Summary
Background
Gliomas are the most lethal type of primary brain tumor in adult. Long noncoding RNAs (lncRNAs), which are involved in the progression of various cancers, may offer a potential gene therapy target in glioma.
Methods and Findings
We first classified gliomas into three molecular subtypes (namely LncR1, LncR2 and LncR3) in Rembrandt dataset using consensus clustering. Survival analysis indicated that LncR3 had the best prognosis, while the LncR1 subtype showed the poorest overall survival rate. The results were further validated in an independent glioma dataset GSE16011. Additionally, we collected and merged data of the two databases (Rembrandt and GSE16011 dataset) and analyzed prognosis of each subtype in WHO II, III and IV gliomas. The similar results were obtained. Gene Set Variation Analysis (GSVA) demonstrated that LncR1 subtype enriched cultured astroglia's gene signature, while LncR2 subtype was characterized by neuronal gene signature. Oligodendrocytic was rich in LncR3. In addition, IDH1 mutation and 1p/19q LOH were found rich with LncR3, and EGFR amplification showed high percentage in LncR1 in GSE16011 dataset.
Conclusions
We report a novel molecular classification of glioma based on lncRNA expression profiles and believe that it would provide a potential platform for future studies on gene treatment for glioma and lead to more individualized therapies to improve survival rates.
Keywords: Glioma, LncRNA, Molecular subtypes, Prognosis
Introduction
Glioma is known as the most common and lethal type of intracranial tumor in adult 1. Advances in the conventional treatments of gliomas, including of surgery, radiotherapy and chemotherapy, have not yet obtained the satisfied therapeutic efficacy or improved prognosis 2. The development of molecular biology and its application in study of tumorigenesis provide an alternative approach to glioma treatment 3. High efficiency of this approach relies heavily on the right classification of patients for each type of treatment. The current histopathological classification system has offered a valuable basis for defining groups of patients for clinical assessment, and predicts the clinical behavior of the respective neoplasm with direct impact on the applied treatment regimes; however, there is a high rate of divergent diagnoses, inexact prognostic capabilities, and poor therapeutic predictive properties, indicating an urgent need for an objective, molecular‐based classification system 4. Several molecular‐based classification systems, such as mRNA expression based and DNA methylation based, have been established 5, 6. lncRNAs, with the length between 200 nucleotides to several kilobases, are a novel class of transcripts that regulate cellular processes 7, the perturbation of which can result in the development and progression of cancer, including of gliomas 8, 9, 10, yet few approaches take the lncRNA molecular abnormalities into consideration. In our study, we screen different lncRNA expression profiling in REMBRANDT database (training set) and find three underlying subtypes in glioma. Mutation in isocitrate dehydrogenase 1(IDH1), loss of heterozygosity on short arm of chromosome 1 and long arm of chromosome 19 (1p/19q LOH), as well as amplification of epithelial growth factor receptor (EGFR), are detected in each subtype; we then estimated the situation of overall survival of three subtypes using Kaplan–Meier. Independent dataset GSE16011 was used for validation.
Materials and methods
LncRNA Expression Profiles on Microarrays and Consensus Clustering
Microarray data from Rembrandt and GSE16011 databases were gathered from published studies 11, 12. The CEL files for the Rembrandt and GSE16011 dataset (Affymetrix GeneChip Human Genome U133 Plus 2.0 Array) were separately merged and computed with Matlab software. The expression data were normalized according to the robust multiarray average (RMA) normalization and expressed in a natural scale. LncRNA expression profiles on Affymetrix HG‐U133 Plus 2.0 arrays were identified based on the NetAffx annotation of the probe sets and the Refseq and Ensembl annotations of lncRNAs as described previously in ref 13. Total 2448 probe sets (corresponding to 1970 lncRNAs genes) that were represented on Affymetrix GeneChip Human Genome U133 Plus 2.0 Array were included in our analysis.
In the present study, we followed the strategy of using the larger dataset (Rembrandt set) as training set, and the smaller one (GSE16011) as the validation set. Firstly, 2448 lncRNA probe sets were first filtered to select for a coefficient of variation (CV) >0.5, allowing us to have most of the variations in lncRNA expression across the samples in the training set (Rembrandt set). Probes showing highly variable expression (CV > 0.5; probe number = 525; gene number = 395) were further mean centered and normalized by Cluster 3.0 and used for consensus clustering 14. Secondly, consensus clustering was performed using the hierarchical clustering method with average linkage and a distance metric equal to one minus the Pearson correlation coefficient. A total of 100 permutation tests were performed with a subsampling ratio of 0.8. The optimal number of glioma subgroups was determined using a consensus clustering cumulative distribution function (CDF) and consensus matrices.
Bioinformatics Analysis and Statistical Analysis
Prediction Analysis of Microarrays (PAM) was used to annotate the samples in validation set (GSE16011) with LncR1, LncR2 and LncR3 labels 15. Gene set variation analysis with LncR1, LncR2 and LncR3 subtypes was analyzed by GSVA package of R. Gene list was obtained from GSVA data package 16. Kaplan–Meier survival analysis was used to estimate the survival distributions. The log‐rank test was used to assess the statistical significance between stratified survival groups using GraphPad Prism 6.0 statistical software (GraphPad Software, lnc., San Diego, CA, USA). P < 0.05 was considered as significant.
Results
Consensus Clustering Identifies Three Molecular Subtypes in Glioma
In the present study, 2448 probe sets (corresponding to 1970 lncRNAs genes) represented on the Affymetrix HG‐U133 Plus 2.0 arrays were identified as described in ref 13. The profiles of all 2448 lncRNA probes from Rembrandt dataset (475 samples: 148 cases of astrocytoma, 227 cases of glioblastoma (GBM), 67 cases of oligodendroglioma, 11 cases of mixed and 21 cases without tumors, 1 case without histology annotation) were included for further analysis. After filtered by variable coefficient (CV), total 525 probes (C.V. > 0.05) were selected for intrinsic molecular subtype discovery using consensus cluster. As shown in Figure 1, three lncRNA‐based subtypes (LncR1, LncR2 and LncR3)existed in gliomas. The result was visualized in treeview (Figure 2A). In addition, four characteristic gene signatures were clearly identified using consensus cluster, and three of them correspond to each subtype of gliomas, respectively (Figure S1).
Figure 1.
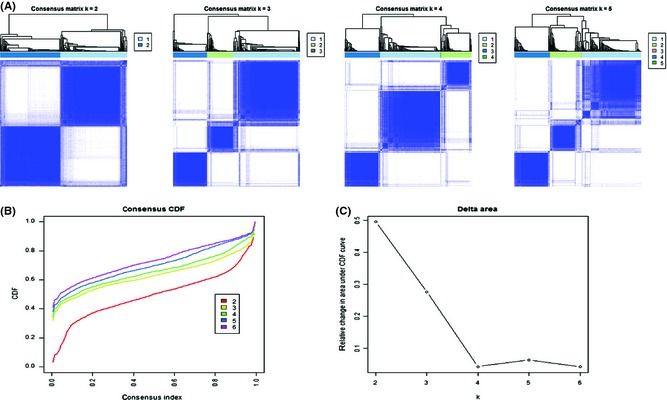
Identification of three molecular subtypes based on the lncRNA profiles in glioma. (A) Consensus clustering matrix of 475 samples from Rembrandt dataset for k = 2 to k = 4. (B) Consensus clustering CDF for k = 2 to k = 6. (C) Relative change in area under CDF curve for k = 2 to k = 6. Total 525 probes (C.V. > 0.05) were selected for consensus cluster, and results are shown in Figure S1.
Figure 2.
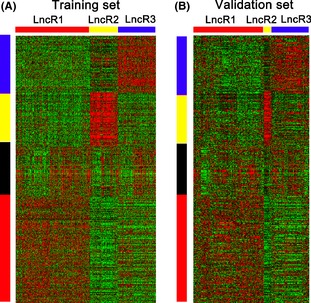
Differentially expressed lncRNA probe sets identify three molecular subtypes in both training and validation set. (A) Using the 525 probes, 475 Rembrandt samples in Rembrandt database (training set) were clustered based on the differentially expressed lncRNA probe sets. (B) The same gene order from the Rembrandt samples was maintained in the GSE16011 dataset (validation set) (n = 284). Gene expression levels are indicated as follows: red, high expression; green, low expression. The bar colors represent the sample types as indicated: red, LncR1 subtype; yellow, LncR2 subtype; blue, LncR3 subtype. Ordered gene expressions for all samples with different color bars are displayed for each gene expression subtype.
LncRNA‐Based Molecular Subtypes in Validation Dataset
An independent dataset of 284 samples (159 cases of glioblastoma (GBM), 52 cases of oligodendrocytoma (OD), 8 cases of pilocytic astrocytoma (PA), 28 cases of oligoastrocytoma (OA), 29 cases of astrocytoma(A), and 8 cases without tumors) on the Affymetrix HG‐U133 Plus 2.0 arrays, GSE16011, was downloaded from GEO as validation set. The molecular subtype of all samples was predicted using Prediction analysis of microarray (PAM). Gene order from the REMBRANDT samples was maintained in the validation dataset, and data were visualized in treeview (Figure 2B). By keeping the same gene order as sequence in training set, we clearly regained the glioma sample groups.
Functional Annotation and Clinical Characteristics of Each Subtype
Overall survival of each subtype estimated by Kaplan–Meier was found to be significantly different from each other. As shown in Figure 3A, LncR3 had the best prognosis, LncR1 subtype showed the poorest overall survival, and LncR2 obtained the intermediate clinical outcome in both training set and validation set. The results were further validated in GSE16011 dataset (Figure 3B). We also analyzed prognosis of each subtype in WHO II, III and IV stage gliomas and obtained the similar results (Figure S2). In addition, IDH1 mutation and 1p/19q LOH were enriched in LncR3, and other subtypes did, however, display small foci expression of these two markers in nonoverlapping cellular elements. EGFR amplification showed high percentage in LncR1, which is observed almost exclusively in this subtype (Figure 4).
Figure 3.
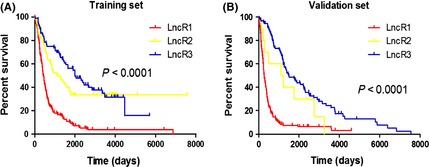
Kaplan–Meier estimates of overall survival in patients with three subtypes. (A) Training set; (B) validation set.
Figure 4.

Integrated view of IDH1 mutation, 1p/19q LOH and EGFR amplification status across three LncRNA based subtypes.
To discern the biological meaning of the subtypes, we used GSVA to calculate all samples for analyzing the association between subtypes and gene expression profile of neurons, oligodendrocytes, astrocytes, and cultured astroglias. The enrichment score was calculated using GSVA package in R software and suggested the similarity between these expression pattern of these geneset and the samples reflect. As shown in Figure 5 with this exploratory analysis, the LncR1 subtype was highly enriched with the cultured astroglia geneset, while the LncR2 subtype shows a high correlation with the neuronal signature. The LncR3 seems to have link with oligodendrocytic geneset differentiation.
Figure 5.

GSVA scores of each subtype shows a relation to specific cell type. Gene expression signatures of oligodendrocytes, astrocytes, neurons and cultured astroglial cells were used to indicate how closely the expression in a sample reflects the expected expression pattern of the geneset. Red unit indicates a positive correlation between specific cell type and the tumor sample expression profile, while the blue unit indicates the reverse.
Discussion
Glioma, the most common and intractable type of intracranial tumor in adults, is a clinically heterogeneous disease with an extremely poor prognosis in spite of multimodal treatment approaches. The advance in molecular biology provides an important novel approach to glioma treatments 17. The key of this method calls for an objective and detailed classification in patient according with the inherent gene expression pattern. Development of molecular classification in gliomas is very rapid in recent years 18, 19, 20. Kinds of classification systems, based on mRNA expression 21, 22, 23, microRNA expression 24, or methylation 25, have been reported by several groups. In Roel G.W. Verhaak's study, they divided GBM into proneural, neural, classical, and mesenchymal subtypes by robust genes (EGFR, NF1, and PDGFRA/IDH1) expression‐based molecular classification 26. Houtan Noushmehr et al. characterized a distinct subgroup of 272 GBM tumors using the DNA methylation analysis and exhibiting CIMP. In the last few years, biological and physiological importance of the lncRNA has been exposed under limelight 27, 28. It is now well documented that many lncRNAs take part in many key biological processes, including dosage compensation, genomic imprinting, chromatin regulation, alternative splicing of pre‐mRNA, nuclear organization and are dysregulated in the development and progression of glioma29, 30. Thus, the molecular classification of gliomas based on lncRNA expression might provide a better system for gene therapy target selection. Our study reveals the subtypes of glioma by screening different lncRNA expression profiling in 475 glioma samples and normal brain samples from the REMBRANDT and finds three underlying subtypes in glioma. Independent dataset GSE16011 was used for validation, and the similar result was obtained.
To reveal the biological meaning of the LncR subtypes, we used GSVA to calculate the relationship between all samples of subtypes and gene signatures of oligodendrocytes, astrocytes, neurons, and cultured astroglial cells from the brain transcriptome database presented by Cahoy et al.31. The results showed that the LncR1 subtype was highly enriched with the cultured astroglia signature but rarely the neuronal signature, which is strongly associated with the LncR2 subtype. With regard to the relationship between each subtype and specific cell type, results showed that oligodendrocytic characteristics were distributed in the LncR1 and LncR3, but rare in LncR2. Additionally, astrocytoma and neuronal are concentrated in the LncR3 and LncR2 subtypes, respectively. The results provide a bridge for guidance in combination with clinical treatment and gliomas' molecular biology research.
Predicting the prognosis of glioma is also an advantage of the molecular classification, Bao ZS, and his coworkers identified mRNA expression signature to improve outcome prediction for patients with mesenchymal GBM4. It is generally agreed upon that mutations in isocitrate dehydrogenase 1(IDH1) 32, 33, loss on 1p/19q 34, 35, 36, as well as amplification of EGFR 37, 38, have clinical prognostic value and are strongly associated with various subtypes of diffuse gliomas. With increasing knowledge of both the predetermine and prognostic significance of these markers, their role in classification is emerging. In our study, data showed that LncR1 subtype is rich in amplification of EGFR and has the worst clinical outcome. Patients in LncR3 subtype, displaying both strong IDH1 mutations and loh 1p/19q, had best survival of all. On the other hand, LncR2 subtype, without obvious distribution of the three markers, shows an intermediate survival in three subtypes.
In summary, our results indicate that there are three molecular subtypes in glioma based on the lnc RNA profiles. Although the possible functional pathways of many identified lncRNA genes are still little understood, researches on lncRNA are in full swing around the world. Our results of the classification based on the lncRNA profiles may provide an efficient classification tool for clinical prognosis evaluation and selection of the target of gene therapy of human gliomas.
Funding
This work was supported by National High Technology Research and Development Program 863 (2012AA02A508), Jiangsu Provincial Special Program of Medical Science (BL2012028), and National Natural Science Foundation of China (91229121, 81272792).
Conflict of Interest
None of the authors have any conflict of interest to disclose.
Supporting information
Figure S1. Total 525 probes (C.V. > 0.05) were selected for Consensus cluster and classified to four subtypes.
Figure S2. Kaplan–Meier estimates of overall survival in patients with three histological subtypes. (A) WHO II stage gliomas. (B) WHO III stage gliomas. (C) WHO IV stage gliomas.
The first two authors contributed equally to this work.
References
- 1. Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol 2007;114:97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mulhern RK, Merchant TE, Gajjar A, et al. Late neurocognitive sequelae in survivors of brain tumours in childhood. Lancet Oncol 2004;5:399–408. [DOI] [PubMed] [Google Scholar]
- 3. Argyriou AA, Kalofonos HP. Molecularly targeted therapies for malignant gliomas. Mol Med 2009;15:115–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bao ZS, Zhang CB, Wang HJ, et al. Whole‐genome mRNA expression profiling identifies functional and prognostic signatures in patients with mesenchymal glioblastoma multiforme. CNS Neurosci Ther 2013;19:714–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mischel PS, Shai R, Shi T, et al. Identification of molecular subtypes of glioblastoma by gene expression profiling. Oncogene 2003;22:2361–2373. [DOI] [PubMed] [Google Scholar]
- 6. Martinez R, Schackert G. Epigenetic aberrations in malignant gliomas: An open door leading to better understanding and treatment. Epigenetics 2007;2:147–150. [DOI] [PubMed] [Google Scholar]
- 7. Ponting CP, Belgard TG. Transcribed dark matter: Meaning or myth? Hum Mol Genet 2010;19:R162–R168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Spizzo R, Almeida MI, Colombatti A, et al. Long non‐coding RNAs and cancer: A new frontier of translational research? Oncogene 2012;31:4577–4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cheetham SW, Gruhl F, Mattick JS, et al. Long noncoding RNAs and the genetics of cancer. Br J Cancer 2013;108:2419–2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Maruyama R, Suzuki H. Long noncoding RNA involvement in cancer. BMB Rep 2012;45:604–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gravendeel LA, Kouwenhoven MC, Gevaert O, et al. Intrinsic gene expression profiles of gliomas are a better predictor of survival than histology. Cancer Res 2009;69:9065–9072. [DOI] [PubMed] [Google Scholar]
- 12. Madhavan S, Zenklusen JC, Kotliarov Y, et al. Rembrandt: Helping personalized medicine become a reality through integrative translational research. Mol Cancer Res 2009;7:157–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang X, Sun S, Pu JK, et al. Long non‐coding RNA expression profiles predict clinical phenotypes in glioma. Neurobiol Dis 2012;48:1–8. [DOI] [PubMed] [Google Scholar]
- 14. Wilkerson MD, Hayes DN. ConsensusClusterPlus: A class discovery tool with confidence assessments and item tracking. Bioinformatics 2010;26:1572–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tibshirani R, Hastie T, Narasimhan B, et al. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A 2002;99:6567–6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hanzelmann S, Castelo R, Guinney J. GSVA: Gene set variation analysis for microarray and RNA‐seq data. BMC Bioinformatics 2013;14:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mischel PS, Cloughesy TF. Targeted molecular therapy of GBM. Brain Pathol 2003;13:52–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huang H, Okamoto Y, Yokoo H, et al. Gene expression profiling and subgroup identification of oligodendrogliomas. Oncogene 2004;23:6012–6022. [DOI] [PubMed] [Google Scholar]
- 19. Rao SA, Santosh V, Somasundaram K. Genome‐wide expression profiling identifies deregulated miRNAs in malignant astrocytoma. Mod Pathol 2010;23:1404–1417. [DOI] [PubMed] [Google Scholar]
- 20. Rickman DS, Bobek MP, Misek DE, et al. Distinctive molecular profiles of high‐grade and low‐grade gliomas based on oligonucleotide microarray analysis. Cancer Res 2001;61:6885–6891. [PubMed] [Google Scholar]
- 21. Loew S, Schmidt U, Unterberg A, et al. The epidermal growth factor receptor as a therapeutic target in glioblastoma multiforme and other malignant neoplasms. Anticancer Agents Med Chem 2009;9:703–715. [DOI] [PubMed] [Google Scholar]
- 22. Pomeroy SL, Tamayo P, Gaasenbeek M, et al. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature 2002;415:436–442. [DOI] [PubMed] [Google Scholar]
- 23. Eberhart CG. Molecular diagnostics in embryonal brain tumors. Brain Pathol 2011;21:96–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim TM, Huang W, Park R, et al. A developmental taxonomy of glioblastoma defined and maintained by MicroRNAs. Cancer Res 2011;71:3387–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 2010;17:510–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010;17:98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kaikkonen MU, Lam MT, Glass CK. Non‐coding RNAs as regulators of gene expression and epigenetics. Cardiovasc Res 2011;90:430–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ellis BC, Molloy PL, Graham LD. CRNDE: A long non‐coding RNA involved in cancer, neurobiology, and development. Front Genet 2012;3:270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Qi P, Du X. The long non‐coding RNAs, a new cancer diagnostic and therapeutic gold mine. Mod Pathol 2013;26:155–165. [DOI] [PubMed] [Google Scholar]
- 30. Harries LW. Long non‐coding RNAs and human disease. Biochem Soc Trans 2012;40:902–906. [DOI] [PubMed] [Google Scholar]
- 31. Cahoy JD, Emery B, Kaushal A, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: A new resource for understanding brain development and function. J Neurosci 2008;28:264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jiao Y, Killela PJ, Reitman ZJ, et al. Frequent ATRX, CIC, FUBP1 and IDH1 mutations refine the classification of malignant gliomas. Oncotarget 2012;3:709–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cohen AL, Holmen SL. Colman H IDH1 and IDH2 mutations in gliomas. Curr Neurol Neurosci Rep 2013;13:345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hatanpaa KJ, Burger PC, Eshleman JR, et al. Molecular diagnosis of oligodendroglioma in paraffin sections. Lab Invest 2003;83:419–428. [DOI] [PubMed] [Google Scholar]
- 35. Durand KS, Guillaudeau A, Weinbreck N, et al. 1p19q LOH patterns and expression of p53 and Olig2 in gliomas: Relation with histological types and prognosis. Mod Pathol 2010;23:619–628. [DOI] [PubMed] [Google Scholar]
- 36. Pinto LW, Araujo MB, Vettore AL, et al. Glioblastomas: Correlation between oligodendroglial components, genetic abnormalities, and prognosis. Virchows Arch 2008;452:481–490. [DOI] [PubMed] [Google Scholar]
- 37. Burel‐Vandenbos F, Turchi L, Benchetrit M, et al. Cells with intense EGFR staining and a high nuclear to cytoplasmic ratio are specific for infiltrative glioma: A useful marker in neuropathological practice. Neuro Oncol 2013;15:1278–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schulte A, Liffers K, Kathagen A, et al. Erlotinib resistance in EGFR‐amplified glioblastoma cells is associated with upregulation of EGFRvIII and PI3Kp110delta. Neuro Oncol 2013;15:1289–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Total 525 probes (C.V. > 0.05) were selected for Consensus cluster and classified to four subtypes.
Figure S2. Kaplan–Meier estimates of overall survival in patients with three histological subtypes. (A) WHO II stage gliomas. (B) WHO III stage gliomas. (C) WHO IV stage gliomas.


