Abstract
Chemical signals are produced by aquatic organisms following predatory attacks or perturbations such as parasitic infection. Ectoparasites feeding on fish hosts are likely to cause release of similar alarm cues into the environment due to the stress, wounding, and immune response stimulated upon infection. Alarm cues are often released in the form of proteins, antimicrobial peptides, and immunoglobulins that provide important insights into bodily function and infection status. Here we outline a noninvasive method to identify potential chemical cues associated with infection in fish by extracting, purifying, and characterizing proteins from water samples from cultured fish. Gel free proteomic methods were deemed the most suitable for protein detection in saline water samples. It was confirmed that teleost proteins can be characterized from water and that variation in protein profiles could be detected between infected and uninfected individuals and fish and parasite only water samples. Our novel assay provides a noninvasive method for assessing the health condition of both wild and farmed aquatic organisms. Similar to environmental DNA monitoring methods, these proteomic techniques could provide an important tool in applied ecology and aquatic biology.
Keywords: alarm cues, gel free MS, Kryptolebias marmoratus, odor, parasitic infection
1. Introduction
Chemical cues released into the environment following natural perturbations, predator attacks, or social threats allow animals to assess potential dangers and may lead to increased survival chances.1 In the aquatic environment, cues released in the form of soluble pheromones or as chemicals following physical trauma to the epidermis are described as alarm cues.2 These cues can signal the presence of danger, including the scent of predatory species in the environment (disturbance cues)3,4 or the odor of parasitized conspecifics.5 Additionally, innate immune responses activated upon pathogen infection can cause variation in body odor; this has been documented in humans, for example, whereby endotoxin-exposed individuals smelled significantly more unpleasant than their control counterparts, indicating a social cue of sickness.6 Changes in body odor have also been seen in infected mice, where females prefer the urinary odors of uninfected male individuals.7 Yet, studies that describe infection odors in aquatic environments are scarce,8,9 despite the importance of chemical communication, particularly in potentially turbid conditions.10
Chemical cues also allow individuals to recognize neighbors, assess social hierarchies, and determine mating ability by genotype matching, which may translate into attraction or conspecific avoidance.11,12 Odor based mate choices have been linked to the Major Histocompatibility Complex (MHC) and its associated peptides13 and allow individuals to discriminate between MHC genotypes on the basis of variation in immune-related peptides released into the water.13 This enables mate choice to maximize the immunocompetence of offspring,14 and MHC-related peptides are likely used for relatedness discrimination in several species, e.g., Salvelinus alpinus(15) and Gasterosteus aculeatus.13 Analyses of social communication in mice has also provided evidence that a variety of polymorphic proteins in urine (major urinary proteins) provide a unique identity to the owner and are used as scent signals.16
Many teleost fish can produce and perceive water-borne cues released by stressed conspecifics, including free cortisol17 and alarm cues released by specialized skin cells when individuals are injured.18 Zebrafish (Danio rerio), for example, are able to detect cues in the water from physically stressed individuals and avoid them,1 and Atlantic salmon can detect when a predator has consumed conspecifics.19 Similarly, Nile tilapia (Oreochromis niloticus) display antipredatory behaviors (including freezing and dorsal fin erection) in response to water conditioned with conspecific skin extract that mimics a predatory attack.20 Alarm cues also arise from parasitic infections, particularly ectoparasites that cause trauma to the epidermis initiating immune responses in a similar way to predator attacks.21,22 Argulid parasites attach to host fish by large maxillary suckers and feed on host blood and skin,23 this type of parasite feeding causes skin wounding, ulcers and increases the susceptibility of the host to secondary infections.24 Immune responses in fish skin, largely identified in the mucosal layer, include several components that reduce pathogen and allergen entry.25 Such components consist of antimicrobial peptides and enzymes (such as lysozymes, phosphatases, and proteases) for enhanced protection and wound healing,26−28 lectins and proteins that enhance pathogen expulsion and immune activation,25,29,30 and immunoglobulin antibodies for protection against surface infections.31 It is on this basis that the current study aimed to investigate excretory-secretory proteins released by hosts and parasites upon infection.
Gel based and gel free proteomic techniques are often used to detect proteins from body tissues of an organism32,33 and are also useful for studying the excretory-secretory proteins produced by endoparasitic helminths.34 For example, Fasciola hepatica, an economically important helminth of livestock is well studied, partly as it can easily be cultured in media, providing a clearer understanding of host–parasite communication and host manipulation.35−37 Fewer studies use in vitro culturing techniques to obtain proteins from ectoparasitic organisms; culturing of such parasites commonly involves rearing host teleost species under laboratory conditions followed by subsequent infection of these hosts via anesthesia.46 Despite difficulties associated with culturing these parasites,38 such systems provide a unique chance to collect and analyze protein data from host–parasite interactions. Furthermore, there is a distinct lack of knowledge surrounding environmental proteins from fresh and marine water bodies and the potential use of proteins as a noninvasive detection method for infection. Research into meta-proteomics of water samples to date has focused on methods for marine sediments (e.g., ref (39)). Yet, these methods have never been applied to teleost fish, particularly in brackish water samples or in relation to infection or parasite presence, which can be critical for the early detection of infection in aquatic species.
Kryptolebias marmoratus (the mangrove killifish) is a small, naturally inbred, cyprinid fish species that is easily maintained under laboratory conditions. Isogeneity in this species via constant self-ing makes it a useful model organism for the study of physiological plasticity and response to environmental stressors.40,41Argulus foliaceus, is a generalist fish parasite found throughout Europe that is known to cause epizootics and problems in both brackish and freshwater fish farms.42−45 Here we assessed the use of a novel noninvasive proteomic approach to identify fish infection by comparing proteins released into water by infected and uninfected K. marmoratus. Additionally, we used this study system to develop a proteomic method of potential application to assess health status and stress response in fish populations.
2. Experimental Procedures
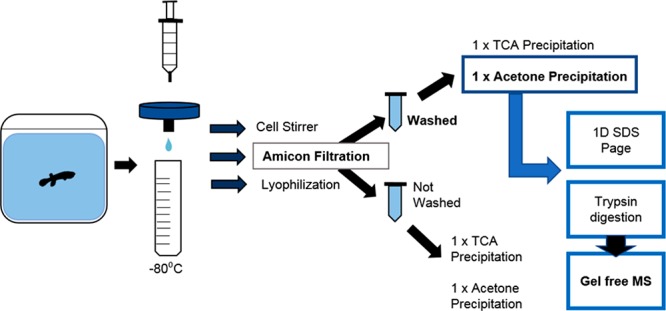
2.1. Water Sample Collection, Preparation, and Visualization
The study species, K. marmoratus, originated in Belize from two self-fertilizing lines (DAN and R) that have undergone >30 generations of self-ing.46 Eighty size-matched K. marmoratus (12–14 months old) reared individually from hatching were selected from the two lines (40 DAN and 40 R) and kept in individual aquaria (12 L × 8 W × 8.5 H cm) containing 750 mL of brackish water (15 ppt salinity, constituted from dechlorinated water and marine filtered water) under controlled conditions (12L:12D photoperiod, 24 ± 1 °C).
Twenty fish from each line were individually infected with one adult individual of the ectoparasitic crustacean Argulus foliaceus. The culture of A. foliaceus originated from carp (Cyprinus carpio) caught in a still water fishery in North Lincolnshire, July 2014, and thereafter was maintained on Gasterosteus aculeatus (three-spined sticklebacks) at Cardiff University as detailed in Stewart et al. (2017).47 The other 20 fish from each line were kept as control individuals, as described in Pawluk.48 Two water samples for proteomic analyses (50 mL each) were collected from each aquarium (80 aquaria total) prior to filtration (Minisart 25 mm Pore size 0.2 μm filter) and storage at −80 °C, the second sample acting as a spare. Twenty water samples (50 mL each) were also taken from containers of the same size containing a single A. foliaceus as a control for parasite proteins released into the water. Amicon filter units were used to reduce water samples from 36 mL (3 subsamples of 12 mL) to approximately 2 mL (10 KDa cut off, 5000g for 10 min, with two filter washes); subsequent quantification (Bradford49) using Sigma Bradford Reagent (according to the manufacturer’s instructions) and a Cary 50 Bio UV–visible spectrophotometer at 595 nm was completed. All samples were precipitated using the addition of 4 volumes of ice cold acetone followed by 1 h incubation at −20 °C. Proteins were then pelleted by centrifugation at 4 °C and 21 000g for 15 min. Pellets were then dried for 30 min before being resuspended in 30 μL of Buffer Z (8 M Urea, 2% w/v CHAPS, 33 mM DTT, 0.5% ampholytes pH range 3–10). Subsequent 1D SDS-page was performed using polyacrylamide gels (12.5%); they were then fixed overnight (in 10% ethanol, 40% acetic acid) and silver stained.50 The molecular weight of protein bands identified on 1D gels were calculated using a standard curve of log(Mw) versus the mobility shift (Rf).
2.2. Trypsin Digestion and Gel Free Mass Spectrophotometry
Fifteen water samples were prepared using Amicon filtration, as described previously, prior to trypsin digest and mass spectrometry analysis (3 individual fish replicates per group: DAN infected, DAN control, R infected, R control, and Argulus only). Sample aliquots of 100 μL each were added to 6 M urea, in 100 mM tris buffer, and then reduced using 5 μL reducing agent containing DTT and Tris stock (200 mM DTT, 100 mM Tris) for 1 h. Subsequently, 20 μL alkylating agent (200 mM iodoacetamide and 100 mM Tris) was added to each sample and vortexed before 1 h incubation at room temperature. A further 20 μL of reducing agent was added per sample and incubated at room temperature for 1 h. Sample urea concentration was reduced (to approximately 0.6 M) by diluting each sample with 75 μL of water before mixing. Trypsin digestion began following the addition of trypsin solution to a final concentration of 50 ng/μL, which was subsequently mixed and then incubated overnight at 37 °C. The reaction was stopped by adjusting the pH to >6 by adding concentrated acetic acid. All 15 samples were reduced to 100 μL aliquots using a speed vacuum prior to analysis liquid Chromatography tandem mass spectrometry using the Agilent 6550 iFunnel Q-TOF mass spectrometer with Dual AJS ESI source coupled to a 1200 series HPLC-Chip system (Agilent, Cheshire, UK).
The HPLC-Chip/Q-TOF system was equipped with a capillary loading pump (1200 series, Agilent Technologies) and a nano pump (1200 series, Agilent Technologies). Sample injection was conducted with a micro-auto-sampler (1100 series, Agilent Technologies), where 1 μL of sample in 0.1% formic acid was loaded on to the enrichment column at a flow of 2.5 μL/min followed by separation at a flow of 300 nL/min. A Polaris Chip was used (G4240–62030, Agilent Technologies), comprising a C18 enrichment/trap column (360 nL) and a C18 separation column (150 mm × 75 μm), where ions were generated at a capillary voltage of 1950 V. The solvent system was as follows: solvent A (ultrapure water with 0.1% formic acid), and solvent B (90% acetonitrile with 0.1% formic acid). The liquid chromatography was performed with a piece-linear gradient using 3–8% of solvent B over 0.1 min, 8–35% solvent B over 14.9 min, 35–90% solvent B over 5 min, and hold at 90% solvent B for 2 min. Tandem mass spectrometry was performed in AutoMS2 mode in the 300–1700 Da range, at a rate of 5 spectra per second, performing MS2 on the 5 most intense ions in the precursor scan. Masses were excluded for 0.1 min after MS/MS was performed. Reference mass locking was used for internal calibration using the mass of 391.2843 Da. Peak lists were generated with Mass Hunter Qualitative Analysis software (V B.06, Agilent Technologies) and exported as Mascot Generic Files.
2.3. Data Submission and Assessment Based on MASCOT Scoring
Mass spectral data from all 15 samples were submitted for database searching using the MASCOT program (Matrix Science Ltd., version 2.1). Search parameters allowed a maximum of one missed cleavage. Additionally, a fixed modification was set for cysteine at 161 Da and variable modifications tested for matches 0, 1, 2, or 3 oxidized methionine residues. Lastly, a set peptide tolerance of 1.2 Da and MS/MS tolerance of 0.6 Da were stipulated. Spectra were searched against the K. marmoratus transcriptome48 as well as a recent version of the NCBI nonredundant protein database sequences. Both analyses resulted in a total of 30 protein lists that were subsequently sorted into treatment groups; each sample from each group was filtered for protein number above the MASCOT significance threshold at p = 0.05 (Transcriptome data ≥50, NCBI data ≥59), protein identified in all three replicates and proteins identified in two of three individuals for each treatment (Table S2 and S3). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE51 partner repository with the data set identifier PXD010987.
2.4. GO Annotation, Pathway Analysis, and Statistical Testing
All proteins for each sample and treatment were considered to compare data searches and to complete a functional analysis (GO annotation, level GO ALL) of the results using the Database for Annotation, Visualization and Integrated Discovery, version 6.8 (DAVID).52 Pathway analysis was completed using REACTOME pathways53 for the three biological replicates of each group. Lastly, protein release for treatment groups was compared statistically using t tests. MASCOT scores for samples from the two respective groups (Infected and control) were compared for specific proteins (OSER1, KRT94). Additionally the number of proteins related to specific functional groups (identified from DAVID) were compared for methylation and immune functions. All figures produced for data visualization were created using R version 3.4.0.54
3. Results
3.1. Protein Visualization and Functional Analyses
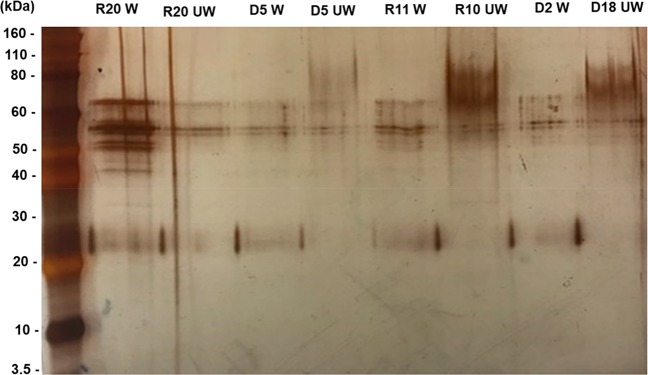
Results from quantification and one-dimensional SDS-page indicated the presence of proteins at approximately 61.6, 47.9, 42.3, and 24.8 kDa (Figure 1).
Figure 1.
SDS-PAGE gel of the expression of proteins concentrated using Amicon filtration steps, from two strains (D: DAN and R: R) for unwashed (UW) and washed (W) filters.
As 1D gel electrophoresis successfully indicated purified proteins a gel free approach was taken to identify all proteins within these samples. Soluble proteins from water samples were identified using both the K. marmoratus transcriptome48 (MASCOT score cut of ≥50) and from a search of the NCBI database (MASCOT score cut of >59). The total number of proteins identified for the K. marmoratus transcriptome and NCBI database ranged from 28 to 53 per sample. The percentage of identified proteins over the MASCOT cut off score for each data set ranged from 2.9–16.3% from transcriptome sourced data and 7.7–20.9% from NCBI sourced data (Table S1). Trypsin (used for digestion) and keratin, mostly deemed to be from human origin and thus contaminants were highly abundant (had a MASCOT score on or above the cut off) and overlapped between search methods (Transcriptome and NCBI) (Table S2 and S3).
Gene ontology analysis indicated that functional proteins involved with immune response and development (Figure S2 and S3) were identified in both data sets; however, on inspection of these proteins there was little overlap between search methods. Pathway analysis (Table 1, Table S5 and Figure S3) was completed for a comparison of enriched protein groups for all 5 groups (DAN control and infected, R control and infected, and Argulus only water). For all fish water samples (DAN and R, infected and control) proteins were mainly involved in metabolism, in three of these groups these proteins had the highest number of hits. Proportionally fewer proteins were reported in Argulus only samples when compared with the others and proteins from this group primarily had roles in transcription and homeostasis. Proteins with roles in cytokine signaling were shown to be in infected fish water samples but not in DAN.
Table 1. Top Five Entities Identified from the REACTOME Pathway Analysis of 5 Groups of Water Samplesa.
| Group | Pathway name | Entities found | Entities total | Entities p-value | Entities FDR | Reactions found |
|---|---|---|---|---|---|---|
| DAN infected | metabolism | 10 | 2116 | 0.84 | 8.35 × 10–01 | 34 |
| immune system | 6 | 2226 | 1.00 | 9.95 × 10–01 | 17 | |
| adaptive immune system | 3 | 944 | 0.93 | 9.28 × 10–01 | 6 | |
| generic transcription pathway | 3 | 1152 | 0.97 | 9.73 × 10–01 | 7 | |
| RNA polymerase II transcription | 3 | 1274 | 0.99 | 9.86 × 10–01 | 18 | |
| DAN control | metabolism | 6 | 2116 | 1.00 | 9.97 × 10–01 | 9 |
| disease | 4 | 1148 | 0.95 | 9.52 × 10–01 | 25 | |
| innate immune system | 4 | 1180 | 0.96 | 9.59 × 10–01 | 12 | |
| post-translational protein modification | 3 | 1415 | 1.00 | 9.97 × 10–01 | 21 | |
| signaling by receptor tyrosine kinases | 2 | 471 | 0.82 | 8.22 × 10–01 | 2 | |
| R infected | metabolism | 8 | 2116 | 0.99 | 9.92 × 10–01 | 23 |
| immune system | 8 | 2226 | 1.00 | 9.96 × 10–01 | 17 | |
| metabolism of proteins | 6 | 2111 | 1.00 | 9.99 × 10–01 | 11 | |
| signal transduction | 6 | 2738 | 1.00 | 1.00 × 1000 | 8 | |
| innate immune system | 5 | 1180 | 0.94 | 9.41 × 10–01 | 8 | |
| R control | signal transduction | 10 | 2738 | 0.98 | 9.84 × 10–01 | 40 |
| metabolism | 8 | 2116 | 0.96 | 9.62 × 10–01 | 24 | |
| metabolism of proteins | 5 | 2111 | 1.00 | 9.98 × 10–01 | 4 | |
| immune system | 4 | 2226 | 1.00 | 1.00 × 1000 | 15 | |
| GPCR downstream signaling | 3 | 1146 | 0.98 | 9.77 × 10–01 | 4 | |
| Argulus | signal transduction | 7 | 2738 | 1.00 | 9.99 × 10–01 | 27 |
| metabolism of proteins | 6 | 2111 | 0.99 | 9.91 × 10–01 | 31 | |
| metabolism | 5 | 2116 | 1.00 | 9.97 × 10–01 | 7 | |
| immune system | 5 | 2226 | 1.00 | 9.98 × 10–01 | 21 | |
| gene expression (transcription) | 3 | 1416 | 0.99 | 9.92 × 10–01 | 5 |
From DAN infected, DAN control, R infected, and R control Kryptolebias marmoratus as well as Argulus only samples.
3.2. Protein Identification and Comparisons
Analysis of proteins identified using the killifish transcriptome identified a similar number of proteins released (MASCOT ≥ 50) by all experimental groups (Table S1). Actin family beta (for cell motility, structure, and integrity) and fish keratin (from host tissue) proteins were expressed in all treatment groups. OSER1, a protein with roles in oxidative stress response, was abundant in all fish water samples, but not in Argulus only samples. Despite this, no significant variations in MASCOT scores between control and infected water samples were identified for OSER1 (t6.96 = 1.521, P = 0.172). In addition to proteins detected from the killifish transcriptome, results were searched against the NCBI database to identify proteins from fish and parasite origins. Of the 47 proteins with a MASCOT score on or above the 59 cut off, 49% were deemed as human contamination (Table S3, in red) but 13% of proteins included claw keratin and proteins associated with teleost fish (polyserase-2-like proteins and Chymotrypsin-like protease proteins).
Variations between infected and uninfected fish samples were identified for both transcriptome and NCBI data sets. Proteins identified from the transcriptome indicated that Keratin 94 (of fish origin), associated with host tissues, was only identified in infected fish water samples. However, these samples did not differ significantly when MASCOT scores from each group were compared (t6.9 = 0.88, P = 0.396). Zinc finger proteins (a class of protein involved with nucleic acid binding and protein dimerization) were highly abundant in DAN infected and Argulus only samples.
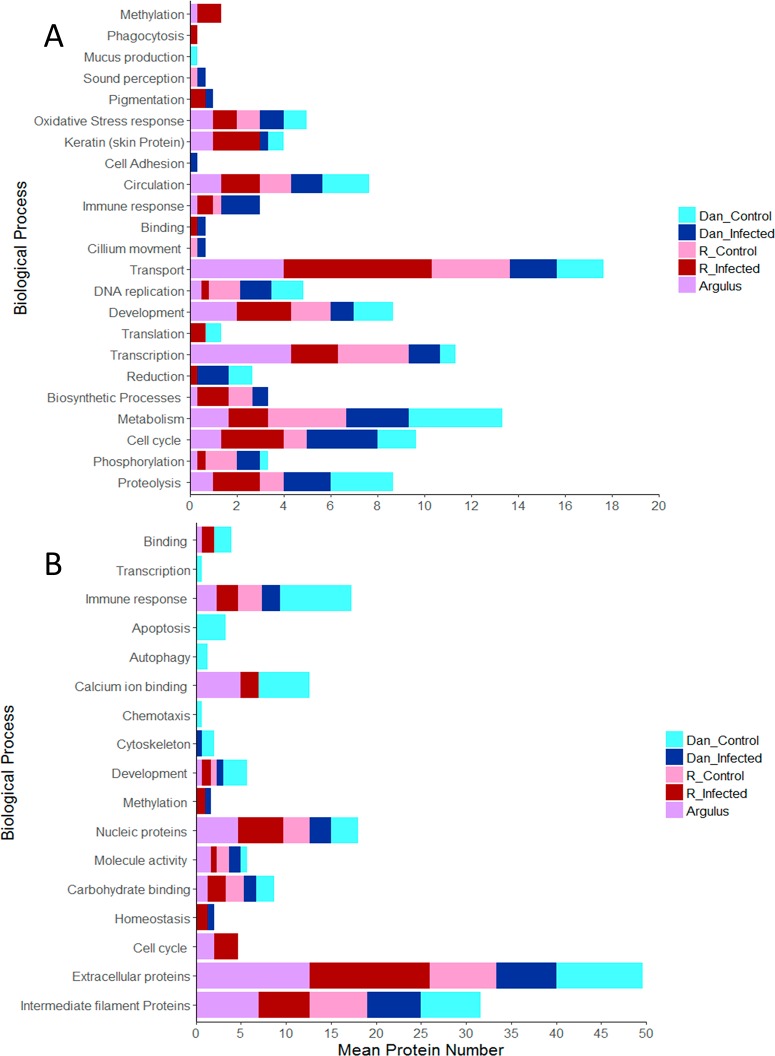
Gene ontology analysis of proteins from the transcriptome was conducted and compared between treatments (Figure 2). This analysis indicated that proteins involved in pigmentation (TSPAN36 and SLC45A2) and binding were only detected among infected fish and no protein group was identified solely in the control groups. Previous work has demonstrated links between infection and pigmentation;55,56 therefore, proteins related to color in K. marmoratus are likely promising indicators of immune response. All groups contained proteins related to metabolism, proteolysis, and circulation. Finally, proteins for sound perception, cilium movement, and mucus production (Figure 2A) were only identified in water from fish origin. Statistical analysis of gene ontology data from NCBI indicated that a significantly greater abundance of proteins for methylation were identified in infected fish water samples when compared with controls (t5 = 2.397, P = 0.042) as can be seen in Figure 2. Conversely despite variation indicated in Figure 2, the number of proteins with roles in immune response did not differ significantly between groups (t5 = −0.682, P = 0.522). For control groups, functional proteins identified were involved in apoptosis and autophagy. Lastly protein families observed in all groups included extracellular proteins, as well as proteins involved in the immune response, and in carbohydrate binding (for all see Figure 2A).
Figure 2.
Go annotation of all proteins identified from infected (with Argulus foliaceus), uninfected and parasite only water samples for two lines (DAN and R) of Kryptolebias marmoratus transcriptome (A) and NCBI data (B).
4. Discussion
Current, nonlethal fish health monitoring involves collection and subsequent analysis of the external surface mucus of teleost species.57,58 Fish skin mucosa acts as an important barrier against pathogens,59,60 and within this barrier proteins may be secreted, synthesized, and released (in the form of dead cells) leading to the production of identifiable immune related molecules.61 Release of molecules into the surrounding aquatic medium is thought to act as chemical stimulus that enable teleost fish to assess the health, genetic background, and identity of conspecifics.11,15 The methods used in the current study indicate that these proteins can be extracted, purified, and characterized from teleost water samples and have the potential to inform infection status noninvasively, without the need for invasive sampling. In the current study, initial gel based methods demonstrated a number of observable protein bands that may be indicative of heat stress and heat shock proteins,62,63 binding proteins,64 and keratins.65 Furthermore, gel-free methods following trypsin digestion of samples greatly increased the number of proteins identified and confirmed the presence of fish keratins, binding proteins, and immune related molecules.
Skin extracts are thought to be present in the mucosa following skin rupture, leading to the release of alarm cues, identifiable by conspecifics.59,66 Fish keratin proteins (and many actin proteins below the MASCOT cutoff) were identified in samples from infected fish only; these proteins are likely to originate from the skin surface (due to the structural function of keratin) in the form of ruptured or dead cells.67 Additionally, keratins from trout have been shown to display antibacterial activity,68 and these proteins will likely indicate danger to surrounding conspecifics. In addition to keratins, binding proteins were highly expressed in the current water samples from infected R line fish. Binding proteins, identified in the mucosa of fish, have been shown to serve many functions including immune response and lipid metabolism,69,70 and it is plausible that these proteins from the mucus are being detected in current samples. Importantly, fish mucus has been shown to contain a plethora of immune related proteins including those from the complement,71 immunoglobulins,72 and antimicrobial peptides.73 Both NCBI and transcriptome based databases indicated immune related proteins in current fish water samples; these were identified for individual characterized proteins (e.g., cytokines) but also at the level of GO annotation and pathway analyses, whereby immune annotations and pathways were highly enriched. One current example of an immune protein identified by MASCOT and GO annotation scoring, from infected R line individuals, is interleukin 6 signal transducer (IL6st). Interleukin 6 receptor was previously identified in the mucus of gilthead sea bream (Sparus aurata) that were chronically stressed,74 and several other interleukin molecules with well-known roles in inflammatory response and protection from pathogens have been described in the skin of Nile tilapia (Oreochromis niloticus).75,76 As fish mucus is continually secreted and replaced,75 it is likely that immune molecules, such as IL6, are transferred to the media on a constant basis. As demonstrated in the current study, the regeneration and movement of mucosal molecules into the aquatic media allows for collection and analysis of these proteins in low water volumes.
Further to specific immune or structural proteins, the mucosa of chronically stressed fish has allowed for research into a variety of stress related proteins thought to influence the response of fish to environmental fluctuations.67,74 Proteins associated with oxidative stress response are commonly studied in fish exposed to toxic environmental pollutants, for example, metals,77 and associated biomarkers have been suggested as a potential target for environmental monitoring and pollutant detection.78,79 Additionally, these proteins have been identified in fish skin mucus and have roles in regulation of cell death.74,80 Here we provide evidence that the OSER1 protein, of fish origin, was detectable from all samples of aquaria water and has the potential to indicate environmental stress response in this species. In this instance OSER1 could be linked with stress related to experimental testing such as sham infection (for control fish) or louse attachment for infected individuals. This warrants further investigation but confirms the ability of current methods to detect a well-known marker.
Many more proteins of interest appeared in two or three individual fish below the MASCOT cutoff. Presence of these proteins might have increased with a larger number of samples at a higher concentration and/or larger volumes of water. Briefly, infected fish samples and Argulus only samples contained proteins (below the MASCOT significance cut off) from mite and lice species; these included PHUM400690 (for immune response to ticks), capicua protein (a transcriptional repressor), and IscW (an immune suppressor). As only one parasite and a small host were tested in the current study, it is likely that larger fish with several parasites would reveal more clues about these proteins. If these proteins were discovered to increase with parasite number they would likely be a strong target for environmental protein detection methods. Proteins with roles in oxidative stress response, response to lice, pigmentation, and immune response identified here have the potential to reveal infection and/or stress status in fish. Proteins associated with Argulus foliaceus were observed in water from infected fish, yet several proteins were only identified in the Argulus control group. These proteins (zinc finger protein and proteins involved in the immune response to lice and ticks) demonstrated the possibility of using proteomic techniques to detect the presence of parasitic organisms within the environment, as with environmental DNA.81−83 Lastly, proteins with roles in oxidative stress response, response to lice, pigmentation, and immune response identified here have the potential to indicate infection and/or stress status in fish, an area that requires further investigation. We propose that the proteomic methods outlined here have the potential to provide a similar tool for evaluating stress and health status in individuals, with the added benefit of functional analysis of expressed proteins. Lastly, we suggest that proteomic and environmental DNA methods could be used in tandem to gain noninvasive data on farmed and wild fish populations.
5. Conclusions
This is the first study outlining protein characterization from brackish water samples without the need for gel electrophoresis and represents a major step forward in the field of metaproteomic techniques. Our novel assay provides a noninvasive method for detecting infection status in fish, which will be useful for assessing the health condition of both wild and farmed aquatic organisms. With further investigation and optimization, protein detection methods represent a promising new method for environmental monitoring and could become an important tool in applied and aquatic biology.
Acknowledgments
This study was supported by a Natural Environment Research Council Industrial CASE studentship (NE/L00948X) and the BCHC (Aberystwyth University) Access Fellowship to RJP. All the experiments in this study have been conducted following Home Office regulations, approved by both Swansea and Cardiff University Ethics Committees and under Home Office license number PPL 302357.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jproteome.8b00953.
Details S1; Tables S1–S5; Figures S1–S3 (PDF)
Author Contributions
RJP, SC, and RS designed the study with help from PMB and RMM. RJP, SC, and RMM wrote the manuscript. PMB provided the lab for experimental procedures. JC provided expertise and facilities for all experimental infections. CGL and JC provided manuscript corrections and advice.
The authors declare no competing financial interest.
Notes
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE51 partner repository with the data set identifier PXD010987.
Supplementary Material
References
- Abreu M. S.; Giacomini A. C. V.; Gusso D.; Koakoski G.; Oliveira T. A.; Marqueze A.; Barreto R. E.; Barcellos L. J. Behavioral responses of zebrafish depend on the type of threatening chemical cues. J. Comp. Physiol., A 2016, 202 (12), 895–901. 10.1007/s00359-016-1129-5. [DOI] [PubMed] [Google Scholar]
- Wyatt T. D. Proteins and peptides as pheromone signals and chemical signatures. Anim. Behav. 2014, 97, 273–280. 10.1016/j.anbehav.2014.07.025. [DOI] [Google Scholar]
- Brown G. E. Learning about danger: chemical alarm cues and local risk assessment in prey fishes. Fish and Fisheries 2003, 4 (3), 227–234. 10.1046/j.1467-2979.2003.00132.x. [DOI] [Google Scholar]
- Chivers D. P.; Mirza R. S.; Johnston J. G. Learned recognition of heterospecific alarm cues enhances survival during encounters with predators. Behaviour 2002, 139 (7), 929–938. 10.1163/156853902320387909. [DOI] [Google Scholar]
- Sharp J. G.; Garnick S.; Elgar M. A.; Coulson G. Parasite and Predator Risk Assessment: Nuanced Use of Olfactory Cues. Proc. R. Soc. London, Ser. B 2015, 282, 20151941. 10.1098/rspb.2015.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson M. J.; Lundström J. N.; Kimball B. A.; Gordon A. R.; Karshikoff B.; Hosseini N.; Sorjonen K.; Olgart Höglund C.; Solares C.; Soop A. The scent of disease: human body odor contains an early chemosensory cue of sickness. Psychol. Sci. 2014, 25 (3), 817–823. 10.1177/0956797613515681. [DOI] [PubMed] [Google Scholar]
- Ehman K.; Scott M. Urinary odour preferences of MHC congenic female mice, Mus domesticus: implications for kin recognition and detection of parasitized males. Anim. Behav. 2001, 62 (4), 781–789. 10.1006/anbe.2001.1805. [DOI] [Google Scholar]
- Lehtonen T. K.; Kvarnemo C. Odour cues from suitors’ nests determine mating success in a fish. Biol. Lett. 2015, 11 (5), 20150021. 10.1098/rsbl.2015.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone C. F.; Moore J. Parasite-induced alteration of odour responses in an amphipod–acanthocephalan system. Int. J. Parasitol. 2014, 44 (13), 969–975. 10.1016/j.ijpara.2014.06.012. [DOI] [PubMed] [Google Scholar]
- Webster M.; Atton N.; Ward A.; Hart P. Turbidity and foraging rate in threespine sticklebacks: the importance of visual and chemical prey cues. Behaviour 2007, 144 (11), 1347–1360. 10.1163/156853907782418222. [DOI] [Google Scholar]
- Boehm T.; Zufall F. MHC peptides and the sensory evaluation of genotype. Trends Neurosci. 2006, 29 (2), 100–107. 10.1016/j.tins.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Charpentier M.; Prugnolle F.; Gimenez O.; Widdig A. Genetic heterozygosity and sociality in a primate species. Behav. Genet. 2008, 38 (2), 151–158. 10.1007/s10519-008-9191-6. [DOI] [PubMed] [Google Scholar]
- Milinski M.; Griffiths S.; Wegner K. M.; Reusch T. B. H.; Haas-Assenbaum A.; Boehm T. Mate choice decisions of stickleback females predictably modified by MHC peptide ligands. Proc. Natl. Acad. Sci. U. S. A. 2005, 102 (12), 4414–4418. 10.1073/pnas.0408264102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consuegra S.; Garcia de Leaniz C. MHC-mediated mate choice increases parasite resistance in salmon. Proc. R. Soc. London, Ser. B 2008, 275 (1641), 1397–1403. 10.1098/rspb.2008.0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsén K. H.; Grahn M.; Lohm J. Influence of MHC on sibling discrimination in Arctic char, Salvelinus alpinus (L.). J. Chem. Ecol. 2002, 28 (4), 783–795. 10.1023/A:1015240810676. [DOI] [PubMed] [Google Scholar]
- Beynon R. J.; Hurst J. Multiple roles of major urinary proteins in the house mouse, Mus domesticus. Biochem. Soc. Trans. 2003, 31, 142–146. 10.1042/bst0310142. [DOI] [PubMed] [Google Scholar]
- Fischer E. K.; Harris R. M.; Hofmann H. A.; Hoke K. L. Predator exposure alters stress physiology in guppies across timescales. Horm. Behav. 2014, 65 (2), 165–172. 10.1016/j.yhbeh.2013.12.010. [DOI] [PubMed] [Google Scholar]
- Mathuru A. S.; Kibat C.; Cheong W. F.; Shui G.; Wenk M. R.; Friedrich R. W.; Jesuthasan S. Chondroitin fragments are odorants that trigger fear behavior in fish. Curr. Biol. 2012, 22 (6), 538–544. 10.1016/j.cub.2012.01.061. [DOI] [PubMed] [Google Scholar]
- Roberts L. J.; Garcia de Leaniz C. Something smells fishy: predator-naïve salmon use diet cues, not kairomones, to recognize a sympatric mammalian predator. Anim. Behav. 2011, 82 (4), 619–625. 10.1016/j.anbehav.2011.06.019. [DOI] [Google Scholar]
- Barreto R. E.; Júnior A. B.; Giassi A. C. C.; Hoffmann A. The ‘club’cell and behavioural and physiological responses to chemical alarm cues in the Nile tilapia. Mar. Freshwater Behav. Physiol. 2010, 43 (1), 75–81. 10.1080/10236241003654139. [DOI] [Google Scholar]
- Forlenza M.; Walker P. D.; De Vries B. J.; Bonga S. E. W.; Wiegertjes G. F. Transcriptional analysis of the common carp (Cyprinus carpio L.) immune response to the fish louse Argulus japonicus Thiele (Crustacea: Branchiura). Fish Shellfish Immunol. 2008, 25 (1–2), 76–83. 10.1016/j.fsi.2007.12.013. [DOI] [PubMed] [Google Scholar]
- Fast M. D. Fish immune responses to parasitic copepod (namely sea lice) infection. Dev. Comp. Immunol. 2014, 43 (2), 300–312. 10.1016/j.dci.2013.08.019. [DOI] [PubMed] [Google Scholar]
- Walker P. D.; Flik G.; Bonga S. W. The biology of parasites from the genus Argulus and a review of the interactions with its host. Host-Parasite Interactions 2004, 55, 107–129. 10.4324/9780203487709-6. [DOI] [PubMed] [Google Scholar]
- Bandilla M.; Valtonen E.; Suomalainen L.-R.; Aphalo P.; Hakalahti T. A link between ectoparasite infection and susceptibility to bacterial disease in rainbow trout. Int. J. Parasitol. 2006, 36 (9), 987–991. 10.1016/j.ijpara.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Ángeles Esteban M. An overview of the immunological defenses in fish skin. ISRN Immunol. 2012, 1–29. 10.5402/2012/853470. [DOI] [Google Scholar]
- McGuckin M. A.; Lindén S. K.; Sutton P.; Florin T. H. Mucin dynamics and enteric pathogens. Nat. Rev. Microbiol. 2011, 9 (4), 265. 10.1038/nrmicro2538. [DOI] [PubMed] [Google Scholar]
- Ellis A. Immunity to bacteria in fish. Fish Shellfish Immunol. 1999, 9 (4), 291–308. 10.1006/fsim.1998.0192. [DOI] [Google Scholar]
- Sarmaşik A. Antimicrobial peptides: a potential therapeutic alternative for the treatment of fish diseases. Turkish J. Biol. 2002, 26 (4), 201–207. [Google Scholar]
- Ingram G. Substances involved in the natural resistance of fish to infection–a review. J. Fish Biol. 1980, 16 (1), 23–60. 10.1111/j.1095-8649.1980.tb03685.x. [DOI] [Google Scholar]
- González-Chávez S. A.; Arévalo-Gallegos S.; Rascón-Cruz Q. Lactoferrin: structure, function and applications. Int. J. Antimicrob. Agents 2009, 33 (4), 301. e1–301. e8. 10.1016/j.ijantimicag.2008.07.020. [DOI] [PubMed] [Google Scholar]
- Zhang Y.-A.; Salinas I.; Li J.; Parra D.; Bjork S.; Xu Z.; LaPatra S. E.; Bartholomew J.; Sunyer J. O. IgT, a primitive immunoglobulin class specialized in mucosal immunity. Nat. Immunol. 2010, 11 (9), 827. 10.1038/ni.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Ambrosio C.; Arena S.; Talamo F.; Ledda L.; Renzone G.; Ferrara L.; Scaloni A. Comparative proteomic analysis of mammalian animal tissues and body fluids: bovine proteome database. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2005, 815 (1–2), 157–168. 10.1016/j.jchromb.2004.07.017. [DOI] [PubMed] [Google Scholar]
- Zargar S.; Gupta N.; Mir R.; Rai V. Shift from gel based to gel free proteomics to unlock unknown regulatory network in plants: a comprehensive review. J. Adv. Res. Biotechnol. 2016, 1, 19. 10.15226/2475-4714/1/2/00107. [DOI] [Google Scholar]
- Brophy P. M.; Mackintosh N.; Morphew R. M. Anthelmintic metabolism in parasitic helminths: proteomic insights. Parasitology 2012, 139 (9), 1205–1217. 10.1017/S003118201200087X. [DOI] [PubMed] [Google Scholar]
- Marcilla A.; Trelis M.; Cortés A.; Sotillo J.; Cantalapiedra F.; Minguez M. T.; Valero M. L.; Del Pino M. M. S.; Muñoz-Antoli C.; Toledo R. Extracellular vesicles from parasitic helminths contain specific excretory/secretory proteins and are internalized in intestinal host cells. PLoS One 2012, 7 (9), e45974 10.1371/journal.pone.0045974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. W.; Menon R.; Donnelly S. M.; Dalton J. P.; Ranganathan S. An integrated transcriptomics and proteomics analysis of the secretome of the helminth pathogen Fasciola hepatica proteins associated with invasion and infection of the mammalian host. Mol. Cell. Proteomics 2009, 8 (8), 1891–1907. 10.1074/mcp.M900045-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morphew R. M.; Wright H. A.; LaCourse E. J.; Woods D. J.; Brophy P. M. Comparative proteomics of excretory-secretory proteins released by the liver fluke Fasciola hepatica in sheep host bile and during in vitro culture ex host. Mol. Cell. Proteomics 2007, 6 (6), 963–972. 10.1074/mcp.M600375-MCP200. [DOI] [PubMed] [Google Scholar]
- Hutson K. S.; Cable J.; Grutter A. S.; Paziewska-Harris A.; Barber I. Aquatic Parasite Cultures and Their Applications. Trends Parasitol. 2018, 34, 1082. 10.1016/j.pt.2018.09.007. [DOI] [PubMed] [Google Scholar]
- Wöhlbrand L.; Feenders C.; Nachbaur J.; Freund H.; Engelen B.; Wilkes H.; Brumsack H. J.; Rabus R. Impact of Extraction Methods on the Detectable Protein Complement of Metaproteomic Analyses of Marine Sediments. Proteomics 2017, 17 (22), 1700241. 10.1002/pmic.201700241. [DOI] [PubMed] [Google Scholar]
- Lee J. S.; Raisuddin S.; Schlenk D. Kryptolebias marmoratus (Poey, 1880): a potential model species for molecular carcinogenesis and ecotoxicogenomics. J. Fish Biol. 2008, 72 (8), 1871–1889. 10.1111/j.1095-8649.2008.01818.x. [DOI] [Google Scholar]
- Harrington R. W. Jr; Kallman K. D. The homozygosity of clones of the self-fertilizing hermaphroditic fish Rivulus marmoratus Poey (Cyprinodontidae, Atheriniformes). Am. Nat. 1968, 102 (926), 337–343. 10.1086/282547. [DOI] [Google Scholar]
- Pasternak A. F.; Mikheev V. N.; Valtonen E. T. Life history characteristics of Argulus foliaceus L.(Crustacea: Branchiura) populations in Central Finland. Ann. Zool. Fenn. 2000, 25–35. [Google Scholar]
- Bower-Shore C. An investigation of the common fish louse, Argulus foliaceus (Linn.). Parasitology 1940, 32 (4), 361–371. 10.1017/S0031182000015869. [DOI] [Google Scholar]
- Mirzaei M.; Khovand H. Prevalence of Argulus foliaceus in ornamental fishes [goldfish (Carassius auratus) and Koi (Cyprinus carpio)] in Kerman, southeast of Iran. J. Parasit. Dis. 2015, 39 (4), 780–782. 10.1007/s12639-013-0406-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aalberg K.; Koščová L.; Šmiga L.; Košuth P.; Koščo J.; Oros M.; Barčák D.; Lazar P. A study of fish lice (Argulus sp.) infection in freshwater food fish. Folia Veterinaria 2016, 60 (3), 54–59. 10.1515/fv-2016-0030. [DOI] [Google Scholar]
- Ellison A.; Jones J.; Inchley C.; Consuegra S. Choosy males could help explaining androdioecy in a selfing fish. Am. Nat. 2013, 181, 855–862. 10.1086/670304. [DOI] [PubMed] [Google Scholar]
- Stewart A.; Jackson J.; Barber I.; Eizaguirre C.; Paterson R.; van West P.; Williams C.; Cable J. Hook, line and infection: a guide to culturing parasites, establishing infections and assessing immune responses in the three-spined stickleback. Adv. Parasitol. 2017, 98, 39–109. 10.1016/bs.apar.2017.07.001. [DOI] [PubMed] [Google Scholar]
- Pawluk R. J.; Uren Webster T. M.; Cable J.; Garcia de Leaniz C.; Consuegra S. Immune-related transcriptional responses to parasitic infection in a naturally inbred fish: roles of genotype and individual variation. Genome Biol. Evol. 2018, 10, 319–327. 10.1093/gbe/evx274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72 (1–2), 248–254. 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Shevchenko A.; Wilm M.; Vorm O.; Mann M. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 1996, 68 (5), 850–858. 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Vizcaíno J. A.; Csordas A.; Del-Toro N.; Dianes J. A.; Griss J.; Lavidas I.; Mayer G.; Perez-Riverol Y.; Reisinger F.; Ternent T. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016, 44 (D1), D447–D456. 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G.; Sherman B. T.; Hosack D. A.; Yang J.; Gao W.; Lane H. C.; Lempicki R. A. DAVID: database for annotation, visualization, and integrated discovery. Genome biology 2003, 4 (9), R60. 10.1186/gb-2003-4-9-r60. [DOI] [PubMed] [Google Scholar]
- Fabregat A.; Sidiropoulos K.; Garapati P.; Gillespie M.; Hausmann K.; Haw R.; Jassal B.; Jupe S.; Korninger F.; McKay S. The reactome pathway knowledgebase. Nucleic Acids Res. 2016, 44 (D1), D481–D487. 10.1093/nar/gkv1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . R: A Language and Environment for Statistical Computing; 2013, Vol. 201.
- Price A. C.; Weadick C. J.; Shim J.; Rodd F. H. Pigments, patterns, and fish behavior. Zebrafish 2008, 5 (4), 297–307. 10.1089/zeb.2008.0551. [DOI] [PubMed] [Google Scholar]
- Seppälä O.; Karvonen A.; Valtonen E. T. Impaired crypsis of fish infected with a trophically transmitted parasite. Anim. Behav. 2005, 70 (4), 895–900. 10.1016/j.anbehav.2005.01.021. [DOI] [Google Scholar]
- Bergsson G.; Agerberth B.; Jörnvall H.; Gudmundsson G. H. Isolation and identification of antimicrobial components from the epidermal mucus of Atlantic cod (Gadus morhua). FEBS J. 2005, 272 (19), 4960–4969. 10.1111/j.1742-4658.2005.04906.x. [DOI] [PubMed] [Google Scholar]
- Salles C.; Gagliano P.; Leitão S.; Salles J.; Guedes H.; Cassano V.; De-Simone S. G. Identification and characterization of proteases from skin mucus of tambacu, a Neotropical hybrid fish. Fish Physiol. Biochem. 2007, 33 (2), 173. 10.1007/s10695-007-9128-7. [DOI] [Google Scholar]
- Reverter M.; Tapissier-Bontemps N.; Lecchini D.; Banaigs B.; Sasal P. Biological and Ecological Roles of External Fish Mucus: A Review. Fishes 2018, 3 (4), 41. 10.3390/fishes3040041. [DOI] [Google Scholar]
- Gomez D.; Sunyer J. O.; Salinas I. The mucosal immune system of fish: the evolution of tolerating commensals while fighting pathogens. Fish Shellfish Immunol. 2013, 35 (6), 1729–1739. 10.1016/j.fsi.2013.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinchmann M. F. Immune relevant molecules identified in the skin mucus of fish using-omics technologies. Mol. BioSyst. 2016, 12 (7), 2056–2063. 10.1039/C5MB00890E. [DOI] [PubMed] [Google Scholar]
- Iwama G. K.; Vijayan M. M.; Forsyth R. B.; Ackerman P. A. Heat shock proteins and physiological stress in fish. Am. Zool. 1999, 39 (6), 901–909. 10.1093/icb/39.6.901. [DOI] [Google Scholar]
- Kiang J. G.; Tsokos G. C. Heat shock protein 70 kDa: molecular biology, biochemistry, and physiology. Pharmacol. Ther. 1998, 80 (2), 183–201. 10.1016/S0163-7258(98)00028-X. [DOI] [PubMed] [Google Scholar]
- Siharath K.; Kelley K. M.; Bern H. A. A low-molecular-weight (25-kDa) IGF-binding protein is increased with growth inhibition in the fasting striped bass, Morone saxatilis. Gen. Comp. Endocrinol. 1996, 102 (3), 307–316. 10.1006/gcen.1996.0074. [DOI] [PubMed] [Google Scholar]
- Conrad M.; Lemb K.; Schubert T.; Markl J. Biochemical identification and tissue-specific expression patterns of keratins in the zebrafish Danio rerio. Cell Tissue Res. 1998, 293 (2), 195–205. 10.1007/s004410051112. [DOI] [PubMed] [Google Scholar]
- Chivers D. P.; Zhao X.; Ferrari M. C. Linking morphological and behavioural defences: prey fish detect the morphology of conspecifics in the odour signature of their predators. Ethology 2007, 113 (8), 733–739. 10.1111/j.1439-0310.2006.01385.x. [DOI] [Google Scholar]
- Jurado J.; Fuentes-Almagro C. A.; Guardiola F. A.; Cuesta A.; Esteban M. Á.; Prieto-Álamo M.-J. Proteomic profile of the skin mucus of farmed gilthead seabream (Sparus aurata). J. Proteomics 2015, 120, 21–34. 10.1016/j.jprot.2015.02.019. [DOI] [PubMed] [Google Scholar]
- Molle V.; Campagna S.; Bessin Y.; Ebran N.; Saint N.; Molle G. First evidence of the pore-forming properties of a keratin from skin mucus of rainbow trout (Oncorhynchus mykiss, formerly Salmo gairdneri). Biochem. J. 2008, 411 (1), 33–40. 10.1042/BJ20070801. [DOI] [PubMed] [Google Scholar]
- Rajan B.; Fernandes J. M.; Caipang C. M.; Kiron V.; Rombout J. H.; Brinchmann M. F. Proteome reference map of the skin mucus of Atlantic cod (Gadus morhua) revealing immune competent molecules. Fish Shellfish Immunol. 2011, 31 (2), 224–231. 10.1016/j.fsi.2011.05.006. [DOI] [PubMed] [Google Scholar]
- Cordero H.; Brinchmann M. F.; Cuesta A.; Meseguer J.; Esteban M. A. Skin mucus proteome map of European sea bass (Dicentrarchus labrax). Proteomics 2015, 15 (23–24), 4007–4020. 10.1002/pmic.201500120. [DOI] [PubMed] [Google Scholar]
- Shen Y.; Zhang J.; Xu X.; Fu J.; Li J. Expression of complement component C7 and involvement in innate immune responses to bacteria in grass carp. Fish Shellfish Immunol. 2012, 33 (2), 448–454. 10.1016/j.fsi.2012.05.016. [DOI] [PubMed] [Google Scholar]
- Salinas I.; Zhang Y.-A.; Sunyer J. O. Mucosal immunoglobulins and B cells of teleost fish. Dev. Comp. Immunol. 2011, 35 (12), 1346–1365. 10.1016/j.dci.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.-J.; Wang P.; Zhang N.; Chen D.-D.; Nie P.; Li J.-L.; Zhang Y.-A. B cell Functions can Be Modulated by antimicrobial Peptides in rainbow Trout Oncorhynchus mykiss: novel insights into the innate nature of B cells in Fish. Front. Immunol. 2017, 8, 388. 10.3389/fimmu.2017.00388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Sánchez J.; Terova G.; Simó-Mirabet P.; Rimoldi S.; Folkedal O.; Calduch-Giner J. A.; Olsen R. E.; Sitjà-Bobadilla A. Skin mucus of gilthead sea bream (Sparus aurata L.). Protein mapping and regulation in chronically stressed fish. Front. Physiol. 2017, 8, 34. 10.3389/fphys.2017.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban M. Á.; Cerezuela R.. Fish mucosal immunity: skin. In Mucosal Health in Aquaculture; Elsevier, 2015; pp 67–92. [Google Scholar]
- Wei X.; Li B.; Wu L.; Yin X.; Zhong X.; Li Y.; Wang Y.; Guo Z.; Ye J. Interleukin-6 gets involved in response to bacterial infection and promotes antibody production in Nile tilapia (Oreochromis niloticus). Dev. Comp. Immunol. 2018, 89, 141–151. 10.1016/j.dci.2018.08.012. [DOI] [PubMed] [Google Scholar]
- Valavanidis A.; Vlahogianni T.; Dassenakis M.; Scoullos M. Molecular biomarkers of oxidative stress in aquatic organisms in relation to toxic environmental pollutants. Ecotoxicol. Environ. Saf. 2006, 64 (2), 178–189. 10.1016/j.ecoenv.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Pandey S.; Parvez S.; Sayeed I.; Haque R.; Bin-Hafeez B.; Raisuddin S. Biomarkers of oxidative stress: a comparative study of river Yamuna fish Wallago attu (Bl. & Schn.). Sci. Total Environ. 2003, 309 (1–3), 105–115. 10.1016/S0048-9697(03)00006-8. [DOI] [PubMed] [Google Scholar]
- McCarthy J. F.; Shugart L. R.. Biomarkers of Environmental Contamination; CRC Press, 1990. [Google Scholar]
- Dzul-Caamal R.; Salazar-Coria L.; Olivares-Rubio H. F.; Rocha-Gómez M. A.; Girón-Pérez M. I.; Vega-López A. Oxidative stress response in the skin mucus layer of Goodea gracilis (Hubbs and Turner, 1939) exposed to crude oil: A non-invasive approach. Comp. Biochem. Physiol., Part A: Mol. Integr. Physiol. 2016, 200, 9–20. 10.1016/j.cbpa.2016.05.008. [DOI] [PubMed] [Google Scholar]
- Thomsen P. F.; Kielgast J. O. S.; Iversen L. L.; Wiuf C.; Rasmussen M.; Gilbert M. T. P.; Orlando L.; Willerslev E. Monitoring endangered freshwater biodiversity using environmental DNA. Mol. Ecol. 2012, 21 (11), 2565–2573. 10.1111/j.1365-294X.2011.05418.x. [DOI] [PubMed] [Google Scholar]
- Ficetola G. F.; Miaud C.; Pompanon F.; Taberlet P. Species detection using environmental DNA from water samples. Biol. Lett. 2008, 4 (4), 423–425. 10.1098/rsbl.2008.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson C. V.; Uren Webster T. M.; Cable J.; James J.; Consuegra S. Simultaneous detection of invasive signal crayfish, endangered white-clawed crayfish and the crayfish plague pathogen using environmental DNA. Biological Conservation 2018, 222 (222), 241–252. 10.1016/j.biocon.2018.04.009. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.