Summary
Aims
Mcl‐1, an antiapoptotic member of the Bcl‐2 family, is overexpressed in human glioblastoma, conferring a survival advantage to tumor cells. The mechanisms underlying its dysregulation have not been clarified. In this study, we explored the involvement of micro‐RNAs that acted as endogenous sequence‐specific suppressors of gene expression.
Methods and results
Using computational and TCGA analysis, we identified miR‐139 as being downregulated in glioblastoma in comparison with human brain tissue, as well as possessing a putative target site in Mcl‐1 mRNA. Overexpression of miR‐139 led to a clear decrease in Mcl‐1 expression in gliomas. Reporter assays revealed direct post‐transcriptional regulation involving miR‐139 and the 3′‐untranslated region of Mcl‐1. Human glioma tissues with low expression of miR‐139 displayed higher expression of Mcl‐1 protein than those with high expression, suggesting that low miR‐139 contributes to Mcl‐1 overexpression. In addition, upregulation of miR‐139 suppressed the proliferation and enhanced temozolomide (TMZ)‐induced apoptosis. Finally, we observed that Mcl‐1 knockdown resulted in similar effects compared with miR‐139 transfection.
Conclusion
Our results suggested that miR‐139 negatively regulated Mcl‐1 and induced apoptosis in cooperation with an anticancer drug TMZ in glioma.
Keywords: Apoptosis, Glioma, Mcl‐1, miR‐139, Temozolomide
Introduction
Micro‐RNAs are a group of small noncoding RNAs that play critical regulatory roles in diverse biological processes, such as development, differentiation, and apoptosis. Mature miRNAs negatively regulate their gene targets through complementary sequence pairing to the 3′UTR of mRNA targets by inducing transcript degradation or translational repression 1. Furthermore, each miRNA can potentially regulate hundreds of mRNAs, and it is predicted that more than one‐third of human genes are miRNA targets 2. Accumulating evidence has revealed the aberrant expression of miRNAs in human glioblastoma. miR‐137 has been reported to be downregulated in glioma, in turn, leading to upregulation of Cox‐2 3. On the other hand, recent reports have demonstrated that miR‐21 4, 5, 6, 7, 8, miR‐221 9, 10, 11, 12, 13, and miR‐23b 14 are upregulated in glioma, leading to downregulation of PTEN, PUMA, and VHL, respectively. Furthermore, the miRNA expression signature was reported to be related to the clinical outcome of patients with glioma 15, 16, 17. Thus, miRNAs may play an important role in glioma development and progression by modulating a variety of gene expression and cellular processes.
Apoptosis resistance is an important characteristic of tumor cells, in addition to dysregulated proliferation and aberrant differentiation. Apoptosis is regulated by a fine balance of Bcl‐2 family proteins, such as antiapoptotic factors and pro‐apoptotic factors. Increased Mcl‐1 expression certainly has the capacity to confer a malignant phenotype 18 and is often associated with increased resistance to apoptosis 19, 20, 21. In addition, Bryan et al. 22 demonstrated that Mcl‐1 is overexpressed in the majority of malignant gliomas and neutralization of Mcl‐1 induced apoptosis and increased chemotherapy‐induced apoptosis. However, the underlying mechanisms of Mcl‐1 overexpression in gliomas are not clearly understood. In the present study, we demonstrate that miR‐139 negatively regulates Mcl‐1 in glioma. MiR‐139 was downregulated in glioma cells and tissues in association with enhanced expression of Mcl‐1. Overexpression of miR‐139 downregulates Mcl‐1 and enhances apoptosis induced by TMZ, a famous anticancer drug for glioma. The present study demonstrates for the first time that miR‐139 directly targets Mcl‐1 and induces apoptosis in cooperation with TMZ targeting Mcl‐1 in glioma.
Materials and Methods
Tissue Samples and Clinical Data
Fifty paraffin‐embedded glioma specimens with clinical data were collected from January 2008 to June 2010, including 14 grade I–II tumors, 18 grade III tumors, and 18 grade IV tumors. This study was approved by the hospital institutional review board, and written informed consent was obtained from all patients.
Cell Culture and Transfection
Human U87, LN229, SNB19, U241, and LN308 glioblastoma cells and low‐grade glioma cell H4 were obtained from the China Academia Sinica Cell Repository, Shanghai, China. The cells were maintained in Dulbecco's modified Eagle's medium (Gibco, Los Angeles, CA, USA) supplemented with 10% fetal bovine serum (Gibco) and were incubated at 37°C in a 4% CO2 atmosphere. Cell transfection was performed using Lipofectamine 2000 (Invitrogen, Camarillo, CA, USA) according to the manufacturer's instructions.
Plasmids and Oligonucleotides
Hsa‐miR‐139 mimics, Mcl‐1 siRNA, were chemically synthesized and purified by high‐performance liquid chromatography (GenePharma, Shanghai, China). Cells were transfected with miR‐139 mimics, Mcl‐1 siRNA oligonucleotides (200 pmol each), using Lipofectamine 2000 (Invitrogen). Cells transfected with scrambled 2′‐OMe oligonucleotides (scramble) were used as control.
qRT‐PCR
For qRT‐PCR assays, total RNA was extracted from cells by RNAiso reagent (Takara, Otsu, Japan). Reverse transcription was performed with PrimeScript RT reagent kit with gDNA eraser (Takara) according to the manufacturer's instructions. The expression of miR‐139 was verified by the altered stem‐loop RT‐PCR with specific RT and PCR primers. U6 snRNA was used as an endogenous control. RT reactions were carried out using RNA fraction with Promega RT kit following the manufacturer's protocol. The PCR conditions were performed by denaturing the DNA at 94°C for 4 min, followed by 40 cycles of amplification: 94°C C for 40 s, 42°C C for 40 s, and 72°C C for 40 s for data collection. Quantitative PCR was performed on an ABI 7400 thermocycler (Applied Biosystems, Foster City, CA, USA) using SYBR Premix Ex TaqTM (Perfect Real Time) kits (TaKaRa) according to the manufacturer's instructions.
Proliferation Assay
Following transfection with miR‐139 mimics, MTT assay was used to quantitate cell viability of human MB cells, as previously described 14. Each experiment was performed in triplicate. The absorbance values of each well were measured with a microplate spectrophotometer (Molecular Devices; Sunnyvale, CA, USA) at 490 nm. All proliferation assays were repeated as independent experiments at least twice.
Apoptosis Assays
Apoptosis was quantitated 48 h after transfection, using annexin V labeling and caspase 3/7 activity. For the annexin V assay, an annexin V–FITC‐labeled Apoptosis Detection Kit (Abcam, Cambridge, MA, USA) was used according to the manufacturer's protocol. Caspase 3/7 activity was measured using Caspase‐Glo 3/7 reagent (Promega, Madison, WI, USA).
Luciferase Reporter Assay
The Mcl‐1 3′UTR‐Luc reporter was created, respectively, by the ligation of Mcl‐1 3′UTR PCR product into the XbaI site of the pGL3 control vector (Promega). The mutant reporter was generated from pGL3‐WT‐Mcl‐1‐3′UTR‐Luc by replacing the binding site of miR‐139 with restriction enzyme cutting site CGGATCCG. For the reporter assay, cells were cultured in 96‐well plates and transfected with wild or mutant luciferase reports and miR‐139. Following 48‐h incubation, luciferase activity was measured using a dual‐luciferase reporter system (Promega). Luciferase activity was measured 48 h after transfection with the dual‐luciferase reporter assay system. The Renilla luciferase activity was utilized as an internal control.
Western Blot and Immunohistochemistry
Western blot and immunohistochemistry assay were performed as previously described 3. Immunoblot and immunohistochemistry assays were performed using antibodies against Mcl‐1 and cytochrome C (1:1000 dilution, Santa Cruze, Dallas, TX, USA) and GAPDH (1:1000 dilution). IHC scores were performed using a semiquantitative grading system as previous study 23.
Nude Mouse Tumor Xenograft Model and miR‐139 Treatment
Nude mouse tumor xenograft model was made as previous study. LN229 glioma cells were subcutaneously injected into 4‐week‐old female nude mice (Cancer Institute of the Chinese Academy of Medical Science). When the tumor volume reached 40 mm3, mice were randomly divided into two groups. Each group was treated with miR‐139 or scrambled oligo in 10‐μL Lipofectamine through local injection of the xenograft tumor at multiple sites. The treatment was performed once every 3 days for 14 days. The tumor volume was measured with a caliper twice a week, using the formula: volume = length × width2/2.
TCGA GBM Data and Statistical Analysis
The TCGA miRNA and mRNA expression microarray data and Mcl‐1 data including survival information for patients with glioblastoma (GBM) were downloaded from the following portal: http://tcga-data.nci.nih.gov/tcga/homepage.htm. The significance of differences between two groups was estimated with the Student's t‐test and χ 2‐test. Statistical technique for comparing means for multiple groups was estimated with ANOVA. All differences were considered statistically significant at the level of P < 0.05. Statistics were performed using the SPSS Graduate Pack 11.0 statistical software (SPSS, Chicago, IL, USA).
Results
Identification of miRNA Binding Elements in the 3′ UTR of Mcl‐1 gene
To identify that miRNAs are capable to negatively regulating Mcl‐1, we performed bioinformatics and TCGA analysis (Figure S1). After this analysis approach, we found that 3′UTR of Mcl‐1 contained the potential binding site for miR‐139 at nt 376 (Figure 1A). Furthermore, TCGA analysis demonstrated that miR‐139 is low expressed in GBM compared with normal brain tissue (Figure S2; Table S1). These observations shed light that Mcl‐1 highly expressed in glioma might be owing to the suppression of miR‐139.
Figure 1.

Mcl‐1 inversely correlated with miR‐139 expression in glioma. (A) Diagram of seed sequence of miR‐139 matched the 3′ UTR of the Mcl‐1 gene. (B) Expression of Mcl‐1 in resected glioma specimen was assessed by IHC assay. FISH assay showed the expression of miR‐139 in the homologous specimens. (C) A statistically significant inverse correlation between miR‐139 and Mcl‐1 protein levels in clinical specimens (Spearman's correlation analysis, r = −0.79097; P < 0.001). (D) Western blot assays of Mcl‐1 in glioma cells. (E) qRT‐PCR assay of miR‐139 in glioma cells.
To verify our hypothesis, we evaluated the expression of miR‐139 and Mcl‐1 by FISH and IHC assay in 40 glioma samples. High‐grade glioma expressed comparatively higher Mcl‐1 expression and lower miR‐139, while low‐grade glioma with lower Mcl‐1 and higher miR‐139 (Figure 1B). Spearman's correlation analysis demonstrated that Mcl‐1 in tumor tissues inversely correlated with miR‐139 expression (Figure 1C). Western blot and qRT‐PCR assay demonstrated that expression of miR‐139 was negatively associated with Mcl‐1 in glioma cells as well (Figure 1D–E).
MiR‐139 Binds to the 3′ UTR of Mcl‐1
To evaluate whether miR‐139 can modulate the levels of Mcl‐1, we transfected LN229 and U87 cells with miR‐139 mimics. Two days after transfection, immunofluorescence assay showed a decrease in Mcl‐1 protein levels in the miR‐139‐transfected group compared with negative group (Figure 2A). Western blot assay demonstrated that miR‐139 overexpression suppressed Mcl‐1 protein levels as well (Figure 2B).
Figure 2.
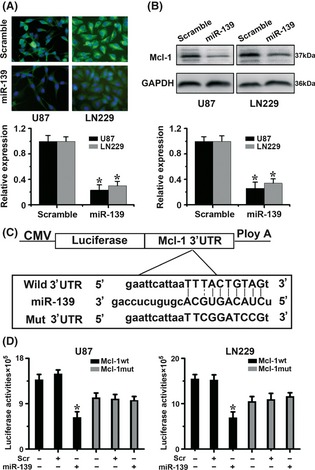
Mcl‐1 is direct target of miR‐139 in glioma cells. (A) U87 and LN229 cells were transfected with the miR‐139 mimics or with a scramble. Expression of the Mcl‐1 protein was evaluated 48 h later by immunofluorescence assay. As shown, the levels of Mcl‐1 were significantly decreased in the presence of miR‐139, but not in the presence of scramble. (B) Western blot analysis of total cell lysates of U87 and LN229 cells transfected with the indicated miRNAs. As shown, transfection with miR‐139 induced efficient Mcl‐1 decrease. (C) Diagram of seed sequence of miR‐139 matched the 3′ UTR of the Mcl‐1 gene. (D) Luciferase reporter assays in glioma cells, following cotransfection of cells with wild‐type or mutant 3′UTR Mcl‐1 and miRNA, as indicated. Data represent fold change in expression (mean ± SE) of three replicates.
To examine whether the downregulation of Mcl‐1 by miR‐139 is caused by direct binding to the putative targeting site in the Mcl‐1, we constructed both wild and mutant luciferase reporter plasmid pMIR‐Mcl‐1‐3′UTR containing the putative miR‐139 binding site of Mcl‐1 3′UTR downstream of the luciferase open reading frame (Figure 2C). Next, we transiently expressed this construct in glioma cells, in the presence or absence of the miR‐139 mimics. And we observed a significant 40–60% decrease in luciferase activity in pGL3‐WT‐Mcl‐1‐3′UTR in the presence of miR‐139 mimics, while no significant change in pGL3‐mut‐Mcl‐1‐3′UTR (Figure 2D). These data suggest that miR‐139 binds to the 3′ UTR of Mcl‐1 and impairs Mcl‐1 mRNA translation.
MiR‐139 Affects Glioma Growth both in vitro and in vivo
To assess the tumor suppressor potential of miR‐139, we first reexpressed miR‐139 in two glioma cells followed by functional assays. Transient‐transfected miR‐139 mimics led to decreased proliferation compared with scramble (Figure 3A, Figure S3) in U87 and LN229 cells.
Figure 3.

miR‐139 suppresses glioma growth both in vitro and in vivo. (A) Representative cartogram showing the proliferation regulated by miR‐139‐treated cells and scramble. (B), (C) Tumor growth curves and mass for miR‐139 mimics‐treated LN229 tumors vs scramble xenograft. Scramble refers to scramble oligonucleotides and uses for negative control. (D), (F) qRT‐PCR and FISH assay for miR‐139 expression in miR‐139 mimics‐treated LN229 tumors vs. scramble xenograft. (E) Western blot assay for Mcl‐1 expression in miR‐139 mimics‐treated LN229 tumors vs. scramble xenograft. (F) IHC assay for Mcl‐1 and caspase‐3 expression in miR‐139 mimics‐treated LN229 tumors vs. scramble xenograft.
To investigate the potential impact of miR‐139 expression in vivo, a LN229 xenograft model was utilized. MiR‐139‐treated group displayed a significant growth and tumor reduction, whereas tumor growth was not impacted by scramble (Figure 3B,C). qRT‐PCR and FISH analysis confirmed the reexpression of miR‐139 in mimics‐transfected tumor (Figure 3D,F). Moreover, miR‐139‐treated glioma model displayed decreased expression of Mcl‐1 (Figure 3E,F) and increased caspase‐3 (Figure 3F) compared with scramble. These data indicated that miR‐139 reintroduction suppressed Mcl‐1 and tumor survival in vitro and vivo.
miR‐139 miRNA Sensitizes Human Glioma cells to TMZ, Which Downregulates Mcl‐1 Expression
To investigate the effect of miR‐139 in the resistance to apoptosis, we transfected LN229 and U87 cells with miR‐139 and then subjected to apoptosis analysis. Overexpression of miR‐139 led to increase apoptosis, caspase‐3/7 activities, and expression of cytoplasm cytochrome C compared with scramble (Figure 4A–C). Next, we exposed miRNA‐transfected cells to TMZ, a well‐known anticancer drug for glioma. TMZ treatment of glioma cells led to a slight increase in the apoptosis rate. Of importance is the finding that TMZ‐induced apoptosis and cytoplasm cytochrome C expression were markedly enhanced in miR‐139 miRNA‐transfected groups (Figures 4B, S4B). In addition, TMZ treatment increased the caspase 3/7 activities of glioma cells and these activities were markedly enhanced in cells transfected with miR‐139 miRNA (Figure 4C). This finding implies that miR‐139 transfection potentiates TMZ‐induced apoptosis in glioma cells.
Figure 4.

Sensitivity to apoptosis is increased after transfection of miR‐139. (A) Western blot assay showed that ectopic expression of miR‐139 promotes mitochondrion release of cytochrome c. (B) U87 and LN229 cells were transfected with miR‐139 mimics or scramble at 40 nM. After 24 h, TMZ was added in fresh media and the cells were incubated for 4 h. Cells were then analyzed by apoptosis assay. (C) In parallel, cells were transfected and treated with TMZ as in panel B, and caspase 3/7 activity was measured.
Mcl‐1 is Involved in the Antitumor Effects of miR‐139
To determine whether the effects of miR‐139 on the cell proliferation and apoptosis are mediated by Mcl‐1, we first silenced Mcl‐1 expression by RNAi (Figure 5A). As shown in Figure 5(B–C), specific knockdown of Mcl‐1 by RNAi inhibited glioma cell proliferation and increased TMZ‐induced apoptosis and expression of cytoplasm cytochrome C (Figure S4A), which phenocopied the effects of miR‐139. Next, we examined whether the restoration of Mcl‐1 could rescue the antitumor effects of miR‐139. We introduced a Mcl‐1 expression construct into glioma cells to recover Mcl‐1 expression (Figure 5D). This construct contains the entire Mcl‐1 coding sequence, but no 3′UTR fragment. Therefore, it is insensitive to miR‐139‐mediated repression. The results showed that Mcl‐1 overexpression could significantly abrogate miR‐139‐dependent effects on glioma proliferation and TMZ‐induced apoptosis (Figure 5E,F). Altogether, these data suggest that the effects of miR‐139 on the cell proliferation and apoptosis are partially mediated by Mcl‐1.
Figure 5.
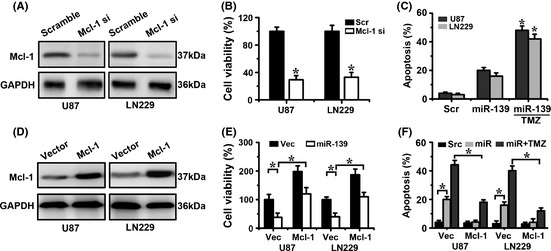
Mcl‐1 impacts proliferation, apoptosis of glioma cells. (A) Mcl‐1 expression levels in U87 and LN229 cells transfected with Mcl‐1 si were assessed by Western blot. (B–C) Representative cartogram showing cell proliferation and apoptosis regulated by Mcl‐1 si. (D) Mcl‐1 expression levels in U87 and LN229 cells transfected with Mcl‐1 were assessed by Western blot. (E–F) Representative cartogram showing proliferation and apoptosis regulated by miR‐139 or/and Mcl‐1.
Discussion
In this study, we emphasized on the role of miR‐139 in glioma. We found that miR‐139 inhibits Mcl‐1 in glioma cells via binding to the 3′UTRs. We also found a significant inverse correlation between miR‐139 expression and Mcl‐1 expression in glioma tissues. Forced miR‐139 expression in glioma cells strongly inhibits cell proliferation and potentiates TMZ‐induced apoptosis. We also showed that the effects of miR‐139 on glioma cells are partially mediated by Mcl‐1 downregulations. Therefore, we are the first comprehensive study of the role of miR‐139 in human gliomas.
In the last 2 years, a few studies have investigated the role of miR‐139 and its targets in cancer. Fan et al. 24 demonstrated that miR‐139 expression is downregulated in human HCC cell lines and derepression of c‐Fos caused by miR‐139 downregulation contributes to the metastasis of HCC. Guo et al. 25 identified that miR‐139 decreases proliferation by directly targeting RAP1B, which has been shown to regulate integrin‐mediated cell signaling. Shen et al. 26 provide evidence that miR‐139 might function as a metastasis suppressor in CRC by targeting type I insulin‐like growth factor receptor. In additional, miR‐139 could target FoxO1 to preserve homeostatic regulation of its transcriptional activity in response to environmental stimuli in mouse hepatocytes 27. Whether these targets are operative in sensitization of glioma cells with anticancer drugs remains to be determined. Our study firstly showed that miR‐139 expression was reduced in human glioma samples and miR‐139 overexpression suppressed the proliferation and enhanced TMZ‐induced apoptosis of glioma cells. Thus, our findings, together with those from other groups, suggest a fundamental role of miR‐139 in tumorigenesis as well as in the phenotypes of cancer cells. Furthermore, we only concluded that Mcl‐1 was one of interesting targets of miR‐139, not the only target in glioma.
Apoptosis is a major barrier that must be circumvented during malignant transformation and tumor progression. Cancer cells evolve to evade apoptosis so that they can escape from the surveillance system and chemotherapy in the crucial tumor growth environment. We showed that miR‐139 could suppress the proliferation of glioma both in vitro and vivo. Therefore, downregulation of miR‐139 may facilitate the adaptation of cancer cells to the crucial growth environment and, in turn, facilitate the development of glioma. It is noteworthy that recurrent glioma is poorly responsive to TMZ. Interestingly, miR‐139 could also sensitize glioma cells to chemotherapeutic drug‐induced apoptosis. Therefore, reintroduction of miR‐139 into glioma cells may not only limit cancer growth but also sensitize glioma cells to TMZ.
It has been shown that in many tumors, the Mcl‐1 protein is expressed at levels much higher than that in its normal counterpart. Some factors have been identified to be able to contribute to the increasing gene transcription, such as activation of transcription factor STAT3, and phosphatidylinositol 3‐kinase (PI3K) pathway 28, 29. The inhibition of Mcl‐1 amplified signaling leads to the inhibition of cell proliferation and metastasis. Hence, it is a promising project for tumor therapy. Mcl‐1‐targeted RNAi involving miRNA, small hairpin RNA (shRNA), or small interference RNA (siRNA) has also been successfully used in human cancer cell lines, including CRC, breast cancer, prostate cancer, lung cancers, hepatocellular carcinoma, and gastric cancer cells 18, 30, 31, 32, 33, 34, 35. In this regard, it is important to note that one of the targets of miR‐139 is Mcl‐1. Our results provide the first insight that forced Mcl‐1 expressions partially rescue cell proliferation and induced apoptosis by miR‐139. In addition, the target of the Mcl‐1 3′‐UTR was confirmed using a reporter gene luciferase that carried its putative binding site for miR‐139.
So far as we know, Mcl‐1 activation, as the consequence of transcriptional or post‐transcriptional regulation, evokes pleiotropic biological responses. Its activation could block the progression of apoptosis by binding and sequestering the pro‐apoptotic proteins Bcl‐2 homologous antagonist killer (Bak) and Bcl‐2‐associated protein X (Bax), which are capable of forming pores in the mitochondrial membrane, allowing the release of cytochrome c into the cytoplasm. In our present study, we investigated whether the Mcl‐1 inhibition mediated by miR‐139 could affect mitochondrial apoptosis pathways. Cytochrome c was increased after miR‐139 cell transduction. Thus, we conclude that miR‐139 could regulate mitochondrial apoptosis signaling by directly targeting MCL‐1 in glioma.
In conclusion, we demonstrate for the first time that miR‐139 exerts effects as tumor suppressor in gliomas through downregulating the expression of MCL‐1. These data suggest a novel function and a therapeutic application of miR‐139 in gliomas.
Conflict of Interest
The authors declare no conflict of interest.
Supporting information
Figure S1. Bioinformatics and TCGA analysis approach.
Figure S2. Relative expression of miR‐139 in TCGA and different glioma cells.
Figure S3. miR‐139 suppresses glioma proliferation.
Figure S4. Western blot assay of cytochrome c in different treatment.
Acknowledgments
This project was supported by the National High Technology Research and Development Program 863 (2012AA02A508), National 11th five years supporting project (2007BA105B08), International Science and Technology Cooperation Program of China (2012DFA30470), Research fund for the doctoral program of higher school (20102307110008), and Heilongjiang natural science Key projects (C03030307).
The first three authors contributed equally to this work.
References
- 1. Cai Y, Yu X, Hu S, et al. A brief review on the mechanisms of miRNA regulation. Genomics Proteomics Bioinformatics 2009;7:147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 2006;6:857–866. [DOI] [PubMed] [Google Scholar]
- 3. Chen L, Wang X, Wang H, et al. miR‐137 is frequently down‐regulated in glioblastoma is a negative regulator of Cox‐2. Eur J Cancer 2012;48:3104–3111. [DOI] [PubMed] [Google Scholar]
- 4. Moore LM, Zhang W. Targeting miR‐21 in glioma: a small RNA with big potential. Expert Opin Ther Targets 2010;14:1247–1257. [DOI] [PubMed] [Google Scholar]
- 5. Ren Y, Kang CS, Yuan XB, et al. Co‐delivery of as‐miR‐21 and 5‐FU by poly(amidoamine) dendrimer attenuates human glioma cell growth in vitro. J Biomater Sci Polym Ed 2010;21:303–314. [DOI] [PubMed] [Google Scholar]
- 6. Shi L, Cheng Z, Zhang J, et al. The mechanism of apoptosis in human U87 glioma cells induced by miR‐21 antisense oligonucleotide. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 2008;25:497–501. [PubMed] [Google Scholar]
- 7. Zhou JY, Zhou C, Wang LL, et al. Influence of knock‐down of miR‐21 expression on the radiosensitivity of glioma SHG‐44 cells. Zhonghua Zhong Liu Za Zhi 2011;33:747–751. [PubMed] [Google Scholar]
- 8. Zhou X, Zhang J, Jia Q, et al. Reduction of miR‐21 induces glioma cell apoptosis via activating caspase 9 and 3. Oncol Rep 2010;24:195–201. [DOI] [PubMed] [Google Scholar]
- 9. Chen L, Zhang J, Han L, et al. Downregulation of miR‐221/222 sensitizes glioma cells to temozolomide by regulating apoptosis independently of p53 status. Oncol Rep 2012;27:854–860. [DOI] [PubMed] [Google Scholar]
- 10. Zhang C, Kang C, You Y, et al. Co‐suppression of miR‐221/222 cluster suppresses human glioma cell growth by targeting p27kip1 in vitro and in vivo . Int J Oncol 2009;34:1653–1660. [DOI] [PubMed] [Google Scholar]
- 11. Zhang C, Zhang J, Hao J, et al. High level of miR‐221/222 confers increased cell invasion and poor prognosis in glioma. J Transl Med 2012;10:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science 2008;321:1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang J, Han L, Ge Y, et al. miR‐221/222 promote malignant progression of glioma through activation of the Akt pathway. Int J Oncol 2010;36:913–920. [DOI] [PubMed] [Google Scholar]
- 14. Chen L, Han L, Zhang K, et al. VHL regulates the effects of miR‐23b on glioma survival and invasion via suppression of HIF‐1alpha/VEGF and beta‐catenin/Tcf‐4 signaling. Neuro Oncol 2012;14:1026–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ma R, Yan W, Zhang G, et al. Upregulation of miR‐196b Confers a Poor Prognosis in Glioblastoma Patients via Inducing a Proliferative Phenotype. PLoS ONE 2012;7:e38096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gabriely G, Yi M, Narayan RS, et al. Human glioma growth is controlled by microRNA‐10b. Cancer Res 2011;71:3563–3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jiang L, Mao P, Song L, et al. miR‐182 as a prognostic marker for glioma progression and patient survival. Am J Pathol 2010;177:29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sieghart W, Losert D, Strommer S, et al. Mcl‐1 overexpression in hepatocellular carcinoma: a potential target for antisense therapy. J Hepatol 2006;44:151–157. [DOI] [PubMed] [Google Scholar]
- 19. Nguyen M, Marcellus RC, Roulston A, et al. Small molecule obatoclax (GX15‐070) antagonizes MCL‐1 and overcomes MCL‐1‐mediated resistance to apoptosis. Proc Natl Acad Sci USA 2007;104:19512–19517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hussain SR, Cheney CM, Johnson AJ, et al. Mcl‐1 is a relevant therapeutic target in acute and chronic lymphoid malignancies: down‐regulation enhances rituximab‐mediated apoptosis and complement‐dependent cytotoxicity. Clin Cancer Res 2007;13:2144–2150. [DOI] [PubMed] [Google Scholar]
- 21. Paoluzzi L, Gonen M, Gardner JR, et al. Targeting Bcl‐2 family members with the BH3 mimetic AT‐101 markedly enhances the therapeutic effects of chemotherapeutic agents in in vitro and in vivo models of B‐cell lymphoma. Blood 2008;111:5350–5358. [DOI] [PubMed] [Google Scholar]
- 22. Day BW, Stringer BW, Spanevello MD, et al. ELK4 neutralization sensitizes glioblastoma to apoptosis through downregulation of the anti‐apoptotic protein Mcl‐1. Neuro Oncol 2011;13:1202–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang J, Han L, Zhang A, et al. AKT2 expression is associated with glioma malignant progression and required for cell survival and invasion. Oncol Rep 2010;24:65–72. [DOI] [PubMed] [Google Scholar]
- 24. Fan Q, He M, Deng X, et al. Derepression of c‐Fos caused by MicroRNA‐139 down‐regulation contributes to the metastasis of human hepatocellular carcinoma. Cell Biochem Funct 2012; doi: 10.1002/cbf.2902. [DOI] [PubMed] [Google Scholar]
- 25. Guo H, Hu X, Ge S, et al. Regulation of RAP1B by miR‐139 suppresses human colorectal carcinoma cell proliferation. Int J Biochem Cell Biol 2012;44:1465–1472. [DOI] [PubMed] [Google Scholar]
- 26. Shen K, Liang Q, Xu K, et al. MiR‐139 inhibits invasion and metastasis of colorectal cancer by targeting the type I insulin‐like growth factor receptor. Biochem Pharmacol 2012;84:320–330. [DOI] [PubMed] [Google Scholar]
- 27. Hasseine LK, Hinault C, Lebrun P, et al. miR‐139 impacts FoxO1 action by decreasing FoxO1 protein in mouse hepatocytes. Biochem Biophys Res Commun 2009;390:1278–1282. [DOI] [PubMed] [Google Scholar]
- 28. Vega F, Medeiros LJ, Leventaki V, et al. Activation of mammalian target of rapamycin signaling pathway contributes to tumor cell survival in anaplastic lymphoma kinase‐positive anaplastic large cell lymphoma. Cancer Res 2006;66:6589–6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Araki T, Hayashi M, Watanabe N, et al. Down‐regulation of Mcl‐1 by inhibition of the PI3‐K/Akt pathway is required for cell shrinkage‐dependent cell death. Biochem Biophys Res Commun 2002;290:1275–1281. [DOI] [PubMed] [Google Scholar]
- 30. Wu Y, Crawford M, Yu B, et al. MicroRNA delivery by cationic lipoplexes for lung cancer therapy. Mol Pharm 2011;8:1381–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Crawford M, Batte K, Yu L, et al. MicroRNA 133B targets pro‐survival molecules MCL‐1 and BCL2L2 in lung cancer. Biochem Biophys Res Commun 2009;388:483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Su H, Yang JR, Xu T, et al. MicroRNA‐101, down‐regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res 2009;69:1135–1142. [DOI] [PubMed] [Google Scholar]
- 33. Steele R, Mott JL, Ray RB. MBP‐1 upregulates miR‐29b that represses Mcl‐1, collagens, and matrix‐metalloproteinase‐2 in prostate cancer cells. Genes Cancer 2010;1:381–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Braconi C, Valeri N, Gasparini P, et al. Hepatitis C virus proteins modulate microRNA expression and chemosensitivity in malignant hepatocytes. Clin Cancer Res 2010;16:957–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Saito Y, Suzuki H, Tsugawa H, et al. Chromatin remodeling at Alu repeats by epigenetic treatment activates silenced microRNA‐512‐5p with downregulation of Mcl‐1 in human gastric cancer cells. Oncogene 2009;28:2738–2744. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Bioinformatics and TCGA analysis approach.
Figure S2. Relative expression of miR‐139 in TCGA and different glioma cells.
Figure S3. miR‐139 suppresses glioma proliferation.
Figure S4. Western blot assay of cytochrome c in different treatment.


