Abstract
Despite the availability of antiretroviral therapy (ART) that fully suppresses human immunodeficiency virus type-1 (HIV), markers of inflammation and minor neurocognitive impairment are frequently identified in HIV-infected persons. Increasing data support that low level replication defective viral RNA is made by infected cells despite the absence of infectious virus. Specific GU-rich single-stranded RNA from the HIV long terminal repeat region (ssRNA40) signaling through toll-like receptor (TLR)-7 and -8 has been shown to induce the secretion of interleukin-1β (IL-1β) in primary monocytes. Here, we examined the activation of microglial cells by HIV ssRNA40, and the potential subsequent neurotoxicity. Our findings show that exposure of human primary microglia to ssRNA40 activates the NLR family pyrin domain containing 3 (NLRP3) inflammasome. Following exposure to ssRNA40, pro-inflammatory cytokines IL-1β, IL-18 and neurotoxic cytokines TNF-α, IL-1α and C1q expression and extracellular secretion are increased. The released cytokines are functional since culture supernatants from ssRNA40 exposed microglia induced toxicity of human primary neurons. Moreover, inflammasome activation of microglia increased ROS generation with a loss of mitochondrial membrane potential and mitochondrial integrity. Treatment with ssRNA40 resulted in a blockade of autophagy/mitophagy mediated negative regulation of NLRP3 inflammasome activity with release of inflammatory cytokines, caspase-1 activation and pyroptotic microglial cell death. Thus, HIV ssRNA mediated activation of microglial cells can contribute to neurotoxicity and neurodegeneration via secretion of inflammatory and neurotoxic cytokines. These findings provide a potential mechanism that explains the frequent minor cognitive deficits and chronic inflammation that persist in HIV-infected persons despite treatment with suppressive ART.
Keywords: HIV ssRNA40, inflammasome, mitophagy, microglial cells, neurotoxicity
INTRODUCTION
Combination antiretroviral therapy (ART) has greatly reduced the burden of illness associated with human immunodeficiency virus type-1 (HIV) infection and led to strategies capable of significantly reducing transmission (Cohen et al., 2016). An important contributor to the improvement in the overall outcome of HIV-infected persons has been the marked decline of HIV-associated dementia. However, although severe neurological disease is now uncommon with ART, minor cognitive impairment persists and may occur in from 10–20% of infected patients on treatment (Farhadian, Patel, & Spudich, 2017; Heaton et al., 2015)
HIV enters the central nervous system (CNS) during primary infection predominantly infecting microglia and macrophages (Burdo, Lackner, & Williams, 2013; Garden, 2002). Although actively replicating virus is detected within the CNS in those with HIV-associated dementia, infectious virus is uncommonly detected in cerebrospinal fluid in patients fully suppressed on ART (Anderson et al., 2017). However, despite the ability of ART to provide sustained viral suppression, markers of chronic inflammation remain elevated suggesting that low level HIV replication and/or release of viral RNA and proteins may be driving the persistent inflammatory response (Baxter, O’Doherty, & Kaufmann, 2018; Eden et al., 2016; Garden, 2002; Golumbeanu et al., 2018; Pollack et al., 2017; Shan et al., 2017)
Much research has examined the effects of HIV on the CNS. However, the mechanistic causes of HIV-associated neurological disorders (HAND) remain mostly unknown. Interleukin-1β (IL-1β) has been associated with HIV disease progression, and is released during NLR family pyrin domain containing 3 (NLRP3) inflammasome activation (Chattergoon et al., 2014; Chen, Partridge, Sell, Torres, & Martin-Garcia, 2017; Guo, Gao, Taxman, Ting, & Su, 2014). Recent reports suggest that NLRP3 inflammasome activation is associated with the enhanced appearance of mitochondria damage signals and subsequent mitochondrial destabilization in host cells (Gurung, Lukens, & Kanneganti, 2015; Misawa et al., 2013; Nakahira et al., 2011; Shimada et al., 2012; Yu et al., 2014). To minimize the deleterious effects of damaged mitochondria, cells employ selective autophagy of mitochondria, mitophagy, which has a critical role in maintaining the health of cells by promoting the turnover and preventing the accumulation of damaged mitochondria that can lead to cellular degeneration. The mitochondrial serine/threonine kinase PTEN induced putative kinase 1 (PINK1) and the cytolytic E3 ubiquitin ligase Parkin, play an important role in mitophagy (Lazarou, Jin, Kane, & Youle, 2012; Matsuda et al., 2010; Narendra, Tanaka, Suen, & Youle, 2008; D. P. Narendra et al., 2010). In addition, autophagy receptors including p62/SQSTM1, Optineurin (OPTN), neighbor of BRCA1 (NBR1), and nuclear DOT protein 52 kDa (NDP52) bind polyubiquitinated cargo through the ubiquitin binding domain (UBD) and target them to the autophagy/mitophagy pathway by interacting with LC3 through their LC3 interacting region (LIR) domain (Deng et al., 2017). Recent studies suggest that NDP52 and OPTN are the primary receptors for PINK1 and Parkin mediated mitophagy (Deng et al., 2017; Lazarou et al., 2015). PINK1 can specifically recruit these receptors directly on the damaged mitochondria to mediate both Parkin dependent and Parkin independent mitophagy (Lazarou et al., 2015).
Several studies have implicated NLRP3 inflammasome activation to be associated with neuroinflammation and HAND (Bandera et al., 2018; Feria, Taborda, Hernandez, & Rugeles, 2018; Marais et al., 2017; Walsh et al., 2014). However, the mechanism(s) driving the inflammatory process during sustained HIV suppression have not been identified. Here, we show that GU-rich RNA from the HIV long terminal repeat region (LTR) signaling through TLR-7 and −8 on microglial cells results in activation of the NLRP3 inflammasome with release of neurotoxic cytokines resulting in degeneration of primary human neurons (HPN).
MATERIALS AND METHODS
Ethics Statement
Healthy HIV seronegative donors were enrolled for venous blood draw using a protocol that was reviewed and approved by the Human Research Protection Program of the University of California, San Diego in accordance with the requirements of the Code of Federal Regulations on the Protection of Human subjects (45 CFR 46 and 21 CFR 50 and 56) and was in full compliance with the principles expressed in the Declaration of Helsinki. Written informed consent was obtained from all blood donors prior to their participation. Normal human fetal brain tissue was obtained from the University of Washington and approved by the Human Research Protection Program of the University of California, San Diego in accordance with the requirements of the Code of Federal Regulation on the Protection of Human subjects (45 CFR§46.102 (f)).
Cell culture
Peripheral blood mononuclear cells (PBMCs) were isolated from whole blood of HIV seronegative donors using density gradient centrifugation over Ficoll–Paque Plus (GE Healthcare Cat# 114402). Primary human monocytes were isolated from PBMCs using monocyte isolation kit II, human (Miltenyi Biotech Cat# 130–091-103). Human microglia cells (HMG) were differentiated from primary human monocytes using methods as described previously (Etemad, Zamin, Ruitenberg, & Filgueira, 2012; Rawat & Spector, 2017). Briefly, monocytes were seeded at 2.5×105 cells/well in a 48-well plate in serum free RPMI Glutamax medium (Gibco Cat# 61870036) supplemented with MCSF (10ng/ml; Peprotech Cat# 300–25), MCP1 (100ng/ml; Peprotech Cat# 300–04), GMCSF (10ng/ml; Peprotech Cat# 300–03) and NGF-β (10ng/ml; R&D Systems Cat# 256-GF) with media changed every 3 days.
For autophagy induction in HMG cells, culture medium was supplemented with trehalose (Sigma, Cat# T0167) at 100mM concentration. The lysosomal degradation inhibitor bafilomycin A1 (Cell Signaling Cat# 54645) was prepared in dimethyl sulfoxide (DMSO) and added to the medium at a concentration of 10nM to measure the autophagic flux. DMSO was added to the medium of untreated cells as vehicle control.
Human microglia isolation and cell culture
Human fetal brain derived microglia cells (HFMG) were isolated as previously described (Rawat & Spector, 2017). Briefly, human fetal brain derived cells were plated at a density of ~7×106 cells/ml in a T75 tissue culture flask in DMEM high glucose (Gibco Cat# 11965084) supplemented with 10% AB-human serum (MP Biomedicals Cat# 092938249) and M-CSF (10ng/ml). After 10–14 days, flasks were shaken and floating microglia were collected and cultured at a density of 2.5×105 cells/well in a 48 well plate.
Human Primary Neuron Isolation and Cell Culture
For isolation of human primary neurons (HPN), forebrain fetal tissue was obtained from Advanced Bioscience Resources (Alameda, CA) and from the University of Washington, School of Medicine according to the University of California San Diego Institutional Review Board guidelines and processed as previously described (Avramut, Zeevi, & Achim, 2001; Teodorof et al., 2014). Briefly, neurons were seeded at 105 cells per well on Poly-D-Lysine/Laminin coated glass coverslips (Fisher Scientific Cat# 08–774-385) in 24-well plates and kept in neurobasal media (Gibco Cat# 21103049) enriched with B27 supplement (Gibco Cat# 17504044), Glutamax supplement (2 mM; Gibco Cat# 35050061), gentamicin (10 μg/mL; Gibco Cat# 15750078) for 2 weeks with media change every 3 days.
Inflammasome activation assay
We seeded HMG or HFMG at a density of 2.5×105 cells /well in a 48 well plate. At day 6, cells were transfected with GU-rich RNA within the HIV LTR (ssRNA40)/lyovec (Invivogen Cat# tlrl-lrna40) and ssRNA41/lyovec (identical to ssRNA40 with the exception that uridines are replaced with adenosines) (Invivogen Cat# tlrl-lrna41) for 24h and 48h. Both cell supernatants for ELISA and cell lysates for western blot analysis were collected. For inhibitor treatments, caspase-1 inhibitor, Ac-YVAD-cmk (Invivogen Cat# inh-yvad) or Z-WEHD-FMK (R&D Systems Cat# FMK002) or MCC950 (Adipogen Cat# AG-CR1–3615), cells were pre-incubated for 2h at 37°C prior to addition of ssRNA40. In parallel, untreated and ssRNA41 treated cells were also incubated at 37°C.
Immunoblotting
Cell lysates were prepared using RIPA lysis buffer (Pierce, Cat# 89900) containing protease and phosphatase inhibitor cocktail (Thermo Scientific Cat# 78441) according to the manufacturer’s instructions. The protein samples were resolved on Bolt™ 12% Bis-Tris Plus Gels (Novex Cat# NW00125BOX) and transferred to polyvinylidene difluoride membranes or nitrocellulose membrane (Thermo Scientific Cat# 88585, 88081), followed by detection with the WesternBreeze chemiluminescence kit (Novex Cat# WB7106, WB7104) according to the manufacturer’s instructions. Primary antibodies used were IL-18 (Abcam Cat# ab191152, RRID:AB_2737346), TNF-α (Abcam Cat# ab9635, RRID:AB_2203950), IL-1α (Abcam Cat# ab9614, RRID:AB_308732), PINK1 (Abcam Cat# ab23707, RRID:AB_447627), SQSTM1 (Abcam Cat# ab56416, RRID:AB_945626), Optineurin (Abcam Cat# ab89820, RRID:AB_2042519), BECN1 (Novus Cat# NB110–87318, RRID:AB_1201249), LC3B (Novus Cat# NB100–2220, RRID:AB_10003146), TBK1 (Cell Signaling Technology Cat# 3504S, RRID:AB_2255663), phospho-TBK1 (Ser172) (Cell Signaling Technology Cat# 5483, RRID:AB_10693472), NDP52 (Cell Signaling Technology Cat# 9036S, RRID:AB_11127597), IL-1β (Cell Signaling Technology Cat# 9036S, RRID:AB_2737350), NLRP3/NALP3 (AdipoGen Cat# AG-20B-0014, RRID:AB_2490202), Parkin (Santa Cruz Biotechnology Cat# sc-30130, RRID:AB_653855) and β-Actin (Sigma-Aldrich Cat# A5316, 35 RRID:AB_476743).
siRNA transfections
siRNA transfections of HMG cells with silencer select siRNA (Thermo Fisher Scientific Cat# 4392420) targeting NLRP3 (Assay ID s534397) or negative control siRNA (Thermo Fisher Scientific Cat# 4390843) were performed according to manufacturer’s protocol. Two days later cells were analyzed for target gene silencing by qPCR analysis and used in experiments.
Cytokine and chemokine assay
Cell culture supernatants collected from microglial cells at 24h and 48h post ssRNA40, ssRNA41 or vehicle treatment were used for quantification of cytokines using ELISA. Human IL-1β (R&D systems Cat# DLB50), human IL-18 (eBioscience Cat# BMS267–2), humanIL-1 alpha (eBioscience Cat# BMS243–2), human TNF-alpha (eBioscience Cat# BMS223–4), human complement C1q (Abcam Cat# ab170246) production were quantified by ELISA in these culture supernatants. These cell culture supernatants were also analyzed for relative levels of selected cytokines and chemokines using a membrane-based antibody array (R&D Systems Cat# ARY005B) following manufacturer’s instructions.
Mitochondrial assay
Following incubation with ssRNA40 or ssRNA41 for 24h or 48h, HMG cells were washed with 1X PBS and incubated with MitoSOX Red (Molecular Probes Cat# M36008) for ROS measurement; TMRE (Molecular Probes Cat# T669) or Mitotracker Green and Deep Red (Molecular Probes Cat# M7514, M22426) for measuring mitochondrial membrane potential. Following 10–20 min incubation, HMG cells were washed and collected in PBS for analysis by flow cytometry using BD FACSCanto RUO-ORANGE analyzer. Data were analyzed using FlowJo v10 software (Tree Star).
Cytotoxicity and cell death detection assay
Quantitative measurement of cytotoxicity and cell death in ssRNA40 exposed HMG cells was performed using Cell Death Detection ELISAPlus Kit (Roche Cat# 11774425001) and LDH Cytotoxicity Detection Kit (Takara Bio Inc. Cat# MK401). Briefly, following 24h and 48h treatment with ssRNA40 or ssRNA41, LDH release was measured in the culture supernatants by reading the absorbance at 490nM and cells were lysed with 200µl lysis buffer for 30 min. Cytoplasmic fractions were collected from lysates following centrifugation and analyzed for nucleosomal DNA release by ELISA using antibodies against DNA and histones. Neuronal cytotoxicity and cell death was also measured using LDH Cytotoxicity Detection Kit (Takara Bio Inc. Cat# MK401) and Cell Death Detection ELISAPlus Kit (Roche Cat# 11774425001) as described above. Briefly, culture supernatants collected from ssRNA40 exposed HMG cells were used to treat HPN for 24h at 37°C. Following incubation, cytoplasmic fractions were collected as described above and analyzed for nucleosomal DNA release by ELISA. Culture supernatants were analyzed for LDH release by ELISA.
Active caspase-1 measurement assay
The levels of active caspase-1 were quantified in live cells using FLICA 660 Caspase-1 Assay Kit (ImmunoChemistry Technologies Cat# 9122). This assay employs a fluorescent inhibitor probe 660-YVAD-fmk to label active caspase-1 in living cells. Briefly, following 24h treatment with ssRNA40 or ssRNA41, HMG cells were washed with PBS and incubated with FLICA™ 660-YVAD-fmk (1:60 dilution) for 30min. After two washes with PBS, cells were further stained with Aqua stain (Live/Dead™ Fixable Dead cell Stain Kit; Molecular Probes Cat# L34957). Cells were resuspended in PBS for flow cytometry analysis using BD FACSCanto RUO-ORANGE analyzer. Data were analyzed using FlowJo v10 software (Tree Star). The levels of active caspase-1 were also measured in the culture supernatants using the Human Caspase-1/ICE Quantikine ELISA Kit (R&D Systems Cat# DCA100). This assay specifically measures the active caspase-1 by using an antibody against p20 subunit of caspase-1.
Cell death detection by Annexin V/propidium iodide (PI) staining
For cell death detection by Annexin V/PI staining, HMG cells incubated with ssRNA40 or ssRNA41 were washed with PBS and then stained with Annexin V and PI (Molecular Probes Cat# V13242) according to the manufacturer’s instructions. Following staining, cells were immediately obtained for flow cytometry analysis. Data were analyzed using FlowJo v10 software (Tree Star).
Immunofluorescence Microscopy
Following treatment with ssRNA40 or ssRNA41 cells were fixed with 4% (w/v) paraformaldehyde (Sigma-Aldrich Cat# P6148) and permeabilized with 0.25% (v/v) TritonX-100 (Sigma-Aldrich Cat# T9284). For immunofluorescence experiments cells were incubated with appropriate primary antibodies: Parkin (Santa Cruz Biotechnology Cat# sc-30130, RRID:AB_653855), PINK1 (Abcam Cat# ab23707, RRID:AB_447627), SQSTM1/p62 (Abcam Cat# ab56416, RRID:AB_945626), Optineurin (Abcam Cat# ab23666, RRID:AB_447598), MAP2 (Novus Cat# NB300–213, RRID:AB_2138178), TOM20 (Santa Cruz Biotechnology Cat# sc-17764, RRID:AB_628381), TOM20 (Santa Cruz Biotechnology Cat# sc-11415, RRID:AB_2207533), ASC (Santa Cruz Biotechnology Cat# sc-30130, RRID: AB_2737351) followed by incubation with Alexa Fluor-conjugated secondary antibodies (Molecular Probes). Cells were nuclear stained and mounted using Prolong Gold Antifade mountant with 4’6-diamidino-2-phenylindole (DAPI) (Molecular Probes Cat# P36935). Stained cells were visualized using Olympus Fluoview FV-1000 confocal imaging system and minimally processed using Adobe Photoshop CS6.
RNA Isolation and Real-Time PCR
Total RNA was isolated using RNeasy Mini kit (Qiagen Cat# 74104) according to the manufacturer’s protocol and 150–300 ng RNA was used in 20–40 μl of reverse transcription reaction using high-capacity cDNA Reverse Transcription Kit (Applied Biosystems Cat# 4368814). TaqMan Gene expression Assay (Applied Biosystems Cat# 4331182) were used for IL1B (Hs01555410_m1), IL1A (Hs00174092_m1), TNF (Hs00174128_m1), IL18 (Hs01038788_m1), C1qA (Hs00381122_m1) and gapdh (Hs02786624_g1) qPCR analysis. TaqMan Non-coding RNA Assay (Applied Biosystems Cat# 4426961) was used for miR-223 (Hs04333910_s1) qPCR analysis. qPCR reactions were performed in triplicate following manufacturer’s instructions. mRNA or miRNA levels were normalized with gapdh (Hs02786624_g1) and U6snRNA (Assay ID: 001093), respectively, by the formula 2−ΔCt, where ΔCt= CtgeneX/miRX-Ctgapdh/U6. All qRT-PCR assays were performed in triplicate and the data presented as mean ± standard deviation (SD).
Statistical Analysis
Comparison between groups was performed using the paired, two-tailed, Student’s t test, or one-way Anova as appropriate. Differences with a p-value <0.05 were considered statistically significant.
RESULTS
HIV-1 RNA primes expression of inflammasome molecules and neurotoxic cytokines in microglial cells
Inflammation is thought to play a central role in HIV-associated neurological disorders that persist despite sustained viral suppression with ART. Since during suppressive treatment, replication competent virus can be absent but HIV mRNA remains detectable, we designed our initial experiments to examine the effect of GU-rich ssRNA within the HIV LTR (ssRNA40) on human primary monocyte derived microglia (HMG) cell activation of inflammatory markers associated with the NLRP3 inflammasome. We chose ssRNA40 because the mRNAs from the HIV LTR can be made despite ART suppression (Baxter et al., 2018; Golumbeanu et al., 2018; Pollack et al., 2017) and ssRNA40 is known to signal through TLR7/8 (Alter et al., 2007; Guo et al., 2014).
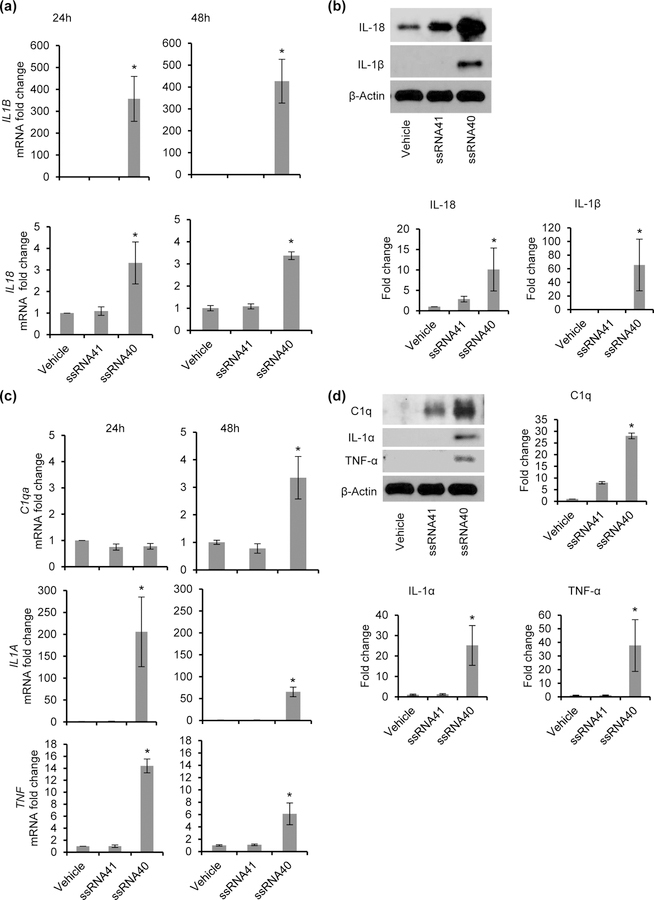
HMG were treated with ssRNA40 or ssRNA41 and harvested at 24h and 48h for inflammatory and neurocytotoxic cytokine expression and release. Following treatment, ssRNA40 compared to ssRNA41- or vehicle-treated controls increased the expression of IL-1β and IL-18 at both the mRNA (Figure 1a) and protein level (Figure 1b). In the presence of ssRNA40, HMG exhibited >5-fold increase in IL-18 protein expression (Figure 1b) and >25-fold increase in IL-1β protein expression compared to the ssRNA41 control (Figure 1b). Thus, ssRNA40 can induce the expression of key inflammasome associated cytokines, IL-18 and IL-1β, in microglia cells.
Figure 1. HIV ssRNA40 increases expression of key inflammasome molecules and neurocytotoxicity inducing cytokine gene expression in microglial cells.
HMG cells were treated with ssRNA40 (5μg/mL), ssRNA41 (5μg/mL) or vehicle (LyoVec) control for 24h and 48h and analyzed for expression of IL-1β, IL-18, C1q, IL-1α and TNFα by qPCR and immunoblotting. (a) Relative fold change in IL1B and IL18 mRNA. (b) Top, Immunoblot showing expression of IL-1β, IL-18 and human β-Actin. Bottom, relative fold change in IL-1β, IL-18 protein normalized to β-Actin and compared to control cells (vehicle) (c) Relative fold change in mRNA expression of neurocytotoxicity inducing cytokines C1qa, IL1A, and TNF mRNA in microglia cells exposed to HIV RNA for 24h and 48h. (d) Left, Immunoblots showing expression of C1q, IL-1α, TNF-α, and β-Actin. Right, relative fold change in C1q, IL-1α, TNF-α protein normalized to β-Actin and compared to control cells (vehicle). Results are presented as mean ± SD, n=3; *P<0.05.
LPS-induced CNS neuroinflammation and neurodegenerative diseases have been found to involve formation of neurotoxic, reactive A1 astrocytes (Sofroniew & Vinters, 2010), and activated microglia cells can induce A1 astrocyte formation through secretion of C1q, IL-1α and TNF-α cytokines (Liddelow et al., 2017). These A1 astrocytes appear to play a central role in neural inflammation through the secretion of neurotoxins that induce rapid death of neurons and oligodendrocytes. Having demonstrated that ssRNA40 induces the expression of key inflammasome molecules, we hypothesized that ssRNA40 would also induce the expression of neurotoxic cytokines C1q, IL-1α and TNF-α in microglial cells (Figures 1c and 1d). Similar to the NLRP3 inflammasome associated cytokines IL-18 and IL-1β, ssRNA40 also induced the expression of neurotoxic A1 astrocyte inducing cytokines at both the mRNA (Figure 1c) and protein levels (Figure 1d). Microglia stimulation with ssRNA40 resulted in >25-fold increase in C1q protein expression (Figure 1d), >15-fold increase in IL-1α (Figure 1d) and >20-fold increase in TNF-α expression by 48h compared to scrambled ssRNA41 treated control microglia (Figure 1d). Thus, ssRNA40 can activate human microglia cells to induce the expression of key cytokines that are inflammatory and neurotoxic (Figures 1a–1d).
HIV-RNA mediated processing and release of inflammatory and neurotoxic cytokines
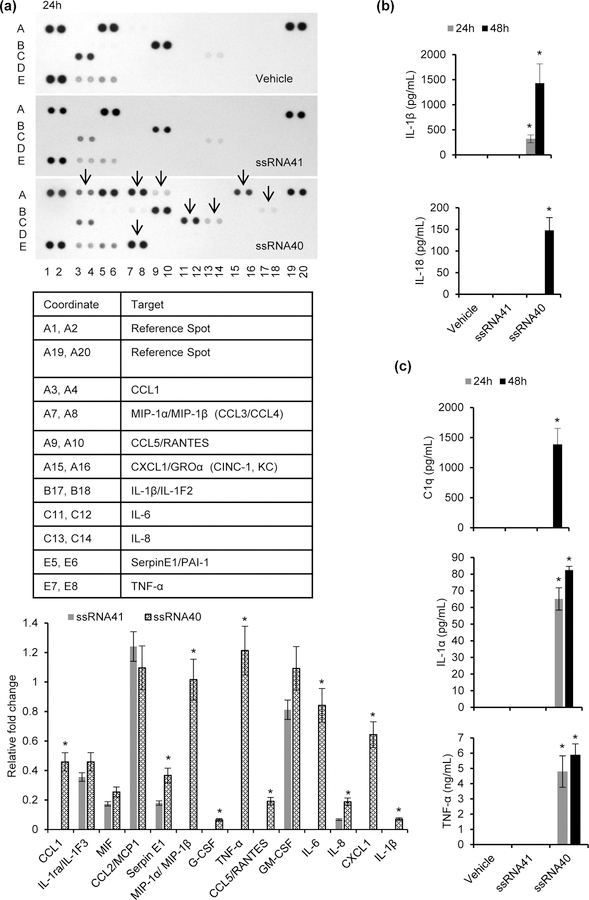
In addition to secreting cytokines that mediate inflammation and neurotoxicity, activated microglia are known to secrete chemokines that can disrupt the blood brain barrier permitting the infiltration of infected monocytes and leukocytes into the CNS. To assess a broad number of potential cytokines secreted by microglia during HIV RNA exposure, HMG were treated with vehicle, ssRNA41 or ssRNA40, and analyzed for expression of different human cytokines and chemokines using a proteome profiler human cytokine array (Figure 2a). Following exposure to ssRNA40, HMG increased expression and secretion of CCL1, CCL3, CCL4, CCL5, IL-6, IL-8, CXCL1, SerpinE1, TNF-α and IL-1β (Figure 2a top and bottom). ssRNA41 treated HMG exhibited minimal or no expression of these cytokines and chemokines in their culture supernatants as observed in the densitometric analysis for ssRNA41 and ssRNA40 cytokine array immunoblots (p<0.05, Figure 2a, bottom). We also performed a quantitative analysis for some of the key NLRP3 inflammasome and neurocytoxicity associated cytokines by ELISA (Figures 2b, 2c). Both IL-1β and IL-18 were exclusively released in culture supernatants derived from ssRNA40 exposed HMG (p<0.001, Figure 2b) and suggest that the NLRP3 inflammasome pathway is induced in microglia following treatment with ssRNA40 for 24h (Figure 2b). In addition, we also confirmed by ELISA the release of C1q, IL-1α and TNF-α in the culture supernatants derived from ssRNA40 exposed HMG (Figure 2c). Similar to the induction of inflammatory cytokines, key neurotoxic cytokines were exclusively released from HMG exposed to ssRNA40 (p<0.01, Figure 2c). None of these inflammatory and neurotoxic cytokines were observed in the ssRNA41 exposed microglial cell culture supernatants (Figures 2b, 2c).
Figure 2. Inflammatory cytokines and neurocytotoxic cytokines released from human microglia following exposure to HIV ssRNA40.
(a) Top, membrane based array immunoblot showing expression profile of human cytokines and chemokines in culture supernatants from HMG cells exposed to ssRNA40, ssRNA41 or vehicle for 24h. Bottom, relative fold change in expression of selective cytokines and chemokines compared to control cells (ssRNA41). (b) Inflammatory cytokines IL-1β and IL18 release in culture supernatants from HMG cells treated with ssRNA40, ssRNA41 or vehicle for 24h and 48h. (c) Neurocytoxicity inducing cytokines C1q, IL-1α and TNF-α release in culture supernatants from HMG cells treated with ssRNA40, ssRNA41 or vehicle for 24h and 48h measured by ELISA. Results are presented as mean ± SD, n=3; *P<0.05.
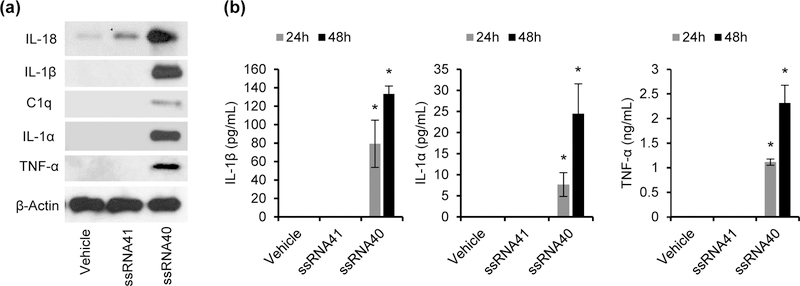
Although we have shown previously that HMG closely resemble HFMG at both the molecular and physiological levels (Rawat & Spector, 2017), we assessed in the limited number of HFMG available if IL-18, IL-1β, C1q, IL-1α and TNF-α were also upregulated following exposure with ssRNA40. Following the same methods used for HMG, each of the key cytokines previously identified were also found to be upregulated by protein expression and secretion in HFMG culture supernatants (Figures 3a and 3b).
Figure 3. HIV ssRNA40 induces inflammatory cytokines, and neurocytotoxicity inducing cytokine expression and release from primary human fetal microglia.
(a) HFMG cells were treated with ssRNA40 (5μg/mL), ssRNA41 (5μg/mL) or vehicle (LyoVec) control for 24h and 48h and analyzed for expression of IL-18, IL-1β, C1q, IL-1α, TNF-α and β-Actin by immunoblotting at 48h. (b) Culture supernatants from HFMG cells treated with ssRNA40, ssRNA41 or vehicle for 48h were analyzed for IL-1β, IL-1α and TNF-α release by ELISA. Representative data from two independent donors repeated in triplicate. Results are presented as mean ± SD, n=3; *P<0.05.
HIV RNA mediated activation of microglial cells is neurotoxic to human primary neurons
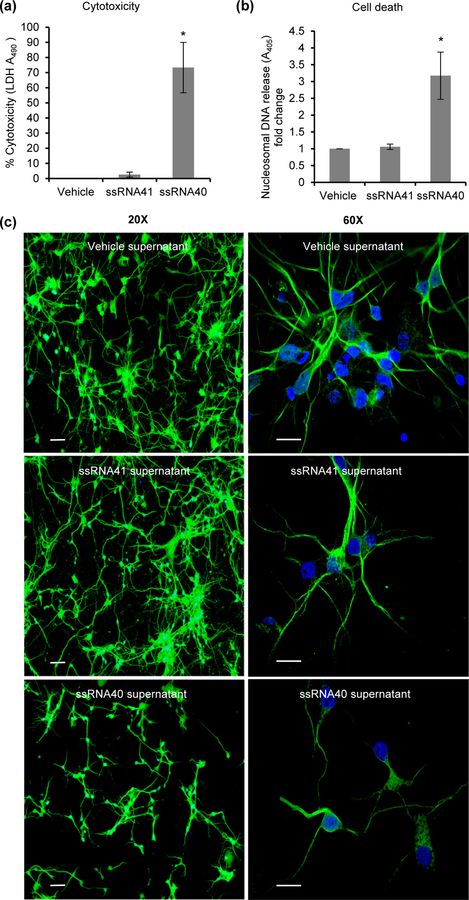
Although to this point, we had shown that specific cytokines and chemokines are released during ssRNA40 activation of microglial cells, it was important to identify the functional activity of these proteins to induce neurotoxicity of HPNs. For these experiments, HPNs were cultured with neuronal media mixed with culture supernatants collected from HMG treated with vehicle, ssRNA41 or ssRNA40 in a 1:10 ratio. After 24h, cells were analyzed for neurotoxicity and viability by LDH assay (Figure 4a) and cell death by nucleosomal DNA release (Figure 4b). Incubation of HPN with culture supernatants from ssRNA40 treated HMG showed increased cytotoxicity as measured by both assays (Figures 4a and 4b). These neurotoxic effects were further confirmed using MAP2 antibody staining and immunofluorescence microscopy (Figure 4c). ssRNA40 activated HMG derived culture supernatants were again found to be cytotoxic having a deleterious effect on HPN number (Figure 4c, 20X view) and the neurite length (Figure 4c, 60X view) compared to control cells.
Figure 4. HIV ssRNA40 mediated activation of microglia can induce cytotoxicity and cell death in human primary neurons.
Cell culture supernatants were collected from ssRNA40, ssRNA41 and vehicle treated HMG cells at 48h. HPN cells were cultured in 10% (v/v) neurons culture medium mixed with supernatants derived from HMG cells for 24h at 37°C. (a) LDH release in HPN culture supernatants at 24h post incubation with HMG derived supernatants. 10% neuronal culture medium was used for background normalization. (b) Cell death analysis in HPN cells incubated with 10% HMG derived culture supernatants using ELISA based determination of nucleosomal DNA in the cytoplasmic fraction. Data are presented as mean ± SD, n=4; *P<0.05. (c) Representative immunofluorescent images of HPN cells treated with 10% culture supernatants from HMG cells and stained with MAP2 (green) and DAPI (blue) showing a decrease in HPN number and neurite length. Scale bar indicates 20µm in 20X view and 10µm in 60X view (n=2).
IL-1β release in response to HIV RNA is dependent on NLRP3 inflammasome activation
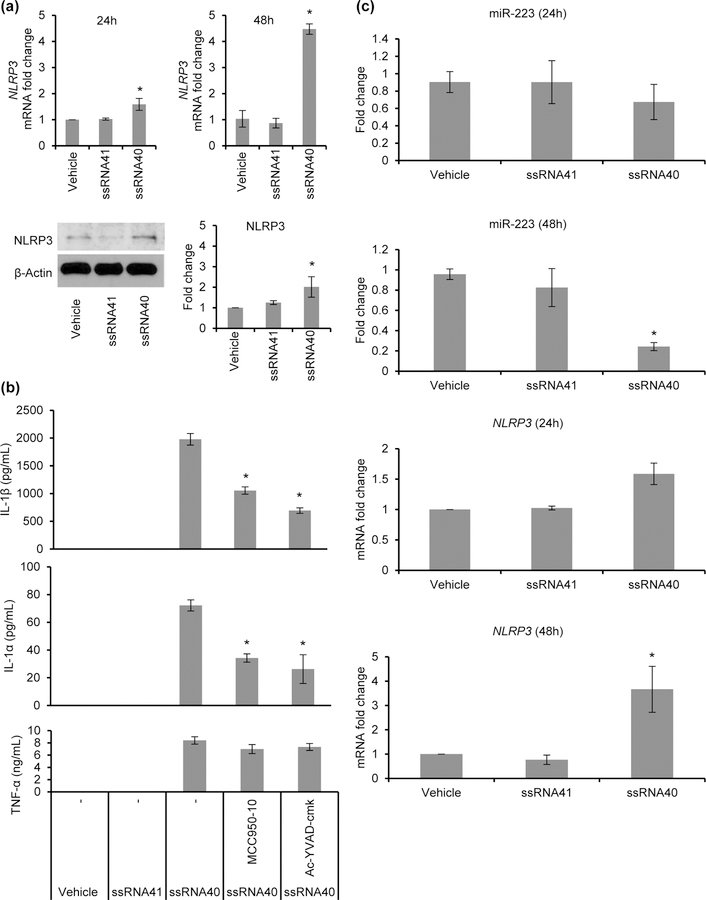
Having shown the release of cytokines that are both pro-inflammatory and induce differentiation of A1 astrocytes when microglial cells are exposed to ssRNA40, we next assessed whether this induction requires NLRP3 inflammasome activation. Activation of the NLRP3 inflammasome involves pro-caspase-1 processing and activation with subsequent caspase-1 dependent maturation of proIL-1β to mature IL-1β. The expression of NLRP3 was evaluated by qPCR and immunoblotting. Following microglial cell treatment with ssRNA40 for 48h, there was a >4-fold increase in NLRP3 mRNA (p=0.02, Figure 5a, top) and >2-fold increase in NLRP3 protein (p=0.03, Figure 5a, bottom) compared to untreated and ssRNA41 treated control cells.
Figure 5. HIV ssRNA40 activates NLRP3 inflammasome and inhibits the expression of its negative regulator, miR-223.
(a) HMG cells were treated with ssRNA40 for 24h and 48h; NLRP3 levels were analyzed with qPCR (top) and immunoblotting (bottom). Left, representative immunoblots for NLRP3 and β-Actin using antibody to NLRP3 and human β-Actin and right, densitometric analysis for fold change in NLRP3 expression from 3 independent donors. Results are presented as mean ± SD, n=3; *P<0.05. (b) Pharmacologic inhibition of NLRP3 by MCC950 inhibits ssRNA40 induced inflammasome activation in microglia cells. HMG cells were pre-treated with NLRP3 inflammasome inhibitor, MCC950 (10μM), or caspase-1-specific inhibitor Ac-YVAD-cmk (50μM), or vehicle control prior to their exposure to ssRNA40, ssRNA41 or vehicle control and production of IL-1β, IL-1α, and TNF-α in culture supernatants were measured at 24h post treatment by ELISA. (c) HMG cells were exposed to ssRNA40 (5μg/mL), ssRNA41 or vehicle control and monitored for expression of miR-223, and NLRP3 mRNA at 24h and 48h post treatment time points. Results are presented as mean ± SD, n=3; *P<0.05.
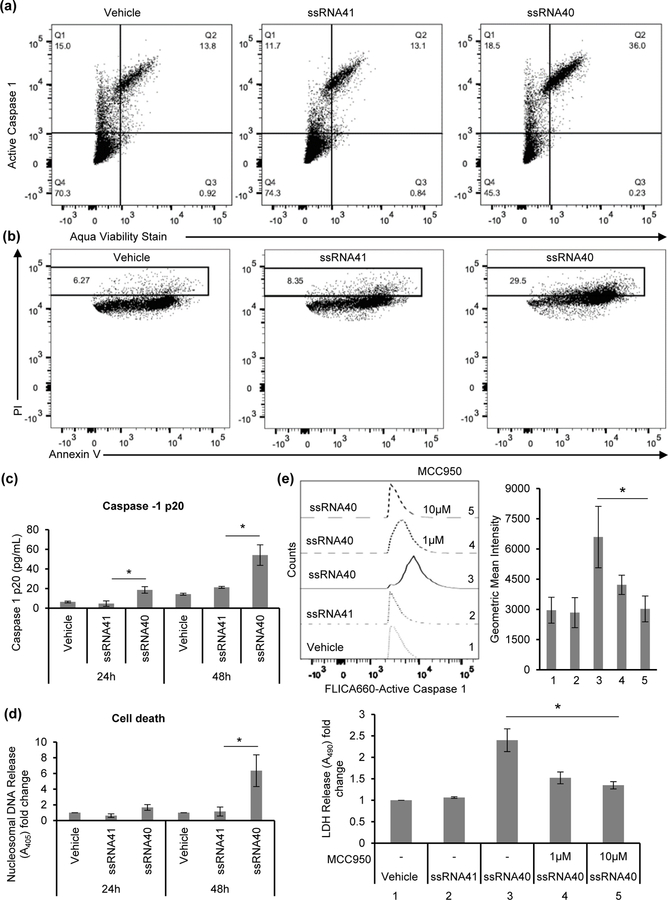
After confirming the NLRP3 inflammasome priming event, we next analyzed its activation in presence of ssRNA40. MCC950 is a known potent and selective small molecule inhibitor of NLRP3 that specifically inhibits activation of the NLRP3 inflammasome and reduces IL-1β production in vivo (Coll et al., 2015). Here, HMG were pre-treated with MCC950 or the caspase-1 inhibitor Ac-YVAD-cmk for 2h, after which cells were treated with ssRNA40 for 24h. Post 24h incubation, cell culture supernatants were analyzed for IL-1β, IL-1α and TNF-α release. NLRP3 inflammasome inhibition by MCC950 significantly reduced the processing and release of IL-1β following ssRNA40 treatment. Overall, there was >50% reduction in IL-1β and IL-1α release in culture supernatants following MCC950 and AcYVAD-fmk pretreatment compared to untreated ssRNA40 exposed microglial cells (p<0.01, Figure 5b). Since TNF-α secretion is not caspase-1 dependent, as expected, no significant difference was observed in TNF-α secretion from MCC950 /AcYVAD-fmk pre-treated and vehicle pre-treated ssRNA40 exposed microglial cells (Figure 5b). Thus, IL-1β release following ssRNA40 treatment is at least in part dependent on the activation of NLRP3 and requires its activation for IL-1β release.
MicroRNA-223 (miR-223) is a myeloid specific miRNA with regulatory effects on NLRP3 inflammasome activity. By binding to a conserved site on the 3’ untranslated region of NLRP3, miR-223 reduces NLRP3 inflammasome activity (Bauernfeind et al., 2012; Haneklaus et al., 2012). Previously Chivero et al reported that Tat-mediated inflammasome activation decreases miR-223 expression (Chivero et al., 2017). Here, we assessed the levels of miR-223 and NLRP3 expression following ssRNA40 treatment of HMG. Quantitation of miR-223 and NLRP3 mRNA was performed by qPCR following treatment of microglia with ssRNA40 for 24h and 48h (Figure 5c). Untreated or ssRNA41 treated cells were used as controls. miR-223 levels were significantly downregulated in ssRNA40 exposed microglia at 48h post-treatment (Figure 5c). Although the exposure to ssRNA40 significantly reduced the expression of miR-223, the expression of NLRP3 mRNA was enhanced in ssRNA40 treated microglia (Figure 5c). These findings suggest that ssRNA40 also promotes NLRP3 inflammasome activity by downregulating the levels of miR-223 in HMG cells.
HIV ssRNA induced inflammasome activation leads to mitochondrial damage in microglial cells
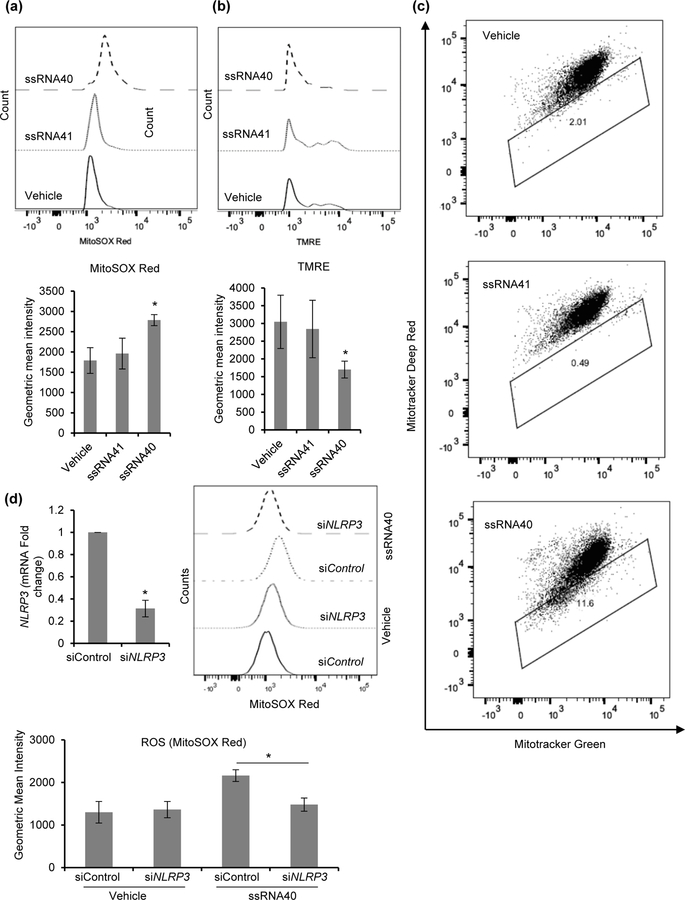
Markers of mitochondrial damage have been observed in macrophages during inflammasome activation (Zhou, Yazdi, Menu, & Tschopp, 2011). Reactive oxygen species (ROS) generating mitochondria provide secondary signaling to the NLRP3 inflammasome. To assess the ssRNA40 mediated mitochondrial damage in microglial cells, we used three types of mitochondria labels that quantify respiring mitochondria (Mitotracker Deep Red), total mitochondria (Mitotracker Green) and ROS (MitoSOX) generating mitochondria using fluorescence-activated cell sorting (Dingley, Chapman, & Falk, 2012). We also used tetramethylrhodamine ethyl ester (TMRE) to measure mitochondrial membrane potential (ΔΨm). HMG were treated with vehicle, ssRNA41 or ssRNA40 for 24h, and analyzed for specific mitochondrial markers. ROS generation is significantly increased during mitochondrial damage in macrophages (Zhou et al., 2011). MitoSOX red staining, a fluorescent label for ROS generating mitochondria, was significantly increased in ssRNA40 treated cells (p=0.007) indicating greater ROS generating damaged mitochondria in these cells compared to control cells (Figure 6a). TMRE staining, a ΔΨm sensitive dye, indicated enhanced mitochondrial depolarization during ssRNA40 stimulation in HMG cells (Figure 6b). ssRNA40 treated HMG exhibited a significant decrease in ΔΨm compared to vehicle and ssRNA41 treated control cells (p = 0.016). Dissipation of ΔΨm was also examined by co-staining with Mitotracker Green and Mitotracker Deep Red stains which the cells take up in a ΔΨm independent and dependent manner, respectively. In the case of ssRNA40 treated cells, initial mitochondrial hyperpolarization was followed by depolarization as indicated by a decrease in Mitotracker Deep Red staining (Figure 6c). Vehicle and ssRNA41 treated control cells did not show any significant changes in mitochondrial polarization (Figure 6c). To further confirm the role of NLRP3 in ssRNA40 mediated mitochondrial damage in microglia cells, we employed RNA interference to knockdown the expression of NLRP3 (Figure 6d, top left). We then quantified the ROS generating damaged mitochondria in control and NLRP3 depleted HMG cells in presence of ssRNA40 using MitoSOX red staining and flow cytometry analysis (Figure 6d). MitoSOX red staining was significantly decreased in NLRP3 depleted cells (p=0.02) indicating lower ROS generating damaged mitochondria in NLRP3 depleted HMG cells in presence of ssRNA40 compared to control siRNA transfected ssRNA40 exposed HMG cells (p=0.02, Figure 6d). These mitochondrial studies further confirm that ssRNA40 mediated NLRP3 inflammasome activation is associated with loss of mitochondrial integrity and ROS generation (Figures 6a–6d).
Figure 6. HIV ssRNA40 mediated NLRP3 inflammasome activation is associated with mitochondrial damage in microglial cells.
HMG cells were primed with ssRNA40, ssRNA41 or vehicle for 24h and stained with (a) MitoSOX Red (b) TMRE (black solid line, Vehicle; black dotted line, ssRNA41; black dashed line, ssRNA40), and (c) Mitotracker Green and Mitotracker Deep Red. (a) Representative histogram indicating increase in ROS generation in ssRNA40 treated HMG cells. (b) Representative histogram showing loss of mitochondrial membrane potential (Δψm) in ssRNA40 stimulated HMG cells. (c) HMG cells were treated with ssRNA40, ssRNA41 or vehicle for 24h followed by staining with Mitotracker Deep Red and Mitotracker Green. An increase in gated cells which were less brightly stained with Mitotracker Deep Red (i.e., downward shift) is indicative of loss of Δψm. Data are representative of three independent donors (n=3, *P<0.05). (d) HMG transfected with non-specific control siRNA (siControl), or NLRP3 siRNA (siNLRP3) were exposed to vehicle (LyoVec) or ssRNA40 (5µg/mL) for 24h. At 24h, cells were harvested and analyzed for NLRP3 expression by qPCR. ROS generating mitochondria were analyzed by intracellular MitoSOX red staining. Left, relative fold change in NLRP3 mRNA. Right, representative histogram showing decrease in ssRNA40 induced ROS generation in NLRP3 silenced HMG cells (top) and the respective geometric mean intensity (bottom). Data are representative of three independent donors (n=3, *P<0.05).
Expression of PINK1 and Parkin is increased following ssRNA40 induced mitochondrial damage in human microglia
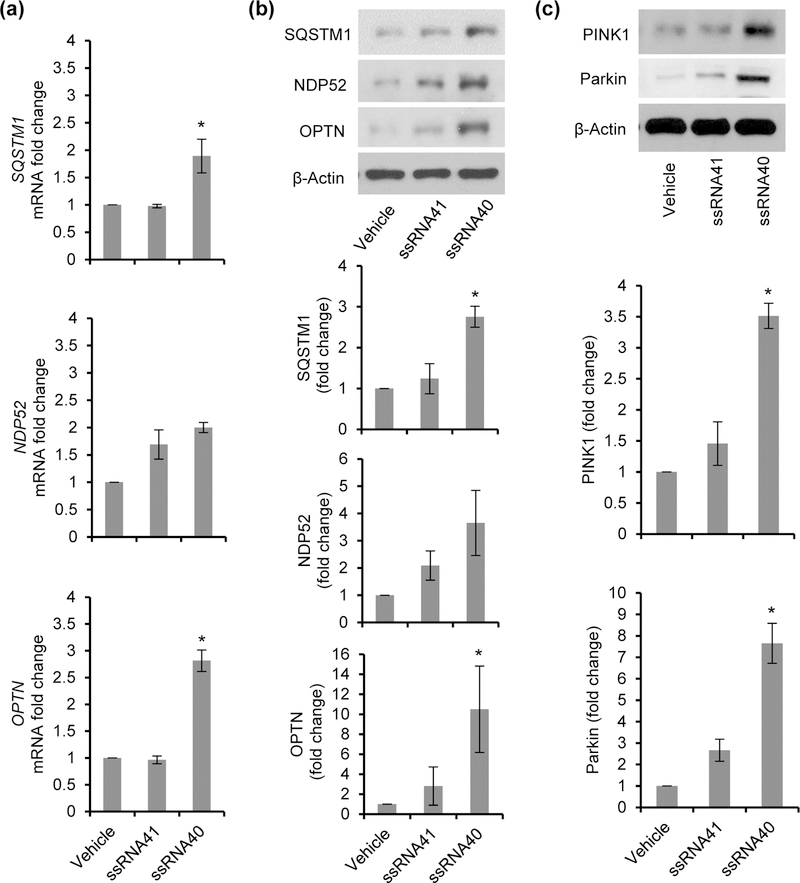
Several neurodegenerative disorders including Parkinson’s disease and Alzheimer’s disease exhibit signs of mitochondrial damage and inflammasome activation (Golpich et al., 2017; Guo, Callaway, & Ting, 2015; Saresella et al., 2016; Sarkar et al., 2017). Mitochondrial destabilization and damage enhances the release of mitochondrial DNA and ROS generation. Additionally, the inhibition of mitophagy/autophagy leads to the accumulation of ROS generating damaged mitochondria leading to the activation of the NLRP3 inflammasome (Nakahira et al., 2011; Saitoh et al., 2008; Zhou et al., 2011). Autophagy receptors, NBR1, OPTN, p62/SQSTM1, NDP52 and TAX1BP1 also play an important regulatory role during inflammasome activity by binding to damaged mitochondria and subsequently targeting them for autophagic degradation by binding to LC3 (Deng et al., 2017; Lazarou et al., 2015). In the case of LPS primed macrophages, p62/SQSTM1 is specifically induced in a NF-κB dependent manner and restricts excessive inflammasome activation (Zhong et al., 2016). Numerous studies also suggest that autophagy receptors play an important role in regulating the inflammasome activity by elimination of these damaged mitochondria and thus prevent excessive inflammasome activation, IL-1β release and caspase-1 induced death (pyroptosis) (Shi et al., 2012; Takahama, Akira, & Saitoh, 2018; Yu et al., 2014). Therefore in our next set of experiments, we analyzed the expression of autophagy receptors, p62/SQSTM1, NDP52 and OPTN in ssRNA40 exposed HMG. HMG cells were treated with vehicle, ssRNA40 or ssRNA41 for 24h as previously described and expression of specific autophagy receptors was analyzed by qPCR and immunoblotting. Following treatment with ssRNA40, expression of p62/SQSTM1 and OPTN was significantly induced at both mRNA (p <0.05) (Figure 7a) and protein level (p <0.005) (Figure 7b) compared to ssRNA41 treated control cells. However, a small but not significant increase was observed in NDP52 expression (p=0.27, Figure 7a) (p=0.22, Figure 7b).
Figure 7. Inflammasome activation by HIV ssRNA40 induces autophagy receptor expression in microglial cells.
(a) Expression analysis of autophagy receptors by qPCR following 24h exposure of HMG cells to ssRNA40. Results are presented as mean ± SD, n=3; *P<0.05. (b) Top, representative immunoblots showing expression of different autophagy receptors in HMG cells at 24h post treatment with ssRNA40. Bottom, densitometric analysis for 3 independent donors showing fold changes in expression of SQSTM1, NDP52 and OPTN in ssRNA40 stimulated cells compared to vehicle treated cells. (c) Top, representative immunoblots for PINK1, Parkin and β-Actin expression in lysates from HMG stimulated with ssRNA40, ssRNA41 or vehicle for 24h. Bottom, densitometric analysis for three independent donors showing fold change in expression of PINK1 and Parkin in ssRNA40 treated cells compared to vehicle treated cells. Results are presented as mean ± SD, n=3; *P<0.05.
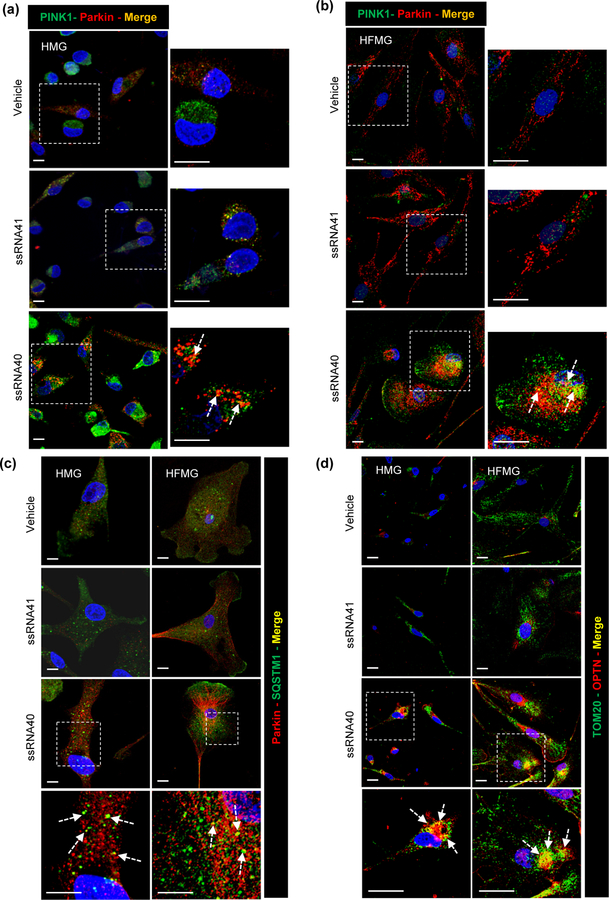
PINK1 recruits Parkin to the depolarized mitochondria to drive mitophagy (Narendra et al., 2008; D. P. Narendra et al., 2010). Previous studies have identified an important role for PINK1 and Parkin proteins in elimination of damaged mitochondria through mitophagy (Matsuda et al., 2010; Narendra et al., 2008; D. P. Narendra et al., 2010). Thus in addition to other autophagy receptors, we also examined PINK1-Parkin mediated mitophagy in ssRNA40 exposed HMG. Microglial cells were treated as described above, and PINK1 and Parkin protein expression was analyzed by immunoblotting at 24h post treatment (Figure 7c). Microglia stimulation with ssRNA40 resulted in a 2-fold increase in PINK1 expression and 2.5-fold increase in Parkin expression compared to control HMG (p<0.05, Figure 7c). Next, we determined the recruitment of these receptors in ssRNA40 stimulated microglial cells by immunofluorescence microscopy. First, we analyzed the localization of the PINK1-Parkin complex in both HMG and HFMG cells (Figures 8a, 8b). Microglial cultures were exposed to ssRNA40 for 24h, fixed and stained with PINK1 (green) and Parkin (red) antibodies. ssRNA41 treated and untreated cells were used as controls. While untreated and ssRNA41 treated control cells showed minimal basal PINK1 and Parkin expression and co-localization (orange-yellow puncta) (Figures 8a, 8b), ssRNA40 treated HMG and HFMG cells exhibited an increase and co-localization of both PINK1 and Parkin (enlarged view, Figures 8a, 8b).
Figure 8. HIV ssRNA40 promotes mitochondrial damage and PINK1-Parkin, and autophagy receptors p62 and OPTN recruitment in microglia cells.
(a and b) Intracellular distribution of PINK1 and Parkin in HMG (a) and HFMG (b) cells stimulated with ssRNA40 for 24h examined by confocal microscopy. Arrow: PINK1 and Parkin co-localization (yellow puncta) in enlarged image (c) Representative immunofluorescent images showing intracellular distribution of SQSTM1 (green) and Parkin (red) in HMG (left) and HFMG (right) cells stimulated with ssRNA40 for 24h. Arrow: SQSTM1 and Parkin co-localization (yellow puncta) in enlarged image (d) Intracellular distribution of OPTN (red) and mitochondria (green, Tom20) following 24h stimulation with ssRNA40 in HMG (left) and HFMG (right) cells. Arrow: Tom20 and OPTN co-localization (yellow puncta) in enlarged image. Scale bar indicates 10µm. Data are representative of three independent donors (n=3).
Though p62/SQSTM1 is reported to play an important role in Parkin dependent mitophagy (Zhong et al., 2016), its exact function is unclear. A recent study showed that Parkin dependent recruitment of p62/SQSTM1 is required for mitochondrial clustering but not for mitophagy in Hela cells (Narendra, Kane, Hauser, Fearnley, & Youle, 2010). In addition to Parkin expression being significantly enhanced in HMG when exposed to ssRNA40 treatment (P=0.03, Figure 7c), we also found increased expression and colocalization of p62/SQSTM1 and Parkin in the perinuclear region compared to control cells (Figure 8c, left). We observed similar expression and co-localization pattern for SQSTM1 and Parkin in ssRNA40 treated HFMG cells (Figure 8c, right).
To further confirm the recruitment of OPTN on damaged mitochondria, HMG and HFMG cells were treated with ssRNA40 for 24h, fixed and stained with Tom20 (green) (a mitochondrial protein) and OPTN (red) (Figure 8d). Mitochondrial recruitment of OPTN was identified by co-localization of Tom20 (green) and OPTN (red) in immunofluorescent images (Figure 8d; yellow puncta). OPTN expression and its recruitment on mitochondria were specifically enhanced in ssRNA40 exposure of HMG (Figure 8d, left) and HFMG cells (Figure 8d, right).
Inflammasome activation and inhibition of mitophagy leads to caspase-1-dependent cell death of microglial cells
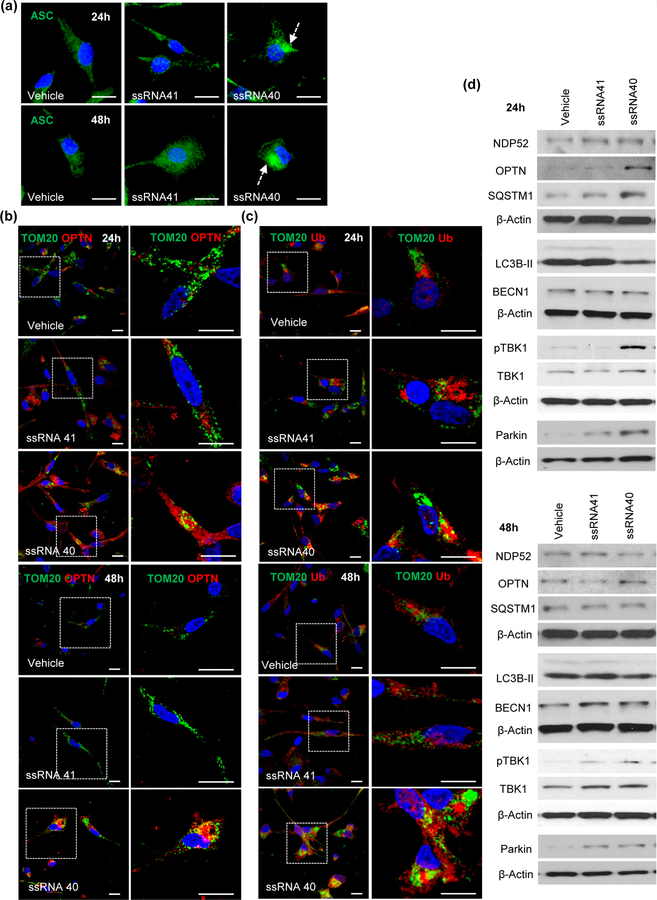
The NLRP3 inflammasome complex typically consists of three major components, the receptor NLRP3, the adaptor ASC and effector caspase-1. In normal resting cells, ASC displays a soluble cytoplasmic localization, but upon inflammasome activation, ASC receptor is mobilized to form a large single perinuclear polymeric structure, “ASC speckle”, an important marker for inflammasome activation (Cai et al., 2014; Franklin et al., 2014; Lu et al., 2014). Thus, in order to further confirm inflammasome activation, we assessed the development of ASC speckle formation in microglia in the presence of ssRNA40 (Figure 9a). For these experiments, HMG were treated with vehicle, ssRNA41 or ssRNA40 for 24h or 48h, and stained with ASC (green) (Figure 9a). NLRP3 inflammasome activation by ssRNA40 induced formation of perinuclear ASC speckle specifically in ssRNA40 treated HMG cells (Figure 9a), while vehicle and ssRNA41 treated cells displayed a diffuse cytoplasmic localization for ASC (Figure 9a).
Figure 9. HIV ssRNA40 mediated inflammasome activation results in accumulation of damaged mitochondria and reduced autophagy in microglial cells.
(a) HMG cells were treated with vehicle (LyoVec), ssRNA41 or ssRNA40. Intracellular ASC expression (green), aggregation (green speckle) and localization are analyzed by confocal immunofluorescence microscopy at 24h (top) and 48h (bottom). Perinuclear ASC speckle (dotted arrow) formation is specifically induced in ssRNA40 treated HMG cells (n=3). Intracellular expression and recruitment of OPTN (red) (b) and Poly-Ub (red) (c) on mitochondria (green, Tom20) in HMG stimulated with ssRNA40 for 24h (Top) and 48h (Bottom). Scale bar indicates 10µm. Data are representative of three independent donors (n=3). (d) Representative immunoblots showing the expression of autophagy receptors NDP52, OPTN, SQSTM1; core autophagy proteins LC3B, BECN1; and mitophagy associated proteins pTBK1, TBK1, Parkin in HMG following incubation with ssRNA40 for 24h (top) and 48h (bottom). Data are representative of three independent donors (n=3).
To this point, our findings demonstrate that ssRNA40 triggers the NLRP3 inflammasome and induces mitochondrial damage in microglial cells. However, in some cell types, activation of autophagy and mitophagy can negatively regulate NLRP3 by targeting the ubiquitinated mitochondria and inflammasome for degradation (Nakahira et al., 2011; Shi et al., 2012). Therefore, our next studies were designed to determine whether ssRNA40 mediated inflammasome activation is regulated by autophagy/mitophagy in microglial cells. HMG cells were exposed to vehicle, ssRNA41 or ssRNA40 for 24h or 48h, and stained with Tom20/OPTN and Tom20/Poly-Ubiquitin (Poly-Ub) to identify damaged mitochondria aggregates in response to ssRNA40 induced inflammasome activation (Figures 9b and 9c). Following ssRNA40 exposure, OPTN and Poly-Ub aggregates localized near perinuclear regions and colocalized with Tom20 (Figures 9b, 9c). These findings suggest that ssRNA40 induced NLRP3 inflammasome activation increases mitochondrial aggregates in microglial cells which are poorly eliminated by autophagy/mitophagy as indicated by the continued presence of these aggregates at 48h post ssRNA40 stimulation (bottom, Figures 9b, 9c).
Having identified that autophagy mediated elimination of damaged mitochondria and regulation of inflammasome activity is impaired in ssRNA40 exposed microglia cells, we next assessed the expression levels of key autophagy and mitophagy proteins in ssRNA40 exposed HMG. Lysates prepared from microglial cells treated with ssRNA40, ssRNA41, and vehicle were analyzed for expression of autophagy related proteins NDP52, OPTN, p62/SQSTM1, LC3B-II and BECN1 and mitophagy associated proteins pTBK1 and Parkin by immunoblotting (Figure 9d and Supporting Information Figure S1). At 24h, autophagy receptors OPTN and p62/SQSTM1 expression increased by 6- and 2.4-fold, respectively. By 48h, OPTN remained elevated while p62/SQSTM1 returned to the level of controls (Figure 9d and Supporting Information Figure S1a). Core autophagy proteins, LC3B-II and BECN1, expression remained unchanged following ssRNA40 treatment of microglia at both time points suggesting that autophagy fails to go to completion (Figure 9d and Supporting Information Figure S1b).
We further confirmed the block in mitophagy by analyzing the turnover of autophagy receptors OPTN, SQSTM1 and NDP52 in a cycloheximide chase experiment (Supporting Information Figure S2). Protein synthesis was inhibited in ssRNA40 treated HMG cells by adding cycloheximide (1μM) at different time points (0h, 6h, 12h and 24h) post ssRNA40 treatment, and expression of OPTN, SQSTM1 and NDP52 was analyzed. Consistent with our previous data (Figure 6), expression of autophagy receptors OPTN and SQSTM1 was increased in ssRNA40 treated cells at 24h and significantly inhibited when cycloheximide was administered between 0–24h post ssRNA40 exposure (p<0.05, Supporting Information Figure S2). However, when cycloheximide treatment was not initiated until 24h post ssRNA40 treatment, there was no significant change in OPTN expression suggesting that following prolonged ssRNA40 exposure there is inhibition of OPTN turnover (Supporting Information Figure S2). SQSTM1 expression was decreased regardless of when cycloheximide treatment was initiated (0h, 6h, 12h, 24h) post ssRNA40 exposure. However, when cycloheximide was administered at 24h post ssRNA40 exposure the inhibition of SQSTM1 was less (Supporting Information Figure S2). These data suggest that autophagy receptor expression is induced in the presence of ssRNA40 to regulate the early inflammasome activity. However, the reduced turnover following continuous exposure to ssRNA40 results in lower autophagy mediated control of late inflammasome activity in microglial cells.
The mitophagy associated protein, pTBK1, which increases recruitment of OPTN on damaged mitochondria, increased 6.5-fold at 24h (Top, Figure 9c), and similar to OPTN and p62/SQSTM1 remained increased but to a lesser extent (2-fold) at 48h post-exposure (Figure 9d and Supporting Information Figure S1c). Similarly, >3-fold increase was observed in mitophagy receptor, Parkin, expression at 24h and remained increased but to a lesser extent (2-fold) at 48h post ssRNA40 treatment (Top, Figure 9d and Supporting Information Figure S1c). Absence of regulatory functions of autophagy during inflammasome activation can activate caspase-1 and induce caspase-1 mediated pro-inflammatory cell death called pyroptosis (Yu et al., 2014). We measured the caspase-1 activation mediated cell death (pyroptosis) in ssRNA40 exposed HMG cells. Cells were stained for active caspase-1 and aqua viability stain, and analyzed by flow cytometry (Figure 10a). HIV ssRNA40 exposed HMG cells showed ~3 fold-increase in active caspase-1 mediated cell death (Figure 10a). We further confirmed the active caspase-1 mediated pyroptotic death in HMG cells during ssRNA40 mediated NLRP3 inflammasome activation by Annexin V/ PI staining (Figure 10b). Consistent with our previous observations, caspase-1 activation in HIV RNA treated cells resulted in >3-fold increase in the percentage of Annexin V/PI dual positive cells compared to the ssRNA41 control cells indicating the induction of pyroptotic cell death (Figure 10b). For quantitative measurement of rate of pyroptotic cell death following ssRNA40 mediated inflammasome activation, active caspase-1 release in culture supernatants and nucleosomal DNA release in the cytoplasm were measured by ELISA at 24h and 48h post ssRNA40 treatment. At 24h and 48h, culture supernatants derived from ssRNA40 exposed HMG contained 2-fold higher and >3-fold of active, cleaved (20 kDa) caspase-1, respectively compared to untreated and ssRNA41 controls (Figure 10c). Similar to casapse-1 activation, ssRNA40 exposed HMG showed 1.5-fold higher and >6-fold increase in nucleosomal DNA release in the cytoplasm compared to ssRNA41 treated cells by 24h and 48h (Figure 10d). Blocking NLRP3 activation with MCC950 resulted in a decrease in the caspase-1 (p<0.05, Figure 10e, top) activation and cell death (p<0.05, Figure 10e, bottom) in ssRNA40 treated cells. This observation is consistent with our previous findings demonstrating upon prolonged exposure (48h) with HIV ssRNA40, there is significant reduction in the turnover of key autophagy receptors OPTN and SQSTM1, decreased expression of mitophagy proteins Parkin and pTBK1, and decreased LC3B lipidation resulting in enhanced NLRP3 inflammasome activity and mitochondrial damage in microglial cells. NLRP3 inflammasome activation in turn increases caspase-1 activity and induces cell death (Figure 10a–10e). Together these studies strongly suggest that autophagy/mitophagy mediated regulation of NLRP3 inflammasome is impaired in ssRNA40 stimulated microglia and induces caspase-1 mediated pyroptosis (Figures 10a–10e).
Figure 10. Induction of caspase-1 mediated cell death in HIV ssRNA40 treated microglial cells.
Activation of caspase-1 in ssRNA40 treated HMG. (a) Representative flow cytometric analysis for active caspase-1 mediated pyroptotic cell death in HMG cells at 24h post ssRNA40 treatment. (b) Measurement of pyroptotic cell death in HMG cells following HIV ssRNA40 mediated NLRP3 inflammasome by Annexin V and propidium iodide (PI) staining using flow cytometric analysis. Data are representative of three independent donors (n=3, *P<0.05). (c and d) Quantitative measurement of rate of pyroptotic cell death in microglia cells following their exposure to HIV ssRNA40. Quantitative measurement of active caspase-1 (p20) expression in culture supernatants (c) and nucleasomal DNA in the cytoplasmic fraction (d) derived from HMG cells stimulated with ssRNA40 for 24h and 48h by ELISA. Results are presented as mean ± SD, n=3; *P<0.05. (e) Pharmacologic inhibition of NLRP3 by MCCP950 inhibits ssRNA40 induced pyroptosis. HMG cells were pre-treated with NLRP3 inflammasome inhibitor, MCC950 (1μM and 10μM) or vehicle control prior to their exposure to ssRNA40 (5μg/mL) for 48h. Cells were analyzed for active caspase-1 by flow cytometry and culture supernatants were analyzed for cell death by LDH release assay. Top, representative histogram (left) and geometric mean intensity measurement (right) from flow cytometric analysis for intracellular active caspase-1 expression. Bottom, quantitative measurement of LDH release in the culture supernatants. Results are presented as mean ± SD, n=3; *P<0.05.
Induction of autophagy reduces ssRNA40 induced inflammasome activity and cell death
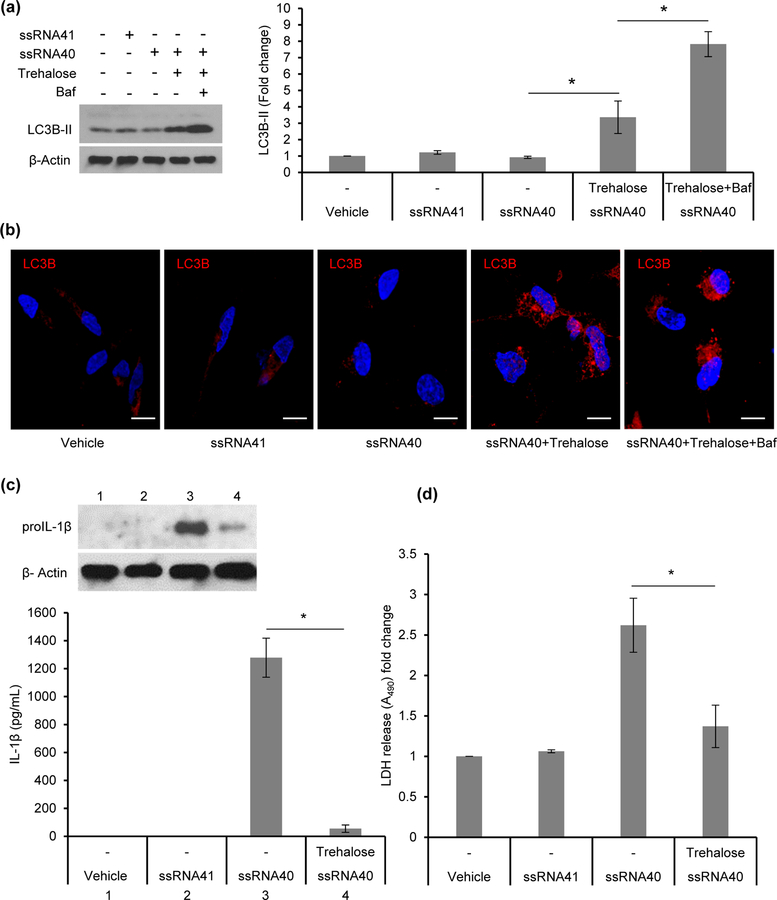
Having found that autophagy mediated regulation of inflammasome activity is impaired in ssRNA40 exposed microglial cells, we next assessed whether induction of autophagy can rescue ssRNA40 induced inflammasome activity and cell death. Although rapamycin is often used as an mTOR dependent inducer of autophagy, it can have a paradoxical effect on inflammation (Dupont et al., 2011; Weichhart et al., 2011; Yang et al., 2015). For this reason, in these studies we have used trehalose, a known mTOR independent inducer of autophagy which has been shown to promote clearance of toxic aggregates in several models of neurodegenerative disease (Du, Liang, Xu, Sun, & Wang, 2013; Sarkar, 2013; Sarkar, Davies, Huang, Tunnacliffe, & Rubinsztein, 2007; Tanaka et al., 2004; Zhang et al., 2014). After pretreatment of HMG with trehalose (100mM) and exposure with ssRNA40 for 24h, LC3B lipidation was 3-fold higher compared to controls (p=0.01, Figure 11a). To confirm that the increased lipidation is due to increased autophagic flux, HMG were treated with bafilomycin A1, a known disrupter of autophagic flux by inhibiting V-ATPase-dependent acidification and CA-P60A/SERCA-dependent autophagosome formation. Bafilomycin A1 co-treatment significantly increased LC3B-II levels in trehalose treated cells indicating that the autophagic flux induced by trehalose was disrupted with bafilomycin A1 treatment leading to accumulation of LC3B-II (p<0.005, Figure 11a). We further confirmed these findings using confocal immunofluorescence microscopy (Figure 11b). Thus, autophagy induction significantly enhanced the LC3B lipidation in ssRNA40 treated cells (Figure 11a and 11b).
Figure 11. Induction of autophagy reduces ssRNA40 mediated inflammasome activity and cell death.
HMG cells were pre-treated with trehalose (100mM) for 6h prior to their exposure to ssRNA40 (5μg/mL). Bafilomycin A1 (Baf) co-treatment with trehalose was used to confirm the increase in autophagic flux in presence of trehalose. LyoVec and ssRNA41 (5μg/mL) treated cells were used as controls. At 24h post ssRNA40 treatment, cells were harvested and analyzed for LC3B lipidation by immunoblotting and confocal immunofluorescence microscopy. (a) Left, representative immunoblot showing expression of LC3B-II and β-Actin. Right, relative fold change (densitometric analysis) in LC3B-II protein normalized to β-Actin. Results are presented as mean ±SD, n=3; *P<0.05. (b) Representative immunofluorescent images of HMG cells treated with ssRNA40 in presence or absence of trehalose. Bafilomycin A1 co-treatment was used to confirm the increase in autophagic flux in the presence of trehalose. Vehicle (LyoVec) and ssRNA41 treated cells were used as controls. At 24h post ssRNA40 treatment cells were stained with LC3B antibody (red) and DAPI (blue). Scale bar indicates 10μm (n=3). (c and d) Trehalose (100mM) pre-treated HMG cells were exposed to ssRNA40 (5μg/mL). Vehicle (LyoVec) and ssRNA41 (5μg/mL) treated cells were used as controls. At 48h post ssRNA40 treatment, cells were harvested and lysates were analyzed for pro-IL-1β expression by immunoblotting and culture supernatants were analyzed for IL-1β and LDH release. (c) Top, representative immunoblot showing expression of pro-IL-1β. Bottom, quantitative measurement of inflammatory cytokine IL-1β release in culture supernatants from vehicle (LyoVec), ssRNA41, ssRNA40, and trehalose and ssRNA40 co-treated HMG cells by ELISA. Results are presented as mean ±SD, n=4; *P<0.05 (d) Cell death analysis by quantitative measurement of LDH release in the culture supernatants from vehicle (LyoVec), ssRNA41 (5μg/mL), ssRNA40 (5μg/mL), and trehalose (100mM) and ssRNA40 co-treated HMG cells by ELISA. Results are presented as mean ±SD, n=4; *P<0.05.
Next, we examined whether induction of autophagy by trehalose reduces ssRNA40 induced inflammasome activity and cell death. HMG were pre-treated with trehalose prior to exposure to ssRNA40 and at 48h cell lysates were analyzed for proIL-1β expression, and supernatants for IL-1β secretion and LDH release. In presence of trehalose, ssRNA40 induced inflammasome activation was significantly reduced as indicated by a decrease in proIL-1β expression (top, Figure 11c) and >20-fold decrease in IL-1β release (bottom, Figure 11c, p<0.005). Trehalose treatment also reduced ssRNA40 induced inflammasome mediated cell death by >50% (p<0.005, Figure 11d).
Having established that reduced autophagy/mitophagy mediated regulatory function in ssRNA40 treated microglia leads to excessive release of inflammatory cytokines, we were interested in identifying the mechanism(s) by which ssRNA40 modulates autophagy. Previous studies have shown that autophagy machinery can deliver pathogen associated molecular patterns (PAMPs) to endosomal TLRs (Ling et al., 2006), and a subset of TLR ligands are known to control autophagy upon recognition by TLRs (Campbell & Spector, 2012; Delgado, Elmaoued, Davis, Kyei, & Deretic, 2008; Heil et al., 2004). These studies demonstrate that HIV ssRNA40 can be recognized through TLR7 and TLR8 expressed in macrophage endosomes. Therefore, we examined whether the ssRNA40 mediated inflammasome activation is dependent on TLR7/8 signaling in microglia (Supporting Information Figure S3). Following knockdown of TLR7 and TLR8 using siRNA (Supporting Information Figure S3a), inflammasome activation and IL-1β release was decreased by 50%, 60% and 70% in TLR7, TLR8 and TLR7/8 depleted microglial cells, respectively (Supporting Information Figure S3b). Next, we examined whether TLR7/8 controls autophagy in ssRNA40 treated microglia. LC3B lipidation and expression of autophagy/mitophagy receptors OPTN and p62/SQSTM1 were assessed in TLR7, TLR8 and TLR7/8 depleted microglial cells in presence of ssRNA40 and compared to controls (Supporting Information Figure S3c). Partial knockdown of TLR7/8 did not produce a significant change in LC3B lipidation in ssRNA40 treated microglia cells (Supporting Information Figure S3c). However, OPTN and p62/SQSTM1 expression were decreased >2-fold in TLR8 and TLR7/8 depleted cells (p<0.05, Supporting Information Figure S3c). This decrease was not observed in TLR7 reduced cells suggesting that TLR8 signaling in microglia may have a greater role in ssRNA40 mediated inflammasome activation (Supporting Information Figure S3c).
DISCUSSION
In the era of effective ART, chronic inflammation is increasingly being identified as an important potential mechanism leading to an increased risk of many disorders including accelerated aging, cardiovascular disease and cognitive impairment. How HIV promotes inflammation despite the lack of detectable replication competent virus remains poorly understood. Recent studies demonstrating the presence of HIV transcripts without the detection of infectious virus (Baxter et al., 2018; Golumbeanu et al., 2018; Pollack et al., 2017) suggested to us that viral RNA that continues to be made in latent reservoirs could be an important driver of inflammation. In the findings reported here, we have identified that GU-rich HIV ssRNA from the LTR known to signal through TLR-7/8 induces IL-1β and IL-18 in human microglial cells through the induction of the NLRP3 inflammasome. Consistent with previous findings that reported induction of neurotoxic reactive astrocytes by activated microglia, ssRNA40 activated human microglia also induced neurotoxicity in primary human neurons through secretion of C1q, IL-1α and TNF-α cytokines (Liddelow et al., 2017).
Prior studies of RNA viruses including influenza, encephalomyocarditis virus, vesicular stomatitis virus and HIV suggest that the NLRP3 inflammasome is a common pathway of the host innate antiviral response against RNA viruses (Chattergoon et al., 2014; Guo et al., 2014; Ichinohe, Pang, & Iwasaki, 2010; Rajan, Rodriguez, Miao, & Aderem, 2011). Influenza virus and HIV were previously reported to induce NLRP3 dependent IL-1β secretion in infected macrophages and monocytes, respectively (Guo et al., 2014; Ichinohe et al., 2010). Here we demonstrate that HIV ssRNA40 alone is sufficient to induce both IL-1β expression and caspase-1 dependent processing and IL-1β secretion in human microglial cells. Unlike influenza virus, HIV ssRNA40 serves as a potent trigger for robust NLRP3 activation in human microglia even in absence of active virus replication.
NLRP3 inflammasome activation is thought to require two signals. Signal 1 primes the expression of proIL-1β, pro-IL-18 and NLRP3, and signal 2 drives the caspase-1 activation and processing of IL-1β and IL-18 (Guo et al., 2015; Kanneganti, 2010). Many RNA viruses activate signal 1 through TL7 mediated recognition of ssRNA and priming of pro IL-1β expression (Chattergoon et al., 2014; Diebold, Kaisho, Hemmi, Akira, & Reis e Sousa, 2004; Lehnardt, 2010; Lund et al., 2004); however, the mechanism of activation is not clearly defined for most of these viruses. In the case of influenza virus, the M2 protein serves as second signal and triggers robust NLRP3 activation and caspase-1 dependent IL-1β secretion (Ichinohe et al., 2010). Similarly, HIV ssRNA sensing via TLR7/TLR8 primes expression of pro-IL-1β, pro-IL-18 and NLRP3 in human primary microglia. However, for microglial cells following ssRNA40 exposure, mitochondrial damage and ROS generation may serve as the second signal to drive NLRP3 inflammasome activation, and IL-1β and IL-18 secretion.
Several previous reports suggest that autophagy is a key regulator of NLRP3 inflammasome activity (Nakahira et al., 2011; Shi et al., 2012). PINK1-Parkin mediated selective mitophagy is known to limit inflammasome activity by targeting the ubiquitinated damaged mitochondria for degradation and prevents their accumulation and ROS generation that can otherwise amplify the NLRP3 inflammasome response (Shi et al., 2012; Tschopp & Schroder, 2010). Autophagy receptors NDP52 and OPTN were previously reported to play a key role in PINK1-Parkin mediated mitophagy and exhibit tissue specific expression (Lazarou et al., 2015). Specifically, the mitochondrial fraction from brain tissue is reported to have the high expression for OPTN and p62 with minimal expression for NDP52 (Lazarou et al., 2015). Similarly, we have identified that OPTN and p62 are increased whereas NDP52 was not induced in microglial cells following exposure to ssRNA40.
PINK1-Parkin dependent TBK1 activation also plays a key role in recruitment of autophagy receptors OPTN and NDP52 on damaged mitochondria following mitochondrial depolarization to promote mitophagy (Heo, Ordureau, Paulo, Rinehart, & Harper, 2015; Richter et al., 2016). Consistent with prior reports, we found that following 24h exposure of microglia to ssRNA40, TBK1 activation is enhanced as indicated by its increased phosphorylation and recruitment of autophagy receptors OPTN and p62 on damaged mitochondria. These findings suggest that microglial cells activate mitophagy to limit early inflammasome activity. However, although there is inflammasome activation following exposure to ssRNA40, there is limited LC3B lipidation and BECN1 expression indicating that autophagic flux fails to go to completion. We found that the turnover of autophagy receptors OPTN and p62 is reduced at 48h post ssRNA40 treatment of microglia. In addition, caspase-1 activity is enhanced, and TBK1 phosphorylation and Parkin expression is reduced following prolonged exposure of microglial cells to ssRNA40, as observed previously with Pseudomonas aeruginosa exposed macrophages, where caspase-1 mediated cleavage of TRIF results in inhibition of autophagy (Jabir et al., 2014). Thus, unlike previously reported macrophage cell lines where inflammasome activation is regulated through autophagy, exposure of microglial cells to HIV ssRNA40 leads to a chronic inflammatory state with elevated levels of caspase-1 and IL-1β secretion (Shi et al., 2012).
Pyroptosis, a highly inflammatory cell death, is characterized by loss of membrane integrity and secretion of large amounts of inflammatory cytokines (Bergsbaken, Fink, & Cookson, 2009). Some earlier studies have shown that autophagy can negatively regulate pyroptosis (Shi et al., 2012; Suzuki et al., 2007; Yu et al., 2014). In agreement with these previous reports, our current findings also suggest that persistent stimulation of microglial cells with HIV ssRNA40 triggers pyroptosis due to a blockade of autophagy mediated regulation of inflammasome activity.
In summary, we showed here that HIV GU-rich ssRNA activates microglial cells via the NLRP3 inflammasome pathway, and can significantly contribute to neurotoxicity and neurodegeneration via secretion of inflammatory and neurotoxic cytokines. Blockade of autophagy/mitophagy mediated negative regulation of inflammasome activity leads to excessive release of inflammatory cytokines, caspase-1 activation, pyroptotic cell death of microglial cells and neurotoxicity that can contribute to the cognitive deficits and chronic inflammation commonly seen in HIV-infected individuals on suppressive antiretroviral treatment.
Supplementary Material
Main Points.
HIV ssRNA40 activates the NLRP3 inflammasome in microglia leading to release of inflammatory and neurotoxic cytokines, caspase-1 activation, inhibition of autophagy/mitophagy mediated regulation of NLRP3 inflammasome, and pyroptotic cell death.
ACKNOWLEDGMENTS
This work was supported, in whole or in part, by the National Institute of Neurological Disorders and Stroke of the NIH (www.ninds.nih.gov) (R01 NS084912 and R01 NS104015 to SAS) and the International Maternal Pediatric Adolescent AIDS Clinical Trials Network (impaactnetwork.org). Overall support for the International Maternal Pediatric Adolescent AIDS Clinical Trials (IMPAACT) Network was provided by the National Institute of Allergy and Infectious Diseases (www.niaid.nih.gov) (NIAID) of the National Institutes of Health (NIH) under Award Numbers UM1AI068632 (IMPAACT LOC), UM1AI068616 (IMPAACT SDMC) and UM1AI106716 (IMPAACT LC), with co-funding from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (www.nichd.nih.gov) (NICHD) and the National Institute of Mental Health (www.nimh.nih.gov) (NIMH). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We thank Rodney Trout for technical assistance.
REFERENCES
- Alter G, Suscovich TJ, Teigen N, Meier A, Streeck H, Brander C, & Altfeld M (2007). Single-stranded RNA derived from HIV-1 serves as a potent activator of NK cells. J Immunol, 178(12), 7658–7666. doi: 10.4049/jimmunol.178.12.7658 [DOI] [PubMed] [Google Scholar]
- Anderson AM, Munoz-Moreno JA, McClernon DR, Ellis RJ, Cookson D, Clifford DB, … Group C (2017). Prevalence and Correlates of Persistent HIV-1 RNA in Cerebrospinal Fluid During Antiretroviral Therapy. J Infect Dis, 215(1), 105–113. doi: 10.1093/infdis/jiw505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avramut M, Zeevi A, & Achim CL (2001). The immunosuppressant drug FK506 is a potent trophic agent for human fetal neurons. Brain Res Dev Brain Res, 132(2), 151–157. doi: 10.1016/S0165-3806(01)00307-8 [DOI] [PubMed] [Google Scholar]
- Bandera A, Masetti M, Fabbiani M, Biasin M, Muscatello A, Squillace N, … Trabattoni D (2018). The NLRP3 Inflammasome Is Upregulated in HIV-Infected Antiretroviral Therapy-Treated Individuals with Defective Immune Recovery. Front Immunol, 9, 214. doi: 10.3389/fimmu.2018.00214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauernfeind F, Rieger A, Schildberg FA, Knolle PA, Schmid-Burgk JL, & Hornung V (2012). NLRP3 inflammasome activity is negatively controlled by miR-223. J Immunol, 189(8), 4175–4181. doi: 10.4049/jimmunol.1201516 [DOI] [PubMed] [Google Scholar]
- Baxter AE, O’Doherty U, & Kaufmann DE (2018). Beyond the replication-competent HIV reservoir: transcription and translation-competent reservoirs. Retrovirology, 15(1), 18. doi: 10.1186/s12977-018-0392-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergsbaken T, Fink SL, & Cookson BT (2009). Pyroptosis: host cell death and inflammation. Nat Rev Microbiol, 7(2), 99–109. doi: 10.1038/nrmicro2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdo TH, Lackner A, & Williams KC (2013). Monocyte/macrophages and their role in HIV neuropathogenesis. Immunol Rev, 254(1), 102–113. doi: 10.1111/imr.12068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Chen J, Xu H, Liu S, Jiang QX, Halfmann R, & Chen ZJ (2014). Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell, 156(6), 1207–1222. doi: 10.1016/j.cell.2014.01.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell GR, & Spector SA (2012). Toll-like receptor 8 ligands activate a vitamin D mediated autophagic response that inhibits human immunodeficiency virus type 1. PLoS Pathog, 8(11), e1003017. doi: 10.1371/journal.ppat.1003017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattergoon MA, Latanich R, Quinn J, Winter ME, Buckheit RW 3rd, Blankson JN, … Cox AL (2014). HIV and HCV activate the inflammasome in monocytes and macrophages via endosomal Toll-like receptors without induction of type 1 interferon. PLoS Pathog, 10(5), e1004082. doi: 10.1371/journal.ppat.1004082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen NC, Partridge AT, Sell C, Torres C, & Martin-Garcia J (2017). Fate of microglia during HIV-1 infection: From activation to senescence? Glia, 65(3), 431–446. doi: 10.1002/glia.23081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivero ET, Guo ML, Periyasamy P, Liao K, Callen SE, & Buch S (2017). HIV-1 Tat Primes and Activates Microglial NLRP3 Inflammasome-Mediated Neuroinflammation. J Neurosci, 37(13), 3599–3609. doi: 10.1523/JNEUROSCI.3045-16.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen MS, Chen YQ, McCauley M, Gamble T, Hosseinipour MC, Kumarasamy N, … Team HS (2016). Antiretroviral Therapy for the Prevention of HIV-1 Transmission. N Engl J Med, 375(9), 830–839. doi: 10.1056/NEJMoa1600693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll RC, Robertson AA, Chae JJ, Higgins SC, Munoz-Planillo R, Inserra MC, … O’Neill LA (2015). A small-molecule inhibitor of the NLRP3 inflammasome for the treatment of inflammatory diseases. Nat Med, 21(3), 248–255. doi: 10.1038/nm.3806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado MA, Elmaoued RA, Davis AS, Kyei G, & Deretic V (2008). Toll-like receptors control autophagy. EMBO J, 27(7), 1110–1121. doi: 10.1038/emboj.2008.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z, Purtell K, Lachance V, Wold MS, Chen S, & Yue Z (2017). Autophagy Receptors and Neurodegenerative Diseases. Trends Cell Biol, 27(7), 491–504. doi: 10.1016/j.tcb.2017.01.001 [DOI] [PubMed] [Google Scholar]
- Diebold SS, Kaisho T, Hemmi H, Akira S, & Reis e Sousa C (2004). Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science, 303(5663), 1529–1531. doi: 10.1126/science.1093616 [DOI] [PubMed] [Google Scholar]
- Dingley S, Chapman KA, & Falk MJ (2012). Fluorescence-activated cell sorting analysis of mitochondrial content, membrane potential, and matrix oxidant burden in human lymphoblastoid cell lines. Methods Mol Biol, 837, 231–239. doi: 10.1007/978-1-61779-504-6_16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Liang Y, Xu F, Sun B, & Wang Z (2013). Trehalose rescues Alzheimer’s disease phenotypes in APP/PS1 transgenic mice. J Pharm Pharmacol, 65(12), 1753–1756. doi: 10.1111/jphp.12108 [DOI] [PubMed] [Google Scholar]
- Dupont N, Jiang S, Pilli M, Ornatowski W, Bhattacharya D, & Deretic V (2011). Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1beta. EMBO J, 30(23), 4701–4711. doi: 10.1038/emboj.2011.398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden A, Marcotte TD, Heaton RK, Nilsson S, Zetterberg H, Fuchs D, … Gisslen M (2016). Increased Intrathecal Immune Activation in Virally Suppressed HIV-1 Infected Patients with Neurocognitive Impairment. PLoS One, 11(6), e0157160. doi: 10.1371/journal.pone.0157160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etemad S, Zamin RM, Ruitenberg MJ, & Filgueira L (2012). A novel in vitro human microglia model: characterization of human monocyte-derived microglia. J Neurosci Methods, 209(1), 79–89. doi: 10.1016/j.jneumeth.2012.05.025 [DOI] [PubMed] [Google Scholar]
- Farhadian S, Patel P, & Spudich S (2017). Neurological Complications of HIV Infection. Curr Infect Dis Rep, 19(12), 50. doi: 10.1007/s11908-017-0606-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feria MG, Taborda NA, Hernandez JC, & Rugeles MT (2018). HIV replication is associated to inflammasomes activation, IL-1beta, IL-18 and caspase-1 expression in GALT and peripheral blood. PLoS One, 13(4), e0192845. doi: 10.1371/journal.pone.0192845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin BS, Bossaller L, De Nardo D, Ratter JM, Stutz A, Engels G, … Latz E (2014). The adaptor ASC has extracellular and ‘prionoid’ activities that propagate inflammation. Nat Immunol, 15(8), 727–737. doi: 10.1038/ni.2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garden GA (2002). Microglia in human immunodeficiency virus-associated neurodegeneration. Glia, 40(2), 240–251. doi: 10.1002/glia.10155 [DOI] [PubMed] [Google Scholar]
- Golpich M, Amini E, Mohamed Z, Azman Ali R, Mohamed Ibrahim N, & Ahmadiani A (2017). Mitochondrial Dysfunction and Biogenesis in Neurodegenerative diseases: Pathogenesis and Treatment. CNS Neurosci Ther, 23(1), 5–22. doi: 10.1111/cns.12655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golumbeanu M, Cristinelli S, Rato S, Munoz M, Cavassini M, Beerenwinkel N, & Ciuffi A (2018). Single-Cell RNA-Seq Reveals Transcriptional Heterogeneity in Latent and Reactivated HIV-Infected Cells. Cell Rep, 23(4), 942–950. doi: 10.1016/j.celrep.2018.03.102 [DOI] [PubMed] [Google Scholar]
- Guo H, Callaway JB, & Ting JP (2015). Inflammasomes: mechanism of action, role in disease, and therapeutics. Nat Med, 21(7), 677–687. doi: 10.1038/nm.3893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Gao J, Taxman DJ, Ting JP, & Su L (2014). HIV-1 infection induces interleukin-1beta production via TLR8 protein-dependent and NLRP3 inflammasome mechanisms in human monocytes. J Biol Chem, 289(31), 21716–21726. doi: 10.1074/jbc.M114.566620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurung P, Lukens JR, & Kanneganti TD (2015). Mitochondria: diversity in the regulation of the NLRP3 inflammasome. Trends Mol Med, 21(3), 193–201. doi: 10.1016/j.molmed.2014.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haneklaus M, Gerlic M, Kurowska-Stolarska M, Rainey AA, Pich D, McInnes IB, … Masters SL (2012). Cutting edge: miR-223 and EBV miR-BART15 regulate the NLRP3 inflammasome and IL-1beta production. J Immunol, 189(8), 3795–3799. doi: 10.4049/jimmunol.1200312 [DOI] [PubMed] [Google Scholar]
- Heaton RK, Franklin DR Jr., Deutsch R, Letendre S, Ellis RJ, Casaletto K, … Group C (2015). Neurocognitive change in the era of HIV combination antiretroviral therapy: the longitudinal CHARTER study. Clin Infect Dis, 60(3), 473–480. doi: 10.1093/cid/ciu862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, … Bauer (2004). Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science, 303(5663), 1526–1529. doi: 10.1126/science.1093620 [DOI] [PubMed] [Google Scholar]
- Heo JM, Ordureau A, Paulo JA, Rinehart J, & Harper JW (2015). The PINK1-PARKIN Mitochondrial Ubiquitylation Pathway Drives a Program of OPTN/NDP52 Recruitment and TBK1 Activation to Promote Mitophagy. Mol Cell, 60(1), 7–20. doi: 10.1016/j.molcel.2015.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinohe T, Pang IK, & Iwasaki A (2010). Influenza virus activates inflammasomes via its intracellular M2 ion channel. Nat Immunol, 11(5), 404–410. doi: 10.1038/ni.1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabir MS, Ritchie ND, Li D, Bayes HK, Tourlomousis P, Puleston D, … Evans TJ (2014). Caspase-1 cleavage of the TLR adaptor TRIF inhibits autophagy and beta-interferon production during Pseudomonas aeruginosa infection. Cell Host Microbe, 15(2), 214–227. doi: 10.1016/j.chom.2014.01.010 [DOI] [PubMed] [Google Scholar]
- Kanneganti TD (2010). Central roles of NLRs and inflammasomes in viral infection. Nat Rev Immunol, 10(10), 688–698. doi: 10.1038/nri2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarou M, Jin SM, Kane LA, & Youle RJ (2012). Role of PINK1 binding to the TOM complex and alternate intracellular membranes in recruitment and activation of the E3 ligase Parkin. Dev Cell, 22(2), 320–333. doi: 10.1016/j.devcel.2011.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, … Youle RJ (2015). The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature, 524(7565), 309–314. doi: 10.1038/nature14893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnardt S (2010). Innate immunity and neuroinflammation in the CNS: the role of microglia in Toll-like receptor-mediated neuronal injury. Glia, 58(3), 253–263. doi: 10.1002/glia.20928 [DOI] [PubMed] [Google Scholar]
- Liddelow SA, Guttenplan KA, Clarke LE, Bennett FC, Bohlen CJ, Schirmer L, … Barres BA (2017). Neurotoxic reactive astrocytes are induced by activated microglia. Nature, 541(7638), 481–487. doi: 10.1038/nature21029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling YM, Shaw MH, Ayala C, Coppens I, Taylor GA, Ferguson DJ, & Yap GS (2006). Vacuolar and plasma membrane stripping and autophagic elimination of Toxoplasma gondii in primed effector macrophages. J Exp Med, 203(9), 2063–2071. doi: 10.1084/jem.20061318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A, Magupalli VG, Ruan J, Yin Q, Atianand MK, Vos MR, … Egelman EH (2014). Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell, 156(6), 1193–1206. doi: 10.1016/j.cell.2014.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund JM, Alexopoulou L, Sato A, Karow M, Adams NC, Gale NW, … Flavell RA (2004). Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc Natl Acad Sci U S A, 101(15), 5598–5603. doi: 10.1073/pnas.0400937101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marais S, Lai RPJ, Wilkinson KA, Meintjes G, O’Garra A, & Wilkinson RJ (2017). Inflammasome Activation Underlying Central Nervous System Deterioration in HIV-Associated Tuberculosis. J Infect Dis, 215(5), 677–686. doi: 10.1093/infdis/jiw561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda N, Sato S, Shiba K, Okatsu K, Saisho K, Gautier CA, … Tanaka K (2010). PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol, 189(2), 211–221. doi: 10.1083/jcb.200910140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misawa T, Takahama M, Kozaki T, Lee H, Zou J, Saitoh T, & Akira S (2013). Microtubule-driven spatial arrangement of mitochondria promotes activation of the NLRP3 inflammasome. Nat Immunol, 14(5), 454–460. doi: 10.1038/ni.2550 [DOI] [PubMed] [Google Scholar]
- Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, … Choi AM (2011). Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol, 12(3), 222–230. doi: 10.1038/ni.1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D, Kane LA, Hauser DN, Fearnley IM, & Youle RJ (2010). p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy, 6(8), 1090–1106. doi: 10.4161/auto.6.8.13426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, & Youle RJ (2008). Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol, 183(5), 795–803. doi: 10.1083/jcb.200809125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra DP, Jin SM, Tanaka A, Suen DF, Gautier CA, Shen J, … Youle RJ (2010). PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol, 8(1), e1000298. doi: 10.1371/journal.pbio.1000298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack RA, Jones RB, Pertea M, Bruner KM, Martin AR, Thomas AS, … Ho YC (2017). Defective HIV-1 Proviruses Are Expressed and Can Be Recognized by Cytotoxic T Lymphocytes, which Shape the Proviral Landscape. Cell Host Microbe, 21(4), 494–506 e494. doi: 10.1016/j.chom.2017.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan JV, Rodriguez D, Miao EA, & Aderem A (2011). The NLRP3 inflammasome detects encephalomyocarditis virus and vesicular stomatitis virus infection. J Virol, 85(9), 4167–4172. doi: 10.1128/JVI.01687-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat P, & Spector SA (2017). Development and characterization of a human microglia cell model of HIV-1 infection. J Neurovirol, 23(1), 33–46. doi: 10.1007/s13365-016-0472-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter B, Sliter DA, Herhaus L, Stolz A, Wang C, Beli P, … Dikic I (2016). Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria. Proc Natl Acad Sci U S A, 113(15), 4039–4044. doi: 10.1073/pnas.1523926113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, … Akira S (2008). Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature, 456(7219), 264–268. doi: 10.1038/nature07383 [DOI] [PubMed] [Google Scholar]
- Saresella M, La Rosa F, Piancone F, Zoppis M, Marventano I, Calabrese E, … Clerici M (2016). The NLRP3 and NLRP1 inflammasomes are activated in Alzheimer’s disease. Mol Neurodegener, 11, 23. doi: 10.1186/s13024-016-0088-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S (2013). Regulation of autophagy by mTOR-dependent and mTOR-independent pathways: autophagy dysfunction in neurodegenerative diseases and therapeutic application of autophagy enhancers. Biochem Soc Trans, 41(5), 1103–1130. doi: 10.1042/BST20130134 [DOI] [PubMed] [Google Scholar]
- Sarkar S, Davies JE, Huang Z, Tunnacliffe A, & Rubinsztein DC (2007). Trehalose, a novel mTOR-independent autophagy enhancer, accelerates the clearance of mutant huntingtin and alpha-synuclein. J Biol Chem, 282(8), 5641–5652. doi: 10.1074/jbc.M609532200 [DOI] [PubMed] [Google Scholar]
- Sarkar S, Malovic E, Harishchandra DS, Ghaisas S, Panicker N, Charli A, … Kanthasamy AG (2017). Mitochondrial impairment in microglia amplifies NLRP3 inflammasome proinflammatory signaling in cell culture and animal models of Parkinson’s disease. NPJ Parkinsons Dis, 3, 30. doi: 10.1038/s41531-017-0032-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan L, Deng K, Gao H, Xing S, Capoferri AA, Durand CM, … Siliciano RF (2017). Transcriptional Reprogramming during Effector-to-Memory Transition Renders CD4(+) T Cells Permissive for Latent HIV-1 Infection. Immunity, 47(4), 766–775 e763. doi: 10.1016/j.immuni.2017.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi CS, Shenderov K, Huang NN, Kabat J, Abu-Asab M, Fitzgerald KA, … Kehrl JH (2012). Activation of autophagy by inflammatory signals limits IL-1beta production by targeting ubiquitinated inflammasomes for destruction. Nat Immunol, 13(3), 255–263. doi: 10.1038/ni.2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada K, Crother TR, Karlin J, Dagvadorj J, Chiba N, Chen S, … Arditi M (2012). Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity, 36(3), 401–414. doi: 10.1016/j.immuni.2012.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofroniew MV, & Vinters HV (2010). Astrocytes: biology and pathology. Acta Neuropathol, 119(1), 7–35. doi: 10.1007/s00401-009-0619-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Franchi L, Toma C, Ashida H, Ogawa M, Yoshikawa Y, … Nunez G (2007). Differential regulation of caspase-1 activation, pyroptosis, and autophagy via Ipaf and ASC in Shigella-infected macrophages. PLoS Pathog, 3(8), e111. doi: 10.1371/journal.ppat.0030111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahama M, Akira S, & Saitoh T (2018). Autophagy limits activation of the inflammasomes. Immunol Rev, 281(1), 62–73. doi: 10.1111/imr.12613 [DOI] [PubMed] [Google Scholar]
- Tanaka M, Machida Y, Niu S, Ikeda T, Jana NR, Doi H, … Nukina N (2004). Trehalose alleviates polyglutamine-mediated pathology in a mouse model of Huntington disease. Nat Med, 10(2), 148–154. doi: 10.1038/nm985 [DOI] [PubMed] [Google Scholar]
- Teodorof C, Divakar S, Soontornniyomkij B, Achim CL, Kaul M, & Singh KK (2014). Intracellular mannose binding lectin mediates subcellular trafficking of HIV-1 gp120 in neurons. Neurobiol Dis, 69, 54–64. doi: 10.1016/j.nbd.2014.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschopp J, & Schroder K (2010). NLRP3 inflammasome activation: The convergence of multiple signalling pathways on ROS production? Nat Rev Immunol, 10(3), 210–215. doi: 10.1038/nri2725 [DOI] [PubMed] [Google Scholar]
- Walsh JG, Reinke SN, Mamik MK, McKenzie BA, Maingat F, Branton WG, … Power C (2014). Rapid inflammasome activation in microglia contributes to brain disease in HIV/AIDS. Retrovirology, 11, 35. doi: 10.1186/1742-4690-11-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichhart T, Haidinger M, Katholnig K, Kopecky C, Poglitsch M, Lassnig C, … Saemann MD (2011). Inhibition of mTOR blocks the anti-inflammatory effects of glucocorticoids in myeloid immune cells. Blood, 117(16), 4273–4283. doi: 10.1182/blood-2010-09-310888 [DOI] [PubMed] [Google Scholar]
- Yang P, Zhao Y, Zhao L, Yuan J, Chen Y, Varghese Z, … Ruan XZ (2015). Paradoxical effect of rapamycin on inflammatory stress-induced insulin resistance in vitro and in vivo. Sci Rep, 5, 14959. doi: 10.1038/srep14959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Nagasu H, Murakami T, Hoang H, Broderick L, Hoffman HM, & Horng T (2014). Inflammasome activation leads to Caspase-1-dependent mitochondrial damage and block of mitophagy. Proc Natl Acad Sci U S A, 111(43), 15514–15519. doi: 10.1073/pnas.1414859111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Chen S, Song L, Tang Y, Shen Y, Jia L, & Le W (2014). MTOR-independent, autophagic enhancer trehalose prolongs motor neuron survival and ameliorates the autophagic flux defect in a mouse model of amyotrophic lateral sclerosis. Autophagy, 10(4), 588–602. doi: 10.4161/auto.27710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z, Umemura A, Sanchez-Lopez E, Liang S, Shalapour S, Wong J, … Karin M (2016). NF-kappaB Restricts Inflammasome Activation via Elimination of Damaged Mitochondria. Cell, 164(5), 896–910. doi: 10.1016/j.cell.2015.12.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Yazdi AS, Menu P, & Tschopp J (2011). A role for mitochondria in NLRP3 inflammasome activation. Nature, 469(7329), 221–225. doi: 10.1038/nature09663 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.