Abstract
The field of microfluidics or lab-on-a-chip technology aims to improve and extend the possibilities of bioassays, cell biology and biomedical research based on the idea of miniaturization. Microfluidic systems allow more accurate modelling of physiological situations for both fundamental research and drug development, and enable systematic high-volume testing for various aspects of drug discovery. Microfluidic systems are in development that not only model biological environments but also physically mimic biological tissues and organs; such ‘organs on a chip’ could have an important role in expediting early stages of drug discovery and help reduce reliance on animal testing. This Review highlights the latest lab-on-a-chip technologies for drug discovery and discusses the potential for future developments in this field.
Drug discovery typically investigates interactions between a lead compound (for example, a potential drug) and a target (for example, a protein, cell membrane or whole cell)1. It usually requires the adaptation of chemical environments to allow the interaction between a lead compound and target to take place; that is, it ensures the accessibility of interaction sites in addition to preventing unwanted adsorption. Effective drug discovery relies on multiple levels of qualitative and quantitative results, including simple ‘yes’ and ‘no’ characterizations, affinity constants for drug–target interactions and kinetic rate constants. The requirement for multiple read-outs places a heavy burden on the testing methods used, as drug discovery typically requires the testing of millions of different chemical combinations. A high-throughput system for handling this large number of samples must be parallel and utilize small chemical volumes to keep the cost of development at an affordable level. The current trends to miniaturize, automatize and parallelize assays address these issues directly while simultaneously increasing resolution and accuracy. These improvements are fuelled by the rapid compound development in combinatorial chemistry, thus demanding new methods with even higher throughput.
In this context, microfluidic devices are showing promise as new and influential players2. These small platforms are also termed micro total analysis systems (μTAS)3,4 or ‘labs on a chip’. They have historically been made from silicon and/or glass using semiconductor processing techniques; soft lithography has recently enabled their fabrication from polymers. Microfluidic systems may contain channels, valves, mixers and other building blocks with typical sizes on the order of tens of micrometres.
Microfluidic technologies have the unique ability to integrate biosensor technology with microscopy-based read-outs. In combination with automated imaging systems possessing high-throughput capabilities and new data processing and storage strategies, microfluidics provides new tools for highly parallel, multiplexed assays with a higher information quality (BOX 1). Furthermore, microfluidic devices provide the possibility to isolate, purify, manipulate and transport particles, biomolecules, bacteriophages, cells or even organisms for a simplified, parallel analysis. Although microfluidics may still be a maturing discipline, microfluidic devices and systems are already being used in many different stages of drug discovery and development5,6.
Box 1 |. Origin of microfluidics.
In a broader setting, the rise of microfluidics is linked to the development of integrated circuit technology and wafer fabrication facilities. The integrated circuit industry experienced rapid growth over the past two decades; the effort to keep up with Moore’s law led to constantly increasing wafer sizes and shrinking minimum feature dimensions. This development led to smaller and faster electronic devices but left older generations of fabrication facilities outdated within the electronics industry. Using these older integrated circuit fabrication facilities to develop microfluidic devices and systems appears to be obvious for the following reasons: requirements for microfluidic manufacturing are much less stringent, as the size of microchannels is rarely below 10 micrometres; simple contact lithography is sufficient at this scale; and there is also no pressure to push microfluidic channel sizes into submicrometre dimensions because the Hagen–Poiseuille equation dictates that the pressure drop across a pipe is inversely proportional to the fourth power of the tube diameter at a constant flow rate, thus significantly higher pressures are needed to maintain the same flow rates in smaller channels. Owing to these three facts, nearly all abandoned integrated circuit fabrication facilities are suitable for microfluidic fabrication. There are only a few building blocks that are foreign to the integrated circuit industry that still need to be added, such as valves, pumps, mixers and connectors between the chips and supporting systems.
Microfluidic fabrication has been developed with two sets of materials: silicon or glass and polymers. Silicon and glass have well-controlled mechanical and chemical properties but they also have high manufacturing costs and high processing complexity, particularly for disposable devices. By contrast, polymers can easily be fabricated via soft lithography or hot embossing, where a single mould can serve as a template for many devices. These processes are more suitable for high-volume disposable devices. However, the mechanical and chemical properties of polymers are less reliable, making surface modification a vital step for robust device functionality. Consequently, recent innovations in surface treatments have mainly been focused on polymeric materials. Although many researchers still use the more expensive silicon or glass chips for research, there is a substantial amount of work targeted towards making reliable polymeric devices.
Adapting and developing these new systems for the pharmaceutical industry introduces several unexpected problems. The first challenge is to convince the industry to replace existing multimillion dollar systems with tiny chips. This is not as trivial as it might seem. Although microfluidic devices can replicate the performance of conventional robotic systems at a much lower cost, commercial companies are often not interested in lowering costs unless this also increases production volume to maintain market share. In established markets it appears to be easier to succeed by creating complementary technologies rather than transformative technologies. The second challenge facing lab-on-a-chip development is determining the effectiveness of current integrated circuit laboratories for fabrication. Integrated circuit laboratories have been a convenient resource for microfluidic development but they were designed without an awareness of the vital issues surrounding microfluidics — for example, material compatibility and biocompatibility, surface chemistry and fluid mechanics. Finally, microfluidic technologies face a scaling limit where it will become less desirable to move to smaller length scales. With lithographic limits being pushed deeper into the submicrometre range, what prevents fluidic systems from following suit? Apart from the above discussion of fluid mechanics, every system has a limit of detection that dictates the minimum signal required for a positive result.
As most of these systems are far from the single-molecule limit of detection, the transition to smaller length scales might jeopardize data integrity. Although lab-on-a-chip systems have shown remarkable results, their integration into commercial systems might be considerably slower than expected.
More fundamentally, scaling laws predict that molecular assays with very small volumes are advantageous in terms of throughput. This is related to molecular diffusion mass transfer (Fick’s law), heat dissipation, multiplexing (arrays) or the surface-to-volume ratio. A tenfold reduction in size should lead to a 100-fold increase in the time to result in molecular binding assays. In addition, this leads to a 100-fold increase in the number of assays that can be placed on a given area, resulting in a throughput that is 10,000 times higher and should give identical quality results. Fundamentally, a microwell plate-based assay format at individual volumes of 1 picolitre should yield a throughput of up to 2,500,000 independent chemical reactions per second and square centimetre. Academic research has provided numerous results to prove such scaling laws; however, just a small fraction of proven methods have made it into commercially available instrumentation. It seems obvious that conventional robotics will not yet be able to provide millions of compounds per second, and it is questionable how urgently such a high throughput is actually needed.
An earlier review5 on the same topic in 2006 concluded that “some challenges still remain... before microfluidic platforms can be used to adapt or replace existing assays”, especially mentioning standardization and adaptation of biological assays to the micron scale. Apparently this has not happened yet, and it is interesting to speculate on the reasons for this. We can identify four possible reasons: economic, psychological, legal and technology-related.
First, previous investments in large-scale robotics and the microwell plate format make it necessary to adapt chip technology to it rather than replace it. High-efficiency chip instrumentation would make instrument manufacturers see their market volume (and profits) shrink, and therefore they are not inclined to adopt it. Second, end users are generally conservative and a low risk of failure is a high priority, therefore they rely on well-established instrumentation. In this regard, it is no surprise that it will take another generation of biologically trained scientists to get ahead in this field. Likewise, it is worth considering how long it took for the microwell plate, the XY robot or fluorescence resonance energy transfer (FRET) detection to move from first being mentioned in the scientific literature to being accepted in high-throughput screening laboratories. Third, a few important patents were in the hands of only a few companies and were aggressively enforced. For example, any array of microfluidic manifolds and any kind of ‘PCR on chip’ or ‘electrophoresis on chip’ technologies were broadly covered by such patents, and prevented a widespread application of, for example, a 96-well adaptation of microfluidic electrophoresis assays by other companies. Last, of course there are still a few bottlenecks left with regard to technology and science. Challenges include interfacing submicrolitre volumes from the conventional laboratory to the chip. The smallest volume that can be manually pipetted is 100 nl, whereas some microfluidic procedures require a volume as small as 1 pl. Another problem is avoiding air bubble formation on reusable chips, or the fundamental incompatibility of very low (and very potent) molecule concentrations and very small volumes.
Here, we focus on a set of recently developed microfluidic techniques that facilitate high-throughput analysis and review their applications in biological systems that are relevant to drug development. We also provide an outlook into microfluidic approaches to modelling biological environments and systems, including specific examples of tissue culture techniques, ‘organs on a chip’ and ‘organisms on a chip’
Microfluidic techniques
For applications in chemistry or biochemistry, the main microfluidic standard operations include sample preparation, injection (metering), valving, pumping, fluid mixing and the use of reactors, separators and detectors for the identification and quantification of analytes. However, more recently the focus has shifted towards biological applications. A few notable examples are discussed below.
Microfluidic patch-clamp techniques.
Patch-clamp techniques for measuring currents across ion channels in cell membranes are essential tools in drug discovery. However, the conventional patch-clamp techniques using glass micropipettes are cumbersome and have a low throughput. To overcome these drawbacks, Tang et al.7 and Chen et al.8 presented chip-based patch-clamp devices that were designed to study ion channels in living cells. Tang et al. used a lateral patch-clamping approach in a standard 1,536-well microplate format that is compatible with standard high-throughput systems. Moreover, the lateral approach allows the method to be easily integrated into robust microfluidic devices that can administer the unique spatiotemporal doses of a drug. The hybrid device consisted of a silicon chip integrating 12 glass capillaries to immobilize and patch the cells, and a polydimethylsiloxane (PDMS) layer bonded to the chip with an array of defined holes. Electrophysiological measurements were carried out in parallel using up to 12 captured cells of a rat basophilic leukaemia cell line.
Droplet microfluidics.
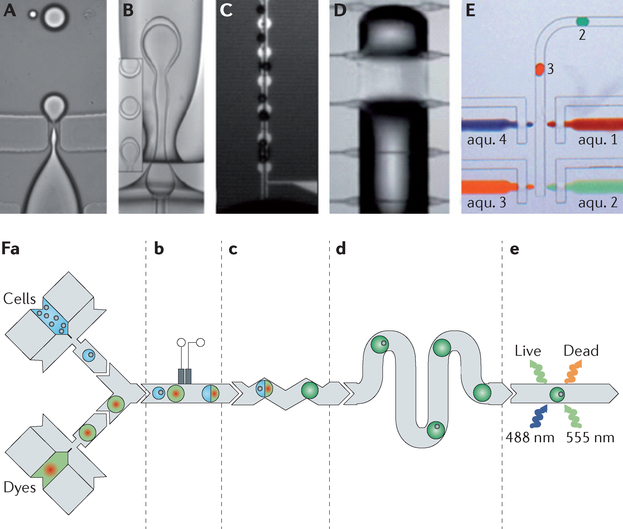
Among many microfluidic techniques used for drug discovery, droplet microfluidics9 has emerged as a powerful tool in biochemical operations because it requires only pico- to nanolitre sample volumes. In these systems, droplets or plugs can be generated by immersing aqueous samples in a water-immiscible carrier medium, such as a hydrophobic compound or gas. However, fine control of the size, shape, frequency and monodispersity of the droplets still attracts a considerable amount of attention for reliable, high-throughput assays. To improve droplet formation, hydrodynamic methods10–14 (FIG. 1A,1B) use channel geometry and fluid flow characteristics to control the generation of droplets. These designs all depend on shear forces in fluids, which are dictated by the channel’s hydrodynamic properties (such as the geometry, flow rate and viscosity), to generate droplets. Electrohydrodynamic technologies have also been used for droplet formation, where an electrical signal through integrated electrodes is used to control droplet properties. Electrowetting, for example, uses an electrical signal to induce fluidic wetting, causing the formation of a short liquid cylinder. After the signal is switched off, the channel reverts back to its hydrophobic nature and the cylinder forms a droplet that breaks off from the reservoir15. Dielectrophoresis can similarly pull droplets from a fluid reservoir16–18 (FIG. 1C). Thermocapillary actuation19–20 (FIG. 1D), acoustic actuation21–24 and valve-based systems25–26 (FIG. 1E) can also be used to create and manipulate droplets.
Figure 1 |. Droplet-based microfluidics.
A | Experimental image depicting droplet generation sequences inside the flow-focusing orifice11. B | Steady-state droplet formation mechanisms that result in monodispersed double emulsions with a single internal droplet12. C | Experimental demonstration of feedback-controlled droplet dispensing16. D | Thermocapillary-actuated droplet generation19. E | Generation of droplets with distinct compositions using mechanical valves (where ‘aqu.’ means aqueous solution)25. F | Schematic illustration of droplet-based cellular drug screening46. Fa | A set of two nozzles encapsulates cells and fluorescent dyes. A fork enables the interdigitation of the streams, resulting in cell-containing droplets alternating with dye-containing droplets. Fb | A fusion module delivers an alternating electrical current, which permits electrically controlled droplet merging. Fc | The dyes are then thoroughly mixed with the cells in a passive micromixer. Fd | A delay line optimizes cell staining by enabling on-chip incubation of the droplet. Fe | A detection module confines the droplet laterally and vertically to collect the laser-excited fluorescent signals. Image A is reproduced, with permission, from REF. 11 © (2003) American Institute of Physics. Image B is reproduced, with permission, from REF. 12 © (2005) The American Association for the Advancement of Science. Image C is reproduced, with permission, from REF. 16 © (2009) Royal Society of Chemistry. Image D is reproduced, with permission, REF. 19 © (2010) Royal Society of Chemistry. Image E is reproduced, with permission, from REF 25 © (2009) Royal Society of Chemistry. Image F is reproduced, with permission, from REF. 46 © (2009) National Academy of Sciences.
Droplet microfluidics has many attractive characteristics for drug discovery, including minimal sample consumption, low cross-contamination, fast mixing, miniaturized space, high-throughput capabilities and multiplexed detection27. In molecular biology, droplets with homogeneous diameters and controlled content that does not evaporate are important for screening experiments that rely on high reproducibility, such as protein crystallization28, gene mutation29, DNA assays30 and molecular evolution31.
Applications of microfluidic techniques
For applications in chemistry or biochemistry, the main microfluidic techniques include electrophoresis, chromatography and binding assays. These techniques facilitate genetic, protein, glycan and metabolite analysis and are used for genome sequencing, clinical diagnostics, drug discovery and basic research support. Again, the recent trend towards applications in cell biology, embryology or tissue engineering is dramatically changing the situation. The examples given below cover only a few of the relevant applications.
Enzyme activity and kinetics.
Enzymes are important targets in early-stage toxicity screens32. Furthermore, kinetic data on the reaction of enzymes with small molecules are gaining significance for drug discovery and development33. Several microfluidic platforms have been developed to meet the resulting demand for the rapid determination of enzyme activity. In a so-called immobilized microfluidic enzyme reactor (IMER), the enzyme is typically immobilized on a solid substrate and supplied with a continuous flow of reagents. Microreactors allow essentially the same operating modes as macroscale reactors — batch and continuous modes — where the latter has proven to be most advantageous. These systems have been increasingly utilized for the determination of enzyme kinetics. An overview of the fundamental features, applications and current state of IMER systems has recently been published33.
Another microfluidic approach has investigated the dynamics of enzyme inhibition using an inhibitor gradient generated in a microchannel. Kinetic rate constants and inhibition constants were calculated from the fluorescence data in the microfluidic system as well as in a plate approach used as a reference. This microfluidic approach was faster (2 minutes instead of at least 15 minutes) and simpler to operate, and the standard deviation data for the kinetic constants were lower than the data obtained with the plate approach34.
Further developments include the replacement of covalent enzyme immobilization to avoid potential damage to the enzyme caused by chemical procedures. Pore-limit electrophoresis with enzyme assay, for instance, is a two-step process that combines pore-limit electrophoresis with an in situ enzyme activity assay. The experiment is performed in a microchannel containing a polyacrylamide pore-size gradient gel using fluorescence detection. This zymographic platform successfully allowed the determination of the molecular weight, quantity and kinetic rate constants of enzymes that are present in heterogeneous protein samples32.
Drug-protein interactions using droplet microfluidics: binding of the anticoagulant drug warfarin.
The investigation of drug-protein interactions using conventional bioassays typically requires the immobilization of a drug or protein in the static well of a microtitre plate. Recently, researchers used droplet microfluidics to study the interaction between human serum albumin (HSA) and the anticoagulant drug warfarin. First, magnetic beads were coated with HSA in a bulk solution. Next, warfarin was added and the system was allowed to reach equilibrium. The samples were then injected into a microfluidic device where they were encapsulated in droplets of mineral oil. The droplets containing magnetic beads (and thus HSA and warfarin complexes) were separated from the solution containing the free drug at a T-junction using a magnet on one side. As the experiments were performed using radiolabelled 14C-warfarin, the concentration of warfarin bound to HSA on the beads and the concentration of free warfarin could easily be determined by a scintillation counter. The droplet separation allowed for a relatively simple measurement of the concentration ratio between bound and free warfarin, allowing researchers to calculate the affinity constant for the binding between the drug (warfarin) and the target (HSA)35.
Plaque formation in Alzheimer’s disease.
Alzheimer’s disease is characterized by the formation of plaques composed of amyloid-β (Aβ) aggregates within brain tissue. To study the inhibitory effects of small molecules and metal ions on Aβ aggregation, the Aβ42 peptide was studied in a microfluidic system. First, the Aβ42 peptide was covalently immobilized into microfluidic channels via silanization of PDMS and subsequent reaction with N-hydroxysuccinimide and carbodiimide. After a blocking step with bovine serum albumin (BSA), Aβ aggregation was studied by incubating the system with solutions of Aβ42 peptide with or without a potential inhibitor (small molecules or metal ions). The results were extracted via fluorescence microscopy and atomic force microscopy; they verified previous results and were obtained faster and with much lower reagent consumption36.
Membrane protein studies: inhibitors of a hepatitis C virus membrane protein.
Membrane proteins are typically embedded in the lipid bilayers that comprise most biological membranes. These proteins allow the cell to interact with its surrounding environment, and generally regulate cellular processes in response to external influences. Their influence on cell behaviour is the basis for their significance in drug discovery. These proteins can be immobilized onto artificial lipid bilayer membranes to ensure their functionality in subsequent investigations, which may offer an alternative to cell-based studies. An overview of the microtechnologies used for membrane protein studies and applications in this field has recently been published37. In 2008, Einav et al.38 used a lab-on-a-chip device to reveal inhibitors of a key membrane protein of the hepatitis C virus (HCV). In this study, a high-throughput microfluidic platform based on “mechanically induced trapping of molecular interactions”, which actively trapped surface-bound molecules while washing away unbound molecules39, was used for affinity measurements. Einav et al. labelled the HCV transmembrane protein NS4B with green fluorescent protein (GFP) and immobilized it within a microchannel via an anti-GFP antibody that was bound to the surface of the microchannel. The affinity of Cy5-labelled HCV RNA probes to the GFP-labelled NS4B was detected by fluorescence measurements. After identifying the part of the RNA that specifically binds NS4B, a compound library was screened for inhibitors of this binding. In total, 1,280 compounds were tested, resulting in the identification of 18 compounds for further analysis. The high sample throughput, in combination with low sample consumption, demonstrated the utility of the microfluidic platform used in this study38.
Detection of exosomes in cancer.
Exosomes are shed from both normal and cancer cells. Initially, these nanoscale vesicles were only considered as inert extracellular debris but now it is believed that they have an important role in long distance cell-cell communication and horizontal transfer of information — for example, during tumour progression. Recent findings have shown that they contain and serve as carriers for bioactive molecules (such as proteins, mRNAs, small RNAs or microRNAs) and might be involved in important steps of tumour progression, such as the formation of the pre-metastatic niche. Thus, they would be interesting targets for new drugs: as biomarkers for diagnosis and/or prognosis of cancer, or as natural transport vehicles for drug delivery40,41. Unfortunately, these nanosized vesicles are difficult to isolate using standard centrifugation protocols. They can be isolated from blood, however, using microfluidic channels that are equipped with herringbone patterns; a similar concept is also used to isolate rare circulating tumour cells from blood42. We expect that microfluidic systems with features and functional elements that have similar dimensions to small vesicles will also allow improved handling, analysis and manipulation of exosomes.
Measurements of cell viability.
Perhaps one of the most noteworthy developments in droplet technology for drug discovery was the encapsulation of single cells or even animals within droplets43–48. Single-cell droplets are compatible with high-throughput screening and sorting45. Brouzes et al.46 developed a droplet viability assay that permits the quantitative scoring of cell viability and growth within intact droplets, thus allowing an entire drug library to be screened for cytotoxic effects against U937 cells. The assay merged a cell-containing droplet with a fluorescently encoded droplet containing various concentrations of the drug mitomycin C, incubated them for approximately 15 minutes and then characterized cell viability by fluorescence imaging46 (FIG. 1F).
DNA synthesis.
Synthetic biological building blocks such as DNA oligomers or DNA analogues can be used to design and manufacture synthetic genetic networks49, metabolic pathways for the production of small molecules and synthetic genomes for bacteriophages or bacteria (see also BOX 2). Kong et al.50 and Lee et al.51 have successfully demonstrated synthetic DNA constructs that can be used for these purposes, but the substantial costs related to synthesizing larger DNA molecules have prevented the use of these building blocks on a larger scale. Kong et al. used a multichamber microfluidic device consisting of four 500 nl reactors to carry out polymerase construction and amplification (PCA)-based synthesis of 1-kb-long DNA constructs from very low concentrations of oligonucleotides.
Box 2 |. Systems biology and synthetic biology.
Systems biology and synthetic biology are two emerging biological disciplines with the potential to have a substantial impact on drug discovery and, in particular, on process steps such as target identification, elucidation of new (molecular) mechanisms of action of small molecules, predictive toxicology and drug production94,95. The aim of systems biology is to describe and understand biology and biological systems from the molecular level up to the systems level. Systems biology attempts to gain further insight into complex genetic and physiological networks by systematically integrating and mining large-scale data sets obtained from ‘omics’ experiments and computational modelling of regulatory networks and interplay between pathways on molecular, cellular, tissue, organ and whole-organism levels. However, the innate complexity of biological systems reveals a seemingly infinite number of targets and mechanisms for potential therapeutic intervention. Multiplexed high-throughput methods as well as methods that are capable of generating high levels of content are required to meet the challenges of systems biology.
The young and often controversial field of synthetic biology has drawn a lot of attention from the scientific community because of notable recent achievements such as the programming of cells by multiplex genome engineering96 or the creation of a bacterium with a synthetic genome97. This field applies engineering principles to develop designs and strategies to create synthetic biological parts, devices and complex systems. The success of the field will rely on two factors; the first factor is the ability to create comprehensive libraries of well-characterized and standardized synthetic biological parts, such as synthesized DNA, phospholipids, membranes and vesicles. These components will become the building blocks for designing and constructing complex synthetic circuits or even synthetic cells. The second factor will be the development of new tools to facilitate the de novo synthesis and assembly of synthetic biological materials with a high resolution and accuracy — requirements that are most likely to be met by nano- and microfluidics technology98. These synthesized constructs will have a reduced complexity compared to living cells and organisms, and may help to elucidate disease mechanisms. Lab-on-a-chip devices can be used to generate new knowledge on biological building blocks or to immobilize, transport and manipulate particles, biomolecules, bacteriophages, cells or even organisms99.
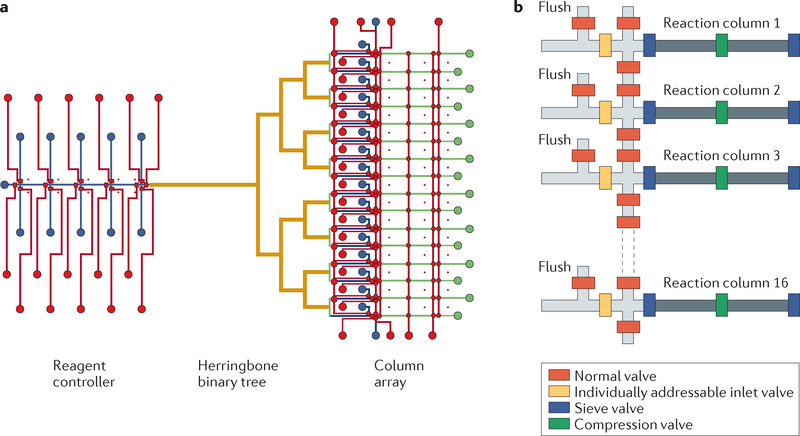
Microfluidic systems use less reagent for more cost-effective synthesis, and can create more complex genes because of the ability to maintain reagents at required concentrations even in complex oligonucleotide pools. Lee et al. parallelized the synthesis of genes on a chip using a multilayer microfluidic device. Conventional solid-phase chemistry was used to synthesize a gene fragment of Bacillus cereus in a design that encompassed valves, purge lines, reagent controllers, a herringbone binary tree for proper mixing of reagents and an array of 16 nl reaction columns (FIG. 2). The multiple reaction chambers allowed the researchers to harvest the constructs separately to obtain homogenous samples, as opposed to the mixtures obtained from common microarray technologies.
Figure 2 |. Parallelized synthesis of oligonucleotides on a chip.
a | Schematic illustration of a 16-column microfluidic DNA synthesizer. The device consists of control lines (shown in red), fluidic lines (shown in blue), a herringbone mixer (shown in yellow) and a square profiled binary tree and the reactor columns (shown in green). b | A diagram of the column array with valves for a controlled fluid inlet, fluid flow and washing of beads51.
Protein expression.
A better understanding of signalling pathways, key regulatory nodes and/or crosstalk between different pathways will allow us to better predict the target effects of drugs and to profile the effects of drugs on the level of metabolic pathways52. Khnouf et al.53 recently presented a module for the cell-free expression of functional soluble and membrane proteins. Cell-free screening systems are indispensable to drug discovery processes for gaining complementary information about drug-target effects. The disposable, three-layer device used by Khnouf et al. has the footprint and pitch of a standard 96-well microtitre plate. The upper and lower layers were mechanically machined polypropylene plates; the former contained a reaction chamber and three adjacent ports for loading solutions, and the latter provided the feeding chamber. Dialysis membranes were used to separate all three layers of the channel, making standard dispensing devices and microplate readers applicable for the reactions and read-outs. Two membrane-associated proteins (bacteriorhodopsin and apolipoprotein A) were co-expressed in a single reaction, and two soluble proteins (luciferase and β-lactamase) were synthesized using this device. The functionality of the produced proteins was demonstrated in different enzyme inhibition assays. The device significantly reduced reagent consumption compared to commercially available cell-free protein synthesis kits and used a straightforward strategy for parallelized high-throughput applications using standardized equipment in drug discovery. The development of such platforms is an important initial step towards standardized and scalable cell-free microfluidic screening systems and will help us to gain deeper insights into important drug targets such as membrane proteins and enzymes, as well as helping us to better understand their effects on biological systems54 (see also BOX 2).
Microfluidic model systems of disease or toxicity
A growing number of in vitro and in vivo models based on lab-on-a-chip technology are being developed for disease or toxicity studies. These range from cell-free assays to cell-based systems (including single-cell arrays and three-dimensional cell culture systems), in addition to small multicellular ‘organisms on a chip’. As described above, on-chip devices have shown the capability to immobilize, manipulate and transport particles, biological parts or cells; they have generated stable static or dynamic gradients of chemical and biochemical compounds; they have provided a constant or intermittent supply of nutrients and removal of metabolites; and they have allowed multiplexed analyses including parallel read-outs of cell culture parameters by on-chip integrated sensors55. Integrating these tools with cell culture facilities and cell-based assays will lead to more advanced culture techniques, highly sophisticated and integrated devices as well as higher-throughput and multiplexed assays.
Cell-based microfluidic model systems.
Cell-based assays are highly dependent on how cells behave in vitro and on disease-relevant cell culture models that accurately represent the pathophysiological states found in vivo56. For in vitro models, the most important factors influencing successful in vivo reproduction are the cell type and source. The correct choice of suitable and stable cell sources is decisive for the reliability and validity of in vitro studies; the cells must have the right phenotypic expression and should come from appropriate lines — that is, normal, mutant, transfected or malignantly transformed cell lines. Primary cells, particularly those derived from the liver, heart or brain, are frequently used as a cell source. Primary cells are initially characterized by their phenotypic behaviour and are therefore ideal candidates for drug discovery and toxicology studies. A major drawback of primary cells, however, is their early loss of organotypic functions. Even short-term culturing generally leads to a reduction in specificity owing to their isolation, drastic alteration of the cellular environment and their transfer to standard plastic cell culture vessels57. Microfluidic tools and platforms offer new opportunities to better control the essential spatiotemporal cues of the cellular microenvironment to maintain organotypic functions for a longer period of time. Moreover, microfluidic techniques allow for the improved handling, positioning, manipulation and analysis of living cells.
For example, microfluidic approaches might be useful in overcoming the limitations of current in vitro techniques for using stem cells in drug discovery. Reprogrammed patient-derived stem cells were recently proclaimed as the beginning of a new era for drug discovery58. One central aim is to differentiate stem cells from patients with a specific disease towards disease-relevant cell types in order to produce in vitro models of the disease or even patient-related models for drug-target evaluation and/or toxicity studies59. However, current in vitro techniques provide a very simplistic and synthetic environment for stem cells and thus are not able to support efficient and terminal differentiation of stem cells. Microfluidic assays might enable better control of cell fate in vitro — for example, by improving the spatiotemporal control of biochemical and biophysical factors60–62. However, the development of such assays is still in its infancy, as our understanding of the control of cell fate by environmental factors is still in the early stages.
Another application of microfluidic cell culture systems is the analysis of DNA damage in cancer models. The comet assay, also referred to as a single-cell gel electrophoresis assay, is a standard technique for analysing DNA damage and repair on a single-cell level. Wood et al.63 recently fabricated a microwell array patterned in an agarose layer that allowed gravity-driven single-cell trapping and high-throughput analysis of DNA damage. The microstructured device solved some of the inherent problems of the widely used standard comet assay, such as difficulties in the retrieval of cell locations and impeded cell analysis because of overlapping cells. The device thus facilitates fully automated and robust imaging and analysis on the basis of spatially patterned cells by capturing them into microwells, and reduces failures of cell analysis because cells are in a single focal plane and are not overlapping. The microwell array was fabricated with a simple stamping procedure into a molten agarose layer that was directly compatible with the original comet assay. To study DNA damage provoked by chemicals or the inhibition of DNA repair mechanisms (mediated by small molecules), the patterned array of agarose microwells can be sandwiched between a glass substrate and a bottomless microtitre plate. This new device, offering the possibility to measure DNA damage and repair kinetics on a platform that is capable of a high throughput, is a new and powerful tool for drug screening on a single-cell level.
Chen et al.8 devised another approach for characterizing the response of single cells to drugs based on electrophysiological measurements in a microfluidic device. In contrast to the approach described above, a planar patch-clamp technique was proposed, in which cells were immobilized on a small aperture integrated into a microfluidic mixer. This three-layer patch-clamp device used PDMS in the upper and lower layers for fluid transport and electrical connection, and a poly(methylmethacrylate) (PMMA) layer or glass chip in the middle with a through-hole to trap the cells. A passive zig-zag micromixer in the top layer was used to precisely control drug concentrations. Although the total channel length was around 8 mm (mainly owing to the micromixer), a complete exchange of solution in the device was possible within 20 seconds; a fast exchange of solutions is a key parameter for studying dynamic effects on a single-cell level. As a proof of concept, volume-regulated chloride channels in human embryonic kidney (HEK) 293T cells were electrophysiologically characterized while being exposed to different osmolarities. Furthermore, researchers studied the impact of different concentrations of tetraethyl-ammonium chloride on potassium channel blocking, in addition to the response of volume-regulated chloride channels to sequential solution exchange. The latter protocol will gain significance in drug testing for elucidating dynamic effects.
Researchers have raised an important question: can these devices or aspects of these devices be used multiple times, or can they only be used safely and reliably as disposables? In the latter case, the future focus of this work will inevitably turn to the cost-effective production of such devices. Yet another important question remains unanswered: do cells in such devices still behave in an organotypic or disease-relevant manner, or do they rapidly change their physiology as a result of the limited confines of their now unfamiliar extracellular environment? Answers to these particular questions will have a substantial impact on the ultimate utility of these devices.
Three-dimensional cell culture.
To date, the dominant method for culturing cells on glass or plastic surfaces involves using a flat layer, regardless of the application. However, three-dimensional cultured cells have become increasingly popular because they more closely resemble cell behaviour in vivo64. The shortcomings of current cell culture methods are thought to be one of the decisive reasons for the late-stage failures and high attrition rates of new drug candidates65–66. Several studies, for example, have shown that there are different mechanisms of drug resistance in three-dimensional cultured cancer cells compared to two-dimensional cultured cells67–68. The idea of three-dimensional cell culture was first formulated by Alexis Carrel69, who observed that the organotypic functions of fragments of a chicken heart could be maintained over 3 months if they were three-dimensionally cultured; following this study, some of the now widely used three-dimensional cell culture techniques were developed70,71. The long-expected paradigm shift has been decelerated mainly because of the elaborate and costly techniques associated with three-dimensional cell culture. Nonetheless, new developments in chip-based technologies and microfluidic platforms are now providing more convenient access to three-dimensional cell culture techniques and experiments, thus reviving the field of three-dimensional cell culture for a broader research community.
The supply of nutrients to cells and the removal of metabolites are crucial factors for successful three-dimensional cell culture. If there is no active fluid flow through the artificial tissue, the size of the aggregates is strongly limited by the diffusion of gases, nutrients and metabolites; thus, the size of cell aggregates is typically limited to approximately a few hundred micrometres72. Techniques to produce cell aggregates date back to the early twentieth century; Holtfreter70 created spherically shaped multicellular aggregates (also known as spheroids) using a non-adhesive agarose layer as a cell culture substrate. Today, micropatterned substrates can be used to precisely control the size and shape of these aggregates — also referred to as microtissues. Tekin et al.73 fabricated stimuli-responsive microgroove arrays to form artificial tissue constructs with a defined size and shape that were used as building blocks for modular tissue engineering approaches or as tissue models for drug discovery processes. Their method relied on a conformal coating of poly(N-isopropylacrylamide) (PNIPAAm) on top of the microgroove arrays; as a thermoresponsive polymer, PNIPAAm could be reversibly switched between a cell-attractive state and a cell-repellent state based on the temperature of the solution. A low temperature (24 °C) during inoculation, combined with gravity, guaranteed that cells selectively seeded in the base of the grooves. Raising the temperature during culture then allowed the cells to anchor to the groove walls and adopt the shape of the grooves. Finally, a lowered temperature was again applied to detach cells in a headfirst conformation in order to retrieve the geometrically defined tissue fibres.
Geometric control of the shape of the aggregates is one possible way to passively control the formation of spatiotemporal gradients in tissue constructs. Alternatively, microtissues can be processed in microfluidic devices to test their dynamic responses to a drug or their responses to a series of different drugs. Many important biological processes, including cell proliferation, migration and differentiation, are controlled in vivo by spatiotemporal gradients of soluble factors (for example, paracrine and/or endocrine factors). Drug development, however, is highly dependent on high-throughput methods, which generally do not support such fine spatiotemporal control. Therefore, methods must be developed to produce large quantities of identical, traceable microtissues, which can be processed in parallel to provide fast and reliable results on the behaviour and response of three-dimensional cultured cells.
Chen et al.67 developed a method to mechanically stabilize multicellular constructs via photo-encapsulation of 500–1,000 cells in encoded polyethylene-glycol-based hydrogels, so that the microtissues could be studied with a multiplexed microfluidic approach. The method was tested with mouse embryonic stem cells, bipotential hepatic progenitors, mature hepatocytes and hepatoma cells. The stabilized and encoded microtissues had a size of 250–350 μm and were used in high-throughput analytical processes, such as flow sorting and analysis, to provide a solid basis for quantitative statistical analysis. The authors developed two different methods for multiplexing the process: the first was based on embedding fluorescent micro- or nanoparticles into the cell-containing capsules. The second was based on an orthogonal detection method using embedded biotinylated particles that were labelled after the encapsulation process by diffusion into the hydrogels with different streptavidin-conjugated near-infrared-emitting molecules. The assay was used to assess the chemotherapeutic effect of different drug-gene combinations in an RNA interference experiment. First, pooled and encapsulated HepG2 cells were treated with small interfering RNA to silence the anti-apoptotic gene B cell lymphoma XL (BCL-XL), and were then exposed to two different doses of doxorubicin. The apoptotic effect was measured by a combined flow analysis and near-infrared scanning process. New strategies using continuous-flow lithographic DNA-based encoding could be used in such approaches to further improve the multiplexing capabilities74. Using multiple cell types, heterogenic microtissues with a more complex and natural architecture mimicking liver or cardiac tissues could also be produced and used as in vitro models for drug testing. The aforementioned methods will lay the foundation for standardized high-throughput tests to evaluate the response of organotypic three-dimensional cultured (human) cells to different drugs and drug combinations. These types of assays will help to refine drug development strategies and will eventually reduce costs by reducing the reliance on animal testing.
Microfluidic platforms with an increased functionality are currently being developed to improve data quality from in vitro assays, as well as to shed light on the delicate relationship between microenvironments and cellular development60,75–77. In the near future, such tools could have the potential to provide deeper insight into the complex spatiotemporal cues that govern cell fate, but eventually they must also be able to translate this knowledge into a high-throughput format; that is, these microfluidic platforms need to be turned into high-throughput devices. Only when these milestones have been realized will cell-based assays have the accuracy needed for their use as successful drug discovery platforms. To this end, one of our own approaches involves the development of high-throughput fabrication techniques for the large-scale production of biofunctionalized microstructures for three-dimensional cell culture applications. These film-based microstructures are produced by a combination of material modification techniques and a microscale thermoforming process. This technology allows the modification of thin polymer films in their planar state before they are formed because the modifications can be preserved during the solid-state forming as a result of a permanent material coherence. It enables the production of microcavity arrays for single-cell and/or three-dimensional cell aggregates and microfluidic structures with high-resolution, site-specific surface modifications. The underlying concept encompasses not only the possibility of film-based continuous manufacturing of very flexible, functionalized cell culture platforms but also provides access to continuous reel-to-reel processes for these applications78.
Organs on a chip.
Three-dimensional cell culture techniques have great potential to assist the transition of drug testing from animal-based assays to in vitro assays. Currently, however, the results obtained with new in vitro systems cannot replace animal testing because they do not take into account the complex interactions between different tissues and organs. Animal testing is not ideal either, as the predictive value of such tests is limited owing to metabolic differences between humans and animals, and many ethical issues are raised by the testing. In efforts to bypass animal testing, co-culture or multitissue-based microfluidic devices integrate two or more cell types in two-dimensional or three-dimensional cell cultures to simulate various human organs on a single chip79–81. These systems aim to mimic the tissue-tissue interfaces in the body or at least some of the physiologically relevant processes that are part of the so-called ADME (absorption, distribution, metabolism and elimination) processes in the body82.
In this effort, Sung et al.83 developed a microfluidic device to test different cell types in three-dimensional cultures using separate chambers connected by a microfluidic network to reproduce the pharmacokinetic profiles of drugs in the body. The chip mimicked a cancerous colon, cancerous liver and healthy bone marrow using three separate compartments for the matrigel-encapsulated colon cancer cells (HCT-116), hepatoma cells (HepG2/C3A) and alginate-encapsulated myeloblasts (Kasumi-1) in three-dimensional culture. The geometry of the compartments and channels as well as the applied flow rates were adjusted so that the residence times of the fluid in the various microfluidic compartments matched the physiological residence times of blood in the corresponding organs. Using this device, Sung et al. demonstrated that the complex interactions of the different tissues yielded cytotoxic effects (with a common anticancer drug) that were different to the effects that have been observed in a conventional 96-well-plate-based assay.
One of the most prominent organ-on-a-chip devices was recently presented by Huh and colleagues84. The authors developed a microfluidic device that was able to mimic the functional alveolar-capillary interface of a human lung by integrating mechanical cell actuation into their co-culture model. A compartmental, two-layer PDMS structure was used to co-culture cells in a middle channel while applying a vacuum from two channels on either side to generate the mechanical actuation. The upper main channel contained human alveolar epithelial cells cultured in air on the upper side of a thin PDMS membrane, whereas the lower channel contained a fluid medium with microvascular endothelial cells attached to the opposite surface of the membrane. Applying a vacuum to the side channels resulted in a cyclic stretching of the PDMS membrane in the main channel. The applied mechanical stimulus had an impact on tissue inflammatory responses in a nanoparticle-based toxicology test and showed enhanced cellular uptake of nanoparticles that was similar to what is observed in in vivo tests.
Devices such as the one presented by Huh et al., which deliver results of drug efficacy and toxicity on an organ level, may provide valuable information for early-phase decisions in future drug development. Patient-derived cells could even lay the groundwork for personalized medicine, providing the ability to optimize drug concentrations and compositions for different patient groups or even individual patients. To continue this development, researchers have embarked on unconventional, innovative approaches for overcoming the current challenges, including the use of ‘more authentic’ cells (cells that behave more naturally and are therefore more representative of the tissue in question) and the reconstitution of more complex biological structures or entities on a chip. Günther et al.85 presented a method for the fixation and long-term culture of an isolated blood vessel on a microfluidic chip. This chip used three microchannel systems for the fixation, perfusion and superfusion of the vessel. The configuration created a well-defined environment for the functional testing of the vessel, showing smooth muscle and endothelial function under the influence of phenylephrine or acetylcholine. In the future, such devices may enable the use of automated, standardized, artery-based pharmacological and/or toxicological screens, and are likely to deliver valuable information about the underlying mechanisms of the blood-tissue interface relating to the bioavailability of new drugs.
Whole organisms on a chip.
Many small vertebrate animals, such as the clawed frog Xenopus laevis and the zebrafish Danio rerio, are gaining increased attention for drug discovery because they allow the testing of substances and chemical compounds on a systemic level. Despite their physiological differences to humans, whole-organism-based screens can provide deep insights into the effects of drug candidates on developmental processes, tissue-tissue interactions and metabolism. Although these phenotypic screens can provide valuable pharmacokinetic and pharmacodynamic information, which is nearly impossible with protein and/or cell-based assays, they are usually more elaborate and time-consuming86. Recent developments in automated microscopy87,88 and microfluidics, however, have simplified and accelerated this process. Microfluidic platforms have been created to manipulate and analyse the roundworm Caenorhabditis elegans89,90, Drosophila melanogaster embryos91, X. laevis eggs92 and D. rerio93 in highly spatiotemporally controlled environments. These devices were designed to fulfil tasks that were previously difficult to accomplish with other methods, such as handling, positioning, orienting and manipulating entire organisms.
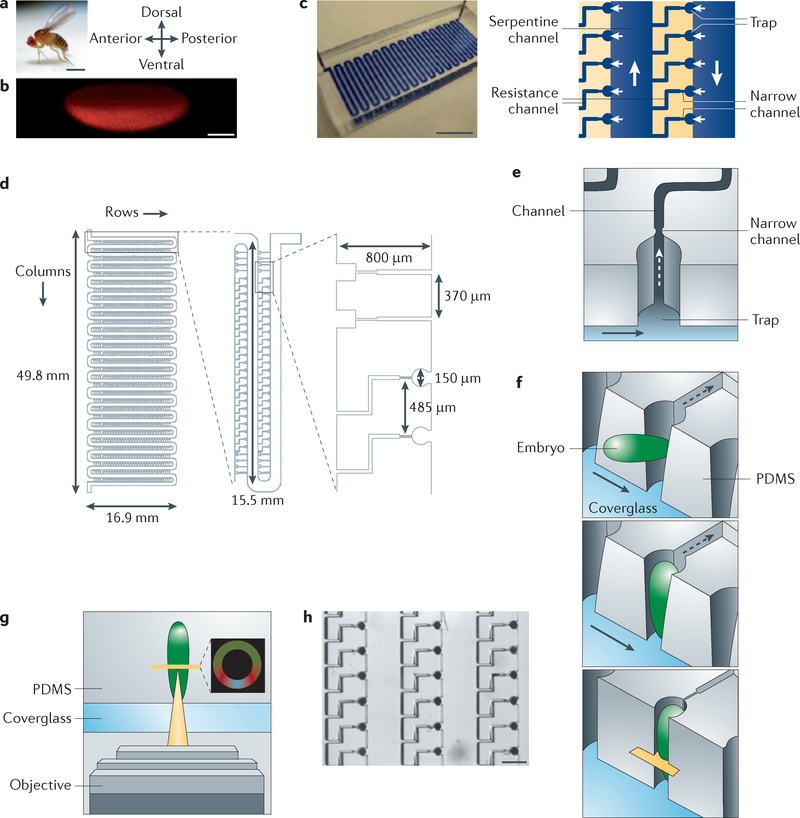
Recently, Chung et al.91 introduced a microfluidic array with the ability to order and vertically orient D. melanogaster embryos on a large scale. This enabled the successful quantitative imaging of the embryos based on the alignment of their dorsoventral axis, which had previously been a major drawback hampering coverslip-based approaches. The microfluidic device consisted of a meandering manifold (700 μm in width) to inject and transport the embryos to intersections with cross-flow channels (FIG. 3). The live embryos (500 μm in length) were vertically trapped in a cylindrical cut-out at the intersection of the main channel and cross-flow channels. The authors used the elastic behaviour of PDMS to open and/or widen the trap with the initial positive injection pressure. As soon as the embryos were vertically oriented within the trap, the pressure was released and the trap was contracted to fix the embryos and their vertical orientation. This lock-in feature allowed the researchers to securely transport the embryos in their fixed position; this orientation is a crucial prerequisite for high-throughput, robotic screening systems. This device allowed a quantitative analysis of the dorsoventral pattern formation of morphogens.
Figure 3 |. Microfluidic array to order and vertically orient Drosophila melanogaster embryos with a high throughput.
a | Image of an adult Drosophila melanogaster with dorsal, posterior, ventral and anterior directions. Scale bar: 1 mm. b | Image of an early embryo stained using an antibody against the protein Dorsal. Scale bar: 100 μm. c | Photograph of the microfluidic device (left) and a schematic diagram indicating the fluid flow (right). Scale bar: 1 cm (left) and 500 μm (right). d | Design of the array (top view). e | Scanning electron micrograph of the truncated cylindrical embryo trap. Scale bar: 100 μm. f | Schematic process sequence for the trapping of the embryo (imaging focal plane shown in yellow) and bulk flow in the meander-shaped manifold (indicated by the arrows). g | Schematic illustration of the imaging set-up with an inset showing a confocal image of a D. melanogaster embryo stained with antibodies to Dorsal, Twist and Rolled (also known as dpERK). h | Details of the array with trapped embryos. Scale bar: 500 μm. PDMS, polydimethylsiloxane. This figure is modified, with permission, from REF. 91 © (2011) Macmillan Publishers Ltd. All rights reserved.
C. elegans is a widely used model organism in developmental biology and drug discovery. Its high transparency, fully sequenced genome, fully mapped cell fate pattern and ease of culture make this organism a prime candidate for systematic studies. Cultivating and studying these organisms over the course of their entire life cycle can provide valuable insight into age-related phenotypic and behavioural screening89. Hulme et al.89 developed a microfluidic device consisting of an array of 16 dome-shaped chambers for housing individual worms, a microchannel network for exchanging fluid, integrated screw valves and ‘worm clamps’ strictly for this purpose. Wedge-shaped channels acted as the worm clamps to immobilize C. elegans for high-resolution imaging and laser-based manipulation. The chambers were designed with a diameter of 1.5 mm in order to be small enough to prevent the worms from exiting the field of view but large enough to prevent interference with their swimming behaviour.
For in vivo chemical and genetic screening of zebrafish (D. rerio) larvae, Pardo-Martin et al.93 developed a high-throughput system denoted ‘vertebrate automated screening technology’ The system had: an automatic loading step of the zebrafish larvae, using simple suction into a tube from a reservoir or microtitre plate; an optical detection step composed of light-emitting diodes and a photodiode; a positioning and rotation step in a rotatable capillary with a diameter of 800 μm; image focusing and acquisition; an optional laser manipulation step; and a final dispensing step back into a multi-well plate. The highly automated system utilized image processing software to control a three-axis stage with two stepper motors in order to position and orient the zebrafish larvae inside the capillary. The system allowed confocal imaging and laser-based microsurgery of oriented zebrafish larvae within 19 seconds.
The use of microfluidic platforms to obtain pharmacokinetic and pharmacodynamic data at the organism level is a very recent development. More complex devices are expected to fuel this momentum and deliver valuable large-scale data sets for drug discovery processes.
Conclusions
Despite the lack of commercially available chip-based drug discovery instruments, the maturation of lab-on-a-chip devices and microfluidic platforms has had an increasing impact on nearly all aspects and steps of the drug discovery process. Lab-on-a-chip-based in vitro and in vivo models, which are mainly used for phenotypical screenings in the preclinical phase, allow for higher reliability and better control than traditional in vitro assays, to better model the human system. Microfluidic techniques have well-established benefits for miniaturization, automatization and parallelization; however, they also hold great promise for long-term, functional cell-based models as a result of their ability to spatiotemporally control cellular microenvironments with a higher resolution and accuracy. We believe that improved cell culture techniques will have a vital role in all future cell-based assays, especially those using stem, progenitor or induced pluripotent cells. Current devices already display impressive complexity — for example, creating gradients of soluble factors over three-dimensional tissue constructs — but current systems are still far from being capable of precisely controlling cell fate and behaviour via artificial instructive environments. In this regard, there is an urgent need to develop new and innovative strategies for integrating spatiotemporally patterned biochemical and biophysical cues into microfluidic devices. The ability to reconstruct physiological or even patient-relevant models in vitro is a major step towards producing more reliable and predictive data in early phases of drug discovery. These models will eventually reduce the need for animal testing and facilitate the development of safer and more effective drugs.
In summary, we believe that microfluidics will not immediately revolutionize drug discovery. However, it will increasingly contribute to the development of novel drug discovery tools in combination with well-established methods. Paradigm shifts may occur in the longer-term future, and will probably concern the use of microfluidic techniques in cell biology and tissue engineering. In particular, the use of multiphase flow (for example, droplets), applications in cell biology (for example, three-dimensional cell culture) and tissue engineering will be the focus of interest in the coming few years. The expiry of a small number of very broad patents will allow many companies to benefit from microfluidic and chip technology as an integrative element to their proprietary technology and instrumentation. This is likely to have a positive impact on the application of this technology by end users.
Acknowledgements
This research was supported by the Korea Institute of Science and Technology and the US National Institutes of Health (Director’s New Innovator Award: 1DP2OD007209-01). The authors thank B. Kiraly, F. Guo and an anonymous peer reviewer for helpful discussions.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
References
- 1.Keseru GM & Makara GM The influence of lead discovery strategies on the properties of drug candidates. Nature Rev. Drug Discov 8, 203–212 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Manz A, Graber N & Widmer HM Miniaturized total chemical analysis systems: a novel concept for chemical sensing. Sens. Actuators B Chem 1, 244–248 (1990). [Google Scholar]
- 3.Squires TM & Quake SR Microfluidics: fluid physics at the nanoliter scale. Rev. Mod. Phys 77, 977–1026 (2005). [Google Scholar]
- 4.Arora A, Simone G, Salieb-Beugelaar G, Kim JT & Manz A Latest developments in micro total analysis systems. Anal. Chem 82, 4830–4847 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Dittrich PS & Manz A Lab-on-a-chip: microfluidics in drug discovery. Nature Rev. Drug Discov. 5, 210–218 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Kang L, Chung BG, Langer R & Khademhosseini A Microfluidics for drug discovery and development: from target selection to product lifecycle management. Drug Discov. Today 13, 1–13 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang KC, Reboud J, Kwok YL, Peng SL & Yobas L Lateral patch-clamping in a standard 1536-well microplate format. Lab Chip 10, 1044–1050 (2010). [DOI] [PubMed] [Google Scholar]
- 8.Chen C-Y, Tu T-Y, Jong D-S & Wo AM Ion channel electrophysiology via integrated planar patch-clamp chip with on-demand drug exchange. Biotechnol. Bioengineer. 108, 1395–1403 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Teh SY, Lin R, Hung LH & Lee AP Droplet microfluidics. Lab Chip 8, 198–220 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Thorsen T, Roberts RW, Arnold FH & Quake SR Dynamic pattern formation in a vesicle-generating microfluidic device. Phys. Rev. Lett 86, 4163–4166 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Anna SL, Bontoux N & Stone HA Formation of dispersions using “flow focusing” in microchannels. Appl. Phys. Lett 82, 364–366 (2003). [Google Scholar]
- 12.Utada AS et al. Monodisperse double emulsions generated from a microcapillary device. Science 308, 537–541 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Mao X, Waldeisen JR & Huang TJ “Microfluidic drifting” — implementing three-dimensional hydrodynamic focusing with a single-layer planar microfluidic device. Lab Chip 7, 1260–1262 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Mao X, Lin S-CS, Dong C & Huang TJ Single-layer planar on-chip flow cytometer using microfluidic drifting based three-dimensional (3D) hydrodynamic focusing. Lab Chip 9, 1583–1589 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Cho SK, Moon HJ & Kim CJ Creating, transporting, cutting, and merging liquid droplets by electrowetting-based actuation for digital microfluidic circuits. J. Microelectromechan. Systems 12, 70–80 (2003). [Google Scholar]
- 16.Wang KL, Jones TB & Raisanen A DEP actuated nanoliter droplet dispensing using feedback control. Lab Chip 9, 901–909 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Ahmed R & Jones TB Dispensing picoliter droplets on substrates using dielectrophoresis. J. Electrostat 64, 543–549 (2006). [Google Scholar]
- 18.He MY, Kuo JS & Chiu DT Electro-generation of single femtoliter- and picoliter-volume aqueous droplets in microfluidic systems. Appl. Phys. Lett 87, 031916 (2005). [Google Scholar]
- 19.Darhuber AA, Valentino JP & Troian SM Planar digital nanoliter dispensing system based on thermocapillary actuation. Lab Chip 10, 1061–1071 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Darhuber AA, Valentino JP, Troian SM & Wagner S Thermocapillary actuation of droplets on chemically patterned surfaces by programmable microheater arrays. J. Microelectromech. Syst 12, 873–879 (2003). [Google Scholar]
- 21.Lee C-Y, Pang W, Yu H & Kim ES Subpicoliter droplet generation based on a nozzle-free acoustic transducer. Appl. Phys. Lett 93, 034104 (2008). [Google Scholar]
- 22.Franke T, Abate AR, Weitz DA & Wixforth A Surface acoustic wave (SAW) directed droplet flow in microfluidics for PDMS devices. Lab Chip 9, 2625–2627 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Shi J, Ahmed D, Mao X, Lin S-CS & Huang TJ Acoustic tweezers: patterning cells and microparticles using standing surface acoustic waves (SSAW). Lab Chip 9, 2890–2895 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Shi J et al. Continuous particle separation in a microfluidic channel via standing surface acoustic waves (SSAW). Lab Chip 9, 3354–3359 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Zeng SJ, Li BW, Su XO, Qin JH & Lin BC Microvalve-actuated precise control of individual droplets in microfluidic devices. Lab Chip 9, 1340–1343 (2009). [DOI] [PubMed] [Google Scholar]
- 26.Unger MA, Chou HP, Thorsen T, Scherer A & Quake SR Monolithic microfabricated valves and pumps by multilayer soft lithography. Science 288, 113–116 (2000). [DOI] [PubMed] [Google Scholar]
- 27.Fidalgo LM et al. From microdroplets to microfluidics: selective emulsion separation in microfluidic devices. Angew. Chem. Int. Ed. Engl 47, 2042–2045 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Chen DL, Gerdts CJ & Ismagilov RF Using microfluidics to observe the effect of mixing on nucleation of protein crystals. J. Am. Chem. Soc 127, 9672–9673 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pekin D et al. Quantitative and sensitive detection of rare mutations using droplet-based microfluidics. Lab Chip 11, 2156–2166 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Niu XZ, Gielen F, Edel JB & deMello AJ A microdroplet dilutor for high-throughput screening. Nature Chem. 3, 437–442 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Guttenberg Z et al. Planar chip device for PCR and hybridization with surface acoustic wave pump. Lab Chip 5, 308–317 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Hughes AJ & Herr AE Quantitative enzyme activity determination with zeptomole sensitivity by microfluidic gradient-gel zymography. Anal. Chem 82, 3803–3811 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Matosevic S, Szita N & Baganz F Fundamentals and applications of immobilized microfluidic enzymatic reactors. J. Chem. Technol. Biotechnol 86, 325–334 (2011). [Google Scholar]
- 34.Garcia E, Hasenbank MS, Finlayson B & Yager P High-throughput screening of enzyme inhibition using an inhibitor gradient generated in a microchannel. Lab Chip 7, 249–255 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Lombardi D & Dittrich PS Droplet microfluidics with magnetic beads: a new tool to investigate drug–protein interactions. Anal. Bioanal. Chem 399, 347–352 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Lee JS, Ryu J & Park CB High-throughput analysis of Alzheimer’s (β-amyloid aggregation using a microfluidic self-assembly of monomers. Anal. Chem 81, 2751–2759 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Suzuki H & Takeuchi S Microtechnologies for membrane protein studies. Anal. Bioanal. Chem 391, 2695–2702 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Einav S et al. Discovery of a hepatitis C target and its pharmacological inhibitors by microfluidic affinity analysis. Nature Biotech. 26, 1019–1027 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maerkl SJ & Quake SR A systems approach to measuring the binding energy landscapes of transcription factors. Science 315, 233–237 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Nazarenko I et al. Cell surface tetraspanin Tspan8 contributes to molecular pathways of exosome-induced endothelial cell activation. Cancer Res. 70, 1668–1678 (2010). [DOI] [PubMed] [Google Scholar]
- 41.Alvarez-Erviti L et al. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nature Biotech. 29, 341–345 (2011). [DOI] [PubMed] [Google Scholar]
- 42.Stott SL et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc. Natl Acad. Sci. USA 107, 18392–18397 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Edd JF et al. Controlled encapsulation of singlecells into monodisperse picolitre drops. Lab Chip 8, 1262–1264 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.He M et al. Selective encapsulation of single cells and subcellular organelles into picoliter- and femtoliter-volume droplets. Anal. Chem 77, 1539–1544 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Baret JC et al. Fluorescence-activated droplet sorting (FADS): efficient microfluidic cell sorting based on enzymatic activity. Lab Chip 9, 1850–1858 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Brouzes E et al. Droplet microfluidic technology for single-cell high-throughput screening. Proc. Natl Acad. Sci. USA 106, 14195–14200 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clausell-Tormos J et al. Droplet-based microfluidic platforms for the encapsulation and screening of mammalian cells and multicellular organisms. Chem. Biol 15, 427–437 (2008). [DOI] [PubMed] [Google Scholar]
- 48.Shi WW, Qin JH, Ye NN & Lin BC Droplet-based microfluidic system for individual Caenorhabditis elegans assay. Lab Chip 8, 1432–1435 (2008). [DOI] [PubMed] [Google Scholar]
- 49.Friedland AE et al. Synthetic gene networks that count. Science 324, 1199–1202 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kong DS, Carr PA, Chen L, Zhang S & Jacobson JM Parallel gene synthesis in a microfluidic device. Nucleic Acids Res. 35, e61 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee C-C, Snyder TM & Quake SR A microfluidic oligonucleotide synthesizer. Nucleic Acids Res. 38, 2514–2521 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Butcher EC, Berg EL & Kunkel EJ Systems biology in drug discovery. Nature Biotech. 22, 1253–1259 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Khnouf R, Olivero D, Jin S, Coleman MA & Fan ZH Cell-free expression of soluble and membrane proteins in an array device for drug screening. Anal. Chem 82, 7021–7026 (2010). [DOI] [PubMed] [Google Scholar]
- 54.Overington JP, Al-Lazikani B & Hopkins AL How many drug targets are there? Nature Rev. Drug Discov 5, 993–996 (2006). [DOI] [PubMed] [Google Scholar]
- 55.Wu M-H, Huang S-B & Lee G-B Microfluidic cell culture systems for drug research. Lab Chip 10, 939–956 (2010). [DOI] [PubMed] [Google Scholar]
- 56.Pruss RM Phenotypic screening strategies for neurodegenerative diseases: a pathway to discover novel drug candidates and potential disease targets or mechanisms. CNS Neurol. Disord. Drug Targets 9, 693–700 (2010). [DOI] [PubMed] [Google Scholar]
- 57.Beigel J, Fella K, Kramer P-J, Kroeger M & Hewitt P Genomics and proteomics analysis of cultured primary rat hepatocytes. Toxicol. In Vitro 22, 171–181 (2008). [DOI] [PubMed] [Google Scholar]
- 58.Rubin LL Stem cells and drug discovery: the beginning of a new era? Cell 132, 549–552 (2008). [DOI] [PubMed] [Google Scholar]
- 59.Rubin LL & Haston KM Stem cell biology and drug discovery. BMC Biol. 9, 42 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lutolf MP & Blau HM Artificial stem cell niches. Adv. Mater 21, 3255–3268 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kobel S & Lutolf MP Biomaterials meet microfluidics: building the next generation of artificial niches. Curr. Opin. Biotechnol 22, 690–697 (2011). [DOI] [PubMed] [Google Scholar]
- 62.Edalat F, Bae H, Manoucheri S, Cha J & Khademhosseini A Engineering approaches toward deconstructing and controlling the stem cell environment. Ann. Biomed. Eng 2011, 1–15 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wood DK, Weingeist DM, Bhatia SN & Engelward B P Single cell trapping and DNA damage analysis using microwell arrays. Proc. Natl Acad. Sci. USA 107, 10008–10013 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Abbott A Cell culture: Biology’s new dimension. Nature 424, 870–872 (2003). [DOI] [PubMed] [Google Scholar]
- 65.Bickle M The beautiful cell: high-content screening in drug discovery. Anal. Bioanal. Chem 398, 219–226 (2010). [DOI] [PubMed] [Google Scholar]
- 66.Paul SM et al. How to improve R&D productivity: the pharmaceutical industry’s grand challenge. Nature Rev. Drug Discov 9, 203–214 (2010). [DOI] [PubMed] [Google Scholar]
- 67.Chen AA, Underhill GH & Bhatia SN Multiplexed, high-throughput analysis of 3D microtissue suspensions. Integr. Biol 2, 517–527 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tung Y-C et al. High-throughput 3D spheroid culture and drug testing using a 384 hanging drop array. Analyst 136, 473–478 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Carrel A On the permanent life of tissues outside of the organsism. J. Exp. Med 15, 516–528 (1912). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Holtfreter J A study of the mechanics of gastrulation. J. Exp. Zool 95, 171–212 (1944). [Google Scholar]
- 71.Leighton J A sponge matrix method for tissue culture; formation of organized aggregates of cells in vitro. J. Natl Cancer Inst 12, 545–561 (1951). [PubMed] [Google Scholar]
- 72.Kunz-Schughart LA, Freyer JP, Hofstaedter F & Ebner R The use of 3D cultures for high-throughput screening: the multicellular spheroid model. J. Biomol. Screen 9, 273–285 (2004). [DOI] [PubMed] [Google Scholar]
- 73.Tekin H et al. Responsive microgrooves for the formation of harvestable tissue constructs. Langmuir 27, 5671–5679 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pregibon DC, Toner M & Doyle PS Multifunctional encoded particles for high-throughput biomolecule analysis. Science 315, 1393–1396 (2007). [DOI] [PubMed] [Google Scholar]
- 75.Freytes DO, Wan LQ & Vunjak-Novakovic G Geometry and force control of cell function. J. Cell. Biochem 108, 1047–1058 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kobel S & Lutolf MP High-throughput methods to define complex stem cell niches. Biotechniques 48, IX–XXII (2010). [DOI] [PubMed] [Google Scholar]
- 77.Lii J et al. Real-time microfluidic system for studying mammalian cells in 3D microenvironments. Anal. Chem 80, 3640–3647 (2008). [DOI] [PubMed] [Google Scholar]
- 78.Truckenmüller R et al. Thermoforming of film-based biomedical microdevices. Adv. Mater 23, 1311–1329 (2011). [DOI] [PubMed] [Google Scholar]
- 79.Baker M Tissue models: a living system on a chip. Nature 471, 661–665 (2011). [DOI] [PubMed] [Google Scholar]
- 80.van Midwoud PM, Merema MT, Verpoorte E & Groothuis GMM A microfluidic approach for in vitro assessment of interorgan interactions in drug metabolism using intestinal and liver slices. Lab Chip 10, 2778–2786 (2010). [DOI] [PubMed] [Google Scholar]
- 81.van Midwoud PM, Verpoorte E & Groothuis GMM Microfluidic devices for in vitro studies on liver drug metabolism and toxicity. Integr. Biol 3, 509–521 (2011). [DOI] [PubMed] [Google Scholar]
- 82.Esch MB, King TL & Shuler ML The role of body-on-a-chip devices in drug and toxicity studies. Annu. Rev. Biomed. Eng 13, 55–72 (2011). [DOI] [PubMed] [Google Scholar]
- 83.Sung JH & Shuler ML A micro cell culture analog (μCCA) with 3-D hydrogel culture of multiple cell lines to assess metabolism-dependent cytotoxicity of anticancer drugs. Lab Chip 9, 1385–1394 (2009). [DOI] [PubMed] [Google Scholar]
- 84.Huh D et al. Reconstituting organ-level lung functions on a chip. Science 328, 1662–1668 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Günther A et al. A microfluidic platform for probing small artery structure and function. Lab Chip 10, 2341–2349 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Baker M Screening: the age of fishes. Nature Methods 8, 47–51 (2011). [DOI] [PubMed] [Google Scholar]
- 87.Bullen A Microscopic imaging techniques for drug discovery. Nature Rev. Drug Discov 7, 54–67 (2008). [DOI] [PubMed] [Google Scholar]
- 88.Conrad C et al. Micropilot: automation of fluorescence microscopy-based imaging for systems biology. Nature Methods 8, 246–249 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hulme SE et al. Lifespan-on-a-chip: microfluidic chambers for performing lifelong observation of C. elegans. Lab Chip 10, 589–597 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kim N, Dempsey CM, Zoval JV, Sze J-Y & Madou MJ Automated microfluidic compact disc (CD) cultivation system of Caenorhabditis elegans. Sens. Actuators B Chem 122, 511–518 (2007). [Google Scholar]
- 91.Chung K et al. A microfluidic array for large-scale ordering and orientation of embryos. Nature Methods 8, 171–176 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jimenez AM et al. Towards high throughput production of artificial egg oocytes using microfluidics. Lab Chip 11, 429–434 (2011). [DOI] [PubMed] [Google Scholar]
- 93.Pardo-Martin C et al. High-throughput in vivo vertebrate screening. Nature Methods 7, 634–636 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pujol A, Mosca R, Farres J & Aloy P Unveiling the role of network and systems biology in drug discovery. Trends Pharmacol. Sci 31, 115–123 (2010). [DOI] [PubMed] [Google Scholar]
- 95.Weber W & Fussenegger M The impact of synthetic biology on drug discovery. Drug Discov. Today 14, 956–963 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang HH et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature 460, 894–898 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gibson DG et al. Creation of a bacterial cell controlled by a chemically synthesized genome. Science 329, 52–56 (2010). [DOI] [PubMed] [Google Scholar]
- 98.Gulati S et al. Opportunities for microfluidic technologies in synthetic biology. J. R. Soc. Interface 6 (Suppl. 4), 493–506 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vinuselvi P et al. Microfluidic technologies for synthetic biology. Int. J. Mol. Sci 12, 3576–3593 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]