Summary
Objectives
This study sought to evaluate the influence of the genetic polymorphisms on platelet reactivity and clinical outcomes in acute ischemic stroke patients taking clopidogrel.
Background
Little research has been published on relationships between genetic polymorphisms, platelet reactivity, and clinical outcomes in stroke patients treated with clopidogrel.
Methods
Patients hospitalized in Changhai Hospital with acute ischemic stroke were randomly enrolled into treatment with a 75‐mg daily maintenance dose of clopidogrel. Genotyping was detected by the MassARRAY iPLEX genotyping system (Sequenom Inc, San Diego, CA), and platelet reactivity was evaluated by the VerifyNow P2Y12 test (Accumetrics Inc., San Diego, CA). Sixteen single nucleotide polymorphisms (SNPs) within 9 genes were selected and high on‐clopidogrel platelet reactivity (HPR) was defined as P2Y12 reaction units (PRU) value ≥230. The primary endpoint was ischemic events, including major adverse cardiac events (MACE), recurrence of stroke, transient ischemic attack (TIA), and the composite of vascular death, and the secondary endpoint was bleeding.
Results
Of the 345 patients recruited, 275 (79.7%) patients were followed up for 1 year and 122 (35.4%) patients were categorized as HPR. Among the SNPs selected, only the CYP2C19*2 allele and the CYP2C19*3 allele were statistically significantly associated with PRU (P < 0.001 and P = 0.003, respectively). Similarly, the prevalence of HPR was associated with CYP2C19*2 and CYP2C19*3 (P < 0.001 and P = 0.001, respectively). During the 1 year of follow‐up, a total of 64 (23.3%) cases of clinical events occurred, including 60 ischemic events and 4 bleeding events. There were no correlation between CYP2C19 variant alleles and clinical outcomes (P > 0.05), but a statistically significant relevance was found between the HPR and the ischemic events in 1 year of follow‐up (P = 0.001).
Conclusions
CYP2C19*2 and CYP2C19*3 had a significant impact on clopidogrel response, but was not associated with ischemic events during 1 year of follow‐up in patients with acute ischemic stroke. HPR was an independent risk factor for ischemic events, and the VerifyNow P2Y12 test may be available to guide individualized antiplatelet therapies in stroke patients in China.
Keywords: Clopidogrel, CYP2C19, Ischemic stroke, The VerifyNow P2Y12 test
Introduction
Stroke is common and often leads to disabling events worldwide, and it has become the first cause of morbidity and mortality in China 1. Clopidogrel is one of the most commonly used antiplatelet agents, which is often recommended as the primary treatment and the secondary prevention for ischemic stroke in guidelines 2, 3. However, there is a wide range of interindividual variations in platelet response to clopidogrel therapy, and increasing evidence has supported the relation between high on‐clopidogrel platelet reactivity (HPR) and an increased risk of recurrent atherosclerotic events 4, 5, 6.
As a thienopyridine prodrug, clopidogrel requires hepatic transformation before it can achieve its antiplatelet effect and numerous genes affect the pharmacokinetic and pharmacodynamic response of clopidogrel 7. Among these genes, cytochrome P450 (CYP) 2C19 is the gene most firmly believed involved in the interindividual variations in clopidogrel response as confirmed by many studies 8, 9, 10. Asians seem to have a higher probability of carrying a loss‐of‐function (LOF) CYP2C19 allele (e.g., CYP2C19*2 or CYP2C19*3) and a lower probability of carrying a gain‐of‐function (GOF) CYP2C19 allele (e.g., CYP2C19*17) than persons of either African or Caucasian ethnicity 11; thus, Asians are much more likely to experience HPR and are at higher rate of recurrent ischemic events. In addition, related studies have shown that other genetic variants, such as ABCB1 12, CYP2C9 13, PON1 14, ITGB3 15, P2RY12 16, CYP2B6 17, P2Y12 18, and CES1 19, may also affect the antiplatelet effect of clopidogrel; however, an overall and convincing agreement for the impact of these genetic variants on response to clopidogrel and adverse clinical events has not been reached.
In recent decades, compared with numerous studies that explore the pharmacogenomics of clopidogrel in patients with cardiovascular disease, there have been few studies well researched in the area of stroke and little data have been available to guide effective antiplatelet therapy in stroke patients with HPR. In this prospective, observational study, a set of genetic variants, including CYP2C19 (*2, *3, *4, *5, *7, *8, *17), ABCB1, CYP2C9*3, ITGB3, PON1, P2RY12, CYP2B6(*1B, *9), P2Y12, and CES1, were selected to evaluate the variations in clopidogrel response and clinical outcomes of ischemic stroke patients in China.
Materials and Methods
Study Populations
This study was a single‐center, prospective, observational trial. Patients with acute ischemic stroke within 14 days of onset and who had been hospitalized at Changhai Hospital, Shanghai, China, were selected randomly for the study. The diagnosis of new onset cerebral infarction was confirmed by computer tomography or magnetic resonance imaging. Exclusion criteria were as follows: valvular heart diseases, tumors, or hemorrhagic diseases that may affect platelet count, the taking of other prohibited drugs (e.g., warfarin, proton pump inhibitors), and persons with an allergic or intolerant to clopidogrel. All eligible patients received a daily clopidogrel maintenance dose of 75 mg during 1 year of follow‐up.
Informed written consent was obtained from each patient before study enrollment, and the study protocol was approved by the Committee on Ethics of Second Military Medical University in China.
Genetic Testing
DNA samples were obtained from peripheral blood and sixteen single nucleotide polymorphisms (SNPs) in 9 genes were genotyped as follows: CYP2C19*2(rs4244285), CYP2C19*3(rs4986893), CYP2C9*3 (rs1057910), CYP2C19*4(rs28399504), CYP2C19*5(rs56337013), CYP2C19*7(rs72558186), CYP2C19*8(rs41291556), CYP2C19*17(rs12248560), ABCB1(rs1045642), ITGB3(rs5918), PON1(rs662), P2RY12(rs6785930), CYP2B6*1B(rs7254579), CYP2B6*9(rs8192719), P2Y12(rs2046934), and CES1(rs71647871). We used the genotyping system (MassARRAY iPLEX; Sequenom Inc, San Diego, CA, USA) for SNP genotyping.
Platelet Function Testing
Platelet reactivity was detected by the VerifyNow P2Y12 test (Accumetrics, San Diego, CA, USA). Blood samples were collected from patients’ peripheral veins on the 5th–7th day of clopidogrel treatment and put into Vacutainer tubes (BD Medical Systems) containing EDTA and sodium citrate (3.2%). Platelet function testing was assessed within 2 h after blood collection and the first 3 mL of blood was always discarded. The results were expressed as P2Y12 reaction units (PRU), base units (BASE), and percentage of inhibition (%Inhibition).
Main Endpoints
The main endpoints of this study included HPR and the incidence of ischemic events and bleeding events during 1 year of follow‐up. The threshold for defining HPR was PRU value ≥230 based on the previous recommendation with the VerifyNow P2Y12 test 4. The primary endpoint was ischemic events, including MACE, recurrence of stroke, TIA, and the composite of vascular death. The secondary endpoint was bleeding. The presence of endpoints was confirmed by event‐related hospital discharge reports.
Statistical Analysis
Data were presented as mean ± standard deviation or expressed in terms of frequencies and percentages. The selected SNPs were evaluated by Pearson's chi‐squared test for the Hardy–Weinberg equilibrium. Association between platelet reactivity and clinical endpoints was analyzed using multiple logistic regression analysis. To assess the cumulative risk of primary endpoint, a Kaplan–Meier survival curve was carried out. A P‐value <0.05 denoted statistical significance. All statistical analyses were performed with the Statistical Analysis System 9.2 (SAS Institute Inc, Cary, NC, USA) or the SPSS version 17 (SPSS Inc., Chicago, IL, USA).
Results
Study Population
A total of 345 patients were admitted to this study from August 2012 to July 2013; 275 patients completed the follow‐up for 1 year and 64 (23.3%) patients had at least one of the predefined endpoints during the follow‐up, including 60 ischemic events and 4 bleeding events. Overall, the characteristics of the study population and the respective clinical data of patients with and without primary endpoint are shown in Table 1. Included in the cohort were 234 (67.8%) men and 111 (32.2%) women with a mean age of 68.1 ± 11.5 years and a mean BMI of 23.8 ± 3.4. In total, 235 (68.1%) patients had total anterior circulation infarction (TACI) or partial anterior circulation infarction (PACI); 149 (43.2%) patients had posterior circulation infarction (POCI); and 39 (11.3%) patients had lacunar infarction (LACI). Among the risk factors for cerebrovascular disease, only a history of heart disease was related to the primary endpoint (P = 0.014).
Table 1.
Characteristics of the study population
| Variables | All patients (N = 345) | Endpoints (N = 60) | No endpoints (N = 211) | P‐value |
|---|---|---|---|---|
| Age | 68.1 ± 11.5 | 70.4 ± 10.5 | 67.3 ± 11.7 | 0.069 |
| BMI | 23.8 ± 3.4 | 23.4 ± 3.1 | 24.0 ± 3.4 | 0.264 |
| Gender | ||||
| Male (%) | 234 (67.8) | 41 (68.3) | 147 (69.7) | 0.843 |
| Female (%) | 111 (32.2) | 19 (31.7) | 64 (30.3) | |
| Hypertension (%) | 245 (71.2) | 43 (71.7) | 154 (73.0) | 0.840 |
| Diabetes (%) | 133 (38.6) | 23 (38.3) | 86 (40.8) | 0.735 |
| Dyslipidemia (%) | 269 (78.0) | 50 (83.3) | 161 (76.3) | 0.247 |
| Heart disease history (%) | 78 (22.6) | 22 (36.7) | 41 (19.4) | 0.005 |
| Stroke history (%) | 106 (30.7) | 25 (41.7) | 65 (30.8) | 0.115 |
| TACI/PACI (%) | 235 (68.1) | 44 (73.3) | 150 (71.1) | 0.734 |
| POCI (%) | 149 (43.2) | 28 (46.7) | 83 (39.3) | 0.308 |
| LACI (%) | 39 (11.3) | 9 (15.0) | 25 (11.8) | 0.478 |
| Base | 275 ± 55.6 | 286 ± 59.0 | 272 ± 53.8 | 0.078 |
| PRU | 197 ± 71.6 | 213 ± 74.8 | 191 ± 68.8 | 0.032 |
| PRU>=230(%) | 122 (35.4) | 29 (48.3) | 61 (28.9) | 0.005 |
| %Inhibition | 28.5 ± 23.5 | 26.5 ± 23.6 | 29.6 ± 23.6 | 0.363 |
Data are shown as mean ± SD or absolute and relative frequencies. BMI, body mass index; TACI, total anterior circulation infarction; PACI, partial anterior circulation infarction; POCI, posterior circulation infarction; LACI, lacunar infarction.
Genotype, Platelet Reactivity, and Clinical Endpoints
Of the 339 eligible blood samples, CES1 and CYP2C19 (*4, *5, *7, *8) were not detected in the population. The results of genetic testing are summarized in Table 2. In our study, ADP‐induced platelet aggregation, indicated by PRU value, was only significantly associated with the presence of the CYP2C19*2 allele and the CYP2C19*3 allele among all the SNPs tested (P < 0.01). Similarly, these two alleles were also associated with the prevalence of HPR (P < 0.001 and P = 0.001, respectively). However, none of the selected SNPs were correlated with clinical endpoints (P > 0.05).
Table 2.
Associations between genetic polymorphisms, platelet reactivity, and clinical endpoints
| Gene | SNP | Alleles | Endpoint p | PRU p | HPR p |
|---|---|---|---|---|---|
| CYP2C19*2 | rs4244285 | G>A | 0.679 | <0.001 | <0.001 |
| CYP2C19*3 | rs4986893 | G>A | 0.644 | 0.003 | 0.001 |
| CYP2C19*17 | rs12248560 | C>T | 0.215 | 0.730 | 1.000 |
| ABCB1 | rs1045642 | T>A | 0.680 | 0.608 | 0.715 |
| ITGB3 | rs5918 | T>C | 1.000 | 0.826 | 0.554 |
| PON1 | rs662 | A>G | 0.236 | 0.194 | 0.448 |
| P2RY12 | rs6785930 | G>A | 0.935 | 0.331 | 0.297 |
| CYP2C9*3 | rs1057910 | A>C | 0.424 | 0.120 | 0.296 |
| CYP2B6*1B | rs7254579 | T>C | 0.842 | 0.361 | 0.313 |
| CYP2B6*9 | rs8192719 | C>T | 0.500 | 0.913 | 0.683 |
| P2Y12 | rs2046934 | C>T | 0.583 | 0.933 | 0.544 |
| CES1 | rs71647871 | G>A | NA | NA | NA |
| CYP2C19*4 | rs28399504 | A>G | NA | NA | NA |
| CYP2C19*5 | rs56337013 | C>T | NA | NA | NA |
| CYP2C19*7 | rs72558186 | T>A | NA | NA | NA |
| CYP2C19*8 | rs41291556 | T>C | NA | NA | NA |
SNP, single nucleotide polymorphism; NA, not available.
CYP2C19 Genotype and PRU
All eligible patients were classified into 7 groups based on CYP2C19 genotype and frequencies of different CYP2C19 genotypes are shown in Table 3. About 201 (59.3%) patients carried at least one LOF CYP2C19 allele. The PRU value rose in accordance with the number of CYP2C19*2 and CYP2C19*3, and the results were displayed in Figures 1 and 2. Compared with noncarriers of LOF CYP2C19 allele, carriers increased the risk of HPR by the presence of the CYP2C19*2 allele (ORadj: 2.248; 95% CI: 1.317–3.837; P = 0.003) and the CYP2C19*3 allele (ORadj: 6.008; 95% CI: 1.645–21.940; P = 0.007) after adjustment for demographics. As only 5 (1.5%) patients carried GOF CYP2C19 allele, we failed to find a statistically significant relationship between CYP2C19*17 and PRU.
Table 3.
Frequencies of CYP2C19 genotype and PRU
| CYP2C19 | N (%) | PRU (mean ± SD) |
|---|---|---|
| *1*1 | 136 (40.1) | 176 ± 73.0 |
| *1*2 | 124 (36.6) | 198 ± 66.3 |
| *1*3 | 20 (5.9) | 229 ± 80.3 |
| *1*17 | 3 (0.9) | 144 ± 75.7 |
| *2*2 | 42 (12.4) | 236 ± 55.8 |
| *2*3 | 12 (3.5) | 243 ± 31.7 |
| *2*17 | 2 (0.6) | 250 ± 21.9 |
Figure 1.
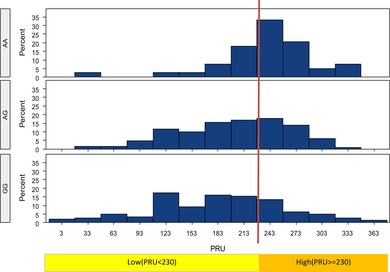
PRU value in different genotypes for CYP2C19*2 (c.681G>A; rs4244285) GG, CYP2C19 *1*1; AG: CYP2C19 *1*2; AA: CYP2C19 *2*2.
Figure 2.
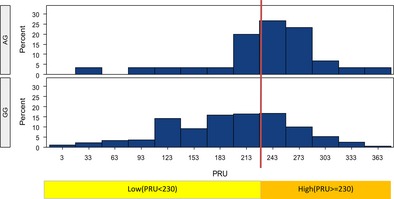
PRU value in different genotypes for CYP2C19*3 (c.636G>A; rs4986893) GG, CYP2C19 *1*1; AG: CYP2C19 *1*3.
PRU and Clinical Endpoints
There was a great deal of variation among the PRU values, ranging from 3 to 382. There were 122 (35.4%) patients categorized as HPR, with an average value of 269.8. The PRU values in the group of patients with primary endpoint were much higher than in patients without primary endpoint (P = 0.032). In multiple logistic regression analysis, HPR was correlated with an increased rate of ischemic events after adjustment for demographics (OR 2.149 [1.183, 3.902], P = 0.012), which corresponded to the result of Kaplan–Meier analysis (Figure 3, P = 0.002).
Figure 3.
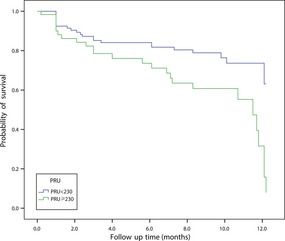
Kaplan–Meier cumulative event rates of the primary endpoint (MACE, recurrence of stroke, TIA, and the composite of vascular death) based on the presence of HPR during the follow‐up period (P = 0.003).
Discussion
In this observational study, we only found that the CYP2C19 LOF alleles (CYP2C19*2 and CYP2C19*3) were significantly correlated with HPR in stroke patients, which was consistent with the results obtained from healthy volunteers and patients with cardiovascular disease 8, 17, 20, 21. Furthermore, the frequencies of CYP2C19*2 and CYP2C19*3 were approximately 32.7% and 4.7% in this study cohort, which were much higher than recent updated data from CPIC guidelines for CYP2C19 genotype and clopidogrel therapy in Africans or Caucasians 11. Therefore, Chinese might have a higher probability of being carriers of CYP2C19 LOF allele and poor metabolizers of clopidogrel than Africans or Caucasians. However, we failed to demonstrate the relationship between CYP2C19 LOF allele and ischemic events, which challenged the literature that CYP2C19 LOF allele impacted cardiovascular outcomes in clopidogrel‐treated patients, especially in patients undergoing PCI 9, 15, 22, 23. It is notable that our findings are also different from a similar study conducted by Jia et al. 24 in China. However, they adopted National Institute of Health Stroke Scale and modified Rankin scale scores to assess the antiplatelet effect of clopidogrel, which may not be appropriate. As the scores were used to assess functional recovery of stroke patients, they might be hardly related to clopidogrel efficacy. In another similar study, Qiu et al. 25 just found an association of CYP2C19 LOF allele with neurological function assessed by modified Rankin scale, but failed to demonstrate a correlation between CYP2C19 LOF allele and ischemic events. Considering the limited number of patients enrolled and short time of follow‐up, three of the studies, including this present study, failed to find significant difference in adverse clinical events between carriers of CYP2C19 LOF allele and noncarriers. On the basis of a large number of clinical trials and meta‐analyses of CYP2C19 and clopidogrel, some investigators 26, 27, 28 pointed out that the clinical relevance of CYP2C19 LOF allele with clopidogrel was indication specific, which means the effect size of CYP2C19 LOF allele on clopidogrel pharmacodynamics was parallel with the effect of clopidogrel for a given indication. Clinical trial data suggest clopidogrel was associated with 35–80% risk reduction in patients undergoing PCI 26, but only with an 22% risk reduction in the ischemic stroke population 29. Thus, it was not surprising that the impact of CYP2C19 LOF allele on recurrent ischemic events in patients with stroke was less evident than that in patients undergoing PCI.
Among the 339 eligible blood samples, no homozygotes of CYP2C19 *17 allelic variant were found, except for a total of 5 (1.5%) heterozygotes (CYP2C19 *1*17 and CYP2C19*2*17). The frequency of the CYP2C19*17 allele was about 0. 7% in our study population, which was lower than 2.7% in East Asians and much lower than 21% in Europeans reported by the CPIC guidelines 11. Our result showed that CYP2C19*17 was not associated with enhanced platelet response to clopidogrel or with an increased risk for bleeding, which was also different from several studies in Western populations 30, 31. The discrepancy of the results could be partially explained by the lower prevalence of the CYP2C19 *17 allele and no prevalence of homozygotes in our study cohort compared with those documented in Caucasians. In addition, it could be ascribed to the relatively low rate of bleeding events and short time of follow‐up in this study.
It was notable that our study demonstrated a statistically significant association between HPR and poorer clinical outcomes in clopidogrel‐treated patients with ischemic stroke. HPR was shown to be an independent risk factor for ischemic events in stroke patients during clopidogrel treatment. Consequently, these findings raised the importance of platelet function tests to predict clinical endpoints in patients treated with clopidogrel. As platelet function tests reflected the influence of all genetic and nongenetic variables that acted on the pharmacokinetics and pharmacodynamics of clopidogrel, we thought platelet function test might be more reliable and helpful than CYP2C19 genotyping to guide individualized clopidogrel therapy based on our findings. The VerifyNow P2Y12 test, a really point‐of‐care platelet function assay, had been commonly used worldwide, but rarely applied in China. To the best of our knowledge, this study was the first to implement the VerifyNow P2Y12 test to assess clopidogrel response in stroke patients in China and the results showed that it had promising applications.
We acknowledge that there are several limitations to this study. First, this investigation was a single‐center study and only 79.7% patients completed the 1 year of follow‐up mainly due to economic reasons. Second, our study was limited by the use of only one test to evaluate HPR. Finally, we assumed that the relationship between CYP2C19 polymorphisms and clinical endpoints might be revealed by larger sample size and longer follow‐up times.
Conclusion
We conclude that CYP2C19*2 and CYP2C19*3 had significant impact on clopidogrel response, but was not associated with ischemic events during 1 year of follow‐up in patients with acute ischemic stroke. CYP2C19*17 was associated neither with an enhanced platelet response to clopidogrel nor with increased risk for bleeding events due to its low prevalence in Chinese. HPR was an independent predictor of ischemic events, and the VerifyNow P2Y12 test may be available to instruct antiplatelet strategies in stroke patients in China.
Conflicts of Interest
The authors declare no conflict of interest.
Acknowledgments
We thank all the patients enrolled in the study. This study was supported by Science and Technology Commission Foundation of Shanghai Municipality (No. 14430721200), Shanghai Pujiang Program (No. 14PJC002) and Shanghai Municipal Commission of Health and Family Planning (No. 20144Y0226, 20124035).
The first two authors contributed equally to the work.
References
- 1. Yang G, Wang Y, Zeng Y, et al. Rapid health transition in China, 1990‐2010: findings from the Global Burden of Disease Study 2010. Lancet 2013;381:1987–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Braunwald E, Antman EM, Beasley JW, et al. ACC/AHA Guideline Update for the Management of Patients With Unstable Angina and Non–ST‐Segment Elevation Myocardial Infarction—2002: Summary Article A Report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines (Committee on the Management of Patients With Unstable Angina). Circulation 2002;106:1893–1900. [DOI] [PubMed] [Google Scholar]
- 3. Furie KL, Kasner SE, Adams RJ, et al. Guidelines for the prevention of stroke in patients with stroke or transient ischemic attack: A guideline for healthcare professionals from the american heart association/american stroke association. Stroke 2011;42:227–276. [DOI] [PubMed] [Google Scholar]
- 4. Parodi G, Marcucci R, Valenti R, et al. High residual platelet reactivity after clopidogrel loading and long‐term cardiovascular events among patients with acute coronary syndromes undergoing PCI. JAMA 2011;306:1215–1223. [DOI] [PubMed] [Google Scholar]
- 5. Price MJ, Angiolillo DJ, Teirstein PS, et al. Platelet reactivity and cardiovascular outcomes after percutaneous coronary intervention: A time‐dependent analysis of the Gauging Responsiveness with a VerifyNow P2Y12 assay: Impact on Thrombosis and Safety (GRAVITAS) trial. Circulation 2011;124:1132–1137. [DOI] [PubMed] [Google Scholar]
- 6. Stone GW, Witzenbichler B, Weisz G, et al. Platelet reactivity and clinical outcomes after coronary artery implantation of drug‐eluting stents (ADAPT‐DES): A prospective multicentre registry study. Lancet 2013;382:614–623. [DOI] [PubMed] [Google Scholar]
- 7. Kazui M, Nishiya Y, Ishizuka T, et al. Identification of the human cytochrome P450 enzymes involved in the two oxidative steps in the bioactivation of clopidogrel to its pharmacologically active metabolite. Drug Metab Dispos 2010;38:92–99. [DOI] [PubMed] [Google Scholar]
- 8. Collet JP, Hulot JS, Pena A, et al. Cytochrome P450 2C19 polymorphism in young patients treated with clopidogrel after myocardial infarction: A cohort study. Lancet 2009;373:309–317. [DOI] [PubMed] [Google Scholar]
- 9. Mega JL, Close SL, Wiviott SD, et al. Cytochrome p‐450 polymorphisms and response to clopidogrel. N Engl J Med 2009;360:354–362. [DOI] [PubMed] [Google Scholar]
- 10. Steinhubl SR. Genotyping, clopidogrel metabolism, and the search for the therapeutic window of thienopyridines. Circulation 2010;121:481–483. [DOI] [PubMed] [Google Scholar]
- 11. Scott SA, Sangkuhl K, Stein CM, et al. Clinical pharmacogenetics implementation consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin Pharmacol Ther 2013;94:317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mega JL, Close SL, Wiviott SD, et al. Genetic variants in ABCB1 and CYP2C19 and cardiovascular outcomes after treatment with clopidogrel and prasugrel in the TRITON‐TIMI 38 trial: A pharmacogenetic analysis. Lancet 2010;376:1312–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gladding P, White H, Voss J, et al. Pharmacogenetic testing for clopidogrel using the rapid INFINITI analyzer: A dose‐escalation study. JACC Cardiovasc Interv 2009;2:1095–1101. [DOI] [PubMed] [Google Scholar]
- 14. Bouman HJ, Schomig E, van Werkum JW, et al. Paraoxonase‐1 is a major determinant of clopidogrel efficacy. Nat Med 2011;17:110–116. [DOI] [PubMed] [Google Scholar]
- 15. Cayla G, Hulot JS, O'Connor SA, et al. Clinical, angiographic, and genetic factors associated with early coronary stent thrombosis. JAMA 2011;306:1765–1774. [DOI] [PubMed] [Google Scholar]
- 16. Rudez G, Bouman HJ, van Werkum JW, et al. Common variation in the platelet receptor P2RY12 gene is associated with residual on‐clopidogrel platelet reactivity in patients undergoing elective percutaneous coronary interventions. Circ Cardiovasc Genet 2009;2:515–521. [DOI] [PubMed] [Google Scholar]
- 17. Price MJ, Murray SS, Angiolillo DJ, et al. Influence of genetic polymorphisms on the effect of high‐ and standard‐dose clopidogrel after percutaneous coronary intervention: The GIFT (Genotype Information and Functional Testing) study. J Am Coll Cardiol 2012;59:1928–1937. [DOI] [PubMed] [Google Scholar]
- 18. Fontana P, Dupont A, Gandrille S, et al. Adenosine diphosphate‐induced platelet aggregation is associated with P2Y12 gene sequence variations in healthy subjects. Circulation 2003;108:989–995. [DOI] [PubMed] [Google Scholar]
- 19. Lewis JP, Horenstein RB, Ryan K, et al. The functional G143E variant of carboxylesterase 1 is associated with increased clopidogrel active metabolite levels and greater clopidogrel response. Pharmacogenet Genomics 2013;23:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim KA, Park PW, Hong SJ, Park JY. The effect of CYP2C19 polymorphism on the pharmacokinetics and pharmacodynamics of clopidogrel: A possible mechanism for clopidogrel resistance. Clin Pharmacol Ther 2008;84:236–242. [DOI] [PubMed] [Google Scholar]
- 21. Umemura K, Furuta T, Kondo K. The common gene variants of CYP2C19 affect pharmacokinetics and pharmacodynamics in an active metabolite of clopidogrel in healthy subjects. J Thromb Haemost 2008;6:1439–1441. [DOI] [PubMed] [Google Scholar]
- 22. Liu Y, Liu N, Li W, Shao H, Zhi H, Li J. Relationship of CYP2C19*2 and CYP2C19*3 gene polymorphism with clopidogrel response variability and recurrent cardiovascular events in Chinese patients undergoing percutaneous coronary intervention. Pharmacology 2013;91:165–172. [DOI] [PubMed] [Google Scholar]
- 23. Shuldiner AR, O'Connell JR, Bliden KP, et al. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. JAMA 2009;302:849–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jia DM, Chen ZB, Zhang MJ, et al. CYP2C19 polymorphisms and antiplatelet effects of clopidogrel in acute ischemic stroke in China. Stroke 2013;44:1717–1719. [DOI] [PubMed] [Google Scholar]
- 25. Qiu LN, Sun Y, Wang L, et al. Influence of CYP2C19 polymorphisms on platelet reactivity and clinical outcomes in ischemic stroke patients treated with clopidogrel. Eur J Pharmacol 2015;747:29–35. [DOI] [PubMed] [Google Scholar]
- 26. Johnson JA, Cavallari LH. Pharmacogenetics and cardiovascular disease–implications for personalized medicine. Pharmacol Rev 2013;65:987–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shuldiner AR, Vesely MR, Fisch A. CYP2C19 genotype and cardiovascular events. JAMA 2012;307:1482; author reply 4‐5. [DOI] [PubMed] [Google Scholar]
- 28. Siasos G, Tousoulis D, Stefanadis C. CYP2C19 genotype and cardiovascular events. JAMA 2012;307:1483–1484; author reply 4‐5. [DOI] [PubMed] [Google Scholar]
- 29. Antithrombotic Trialists C . Collaborative meta‐analysis of randomised trials of antiplatelet therapy for prevention of death, myocardial infarction, and stroke in high risk patients. BMJ 2002;324:71–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li Y, Tang HL, Hu YF, Xie HG. The gain‐of‐function variant allele CYP2C19*17: A double‐edged sword between thrombosis and bleeding in clopidogrel‐treated patients. J Thromb Haemost 2012;10:199–206. [DOI] [PubMed] [Google Scholar]
- 31. Sibbing D, Koch W, Gebhard D, et al. Cytochrome 2C19*17 allelic variant, platelet aggregation, bleeding events, and stent thrombosis in clopidogrel‐treated patients with coronary stent placement. Circulation 2010;121:512–518. [DOI] [PubMed] [Google Scholar]


