deacetylase 4 (HDAC4) phosphorylation which facilitates HDAC4 recruitment to FOXP1 promoter during Mac-1 clustering.
(A) Immunoblot data of cytoplasmic and nuclear protein extracts from co-transfected HEK293 cells showing IRAK1-WT inhibits HDAC4 Ser632 phosphorylation. Two experiments were shown as exp 1 and exp 2. (B) Graph showing quantification of phosphor S632 HDAC4 bands from the blots shown in A. by ImageJ normalized to whole HDAC4. (C) Downregulation of HDAC4 Ser632 phosphorylation in nuclear extracts from THP-1 cells after 2 or 4 hours Mac-1 clustering. (D) Protein expression of CaMKIIδ in Mac-1 clustered THP-1 cells. (E) Mac-1 clustering inhibits Thr286 phosphorylation of CaMKIIδ. (F) IRAK1-WT inhibits Thr286 phosphorylation of CaMKIIδ in co-transfected HEK293 cells. (G) Summarized results of separate immunoblot experiments represented by F as graph showing quantification of phosphor-Thr286 CaMKIIδ bands on blots images analyzed by ImageJ normalized to total CaMKIIδ controls. (H) Enhanced recruitment of HDAC4 to P1 chromatin 2 hours after Mac-1 clustering detected by ChIP. Enriched amplicons were detected by quantitative PCR with 44/ak, 45/al, 46/am, 47/an and 48/aq (Supplementary Table). Immunoblot results of 2 hours and 4 hours after Mac-1 clustering in C represent 2 and 4 separate Mac-1 clustering/nuclear protein extraction/immunoblot experiments respectively. Results are n = 3 separate co-transfection/immunoblots experiments for G. Data represent the mean ± SD. *, P<0.05; **, P < 0.01; ***, P<0.001. Two tailed T-test.
We next sought to unpack the mechanism by which IRAK1-WT represses HDAC4 phosphorylation. IRAK1 is a kinase, so IRAK1 itself may only act as an indirect regulator in this process. Instead, several kinase families are responsible for phosphorylation of HDAC4 [as reviewed in (24)]. To identify the intermediate kinase which may be involved in HDAC4 phosphorylation during Mac-1 clustering, we employed Phospho Explorer Antibody Array (Full Moon BioSystems), which consists of 1318 antibodies for broad-scope protein phosphorylation profiling screening, using protein extracts from Mac-1-clustered compared to non-clustered control cells. The results displayed that phosphorylation and activation status of multiple kinases including known and potential candidates were affected by Mac-1 clustering (data not shown). Expression constructs of 18 candidate genes selected from the kinase array were co-transfected with HDAC4 into HEK293 cells followed by immunoblotting with HDAC4-Ser632 phosphorylation antibody. We were not able to identify new candidate kinase(s) capable of phosphorylating HDAC4. We also analyzed a few known kinases which showed marginally low reduction of phosphorylation/inactivation in Phospho Explorer Antibody Array hybridization (data not shown). Among them, calcium/calmodulin dependent protein kinase II (CaMKII) appeared to be an interesting candidate.
CaMKII is a multimeric enzyme formed by its four subunits α, β, δ and γ. While expression of α and β are generally restricted in brain, δ and γ are expressed in most of other tissues (25). CaMKIIδ and CaMKIIγ have been well characterized to phosphorylate HDAC4 Ser632 site in cardiomyocytes (22). To probe which CaMKII(s) function in THP-1 cells, we carried out Mac-1 clustering time course experiments and protein lysates at each time point were immunoblotted by Pan-CaMKII antibody. As shown in Figure 7.D, all samples from 0, 0.5, 1, 2 and 3 hours possessed a dominant ~52 kd protein band, corresponding to δ or α subunits. Potential β and γ subunits with higher molecular weights were not evident. Re-blotting of same filter blot by a knockout validated CaMKIIδ-specific antibody (ab181052, Abcam) exhibited the same pattern of the ~52 kd band detected by Pan-CaMKII (Figure 7.D, middle image panel). CaMKIIα expression was undetectable with CaMKIIα-specific antibodies. Thus, we concluded that CaMKIIδ is the dominant subunit expressed in THP-1 cells.
Phosphorylation of Threonine 286 of CaMKII activates the kinase (for CaMKIIδ the corresponding site is Thr287). We speculated that if CaMKII is involved in downregulation of HDAC4 phosphorylation in THP-1 cells, reduced phosphorylation of this site may be expected during Mac-1 clustering. Indeed, similar Mac-1 clustering time course experiments with Phosphor-Thr286 antibodies from different sources (ab32678, Abcam, Exp 1; and CST12716, Cell Signaling Tech, Exp 2; Figure 7.E) demonstrated that within only 30 min of inducing clustering, Thr286 phosphorylation of CaMKII was markedly reduced. Importantly, this alteration in CaMKII phosphorylation status preceded changes in HDAC4 phosphorylation (Figure 7. C) and FOXP1 expression (Figure 2.D), strongly suggesting a functional role for CaMKIIδ in signaling triggered by Mac-1 clustering.
We further hypothesized that IRAK1 may be able to inhibit HDAC4 phosphorylation by downregulating phosphorylation of CaMKIIδ. To test this hypothesis, we co-transfected constructs expressing IRAK1-WT and CaMKIIδ in HEK293 cells. As shown in Figure 7.F, IRAK1-WT significantly reduced phosphorylation of CaMKIIδ as detected by phosphor-Thr286 antibody. Results of multiple transfections/immunoblots quantified in Figure 7.G graph exhibited average 57.1 ± 15.4 % reduction of phosphor-Thr286 level by IRAK1-WT (P<0.05). The mechanism by which IRAK1 inhibits CaMKIIδ phosphorylation is unknown and the focus of ongoing studies.
Next, we sought to determine whether Mac-1 signaling influenced HDAC4 recruitment to FOXP1 gene chromatin by performing ChIP experiments. As shown in Figure 7.H, Mac-1 clustering significantly increased the binding of HDAC4 to five amplicons of P1 promoter (4.9 ± 1.3, 4.7 ± 0.5, 3.5 ± 0.1, 5.5 ± 0.3, 3.1 ± 0.3-fold, respectively, in Mac-1 clustered compared to 1.4 ± 0.4, 1.1 ± 0.4, 0.8 ± 0.2, 1.4 ± 0.3, 1.3 ± 0.3-fold, respectively, in non-clustered cells (negative group IgG control as 1.0; P<0.05, <0.001, <0.001, <0.001, <0.01, clustered vs. non-clustered, respectively).
Taken together, these observations support a new “outside-in” Mac-1 signaling cascade involving IRAK1, CaMKIIδ, and HDAC4. IRAK1 reduces phosphorylation/activation of CaMKIIδ, which subsequently in turn results in reduced phosphorylation and increased chromatin recruitment of HDAC4 to FOXP1 gene promoter, thereby positioning HDAC4 appropriately for suppression of FOXP1 expression.
3.8. FOXP1-IT1 lncRNA binds directly to HDAC4 and mediates IRAK1-HDAC4 pathway
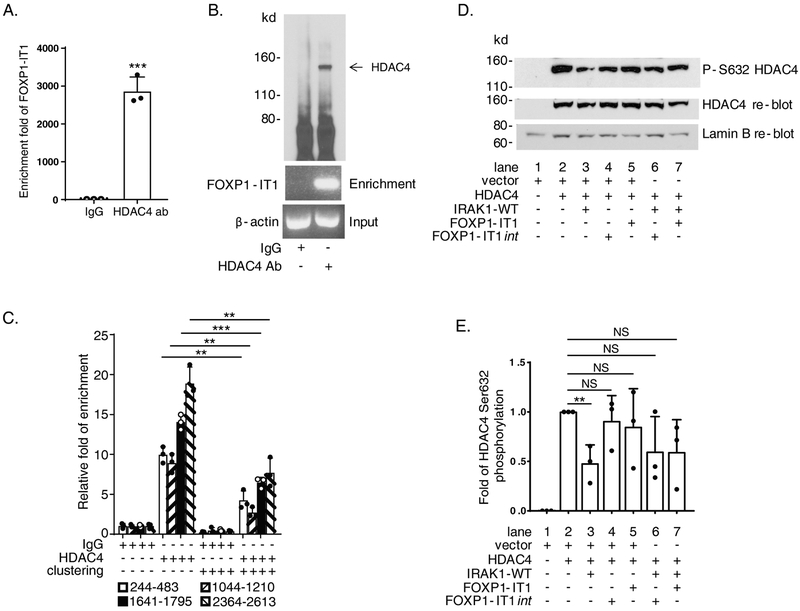
We have shown that overexpression of FOXP1-IT1 lncRNA significantly reduced Mac-1 clustering dependent FOXP1 downregulation, CD11b expression and Mac-1 ligand binding (Figure 5.B, C, D, and E). Next, we examined whether the mechanism of action of FOXP1-IT1 intersected with the Mac-1/IRAK1/HDAC4 pathway. RNA immunoprecipitation (RIP), performed with nuclear extracts of proliferating THP-1 cells and detected by splicing-specific primer pair which crosses two exons boundary of the lncRNA (see Supplementary Table, no.31/ad), demonstrated enriched FOXP1-IT1 lncRNA by HDAC4 antibody (2,866 ± 376 fold enrichment, P<0.001 IgG control vs. anti-HDAC4 antibody; Figure 8.A) and immunoprecipitated HDAC4 protein (Figure 8.B), indicating FOXP1-IT1 lncRNA is capable of binding to HDAC4. We then carried out RIP with nuclear extracts of Mac-1-clustered THP-1 cells. Enriched nuclear RNA was DNase I treated, reverse transcribed, and then detected by qPCR using four pairs of FOXP1-IT1 primers. Mac-1 clustering was accompanied by significant reduction of FOXP1-IT1 lncRNA enrichment by HDAC4 antibody compared to non-clustered cells in all 4 amplicons. As Figure 8.C shows, enrichment of four areas spanning nt 244..483, 1044..1210, 1641..1795, and 2364..2613 respectively of the 3408 nt FOXP1-IT1 cloned was reduced markedly after clustering of Mac-1 (% reduction = 57.7 ± 8.7%, 68.6 ± 11.6%, 54.0 ± 2.8% and 58.3 ± 13.6%; P<0.01, P<0.01, P<0.001, and P<0.01, respectively, clustered vs. non-clustered). These observations are consistent with a working model that FOXP1-IT1 binds directly to HDAC4 and may “trap” more HDAC4 molecules away from P1 chromatin in Mac-1 non-clustered cells; in contrast, in Mac-1clustered cells, reduced FOXP1-ITI/HDAC4 binding may “free” HDAC4 to be recruited to P1 chromatin microenvironment.
Figure 8. FOXP1-IT1 long non-coding RNA (lncRNA) binds directly to histone deacetylase 4 (HDAC4) and mediates IL-1 receptor-associated kinase 1 (IRAK1) downregulation of HDAC4 phosphorylation.
(A) & (B) FOXP1-IT1 lncRNA binding to HDAC4 detected by RNA immunoprecipitation (RIP) with splicing-specific primer pair no.31/ad (Supplementary Table) in proliferating THP-1 cells. (C) Reduced FOXP1-IT1 + HDAC4 binding in 4 hours Mac-1 clustered THP-1 cells detected by RIP with quantitative PCR (qPCR) primer pairs no. w/y, 18/r, 37/aj and 36/ai (Supplementary Table). (D) Representative immunoblot of nuclear protein extracts from co-transfected HEK293 cells showing FOXP1-IT1 mediates IRAK1-WT inhibition of HDAC4 Ser632 phosphorylation. (E) Summarized results of 3 separate co-transfection/nuclear protein extraction/immunoblot experiments represented by D as graph showing quantification of phosphor-Ser632 of HDAC4 bands on blots images analyzed by ImageJ normalized to total HDAC4 protein controls. Data represent the mean ± SD. **, P < 0.01; ***, P<0.001. Two tailed T-test.
The findings that FOXP1-IT1 lncRNA physically interacted with HDAC4 (Figure 8.A, B and C) and IRAK1 modulated HDAC4 phosphorylation (Figure 7.A and B) led us to hypothesize that HDAC4 might be the intermediate target of IRAK1 and FOXP1-IT1 lncRNA. To investigate this possibility, IRAK1, HDAC4 and FOXP1-IT1 were co-transfected into HEK293 cells and HDAC4 Ser632 phosphorylation in nuclear protein extracts was analyzed by immunoblotting. Results represented by Figure 8.D and quantified in Figure 8.E demonstrate that, while co-transfection of IRAK1-WT and HDAC4 inhibited HDAC4 Ser632 phosphorylation (fold inhibition = 0.52 ± 0.19; lane 2 HDAC4 vs. lane 3 HDAC4 + IRAK1-WT, P<0.01), co-transfection of either FOXP1-IT1 or FOXP1-IT1int and HDAC4 had no inhibitory effect on HDAC4 phosphorylation (0.91± 0.26 fold, lane 2 HDAC4 vs. lane 4 HDAC4 + FOXP1-IT1int, P =0.56; and 0.85 ± 0.39 fold, lane 2 HDAC4 vs. lane 5 HDAC4 + FOXP1-IT1, P =0.53). Importantly, co-transfection of either FOXP1-IT1 or FOXP1-IT1int with HDAC4 and IRAK1-WT (lane 6 and 7) significantly rescued the reduced HDAC4-Ser632 phosphorylation mediated by IRAK1 (from 0.48 ± 0.19 fold, lane 3 back to 0.60 ± 0.36 fold, lane 6; lane 6 HDAC4 + IRAK1-WT + FOXP1-IT1int vs. lane 2 HDAC4, P = 0.12; and similarly, back to 0.60 ± 0.33 fold, lane 7; lane 7 HDAC4 + IRAK1-WT + FOXP1-IT1 vs. lane 2 HDAC4, P = 0.10). These results imply similar function of FOXP1-IT1 and FOXP1-IT1int to positively regulate FOXP1 expression, as supported by Figure 4.C-D.
3.9. Proposed model for regulation of FOXP1 gene expression
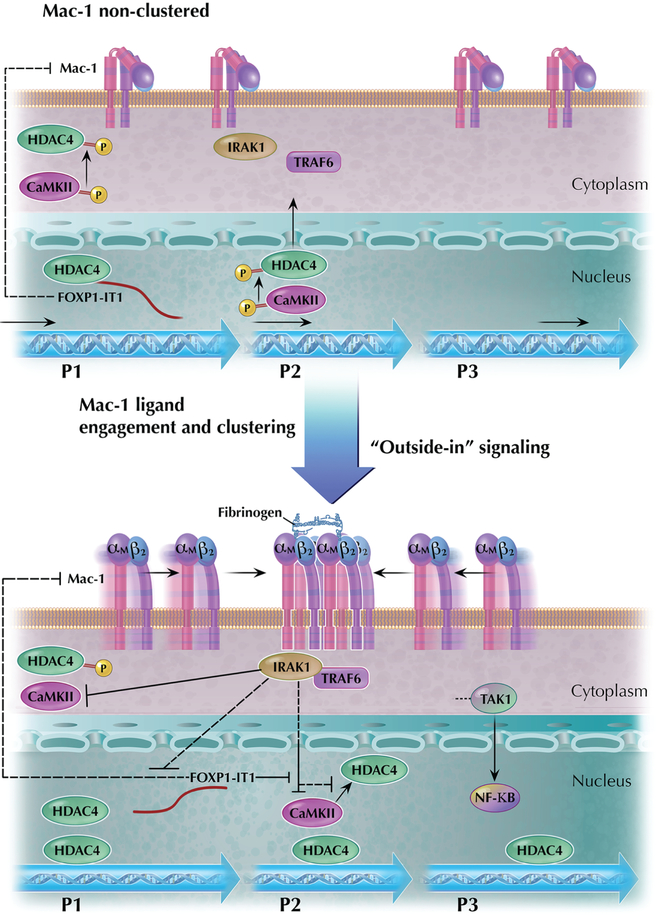
Based upon the findings outlined above, we propose a model for the regulation of FOXP1 gene expression by Mac-1 integrin signaling (Figure 9). FOXP1 is a multi-promoter gene with both positive (FOXP1-IT1 lncRNA, CaMKIIδ) and negative (IRAK1 and HDAC4) regulators of gene expression. Ligand engagement and integrin clustering initiates “outside-in” signaling by Mac-1 that is accompanied by physical interaction between Mac-1 and IRAK1 (12). This complex formation triggers a novel IRAK1 pathway identified here, which is distinct from canonical NFκB signaling, bifurcating at the level of IRAK1/TRAF6 and independent of TAK1-related downstream p65/p50 intermediates. Instead, IRAK1 appears to reduce HDAC4 phosphorylation in an indirect manner mediated via inhibition of phosphorylation/activation of CaMKIIδ, which binds directly to HDAC4 (22). This process promotes nuclear retention of hypo-phosphorylated HDAC4 and recruitment of HDAC4 to FOXP1 promoter chromatin that ultimately results in suppression of FOXP1 gene expression. The novel IRAK1-CaMKII-HDAC4 pathway is influenced by the newly cloned FOXP1-IT1 lncRNA. As lncRNA whose gene is embedded within FOXP1 gene locus itself, FOXP1-IT1 works in cis to activate FOXP1 gene promoter, “traps” HDAC4 by physical binding, and counteracts IRAK1-mediated HDAC4 de-phosphorylation. FOXP1-IT1 also downregulates Mac-1 expression in an unknown mechanism, thereby providing a feedback loop to dampen Mac-1 signaling and maintain FOXP1 expression at a higher level. Reciprocally, Mac-1 clustering-dependent signaling represses FOXP1-IT1 expression, thereby favoring IRAK1-directed signaling events for downregulation of FOXP1 expression.
Figure 9. Schematic mechanism of Mac-1 regulation of human FOXP1 gene promoters and expression.
Diagram of the proposed Mac-1 signaling pathway that regulates the human FOXP1 gene expression. FOXP1 is a multi-promoter gene with both positive (FOXP1-IT1 lncRNA, CaMKIIδ) and negative (IRAK1 and HDAC4) regulators of gene expression. Ligand engagement and integrin clustering initiates “outside-in” signaling by Mac-1 that is accompanied by physical interaction between Mac-1 and IRAK1. The figure highlights localization of regulatory factors of FOXP1 gene in different cellular compartments, which change when cell transits from the status of Mac-1 non-clustering (upper panel) to that of Mac-1 ligand engagement and clustering (lower panel).
4. Discussion
In this report, we have identified human FOXP1 is a multi-promoter (P1, P2, and P3) gene regulated by β2-leukocyte integrin Mac-1 through a complex signaling network involving IRAK-1, HDAC4, CaMKIIδ and a novel cloned FOXP1-IT1 long non-coding RNA (lncRNA), whose gene is embedded within FOXP1 itself. This conclusion is supported by the following data: 1) Clustering of the Mac-1 downregulates transcription from these promoters; 2) IRAK1, which is physically associated with Mac-1, is a potent suppressor of human FOXP1 promoter; 3) FOXP1-IT1 lncRNA has been cloned and found to bind to HDAC4 and regulate FOXP1 in cis manner; 4) Overexpression of FOXP1-IT1 counteracted Mac-1 clustering-dependent downregulation of FOXP1, reduced IRAK1 downregulation of HDAC4 phosphorylation, and attenuated differentiation of THP-1 monocytic cells; and 5) Mac-1 clustering inhibited FOXP1-IT1 expression with reduced binding to HDAC4 as well as phosphorylation of CaMKIIδ to activate the IRAK1 signaling pathway. Since FOXP1 has a wide range of biological functions, the investigation of the regulation of FOXP1 expression at the promoter level is likely to have broad scientific implications. Furthermore, the alternative promoters identified in this study also possess rich and diverse transcription factor binding sites (data not shown), thereby providing additional lines of investigation for future studies directed to understanding the functional versatility of FOXP1 gene.
Clustering of Mac-1 has been shown to trigger “outside-in” signaling that regulates gene expression, monocyte differentiation, and monocyte/macrophage function (8-11). In a series of studies, our laboratory reported that ligand engagement and clustering of Mac-1 promotes activation of NFκB through a cascade involving the physical association of Mac-1 with IRAK1 and downstream signaling via TNF receptor associated factor 6 (TRAF6) and TGF-β-activated kinase 1 (TAK1)(12). Mac-1 clustering also downregulated the expression of the transcription factor FOXP1, which operates as a repressor of c-Fms (13). The “outside-in” signaling triggered by Mac-1 clustering involves intracellular signalosome. Xue et al. reported that Mac-1 clustering by immobilized intercellular adhesion molecule-1 (ICAM-1) or μ2-antibody triggered phosphorylation of protein kinase C delta (PKCδ), which was required for downregulation of FOXP1 (26). We recently reported that Mac-1 binding to platelet glycoprotein Ibα (GPIbα) also triggers phosphorylation of PKCδ and downregulates FOXP1 (27). The present work, inspired by our prior observation of physical association of Mac-1 and IRAK1 (12), identified a novel IRAK1-CaMKIIδ-HDAC4 pathway for FOXP1 gene regulation that is independent of downstream events of canonical NFκB signaling, thereby greatly enhancing our understanding of “outside-in” signaling by Mac-1.
The precise mechanism by which IRAK1 downregulates CaMKIIδ phosphorylation is unknown. The kinase activity of IRAK1 is only partially responsible for the ability of IRAK1 to conduct this signaling cascade (Figure 3.E; Figure 7.A). Indeed, there is precedence for such action by IRAK1. Hartupee et al. reported that signals from the IL-1 receptor can separate into two pathways at the level of IRAK1, one of which is TRAF6-independent pathway that is responsible for mRNA stabilization (28). Inoue and coworkers reported that T cells downregulate macrophage TNF production by IRAK1-mediated IL-10 expression (29) involving IRAK1 nuclear translocation without altering the NFκB pathway. The potential function of IRAK1 in the nucleus is particularly interesting. If IRAK1 exerts its effect in nucleus in this study, it may help clarify its regulation of CaMKIIδ. CaMKIIδ possesses two major subtypes, the CaMKIIδB and CaMKIIδC , the former contains an additional 11 amino acids , a nuclear localization sequence, while CaMKIIδC stays in cytoplasm (25). Further investigation of how IRAK1 mediates nuclear CaMKIIδB might be beneficial to understanding shuttling and recruitment of HDAC4 to FOXP1 gene chromatin. As proposed in Figure 9, IRAK1 may also organize a series of kinases or other enzymes to affect chromatin modifying enzymes. We do not rule out the possibility that other HDAC4-phosphorylating enzymes may be involved. Future studies are focusing on characterizing such specific enzyme(s) whose activity is regulated by IRAK1.
The cis regulation of FOXP1 by the FOXP1-IT1 lncRNA, whose gene is embedded within the FOXP1 locus, is a novel finding that provides an unusual example of transcriptional control. lncRNAs have been linked to several human diseases (30). MIAT and ANRIL are two lncRNAs associated with myocardial infarction, coronary artery disease, and diabetes (31, 32). Several lncRNAs including lincRNA-Cox2, THRIL, PACER, Morrbid, and lnc13, have been reported to play modulating roles in innate immunity [as well reviewed by HY Chang in (33)]. lncRNAs act by regulating recruitment of transcriptional activators/repressors and chromatin remodeling (e.g., methylation and acetylation) (20, 34). Our finding that FOXP1-IT1 binds to HDAC4 (Figure 8.A & C) and counteract IRAK1-mediated de-phosphorylation of HDAC4 (Figure 8.D & E) provides evidence for a potential regulatory interaction between FOXP1-IT1 and chromatin modifying enzymes. The ability of FOXP1-IT1 to bind directly to HDAC4 provides an additional mechanism by which FOXP1-IT1 is capable of regulating FOXP1 expression—namely, FOXP1-IT1 sequesters HDAC4 away from P1 chromatin, thereby de-repressing P1 transcriptional activity.
In summary, this study provides evidence for regulation of FOXP1 gene expression by an unreported novel lncRNA-mediated IRAK1 pathway through a newly-identified multi-promoter structure of the gene. The augmentation of FOXP1 expression by small molecule inhibitors of IRAK1 and HDAC4, in addition to increased expression of FOXP1 protein by bortezomib (13), provides a novel anti-inflammatory therapeutic strategy for monocyte/macrophage-related inflammatory diseases.
Supplementary Material
Highlights.
Identification of functional multi-promoter structure of the human FOXP1 gene.
“Outside-in” integrin signaling downregulates promoter activity of FOXP1 via IRAK1.
IRAK1-CaMKIIδ-HDAC4 cascade enhances HDAC4 recruitment to FOXP1 promoter chromatin.
FOXP1-IT1 lncRNA associates with HDAC4 to mediate integrin-triggered IRAK1 pathway.
Integrin-IRAK1-HDAC4 regulation of FOXP1 exists in authentic human blood monocytes.
Acknowledgements
This work was supported by National Institutes of Health grants R37 HL57506 and R01 HL126645 to D.I.S. and National Cancer Institute Grant-U54CA116867 sponsored Transdisciplinary Research on Energetics and Cancer Developmental Project Funding RES503563 and RES504393 to C.S. Cloned alternative transcripts and candidate promoter regions are issued GenBank Accession numbers KC551795, KC551794 and KU304332, for PR25 (PI-exon a), PR17 (P1-exon b) and PR4 (P2-exon); and TPA BK008733, TPA BK009919 and TPA BK005686 for P1, P2 and P3 respectively. We are grateful to Tularik Inc. for the kind supply of wild-type IRAK1 and kinase-inactive mutant of IRAK (K239S) constructs. This research was supported by the Hematopoietic Biorepository and Cellular Therapy Shared Resource of the Case Comprehensive Cancer Center (P30CA043703) for human blood sample collection. We thank Dr. Yuan Lu and Dr. Ganapati H. Mahabaleshwar for their critical review of the manuscript. We are grateful to Dr. Xudong Liao and Dr. Rongli Zhang for help during figure preparation.
Footnotes
Conflict of interest statement: The authors have declared no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Tenen DG, Hromas R, Licht JD, and Zhang DE. Transcription factors, normal myeloid development, and leukemia. Blood. 1997;90(2):489–519. [PubMed] [Google Scholar]
- 2.Friedman AD. Transcriptional regulation of granulocyte and monocyte development. Oncogene. 2002;21(21):3377–90. [DOI] [PubMed] [Google Scholar]
- 3.Hettinger J, Richards DM, Hansson J, Barra MM, Joschko AC, Krijgsveld J, and Feuerer M. Origin of monocytes and macrophages in a committed progenitor. Nature immunology. 2013;14(8):821–30. [DOI] [PubMed] [Google Scholar]
- 4.Rosenbauer F, and Tenen DG. Transcription factors in myeloid development: balancing differentiation with transformation. Nature reviews Immunology. 2007;7(2):105–17. [DOI] [PubMed] [Google Scholar]
- 5.Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, and Ley K. Development of monocytes, macrophages, and dendritic cells. Science (New York, NY). 2010;327(5966):656–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wiktor-Jedrzejczak W, Bartocci A, Ferrante AW Jr., Ahmed-Ansari A, Sell KW, Pollard JW, and Stanley ER. Total absence of colony-stimulating factor 1 in the macrophage-deficient osteopetrotic (op/op) mouse. Proceedings of the National Academy of Sciences of the United States of America. 1990;87(12):4828–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dai XM, Ryan GR, Hapel AJ, Dominguez MG, Russell RG, Kapp S, Sylvestre V, and Stanley ER. Targeted disruption of the mouse colony-stimulating factor 1 receptor gene results in osteopetrosis, mononuclear phagocyte deficiency, increased primitive progenitor cell frequencies, and reproductive defects. Blood. 2002;99(1):111–20. [DOI] [PubMed] [Google Scholar]
- 8.Simon DI. Opening the field of integrin biology to "biased agonism". Circulation research. 2011;109(11):1199–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan ST, and Edgington TS. Coupling of the adhesive receptor CD11b/CD18 to functional enhancement of effector macrophage tissue factor response. The Journal of clinical investigation. 1991;87(1):50–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan SM. The leucocyte beta2 (CD18) integrins: the structure, functional regulation and signalling properties. Bioscience reports. 2012;32(3):241–69. [DOI] [PubMed] [Google Scholar]
- 11.Lim J, and Hotchin NA. Signalling mechanisms of the leukocyte integrin alphaMbeta2: current and future perspectives. Biology of the cell. 2012;104(11):631–40. [DOI] [PubMed] [Google Scholar]
- 12.Shi C, Zhang X, Chen Z, Robinson MK, and Simon DI. Leukocyte integrin Mac-1 recruits toll/interleukin-1 receptor superfamily signaling intermediates to modulate NF-kappaB activity. Circulation research. 2001;89(10):859–65. [DOI] [PubMed] [Google Scholar]
- 13.Shi C, Zhang X, Chen Z, Sulaiman K, Feinberg MW, Ballantyne CM, Jain MK, and Simon DI. Integrin engagement regulates monocyte differentiation through the forkhead transcription factor Foxp1. The Journal of clinical investigation. 2004;114(3):408–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi C, Sakuma M, Mooroka T, Liscoe A, Gao H, Croce KJ, Sharma A, Kaplan D, Greaves DR, Wang Y, et al. Down-regulation of the forkhead transcription factor Foxp1 is required for monocyte differentiation and macrophage function. Blood. 2008;112(12):4699–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koon HB, Ippolito GC, Banham AH, and Tucker PW. FOXP1: a potential therapeutic target in cancer. Expert opinion on therapeutic targets. 2007;11(7):955–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smale ST, and Kadonaga JT. The RNA polymerase II core promoter. Annual review of biochemistry. 2003;72(449–79. [DOI] [PubMed] [Google Scholar]
- 18.Ashburner BP, Westerheide SD, and Baldwin AS Jr. The p65 (RelA) subunit of NF-kappaB interacts with the histone deacetylase (HDAC) corepressors HDAC1 and HDAC2 to negatively regulate gene expression. Molecular and cellular biology. 2001;21(20):7065–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wesche H, Henzel WJ, Shillinglaw W, Li S, and Cao Z. MyD88: an adapter that recruits IRAK to the IL-1 receptor complex. Immunity. 1997;7(6):837–47. [DOI] [PubMed] [Google Scholar]
- 20.Maamar H, Cabili MN, Rinn J, and Raj A. linc-HOXA1 is a noncoding RNA that represses Hoxa1 transcription in cis. Genes & development. 2013;27(11):1260–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459(7243):108–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Backs J, Song K, Bezprozvannaya S, Chang S, and Olson EN. CaM kinase II selectively signals to histone deacetylase 4 during cardiomyocyte hypertrophy. The Journal of clinical investigation. 2006;116(7):1853–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKinsey TA, Zhang CL, Lu J, and Olson EN. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature. 2000;408(6808):106–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Giorgio E, and Brancolini C. Regulation of class IIa HDAC activities: it is not only matter of subcellular localization. Epigenomics. 2016;8(2):251–69. [DOI] [PubMed] [Google Scholar]
- 25.Gray CB, and Heller Brown J. CaMKIIdelta subtypes: localization and function. Frontiers in pharmacology. 2014;5(15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xue ZH, Zhao CQ, Chua GL, Tan SW, Tang XY, Wong SC, and Tan SM. Integrin alphaMbeta2 clustering triggers phosphorylation and activation of protein kinase C delta that regulates transcription factor Foxp1 expression in monocytes. Journal of immunology (Baltimore, Md : 1950). 2010;184(7):3697–709. [DOI] [PubMed] [Google Scholar]
- 27.Wang Y, Gao H, Shi C, Erhardt PW, Pavlovsky A, D AS, Bledzka K, Ustinov V, Zhu L, Qin J, et al. Leukocyte integrin Mac-1 regulates thrombosis via interaction with platelet GPIbalpha. Nature communications. 2017;8(15559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hartupee J, Li X, and Hamilton T. Interleukin 1alpha-induced NFkappaB activation and chemokine mRNA stabilization diverge at IRAK1. The Journal of biological chemistry. 2008;283(23):15689–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inoue M, Arikawa T, Chen YH, Moriwaki Y, Price M, Brown M, Perfect JR, and Shinohara ML. T cells down-regulate macrophage TNF production by IRAK1-mediated IL-10 expression and control innate hyperinflammation. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(14):5295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmitz SU, Grote P, and Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cellular and molecular life sciences : CMLS. 2016;73(13):2491–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishii N, Ozaki K, Sato H, Mizuno H, Saito S, Takahashi A, Miyamoto Y, Ikegawa S, Kamatani N, Hori M, et al. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. Journal of human genetics. 2006;51(12):1087–99. [DOI] [PubMed] [Google Scholar]
- 32.Broadbent HM, Peden JF, Lorkowski S, Goel A, Ongen H, Green F, Clarke R, Collins R, Franzosi MG, Tognoni G, et al. Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Human molecular genetics. 2008;17(6):806–14. [DOI] [PubMed] [Google Scholar]
- 33.Chen YG, Satpathy AT, and Chang HY. Gene regulation in the immune system by long noncoding RNAs. Nature immunology. 2017;18(9):962–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhan A, and Mandal SS. LncRNA HOTAIR: A master regulator of chromatin dynamics and cancer. Biochimica et biophysica acta. 2015;1856(1):151–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Banerji J, Olson L, and Schaffner W. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell. 1983;33(3):729–40. [DOI] [PubMed] [Google Scholar]
- 36.Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, and Leunissen JA. Primer3Plus, an enhanced web interface to Primer3. Nucleic acids research. 2007;35(Web Server issue):W71–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.