SUMMARY
Aims
With developments of etiology of cerebral small vessel disease (CSVD) and genome‐wide association study (GWAS) of stroke, the genetic studies of CSVD are focused on genes related to blood‐brain barrier (BBB) and aging. The project aims to investigate the association between CSVD and susceptibility loci and candidate genes.
Methods
All study subjects admitted Beijing Tiantan Hospital from June 2009 to September 2010 including 197 cerebral small vessel disease patients(S), 198 large artery atherosclerosis control individuals (vascular stenotic rate ≥50% diameter reduction) (L), 200 hypertensive intracerebral hemorrhage control individuals (H) and 197 stroke‐free control individuals (C). 15 SNPs in 4 genes (MYLK, AQP4, NINJ2, and INK4/ARF) were genotyped using Multiplex Snapshot assay. Each SNP was first examined between the groups S and C in different genetic models (codominant, dominant, recessive, overdominant, and log‐additive). Permutation correction was used to adjust for multiple testing. The significant SNP loci were further analyzed in comparing S with L and H, respectively. Subgroup analysis was also performed for each risk‐factor category.
Results
Among the 15 SNPs, rs2222823 and rs2811712 were found to be significantly associated with CSVD after multiple‐testing adjustment. The heterozygote (A/T) of rs2222823 of MYLK has an odds ratio of 0.52 (95% CI =[0.35, 0.79], P= 0.002, adjusted P= 0.031) when compared with homozygotes. The heterozygote (C/T) of rs2811712 of INK4/ARF has an odds ratio of 1.75 (95% CI =[1.13–2.71], P= 0.004, adjusted P= 0.050). The SNP rs2222823 was significant (P= 0.035) in comparing S with H. In comparing S versus L, it is significant for the subgroups of patients without diabetes (P= 0.012) and drinking (P= 0.018). rs2811712 was significant in comparing S with L for the subgroups of patients with hyperlipidemia (P= 0.029) and drinking (P= 0.04).
Conclusion
The heterozygotes (T/A) at the rs2222823 SNP locus of MYLK gene decreases the risk of having cerebral small vessel disease, while the heterozygotes (C/T) at the rs2811712 SNP locus of INK4/ARF gene increases the risk, suggesting that the MYLK and INK4/ARF are the associated genes of cerebral small vessel disease in Han Chinese population.
Keywords: Cerebral small vessel disease, Heterozygote genotype, rs2222823 and rs2811712, SNaPshot array
Introduction
Cerebral small vessel disease (CSVD) is an important cause of silent stroke and vascular dementia. It results from ischemia in the perforating arteries supplying the white matter and deep grey matter nuclei, and it most shows both focal lacunar infarction and diffuse areas of chronic ischemia (white matter changes or leukoaraiosis) in neuroimaging. In China, cerebral small vessel disease is one of the most common stroke subtypes, accounting for more than 30% of ischemic stroke patients [1].
Previous studies of CSVD showed that it might have a background for genetic predisposition [2, 3]. Most of the focus has been on single nucleotide polymorphisms as genetic risk markers for white matter changes or leukoaraiosis, either directly or through their interactions with other genes or medical risk factors [4]. However, no genetic polymorphism had yet shown convincing evidence for an association with CSVD [5]. With developments of etiology study of CSVD, the dysfunction of blood‐brain barrier was considered as one of the important reasons leading to cerebral small vessel disease [6, 7, 8, 9]. Recent studies showed that myosin light chain kinase (MYLK) and aquaporin 4 (AQP4) were the key roles in regulating the permeability of blood brain barrier [10, 11, 12, 13, 14, 15, 16]. The New England Journal of Medicine and Nature recently published two GWAS studies, where the INK4//ARF gene and ninjurin 2 gene (NINJ2) were reported to be associated with stroke [17, 18].
We carried out a genetic association study to examine the relationship between CSVD and the 4 candidate genes, MYLK, AQP4, NINJ2, and INK4/ARF. The results suggest that two tag SNP loci of MYLK and INK4/ARF genes are related with CSVD in Han Chinese population.
Methods
Subjects
The total study population includes 792 Han Chinese, where 197 cerebral small vessel disease patients (S) and 197 stroke‐free control individuals (C) participated the first stage of the study; 198 large artery atherosclerosis control individuals (vascular stenotic rate ≥50% diameter reduction) (L), and 200 hypertensive intracerebral hemorrhage control individuals (H) participated in the second stage of the study. All subjects including cerebral small vessel disease and all control individuals were recruited from consecutive outpatients and inpatients admitted to Beijing Tiantan Hospital from June 2009 to September 2010. This study was an observational study, not focused on geographically matching. All subjects underwent brain MR imaging with angiography or CT scan, carotid artery ultrasound, and Trancranial Doppler. The stroke‐free control individuals were provided imaging examination (MRI or CT) in Cerebrovascular Disease Screening Test department and Health Examination Centre to rule out asymptomatic brain tumor and cerebrovascular disease. The inclusion and exclusion criteria were as follows: CSVD was defined as having one of four imaging features (lacunar infarcts, leukoaraiosis, microbleed, and dilatation of the pervascular spaces), while ruled out the patients with subcortical lesion more than 1.5 cm in diameter, or cortical infarct of any size, or a potential cardioembolic source, or parent artery stenosis, and other large‐vessel diseases; large artery atherosclerosis was defined as being caused by atherosclerosis, defined as carotid, vertebral, or basilar artery stenosis more than 50% by carotid artery ultrasound or MR angiography; hypertensive intracerebral hemorrhage met the cerebral hemorrhage criteria of CT scan or MR image caused by hypertension; stroke‐free was defined as freeing from symptomatic cerebrovascular disease (as TIA) and were normal on MR imaging or CT scan. We asked about the past medical history of the study population and the history of medications to remove medications for lipid lowing, blood sugar lowing, or blood pressure lowers.
The study was approved by the local ethics committees, and patients or their trustees gave written informed consent.
Study Design & Genotyping Analysis
The study design was divided into two stages. To minimize the genotyping labor and cost, the first stage only includes tag SNP loci with minor allele frequency (MAF) >15% in the promoter region and the reported SNP loci in the coding regions of the candidate genes. Comparing group S with group C, fifteen SNP loci on 4 candidate genes (MYLK, AQP4, NINJ2, and INK4/ARF, Table 1) were screened. In the second stage, each significant SNP loci after multiple‐testing adjustment was further examined when S was compared with L and H, respectively.
Table 1.
Properties of the selected gene polymorphisms for genotyping
| Gene & SNP | Chromosome | Chromosome position | SNP property | Functional change | Allele |
|---|---|---|---|---|---|
| INK4/ARF | |||||
| rs3218020 | 9 | 21997872 | 5'‐flanking | / | C/T |
| rs2811712 | – | 21998035 | 5'‐flanking | / | C/T |
| rs10757278 | – | 22124477 | 5'‐flanking | / | C/T |
| AQP4 | |||||
| rs162007 | 18 | 24445847 | 5'‐flanking | / | C/T |
| rs2075575 | – | 24446526 | 5'‐flanking | / | C/T |
| rs162004 | – | 24447936 | 5'‐flanking | / | G/C |
| MYLK | |||||
| rs820463 | 3 | 123357037 | Synon exon 29 | p.Asn1614Asn | C/T |
| rs3732487 | – | 123419573 | Nonsynon exon 18 | p.Asp914Glu | A/C |
| rs3732486 | – | 123419733 | Nonsynon exon 18 | p.Leu861Pro | C/T |
| rs3796164 | – | 123453061 | Nonsynon exon 10 | p.Val261Ala | C/T |
| rs2222823 | – | 123604787 | 5'‐flanking | / | A/T |
| rs1920221 | – | 123606952 | 5'‐flanking | / | C/T |
| NINJ2 | |||||
| rs3809263 | 12 | 773456 | 5'‐flanking | / | G/A |
| rs11833579 | – | 775199 | 5'‐flanking | / | C/T |
| rs7298096 | – | 776471 | 5'‐flanking | / | C/T |
Genomic DNA was extracted from venous blood sample, using QIAamp DNA Blood Mini Kit (Qiagen, CA, USA) according to the manufacturer's instructions. The primers were designed for each SNP locus using Primer3 (http://frodo.wi.mit.edu/), and they were listed in supplementary Table 1. Multiplex PCR reactions were performed to amplify target regions containing the selected SNPs. The PCR amplifications were carried out in a final volume of 20 μl containing 20 ng/μl DNA, 2.4 μl of 2.5 μM dNTP mix (TaKaRa Biotehnology Co.Ltd. TaKaRa Dalian, China), 2 μl of each primer (1 μM), 1.2 μl of 25 mM MgCl2 (Qiagen CA, USA), 0.15 μl of Taq polymerase 5 U/μl (TaKaRa Biotehnology Co.Ltd. TaKaRa Dalian, China), 2 μl of 10×buffer (TaKaRa Biotehnology Co.Ltd. TaKaRa Dalian, China) supplied with 15 mM MgCl2.The PCR conditions were as follows: 1 cycle of 95°C for 2 min, 35 cycles of 94°C for 30 second, primer annealing(Tm‐6) for 30 second, 72°C extension for 1.5 min, followed by 15 min of final extension at 72°C, and a 4°C holding step. After amplification, 10 μl of PCR products was purified by incubating at 37°C for 60 min with 0.2 μl of 5 U/μl Exonuclease (TaKaRa Biotehnology Co.Ltd. TaKaRa Dalian,China) and 1 μl of 1 U/μl SAP (USB), followed by enzymedeactivation at 80°C for 15 min. Next, 3 μl of the purified PCR product was mixed with 3 μl of SNaPshot reaction mix (Applied Biosystems, Foster City, CA, USA), 3.8 μl of ddH2O, and 0.2 μl of the SNaPshot primer (10 μM) in a 10 μl total volume. Primer extension was performed on the GeneAmp PCR system (Applied Biosystems, Foster City, CA, USA) for 28 cycles at 96°C for 10 second, 50°C for 5 second, and 60°C for 30 second, followed by a 4°C hold. After the SNaPshot PCR, the unincorporated ddNTPs were removed enzymatically by incubating samples with 1 U of SAP (TaKaRa Biotehnology Co.Ltd. TaKaRa Dalian,China) for 60 min at 37°C, followed by 15 min at 80°C for enzyme inactivation. All products were separated by capillary electrophoresis in the ABI Prism 3130XL Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) and evaluated using the GeneScan software, version 3.7 (Applied Biosystems, Foster City, CA, USA).
Statistical Analysis
Baseline characteristics, including age, gender, diabetes, hyperlipidemia, smoking, and drinking conditions, were summarized as means and standard deviations or frequencies and percentages as appropriate by the disease groups. ANOVA or the Chi‐squared test was used to compare each characteristic among the groups. Hardy–Weinberg equilibrium was evaluated in all subjects using QuickChi.
For the first stage of the study, logistic regression models were used to examine the effect of each SNP in comparing the group of CSVD patients (S) with the controls (C). For each SNP, 5 genetic models were considered, including the codominant model, dominant model, recessive model, overdominant model, and log‐additive model. All models controlled for age and gender. The Akaike Information Criterion (AIC) was calculated for each genetic model and the genetic model with the smallest AIC was selected. The permutation correction method was used to adjust the results for multiple testing. The significant SNP loci (adjusted P values less than 0.05) were further analyzed in comparing S with L and H, respectively. Subgroup analysis was also carried out to examine each risk factor. All the statistical analyses were performed using SAS 9.2 software (Cary, NC, USA).
Results
The mean age of the study subjects was 58.05 ± 11.5 year (Table 2). The stroke‐free control subjects were younger (55.18 ± 11.7 year) than the other three groups (P < 0.01, Table 2). 62% of the subjects were male, and the control group (C) has the smallest proportion (47.2%) of male subjects (vs. 63.5%, 65.5%, and 71.7% for S, H and L, respectively; P < 0.001). About 40% of the L group subjects had diabetes, and the proportion was 30% for the S group, and 19% for the H group (P < 0.001). The L group also had the highest proportion of hyperlipidemia (P < 0.001) and most smokers (P= 0.002). All three groups had about 40% of the subjects who drink routinely (P= 0.620).
Table 2.
Summary to baseline characteristics by all study subjects
| Cerebral small vessel disease (n = 197) | Stroke‐free control subject (n = 197) | Large artery atherosclerosis control subject (n = 198) | Intracranial hemorrhage control subject (n = 200) | P value | |
|---|---|---|---|---|---|
| Gender (Male), n (%) | 125 (63.45%) | 92 (47.21%) | 142 (71.72%) | 131 (65.50%) | <0.001 |
| Age, median ±SD | 57.85 ± 10.87 | 55.18 ± 11.70 | 61.21 ± 9.83 | 58.01 ± 12.73 | <0.001 |
| Diabetes, n (%) | 57 (29.53%) | N/Aa | 80 (40.26%) | 38 (19.29%) | <0.001 |
| Hyperlipidemia, n (%) | 106 (54.36%) | N/A | 151 (76.62%) | 94 (47.24%) | <0.001 |
| Smoking, n (%) | 82 (41.84%) | N/A | 117 (59.39%) | 95 (47.74%) | 0.002 |
| Drinking, n (%) | 72 (37.70%) | N/A | 83 (42.13%) | 83 (41.71%) | 0.620 |
aN/A, No data are available.
Genotype frequencies of all fifteen SNPs were in Hardy–Weinberg equilibrium for both S and C groups (P > 0.05, Table 3. Controlling for age and gender, only two SNP loci, rs2222823 and rs2811712, in comparing S with C passed the permutation correction for multiple testing, where the adjusted P values were, respectively, 0.031 and 0.050 (Table 3). The AIC values of the overdominant model were the smallest for these two SNP loci (Figure 1). For SNP 2222823, age (OR = 1.13 per 5 years older, 95% CI =[1.03, 1.24], P= 0.011), and gender (OR = 1.98, male vs. female, 95% CI =[1.31, 2.98], P= 0.001) were both significantly in the overdominant model. For SNP 2811712, age (OR = 1.13 per 5 years older, 95% CI =[1.03, 1.24], P= 0.013) and gender (OR = 2.00, male vs. female, 95% CI =[1.33, 3.02], P= 0.001). For rs2222823, the odds ratio was 0.52 (95% CI, 0.35–0.79) for the heterozygous genotype (A/T) when compared with the homozygous genotypes (A/A and T/T). For rs2811712, the heterozygous genotype (C/T) was associated with a higher likelihood of having CVSD than the homozygotes (OR = 1.95, 95% CI =[1.13, 2.71]). Power analysis was performed based on the logistic regression models (overdominant) used in the first stage of the study. The sample size of 394 (197 per group) had a power of 80% to detect an odds ratio of 0.57 or 1.83, assuming that the prevalence rate is 60% for the homozygotes or heterozygotes, and gender and age are not associated with the genotypes.
Table 3.
Association of 15 SNPs in comparing cerebral small vessel disease subjects with stroke‐free controls in overdominant modelsa
| SNP | Genotype | Stroke‐free control subjects (%) | Cerebral small vessel disease subjects (%) | H‐W P value | Overdominant model | ||
|---|---|---|---|---|---|---|---|
| OR | P value | Adjusted P value | |||||
| rs10757278 | C/C‐T/T | 103 (50%) | 103 (50%) | 0.251 | 1.04 | 0.844 | 1.000 |
| C/T | 94 (50%) | 94 (50%) | – | (0.69–1.56) | – | – | |
| rs11833579 | C/C‐T/T | 95 (46%) | 112 (54%) | 0.993 | 1.41 | 0.095 | 0.901 |
| C/T | 102 (55%) | 85 (45%) | – | (0.94–2.13) | – | – | |
| rs162004 | C/C‐G/G | 107 (50%) | 108 (50%) | 0.834 | 1.08 | 0.727 | 1.000 |
| C/G | 90 (50%) | 89 (50%) | – | (0.71–1.61) | – | – | |
| rs162007 | C/C‐T/T | 104 (51%) | 98 (49%) | 0.901 | 0.88 | 0.531 | 0.693 |
| C/T | 93 (48%) | 99 (52%) | – | (0.58–1.32) | – | – | |
| rs1920221 | C/C‐T/T | 118 (55%) | 98 (45%) | 0.062 | 0.68 | 0.065 | 0.904 |
| C/T | 79 (44%) | 99 (56%) | – | (0.45–1.02) | – | – | |
| rs2075575 | C/C‐T/T | 141 (54%) | 121 (46%) | 1.000 | 0.65 | 0.050 | 0.572 |
| C/T | 56 (42%) | 76 (58%) | – | (0.42–1.00) | – | – | |
| rs2222823 | A/A‐T/T | 126 (57%) | 94 (43%) | 0.872 | 0.52 | 0.002 | 0.031a |
| A/T | 71 (41%) | 103 (59%) | – | (0.35–0.79) | – | – | |
| rs2811712 | C/C‐T/T | 114 (45%) | 142 (55%) | 0.240 | 1.75 | 0.004 | 0.050a |
| C/T | 83 (60%) | 55 (50%) | – | (1.13–2.71) | – | – | |
| rs3218020 | C/C‐T/T | 104 (49%) | 110 (51%) | 0.061 | 1.12 | 0.585 | 1.001 |
| C/T | 93 (52%) | 87 (48%) | – | (0.75–1.32) | – | – | |
| rs3732486 | C/C‐T/T | 97 (51%) | 92 (49%) | 0.564 | 0.86 | 0.468 | 1.000 |
| C/T | 100 (49%) | 105 (51%) | – | (0.57–1.30) | – | – | |
| rs3732487 | A/A‐C/C | 96 (51%) | 93 (49%) | 0.762 | 0.91 | 0.632 | 1.001 |
| A/C | 101 (49%) | 104 (51%) | – | (0.60–1.39) | – | – | |
| rs3796164 | C/C‐T/T | 109 (51%) | 103 (49%) | 0.583 | 0.85 | 0.435 | 1.002 |
| C/T | 88 (48%) | 94 (52%) | – | (0.56–1.28) | – | – | |
| rs3809263 | A/A‐G/G | 100 (48%) | 110 (52%) | 0.642 | 1.18 | 0.448 | 1.000 |
| A/G | 97 (53%) | 87 (47%) | – | (0.78–1.75) | – | – | |
| rs7298096 | C/C‐T/T | 91 (44%) | 114 (56%) | 0.681 | 1.64 | 0.018 | 0.523 |
| C/T | 106 (56%) | 83 (44%) | – | (1.09–2.44) | – | – | |
| rs820463 | C/C‐T/T | 102 (48%) | 109 (52%) | 0.733 | 1.18 | 0.425 | 0.842 |
| C/T | 95 (52%) | 88 (48%) | – | (0.79–1.79) | – | – | |
aModels control for age and gender.
Figure 1.
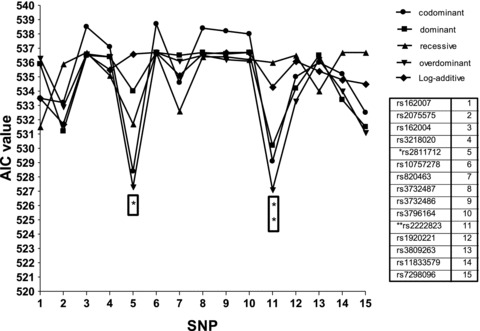
AIC values of five genetic models in relating each SNP loci to the cerebral small vessel disease and control groups. The stars show there are significant differences between cerebral small vessel disease patients and stroke‐free controls, adjusted P value less than 0.05, at SNP loci, rs2811712 and rs2222823, and the AIC values of overdominant model are smaller than other genetic models.
In comparing S with H, the odds ratio was 0.90 (95% CI =[0.81, 0.99], P= 0.035, heterozygotes vs. homrozygotes, Table 4) for the SNP rs2222823 in the overdominant model. This SNP also had a significant effect in patients without diabetes (OR = 0.88, 95% CI =[0.78, 0.99], P= 0.028). In comparing S with L, this SNP does not have an overall significant effect. For the subgroup of patients without diabetes, the odds ratio was 0.85 (95% CI =[0.76, 0.96], P= 0.012). Similarly, for the subgroup of drinking patients, the odds ratio was 0.83 (95% CI =[0.72, 0.97], P= 0.018). For the SNP rs2811712, there were no overall significant differences between the homozygotes and heterozygotes in comparing S with L or H, but the heterozygotes show significantly high risk in two subgroups (Figure 2). For the subgroup of patients with hyperlipidemia, OR was 1.15 (95% CI =[1.02, 1.30], P= 0.029, Table 4); for the drinking subjects, OR was 1.18 (95% CI =[1.01, 1.37], P= 0.040).
Table 4.
Association of two selected SNPs, rs2222823 and rs2811712, in comparing S with H and L, respectivelya
| SNP | Genotype | S | H | L | OR | P value | |||
|---|---|---|---|---|---|---|---|---|---|
| S versus H | S versus L | S versus H | S versus L | ||||||
| rs2222823 | Overall | A/A‐T/T | 126 | 107 | 109 | 0.90 | 0.91 | 0.035a | 0.058 |
| T/A | 71 | 93 | 89 | (0.81–0.99) | (0.82–1.00) | – | – | ||
| No diabetes | A/A‐T/T | 90 | 85 | 60 | 0.88 | 0.85 | 0.028a | 0.012a | |
| T/A | 46 | 74 | 58 | (0.78–0.99) | (0.76–0.96) | – | – | ||
| Diabetes | A/A‐T/T | 32 | 19 | 49 | 0.96 | 1.03 | 0.679 | 0.726 | |
| T/A | 25 | 19 | 31 | (0.79–1.16) | (0.87–1.22) | – | – | ||
| No hyperlipidemia | A/A‐T/T | 59 | 58 | 29 | 0.89 | 0.96 | 0.126 | 0.641 | |
| T/A | 30 | 47 | 18 | (0.78–1.03) | (0.82–1.22) | – | – | ||
| Hyperlipidemia | A/A‐T/T | 66 | 48 | 80 | 0.90 | 0.91 | 0.129 | 0.109 | |
| T/A | 40 | 46 | 71 | (0.78–1.03) | (0.8–1.02) | – | – | ||
| No smoking | A/A‐T/T | 75 | 57 | 48 | 0.90 | 0.95 | 0.151 | 0.481 | |
| T/A | 39 | 45 | 33 | (0.79–1.04) | (0.83–1.08) | – | – | ||
| Smoking | A/A‐T/T | 50 | 48 | 61 | 0.90 | 0.90 | 0.177 | 0.135 | |
| T/A | 32 | 47 | 56 | (0.78–1.04) | (0.79–1.03) | – | – | ||
| No drinking | A/A‐T/T | 76 | 63 | 69 | 0.90 | 0.97 | 0.132 | 0.671 | |
| T/A | 43 | 53 | 45 | (0.79–1.03) | (0.85–1.11) | – | – | ||
| Drinking | A/A‐T/T | 46 | 43 | 40 | 0.88 | 0.83 | 0.105 | 0.018a | |
| T/A | 26 | 40 | 43 | (0.75–1.03) | (0.72–0.97) | – | – | ||
| rs2811712 | Overall | T/T‐C/C | 114 | 121 | 129 | 1.03 | 1.09 | 0.564 | 0.100 |
| C/T | 83 | 79 | 69 | (0.93–1.14) | (0.98–1.20) | – | – | ||
| No diabetes | T/T‐C/C | 85 | 100 | 83 | 1.01 | 1.10 | 0.930 | 0.134 | |
| C/T | 51 | 59 | 35 | (0.83–1.14) | (0.97–1.25) | – | – | ||
| Diabetes | T/T‐C/C | 28 | 18 | 46 | 0.99 | 1.10 | 0.943 | 0.277 | |
| C/T | 29 | 20 | 34 | (0.81–1.23) | (0.93–1.30) | – | – | ||
| No hyperlipidemia | T/T‐C/C | 55 | 64 | 26 | 0.99 | 0.96 | 0.898 | 0.655 | |
| C/T | 34 | 41 | 21 | (0.85–1.15) | (0.82–1.14) | – | – | ||
| Hyperlipidemia | T/T‐C/C | 58 | 56 | 103 | 1.04 | 1.15 | 0.567 | 0.029a | |
| C/T | 48 | 38 | 48 | (0.91–1.20) | (1.02–1.30) | – | – | ||
| No smoking | T/T‐C/C | 69 | 67 | 55 | 1.05 | 1.11 | 0.464 | 0.154 | |
| C/T | 45 | 35 | 26 | (0.92–1.20) | (0.96–1.28) | – | – | ||
| Smoking | T/T‐C/C | 45 | 51 | 74 | 1.00 | 1.11 | 0.957 | 0.154 | |
| C/T | 37 | 44 | 42 | (0.86–1.16) | (0.96–1.28) | – | – | ||
| No drinking | T/T‐C/C | 72 | 73 | 73 | 1.02 | 1.06 | 0.749 | 0.366 | |
| C/T | 47 | 43 | 41 | (0.89–1.16) | (0.93‐ 1.22) | – | – | ||
| Drinking | T/T‐C/C | 38 | 47 | 56 | 1.04 | 1.18 | 0.615 | 0.040a | |
| C/T | 34 | 36 | 27 | (0.89–1.22) | (1.01–1.37) | – | – | ||
aModels control for age and gender.
Figure 2.
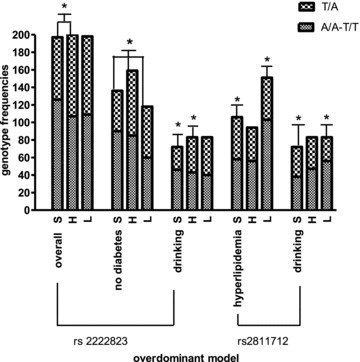
The figure shows the genotype frequencies at rs2222823 and rs2811712 SNP loci in overdominant model among S, L, and H. The stars show there are significant differences in comparisons, P < 0.05.
Discussion
Our study showed that the two SNP loci in two different genes (MYLK and INK4/ARF genes), rs2222823 and rs2811712, were significantly associated with cerebral small vessel disease. The heterozygosis genotype (T/A) at rs2222823 SNP locus had protective effect for cerebral small vessel disease, while the heterozygosis genotype (C/T) at rs2811712 SNP locus had promoting effect for cerebral small vessel disease. In further comparison between S and H or L, these two SNP loci also showed statistically significant association in several risk‐factor categories.
These results suggested that MYLK and INK4/ARF genes were related with cerebral small vessel disease. Firstly, rs2222823 and rs2811712 SNP loci were respectively located the tagSNPs in promoter region of the 5’flanking of MYLK and INK4/ARF genes. Since those SNP loci were important function structures of two genes and had powful associations with MYLK and INK4/ARF genes. Second, many recent studies supported this suggestion. Wang et al. (2007) revealed MYLK gene was related with atherosclerosis disease in their studies [19]. Su et al. found the increased activation of myosin light chain kinase could enhance vascular endothelial permeability [20]. And recent studies about region of INK4/ARF gene found it had closely association with aging and related aging diseases, such as myocardial infarction, stroke, and type 2 diabetes [21, 22, 23]. Thence, the MYLK and INK4/ARF genes were associated with the dysfunction of blood brain barrier and aging atherosclerosis, which was highly correlated with the pathophysiological changes of CSVD. Because CSVD was an aging arteriosclerosis microangiopathy resulted in increased permeability of blood‐brain barrier.
Our study confirmed and refined the results of the GWAS that the INK4/ARF gene and NINJ2 gene were related with stroke. Burton PR et al. and Samani NJ et al. reported the region of INK4/ARF gene on chromosome 9p21.3 was related with coronary artery disease in genomewide association analysis [18, 24], and Jazdzewski K et al. confirmed the region on chromosome 9p21.3 was also increased the risk of stroke [25]. Those studies revealed SNP rs1333049 near the region of INK4/ARF gene was closely associated with both coronary artery disease and stroke. Our results showed SNP rs2811712 locus at the INK4/ARF gene was related with the subtype of cerebral small vessel disease in Chinese population. There was a different SNP locus with the results of Jazdzewski K et al. studies, which might be resulted from ethnical difference, while our result supported the conclusion that the region of INK4/ARF gene on chromosome 9p21.3 was associated with stroke, very likely with cerebral small vessel disease. Ikram MA et al. carried out an analysis of genomewide association data generated from four large cohorts composing the Cohorts for Heart and Aging Research in Genomic Epidemiology consortium, including 19,602 white persons (mean [+/‐SD] age, 63+/‐8 years) in whom 1544 incident strokes (1164 ischemic strokes) developed over an average follow‐up of 11 years. Two intergenic single‐nucleotide polymorphisms (rs11833579 and rs12425791) on chromosome 12p13 and within 11 kb of the gene NINJ2 were associated with stroke (P < 5×10(‐8)) [17]. In Ikram MA's study, the SNP rs11833579 locus at NINJ2 gene were realized that was associated with stroke (all types stroke, including ischemic, hemorrhagic, or of “unknown” type on the basis of clinical and imaging criteria), except the results of an underpowered analysis of the smaller black cohort. However, our result didn't found association between rs 11833579 and CSVD. Tong Y et al. found there was association between two SNPs (rs11833579 and rs12425791) on chromosome 12p13 and ischemic stroke [26]. Matsushita T et al. revealed rs11833579 had no association with atherothrombotic stroke and rs12425791 on chromosome 12p13 was a genetic marker for atherothrombotic stroke in multiethnic population [27]. While Lotta LA et al. found there wasn't association between two single nucleotide polymorphisms (rs11833579 and rs12425791 of the NINJ2gene) on chromosome 12p13 with early‐onset ischemic stroke [28]. Same as Olsson S et al. and Ding H studies [29, 30]. And International Stroke Genetics Consortium showed their well‐powered meta‐analyses did not validate associations between two SNPs (rs12425791 and rs11833579) and stroke [31]. Therefore, we considered that the SNP locus (rs11833579) at NINJ2 gene hadn't an association with CSVD in Chinese population.
In genotype analysis, the AIC of five genetic models was calculated and the data suggest the overdominant model. Although overdominance can be observed for many human traits [32], few cases of stroke susceptibilities due to an overdominant model of inheritance have been reported. However, in some studies of cancer, the polymorphisms turned out to be strongly associated with cancers under the overdominant models [33, 34]. Further research is desired to confirm whether CSVD was in accordance with the overdominant model. There were several reasons that might lead to such bias toward the heterozygotes. Technically, this could appear in using inaccurate restriction enzymes, but was unlikely to occur in the ABI assay for genotyping using SnaPshot technique. Molecularly, it was also possible that the variant allele might have a very strong dominant effect, so that there was little difference between the effects of the variant homozygotes and heterozygotes. However, this needs to be further tested in indepth molecular mechanistic studies in the future. Statistically, because of the relatively small numbers of the variant homozygotes observed in both cases and controls, the effect of the variant homozygotes might be subject to selection bias or other unfavorable genotypes than that of the heterozygotes, or simply there was not enough statistical power to detect the real effect among the variant homozygotes, which only could be corrected by much larger studies in the future. There was no affect to diagnoses the phenotype of cerebral small vessel disease whether the patients were first ever stroke or recurrent stroke, so we didn't get concerned about this condition. But the situation of first ever stroke or recurrent stroke will be helpful to assessing risk factors of CSVD, so we will deeply analyses the risks of CSVD distinguishing these two conditions in the future study,
In conclusion, the heterozygotes (T/A) at rs2222823 SNP locus of the MYLK gene decreased the risk of having cerebral small vessel disease and the heterozygotes (C/T) at rs2811712 SNP locus of INK4/ARF gene increased the risk. These findings suggest that the MYLK and INK4/ARF gene are associated with cerebral small vessel disease in Han Chinese population. Yet, given the small number of patients studied, this observation could be a result of chance. Therefore, our results should be interpreted with caution and need to be validated in a larger cohort of CSVD patients. Stroke is a major cause of disability and death worldwide. Prevention aimed at risk factors of stroke is the most effective strategy to curb the stroke pandemic [35]. Genetic research represents a promising tool for stroke, especially CSVD patients, in both pathway of pathogenesis and the search for new molecular preventive therapy.
Conflict of Interest
The authors declare no conflict of interest.
Supporting information
Table S1. List of the primers for selected SNPs †
Supporting info item
Acknowledgment
This work was supported by a grant from the Major State Basic Research Development Program of China (973 Program) (No.2009CB521905).
The two corresponding authors contributed equally to the work.
References
- 1. Mok VC, Lau AY, Wong A, Lam WW, Chan A, Leung H, et al Long‐term prognosis of Chinese patients with a lacunar infarct associated with small vessel disease: A 5 year longitudinal study. Int J Stroke 2009;4:81–88. [DOI] [PubMed] [Google Scholar]
- 2. Jerrard‐Dunne P, Cloud G, Hassan A, Markus HS. Evaluating the genetic component of ischemic stroke subtypes: A family history study. Stroke 2003;34:1364–1369. [DOI] [PubMed] [Google Scholar]
- 3. Polychronopoulos P, Gioldasis G, Ellul J, Metallinos IC, Lekka NP, Paschalis C, Papapetropoulos T. Family history of stroke in stroke types and subtypes. J Neurol Sci 2002;195:117–122. [DOI] [PubMed] [Google Scholar]
- 4. Assareh A, Mather KA, Schofield PR, Kwok JB, Sachdev PS. The genetics of white matter lesions. CNS Neurosci Ther 2011;17(5):525–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Paternoster L, Chen W, Sudlow CL. Genetic determinants of white matter hyperintensities on brain scans: A systematic assessment of 19 candidate gene polymorphisms in 46 studies in 19,000 subjects. Stroke 2009;40:2020–2026. [DOI] [PubMed] [Google Scholar]
- 6. Knottnerus IL, Ten Cate H, Lodder J, Kessels F, van Oostenbrugge RJ. Endothelial dysfunction in lacunar stroke: A systematic review. Cerebrovasc Dis 2009;27:519–526. [DOI] [PubMed] [Google Scholar]
- 7. Wardlaw JM. What causes lacunar stroke? J Neurol Neurosurg Psychiatry 2005;76:617–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wardlaw JM, Farrall A, Armitage PA, Carpenter T, Chappell F, Doubal F, et al Changes in background blood‐brain barrier integrity between lacunar and cortical ischemic stroke subtypes. Stroke 2008;39:1327–1332. [DOI] [PubMed] [Google Scholar]
- 9. Starr JM, Wardlaw J, Ferguson K, MacLullich A, Deary IJ, Marshall I. Increased blood‐brain barrier permeability in type II diabetes demonstrated by gadolinium magnetic resonance imaging. J Neurol Neurosurg Psychiatry 2003;74:70–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shen Q, Rigor RR, Pivetti CD, Wu MH, Yuan SY. Myosin light chain kinase in microvascular endothelial barrier function. Cardiovasc Res 2010;87:272–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cai S, Pestic‐Dragovich L, O’Donnell ME, Wang N, Ingber D, Elson E, et al Regulation of cytoskeletal mechanics and cell growth by myosin light chainphosphorylation. Am J Physiol 1998;275:C1349–C1356. [DOI] [PubMed] [Google Scholar]
- 12. Yuan SY, Breslin JW, Perrin R, Gaudreault N, Guo M, Kargozaran H, et al Microvascular permeability in diabetes and insulin resistance. Microcirculation 2007;14:363–373. [DOI] [PubMed] [Google Scholar]
- 13. Kumar P, Shen Q, Pivetti CD, Lee ES, Wu MH, Yuan SY. Molecular mechanisms of endothelial hyperpermeability: Implications in inflammation. Expert Rev Mol Med 2009;11:e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Qi LL, Fang SH, Shi WZ, Huang XQ, Zhang XY, Lu YB, et al CysLT2 receptor‐mediated AQP4 up‐regulation is involved in ischemic‐like injury through activation of ERK and p38 MAPK in rat astrocytes. Life Sci 2011;88:50–56. [DOI] [PubMed] [Google Scholar]
- 15. Francesca B, Rezzani R. Aquaporin and blood brain barrier. Curr Neuropharmacol 2010;8:92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nicchia GP, Frigeri A, Liuzzi GM, Svelto M. Inhibition of aquaporin‐4 expression in astrocytes by RNAi determines alteration in cell morphology, growth, and water transport and induces changes in ischemia‐related genes. FASEB J 2003;17:1508–1510. [DOI] [PubMed] [Google Scholar]
- 17. Ikram MA, Seshadri S, Bis JC, Fornage M, DeStefano AL, Aulchenko YS, et al Genomewide association studies of stroke. N Engl J Med 2009;360:1718–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wellcome Trust Case Control Consortium . Genome‐wide association study of 14,000 cases of seven common diseases and 3000 shared controls. Nature 2007;447:661–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang L, Hauser ER, Shah SH, Pericak‐Vance MA, Haynes C, Crosslin D, et al Peakwide mapping on chromosome 3q13 identifies the kalirin gene as a novel candidate gene for coronary artery disease. Am J Hum Genet 2007;80:650–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Su L, Shen L, Clayburgh DR, Nalle SC, Sullivan EA, Meddings JB, et al Targeted epithelial tight junction dysfunction causes immune activation and contributes to development of experimental colitis. Gastroenterology 2009;136:551–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Melzer D. Genetic polymorphisms and human aging: Association studies deliver. Rejuvenation Res 2008;11:523–526. [DOI] [PubMed] [Google Scholar]
- 22. Matarin M, Brown WM, Singleton A, Hardy JA, Meschia JF. Whole genome analyses suggest ischemic stroke and heart disease share an association with polymorphisms on chromosome 9p21. Stroke 2008;39:1586–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sharpless NE, DePinho RA. How stem cells age and why this makes us grow old. Nat Rev Mol Cell Biol 2007;18:703–713. [DOI] [PubMed] [Google Scholar]
- 24. Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, et al Genomewide association analysis of coronary artery disease. N Engl J Med 2007;357(5):443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Karvanen J, Silander K, Kee F, Tiret L, Salomaa V, Kuulasmaa K, et al The impact of newly identified loci on coronary heart disease, stroke, and total mortality in the MORGAM prospective cohorts. Genet Epidemiol 2009;33(3):237–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tong Y, Zhang Y, Zhang R, Geng Y, Lin L, Wang Z, et al Association between two key SNPs on chromosome 12p13 and ischemic stroke in Chinese Han population. Pharmacogenet Genomics 2011;21(9):572–578. [DOI] [PubMed] [Google Scholar]
- 27. Matsushita T, Umeno J, Hirakawa Y, Yonemoto K, Ashikawa K, Amitani H, et al Association study of the polymorphisms on chromosome 12p13 with atherothrombotic stroke in the Japanese population. J Hum Genet 2010;55(7):473–476. [DOI] [PubMed] [Google Scholar]
- 28. Lotta LA, Giusti B, Saracini C, Vestrini A, Volpe M, Rubattu S, et al No association between chromosome 12p13 single nucleotide polymorphisms and early‐onset ischemic stroke. J Thromb Haemost 2010;8(8):1858–1860. [DOI] [PubMed] [Google Scholar]
- 29. Olsson S, Melander O, Jood K, Smith JG, Lövkvist H, Sjögren M, et al Genetic variant on chromosome 12p13 does not show association to ischemic stroke in 3 Swedish case‐control studies. Stroke 2011;42(1):214–216. [DOI] [PubMed] [Google Scholar]
- 30. Ding H, Tu X, Xu Y, Xu C, Wang X, Cui G, Bao X, et al No evidence for association of 12p13 SNPs rs11833579 and rs12425791 within NINJ2 gene with ischemic stroke in Chinese Han population. Atherosclerosis 2011;216(2):381–3812. [DOI] [PubMed] [Google Scholar]
- 31. International Stroke Genetics Consortium; Wellcome Trust Case‐Control Consortium 2 . Failure to validate association between 12p13 variants and ischemic stroke. N Engl J Med 2010;362(16):1547–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Comings DE, MacMurray JP. Molecular heterosis: A review. Mol Genet Metab 2000;71(1–2):19–31. [DOI] [PubMed] [Google Scholar]
- 33. Jazdzewski K, Liyanarachchi S, Swierniak M, Pachucki J, Ringel MD, Jarzab B, et al Polymorphic mature microRNAs from passenger strand of pre‐miR146a contribute to thyroid cancer. Proc Natl Acad Sci USA 2009;106(5):1502–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cui R, Kamatani Y, Takahashi A, Usami M, Hosono N, Kawaguchi T, et al Functional variants in ADH1B and ALDH2 coupled with alcohol and smoking synergistically enhance esophageal cancer risk. Gastroenterology 2009;137(5):1768–1775. [DOI] [PubMed] [Google Scholar]
- 35. Yu JG, Zhou RR, Cai GJ. From hypertension to stroke: Mechanisms and potential prevention strategies. CNS Neurosci Ther 2011;17(5):577–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List of the primers for selected SNPs †
Supporting info item


