ABSTRACT
Post-translational modifications are induced in stressed cells which cause them to be recognised by the system. One such modification is citrullination where the positive charged arginine is modified to a neutral citrulline. We demonstrate most healthy donors show an oligoclonal CD4 response in vitro to at least one citrullinated vimentin or enolase peptide. Unlike rheumatoid arthritis patients, these T cell responses were not restricted by HLA-DRB1 shared epitope (SE) alleles, suggesting they could be presented by other MHC class II alleles. As HLA-DP is less polymorphic than HLA-DR, we investigated whether the common allele, HLA-DP4 could present citrullinated epitopes. The modification of arginine to citrulline enhanced binding of the peptides to HLA-DP4 and induced high-frequency CD4 responses in HLA-DP4 transgenic mouse models. Our previous studies have shown that tumours present citrullinated peptides restricted through HLA-DR4 which are good targets for anti-tumour immunity. In this study, we show that citrullinated vimentin and enolase peptides also induced strong anti-tumour immunity (100% survival, p < 0.0001) against established B16 tumours and against the LLC/2 lung cancer model (p = 0.034) both expressing HLA-DP4. Since most tumours do not constitutively express MHC class II molecules, models were engineered that expressed MHC class II under the control of an IFNγ inducible promoter. Immunisation with citrullinated peptides resulted in 90% survival (p < 0.001) against established B16 HHD tumour expressing IFNγ inducible DP4. These studies show that citrullinated peptides can be presented by a range of MHC class II molecules, including for the first time HLA-DP4, and are strong targets for anti-tumour immunity.
KEYWORDS: Citrullination, cancer, HLA-DP, CD4 T cells, tumour immunotherapy
Introduction
The post-translational conversion of arginine residues to citrulline by peptidylarginine deiminase (PAD) enzymes requires millimolar concentrations of calcium.1,2 This can occur during apoptosis leading to precipitation of proteins and stimulation of CD4 and antibody responses which are associated with autoimmune diseases such as rheumatoid arthritis (RA).3,4 In RA the presentation of citrullinated epitopes is associated with SE alleles such as HLA-DR*0401 (HLA-DR4).5–7 More recently, citrullination has been shown to occur as a result of a degradation and recycling process called autophagy that is induced in stressed cells.8 However, if citrullination is a normal stress response it cannot be restricted to individuals only expressing HLA-DR4. We have previously shown in cancer patients that there is a T cell response to citrullinated peptides that is not restricted through the SE alleles. We have also shown that potent T cell responses to citrullinated vimentin and enolase in HLA-DR4 transgenic mice resulted in strong anti-tumour immunity.9,10 This response was mediated by killer CD4 T cells which secrete high amounts of IFNγ to upregulate MHC class II (MHC-II) and then directly kill the tumour cells, without the need for CD8 T cells.9,11 We have shown that tumour recognition also depends upon autophagy.9,10 In this study, we show CD4 responses in most healthy human donors to at least one citrullinated vimentin and/or enolase peptides that are not restricted to the SE allele. We show enhanced binding of citrullinated compared to wild type peptides to HLA-DP4, an allele expressed by over 70% of the Caucasian population.12 In an HLA-DP4 transgenic mouse model, we confirm that vaccination with the same citrullinated vimentin and enolase peptides can stimulate specific CD4 T cell responses which mediate efficient tumour therapy in aggressive lung and melanoma models. This suggests that there is a repertoire of T cells recognising citrullinated peptides in healthy donors that can be harnessed for cancer therapy.
Results
Citrullinated peptides stimulate responses in healthy donors
In our previous studies, we demonstrated that citrullinated vimentin and enolase peptide are presented by tumour cells since they are targeted by peptide-specific T cells and these can be targeted for tumour therapy.9,10 In these studies we also examined the responses to citrullinated vimentin and enolase peptides in humans and demonstrated that individuals showed a repertoire of responses to these peptides. Analysis of the HLA type of these donors revealed that responders did not share the HLA-DR*0401 type previously shown to be associated with citrullinated peptide-specific responses, suggesting the possibility of restriction through other HLA alleles. To determine if repertoires of T cells responding to citrullinated peptides that are restricted through alleles other than HLA-DR*0401 exist; we examined responses in healthy volunteers to two citrullinated vimentin peptides and one citrullinated enolase peptide. Figure 1Ai shows the presence of significant proliferative responses to citrullinated vimentin and enolase peptides when compared to medium alone in 14/21 (67%) of healthy volunteers. Figure 1Aii shows more detailed examples of responses in some of the donors highlighting that most donors respond to one or more of the citrullinated peptides. Phenotypic analysis of the proliferating responses demonstrated that it was CD4 + T cells proliferating in response to the citrullinated peptides rather than CD4 negative cells (including CD8+ cells). In addition, these responses appear to be predominantly to the citrullinated peptide rather than to the native (wildtype) sequence. Representative data for each peptide is shown in Figure 1(b). The citrullinated enolase and vimentin peptides have previously been shown to be restricted through HLA-DR*0401.9,10,13,14 Analysis of the HLA types of responding donors revealed that of responders only 4/14 (28%) were known to be HLA-DR*0401 positive (Table 1). In contrast 10/14 (71%) of the responding donors were known to be HLA-DP4 positive with three of unknown HLA type. Only one responding donor, BD0011, was known to be HLA-DP4 negative. HLA-DP4 is known to be expressed by up to 70% of the Caucasian population and therefore developing a vaccine relevant to HLA-DP4 positive individuals broadens the use of the vaccine.
Figure 1.
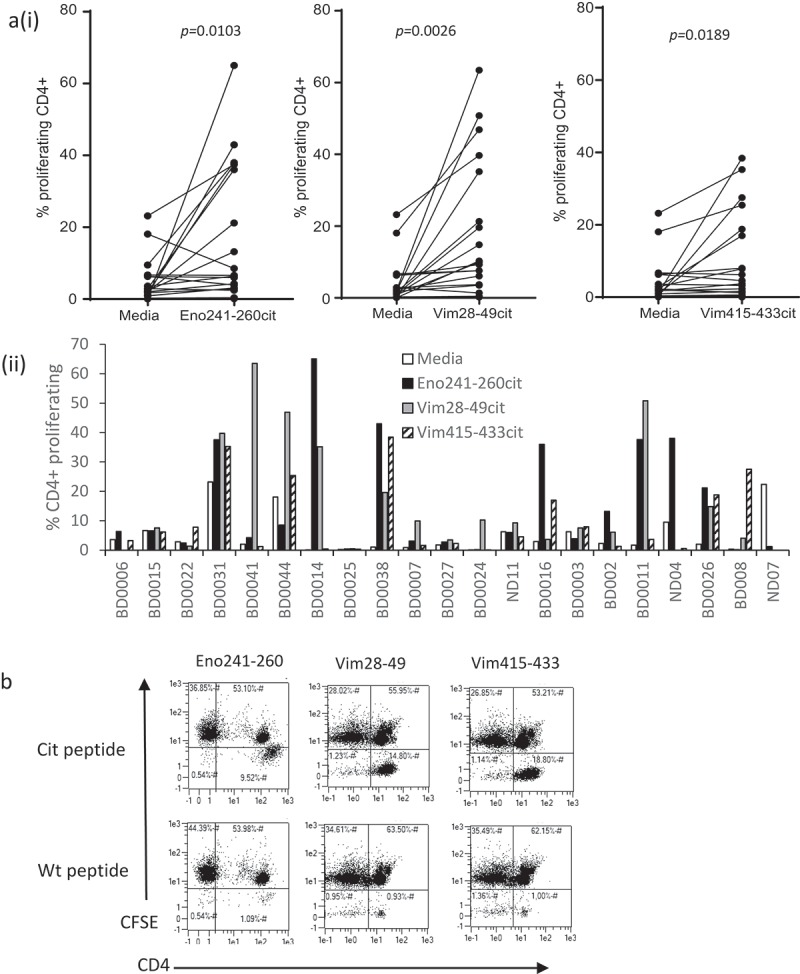
Characterisation of responses to citrullinated peptides in healthy donors. (a), Healthy donor PBMCs were analysed for proliferation in response to citrullinated vimentin or enolase peptides. Results shown as percentage proliferating CD4+ cells shown for each peptide (i) and each donor (ii). (b), Example dot plots showing proliferation of CD4 cells in response to citrullinated (cit) and native (wt) peptides. Results are representative of at least two independent experiments.
Table 1.
HLA typing of healthy donors.
| Donor | Sex | Age | HLA-A | HLA-B | HLA-C | HLA-DR | HLA-DQ | HLA-DP |
|---|---|---|---|---|---|---|---|---|
| BD0025 | F | 40–50 | 2,29 | 7,57 | 6,7 | 1,7,53a | 3,5 | 3,13 |
| BD0016 | M | 50–60 | 1,2 | 8,44 | 5,7 | 3,15,51a,52a | 2,6 | 1,4 |
| BD0008 | M | 50–60 | 1 | 8 | 7 | 3 | 2 | 1,4 |
| BD0002 | M | 40–50 | 2,29 | 44,51 | ND | 7,11 | 2,3 | 4,5 |
| BD0026 | M | 30–40 | 2 | 7,41 | 7,17 | 7,13,52a,53a | 2,3 | 1,4 |
| ND04 | F | 30–40 | 3,24 | 15,27 | 2,3 | 4, 53a | 3 | 4,9 |
| ND07 | F | 20–30 | 2,29 | 15,44 | 3,16 | 4,7,53a | 2,3 | 2 |
| BD0007 | F | 30–40 | 1, 32 | 8, 15 | 7 | 3,13,51a,51c | 2,6 | 4,13 |
| BD0024 | F | 30–40 | 2 | 7,27 | 1,7 | 4,15,53a, 51a | 3,6 | 4 |
| BD0027 | M | 40–50 | 1,11 | 8,57 | 6,7 | 7,8 | 3,4 | 4 |
| BD0011 | M | 20–30 | 33,74 | 18,49 | 07 | 13,15 | 05,06 | 17,18 |
| BD0003 | M | 40–50 | 11,29 | ND | ND | 4,13 | 3,6 | 4 |
| ND11 | M | 40–50 | 02,03 | 07,40 | 03,07 | 12,13 | 03,06 | 02,04 |
| BD0038 | F | 30–40 | 26,33 | 40,58 | 03,03 | 09,11 | 03,03 | 04,05 |
| BD0014 | F | 30–40 | Not available | |||||
| BD0006 | F | 20–30 | Not available | |||||
| BD0015 | F | 40–50 | 03,24 | 07,15 | 03,07 | 04,15 | 03,06 | 04 |
| BD0022 | F | 30–40 | 01,02 | 35,50 | 06,12 | 04,07 | 02,03 | 02,04 |
| BD0031 | M | 20–30 | Not available | |||||
| BD0041 | F | 50–60 | 01,24 | 07,40 | 03,07 | 04,11 | 03,03 | 02,04 |
| BD0044 | F | 20–30 | Not available | |||||
Grey highlight = responding donors, M = male, F = female
Examination of TCR clonality of the responding CD4 T cells revealed a bias of TCR Vβ and Vα sequences among CD4+ proliferating cells (CFSElow) from donor BD0008 to the vimentin 28–49cit peptide and from donor BD0011 to the vimentin 415–433cit peptide on comparison with the non-proliferating CD4 population. The TCRα and β CDR3 tree maps from both donors demonstrate that the non-proliferating CFSEhigh CD4 cells express highly diversified CDR3 nucleotide sequences compared to those expressed in proliferating CFSElow CD4 cells that show a dramatic increase in the relative frequency of a subset of CDR3 sequences, suggesting a more focused (less diverse) repertoire (Figure 2(a–b)). This was confirmed by the much lower diversity index (D50) of the CDR3 sequences from the proliferating CD4 T cells compared to the non-proliferating CD4 T cells (Table 2) which is a reflection of the percent of dominant and unique T clones that account for the cumulative 50% of the total CDR3s counted in the sample, where a more diverse library, exhibits a value close to 50. The oligoclonal nature of the proliferating CD4 T cells, with a more focused TCR repertoire, is further corroborated by the distribution of the TCRα and β V usage, where an increase in frequency of a subset of germline V alleles is apparent, in contrast to the non-proliferating CD4 cells from the same cultures (Figure 2(c–d)).
Figure 2.
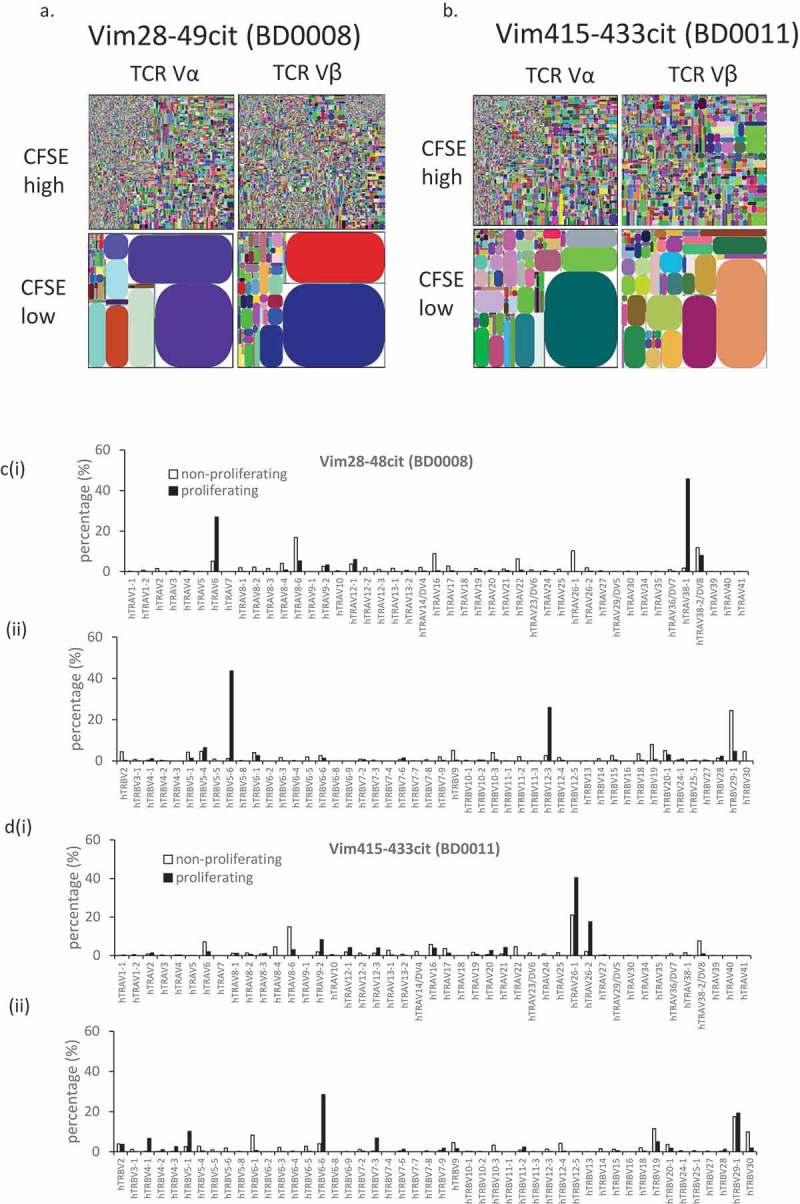
TCR α and β repertoire diversity in CD4+ve CFSE high/low cells responding to citrullinated peptides. Tree maps depicting TCR α and β chain CDR3 clonotype usage in relation to repertoire size in CD4+ve CFSE high/low cells on incubation with citrullinated peptides vimentin 28–49 from donor BD0011 (a) and vimentin 415–433 from donor BD0008(b). Each rectangle in a tree map represents a unique CDR3 nucleotide sequence and the size of each rectangle denotes the relative frequency of an individual sequence. The colours for the individual CDR3 sequences in each tree map plot are chosen randomly and thus do not match between plots. Histogram showing % expression of TCRVα (i) and TCRVβ (ii) chains among proliferating and non-proliferating CD4+ve cells in donors BD0008 (c) and BD0011 (d) to citrullinated peptides vimentin 28–49 and vimentin 415–433.
Table 2.
CDR3 diversity values for the TCR α and β chain from the CD4+ CFSEhigh and CFSElow cells obtained from donors incubated with peptides.
| DONOR | PEPTIDE | SAMPLE | Diversity Index (D50) |
|
|---|---|---|---|---|
| TRA | TRB | |||
| BD0008 | Vimentin 28-49 cit | CD4+ CFSEHigh | 12.8 | 17 |
| CD4+ CFSELow | 0.4 | 0.2 | ||
| BD0011 | Vimentin 415-433 cit | CD4+ CFSEHigh | 9.9 | 5.9 |
| CD4+ CFSELow | 0.7 | 0.5 | ||
Citrullinated peptides bind to HLA-DP4
To assess if responses to the citrullinated (cit) peptides could be restricted through HLA-DP4, the peptides were tested for binding to HLA-DP4. Binding was compared to a known HLA-DP4 restricted peptide from Hepatitis B surface antigen and two peptides from fibrinogen and collagen II that have been shown in the literature15 not to bind to HLA-DP4. In our assay, the biotinylated Hepatitis B peptide showed good binding to HLA-DP4 whereas peptides from fibrinogen and collagen II demonstrated minimal binding over control (Figure 3(a)). The unmodified vimentin 415–433 and 28–49 peptides showed low binding to HLA-DP4 that was not significant over negative peptides but the citrulline modification dramatically increased this binding (p < 0.0001) (Figure 3(a)). The enolase 241–260 peptide showed binding of the unmodified peptide with less difference between binding of the modified and unmodified peptides although the binding of the citrulline-containing peptide was significantly better than the native sequence (p = 0.0244) (Figure 3(a)). To investigate if the addition of biotin to the peptides influenced the binding to HLA-DP4, the binding of the biotinylated Hepatitis B peptide was assessed in the presence of an equal amount of non-biotinylated Hepatitis B peptide. The binding was 50% inhibited by an equal amount of non-biotinylated Hepatitis B peptide (Figure 3(b)), thus demonstrating that the addition of biotin to the peptide had little effect upon the peptide binding to HLA-DP4. To further confirm the binding specificity of the native (un-biotinylated) peptides for the HLA-DP4 allele, the peptides were added in the presence of biotinylated Hepatitis B peptide (previously demonstrated to bind strongly to HLA-DP4) and the degree of inhibition evaluated. The citrullinated vimentin and enolase peptides showed a significant inhibition (p < 0.0001) of binding of the biotinylated Hepatitis B peptide suggesting that they are binding specifically to the HLA-DP4 allele on the membrane prep (Figure 3(c)). The wildtype peptides and negative control peptides from fibrinogen and collagen showed no inhibition of Hepatitis B peptide binding. In addition to this, the citrullinated vimentin and enolase peptides and negative control fibrinogen peptides were titrated against a known concentration of biotinylated Hepatitis B peptide. Figure 3(d) shows that the native Hepatitis B peptide effectively competes with its biotinylated format, with a 1:1 ratio of biotinylated to non-biotinylated peptide causing 50% inhibition in signal. A higher amount of the citrullinated vimentin and enolase peptides are required to achieve 50% inhibition, suggesting these peptides are of an apparent 2–3 fold lower binding affinity compared to the Hepatitis B peptide.
Figure 3.
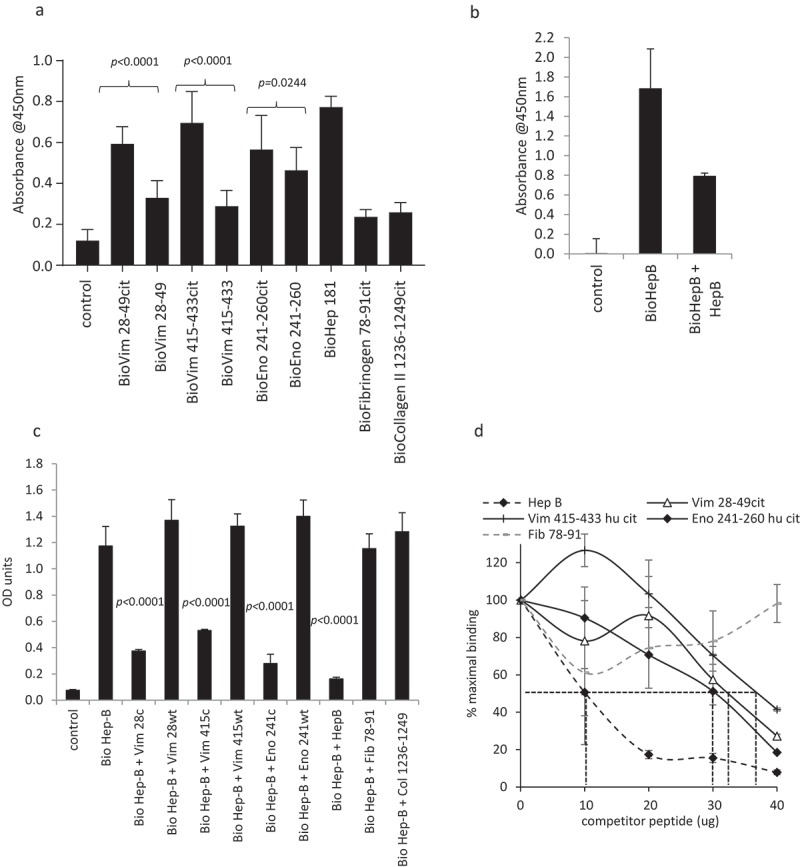
Citrullinated peptides bind to HLA-DP4. Direct binding of biotinylated citrullinated (cit) and native (wt) peptides to HLA-DP4 (a). Binding of 10 µg biotinylated Hepatitis B (HepB) peptide in the presence and absence of 10 µg non-biotinylated HepB 181–193 peptide (b). Competition of 40 µg non-biotinylated competitor peptides in the presence of 10 µg biotinylated HepB 181–193 peptide (c). Titration of non-biotinylated competitor peptide with 10 µg biotinylated HepB 181–193 peptide (d). Results are representative of at least two independent experiments.
The enhanced HLA-DP4 binding as a result of the citrulline modification suggests that peptide/MHC binding affinity may play a role in the induction of HLA-DP4 restricted immune responses to these citrullinated peptides. However, the modified enolase 241–260 sequence demonstrated only a small difference in HLA-DP4 binding affinity over the native sequence, therefore, it is possible that other factors such as TCR contact can play a role in the generation of HLA-DP4 restricted responses.
Responses to citrullinated peptides can be induced in HLA-DP4 transgenic mice
In light of the responses in healthy individuals and HLA binding data, we screened the two citrullinated vimentin and the enolase peptides in a HLA-DP4 transgenic mouse model. HLA-DP4 transgenic mice express human CD4 molecule and lack the expression of endogenous mouse MHC class I and II alleles which are replaced by transgenic HLA-A2 (HHDII) and human HLA-DP4 molecules. HLA-DP4 transgenic mice were vaccinated with the peptides combined with TLR9 and TLR4 agonists, and responses measured ex vivo by IFNγ ELISpot assay. Mice show high-frequency responses to the three citrullinated peptides in HLA-DP4 transgenic mice (p < 0.0001) with minimal cross-reactivity to the wildtype (wt) peptide (Figure 4(a)). Responses are also detected to a known HLA-DP4 peptide from Hepatitis B (Figure 4Aiv). No responses to citrullinated peptides are seen in C57Bl/6 mice or HHDII/DR1 transgenic mice (Supplementary Figure 1) suggesting that these responses were dependent upon the HLA-DP4 allele. To confirm if these responses were CD4 mediated, responses were analysed in the presence of CD4 or CD8 blocking antibodies. Responses were lost in the presence of the CD4 blocking antibody (p < 0.0001 for vimentin 28-49cit and enolase 241–260cit, p = 0.0002 for vimentin 415-433cit) but not affected to the same extent in the presence of the CD8 blocking antibody suggesting these are CD4 mediated responses in the HLA-DP4 transgenic mice (Figure 4(b)). The vimentin 28-49 sequence is homologous between humans and mice, however, the vimentin 415-433 and enolase 241-260 sequences have two and one amino acid differences, respectively, between the human and murine sequences, therefore, responses induced with the human peptide sequences were checked for cross-reactivity to the murine homologues in HLA-DP4 transgenic mice (Figure 4(c)). Responses to both human sequences showed cross-reactivity to the murine counterparts. In addition to this mice were immunised with the murine peptide sequences and showed the generation of responses specific to the citrullinated peptides and not to the wt (Supplementary Figure 2). This confirms that the human peptides behave similarly to the murine peptides and were suitable for study in the mouse model. Peptides used encompassed 19–21 amino acids. In an attempt to map a shorter peptide sequence for each epitope, responses induced with the vimentin 28-49cit, 415-433cit and enolase 241-260cit peptides were tested for reactivity to shorter peptides spanning the longer sequences. Responses showed cross-reactivity with the shorter peptide epitopes vimentin 28–42cit, 418–431cit and enolase 241–255cit in the HLA-DP4 transgenic mice where similar levels of responses were seen (Figure 4(d)). This was consistent with findings in the HLA-DR4 transgenic mice mapping shorter peptide sequences to vimentin 418-431, vimentin 28-42 and enolase 241-255 (Supplementary Figure 3).
Figure 4.
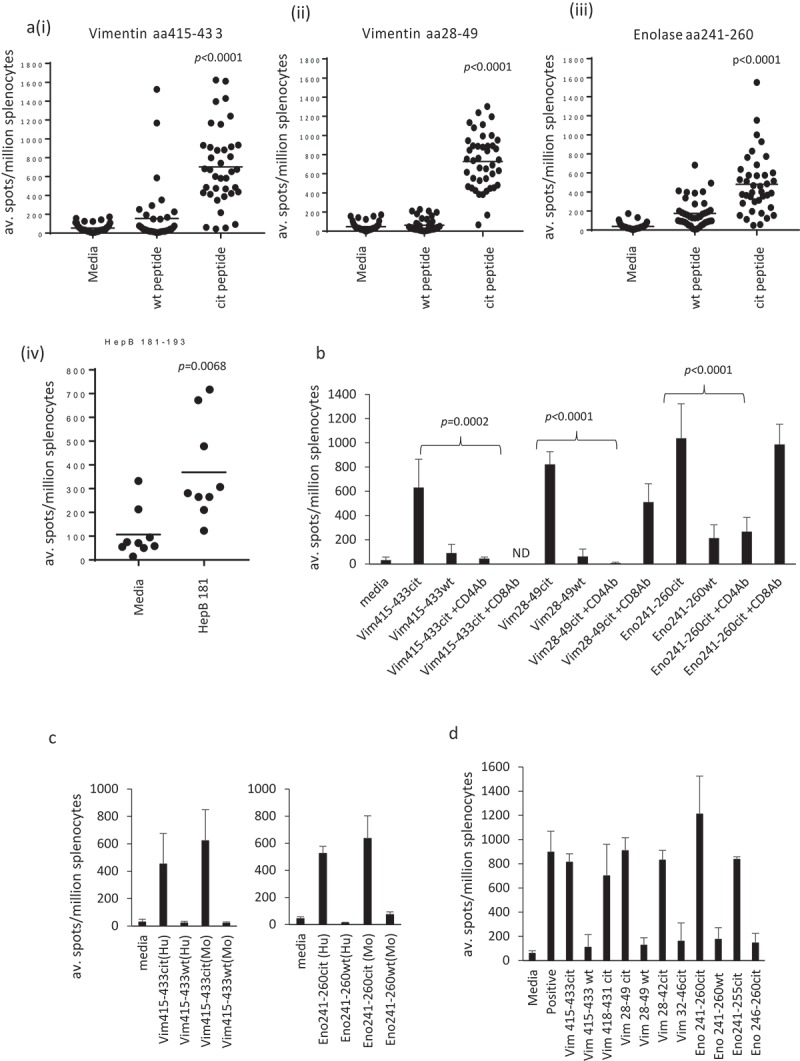
Citrullinated vimentin and enolase peptides stimulate CD4 responses in HLA-DP4 transgenic mice. HLA-DP4 transgenic mice were immunised with citrullinated Vim28–49 (ii), Vim415–433 (i), Eno241-260 peptides (iii) or HepB 181–193 (iv) and immune responses specific to the citrullinated or native (wt) peptides were monitored by IFNγ Elispot assay and compared to control (a). Immune responses were assessed in the presence of CD4 or CD8 blocking antibodies (b). Immune responses to the citrullinated human (Hu) Eno241-260 and Vim415-433 peptides were tested for cross-reactivity to the murine (Mo) peptides (c). Immune responses in HLA-DP4 mice were mapped to shorter peptide sequences (d). Results are representative of at least two independent experiments in which n = 3.
Citrullinated peptides provide efficient tumour therapy against B16 tumours in both HLA-DR4 and HLA-DP4 transgenic mice
We have previously shown that citrullinated vimentin and enolase peptides presented by tumour cells can be good targets for CD4 T cells and these provide good tumour therapy in HLA-DR4 transgenic models. The B16 tumour model lacking endogenous murine MHC class I and II alleles was engineered to express HHDII and HLA-DP4 for use in HLA-DP4 transgenic mice. Cell lines engineered to express inducible HLA-DR4 or HLA-DP4 demonstrated a similar growth rate in the transgenic mice compared the parental B16F1 line in C57Bl/6 mice (p = 0.5673 and p = 0.2166, respectively). The engineered models also demonstrated no significant difference in overall survival (Supplementary Figure 4). To determine if responses to the citrullinated peptides induced similar anti-tumour responses in HLA-DP4 mice to HLA-DR4 mice, we challenged mice with tumour and subsequently vaccinated with citrullinated or wildtype peptide plus adjuvant. Each of the three citrullinated peptides in combination with TLR9/TLR4 ligands provided tumour therapy in a B16 melanoma model constitutively expressing HLA-DP4 or HLA-DR4 that was significantly better than the wildtype peptide (Figure 5(a)), suggesting that these epitopes are presented in this tumour model. A study was also performed with the murine peptide sequences confirming that the similar immune responses also translate to tumour therapy (Supplementary Figure 5). These responses were as effective in the HLA-DP4 model as in the HLA-DR4 model (Figure 5(a) i and ii). The combination of all three citrullinated peptides provided 100% survival in the HLA-DP4 transgenic mouse model (p > 0.0001) (Figure 5(b)). Since most tumours do not constitutively express MHC class II molecules a model was engineered that expressed HLA-DP4 under the control of an IFNγ-inducible promoter. Analysis of the combination of citrullinated peptide-specific responses induced in the presence of TLR9/TLR4 ligands in the tumour model expressing HLA-DP4 under the inducible promoter showed efficient tumour therapy (p < 0.0001) even when expression of MHC class II on the tumour requires induction by IFNγ (Figure 5(c)). Since CD4 T cells can have both direct and indirect (via effects on infiltrating APCs and CD8 T cells) impact on the tumour, the role for direct recognition of the tumour by CD4 T cells was also assessed by the use of a tumour model unable to express HLA-DP4 (Figure 5(d)). No tumour therapy was seen when the tumour cells were unable to express HLA-DP4 suggesting that the tumour cells present the citrullinated peptides on HLA-DP4 which are a direct target for CD4 T cell responses. To examine the efficacy of the immune response against established tumours HLA-DP4 mice were given a single immunisation of the three citrullinated peptides at either 3, 7 or 14 days post tumour implant. Single immunisations at days 3, 7 or 14 were able to efficiently prevent tumour growth and resulted in 90% (p < 0.0001), 60% (p = 0.0023) and 40% (p = 0.025) survival, respectively (Figure 5(e)).
Figure 5.
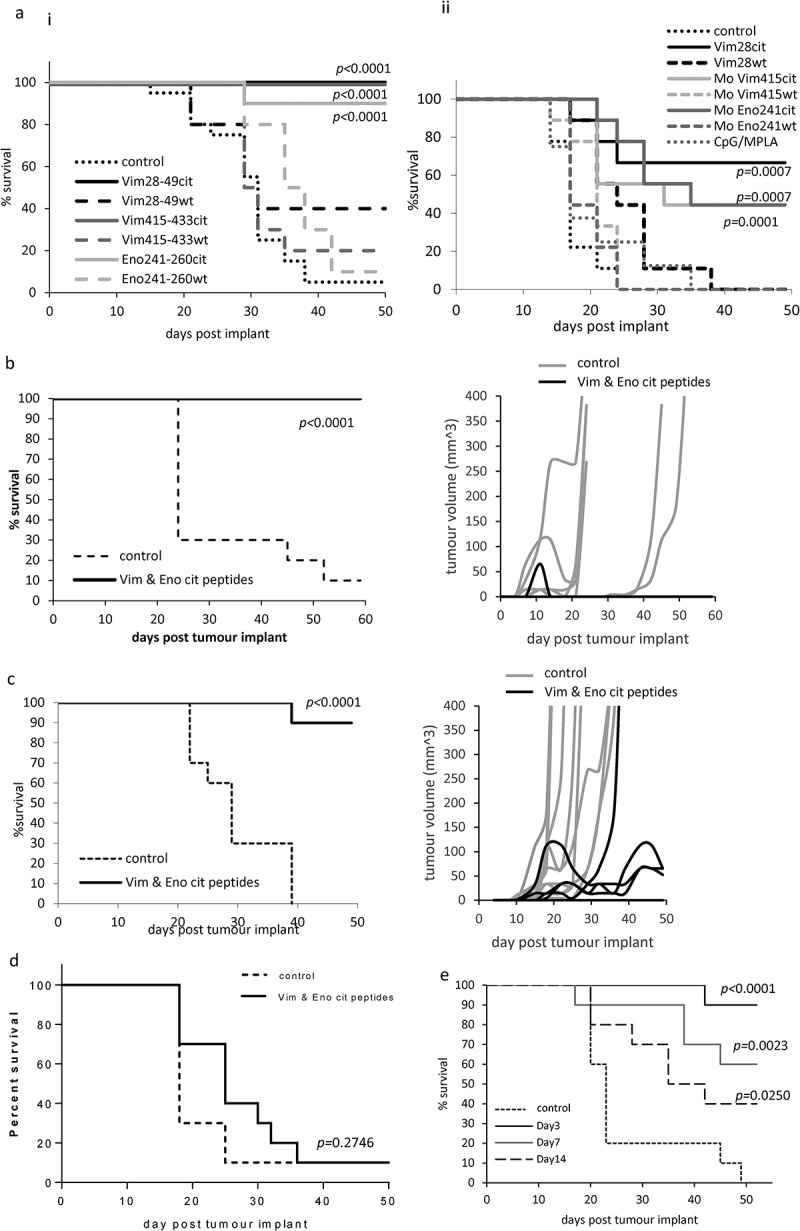
Citrullinated vimentin and enolase peptide vaccination provide tumour therapy in HLA-DR4 and HLA-DP4 transgenic mice. (a), HLA-DP4 (i) or DR4 (ii) transgenic mice were challenged with B16 cells constitutively expressing HLA-DP4 or HLA-DR4 and four days later mice were immunised with citrullinated or native Vim28-49, Vim415-433 or Eno241-260 peptides and tumour growth and survival monitored. HLA-DP4 transgenic mice were challenged with B16 cells constitutively expressing HLA-DP4 (b), expressing HLA-DP4 under an IFNγ inducible promoter (c) or expressing no HLA-DP4 (d) and four days later mice were immunised with combination of citrullinated Vim28-49, Vim415-433 and Eno241-260 peptides and tumour growth and survival monitored. (e), HLA-DP4 transgenic mice were challenged with B16 cells expressing HLA-DP4 under an IFNγ inducible promoter followed by a single vaccination at days 3, 7 or 14. Tumour growth and survival was monitored. N = 10/group.
Citrullinated peptides also provide efficient tumour therapy against lung tumours in both HLA-DP4 and HLA-DR4 transgenic mice
Vimentin and enolase are proteins expressed by many cells and are therefore potential targets for treatments of a wide range of cancers. In addition to the B16 melanoma model, we examined the effect of the citrullinated peptides in combination with TLR9/TLR4 ligands for the therapy of the murine LLC/2 lung tumour line in the HLA-DR4 and HLA-DP4 mouse models. LLC/2 cells were shown to be positive by Western blot for the expression of vimentin and enolase (Supplementary Figure 6). Immunisation of mice in the HLA-DP4 model with the combination of two citrullinated vimentin peptides and the citrullinated enolase peptide showed significant delay in tumour growth over control (p < 0.0001) in the aggressive lung tumour model expressing HLA-DP4 (Figure 6(a)). The two citrullinated vimentin peptides or the citrullinated enolase peptide also showed significant delay in tumour growth (p = 0.034 and p = 0.0456) of the aggressive lung tumour model in HLA-DR4 mice (Figure 6(b)).
Figure 6.
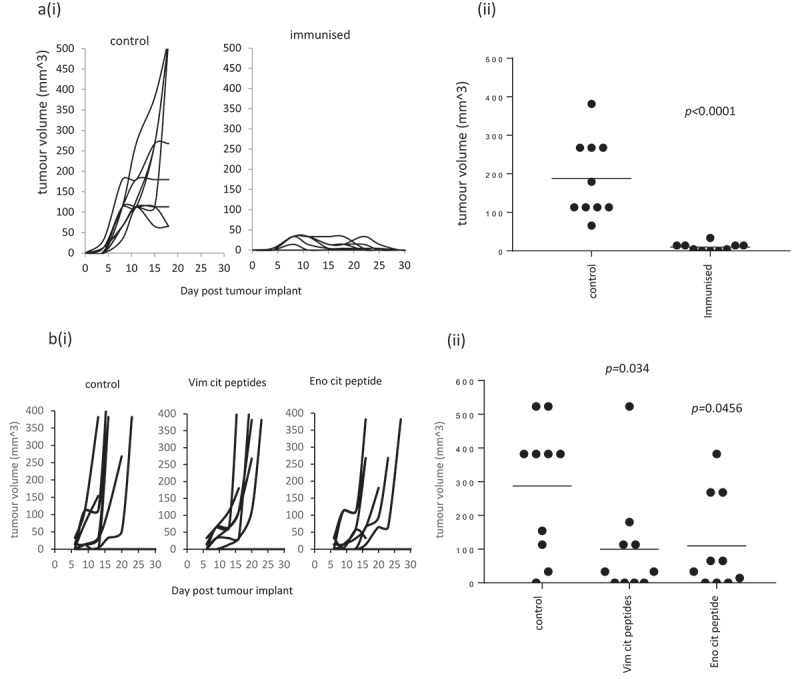
Citrullinated peptides provide efficient therapy of lung tumours. HLA-DP4 (a) or HLA-DR4 (b) transgenic mice were challenged with LLC/2 cells constitutively expressing HLA-DP4 or HLA-DR4. Four days later mice were immunised with citrullinated Vim28-49, Vim415-433 and Eno241-260 peptides and tumour growth monitored. Tumour growth curves (i) and tumour volume (ii) in the HLA-DP4 model at day 15 and in HLA-DR4 model at day 16 are shown. N = 10/group.
Discussion
Citrullinated peptides have been shown to be presented on MHC class II via autophagy in antigen presenting cells.8 We have shown a similar mechanism in tumour cells whereby as a result of autophagy citrullinated peptides are presented in the context of HLA-DR*0401 and recognised by cytotoxic CD4 T cells.9 As autophagy is increased under stressful conditions it seemed plausible that citrullinated peptides could be a general mechanism to alert the immune system to recognise and remove stressed cells. The assumption is therefore that citrullinated peptides must be presented on a range of HLA alleles. In this study, we show that most healthy donors show a CD4 T cell response to one or more citrullinated vimentin and enolase peptides suggesting that this is a common occurrence. The proliferation responses were highly oligoclonal and different between peptides suggesting that these cells were selectively responding to each peptide. In contrast, the non-proliferating cells had a very diverse repertoire suggesting that they had not responded. Indeed, the predominant clones in the proliferating cells were poorly represented in the non-proliferating cells. Citrullinated peptides can be presented in the thymus 16 but responses in healthy donors suggested that the T cells recognising them are positively but not negatively selected. Previous studies in RA patients have shown that citrullinated peptides are predominantly expressed by SE alleles,3,13,17 but in line with our previous studies9,10 the healthy donors in this study did not show this restriction. Indeed, the best correlation with response was expression of the HLA-DP4 restriction with 71% of the responding donors known to express this haplotype and 21% of unknown haplotype. Only one responding donor was known to be SE and HLA-DP4 negative. There have been limited publications suggesting citrullinated peptides can also preferentially bind to HLA-DR9 and HLA-DQ2,7 and 8.18,19 None of our donors expressed HLA-DR9, HLA-DQ7 or HLA-DQ8. 43% of the responding donors expressed HLA-DQ2 with 21% unknown suggesting these alleles could have presented the citrullinated peptides in these donors. There are no previous reports of HLA-DP4 presenting citrullinated peptides.
To confirm this observation, we showed that the citrullinated peptides bound more strongly to HLA-DP4 than the arginine-containing peptides. It has been previously thought that the conversion of arginine to citrulline enhances the binding of some peptides to HLA-DR and DQ alleles.19,20 In contrast it has also been shown that the conversion of arginine to citrulline does not always lead to enhanced peptide MHC class II binding affinity.15 We have also shown that HLA-DP4 transgenic mice make strong Th1 responses to human citrullinated vimentin and enolase peptides that do not cross react with wild type peptides. These responses also cross react with the homologous murine sequences suggesting a breaking of tolerance similar to the scenario that would be encountered in human patients. Increasing evidence is emerging that HLA-DP molecules can present epitope peptides in the context of infectious disease, allergy and cancer.21–24 HLA-DP alleles appear more conserved than DR or DQ alleles, with five alleles frequently expressed in the worldwide population that cover approximately 90% of individuals.25 Recent evidence also suggests that these common HLA-DP alleles can share a HLA supertypic binding specificity.25 It is therefore possible that the citrullinated peptides discussed could show binding to other HLA-DP alleles in addition to HLA-DP4. Since citrullination occurs in times of cellular stress and these ‘stressed cells’ would require clearance by the immune system, the potential to alert the immune system to this through the presentation of citrullinated peptides via MHC class II would be beneficial. Therefore, the reduced polymorphism among HLA-DP alleles suggests these as prime candidates for the presentation of peptides in this universal process and may point to a role of HLA-DP alleles in the clearance of stressed cells. Indeed van Lith et al. have shown that HLA-DP does not require an invariant chain or HLA-DM to form stable dimers making it more accessible to peptides produced during autophagy.26,27 Other studies have shown that HLA-DP does not bind CLIP fragments,28 and known HLA-DP peptide-binding motifs differ from those of (ER-loaded) MHC molecules, so HLA-DP is not likely to compete for classical class II-binding peptides.29 The reported lower expression of HLA-DP molecules30,31 most likely plays a role to avoid autoimmunity and promote self-tolerance. In contrast, co-expression on HLA-DP4 and HLA-DR4 may push T cells over the threshold and result in autoimmune disease.
Our responding healthy donors and our HLA-DP4 transgenic mice immunised with the citrullinated vimentin and enolase peptides induced CD4 T cell responses. These CD4 T cells mediated, anti-tumour immunity in HLA-DP4 transgenic mice against B16 tumours expressing either constitutive or IFNγ inducible HLA-DP4. CD4 T cells are also able to exert indirect effects upon the tumour via the activation of M1 macrophages and provision of help for CD8 T cells.32–34 To address this possibility, a model was used where the tumour cells did not express the relevant MHC class II allele. The loss of tumour therapy in the absence of MHC class II expressed by the tumour implies in this model that the anti-tumour effect is mediated by direct effects of the CD4 T cells upon the tumour. The anti-tumour immunity was similar in HLA-DP4 and HLA-DR4 mice suggesting both alleles can equally present the citrullinated epitopes on tumours. Indeed, a single immunisation with the combination of citrullinated vimentin and enolase peptides induced significant anti-tumour immunity even 14 days after tumours were established. To show that these anti-tumour responses were not restricted to B16 melanoma, similar results were also obtained against the HLA-DP4 expressing Lewis lung carcinoma line, LLC/2. Immunised mice demonstrating strong tumour rejection showed no evidence of toxicity suggesting healthy cells do not present these modified epitopes. Indeed, it has been shown that RA cannot be induced by T cells alone but requires joint erosion, antibody responses and inflammation. This is borne out by studies where no autoimmune symptoms were observed with T cells alone, even in HLA-DR4 transgenic mice which are susceptible to RA.35 Our studies suggest citrullinated vimentin and enolase peptides could be used to stimulate strong anti-tumour immune responses in both HLA-DR4 and HLA-DP4 individuals.
Materials and methods
Laboratory practice
These studies were conducted in a laboratory that operates under exploratory research principles. Standard operating procedures were used for all human and mouse T cell assays. These studies were performed using general research investigative assays. Procedures and raw data can be obtained from Scancell Ltd through corresponding author and may be subject to non-disclosure agreements. Unless otherwise stated all reagents were obtained from Sigma-Aldridge.
Cell lines and culture
The murine melanoma B16F1 cell line (ATCC-CRL-6323) and murine lung carcinoma line LLC/2 (ATCC-CRL-1642) were obtained from the American Type Culture Collection (ATCC). B16F1 cell line was cultured in RPMI medium 1640 (GIBCO/BRL) supplemented with 10% Fetal Calf Serum (FCS), L-glutamine (2mM) and sodium bicarbonate buffered; LLC/2 was cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% FCS. The cell lines utilised were certified mycoplasma free, authenticated by suppliers (STR profiling) and used within 10 passages.
Peptides and adjuvants
Peptides vimentin amino acid (aa)28-42cit (cit-SYVTTST-cit-TYSLGS), aa28-42wt (RSYVTTSTRTYSLGS), aa28-49wt (RSYVTTSTRTYSLGSALRPSTS), aa28-49cit (cit-SYVTTST-cit-TYSLGSAL-cit-PSTS), aa32-46cit (TTST-cit-TYSLGSAL-cit-P), human aa415-433wt (LPNFSSLNLRETNLDSLPL), human aa415-433cit (LPNFSSLNL-cit-ETNLDSLPL), mouse aa415-433wt (LPTFSSLNLRETNLESLPL), mouse aa415-433cit (LPTFSSLNL-cit-ETNLESLPL), human aa418-431cit (FSSLNL-cit-ETNLDSL), human enolase aa241-260cit (VIGMDVAASEFF-cit-SGKYDLD), human aa241-260wt (VIGMDVAASEFFRSGKYDLD), mouse aa241-260cit (VIGMDVAASEFY-cit-SGKYDLD), mouse aa241-260wt (VIGMDVAASEFYRSGKYDLD), human aa 241-255cit (VIGMDVAASEFF-cit-SG), human aa 246-260cit (VAASEFF-cit-SGKYDLD), fibrinogen aa78-91cit (NQDFTN-cit-INKLKNS), collagen II aa1236-1249cit (LQYM-cit-ADQAAGGLR)15 and Hepatitis B surface antigen aa 181–19336 were synthesized at >90% purity by Genscript (USA) and stored lyophilised at −80°C. On the day of use they were reconstituted to the appropriate concentration in phosphate buffered saline (PBS).
Adjuvants used include TLR9 agonist CpG ODN 1826 (Invivogen) and TLR4 agonist monophosphoryl lipid A (MPLA; Sigma); both used at a dose of 5µg/mouse/immunisation.
Plasmids
The construction of the plasmid pVITRO2 Chimeric HLA-DR401 and the IFNγ inducible plasmid pDCGAS chimeric HLA-DR401 have been described previously.9 The HHDII plasmid pCDNA3 HHDII is described in detail elsewhere.37
To generate the plasmid pVITRO2 Human HLA-DP4, the nucleotide sequence encoding the full length human HLA-DPA*0103 α chain flanked by FspI/EcoRI and the HLA-DPB*0401 β chain flanked by BamHI/SalI restriction sites were synthesized (Eurofins MWG). Following sequence confirmation, the HLA-DPA*0103 chain was cloned into the FspI/EcoRI mcs2 of the vector pVITRO2-hygro-mcs (Invivogen). The HLA-DPB*0401 chain was subsequently inserted into the BamHI/SalI mcs1 of the mammalian expression vector alongside the alpha HLA-DPA*0103 chain present within mcs2. To construct the IFNγ inducible plasmid pDCGAS Human HLA-DP4, the HLA-DPA*0103α and HLA-DPB*0401β chains, were sequentially cloned into the pDCGAS chimeric HLA-DR401 plasmid in replacement of the chimeric DR4 chains described elsewhere.9 The IFNγ inducible promoter within this plasmid consists of a TATA box and the GAS (IFNγ activated sequence) direct repeat enhancer element that in the presence of IFNγ drives expression of the HLA-DP401 chains within the pDCOrig vector backbone. After sequence confirmation endotoxin-free plasmid DNA was generated using the endofree Qiagen maxiprep kit (Qiagen, Crawley).
Transfection and flow cytometry
B16F1 melanoma and LLC/2 lung carcinoma cells expressing HLA-DR4 under expression of the constitutive and IFNγ inducible promoters have been described previously.9
The B16F1 cell line previously knocked out for murine MHC class I and II by Zinc finger Technology (Sigma Aldrich) was transfected using Lipofectamine LTX with plus reagent (Invitrogen), with 4 µg of each plasmid, pCDNA3 HHDII in combination with either the pVITRO2 Human HLA-DP4 or pDCGAS Human HLA-DP4 plasmids, where DP4 is under expression of the constitutive or IFNγ inducible promoter, respectively. Transfected cells were selected by growth in the presence of G418 (500µg/ml) with either Hygromycin B (300µg/ml) or Zeocin (300µg/ml). The LLC/2 lung carcinoma cell line was also transfected with the pVITRO2 Human HLA-DP4 plasmid and selected on incubation with media supplemented with Hygromycin B (300µg/ml). Lines were cloned by limiting dilution and expression was confirmed by flow cytometry using the anti-human beta 2 microglobulin FITC (clone TU19, BD Biosciences) and anti-human HLA-DR/DP/DQ (clone WR18, Abcam) PE antibodies. Cells transfected with the IFNγ inducible plasmid were incubated overnight in the absence or presence of murine IFNγ (50ng/ml, Gibco Life Technologies) prior to staining with the antibody.
HLA-DR4 and DP4 binding studies
Binding to HLA-DP4 was assessed by extraction of membrane fractions from B16HHDII/DP4 cells using Mem-PER™ Plus Membrane Protein Extraction Kit (Thermofisher-Scientific) according to manufacturer’s instructions. Membrane preps containing HLA-DP4 were incubated with 10 µg biotinylated peptide for 4hrs at 37°C. For competition assays, the 10 µg biotinylated Hepatitis B peptide was incubated in the presence of specified concentrations of non-biotinylated peptides. Biotinylated peptide bound to HLA-DP4 was visualised by capture onto streptavidin-coated ELISA plates and detected with anti-HLA-DP antibody clone B7/21 (Leinco Technologies Inc, USA) and anti-mouse IgG3 HRP antibody (Invitrogen, UK). Binding was quantified with TMB substrate and absorbance read at 405nm wavelength.
Immunisation protocol
HLA-DR4 mice (Model #4149, Taconic), HLA-A2.1+/+ HLADP4+/+ hCD4+/+ (HLA-DP4) transgenic mice (EM:02221, European Mouse Mutant Archive), HHDII/DR1 mice (Pasteur Institute) or C57Bl/6 mice (Charles River) aged between 8 and 12 weeks were used. All work was carried out under a Home Office approved project licence. For all studies, the mice were randomised into different groups but not blinded to the investigators. Peptides were dissolved in PBS and then emulsified with CpG (ODN 1826) and MPLA and delivered at 25 µg dose unless stated otherwise. Adjuvants were used at 5µg/dose. Peptides in adjuvant were injected subcutaneously at the base of the tail. Mice were immunised on day 0, 7 and 14 and spleens were removed for analysis at day 20.
For tumour challenge experiments mice were challenged with 2.5 × 104 B16 DR4 cells, 4 × 105 B16 HHDII DP4 cells, 1.5 × 106 LLC/2 DR4, 2.5 × 106 LLC/2 DP4, 5 × 104 B16 inducible DR4 or 1 × 105 B16 HHDII inducible DP4 cells subcutaneously on the flank 3 days prior to primary immunisation and then immunised as above unless stated otherwise. Tumour growth was monitored at 3–4 days intervals and mice were humanely euthanised once tumour reached ≥10 mm in diameter. Tumour volume was estimated using the following formula, volume = (π/6)x(LxW2), where L is length and W is width.
Ex vivo ELISpot assay
ELISpot assays were performed using murine IFNγ capture and detection reagents according to the manufacturer’s instructions (Mabtech). In brief, the IFNγ specific antibodies were coated onto wells of 96-well Immobilin-P plate. Synthetic peptides (10µg/ml) and 5 × 105 per well splenocytes were added to the wells of the plate in quadruplicate and plates incubated for 40 h at 37°C in an atmosphere of 5% CO2. Where relevant anti-CD4 (mouse clone GK1.5 or human clone OKT4, BioXcell) or CD8 (mouse clone 2.43, BioXcell) blocking antibodies were added to splenocytes at 20µg/ml for 15 min prior to stimulation with peptide. Lipopolysaccharide (LPS) at 5µg/ml was used as positive control. After incubation, captured IFNγ was detected by biotinylated specific IFNγ antibodies and developed with a streptavidin alkaline phosphatase and chromogenic substrate. Spots were analysed and counted using an automated plate reader (Cellular Technologies Ltd).
Peripheral blood mononuclear cell (PBMC) isolation
PBMC experiments were carried out with ethical approval. Demographics of healthy donors are shown in Table 1. Peripheral blood samples (approx. 50 ml) were drawn into lithium heparin tubes (Becton Dickinson). Samples were maintained at room temperature and processed immediately the following venepuncture. PBMCs were isolated by density gradient centrifugation using Ficoll-Hypaque. Proliferation assays were performed immediately after PBMC isolation. The median number of PBMCs routinely derived from healthy donors was 1.36 × 106 PBMC/ml whole blood (range 0.6 × 106–1.8 x 106/ml). The median viability as assessed by trypan blue exclusion was 90.6% (range 80–97%). For CD25 depletion PBMCs were processed as above and then immediately enriched with anti-CD25 microbeads and MACS cell separation columns (Miltenyi).
Proliferation assay
CD25 depleted PBMCs were subjected to carboxyfluorescein succinimidyl ester (CFSE; Thermofisher) labelling with 5µM CFSE for 5 min at room temperature in PBS 5% FCS followed by a wash in a 10-fold excess of PBS 5% FCS. Cells were resuspended at 1.5-2x106/ml, cultured with 10μg/ml peptide or vehicle (negative control) and analysed at days 7 and 11 for dilution of CFSE by flow cytometry combined with staining for CD4 (efluor 450 clone RPA-T4, Thermofisher) and CD8 (APC clone RPA-T8, Thermofisher).
At day 10 post-stimulation, cells for TCR analysis were stained for CD4 (efluor 450 clone RPA-T4, Thermofisher) and CD8 (APC clone RPA-T8, Thermofisher) and subsequently sorted on a MoFlow Cell Sorter (Beckman Coulter) into CD4+ve/CFSEhigh and CD4+ve/CFSElow populations. Populations were sorted directly into 1.0 ml of RNA protect reagent (Qiagen) diluted 5:1 with FACS sorting buffer (PBS supplemented with 1mM EDTA, 20mM HEPES and 1% FCS). Samples were stored at −80°C until analysis
RT-PCR, NGS bulk sequencing of the TCR α and β chains and repertoire data analysis
Sorted cells (bulk) from CD4+ve/CFSEhigh and CD4+ve/CFSElow populations in RNA protect (Qiagen) were shipped to iRepertoire Inc (Huntsville, AL, USA) for NGS sequencing of the TCR α and TCR β chain to confirm expansion of TCR’s in the CD4+ve/CFSElow cells, proliferating to the peptide in contrast to the non-proliferating CD4+ve/CFSEhigh population. In brief, RNA was purified from sorted cells, RT-PCR was performed, cDNA was then subjected to Amplicon rescued multiplex PCR (ARM-PCR) using human TCR α and β 250 PER primers (iRepertoire Inc., Huntsville, AL, USA). Information about the primers can be found in the United States Patent and Trademark Office (Patent Nos. 7,999,092 and 9,012,148B2). After assessment of PCR/DNA samples, 10 sample libraries were pooled and sequenced using the Illumina MiSeq platform (Illumina, San Diego, CA, USA). The raw data was analysed using IRweb software (iRepertoire). V, D, and J gene usage and CDR3 sequences were identified and assigned D50 diversity values and tree maps generated using iRweb tools. Tree maps show each unique CDR3 as a coloured rectangle, the size of each rectangle corresponds to each CDR3s abundance within the repertoire and the positioning is determined by the V region usage. Diversity was measured using D50 immune repertoire diversity index. The D50 index is a quantitative measure of the degree of diversity of T cells within a sample. The D50 is the percentage of T-cell clones that account for the cumulative 50% of the total CDR3s counted in the sample. The more diverse a library, the closer the value will be to 50. Low diversity values are associated with decreased diversity. Data are presented as non-normalised (which takes into account the frequency of each unique CDR3).
Statistical analysis
Comparative analysis of the ELISpot and peptide binding assay results was performed by applying paired or unpaired ANOVA or Students t-test as appropriate with values of P calculated accordingly. Sidak correction for multiple comparisons was applied where relevant in the analysis of Elispot data and Tukey correction for multiple comparisons was applied where relevant in analysis of peptide binding data. Comparison of tumour survival was assessed by Log Rank (Mantel-Cox) test and tumour size was assessed by Mann–Whitney test using the Graphpad Prism software version 7. P < 0.05 values were considered statistically significant and p < 0.01 values were considered highly significant. The error bars shown in the figures represent the mean + standard deviation.
Funding Statement
This work was funded by Scancell Ltd.
Acknowledgments
The authors would like to thank Barbara Gunn and Stephen Reader for their technical support. The authors would also like to thank Dr Tina Parsons and Dr Mireille Vankemmelbeke for help in proofreading the manuscript.
Disclosure of Potential Conflicts of Interest
VA Brentville, RL Metheringham and LG Durrant have ownership interest in patent WO2017013425 A1. LG Durrant is a director and CSO of Scancell Ltd has ownership interest (including patents) in Scancell Ltd. All authors are employees of Scancell Ltd. Work in this study was funded by Scancell Ltd.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Damgaard D, Senolt L, Nielsen MF, Pruijn GJ, Nielsen CH.. Demonstration of extracellular peptidylarginine deiminase (PAD) activity in synovial fluid of patients with rheumatoid arthritis using a novel assay for citrullination of fibrinogen. Arthritis Res Ther. 2014;16:498. doi: 10.1186/s13075-014-0498-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vossenaar ER, Radstake TR, van der Heijden A, van Mansum MA, Dieteren C, de Rooij DJ, Barrera P, Zendman AJW, van Venrooij WJ.. Expression and activity of citrullinating peptidylarginine deiminase enzymes in monocytes and macrophages. Ann Rheum Dis. 2004;63:373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.James EA, Rieck M, Pieper J, Gebe JA, Yue BB, Tatum M, Peda M, Sandin C, Klareskog L, Malmström V, et al. Citrulline-specific Th1 cells are increased in rheumatoid arthritis and their frequency is influenced by disease duration and therapy. Arthritis Rheumatol. 2014;66:1712–1722. doi: 10.1002/art.38637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sakkas LI, Bogdanos DP, Katsiari C, Platsoucas CD. Anti-citrullinated peptides as autoantigens in rheumatoid arthritis-relevance to treatment. Autoimmun Rev. 2014;13:1114–1120. doi: 10.1016/j.autrev.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 5.Klareskog L, Stolt P, Lundberg K, Kallberg H, Bengtsson C, Grunewald J, Rönnelid J, Harris HE, Ulfgren A-K, Rantapää-Dahlqvist S, et al. A new model for an etiology of rheumatoid arthritis: smoking may trigger HLA-DR (shared epitope)-restricted immune reactions to autoantigens modified by citrullination. Arthritis Rheum. 2006;54:38–46. doi: 10.1002/art.21575. [DOI] [PubMed] [Google Scholar]
- 6.Scally SW, Petersen J, Law SC, Dudek NL, Nel HJ, Loh KL, Wijeyewickrema LC, Eckle SBG, van Heemst J, Pike RN, et al. A molecular basis for the association of the HLA-DRB1 locus, citrullination, and rheumatoid arthritis. J Exp Med. 2013;210:2569–2582. doi: 10.1084/jem.20131241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raychaudhuri S, Sandor C, Stahl EA, Freudenberg J, Lee HS, Jia X, Alfredsson L, Padyukov L, Klareskog L, Worthington J, et al. Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat Genet. 2012;44:291–296. doi: 10.1038/ng.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ireland JM, Unanue ER. Autophagy in antigen-presenting cells results in presentation of citrullinated peptides to CD4 T cells. J Exp Med. 2011;208:2625–2632. doi: 10.1084/jem.20110640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brentville VA, Metheringham RL, Gunn B, Symonds P, Daniels I, Gijon M, Cook K, Xue W, Durrant LG. Citrullinated vimentin presented on MHC-II in tumor cells is a target for CD4+ T-cell-mediated antitumor immunity. Cancer Res. 2016;76:548–560. doi: 10.1158/0008-5472.CAN-15-1085. [DOI] [PubMed] [Google Scholar]
- 10.Cook K, Daniels I, Symonds P, Pitt T, Gijon M, Xue W, Metheringham R, Durrant L, Brentville V. Citrullinated alpha-enolase is an effective target for anti-cancer immunity. Oncoimmunology. 2018;7:e1390642. doi: 10.1080/2162402X.2018.1490854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Durrant LG, Metheringham RL, Brentville VA. Autophagy, citrullination and cancer. Autophagy. 2016;12:1055–1056. doi: 10.1080/15548627.2016.1166326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castelli FA, Buhot C, Sanson A, Zarour H, Pouvelle-Moratille S, Nonn C, Gahery-Ségard H, Guillet J-G, Ménez A, Georges B, et al. HLA-DP4, the most frequent HLA II molecule, defines a new supertype of peptide-binding specificity. J Immunol. 2002;169:6928–6934. [DOI] [PubMed] [Google Scholar]
- 13.Feitsma AL, van der Voort EI, Franken KL, El Bannoudi H, Bg E, Jw D, Huizinga TWJ, de Vries RRP, Toes REM, Ioan-Facsinay A. Identification of citrullinated vimentin peptides as T cell epitopes in HLA-DR4-positive patients with rheumatoid arthritis. Arthritis Rheum. 2010;62:117–125. doi: 10.1002/art.25059. [DOI] [PubMed] [Google Scholar]
- 14.Gerstner C, Dubnovitsky A, Sandin C, Kozhukh G, Uchtenhagen H, James EA, Rönnelid J, Ytterberg AJ, Pieper J, Reed E, et al. Functional and structural characterization of a novel HLA-DRB1*04:01-restricted alpha-enolase T cell epitope in rheumatoid arthritis. Front Immunol. 2016;7:494. doi: 10.3389/fimmu.2016.00494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sidney J, Becart S, Zhou M, Duffy K, Lindvall M, Moore EC, Moore EL, Rao T, Rao N, Nielsen M, et al. Citrullination only infrequently impacts peptide binding to HLA class II MHC. PLoS One. 2017;12:e0177140. doi: 10.1371/journal.pone.0177140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engelmann R, Biemelt A, Cordshagen A, Johl A, Kuthning D, Muller-Hilke B. The prerequisites for central tolerance induction against citrullinated proteins in the mouse. PLoS One. 2016;11:e0158773. doi: 10.1371/journal.pone.0158773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Snir O, Rieck M, Gebe JA, Yue BB, Rawlings CA, Nepom G, Malmström V, Buckner JH. Identification and functional characterization of T cells reactive to citrullinated vimentin in HLA-DRB1*0401-positive humanized mice and rheumatoid arthritis patients. Arthritis Rheum. 2011;63:2873–2883. doi: 10.1002/art.30445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Catalan D, Aravena O, Zuniga R, Silva N, Escobar A, Sabugo F, Wurmann P, Soto L, González R, Alfaro J, et al. Weak CD4+ T-cell responses to citrullinated vimentin in rheumatoid arthritis patients carrying HLA-DR9 alleles. Rheumatol Int. 2012;32:1819–1825. doi: 10.1007/s00296-011-2039-z. [DOI] [PubMed] [Google Scholar]
- 19.Kampstra AS, van Heemst J, Moustakas AK, Papadopoulos GK, Huizinga TW, Toes RE. The increased ability to present citrullinated peptides is not unique to HLA-SE molecules: arginine-to-citrulline conversion also enhances peptide affinity for HLA-DQ molecules. Arthritis Res Ther. 2016;18:254. doi: 10.1186/s13075-016-1153-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.James EA, Moustakas AK, Bui J, Papadopoulos GK, Bondinas G, Buckner JH, Kwok WW. HLA-DR1001 presents “altered-self” peptides derived from joint-associated proteins by accepting citrulline in three of its binding pockets. Arthritis Rheum. 2010;62:2909–2918. doi: 10.1002/art.27594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Waal L, Yuksel S, Brandenburg AH, Langedijk JP, Sintnicolaas K, Verjans GM, Osterhaus ADME, de Swart RL. Identification of a common HLA-DP4-restricted T-cell epitope in the conserved region of the respiratory syncytial virus G protein. J Virol. 2004;78:1775–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fossum B, Gedde-Dahl T 3rd, Hansen T, Eriksen JA, Thorsby E, Gaudernack G. Overlapping epitopes encompassing a point mutation (12 Gly–>arg) in p21 ras can be recognized by HLA-DR, -DP and -DQ restricted T cells. Eur J Immunol. 1993;23:2687–2691. doi: 10.1002/eji.1830231045. [DOI] [PubMed] [Google Scholar]
- 23.Higgins JA, Thorpe CJ, Hayball JD, O’Hehir RE, Lamb JR. Overlapping T-cell epitopes in the group I allergen of dermatophagoides species restricted by HLA-DP and HLA-DR class II molecules. J Allergy Clin Immunol. 1994;93:891–899. [DOI] [PubMed] [Google Scholar]
- 24.Mandic M, Castelli F, Janjic B, Almunia C, Andrade P, Gillet D, Brusic V, Kirkwood JM, Maillere B, Zarour HM. One NY-ESO-1-derived epitope that promiscuously binds to multiple HLA-DR and HLA-DP4 molecules and stimulates autologous CD4+ T cells from patients with NY-ESO-1-expressing melanoma. J Immunol. 2005;174:1751–1759. [DOI] [PubMed] [Google Scholar]
- 25.Sidney J, Steen A, Moore C, Ngo S, Chung J, Peters B, Sette A. Five HLA-DP molecules frequently expressed in the worldwide human population share a common HLA supertypic binding specificity. J Immunol. 2010;184:2492–2503. doi: 10.4049/jimmunol.0903655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Lith M, McEwen-Smith RM, Benham AM. HLA-DP, HLA-DQ, and HLA-DR have different requirements for invariant chain and HLA-DM. J Biol Chem. 2010;285:40800–40808. doi: 10.1074/jbc.M110.148155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crotzer VL, Blum JS. Autophagy and its role in MHC-mediated antigen presentation. J Immunol. 2009;182:3335–3341. doi: 10.4049/jimmunol.0803458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chicz RM, Graziano DF, Trucco M, Strominger JL, Gorga JC. HLA-DP2: self peptide sequences and binding properties. J Immunol. 1997;159:4935–4942. [PubMed] [Google Scholar]
- 29.Falk K, Rotzschke O, Stevanovic S, Jung G, Rammensee HG. Pool sequencing of natural HLA-DR, DQ, and DP ligands reveals detailed peptide motifs, constraints of processing, and general rules. Immunogenetics. 1994;39:230–242. [DOI] [PubMed] [Google Scholar]
- 30.Thomas R, Thio CL, Apps R, Qi Y, Gao X, Marti D, Stein JL, Soderberg KA, Moody MA, Goedert JJ, et al. A novel variant marking HLA-DP expression levels predicts recovery from hepatitis B virus infection. J Virol. 2012;86:6979–6985. doi: 10.1128/JVI.00406-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edwards JA, Durant BM, Jones DB, Evans PR, Smith JL. Differential expression of HLA class II antigens in fetal human spleen: relationship of HLA-DP, DQ, and DR to immunoglobulin expression. J Immunol. 1986;137:490–497. [PubMed] [Google Scholar]
- 32.Borst J, Ahrends T, Babala N, Melief CJM, Kastenmuller W. CD4(+) T cell help in cancer immunology and immunotherapy. Nat Rev Immunol. 2018;18:635–647. doi: 10.1038/s41577-018-0044-0. [DOI] [PubMed] [Google Scholar]
- 33.Fauskanger M, Haabeth OAW, Skjeldal FM, Bogen B, Tveita AA. Tumor killing by CD4(+) T cells is mediated via induction of inducible nitric oxide synthase-dependent macrophage cytotoxicity. Front Immunol. 2018;9:1684. doi: 10.3389/fimmu.2018.01684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haabeth OAW, Fauskanger M, Manzke M, Lundin KU, Corthay A, Bogen B, Tveita AA. CD4(+) T-cell-mediated rejection of MHC class II-positive tumor cells is dependent on antigen secretion and indirect presentation on host APCs. Cancer Res. 2018;78:4573–4585. doi: 10.1158/0008-5472.CAN-17-2426. [DOI] [PubMed] [Google Scholar]
- 35.Taneja V, Behrens M, Basal E, Sparks J, Griffiths MM, Luthra H, David CS. Delineating the role of the HLA-DR4 “shared epitope” in susceptibility versus resistance to develop arthritis. J Immunol. 2008;181:2869–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Celis E, Karr RW. Presentation of an immunodominant T-cell epitope of hepatitis B surface antigen by the HLA-DPw4 molecule. J Virol. 1989;63:747–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xue W, Metheringham RL, Brentville VA, Gunn B, Symonds P, Yagita H, Ramage JM, Durrant LG. SCIB2, an antibody DNA vaccine encoding NY-ESO-1 epitopes, induces potent antitumor immunity which is further enhanced by checkpoint blockade. Oncoimmunology. 2016;5:e1169353. doi: 10.1080/2162402X.2016.1169353. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


