Summary
Background and purpose
As an important oncogenic miRNA, miR‐21 has been reported to play crucial roles in glioblastoma (GBM) carcinogenesis. However, the precise biological function and molecular mechanism of miR‐21 in GBM remain elusive. This study is designed to explore the mechanism of miR‐21 involved in the control of GBM cell growth.
Methods and results
MTT assay, cell cycle analysis, and apoptosis analysis showed that reduction of miR‐21 inhibited cell growth in U87 and LN229 GBM cells. Further, reduction of miR‐21 decreased the expression of human telomerase reverse transcriptase (hTERT) and repressed STAT3 expression and STAT3 phosphorylation. STAT3 inhibition led to a remarkable depletion of hTERT at both mRNA and protein levels by binding to the hTERT gene promoter by performing luciferase reporter assay and chromatin Immunoprecipitation PCR. Finally, knockdown of miR‐21 considerably inhibited tumor growth and diminished the expression of STAT3 and hTERT in xenograft model.
Conclusion
Our findings indicate that miR‐21 regulates hTERT expression mediated by STAT3, therefore controlling GBM cell growth.
Keywords: Glioblastoma, hTERT, miR‐21, STAT3
Introduction
miRNAs, small highly conserved noncoding RNA molecules of approximately 20–22 nucleotides in length, regulate protein expression by cleaving or repressing the translation of targets mRNA. Growing studies have indicated that miRNAs could function as oncogenic miRNAs (oncomiRs) or tumor suppressor miRNAs, playing crucial roles in the transformation and carcinogenesis 1. Among oncomiRs, miR‐21 has been overexpressed in a variety of cancer, including breast, lung, and colon cancer, as well as in glioblastoma (GBM) 2, 3, 4, 5. Previously, we profiled miRNA expression in five GBM cell lines (U251, TJ866, TJ905, TJ899, and A172) and human astrocytoma cell line (H4), and found that miR‐21 exhibited the most significant increase relative to normal brain tissue 6. And downregulation of miR‐21 inhibited epidermal growth factor receptor/protein kinase B (EGFR/AKT) pathway and suppressed the growth of human GBM cells independent of phosphatase and tensin homolog (PTEN) status. Up to date, several genes have been evidenced to be the targets of miR‐21 in GBM cells. Chen et al. 7 identified that reduction of miR‐21 increased programmed cell death 4 (PDCD4) and over‐expression of miR‐21‐inhibited PDCD4‐dependent apoptosis by targeting PDCD4 3′UTR in GBM cells. In addition, miR‐21 regulated matrix metalloproteinase (MMP) activities by targeting MMP inhibitors reversion‐inducing cysteine‐rich protein with kazal motifs (RECK) and tissue inhibitor of metalloproteinases 3 (TIMP‐3) to contribute to glioma malignant phenotype 8. However, the precise biological function and molecular mechanism of miR‐21 in GBM remain elusive.
In this study, we showed that reduction of miR‐21 inhibited cell growth in GBM cells, accompanying a decreased expression of human telomerase reverse transcriptase (hTERT) mediated by signal transducer and activator of transcription 3 (STAT3) transcription. Further, this effect was exerted in a STAT3‐dependent manner. Taken together, these results suggest that modulation of the mechanism responsible for miR‐21 in GBM could be used as a critical therapeutic strategy for GBM intervention and warrants further investigation.
Materials and Methods
Cell Culture and Reagents
The human U87 and LN229 GBM cell lines were purchased from the Cell Bank of Chinese Academy of Sciences (Shanghai, China). Both cells were cultured in Dulbecco's modified Eagle's medium (Gibco, NY, USA), supplemented with 10% bovine serum albumin and maintained at 37°C in an atmosphere of 5% CO2 and routinely passaged at 2‐ to 3‐day intervals. WP1066 and IL‐6 were dissolved in DMSO. For treatment, WP1066 was added to glioma cells at final concentrations of 1.0, 2.5, and 5 μM for 48 h 9. IL‐6 treated cell was referenced to the previous report 10.
Oligonucleotides and Cell Transfection
The 2′‐O‐methy1 (2′‐OMe‐) oligonucleotides were chemically synthesized by GenePharma (Shanghai, China). Sequences used were as follows: miR‐21, 5′‐UAGCUUAUCAGACUGAUGUUGA‐3′; anti‐miR‐21 (As‐miR‐21), 5′‐UCAACAUCAGUCUGAUAAGCUA‐3′; and scrambled miRNA (negative control), 5′‐UUGUACUACACAAAAGUACUG‐3′. For transfection, oligonucleotides were allowed to form transfection complexes with Lipofectamine 2000 (Invitrogen, CA, USA), subsequently added to glioma cells at a final concentration of 50 nM, and left to incubate for 8 h before medium change.
Engineering of Lentiviral Vector‐Based hTERT siRNA
The lentiviral vector mediated for hTERT siRNA was obtained from Shanghai GeneChem (Shanghai, China), consisting of PGC‐LV, pHelper 1.0, and pHelper 2.0. The siRNA sequences targeting hTERT were 5′‐GCA AGT TGC AAA GCA TTG GAA‐3′, and the sequences of negative control were 5′‐TTC TCC GAA CGT GTC ACG T‐3′. The PGC‐LV tagged with green fluorescent protein (GFP) and pHelper 1.0 and pHelper 2.0 were co‐transfected into 293T cells with Lipofectamine 2000. The cultured supernatants were collected, concentrated, and stocked in a refrigerator maintained at −70°C. Viral titers were determined as transduction unit (TU) by infection of 293T cells with dilutions of vector preparation and counting of enhanced GFP. The lentiviral vectors were infected into U87 and LN229 cells with a multiplicity of infection from 5 to 20.
MTT Assay
Cells were seeded into 96‐well plates at 2 × 103 cells per well. Subsequently, 50 μL of MTT dilution (KeyGEN, Nanjing, China) was added into each well at each day of consecutive 3 days after transfection, and the cells were incubated for additional 4 h. Finally, the supernatant was discarded and 150 μL of DMSO was added to each well to dissolve the precipitate. Optical density was measured at the wavelength of 570 nm. These data are presented as the mean ± SD, which are derived from five samples of at least three independent experiments.
Cell Cycle Analysis
Cells were washed with PBS, fixed with 70% ethanol for at least 1 h. After extensive washing, the cells were suspended in Hank's Balanced Salt Solution, containing 50 μg/mL Propidium iodide (PI) and 50 μg/mL RNase A, incubated for 1 h at room temperature, and analyzed by FACScan (Becton Dickinson, CA, USA). Cell cycle analysis was accomplished by ModFit software (Verity Software House, ME, USA). Experiments were performed in triplicate. Results were presented as percentage of cells in a particular phase.
Apoptosis Assay
Annexin V FITC and PI double stain was used to evaluate the percentages of apoptosis. Annexin V− and PI− cells were used as controls. Annexin V+ and PI− cells were designated as apoptotic and Annexin V+ and PI+ cells displayed necrotic. Tests were repeated in triplicate.
Western Blots Analysis
Total proteins were extracted, and the protein concentration was determined by BSA method (keyGEN, China). From each sample, 40 μg of protein lysates was subjected to SDS‐PAGE in 10% acrylamide gel and transferred to PVDF membranes (Millipore Corporation, MA, USA). The membrane was blocked in 5% nonfat milk and incubated with diluted antibodies against hTERT (1:500; Santa Cruz, CA, USA), STAT3, and pSTAT3 (1:500, Cell Signal, USA) overnight at 4°C, followed by incubation with HRP‐conjugated secondary antibody (1:2000; Santa Cruz). After stripping, the membrane was reprobed with GAPDH (1:2000; Shanghai, China) using ultra enhanced chemiluminescence Western blotting detection reagents. All bands obtained from western blot analysis were quantified by densitometry and are presented in the form of a bar graph.
RT‐PCR Analysis
Total RNA was isolated from normal, tumor tissue specimens, and cultured glioma cells using Trizol reagent (Invitrogen) and converted to cDNA using BioRT two‐step RT‐PCR Kit. PCRs were carried out using Ex Taq hot start polymerase (Takara), and β‐actin was used as a control for adjusting the relative of total RNA between the samples. The thermal cycles were carried out under the following conditions: 94°C for 30 seconds, 60°C for 30 seconds, and 72°C for 45 seconds for 35 cycles for hTERT and β‐actin. The primers (Takara, Dalian, China) used for PCR were as follows: hTERT, 5′‐AGG TCA GGC AGC ATC GGG AA‐3′ (forward) and 5′‐AGG CCC TGT GGA TAT CGT CCA G‐3′ (reverse); β‐actin, 5′‐AAG ACC TGT ACG CCA ACA CAG T‐3′ (forward) and 5′‐AGA AGC ATT TGC GGT GGA CGA T‐3′ (reverse). The PCRs for hTERT and β‐actin were fractionated on a 2% agarose gel containing 0.5 mg/mL ethidium bromide. Gels were visualized by Gel Doc™ XR gel documentation system (Bio‐Rad, CA, USA).
Chromatin Immunoprecipitation Analysis (ChIP)
The ChIP assay was performed using reagents commercially obtained from Upstate Biotechnology, and conducted essentially according to the manufacturer's instructions. Briefly, the cells were maintained in 100‐mm cell culture plates and were then fixed with formaldehyde for 10 min. Cells were lysed in SDS lysis buffer, and the chromatin DNA was extracted and sonicated into 200‐ to 1000‐bp fragments. Immunoprecipitation was performed with anti‐STAT3 (Upstate), anti‐RNA Polymerase II (positive control), and IgG (negative control). Purified DNA was used for PCR amplification, and primer sets were designed to flank the putative STAT‐3‐binding sites. Primer sequences used were 5′‐CCAAACCTGTGGACAGAACC‐3′ (forward) and 5′‐AGACTGACTGCCTCCATCGT‐3′ (reverse).
Luciferase Reporter Assay
pGL3‐WT‐hTERT‐promoter reporters were created by ligation of STAT‐3‐binding sites in the hTERT promoter into the BglII site of the pGL3 control vector (Promega, CA, USA). pGL3‐MUT‐hTERT‐promoter reporters were generated from pGL3‐WT‐hTERT‐promoter reporters by deleting the binding sites. Luciferase activity was measured with the Dual‐luciferase reporter assay system.
Xenograft Tumor Assay
The nude mouse subcutaneous LN229 glioma model was constructed as previously described 11. When tumors were established, the mice were divided into three groups (8 mice per group) randomly, which were treated with 200 pmol scramble oligo or As‐miR‐21 in 10 μL Lipofectamine 2000 through local injection of xenograft tumor in multiple sites. The treatment was performed once every 3 days for 21 days. The tumor volume was measured with a caliper once every 3 days, using the formula volume = length × width2/2.
Immunohistochemistry Staining
For immunohistochemical study, the sections were incubated with primary antibodies (1:100 dilution) against hTERT and STAT3 overnight at 4°C, and then incubated with a biotinylated secondary antibody (1:200 dilution) at room temperature for 1 h, followed by incubation with ABC‐peroxidase reagent for 1 h, washed with PBS, stained with 3, 3‐diaminobenzidine (30 mg dissolved in 100 mL Tris buffer containing 0.03% H2O2) for 5 min, and rinsed in water and counterstained with hematoxylin. For the evaluation of hTERT and STAT3 expression, ten randomly selected visual fields per section were examined by light microscope.
Statistical Analysis
Data were analyzed with SPSS 10.0 (SPSS Inc., IL, USA). Statistical evaluation for data analysis was determined using t‐test. Differences with P < 0.05 were considered statistically significant.
Results
As‐miR‐21 Suppresses GBM Cell Growth Accompanying hTERT Downregulation
To evaluate the potential effect of miR‐21 in GBM cells, MTT assay was employed to measure cells viability. As‐miR‐21‐treated cells showed a significant decrease in proliferation relative to untreated cells (Figure 1A). The growth inhibitory effect of As‐miR‐21‐treated cells reached maximum at 3 days post transfection in both U87 and LN229 GBM cells. Next, we analyzed the cell cycle distribution and cell apoptosis. As shown in Figure 1B, As‐miR‐21‐treated cells represented significant ascends in G0/G1 phase in comparison with untreated cells. By Annexin V and PI double staining, significant more apoptotic cells were found in U87 and LN229 cells after transfection of As‐miR‐21 (Figure 1C). These results suggest that knockdown of miR‐21 can induce cells arrest at G0/G1 phases and cell apoptosis, thereby inhibiting cell growth in GBM cells. Further, we found that repression of miR‐21 reduced hTERT protein expression in both U87 and LN229 cells (Figure 1D). And the level of hTERT mRNA expression also was decreased after knockdown of miR‐21 by RT‐PCR. Thus, we reasoned that miR‐21 may regulate hTERT expression at the transcriptional level.
Figure 1.
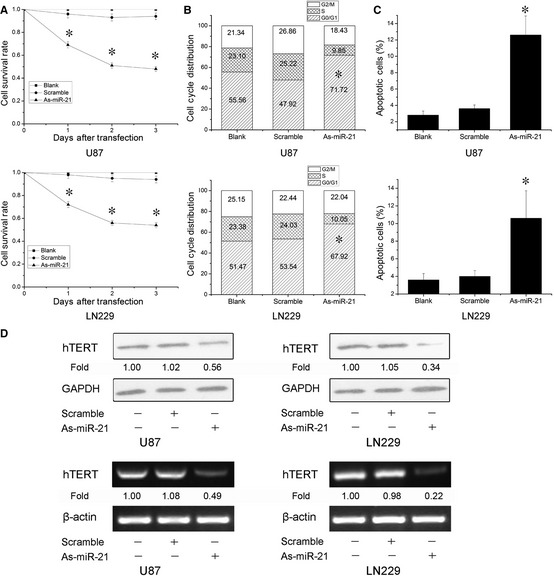
Reduction of miR‐21 suppresses cell growth by human telomerase reverse transcriptase (hTERT) in glioblastoma cells. (A–C) Cells were transfected with As‐miR‐21, and cell survival, proliferation, and apoptosis were measured by MTT assay, cell cycle distribution, and Annexin V FITC and PI double stain. (D) Cells were treated with As‐miR‐21, the levels of hTERT mRNA and protein were detected, and β‐actin and GAPDH were regarded as an endogenous normalizer. n = 3. *P < 0.05 as compared with the control group.
hTERT siRNA Inhibits Cell Growth in GBM Cells
To identify whether the biological effect of hTERT in GBM cells is consistent with the function of miR‐21, we used lentiviral vector–mediated delivery of hTERT siRNA to downregulate hTERT expression in U87 and LN229 cells. MTT assay showed about 29.4 ± 3.4% and 38.6 ± 4.5% survival rates in hTERT siRNA‐treated U87 and LN229 cells compared to the untreated cells, respectively (Figure 2A). Consistently, a marked induction of G1/G0 phase arrest and cell apoptosis was detected after hTERT siRNA treatment (Figure 2B,C). Taken together, hTERT is required for cell survival in GBM cells.
Figure 2.
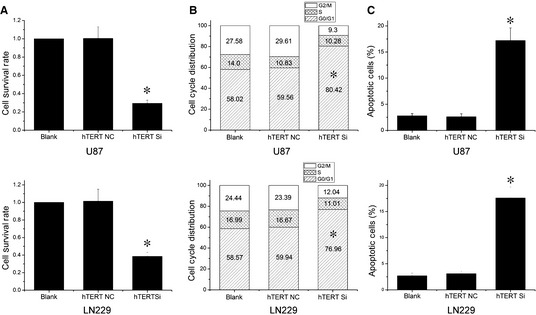
Human telomerase reverse transcriptase (hTERT) siRNA inhibits cell growth in glioblastoma cells. (A–C) Cells were transfected with lentiviral vector hTERT siRNA, and cell survival, proliferation, and apoptosis were measured by MTT assay, cell cycle distribution and Annexin V FITC and PI double stain. n = 3. *P < 0.05 as compared with the control group.
As‐miR‐21 Represses hTERT Expression through STAT3 Transcription
To explore the potential mechanism involved in regulation of hTERT by miR‐21, we identified that STAT3 was an important mediator between miR‐21 and hTERT. Western blot assay showed that reduction of miR‐21 decreased STAT3 expression and phosphorylation in U87 and LN229 cells (Figure 3A). Further, STAT3 inhibitor WP1066 effectively suppressed the phosphorylation of STAT3 compared to the untreated cells. Notably, a remarkable depletion in the mRNA and protein level of hTERT was also found in WP1066‐treated cells (Figure 3B). There data indicate that STAT3 is critical for hTERT regulation of miR‐21.
Figure 3.
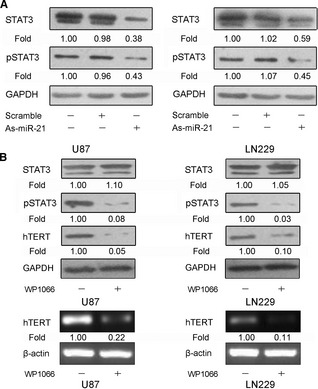
Reduction of miR‐21 represses human telomerase reverse transcriptase (hTERT) expression mediated by STAT3. (A) Cells were transfected with As‐miR‐21, and the levels of STAT3 and pSTAT3 were detected by Western blot analysis. (B) Cells were treated with STAT3 inhibitor WP1066, and STAT3, pSTAT3 and hTERT protein expression was detected by Western blot analysis, and hTERT mRNA expression was tested by PCR assay. β‐Actin and GAPDH were regarded as an endogenous normalizer. n = 3.
To investigate the functional interaction between STAT3 signaling and hTERT gene expression, we scanned the hTERT gene promoter region for regulatory DNA‐binding elements and identified the consensus STAT3‐binding site (TTCNNNGAA) in the hTERT gene promoter at −3308/−3316 bp. To determine in vivo binding of STAT3 to the hTERT promoter, we performed ChIP PCR assays in U87 and LN229 cells. Data showed that hTERT promoter DNA was immunoprecipitated by an anti‐STAT3 antibody (Figure 4A). Moreover, we created pGL3‐WT‐hTERT‐promoter and pGL3‐MUT‐hTERT‐promoter located at −3308/−3316 bp. The wild‐type hTERT promoter demonstrated low activity in dose‐dependent manner after inhibition of STAT3 activity and the mutation could abolish the inhibitory effect of STAT3 inhibition on luciferase activity (Figure 4B). These data indicate that STAT3 regulates hTERT at the transcriptional level.
Figure 4.
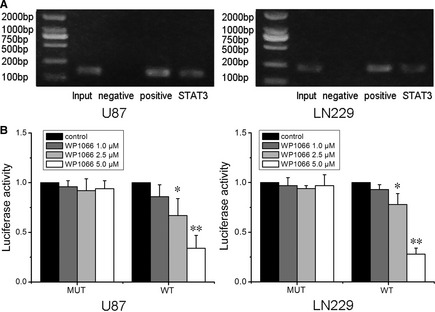
Human telomerase reverse transcriptase (hTERT) is a direct target of STAT3. (A) Chromatin immunoprecipitation analysis was performed on cell lysates using equal portions of anti‐STAT3, anti‐RNA polymerase II (positive control), and IgG (negative control) as described in Materials and methods. Input samples were DNAs amplified from lysates before immunoprecipitation. Results for binding site in the hTERT promoter were shown. (B) pGL3‐WT‐hTERT‐promoter and pGL3‐MUT‐hTERT‐promoter reporters were transfected into cells, which were then treated with WP1066 (1.0, 2.5 and 5.0 μM). Luciferase activity was determined 48 h after transfection. Luciferase activity of control group was normalized as 1 in WT group and MUT group, respectively. Error bars represent standard deviation and were obtained from three independent experiments. n = 3. *P < 0.05 as compared with the control group. **P < 0.01 as compared with the control group.
MiR‐21 Modulates hTERT in a STAT3‐Dependent Manner
Having demonstrated STAT3 as a mediator between miR‐21 and hTERT, we next examined the importance of STAT3 in miR‐21‐mediated hTERT regulation. Enforced expression of miR‐21 by miR‐21 oligonucleotide could increase hTERT expression. However, inhibition of STAT3 activity by WP1066 abrogated miR‐21‐induced hTERT expression in both U87 and LN229 cells (Figure 5A). Furthermore, we treated GBM cells using miR‐21 antisense oligonucleotide and IL‐6 which could obviously activate STAT3. Consequently, STAT3 activation overrode hTERT repression induced by As‐miR‐21 (Figure 5B). These findings demonstrate that STAT3 is a dependent mediator involved in hTERT regulation of miR‐21.
Figure 5.
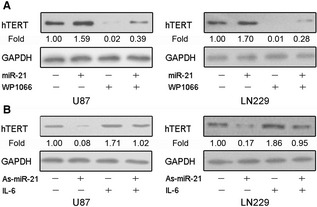
MiR‐21 modulates human telomerase reverse transcriptase (hTERT) in a STAT3‐dependent manner. (A) Cells transfected with As‐miR‐21 were treated with IL‐6 which could elevate the expression of STAT3, and hTERT expression was measured by Western blot analysis. (B) Cells transfected with miR‐21 were treated with WP1066, and hTERT expression was measured by Western blot analysis. GAPDH was regarded as an endogenous normalizer. n = 3.
As‐miR‐21 Inhibits GBM Xenograft Growth
As miR‐21 is frequently elevated in GBM and plays an important role in cell survival, we further examined the effects of knockdown of miR‐21 on tumor growth using a LN229 glioma xenograft model. As shown in Figure 6A, tumors continued growing in both scramble and control groups. However, As‐miR‐21 significantly reduced tumor growth (P < 0.05). Immunohistochemical staining analysis revealed that STAT3 and hTERT levels were downregulated in As‐miR21 group (Figure 6B), confirming the data in vitro.
Figure 6.
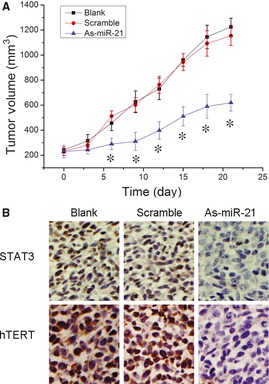
Downregulation of miR‐21 inhibits glioblastoma xenograft growth. (A) When subcutaneous tumors were established, As‐miR‐21 was injected in a multisite injection manner. Tumor volumes were measured every 3 days during treatment. (B) The expression levels of STAT3 and human telomerase reverse transcriptase were tested by immunohistochemistry analyses of xenograft tumors after As‐miR‐21 treatment. *P < 0.05 as compared with the control group.
Discussion
Glioblastoma is one of the most common forms in brain malignant tumors. Despite the comprehensive therapy of surgical resection combined with chemotherapy and radiotherapy, the median survival has been only 9–12 months yet. Thus, there are urgent needs to develop novel therapeutic approaches by targeting the molecules that are altered in this dismal disease. Recently, the discovery of miRNAs has broadened our understanding of the mechanisms involved in tumorigenesis. Here, we showed that reduction of miR‐21 repressed hTERT expression in a STAT3‐dependent fashion, subsequently inhibited GBM cell growth. These findings provided new insights into glioma research and therapeutic strategies for malignant gliomas.
Telomerase activity is essential to maintain the integrity of the replicating tumor cell and establish immortality and thus is required for the survival of the large majority of tumor cells 12, 13. Telomerase expression has been detected in more than 90% of malignant tumors including gliomas, but is absent in most normal somatic tissues 14, 15, 16. Three core components of the telomerase holoenzyme are hTERT, the human telomerase RNA, and the telomerase‐associated protein (TP1). hTERT is catalytic subunit of telomerase, and the telomerase activity is predominantly regulated by hTERT 17, 18. In this study, hTERT siRNA inhibited cell proliferation and induced cell cycle G0/G1 phase arrest and cell apoptosis in GBM cells, consistent with the previous studies. However, little investigations into the correlation between miRNA and telomerase in cancer cells have been previously reported. In thyroid carcinoma cell lines, a tendency for an inverse correlation between miR‐138 and hTERT protein expression was observed, and miR‐138 regulated hTERT expression by targeting hTERT mRNA 3′UTR 19. Our data showed that repression of miR‐21 could inhibit hTERT protein and mRNA expression in GBM cells.
As hTERT expression is specific to cancer cells and tightly associated with telomerase activity, numerous researchers have focused on the cancer‐specific regulation of hTERT 20, 21, 22. By searching the database TFSEARCH, hTERT promoter has several transcription factor binding sites such as NFkB, c‐Myc, SP1, AP1, and STAT3. In human neuroblastoma cells, NFkB becomes functionally activated after ionizing radiation (IR) and mediates telomerase activity upregulation by binding to the gene hTERT promoter region. Consistently, elimination of the NFkB‐recognition site on the telomerase promoter compromises IR‐induced telomerase promoter activation 23. Our data demonstrate that hTERT is directly regulated by STAT3 at the transcriptional level, which is aberrantly activated in human GBM tissues, in line with the previous data 24. Moreover, activation of STAT3 activity abrogates miR‐21‐induced hTERT expression whereas inactivation of STAT3 activity overrides As‐miR‐21‐repressed hTERT expression, suggesting that miR‐21 regulates hTERT expression in a STAT3‐dependent fashion in GBM cells.
In summary, to our knowledge, it is a first time to report that miRNA can modulate hTERT expression in GBM cells. Knockdown of miR‐21 targets hTERT expression in a STAT3‐dependent manner thereby inhibits GBM cell growth. Our findings suggest that modulation of the mechanism involved in hTERT regultaion of miR‐21 could provide a new important therapeutic strategy for GBM treatment and warrants further investigation.
Conflict of Interest
The authors have no conflict of interest.
Acknowledgments
This work was supported by China National Natural Scientific Fund (81101901, 81172389 and 81072078) and National High Technology Research and Development Program 863 (2012AA02A508).
The first two authors contributed equally to this work.
References
- 1. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 2006;6:857–866. [DOI] [PubMed] [Google Scholar]
- 2. Hatley ME, Patrick DM, Garcia MR, Richardson JA, Bassel‐Duby R, van Rooij E, Olson EN. Modulation of K‐Ras‐dependent lung tumorigenesis by MicroRNA‐21. Cancer Cell 2010;18:282–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Terao M, Fratelli M, Kurosaki M, et al. Induction of miR‐21 by retinoic acid in estrogen receptor‐positive breast carcinoma cells: Biological correlates and molecular targets. J Biol Chem 2011;286:4027–4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang P, Zou F, Zhang X, et al. microRNA‐21 negatively regulates Cdc25A and cell cycle progression in colon cancer cells. Cancer Res 2009;69:8157–8165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chan JA, Krichevsky AM, Kosik KS. MicroRNA‐21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res 2005;65:6029–6033. [DOI] [PubMed] [Google Scholar]
- 6. Zhou X, Ren Y, Moore L, et al. Downregulation of miR‐21 inhibits EGFR pathway and suppresses the growth of human glioblastoma cells independent of PTEN status. Lab Invest 2010;90:144–155. [DOI] [PubMed] [Google Scholar]
- 7. Chen Y, Liu W, Chao T, et al. MicroRNA‐21 down‐regulates the expression of tumor suppressor PDCD4 in human glioblastoma cell T98G. Cancer Lett 2008;272:197–205. [DOI] [PubMed] [Google Scholar]
- 8. Gabriely G, Wurdinger T, Kesari S, Esau CC, Burchard J, Linsley PS, Krichevsky AM. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol 2008;28:5369–5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Iwamaru A, Szymanski S, Iwado E, et al. A novel inhibitor of the STAT3 pathway induces apoptosis in malignant glioma cells both in vitro and in vivo. Oncogene 2007;26:2435–2444. [DOI] [PubMed] [Google Scholar]
- 10. Lederle W, Depner S, Schnur S, et al. Mueller MMIL‐6 promotes malignant growth of skin SCCs by regulating a network of autocrine and paracrine cytokines. Int J Cancer 2011;128:2803–2814. [DOI] [PubMed] [Google Scholar]
- 11. Zhang CZ, Zhang JX, Zhang AL, et al. MiR‐221 and miR‐222 target PUMA to induce cell survival in glioblastoma. Mol Cancer 2010;9:229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu JP, Chen W, Schwarer AP, Li H. Telomerase in cancer immunotherapy. Biochim Biophys Acta 2010;1805:35–42. [DOI] [PubMed] [Google Scholar]
- 13. Chen H, Li Y, Tollefsbol TO. Strategies targeting telomerase inhibition. Mol Biotechnol 2009;41:194–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shay JW, Bacchetti S. A survey of telomerase activity in human cancer. Eur J Cancer 1997;33:787–791. [DOI] [PubMed] [Google Scholar]
- 15. You Y, Pu P, Peng Q, Xia Z, Huang Q, Wang C, Wang G. Telomerase activity and regulation in human neuroepithelial tumors. Zhonghua Wai Ke Za Zhi 2002;40:90–93. [PubMed] [Google Scholar]
- 16. Boldrini L, Pistolesi S, Gisfredi S, et al. Telomerase activity and hTERT mRNA expression in glial tumors. Int J Oncol 2006;28:1555–1560. [DOI] [PubMed] [Google Scholar]
- 17. You Y, Geng X, Zhao P, et al. Evaluation of combination gene therapy with PTEN and antisense hTERT for malignant glioma in vitro and xenografts. Cell Mol Life Sci 2007;64:621–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. George J, Banik NL, Ray SK. Combination of hTERT knockdown and IFN‐gamma treatment inhibited angiogenesis and tumor progression in glioblastoma. Clin Cancer Res 2009;15:7186–7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mitomo S, Maesawa C, Ogasawara S, et al. Downregulation of miR‐138 is associated with overexpression of human telomerase reverse transcriptase protein in human anaplastic thyroid carcinoma cell lines. Cancer Sci 2008;99:280–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Strati A, Papoutsi Z, Lianidou E, Moutsatsou P. Effect of ellagic acid on the expression of human telomerase reverse transcriptase (hTERT) alpha+beta+ transcript in estrogen receptor‐positive MCF‐7 breast cancer cells. Clin Biochem 2009;42:1358–1362. [DOI] [PubMed] [Google Scholar]
- 21. Kumari A, Srinivasan R, Wig JD. Effect of c‐MYC and E2F1 gene silencing and of 5‐azacytidine treatment on telomerase activity in pancreatic cancer‐derived cell lines. Pancreatology 2009;9:360–368. [DOI] [PubMed] [Google Scholar]
- 22. Li W, Li L, Liu Z, et al. Expression of the full‐length telomerase reverse transcriptase (hTERT) transcript in both malignant and normal gastric tissues. Cancer Lett 2008;260:28–36. [DOI] [PubMed] [Google Scholar]
- 23. Aravindan N, Veeraraghavan J, Madhusoodhanan R, Herman TS, Natarajan M. Curcumin regulates low‐linear energy transfer gamma‐radiation‐induced NFkappaB‐dependent telomerase activity in human neuroblastoma cells. Int J Radiat Oncol Biol Phys 2011;79:1206–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Konnikova L, Simeone MC, Kruger MM, Kotecki M, Cochran BH. Signal transducer and activator of transcription 3 (STAT3) regulates human telomerase reverse transcriptase (hTERT) expression in human cancer and primary cells. Cancer Res 2005;65:6516–6520. [DOI] [PubMed] [Google Scholar]


