Abstract
Acoustics has a broad spectrum of applications, ranging from noise cancelation to ultrasonic imaging. In the past decade, there has been increasing interest in developing acoustic-based methods for biological and biomedical applications. This Perspective summarizes the recent progress in applying acoustofluidic methods (i.e., the fusion of acoustics and microfluidics) to bioanalytical chemistry. We describe the concepts of acoustofluidics and how it can be tailored to different types of bioanalytical applications, including sample concentration, fluorescence-activated cell sorting, label-free cell/particle separation, and fluid manipulation. Examples of each application are given, and the benefits and limitations of these methods are discussed. Finally, our perspectives on the directions that developing solutions should take to address the bottlenecks in the acoustofluidic applications in bioanalytical chemistry are presented.
Graphical Abstract
INTRODUCTION
Bioanalytical chemistry, which focuses on analyzing biological systems and detecting biomolecules, has become an increasingly important area in analytical chemistry.1 The advance of our knowledge in biology and medicine relies on analytical methods that can extract critical information from complex biological systems. Compared to classical analytical chemistry, one of the major challenges in bioanalysis is the complexity of the biological systems themselves. For example, there are roughly 20 000 protein encoding genes in the complete human genome, and almost 18 000 different proteins from those encoding genes have been identified.2,3 This gives us a glimpse of the incredible complexity of biological systems. Bioanalytical methods must handle complex sample matrices, low abundance of targets, interactions between a myriad of biomolecules, and the dynamics of biological systems (both temporally and spatially). Hence, effective sample processing protocols and tools are indispensable for any bioanalytical methods. Numerous methodologies and technologies have been developed to achieve efficient manipulation of biological samples, such as chromatographic separations, electrophoresis, magnetics, dielectrophoresis, optical tweezers, and acoustics.4−9 In the past decade, we have seen a rapid increase the application of acoustofluidics (i.e., the fusion of acoustics and microfluidics) to bioanalytical methods in these existing technologies.10,11
The first experimental observation of acoustic interactions on particles dates back to the 1860s, when German physicist August Kundt measured the velocity of sound.12Kundt designed an acoustic resonator chamber, later known as Kundt’s tube, to establish a standing acoustic wave field. The position of the pressure nodes was then visualized by adding fine particles to the chamber, which aggregated at the location of the pressure nodes. Early applications of acoustofluidic sample manipulation include concentrating particles and blood cells, and filtering solids from liquid on the macroscale using an acoustic resonator.13−16 Due to their limited resolution and functionality, these early applications did not draw much attention from the bioanalytical community. However, with the advance of microfluidics and microfabrication techniques, the performance and functionality of acoustofluidic sample manipulation techniques have improved substantially in the past decade and continue to rapidly evolve.4,17−24
The unique advantages of acoustofluidic-based sample manipulation have enabled a variety of applications in bioanalysis. First, acoustic waves are mechanical in nature; they are less likely to have a detrimental impact on the cells and biomolecules when controlled at appropriate frequencies and power levels. Generally, this makes acoustofluidic sample manipulation nondestructive and offers superior biocompatibility, making it a viable tool for many biological applications. Second, acoustic waves manipulate samples without physical contact with the biological targets, thus avoiding complications like sample damage and biofouling. Acoustic waves also do not have strict requirements on the composition of the working fluid, so cells and biomolecules can be kept in optimized buffer solutions when using acoustic manipulation. Third, acoustics is an extremely versatile tool in manipulating biological samples. It can be designed to manipulate single cells with micrometer-level precision or separate billions of cells in a few seconds.25
In this Perspective, we categorize the applications of acoustofluidic technology into four major functional areas: sample concentration, fluorescence-activated cell sorter (FACS), label-free cell/particle separation, and fluid manipulation. We discuss the working mechanisms, representations, and advantages and limitations for each application. While we explain most of the working mechanisms for different types of acoustic systems, the detailed physics and corresponding numerical models are not covered to limit the length of this Perspective. We recommend reviews for more in-depth discussions on the specific operating theories and models.10,26−30 This Perspective aims to give readers a comprehensive picture of the state-of-the-art of acoustic technologies in bioanalytical applications, and to facilitate the adoption or selection of acoustic systems for specific applications. In the outlook section, we provide our reflections on the status of acoustofluidic technologies and discuss important future directions in this area.
SAMPLE CONCENTRATION
Concentrating particles or biological cells from a dilute sample is an indispensable step for bioanalysis. The enrichment of a rare species could significantly improve the detection sensitivity and reduce the analysis time. For example, the detection of bacteria in water sample usually requires a cell culture step to provide sufficient bacteria for analysis, which will take between 6 and 24 h. By employing a concentration unit to enrich bacteria, the total analysis time can be significantly reduced. Particle trapping and concentration, as observed in Kundt’s tube, is one of the earliest applications of acoustic waves in bioanalysis. Early works employed a thin vibrating actuator within a glass tube as the resonator to establish a standing acoustic wave field. Particles inside the standing field will experience an acoustic radiation force, which can be expressed as
| (1) |
| (2) |
where ϕ is the acoustic contrast factor; Vp is the volume of the particle; βf, ρf, βp, and ρp are the compressibility and density of the fluid medium and the particles, respectively; and po, λ, and x are the acoustic pressure, the wavelength of the acoustic waves, and the horizontal distance to the pressure node, respectively. Particles with a positive contrast factor (e.g., polystyrene particles and biological cells) will be pushed toward pressure nodes in the standing wave field and will become trapped, resulting in localized particle concentration (Figure 1a).31 This phenomenon was exploited as a means to enhance immunoagglutination assays.32 In an agglutination assay, latex beads are coated with antibodies specific to the target analyte. In the presence of the analyte, latex beads will form aggregates, which can be detected using turbidimetry, nephelometry, or flow cytometry. Using acoustofluidic particle trapping can increase the local concentration of latex beads, thereby improving the probability of analyte-to-particle collisions. Coakley and co-workers demonstrated a 256-fold improvement in sensitivity for the detection of c-reactive proteins using an acoustic-enhanced immunoagglutination assay.33
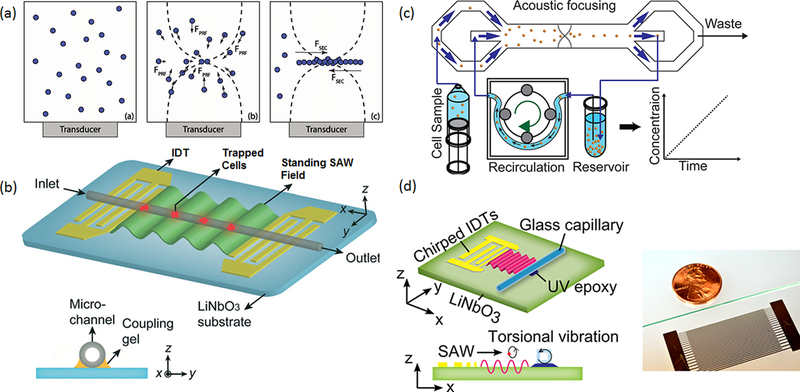
Figure 1.
(a) Illustrations of the trapping process for particles in a standing acoustic wave field. Reproduced with permission from ref 31. Copyright 2012 the Royal Society of Chemistry. (b) A standing surface acoustic wave (SSAW)-based cell concentration device. Cells introduced from the inlet are trapped at the pressure nodes. Reproduced with permission from ref 34. Copyright 2014 the Royal Society of Chemistry. (c) A recirculating cell concentrator using acoustic focusing. Reproduced with permission from ref 35. Copyright 2015 the Royal Society of Chemistry. (d) An acoustic system that can concentrate particles as small as 100 nm in diameter in a glass capillary. Reproduced with permission from ref 24. Copyright 2017 American Chemical Society.
While increasing the local concentration of particles can improve immunoagglutination assays, it is not very useful for many other bioanalytical applications. Most of these applications involving concentrating particles also require an increase in the global concentration for target particles so that it can facilitate downstream analysis, especially for low-abundance samples. In this situation, a particle concentrator that can operate under a continuous-flow condition is desired, as it can process large volumes and transfer the concentrated sample out of the chamber for downstream analysis. Chen et al. reported a standing surface acoustic wave (SSAW) device that can trap particles/cells under a continuous flow (Figure 1b).34 To generate surface acoustic wave (SAWs), a pair of interdigital transducers (IDTs) was fabricated onto a piezo-electric substrate (e.g., LiNbO3). The SAWs were coupled into a polyethylene tubing to establish a SSAW field that is orthogonal to the flow direction. In this way, several pressure node planes were formed along the path of the moving particles/cells, which effectively trapped particles/cells in a continuous flow. The device achieved a concentration factor 100−1000 times for diluted blood cells with a recovery efficiency >90%. The method is highly efficient and does not require a specialized resonance chamber. This makes it suitable for applications that need a rapid concentration of cells from a small sample volume. The major limitation for this type of acoustic-based sample concentration is the saturation of the trapping site, as it could become filled with a large number of trapped cells, making it difficult to handle large-volume samples.
Recently, Soh and co-workers reported an acoustofluidic cell concentrator that can continuously collect concentrated cells, which allows the processing of large sample volumes with ~1000-fold (Figure 1c) enrichment for the particle/cell sample.35 They designed an acoustic resonator that has a pressure node plane parallel to the flow direction. When the cell sample is introduced into the flow chamber, all the cells will be pushed toward the center of the flow chamber (the pressure node plane). By designing two outlets that can collect the center stream and side streams, the cell sample is concentrated as the excess fluid is removed. To further improve the concentration factor, the researchers employed a peristaltic pump to recirculate the cell sample. After a complete enrichment cycle, up to a 1000-fold concentration increase was achieved for red blood cells and human cancer cell lines.
Another important direction in the development of an acoustofluidic particle concentrator involves concentrating nanoparticles. Nanoparticles are used extensively in bioassays due to their large surface-to-volume ratio, and bio-nanoparticles, such as extracellular vesicles, are potential biomarkers for many diseases. If acoustofluidic particle concentration can be applied to nanoparticles, it will certainly expand the application scope of this technology. However, classic acoustic particle concentration using acoustic radiation force does not work for nanoparticles. As shown in eq 1, the amplitude of the acoustic radiation force depends on the volume of the particle. The acoustic radiation force exerted on nanoparticles is much smaller than that exerted on mammalian cells, with a diameter range of 5−30 μm. For nanoparticles, the induced drag force from acoustic streaming is not negligible with respect to the acoustic radiation force. The movement of nanoparticles is then determined by a competition between the drag force and the acoustic radiation force. Thus, several new strategies were reported to achieve nanoparticle concentration. Collins et al. reported a high-frequency SAW device that can locally enrich nanoparticles down to ~300 nm.36 By introducing a high-frequency (193 or 636 MHz) focused SAW on a polydimethylsiloxane (PDMS) microfluidic channel, localized microvortex acoustic streaming can be achieved. Particles between 0.5 and 2 μm can be selectively trapped at the microvortex due to the combined effect of the acoustic radiation force and the microvortex streaming pattern. Although this method provides a simple way to concentrate nanoparticles in a microchannel, using a high-frequency SAW increases the fabrication resolution and the need for specialized equipment in order to obtain the necessary signal generation and amplification.
Mao et al. achieved low-frequency nanoparticle concentration inside a square cross-section glass capillary by synchronizing the effects of the acoustic radiation force and acoustic streaming (Figure 1d).24 They found that, under a certain frequency around 2.6 MHz, a large, single-vortex streaming pattern was formed across the glass capillary. Since the pressure node was also located at the center of the channel, the two forces (i.e., the acoustic radiation force and the drag force) do not compete. Thus, nanoparticles were concentrated to the channel center. Effective concentration of polystyrene particles down to ~80 nm was demonstrated. This method was then used to improve detection signals for a homogeneous nanoparticle-based immunoassay. A 30-fold enhancement in the fluorescence signal was demonstrated with this acoustofluidic nanoparticle concentrator. Hammarstrom et al. also achieved nanoparticle enrichment using a low-frequency, secondary acoustic radiation force, which is generated from particle−particle interactions within close proximity.37 They first trapped a cluster of 10 μm particles as the seed particles. E. coli and nanoparticles were then trapped and enriched as they flowed between these large seed particles in an acoustic field due to the secondary acoustic radiation forces. In a recent report, this method was successfully used to enrich bacteria from the blood of sepsis patients.23 The bacteria enrichment process can be completed in 2 h, which is much shorter than the time for a cell culture (4.6−26.4 h). The future direction of this system is to further improve the sensitivity of this system to identify sepsis with a low bacteria load. Using the similar concept, extracellular vesicles from culture media, urine, and blood have also been enriched, indicating the potential of using acoustofluidic methods to concentrate extracellular vesicles.38,39
In addition to concentrating particles/cells in a chamber, it is also possible to enrich them in an unconfined droplet as result of the acoustic radiation force and acoustic streaming.40 Cooper and colleagues employed a slanted-finger interdigitated transducer to generate SAWs that could actuate a droplet on the SAW substrate.41 Because of the specific streaming patterns generated inside the droplet, cells with different densities settled on different positions in the droplet. They exploited this phenomenon to selectively enrich red blood cells infected with the malaria parasite on the outer ring of the droplet. The enrichment was completed in 3 s, with a 100−1000-fold enrichment factor.
FLUORESCENCE-ACTIVATED CELL SORTER
A FACS is a powerful, high-throughput, single-cell characterization and sorting tool that has revolutionized how cells are studied and purified. One of the fundamental technical requirements for any FACS system is to manipulate cells into a single file (cell focusing) and then select specific cells (cell sorting) after receiving a detection signal. Acoustic waves have been demonstrated to be capable of both cell focusing and cell sorting, with unique advantages compared to classical FACS systems.
As discussed above, the acoustic radiation force can be used to direct particles to the pressure node of a standing acoustic wave field. When the pressure node plane is designed to be parallel to the flow direction, particles can be aligned as they flow through the acoustic field. Thus, acoustic focusing of particles is achieved. Goddard et al. reported the first acoustic-focusing-based flow cytometer in 2006.42 They used a tube with a circular cross section as the flow cell. As a result, when acoustic vibrations with the proper frequency are applied to the side wall of this flow cell, a single-pressure node plane is formed in the center, where particles are aligned in a single file. Acoustic focusing is advantageous over conventional hydrodynamic focusing, as it does not require sheath flow. By removing the need for a sheath flow, as required in traditional flow cytometry, acoustic focusing allows for a much higher sample throughput under the same flow velocity. This prototype system was later developed into the first commercial flow cytometer using acoustic focusing. Another advantage of using acoustic focusing is the ability to generate multiple focused cell (or particle) streams with multiple pressure nodes via a standing acoustic wave field in one device. Graves and colleagues developed a planar flow cell that focuses up to 37 streams of cells with a standard acoustic setup.19 Recently, by combining the flow cell with a line-focused laser beam and a CMOS array detector, this group achieved a throughput of 100 000 events/s.43 While acoustic focusing in a resonator has gained tremendous success in flow cytometry, acoustic focusing can also be achieved using SSAWs to establish a pressure node plane parallel to the flow direction.44 The major advantage of using a SSAW is the flexibility in channel design, which allows for the further miniaturization of the components in future flow cytometers. Chen et al. demonstrated SSAW-based focusing in a microfluidic channel with a length less than 1 cm (Figure 2a).45 The coefficient of variation obtained in their system was comparable to that obtained with a commercial FACS.
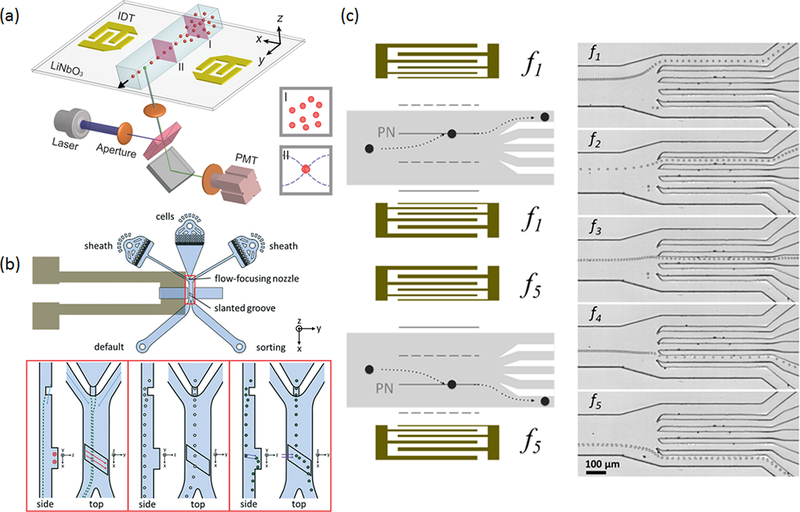
Figure 2.
(a) SSAW-based microfluidic flow cytometer. Cells are focused by establishing a pressure node array in a plane that is parallel to the flow direction. Reproduced with permission from ref 45. Copyright 2014 the Royal Society of Chemistry. (b) A traveling SAW-based cell sorter. A slanted groove structure is fabricated to enhance the acoustic actuation. Reproduced with permission from ref 49. Copyright 2017 the Royal Society of Chemistry. (c) SSAW-based multiplexed cell sorter. The position of the pressure node can be tuned by adjusting the input frequency. Hence, particles can be sorted into five different outlets using five frequencies. Reproduced with permission from ref 46. Copyright 2012 the Royal Society of Chemistry.
Another key functional module in a FACS is the cell-sorting unit. Cell sorting involves directing cells to a designated collection area when a trigger signal is received. Commercial cell sorters typically use a high-voltage electrical field to actuate charged cell droplets. While this sorting mechanism is very efficient and can achieve a throughput up to 70 000 events/s, it stresses the cells during the sorting process, affecting their viability. This sorting process also occurs in an open environment, so pathogen-containing aerosols can be generated and present a practical threat to the operator. Acoustofluidic methods can sort cells with moderate external forces and within an enclosed environment while maintaining a low cost and small device footprint. Both acoustic radiation force and acoustic streaming have been used to actuate cells in a flow cell.46−48
Since cell sorting prefers a highly localized actuation area to ensure single-cell accuracy, the more flexible SAW devices are often favored over resonator-based systems, which have difficulty confining the actuation region to a small area. Franke et al. first reported a traveling SAW microfluidic device for high-throughput cell sorting.47 Only one IDT was fabricated on a piezoelectric substrate to generate the traveling SAWs that propagate orthogonal to the flow direction. When the traveling SAWs propagate into the liquid medium, they will generate acoustic streaming that actuates the fluid, pushing cells (or particles) to the collection outlet. To confine the effective actuation area, the researchers employed a PDMS post to control the position where the traveling SAW enters the liquid medium. A sorting rate of 3000 events/s was achieved with an applied electric voltage of less than 1.8 V. Good cell viability was maintained under these mild sorting conditions. More recently, the group further improved their sorting design by fabricating a slanted groove at the sorting region to replace the PDMS post, which further improves the energy efficiency of the SAW.49 The device achieved 92% purity with a sorting throughput of 1000 events/s (Figure 2b).
Huang and co-workers first reported SSAW-based cell sorting.46 The major advantages of using a SSAW are the high controllability and the flexibility to achieve multiplex sorting. In SSAW-based cell sorting, the direction of cell movement depends on the position of the pressure node. Thus, multiplex sorting can be realized by moving the position of pressure nodes during sorting. Ding et al. employed a pair of chirped IDTs that can resonate at a series of frequencies. In this device, the position of the pressure node can be adjusted by changing the input frequency. As shown in Figure 2c, five-channel sorting was achieved. Based on SSAW-based cell sorting, Nawaz et al. developed a microfluidic FACS that can sort dye-stained HeLa cells from unstained cells with a purity of ~92% at a 1200 events/s throughput.50 Ren et al. further improved the throughput of SSAW cell sorting using focused IDTs.51 Focused IDTs further reduce the actuation area of SSAW-based sorting to ~160 μm, leading to a sorting rate >10 000 events/s. Recently, the same group reported a complete acoustofluidic cell sorter by integrating the SSAW-based cell focusing and SSAW-based cell sorting into one device which is capable of sorting HeLa cells at a throughput of 2500 cells/s.52
LABEL-FREE CELL/PARTICLE SEPARATION
Label-free separation is an important sample-processing function which is routinely performed in bioanalysis to reduce interference from a complex sample and facilitate the detection of targets. As discussed above, acoustic waves can exert forces on particles inside the wave field. The amplitude of this force is dependent on the physical properties of particles, e.g., cell size and cell compressibility. If a sample has a mixture of cells that have different sizes or compressibilities, it is possible to separate them using acoustic waves. To date, most of these acoustofluidic separation methods use standing acoustic waves. In a standing wave field, all the cells will be pushed to the location of the pressure nodes by the acoustic radiation force. Larger cells will migrate to the pressure node faster than smaller ones as they experience a larger acoustic radiation force. Acoustic separation is realized by inserting a bifurcation during the migration process to collect large and small components separately.
In 1995, Johnson and Feke reported the first sized-based acoustic separation system using a half-wavelength resonator as shown in Figure 3a.53,54 A mixture of large and small particles was introduced from one side of the resonator chamber, and then both types of particles were directed toward the channel center (pressure node). Due to the different migration speeds, the particles were separated at the outlet. Later, with the development of microfluidics and microfabrication, Laurell and co-workers further extended this separation scheme to a microfluidic platform, which provides more precise fluidic control and enables integration with downstream analysis modules.17 They used this approach to separate cancer cells from white blood cells. In a recent work, the same team achieved a ~92% separation efficiency with MCF-7 breast cancer cells and white blood cells with a 6 mL/h sample throughput (Figure 3b).55 They integrated an acoustic-based cell concentrator to concentrate the cancer cells after separation, which is essential when dealing with rare cell samples.56 Different components of blood can also be separated using this mechanism. Chen et al. reported a high-throughput platelet separation device using a thin reflective resonator.25 The sample stream was introduced from the bottom layer, while a buffer solution matching the acoustic impedance of whole blood was introduced from the top layer. This setup maximizes the effective use of acoustic pressure nodes, thereby increasing the separation throughput significantly to 10 mL/min, which is almost a 1000-fold improvement over other acoustofluidic platelet separation devices.57 A red blood cell/white blood cell removal rate greater than 85% and a platelet recovery rate greater than 80% were achieved. Compared to centrifugation-based platelet separation, this acoustofluidic method induces less platelet activation and results in better morphology and functionality after separation. Using acoustic impedance matching, Ohlsson et al. separated bacteria from whole blood with a throughput ~ 5 mL/h,58 achieving ~90% bacteria recovery rate and >99% blood cell removal rate. Grenvall et al. demonstrated that subpopulations of white blood cells can be separated using acoustic waves with an improved cell prefocusing technique.59 As the difference between subpopulations of white blood cells is much smaller than other cases, it is crucial to align cells within a narrow range of starting positions. In this work, acoustic focusing and laminar flow focusing were combined to generate a tightly focused stream of cells before they entered the separation region. Lymphocytes and granulocytes were collected with high purity (95.2 ± 0.6% and 98.5 ± 0.7%, respectively) and high recovery (86.5 ± 10.9% and 68.4 ± 10.6%, respectively).
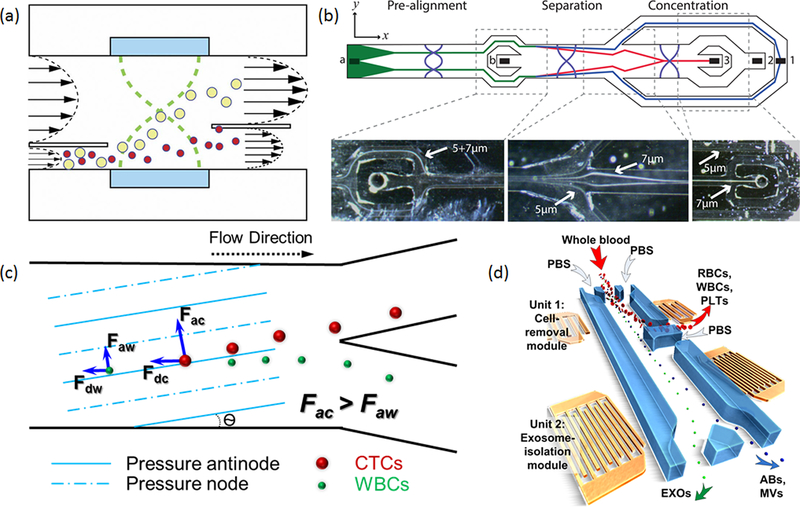
Figure 3.
(a) Typical setup of an acoustofluidic, half-wavelength particle separator. Reproduced with permission from ref 54. Copyright 2012 the Royal Society of Chemistry. (b) An integrated acoustic separator with prefocusing, separation, and concentration units. Reproduced with permission from ref 55. Copyright 2015 American Chemical Society. (c) Working mechanism for taSSAW-based cell separation. Reproduced with permission from ref 22. Copyright 2015 the National Academy of Science. (d) Two-stage separation device using taSSAWs for the isolation of exosomes from whole blood. Two pairs of IDTs with different wavelengths are used to separate cells and vesicles, respectively. Abbreviations: RBCs, red blood cells; WBCs, white blood cells; PLTs, platelets; EXOs, exosomes; Abs, apoptotic bodies; MVs, microvesicles. Reproduced with permission from ref 66. Copyright 2017 the National Academy of Science.
In addition to the resonator setup, SSAWs can also be used to realize size-, density-, or compressibility-based cell separation under a similar working mechanism.18 Moreover, unlike resonator-based separation, where the direction of the pressure node plane is limited by the orientation of the channel wall, the SSAW allows for designs of the acoustic wave field inside the flow cell to be more flexible. Ding et al. developed a tilted-angle standing surface acoustic wave (taSSAW) cell separation device that can separate cancer cells from white blood cells.60The direction of the pressure node in a taSSAW has a small angle to the flow direction, so the separation is the result of the combined effects of acoustic radiation force and hydrodynamic drag force (Figure 3c). In the acoustic resonator setup,17 the maximum separation distance for cells is limited to a quarter of the wavelength (typically 75−300 μm), whereas taSSAW can overcome this separation distance limitation, as the hydrodynamic drag can carry cells over several acoustic wavelengths. As a result, the separation process is more robust and less sensitive to the fluctuations in the laminar fluidic stream and the cells’ initial positions. Li et al. further optimized the taSSAW design and achieved a 20-fold throughput increase over the original design.22 The optimized device was then applied to clinical samples and successfully isolated circulating tumor cells (CTCs) in blood samples from breast cancer patients. Wu et al. increased the separation throughput of the taSSAW device to 7.5 mL/h for a blood sample by employing a PDMS−glass hybrid channel.61 With the increased separation throughput, CTC clusters from prostate cancer patients’ samples were also isolated. Compared to other acoustic methods,62−65 taSSAW allows the use of shorter acoustic wavelengths to improve the separation of smaller particles, such as bacteria and extracellular vesicles. Wu et al. reported a two-stage taSSAW separation device that can directly isolate exosomes from human whole blood samples (Figure 3d).66 The first stage of their device is a low-frequency (19.6 MHz) taSSAW cell separation unit that removes the blood cells while sending plasma and extracellular vesicles to the second-stage separation unit. The second stage is a high-frequency (39.4 MHz) taSSAW separation unit that separates the exosomes (<140 nm) from apoptotic bodies and microvesicles (>140 nm). This integrated, two-stage acoustofluidic separator automates the process of obtaining exosomes from blood samples and, compared to the commonly used multistage centrifugation/ultracentrifugation systems, reduces the time and labor demands.
In addition to the classical standing wave-based cell separation, traveling SAW is an emerging acoustofluidic separation technique. Traveling SAW can also induce the acoustic radiation force which interacts with particles in the liquid medium. The amplitude of the force depends on the relationship between the size of the particles and the wavelength of the traveling SAW. In order to achieve effective separation of microscale particles, a frequency >100 MHz is generally required. Destgeer et al. achieved traveling SAW-based particle separation with an impressive resolution as low as 200 nm.21 Traveling SAW separation does not require strict alignment between the flow cell and the acoustic field, making it easier to fabricate and implement. Furthermore, a disposable traveling SAW separation platform has recently been reported.67 Most traveling SAW separations were carried out using particles. There still needs to be more demonstrations of separating biological samples (e.g., cells) before it becomes more widely considered for bioanalysis applications.
FLUID/DROPLET MANIPULATION
Various fluidic operations are indispensable to most bioassay protocols. Acoustic waves can also manipulate fluids by controlling the streaming patterns via acoustic streaming. The mixing of different analytes is often required for bioassays. However, mixing on the microscale is often problematic because the viscous force dominates the inertial force. Acoustic streaming provides a viable way to disrupt the streamlines and accelerate the mixing of multiple components at the microscale. Both SAW and resonator-based acoustic devices have reportedly achieved effective mixing in a microchamber or in an open droplet.68,69 Yeo and colleagues applied SAW-based mixing to a flow-injection analysis system to enhance the chemiluminescence signal for detection.70 With the assistance of SAW mixing, they achieved a 100-fold enhancement of detection sensitivity without the need for sample preconcentration. Due to the simple nature of the SAW device, it is a suitable platform for portable flow-injection analysis detection.
Pumping for fluidic transport is another basic fluid operation in bioassays. While SAW-based pumping in a microchannel has been reported, the high frequency and power requirements make it less useful for practical bioanalysis.71 A more efficient SAW pumping method involves the transport droplets on a substrate in the direction of the traveling SAW propagation. Bourquin et al. reported a sandwich-bead-based immunoassay for human IFN-g (Figure 4a) using SAW pumping.72 A slanted IDT was used to generate SAWs at three distinct frequencies: 13.2 MHz to mix the beads, 12.12 MHz to transport particles to the droplet center for capture antibody binding, and 13.4 MHz to pump droplets and unbound particles out of the detection area. This method shows the flexibility of SAW-based methods, where one set of electrodes and a control unit can perform three relevant tasks for an immunoassay. The same group further extended the functionality of an open SAW substrate by achieving SAW-induced cell lysis and polymerase chain reaction (PCR) thermal cycling using SAW heating.20 Within 3 s, red blood cells were lysed in a droplet using the higher shear stress generated by the SAW-induced acoustic streaming under high power. By combining these two functional units, a SAW-based PCR device was developed for the rapid detection of the malaria parasite Plasmodium berghei in blood. Recently, Zhang et al. reported the use of SAW-induced acoustic streaming to control aqueous droplets on an oil surface.73 This platform allows for digital microfluidics-like droplet manipulation without the need of a solid surface, thereby reducing the surface nonspecific binding compared to digital microfluidics.
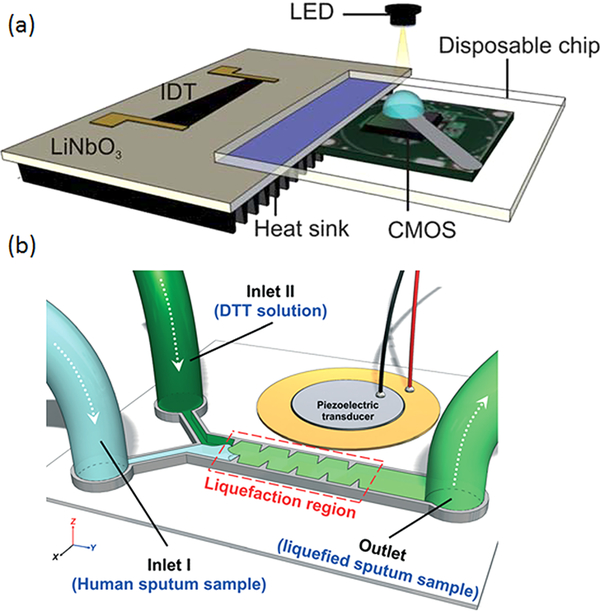
Figure 4.
(a) An integrated SAW immunoassay chip. Reproduced with permission from ref 72. Copyright 2011 the Royal Society of Chemistry. (b) A sputum liquefier based on acoustic sharp-edge mixing. Reproduced with permission from ref 83. Copyright 2015 the Royal Society of Chemistry.
If a fluid droplet on a substrate is actuated with high-power, high-frequency acoustic waves, the droplet will break up into numerous small aerosol droplets. This process is called acoustic nebulization or atomization. Acoustic nebulization has found many applications in aerosol-based drug delivery systems. In bioanalytical chemistry, it is primarily studied as an ambient ionization method for mass spectrometry (MS) analysis. Cooper and co-workers first used SAW-based nebulization (SAWN) as an ionization method for analyzing peptides using a hybrid linear ion trap to develop a Fourier-transform ion cyclotron resonance mass spectrometer.74 This SAWN-based ionization has an operating mechanism similar to that of matrix-assisted laser desorption/ionization (MALDI): pulsed ionization from a chip, while generating multicharged precursor ions. Compared to electrospray ionization (ESI), SAWN produced ions with lower internal energy. SAWN can also be coupled with liquid chromatography and paper microfluidics for rapid MS analysis of biological samples. To date, a wide range of molecules, including heavy metals, small molecules, peptides, and lipids, have been tested using SAWN-MS.75−78 In addition to MS applications, Ashtiani et al. employed SAW-based atomization for cryogenic electron microscopy grid preparation.79
Instead of using SAWs to generate acoustic streaming, acoustic streaming can also be induced using oscillating gas bubbles or sharp edges that are excited by an acoustic energy source.80 Compared to SAW streaming, this induced streaming via an oscillating membrane does not require a piezoelectric substrate or fabricating IDTs, operates at low frequencies (<100 kHz), and can be excited with low-cost and commercially available transducers. Both fluidic pumping and mixing have been achieved by controlling the organization of microvortices generated by the oscillating membranes.81,82 Based on sharp-edge-based fluid mixing, Huang et al. reported the first microfluidic sputum liquefier (Figure 4b).83 Sputum is usually a highly viscous sample and has to be liquefied using a microvortex mixer before any downstream analysis can be carryied out. Common microfluidic systems cannot generate enough body forces to mix the sputum sample and the liquefying reagent. In this work, the streaming generated from oscillating sharp edges is strong enough to induce complete mixing, resulting in a liquefied sample for downstream analysis. The strong streaming generated by oscillating membranes was also used to rotate cells and microorganisms. Ahmed et al. reported the controlled rotation of L4-stage Caenorhabditis elegans in a microchannel with multibubble-induced microvortices.84 This method enables high-quality 3D imaging of the whole body of C. elegans using a normal epifluorescence microscope. Recently, Li et al. demonstrated that sharp-edge vibration can also be used to nebulize liquid samples for MS analysis. They exploited the flexibility of sharp-edge devices and developed the first mechanical-based probe ionization method that can analyze sample surfaces directly.85
PERSPECTIVE AND OUTLOOK
As discussed above, acoustofluidics has demonstrated many important applications in biological sample manipulation. The numerous applications of the acoustofluidic methods in bioanalytical chemistry are summarized in Table 1. For researchers who are interested in embracing acoustofluidic technologies for their own applications, several critical aspects need to be considered. First, one needs to select appropriate acoustofluidic methods based on the targets. Both solids and liquids can be manipulated by acoustofluidic methods, but the underlying manipulation mechanisms are different. Most fluid manipulation methods are achieved through controlling the streaming pattern induced by acoustic waves, while particles are primarily manipulated by using acoustic radiation force. As the size of particles decreases, the impact of streaming will become more and more dominant. Generally, for particles with a diameter less than 1 μm, the streaming effect could dominate the particles’ behavior over the acoustic radiation force. Therefore, special efforts need to be made for dealing with small particles. The second aspect is stopped-flow versus continuous-flow manipulation. Many acoustofluidic methods can be operated under either stopped-flow or continuous-flow conditions. The selection of flow conditions depends on the emphasis of the specific application. If the throughput is a major focus, continuous flow is a better choice, as it generates the desired products continuously. If the manipulation target has a small volume or needs precise manipulation, a stopped-flow condition will be more effective. Third, special care is needed when manipulating living biological objects. To maintain the integrity and normal physiology of living biological objects (e.g., mammalian cells), the input power level should be kept as low as possible for reasonable acoustic radiation pressure level and acoustic streaming velocity. For the experiments requiring high power input, the temperature in the chamber needs to be controlled for optimal physiological temperature. In addition, cavitation effects need to be avoided, as the generation and bursting of microbubbles could damage cell membranes, especially for experiments with frequency less than 1 MHz.
Table 1.
Summary of Applications of the Acoustofluidic Methods in Bioanalytical Chemistry
| target of manipulation | applications | methodsa | references |
|---|---|---|---|
| particle | immunoagglutination assay | standing BAW | 32, 33 |
| microparticle/cell concentration | SSAW | 34 | |
| BAW | 35 | ||
| nanoparticle/bacteria/ vesicle concentration | SAW | 36, 40, 41 | |
| standing wave + acoustic streaming | 22 | ||
| secondary acoustic radiation force | 23, 37–39 | ||
| cell focusing | BAW | 19, 42, 43 | |
| SSAW | 44, 45, 51 | ||
| cell sorting | BAW | 48 | |
| SSAW | 46, 50–52 | ||
| traveling SAW | 47, 49 | ||
| cell/particle separation | BAW | 17, 25, 53–56, 59 | |
| SSAW | 18, 57 | ||
| taSSAW | 22, 60, 61 | ||
| traveling SAW | 21, 67 | ||
| bacteria/vesicle separation | BAW | 58, 63, 65 | |
| SSAW | 62, 64 | ||
| taSSAW | 66 | ||
| fluid | fluid mixing | BAW | 69 |
| SAW | 68, 70 | ||
| oscillating bubble/sharp-edge | 82, 83 | ||
| pumping | SAW | 71–73 | |
| oscillating bubble/sharp-edge | 80, 81 | ||
| cell lysis and PCR | SAW | 20 | |
| nebulization | SAW | 74–79 | |
| vibrating sharp-edge | 85 | ||
| cell/microorganism rotation | oscillating bubble/sharp-edge | 84 |
Abbreviations: BAW, bulk acoustic wave; SAW, surface acoustic wave; SSAW, standing surface acoustic wave; taSSAW, tilted-angle standing surface acoustic wave.
Most acoustofluidic-based manipulation happens instantaneously, so it can serve as an effective sample manipulation tool for real-time analysis of biological samples. For instance, Guo et al. used two pairs of IDTs to generate interfering standing acoustic waves in a microfluidic chamber.86 By controlling the amplitudes and frequencies of the input signals, specific patterns for cell assembly can be formed. Their device enabled the dynamics of intercellular communication to be studied by controlling the gap junction between homotypical cell pairs and heterotypical cell pairs. As the cell-to-cell contact forms, the transfer of dye molecules between neighboring cells can be studied in real time using fluorescence microscopy. In another example, Wiklund et al. employed ultrasonic cell traps to study time-resolved interactions between natural killer cells and cancer cells using fluorescence microscopy.87 These examples demonstrate the potential of applying acoustofluidic methods for real-time analysis when coupled with real-time detection techniques (e.g., microscopy or spectroscopy).
While real-time analysis could provide invaluable time-resolved information for biological events, the combination of acoustic tweezers with information-rich bioanalytical methods (e.g., sequencer, MS, and NMR) is also necessary for bioanalysis. Acoustic tweezers are acoustic systems that can manipulate cells, cell assemblies, or organisms inside a fluidic chamber.27 Most acoustic tweezers are based on standing acoustic waves, which utilize the distribution of the pressure nodes within a given geometry to achieve cell manipulation. Thanks to the robustness and biocompatibility of standing acoustic wave-based manipulation, it has been applied to many biological applications, including cell−cell interaction, controlled cell co-culturing, rapid fabrication of cell spheroids, and generation of single-cell arrays.88−93 In this Perspective, we did not include a discussion of acoustic tweezers since there have not been many bioanalytical applications of this technology, despite it having already shown great potential in the manipulation of single cells, cell assemblies, and micro-organisms with unprecedented precision and versatility.4,94−96 One of the technical hurdles between acoustic tweezers and these information-rich analytical methods is the lack of an interface that can efficiently connect the acoustically manipulated microenvironment to an external analytical instrument for on-chip analysis downstream. For example, acoustic tweezers can assemble specific cell patterns to study intercellular communication with high temporal and spatial resolution, but the information at the gene expression level or at the proteomic level cannot be probed. Therefore, if we can develop interfaces that are compatible with these information-rich analytical methods, this will open new avenues in biological research and reveal new insights into cellular mechanisms.
Another hindrance to the widespread adoption of acoustofluidic methods for in-depth bioanalysis is the lack of systematic characterizations of the influence of acoustic waves on normal cell physiology. Although the existing literature supports the biocompatibility of acoustofluidic cell manipulation under a moderate power input (with respect to cell viability and ability of proliferate), these works are limited to a nonstandardized acoustic system, making it difficult to compare results across different research laboratories.97−99 To address this problem, future studies should attempt to measure the actual force and streaming velocity in the microchannel instead of reporting an input power.100,101 Theoretical models and calibration methods should also be developed to facilitate such characterization. Once a standard is established, an indepth investigation on acoustic parameters and cell responses can be conducted to provide a reference to the research community, allowing the influence of acoustic waves on cells and organisms at the gene expression level to be better understood.
Despite the rapid development of acoustofluidic methods in the past decade, few commercial products are currently available. This is attributable to many reasons, such as the lack of an integration solution, the discrepancies between laboratory-scale device fabrication and mass production, and the limited time in development for many recent acoustofluidic methods. To further facilitate the adoption of acoustofluidic methods for bioanalytical applications, future efforts need to focus on the development of a complete package for certain applications. For example, in addition to cell-focusing and cell-sorting units, an acoustofluidic cell sorter should also integrate pumping and optical detection units and an electronic feedback system to improve the ease of use for end users. In addition, the device material should also be compatible with mass production. Currently, many acoustofluidic methods are based on PDMS and glass materials, which are difficult or expensive for mass production. Plastic-based devices fabricated through injection molding would be far more attractive for commercialization.
Many of the acoustofluidic systems previously mentioned have great potential for point-of-care (POC) applications due to their simplicity, small footprint, low cost, and low power consumption. Acoustofluidic methods for particle concentration, fluid mixing and pumping, and sample nebulization are especially suitable for POC applications. Based on these techniques, acoustofluidic-based immunoassay and PCR assay have been reported. To bring these methods to real POC application scenarios, efforts should be made to improve the quantification capability of existing methods and miniaturize current external components, including signal generators and pumps. Future POC applications of acoustofluidic sample processing systems will also benefit from integrating multiple functional acoustic units into one complete system. The major benefits of an integrated system are standardized control units and streamlined system operation. For example, the integration of acoustic pumping, acoustic cell separation, and acoustic cell concentration will result in a much more compact, automated, and rapid system for rare cell analysis. The key technical challenge in integrating these individual units is how to design the interface to coordinate the different elements without interfering in the normal function of other units.
The common targets for acoustic manipulations include fluids, extracellular vesicles, bacteria, mammalian cells, and small organisms. However, these do not cover the full spectra of biological objects. The potential to acoustically manipulate large biomolecules such as proteins, protein aggregates, and long-chain DNA molecules is not well documented or explored. Recently, Chalasani and co-workers have identified that the TRP-4 protein can sensitize C. elegans neurons’ response to ultrasound and generate behavioral outputs.102 Their research is a possible link to the application of acoustic waves at the molecular level. In addition, the question arises if it is possible for an acoustics method to separate intracellular organelles such as mitochondria. Current standard mitochondria separation involves multiple steps of ultracentrifugation, with a high likelihood of damaging mitochondria. Acoustofluidic separation could potentially reduce the time and cost of this process while improving the overall viability of the separated organelles.
Existing acoustofluidic methods are generally carried out either on a substrate or in a microfluidic chamber. To meet the needs of future in situ or in vivo analysis methods requires acoustofluidic systems to be compact, flexible, self-powered, or powered wirelessly. Recently, Bachman et al. reported an acoustofluidic mixing device that can be operated by a cell phone and a Bluetooth speaker, indicating the potential of powering acoustofluidic device with less power and even wirelessly.103 With the rapid development of flexible electronics, it is also possible to fabricate acoustic electronics on flexible substrates. Luo et al. fabricated SAW transducers on a thin ZnO layer that was deposited on a flexible polyimide and polyethylene terephthalate substrate.104 The flexible SAW device was capable of generating acoustic streaming, which demonstrates the potential of applying acoustofluidic devices for wearable sensors or in vivo analysis.
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (R01HD086325, R01AI120560, and R33CA223908) and West Virginia University startup fund.
Footnotes
The authors declare the following competing financial interest(s): The authors declare the following competing financial interest(s): T.J.H. has four US patents (patent numbers: 8,573,060; 9,608,547; 9,606,086; and 9,757,699) related to acoustofluidics and acoustic tweezers. He has also co-founded a start-up company, Ascent Bio-Nano Technologies Inc., to commercialize technologies involving acoustofluidics and acoustic tweezers.
REFERENCES
- (1).Bowser MT Chem. Rev 2013, 113 (4), 2267–2268. [DOI] [PubMed] [Google Scholar]
- (2).Kim M-S; Pinto SM; Getnet D; Nirujogi RS; Manda SS; Chaerkady R; Madugundu AK; Kelkar DS; Isserlin R; Jain S; Thomas JK; Muthusamy B; Leal-Rojas P; Kumar P; Sahasrabuddhe NA; Balakrishnan L; Advani J; George B; Renuse S; Selvan LDN; Patil AH; Nanjappa V; Radhakrishnan A; Prasad S; Subbannayya T; Raju R; Kumar M; Sreenivasamurthy SK; Marimuthu A; Sathe GJ; Chavan S; Datta KK; Subbannayya Y; Sahu A; Yelamanchi SD; Jayaram S; Rajagopalan P; Sharma J; Murthy KR; Syed N; Goel R; Khan AA; Ahmad S; Dey G; Mudgal K; Chatterjee A; Huang T-C; Zhong J; Wu X; Shaw PG; Freed D; Zahari MS; Mukherjee KK; Shankar S; Mahadevan A; Lam H; Mitchell CJ; Shankar SK; Satishchandra P; Schroeder JT; Sirdeshmukh R; Maitra A; Leach SD; Drake CG; Halushka MK; Prasad TSK; Hruban RH; Kerr CL; Bader GD; Iacobuzio-Donahue CA; Gowda H; Pandey A Nature 2014, 509, 575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Wilhelm M; Schlegl J; Hahne H; Gholami AM; Lieberenz M; Savitski MM; Ziegler E; Butzmann L; Gessulat S; Marx H; Mathieson T; Lemeer S; Schnatbaum K; Reimer U; Wenschuh H; Mollenhauer M; Slotta-Huspenina J; Boese J-H; Bantscheff M; Gerstmair A; Faerber F; Kuster B Nature 2014, 509, 582. [DOI] [PubMed] [Google Scholar]
- (4).Ding X; Lin S-CS; Kiraly B; Yue H; Li S; Chiang I-K; Shi J; Benkovic SJ; Huang TJ Proc. Natl. Acad. Sci. U. S. A 2012, 109 (28), 11105–11109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.() Huang J; Wang F; Ye M; Zou H Journal of Chromatography A 2014, 1372, 1–17. [DOI] [PubMed] [Google Scholar]
- 6.() Schägger H; von Jagow G Anal. Biochem 1987, 166 (2), 368–379. [DOI] [PubMed] [Google Scholar]
- 7.() Pan Y; Du X; Zhao F; Xu B Chem. Soc. Rev 2012, 41 (7), 2912–2942. [DOI] [PubMed] [Google Scholar]
- 8.() Regtmeier J; Duong TT; Eichhorn R; Anselmetti D; Ros A Anal. Chem 2007, 79 (10), 3925–3932. [DOI] [PubMed] [Google Scholar]
- (9).Neuman KC; Nagy A Nat. Methods 2008, 5, 491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Ding X; Li P; Lin S-CS; Stratton ZS; Nama N; Guo F; Slotcavage D; Mao X; Shi J; Costanzo F; Huang TJ Lab Chip 2013, 13 (18), 3626–3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Barani A; Paktinat H; Janmaleki M; Mohammadi A; Mosaddegh P; Fadaei-Tehrani A; Sanati-Nezhad A Biosens. Bioelectron 2016, 85, 714–725. [DOI] [PubMed] [Google Scholar]
- (12).Kundt A Ann. Phys 1866, 203 (4), 497–523. [Google Scholar]
- (13).Yasuda K; Haupt SS; Umemura S; Yagi T; Nishida M; Shibata YJ Acoust. Soc. Am 1997, 102 (1), 642–5. [DOI] [PubMed] [Google Scholar]
- 14.() Cousins CM; Holownia P; Hawkes JJ; Limaye MS; Price CP; Keay PJ; Coakley WT Ultrasound Med. Biol 2000, 26 (5), 881–888. [DOI] [PubMed] [Google Scholar]
- 15.() Trampler F; Sonderhoff SA; Pui PWS; Kilburn DG; Piret JM Bio/Technology 1994, 12, 281. [DOI] [PubMed] [Google Scholar]
- (16).Tolt TL; Feke DL Chem. Eng. Sci 1993, 48 (3), 527–540. [Google Scholar]
- (17).Petersson F; Åberg L; Sward-Nilsson A-M; Laurell T Anal. Chem 2007, 79 (14), 5117–5123. [DOI] [PubMed] [Google Scholar]
- (18).Shi J; Huang H; Stratton Z; Huang Y; Huang TJ Lab Chip 2009, 9 (23), 3354–3359. [DOI] [PubMed] [Google Scholar]
- (19).Piyasena ME; Suthanthiraraj PPA; Applegate RW; Goumas AM; Woods TA; López GP; Graves SW Anal. Chem 2012, 84 (4), 1831–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Reboud J; Bourquin Y; Wilson R; Pall GS; Jiwaji M; Pitt AR; Graham A; Waters AP; Cooper JM Proc. Natl. Acad. Sci. U. S. A 2012, 109 (38), 15162–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Destgeer G; Ha BH; Jung JH; Sung HJ Lab Chip 2014, 14 (24), 4665–72. [DOI] [PubMed] [Google Scholar]
- (22).Li P; Mao Z; Peng Z; Zhou L; Chen Y; Huang P-H; Truica CI; Drabick JJ; El-Deiry WS; Dao M; Suresh S; Huang TJ Proc. Natl. Acad. Sci. U. S. A 2015, 112 (16), 4970–4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Ohlsson P; Evander M; Petersson K; Mellhammar L; Lehmusvuori A; Karhunen U; Soikkeli M; Seppä T; Tuunainen E; Spangar A; von Lode P; Rantakokko-Jalava K; Otto G; Scheding S; Soukka T; Wittfooth S; Laurell T Anal. Chem 2016, 88 (19), 9403–9411. [DOI] [PubMed] [Google Scholar]
- (24).Mao Z; Li P; Wu M; Bachman H; Mesyngier N; Guo X; Liu S; Costanzo F; Huang TJ ACS Nano 2017, 11 (1), 603–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Chen Y; Wu M; Ren L; Liu J; Whitley PH; Wang L; Huang TJ Lab Chip 2016, 16 (18), 3466–3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Yeo LY; Friend JR Annu. Rev. Fluid Mech 2014, 46 (1), 379–406. [Google Scholar]
- (27).Drinkwater BW Lab Chip 2016, 16 (13), 2360–2375. [DOI] [PubMed] [Google Scholar]
- 28.() Ozcelik A; Rufo J; Guo F; Gu Y; Li P; Lata J; Huang TJ Nat. Methods 2018, 15, 1021–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.() Bruus H Lab Chip 2012, 12 (6), 1014–1021. [DOI] [PubMed] [Google Scholar]
- (30).Destgeer G; Sung HJ Lab Chip 2015, 15 (13), 2722–38. [DOI] [PubMed] [Google Scholar]
- (31).Evander M; Nilsson J Lab Chip 2012, 12 (22), 4667–4676. [DOI] [PubMed] [Google Scholar]
- (32).Wiklund M; Radel S; Hawkes JJ Lab Chip 2013, 13 (1), 25–39. [DOI] [PubMed] [Google Scholar]
- (33).Sobanski MA; Tucker CR; Thomas NE; Coakley WT Bioseparation 2000, 9 (6), 351–357. [DOI] [PubMed] [Google Scholar]
- (34).Chen Y; Li S; Gu Y; Li P; Ding X; Wang L; McCoy JP; Levine SJ; Huang TJ Lab Chip 2014, 14 (5), 924–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Jakobsson O; Oh SS; Antfolk M; Eisenstein M; Laurell T; Soh HT Anal. Chem 2015, 87 (16), 8497–8502. [DOI] [PubMed] [Google Scholar]
- (36).Collins DJ; Ma Z; Ai Y Anal. Chem 2016, 88 (10), 5513–5522. [DOI] [PubMed] [Google Scholar]
- (37).Hammarstrom B; Laurell T; Nilsson J Lab Chip 2012, 12 (21), 4296–304. [DOI] [PubMed] [Google Scholar]
- (38).Evander M; Gidlöf O; Olde B; Erlinge D; Laurell T Lab Chip 2015, 15 (12), 2588–2596. [DOI] [PubMed] [Google Scholar]
- (39).Ku A; Lim HC; Evander M; Lilja H; Laurell T; Scheding S; Ceder Y Anal. Chem 2018, 90 (13), 8011–8019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Destgeer G; Cho H; Ha BH; Jung JH; Park J; Sung HJ Lab Chip 2016, 16 (4), 660–667. [DOI] [PubMed] [Google Scholar]
- (41).Bourquin Y; Syed A; Reboud J; Ranford-Cartwright LC; Barrett MP; Cooper JM Angew. Chem., Int. Ed 2014, 53 (22), 5587–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Goddard G; Martin JC; Graves SW; Kaduchak G Cytometry, Part A 2006, 69A (2), 66–74. [DOI] [PubMed] [Google Scholar]
- (43).Kalb DM; Fencl FA; Woods TA; Swanson A; Maestas GC; Juárez JJ; Edwards BS; Shreve AP; Graves SW Anal. Chem 2017, 89 (18), 9967–9975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Shi J; Mao X; Ahmed D; Colletti A; Huang TJ Lab Chip 2008, 8 (2), 221–223. [DOI] [PubMed] [Google Scholar]
- (45).Chen Y; Nawaz AA; Zhao Y; Huang PH; McCoy JP; Levine SJ; Wang L; Huang TJ Lab Chip 2014, 14 (5), 916–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Ding X; Lin S-CS; Lapsley MI; Li S; Guo X; Chan CY; Chiang IK; Wang L; McCoy JP; Huang TJ Lab Chip 2012, 12 (21), 4228–4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Franke T; Braunmuller S; Schmid L; Wixforth A; Weitz DA Lab Chip 2010, 10 (6), 789–94. [DOI] [PubMed] [Google Scholar]
- (48).Jakobsson O; Grenvall C; Nordin M; Evander M; Laurell T Lab Chip 2014, 14 (11), 1943–1950. [DOI] [PubMed] [Google Scholar]
- (49).Ung WL; Mutafopulos K; Spink P; Rambach RW; Franke T; Weitz DA Lab Chip 2017, 17 (23), 4059–4069. [DOI] [PubMed] [Google Scholar]
- (50).Nawaz AA; Chen Y; Nama N; Nissly RH; Ren L; Ozcelik A; Wang L; McCoy JP; Levine SJ; Huang TJ Anal. Chem. 2015, 87 (24), 12051–12058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Ren L; Chen Y; Li P; Mao Z; Huang PH; Rufo J; Guo F; Wang L; McCoy JP; Levine SJ; Huang TJ Lab Chip 2015, 15 (19), 3870–3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Ren L; Yang S; Zhang P; Qu Z; Mao Z; Huang P-H; Chen Y; Wu M; Wang L; Li P; Huang TJ Small 2018, 14 (40), 1801996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Johnson DA; Feke DL Sep. Technol 1995, 5 (4), 251–258. [Google Scholar]
- (54).Lenshof A; Magnusson C; Laurell T Lab Chip 2012, 12 (7), 1210–23. [DOI] [PubMed] [Google Scholar]
- (55).Antfolk M; Magnusson C; Augustsson P; Lilja H; Laurell T Anal. Chem 2015, 87 (18), 9322–8. [DOI] [PubMed] [Google Scholar]
- (56).Antfolk M; Laurell T Anal. Chim. Acta 2017, 965, 9–35. [DOI] [PubMed] [Google Scholar]
- (57).Nam J; Lim H; Kim D; Shin S Lab Chip 2011, 11 (19), 3361–3364. [DOI] [PubMed] [Google Scholar]
- (58).Ohlsson P; Petersson K; Augustsson P; Laurell T Sci. Rep. 2018, 8 (1), 9156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Grenvall C; Magnusson C; Lilja H; Laurell T Anal. Chem 2015, 87 (11), 5596–5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Ding X; Peng Z; Lin S-CS; Geri M; Li S; Li P; Chen Y; Dao M; Suresh S; Huang TJ Proc. Natl. Acad. Sci. U. S. A 2014, 111 (36), 12992–12997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Wu M; Huang P-H; Zhang R; Mao Z; Chen C; Kemeny G; Li P; Lee AV; Gyanchandani R; Armstrong AJ; Dao M; Suresh S; Huang TJ Small 2018, 14 (32), 1801131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Lee K; Shao H; Weissleder R; Lee H ACS Nano 2015, 9 (3), 2321–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Dolatmoradi A; Mirtaheri E; El-Zahab B Lab Chip 2017, 17 (7), 1332–1339. [DOI] [PubMed] [Google Scholar]
- (64).Ai Y; Sanders CK; Marrone BL Anal. Chem 2013, 85 (19), 9126–9134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Dow P; Kotz K; Gruszka S; Holder J; Fiering J Lab Chip 2018, 18 (6), 923–932. [DOI] [PubMed] [Google Scholar]
- (66).Wu M; Ouyang Y; Wang Z; Zhang R; Huang P-H; Chen C; Li H; Li P; Quinn D; Dao M; Suresh S; Sadovsky Y; Huang TJ Proc. Natl. Acad. Sci. U. S. A 2017, 114 (40), 10584–10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Ma Z; Collins DJ; Ai Y Anal. Chem 2016, 88 (10), 5316–5323. [DOI] [PubMed] [Google Scholar]
- (68).Destgeer G; Im S; Ha BH; Jung JH; Ansari MA; Sung HJ Appl. Phys. Lett 2014, 104 (2), No. 023506. [Google Scholar]
- (69).Frommelt T; Kostur M; Wenzel-Schäfer M; Talkner P; Hänggi P; Wixforth A Phys. Rev. Lett 2008, 100 (3), No. 034502. [DOI] [PubMed] [Google Scholar]
- (70).Gracioso Martins AM; Glass NR; Harrison S; Rezk AR; Porter NA; Carpenter PD; Du Plessis J; Friend JR; Yeo LY Anal. Chem 2014, 86 (21), 10812–10819. [DOI] [PubMed] [Google Scholar]
- (71).Cecchini M; Girardo S; Pisignano D; Cingolani R; Beltram F Appl. Phys. Lett 2008, 92 (10), 104103. [Google Scholar]
- (72).Bourquin Y; Reboud J; Wilson R; Zhang Y; Cooper JM Lab Chip 2011, 11 (16), 2725–30. [DOI] [PubMed] [Google Scholar]
- (73).Zhang SP; Lata J; Chen C; Mai J; Guo F; Tian Z; Ren L; Mao Z; Huang P-H; Li P; Yang S; Huang TJ Nat. Commun 2018, 9 (1), 2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Heron SR; Wilson R; Shaffer SA; Goodlett DR; Cooper JM Anal. Chem 2010, 82 (10), 3985–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Astefanei A; van Bommel M; Corthals GL J. Am. Soc. Mass Spectrom 2017, 28 (10), 2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.() Huang Y; Yoon SH; Heron SR; Masselon CD; Edgar JS; Turecek F; Goodlett DR J. Am. Soc. Mass Spectrom 2012, 23 (6), 1062–70. [DOI] [PubMed] [Google Scholar]
- 77.() Yoon SH; Huang Y; Edgar JS; Ting YS; Heron SR; Kao Y; Li Y; Masselon CD; Ernst RK; Goodlett DR Anal. Chem 2012, 84 (15), 6530–6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Tveen-Jensen K; Gesellchen F; Wilson R; Spickett CM; Cooper JM; Pitt AR Sci. Rep 2015, 5 (1), 9736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Ashtiani D; Venugopal H; Belousoff M; Spicer B; Mak J; Neild A; de Marco AJ Struct. Biol 2018, 203 (2), 94–101. [DOI] [PubMed] [Google Scholar]
- (80).Lee AP; Patel MV; Tovar AR; Okabe Y JALA 2010, 15 (6), 449–454. [Google Scholar]
- (81).Huang P-H; Nama N; Mao Z; Li P; Rufo J; Chen Y; Xie Y; Wei C-H; Wang L; Huang TJ Lab Chip 2014, 14 (22), 4319–4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Huang P-H; Xie Y; Ahmed D; Rufo J; Nama N; Chen Y; Chan CY; Huang TJ Lab Chip 2013, 13 (19), 3847–3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Huang PH; Ren L; Nama N; Li S; Li P; Yao X; Cuento RA; Wei CH; Chen Y; Xie Y; Nawaz AA; Alevy YG; Holtzman MJ; McCoy JP; Levine SJ; Huang TJ Lab Chip 2015, 15 (15), 3125–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Ahmed D; Ozcelik A; Bojanala N; Nama N; Upadhyay A; Chen Y; Hanna-Rose W; Huang TJ Nat. Commun 2016, 7 (1), 11085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Li X; Attanayake K; Valentine SJ; Li P Rapid Commun. Mass Spectrom 2018, DOI: 10.1002/rcm.8232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Guo F; Li P; French JB; Mao Z; Zhao H; Li S; Nama N; Fick JR; Benkovic SJ; Huang TJ Proc. Natl. Acad. Sci. U. S. A. 2015, 112 (1), 43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Wiklund M; Christakou A; Ohlin M; Iranmanesh I; Frisk T; Vanherberghen B; Önfelt B Micromachines 2014, 5 (1), 27. [Google Scholar]
- (88).Li S; Glynne-Jones P; Andriotis OG; Ching KY; Jonnalagadda US; Oreffo ROC; Hill M; Tare RS Lab Chip 2014, 14 (23), 4475–4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.() Collins DJ; Morahan B; Garcia-Bustos J; Doerig C; Plebanski M; Neild A Nat. Commun. 2015, 6, 8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.() Alhasan L; Qi A; Al-Abboodi A; Rezk A; Chan PPY; Iliescu C; Yeo LY ACS Biomater. Sci. Eng. 2016, 2 (6), 1013–1022. [DOI] [PubMed] [Google Scholar]
- 91.() Chen K; Wu M; Guo F; Li P; Chan CY; Mao Z; Li S; Ren L; Zhang R; Huang TJ Lab Chip 2016, 16 (14), 2636–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.() Christakou AE; Ohlin M; Vanherberghen B; Khorshidi MA; Kadri N; Frisk T; Wiklund M; Onfelt B Integrative Biology 2013, 5 (4), 712–719. [DOI] [PubMed] [Google Scholar]
- (93).Christakou AE; Ohlin M; Onfelt B; Wiklund M Lab Chip 2015, 15 (15), 3222–3231. [DOI] [PubMed] [Google Scholar]
- (94).Guo F; Mao Z; Chen Y; Xie Z; Lata JP; Li P; Ren L; Liu J; Yang J; Dao M; Suresh S; Huang TJ Proc. Natl. Acad. Sci. U. S. A. 2016, 113 (6), 1522–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.() Baresch D; Thomas J-L; Marchiano R Phys. Rev. Lett 2016, 116 (2), No. 024301. [DOI] [PubMed] [Google Scholar]
- (96).Devendran C; Collins DJ; Ai Y; Neild A Phys. Rev. Lett 2017, 118 (15), 154501. [DOI] [PubMed] [Google Scholar]
- (97).Evander M; Johansson L; Lilliehorn T; Piskur J; Lindvall M; Johansson S; Almqvist M; Laurell T; Nilsson J Anal. Chem 2007, 79 (7), 2984–91. [DOI] [PubMed] [Google Scholar]
- 98.() Wiklund M Lab Chip 2012, 12 (11), 2018–2028. [DOI] [PubMed] [Google Scholar]
- (99).Burguillos MA; Magnusson C; Nordin M; Lenshof A; Augustsson P; Hansson MJ; Elmer E; Lilja H; Brundin P;́ Laurell T; Deierborg T PLoS One 2013, 8 (5), No. e64233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (100).Barnkob R; Augustsson P; Laurell T; Bruus H Lab Chip 2010, 10 (5), 563–570. [DOI] [PubMed] [Google Scholar]
- (101).Wang Y-Y; Yao J; Wu X-W; Wu D-J; Liu X-JJ Appl. Phys 2017, 122 (9), No. 094902. [Google Scholar]
- (102).Ibsen S; Tong A; Schutt C; Esener S; Chalasani SH Nat. Commun 2015, 6 (1), 8264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (103).Bachman H; Huang P-H; Zhao S; Yang S; Zhang P; Fu H; Huang TJ Lab Chip 2018, 18 (3), 433–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (104).Luo J; He X; Zhou J; Wang W; Xuan W; Chen J; Jin H; Xu Y; Dong S Procedia Eng. 2015, 120, 717–720. [Google Scholar]