Summary
Aims
To study the role of curcumin on glioma cells via the SHH/GLI1 pathway in vitro and vivo.
Methods
The effects of curcumin on proliferation, migration, apoptosis, SHH/GLI1 signaling, and GLI1 target genes expression were evaluated in multiple glioma cell lines in vitro. A U87‐implanted nude mice model was used to study the role of curcumin on tumor volume and the suppression efficacy of GLI1.
Results
Curcumin showed cytotoxic effects on glioma cell lines in vitro. Both mRNA and protein levels of SHH/GLI1 signaling (Shh, Smo, GLI1) were downregulated in a dose‐ and time‐dependent manner. Several GLI1‐dependent target genes (CyclinD1, Bcl‐2, Foxm1) were also downregulated. Curcumin treatment prevented GLI1 translocating into the cell nucleus and reduced the concentration of its reporter. Curcumin suppressed cell proliferation, colony formation, migration, and induced apoptosis which was mediated partly through the mitochondrial pathway after an increase in the ratio of Bax to Bcl2. Intraperitoneal injection of curcumin in vivo reduced tumor volume, GLI1 expression, the number of positively stained cells, and prolonged the survival period compared with the control group.
Conclusion
This study shows that curcumin holds a great promise for SHH/GLI1 targeted therapy against gliomas.
Keywords: Apoptosis, Curcumin, GLI1, Malignant glioma, Proliferation
Introduction
To date, the etiology of malignant gliomas is not fully understood. However, more and more evidences indicate that it is associated with multistep process involving accumulation of aberrant activation in signaling pathways such as AKT, NF‐kB, ERK, mTOR, JAK/STAT3, and SHH/GLI1 1, 2, 3. Clement et al. 4 demonstrated that SHH/GLI1 signaling played an important role on tumorigenicity of the human gliomas and their cancer stem cells. In addition, SHH induces the transcription of important transcription factor glioma‐associated oncogene homolog 1 (GLI1). Activation and widespread expression of GLI1 in gliomas are consistent with its isolation from a glioma cell line 5 and its expression in brain tumors in situ 6. Furthermore, with the activation of SHH/GLI1 signaling pathway, the expression of multiple important GLI1‐regulated oncoproteins, such as N‐myc, CyclinD1, Foxm1, and Bcl‐2, changed significantly 7. The oncoproteins played important roles on cell cycle, survival, invasion, and cancer stem cell self‐renewal. Thus, the biological behavior of glioma cells may be suppressed through the inhibition of SHH/GLI1 signaling pathway. Currently, several blockers, including cyclopamine, SANT1, GANT58, and GDC‐0449, have some limited therapeutic effect. However, they also show different levels of side effects. Therefore, it is a pressing need to develop and test of the new alternative chemotherapeutic agents. Curcumin (Figure 1A), the active ingredient of turmeric, has been shown to have multiple anticancer effects including inhibition of cell proliferation, angiogenesis, and induction of apoptosis 8. Both in vivo and in vitro studies have demonstrated that curcumin inhibit cancer growth significantly 9, 10. Treatment of human GBM cells with curcumin in cell cultures or after xenografting in immunocompromised mice resulted in growth suppression, cell cycle arrest, and induction of apoptosis or autophagy 11, 12. These anticancer effects were attributed to curcumin's effect on the NF‐kB, ERK, AKT, or mTOR signaling pathways. However, despite the fact that GLI1 is constitutively activated in the majority of malignant gliomas, no study has been performed to test the possible effect of curcumin on SHH/GLI1 signaling pathway. In this study, we have revealed that the effect of curcumin on various glioma cell lines was through inhibiting the SHH/GLI1 signaling pathway and GLI1‐regulated proteins.
Figure 1.
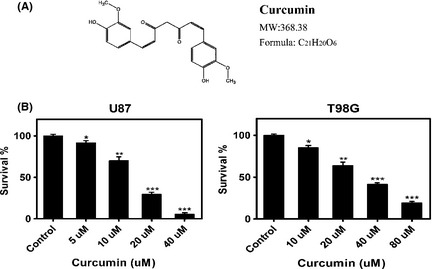
Cytotoxic effect of curcumin on human glioma cells. (A) Chemical structure and molecular weight of curcumin. (B) U87 and T98G cells were cultured in‐96‐well plates, respectively, and treated with increasing concentrations of curcumin for 24 h. Cell death was analyzed using MTT assay. The error bars represent standard error. Data are shown as mean ± SEM for the three replicates. Statistical significance levels are indicated as: *P < 0.05; **P < 0.01 and ***P < 0.001.
Materials and Methods
Cell Lines and Culture Conditions
Human glioma cell lines (U87, T98G) were purchased from Chinese Academy of Sciences Cell Bank. All cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS) and 1% antibiotic at 37°C in a humidified atmosphere with 5% CO2 and 95% air. The cells were routinely passaged at 2‐ to 3‐day intervals.
Cytotoxicity Assay
The cells were seeded into 96‐well plates at 5000–8000 cells/well and incubated overnight. Fresh medium containing various concentrations of curcumin (Sigma‐Aldrich, St. Louis, MO, USA) was used. After 20 h, 20 ul per well of MTT dye (5 mg/mL) was added for another 4 h. Subsequently, the medium was removed completely and added with 150 ul DMSO into each well with gently concussion for 10 min. The amount of formazan was quantified using IMARK microplate reader at 490 nm of absorbance.
Cell Proliferation Assay
The MTT assay was used to determine relative cell growth as above. Fresh medium containing various concentrations of curcumin was used for different time intervals (0, 24, 48, and 72 h). Except for the 0 h, MTT solution (Sigma‐Aldrich) was added. Then, the medium was removed and added with DMSO for each well. The amount of formazan was quantified using IMARK microplate reader at 490 nm of absorbance.
Colony Formation Assay
Briefly, cells were seeded in 6‐well plates (0.5 × 10 3cells per well) and allowed to grow for 24 h. Then, the cells were treated with or without various concentration curcumin for another 24 h. The medium containing curcumin was then removed, and the cells were washed in PBS and cultured for an additional 12 days in complete medium. The colonies obtained were washed with PBS and fixed in 10% formalin for 10 min at room temperature and then stained with Giemsa stain. The colonies were counted and compared with untreated cells. Each treatment was performed in triplicate.
Migration (scratch) Assay
U87 and T98G cells were plated in 6‐well plates (corning). When the cells had reached subconfluency, mitomycinC (10 mg/mL) was added to the medium for 2 h and a scratch was made using a sterile pipette tip. The medium was changed with DMEM supplemented with 2% FBS plus treatment. Photographs of the scratched area were taken at 0 h and after 24 h using a Axiovert 200 microscope (Carl Zeiss), and the number of cells that had migrated over the margin of the scratch was counted. The scratch was captured in six different photographs, and the numbers of migrated cells were averaged. Experiments were repeated three times.
Cell Cycle and Apoptosis Assays
U87 and T98G cells were plated in 6‐well plates, and when the confluence reached 70–80%, curcumin was added to the medium at various concentrations. After 24 h, cells were harvested and fixed in 70% (v/v) ethanol. Then, 50 mg/mL PI (Sigma‐Aldrich) and 10 mg/mL RNase A (Sigma‐Aldrich) were added for 30 min. The cells used for apoptosis analysis were resuspended in binding buffer. Then, 10 ul of FITC annexin V (BD Pharmingen, San Jose, CA, USA) and 5 μL PI (BD Pharmingen) were added and incubated for 15 min. Then, the stained cells were analyzed by flow cytometry (FACSCanto II, BD Biosciences,USA).
RNA Extraction, cDNA Synthesis
Total RNA was extracted from U87 and T98G cell lines using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The first strand cDNAs were synthesized using PrimeScript RT reagents Kit (Perfect Real Time, TaKaRa, Japan) as the manufacturer's instructions. The reverse transcription reaction was carried out at 37°C for 15 min and then inactivated at 85°C for 5 seconds.
Quantitative Real‐time PCR
Quantitative real‐time polymerase chain reaction (qRT‐PCR) was performed in triplicate in LightCycler2.0 (Roche Diagnostics company, CH) and normalized with glyceraldehyde 3‐phosphatedehydrogenase (GAPDH) as endogenous control. Endogenous mRNA levels of SHH, SMO, GLI1 were determined using SYBR® PrimeScript RT‐PCR Kit (TaKaRa, Japan) in accordance with the manufacturer's instructions. The real‐time PCR primers were as follows: Shh: sense 5′‐TGCTGCTAGTCCTCGTCTCCT‐3′; antisense 5′‐TTTTGGGGTGCC‐ TCCTCTT‐3′; Smo: sense 5′CTTCAGCTGCCACTTCTACGACTTC‐3′; antisense: 5′‐TCGGGCGATTCTTGATCTCAC‐3′; GLI1: sense 5′‐GAGCACGAGGGCTGCA GTAA‐3′; antisense 5′‐TCGCAGCGAG CTAGGATCTGTA‐3′; GAPDH: sense5′‐ ACGACCACTTTGTCAAGCTC‐3′; antisense 5′‐ GGTCTACATGGCAACTGAGA ‐3′.
Western Blot Analysis
Treated and untreated U87 and T98G cells were scraped in Thermo scientific RIPA buffer (Pierce, USA) with protease inhibitors. After centrifugation, lysate was separated in 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and the gel was blotted onto PVDF membrane (Millipore, USA). The membrane was blocked in 5% milk–TBST solution and incubated separately with rabbit antihuman Shh (1:500, Bioss Inc, USA), rabbit antihuman GLI1 (1:1000, Cell Signaling Technology, USA), mouse antihuman CyclinD1 antibody (1:300, Santa Cruz, USA),rabbit antihuman Bcl‐2(1:500, Santa Cruz, USA), rabbit antihuman Foxm1(1:1000, Cell Signaling Technology, USA), rabbit antihuman Bax (1:500, Santa Cruz, USA), mouse antihuman pro‐caspase‐9 (1:500, Santa Cruz, USA), rabbit antihuman pro‐caspase‐3 (1:500, Santa Cruz, USA), or mouse antihuman GAPDH antibody (1:1000, Sigma, USA). Following incubation with HRP‐labeled secondary antibody (Introvigen), protein bands were detected with Fujifilm Las‐4000.
Immunofluorescence
The human glioma cell lines T98G (1 × 105 per well) were grown on glass coverslips overnight. Then, they were treated with curcumin (20 uM) or DMSO for 24 h. The cells were fixed in 2% paraformaldehyde, permeabilized, blocked, and incubated with anti‐GLI1 antibody (1:200; Bioss Inc, Woburn, MA, USA). Following washing, the cells were incubated with Alexa594 labeled secondary antibody. The slides were analyzed by confocal laser scanning microscopy (Olympus FV1000 Laser Scanning Microscope, Japan).
GLI1 Reporter Assay
To determine GLI activity, a reporter was kindly provided by Gregory J. Gores, M.D. (Division of liver pathobiology, Mayo Clinic, Rochester, MN) containing eight directly repeated copies of a consensus GLI1 binding site (8×‐GLI1) downstream of the luciferase gene was used. The plasmid was transfected into U87 or T98G cells (0.8 ug/well) using Lipofectamine 2000 reagent (Invitrogen, USA). Cells were cotransfected with 50 ng of a plasmid expressing Renilla luciferase under the control of cytomegalovirus promoter (pGL4.75 Vector; Promega, Madison, WI ,USA). Twenty‐four hours after transfection, cells were treated with and without curcumin for 24 h, as indicated. Then, cell lysates were prepared, and both firefly and Renilla luciferase activities were quantified using the Dual‐Luciferase Reporter Assay System (Promega). Firefly luciferase activity was normalized to Renilla luciferase activity to control for transfection efficiency and cell numbers. Data were expressed as fold increase over solvent‐treated (Con) cells transfected with the 8×‐GLI1/pRL‐CMV reporter constructs.
Tumor Xenograft Study
Briefly, U87 cells(3 × 105 cells per mouse in 3 ul) transfected with luciferase lentivirus (U87‐luc) were injected into the intracranial of 5‐week‐old female nude mice(n = 14) as described earlier 13. The next day, the mice were randomly distributed into two groups following by intraperitoneal injection with curcumin (60 mg/kg, n = 7) or solvent DMSO (Control, n = 7). The curcumin was intraperitoneal injected every day. After 20 days, tumors were measured by bioluminescence of whole mice using an IVIS Lumina Imaging System (Xenogen). When the mice showed typical clinical symptoms or significant weight lost following by frail and in low spirits or at the end of the observation period (40 days), blood samples were collected before being sacrificed and portions of the tumor tissues were used for quantitative real‐time PCR and Western blot assay for GLI1 expression. Liver, lung, heart, and kidney were also removed and fixed in formalin. Cryosections (4 mm) were stained with hematoxylin and eosin and used for immunohistochemical (IHC). These procedures were carried out following approval by the Harbin Medical University Institutional Animal Care and Use Committee.
Immunohistochemical (IHC) Analysis of Xenograft Tumors
Briefly, the tumor tissues were deparaffinaged, rehydrated, treated with 0.3% hydrogen peroxide, and processed for antigen retrieval using heat induction for about 10 min. Then, the samples were processed for staining with anti‐GLI1 antibody (1:200, bs‐1206R, Bioss Inc, USA). GLI1 expression was detected using peroxidase‐conjugated AffiniPure goat anti‐rabbit IgG (ZSGB‐BIO) and 3,3′‐ diaminoben‐zidine (DAB substrate kit, ZSGB‐BIO). Then, the slides were dehydrated, mounted, and visualized using an Olympus CX31 Microscope (Olympus Corporation, Japan). Immunohistochemical analysis was performed as previously described 14.
Statistical Analysis
Results with mean and SEM in this study were calculated using GraphPad Prism software (version 5.0). Comparisons of each treatment data with control were carried out for statistical difference by the paired t‐test. The survival curves were analyzed using the log‐rank test employing the Prism GraphPad software. Statistical significance was assumed at a value of P < 0.05.
Results
Curcumin had Cytotoxic Effects on Malignant Glioma Cells
We investigated the cytotoxic effect of curcumin (Figure 1A) on different malignant glioma cells using the MTT assay. Figure 1B showed dose‐dependent effect of curcumin on two different glioma cell lines (U87 and T98G). The sensitivity to the agent between the two glioma cell lines was different. The medial IC50 was 15 uM and 31 uM for the U87 and T98G, respectively. These results showed that curcumin was cytotoxic against malignant glioma cell lines. Therefore, in subsequently study, we separately chose two doses of curcumin for each cell line (10 uM, 20 uM for U87 and 20 uM, 40 uM for T98G).
Curcumin Suppressed the Core Components and GLI1‐dependent Target Oncoproteins in SHH/GLI1 Signaling Pathway
There are evidence that glioma tumorigenesis has an association with the SHH/GLI1 signaling pathway 4, 15. To investigate whether curcumin influences the SHH/GLI1 pathway and GLI1‐dependent target oncoproteins, glioma cells (T98G) were treated with 40 uM curcumin for different time period (0, 8, 16, 24 h) and then analyzed for SHH, SMO, and GLI1 mRNA expression. It was found that the expression of all three genes decreased significantly in a time‐dependent manner after treatment (Figure 2A). Western blot analysis further verified that curcumin treatment downregulated SHH and GLI1 expressions in a time‐dependent manner (Figure 2B). Likewise, Cyclind1, Bcl‐2, and Foxm1, the downstream factors of GLI1 oncoproteins were inactivated in curcumin‐treated group (Figure 2B). Subsequently, both cell lines were treated with various concentrations of curcumin or with solvent DMSO only (Control) for 24 h. The oncoproteins expression of SHH/GLI1 signaling pathway were then analyzed. These data revealed that almost all the genes showed a dose‐dependent response to the curcumin treatment (Figure 2C), suggesting that curcumin downregulating the core components of the SHH/GLI1 signaling pathway and GLI1‐dependent target oncoproteins in a time‐ and dose‐dependent manner.
Figure 2.
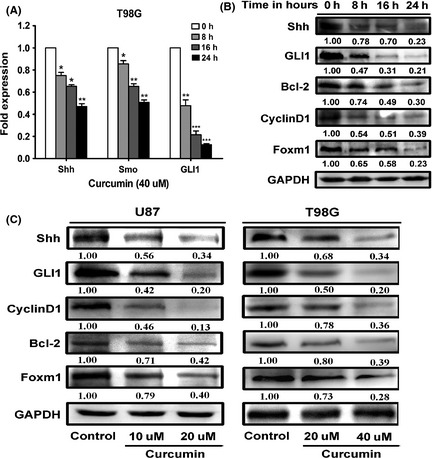
Curcumin inhibited SHH/GLI1 signaling and its downstream targets oncoproteins in time‐ and dose‐dependent manner. (A) Real‐time everse transcription‐PCR analysis of total RNA from T98G cells following 40 uM of curcumin treatment for different time points (0, 8, 16, 24 h) revealed decreasing expression of Shh, Smo, and GLI1 on mRNA level. (B) Western blot assay showed curcumin treatment led to decreased levels of Shh, GLI1, and GLI1‐regulated target (CyclinD1, Bcl‐2, Foxm1) proteins in time‐dependent manner in T98G cells following 40 uM agent exposure for 24 h. (C) U87 and T98G cells were plated and grown for 24 h and then treated with the solvent DMSO (control) or with different (indicated) doses of curcumin dissolved in DMSO for 24 h before harvesting. Western blot assay shows curcumin inhibits core components of the SHH/GLI1 signaling (SHH, GLI1) and its target oncoproteins (CyclinD1, Bcl‐2, Foxm1) in dose‐dependent manner. Data are shown as mean ± SEM for the three replicates. Statistical significance levels are indicated as: *P < 0.05; **P < 0.01 and ***P < 0.001.
Curcumin Treatment Inactivated Transcription Activities of GLI1 and Decreased GLI1 Expression Level at the Cell Nucleus and Cytoplasma
Activating the SHH/GLI1 pathway enriched the GLI1′s level and promoted GLI1 transportation into the cell nucleus. Nuclear staining of GLI1 was a reliable indicator of HH pathway activation as it bound to a consensus GLI1‐binding element in target genes for activation 16. To determine the effect of curcumin on the SHH/GLI1 signaling pathway, T98G cells were treated with curcumin (20 uM), and the GLI1 expression level and localization were analyzed under confocal microscopy. As shown in Figure 3A, curcumin decreased the GLI1's expression level significantly at the cell nucleus and cytoplasma compared with control cells. Subsequently, the 8×‐GLI1 reporter was transfected into U87 and T98G cell lines. After treatment of curcumin, we found that the luciferase activity also decreased significantly compared with control cells (Figure 3B). These data revealed that curcumin suppressed GLI1 activation and prevented its translocation into cell nucleus. Moreover, curcumin treatment reduced the expression of GLI1 in both cytoplasma and nucleus.
Figure 3.
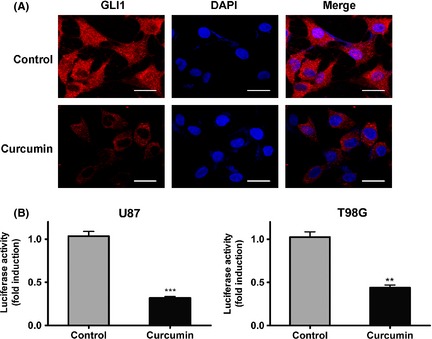
Curcumin treatment decreased GLI1 expression at the cytoplasma,nucleus and inactivated consensus GLI1‐binding sites. (A) T98G cells treated with 20 uM of curcumin for 24 h were subjected to immunofluorescent staining using anti‐GLI1 antibody. Curcumin treatment prevented most of GLI1 translocation into nucleus and resulted in lower expression levels at the cytoplasma. Bars represent 20 um. (B) U87 and T98G cells were transiently transfected (24 h) with a firefly luciferase reporter construct containing eight consecutive consensus GLI1‐binding sites (8×‐GLI1) and cotransfected with pGL4.75. Cells were then treated as indicated with solvent DMSO (control) or curcumin. Both firefly and Renilla luciferase activities were quantified, and the firefly/Renilla luciferase activities were recorded as fold induction over solvent‐treated (Con) cells transfected with the 8×‐GLI1/pGL4.75 vector reporter constructs. Mean ± SEM (n = 3; *P < 0.05).
Curcumin Inhibited Proliferation of Human Glioma Cells and induced G2/M Arrest
This study shows that curcumin is a potent inhibitor of SHH/GLI1 signaling and GLI1‐regulated oncogenes in U87 and T98G glioma cells. To examine the effect of SHH/GLI1 signaling inactivation on cell proliferation, MTT assay was performed to evaluate the potential restraining effect of curcumin on cell growth in the two glioma cell lines (Figure 4A). Due to the different sensitivity to the agent, the two cells were exposed to increasing doses of curcumin (5, 10, 20, 40 umol/L in U87 and 10, 20, 40, 80 umol/L in T98G) for test samples or solvent DMSO only (control) for different time periods (0, 24, 48, 72 h). Both the cell lines responded to the curcumin treatment in a dose‐and time‐dependent manner. Similarly, colony formation was suppressed by curcumin treatment in both glioma cell lines (Figure 4B), suggesting that curcumin's effect on the tumor cells was irreversible. To further understand the mechanism of the growth inhibitory effect of curcumin in human glioma cells, we performed the cell cycle analysis in U87 and T98G cells. The data showed that different doses of curcumin gradually induced a significant G2/M phase arrest in glioma cells following a 24 h exposure (Figure 5A). All these data suggested that curcumin could inhibit glioma cells proliferation, colony formation, and induce a G2/M phase arrest mediated via interfering with SHH/GLI1 signaling.
Figure 4.
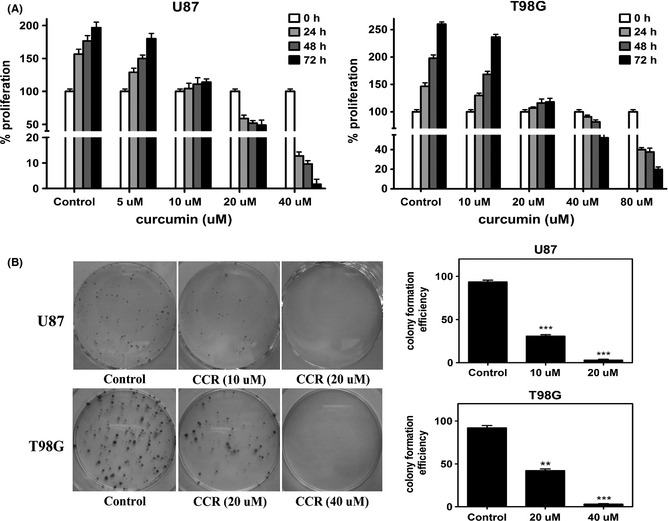
Curcumin suppressed proliferation of U87 and T98G cells. (A) Cells were plated and grown for 24 h and then treated with solvent only (Con) or increasing doses of curcumin as indicated and harvested after 24, 48, and 72 h. Cell viability was examined using the MTT assay. Curcumin treatment resulted in significant decrease dose‐ and time‐dependent manner in cell proliferation in both two cell lines when compared with untreated controls. (B) Curcumin inhibited colony formation. Human glioma cells were incubated with indicated concentrations of curcumin for 24 h and allowed to grow into colonies for 12 days. Incubation with curcumin inhibits colony formation. Data are mean ± SEM for the three replicates. *P < 0.05; **P < 0.01 and ***P < 0.001.
Figure 5.
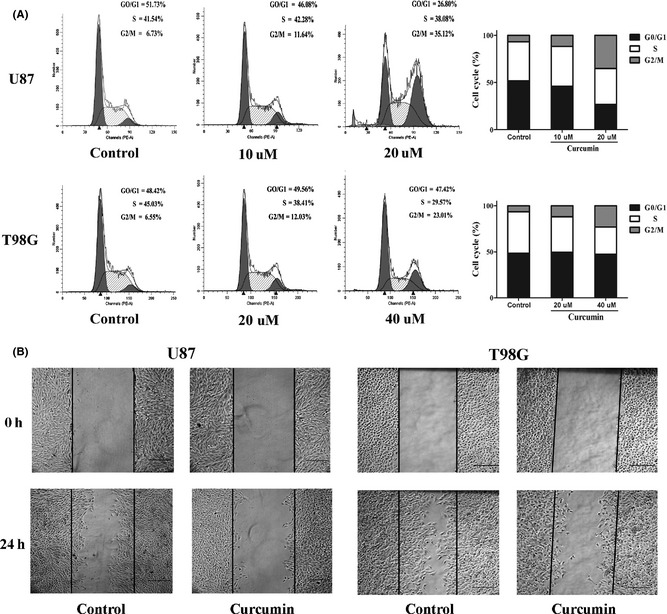
Curcumin‐induced G2/M phase arrest and restrained the migratory potential of glioma cells. (A) Cells were plated and grown in 6‐well plate for 24 h and then treated with solvent only (Con) or the indicated concentrations of curcumin for 24 h. Results were examined by flow cytometry following propidium iodide staining for DNA content. Curcumin treatment induced a significant increase in the percentage of cells in G2/M phase in two cell lines. Histograms and cell cycle distributions shown are representative of three independent experiments. (B) Cell migration was analyzed using the wound‐healing assay. A single scratch was made in the confluent monolayer. The scratch was photographed at 0 h and after 24 h treatment with solvent (Con) or with the indicated doses of curcumin. Cells those migrated into the scratch were counted. Cell migration was restrained by the curcumin treatment compared with control group. *P < 0.05, significant difference between groups. Bars represent 250 um.
Curcumin Restrained the Migration of Glioma Cells
Both U87 and T98G were malignant glioma cell lines which presented highly invasive growth characteristics in vitro and vivo. In accordance with this, downstreams of GLI1‐regulated genes showed a very strong characteristics of promoting motility and invasiveness of cancer cells 17. To test whether the inhibition of SHH/GLI1 signaling by curcumin treatment could influence cell migration in vitro, a wound‐healing assay was performed. The results displayed that curcumin treatment also attenuated the migration of U87 and T98G cells compared with controls (Figure 5B).
Curcumin‐induced Apoptosis of Human Glioma Cells Partly Through the Mitochondrial Pathway
As inhibition of SHH/GLI1 signaling pathway could enhance apoptosis in brain glioma cell lines 18 and the downstream of GLI1‐regulated genes (such as bcl‐2 and Foxm1) also had strong correlation with cell survival 19, 20, we performed the Annexin V/PI staining to verify whether curcumin treatment could induce apoptosis. Flow cytometric analysis revealed that curcumin induced the apoptosis of both glioma cells in a dose‐dependent manner (Figure 6A,B). Subsequently, to confirm the apoptotic route of curcumin‐dependent Shh inhibition activation, the T98G cells were treated with 40 uM of curcumin and harvested after different time periods (0, 8, 16, and 24 h). First, the effect of curcumin on caspase‐3 was examined. Figure 6C shows that the level of caspase‐3 increases significantly after curcumin treatment. Then, we investigated the expressions of Bax and Bcl‐2 proteins and found that both of them altered in a time‐dependent manner. The trend of changes was opposite. The level of Bax gradually increased. The level of Bcl‐2 decreased after curcumin treatment. The Bax/Bcl‐2 ratio (Figure 6D) also gradually increased. To further confirm this, caspase‐9 was also detected. The results show that the expression of caspase‐9 increases in a time‐ dependent manner (Figure 6C). Summing up the above results, we find that inhibition of SHH/GLI1 signaling by curcumin may act as a novel mechanism of the apoptosis.
Figure 6.
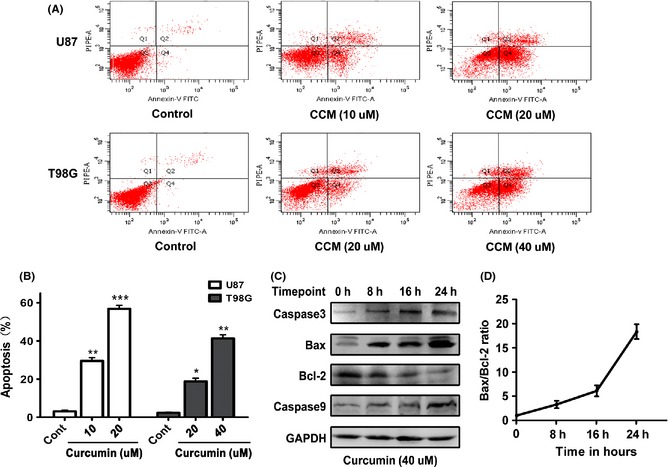
Curcumin triggered apoptosis through the internal pathway in glioma cells. (A) Subconfluent cells were either solvent DMSO‐treated (Con) or challenged with the indicated concentrations of curcumin for 24 h, and then, cell apoptosis was examined by flow cytometry assay using the Annexin V/PI. A representative experiment of the three replicated assays is shown. (B) Histogram representing dose‐dependent apoptosis of the U87 and T98G cell lines. Data are mean ± SEM of three replicates. *P < 0.05; **P < 0.01; ***P < 0.001. (C) The T98G cells were treated with 40 uM curcumin for the indicated periods of time, and then, cell extracts were prepared and used for Western blot assay using the indicated antibodies. Results showed that Bcl‐2 decreased following the increasing time points. Others were increased in a time‐dependent style. (D) Graph showing the Bax/Bcl‐2 ratio. Error bars represent standard error of three different experiments.
Curcumin Inhibited GBM Growth in Vivo through SHH/GLI1 Signaling and Prolonged the Survival Period
To investigate whether antiglioma effect of curcumin can be achieved in vivo, an intracranial glioma model on nude mice was adopted. Mice were intracranial injected with U87‐luc cells with or without the treatment of curcumin; bioluminescence imaging was performed for the whole body to evaluate the effect of curcumin on glioma tumor growth. The curcumin treatment group displayed a marked reduction in the tumor (Figure 7A); Immunohistochemical analysis showed that GLI1 expression and number of positively stained cells decreased significantly in curcumin‐treated samples compared with control; Furthermore, real‐time PCR and Western blot analysis confirmed that both the mRNA and protein level of GLI1 expression significant decreased in curcumin treatment group (Figure 7C). Besides, the treatment of curcumin reduced the influence on general health of nude mice caused by tumor (Table 1). However, the dosage of curcumin did not cause significant tissue or organ metabolic toxicity from checking the serum markers (Table 2) and histological change by H&E analysis (Suppl Figure 1). To further better evaluate the therapeutic effect of curcumin on nude mice, the survival period of each group (n = 7/group) was analyzed by Kaplan–Meier curve; the curcumin‐treated group showed a significant improvement in survival until the end of the observation period compared with the control group (P < 0.05) (Figure 7D).
Figure 7.
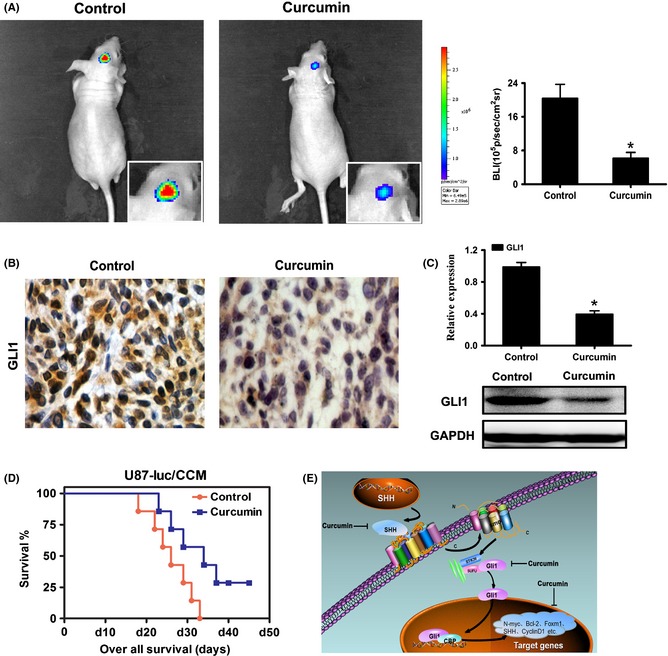
Curcumin inhibited glioma growth in xenograft mouse model through interference with the SHH/GLI1 signaling pathway and prolonged the survival period. (A) Luminescence imaging for curcumin‐treated U87‐luc tumors versus controls. (B) The effect of curcumin on GLI1 expression and localization. Tumor tissues from curcumin or control treated mice were processed for immunohistochemical staining using anti‐GLI1 antibody. Reduced staining of GLI1 was observed in curcumin‐treated mice compared with control. (C) The expression of GLI1 in intracranial graft assessed by real‐time PCR and Western blot. (D) Kaplan–Meier survival curves indicating mice with the treatment of curcumin significantly better outcome compared with controls. (E) Schematic diagram of the effect of curcumin on SHH/GLI1 signaling in glioma. *P < 0.05, significant difference between groups.
Table 1.
Comparison of general health and clinical symptoms between the curcumin treatment group and control group
| Control Group | Curcumin Group | |
|---|---|---|
| Weight loss (%) | 22.0 ± 7.3 | 12.4 ± 8.9 |
| Mental state | listless, less moving | lively, vigorous |
| Food intake (g) | 1.7 ± 0.3 | 2.3 ± 0.5 |
| Skin color | dull, rough, dry | glossy, bright |
| Focal motor deficits | 3/7 | 1/7 |
| Seizure | 2/7 | none |
| Ocular proptosis | 4/7 | 2/7 |
| Skull separating uplift | 3/7 | 1/7 |
Table 2.
Tissue damage serum markers in curcumin‐ and control‐treated U87‐luc implanted nude mice. (t‐test; P > 0.05)
| Control | Curcumin | |
|---|---|---|
| ALT (U/L) | 34 ± 7 | 31 ± 3 |
| AST(U/L) | 131 ± 11 | 138 ± 17 |
| ALP (U/L) | 182 ± 10 | 158 ± 8 |
| TP (g/L) | 55.3 ± 3.2 | 58.6 ± 3.5 |
| CHE(U/L) | 8981 ± 1222 | 7865 ± 1158 |
| BUN(mmol/L) | 6.84 ± 0.80 | 6.56 ± 1.07 |
| Crea(umol/L) | 27.3 ± 5.4 | 20.1 ± 4.1 |
| UA (umol/L) | 134 ± 28 | 116 ± 31 |
| CK (U/L) | 1439 ± 155 | 1352 ± 161 |
| LDH(IU/L) | 1124 ± 246 | 846 ± 169 |
Discussion
Until today, glioblastomas (GBM) are still the most common malignant brain tumors in human. Although neurosurgery, radiotherapy, and chemotherapy have been adopted to treat the condition, even in recent years, new chemotherapy drugs, such as temozolomide and several specific channel blockers, have been used to get better effect to control the tumor 21, 22, 23, the curative efficacy is still unsatisfactory. Therefore, there is a pressing need to search for novel strategies to develop improved therapeutics against GBM. The glioma‐associated oncogene homologue1 (GLI1) isoform was first isolated from a human GBM. GLI1 is overexpressed due to gene amplification or somatic copy number increase in glioma. Clement V et al. 4 demonstrated that the SHH/GLI1 pathway was activated in malignant gliomas. Wang et al. 24 further showed that SHH/GLI1 signaling pathway inhibition restricted cell migration and invasion in human gliomas. Therefore, GLI1 appears to be an attractive target for pharmacologic intervention. There had been several strategies to inhibit the SHH/GLI1 signaling pathway such as using siRNA 18, 25, inhibitors of specific pathway 26, and some chemotherapeutics. However, frequent drug resistance and potent side effects of these agents limited their application in the clinical applications. In contrast, curcumin is a popular dietary supplement that has been shown to have a wide variety of beneficial effects including antioxidant, anti‐inflammatory, anticancer activities, etc. 27. Furthermore, several studies have shown that curcumin could suppress cell proliferation and induce apoptosis of several cancer cells including pancreas, breast, colon, lung, leukemia, prostate carcinoma, and glioblastomas 9, 28, 29, 30, 31, 32. It is also argued that curcumin exerts antiglioma effect by inhibiting NF‐kB, Akt, JAK1,2/STAT3 signaling pathway 33, 34. However, the mechanism of these effects is not clear. In this study, we first find that the action of curcumin is associated with the SHH/GLI1 pathway. Curcumin downregulated the expressions of Shh protein, GLI1 protein, and specific target proteins in the pathways that have important roles on cell proliferation, migration, and apoptosis. Considering the importance of GLI1 in the pathway, we found that curcumin reduces intracellular and intranuclear levels of GLI1 in a dose‐dependent and time‐dependent manner in both GBM cell lines. Furthermore, MTT assay, colony formation, and tumor volume assay showed that GLI1 inhibition by curcumin was correlated with reduced cell growth in vitro and in vivo. Moreover, we found that curcumin induced an accumulation of glioma cells in the G2/M phase of the cell cycle as analyzed by DNA flow cytometer. Because GLI binds to consensus motifs within the promoter or enhancer regions such as N‐myc, CyclinD1, and Foxm1 genes 35, 36, 37, proteins those drive cell proliferation and cell cycle progression were dysregulated. We found that levels of both CyclinD1 and Foxm1 proteins decreased in a dose‐dependent and time‐dependent manner. However, the function of the two proteins is not identical. It is known that CyclinD1 dysregulation by Shh signals drives cell cycle progression at G1/S 37. Meanwhile, Foxm1 promotes cell cycle progression at G2/M phase through direct up‐regulation of CyclinB1 and Cdc25B 38. These facts indicate that SHH/GLI1 signaling promotes cellular progression through upregulating multiple cell cycle regulators during G1/S and G2/M phases. Combined with our study, we suggest that the phase arrest in G2/M after curcumin‐mediated GLI1 inhibition may be the underlying mechanism for the observed growth inhibition in human gliomas. In addition, we also demonstrated that curcumin could promote glioma cells apoptosis which was consistent with previous studies 39, 40, 41. Luthra et al. 42 demonstrated that curcumin induced Bcl‐2 mediated G2/M arrest and apoptosis in human glioma U87 cells. In the current study, we found that after interfering with SHH/GLI1 signaling by curcumin treatment, Bcl‐2 expression level decreased significantly. Meanwhile, Bcl2, an anti‐apoptotic effector, is one of the direct targets of SHH/GLI1 signaling. Subsequently, to verify whether the apoptosis was caused by inhibition of Bcl‐2 through the internal mitochondrial pathway, we also examined the expression of caspase‐3, Bax, and caspase‐9. The results showed that the underlying mechanism of induction apoptosis in human gliomas is most likely related to by curcumin‐mediated GLI1 inhibition. In addition, the efficacy of curcumin to suppress GLI1 activity was also verified by observation the reduced mRNA and protein expression in glioma cells of incranial implanted tumors compared with controls. Curcumin was injected intraperitoneally to increase absorption as suggested previously 13. Moreover, it is also possible that intraperitoneal injection of curcumin may cross the brain blood barrier 43. In our mice model experiment, curcumin significantly reduced the size of incranial tumors in U87‐implanted nude mice and decreased the expression of GLI1 in the cytoplasm and nucleus. Thus, our in vivo results were in accordance with the vitro results. In addition to this, the survival period of the curcumin‐treated mice was prolonged to some extent compared with the control. In conclusion, we demonstrated that curcumin inhibited GBM cells proliferation and induced apoptosis through SHH/GLI1 pathway (Figure 7E), indicating that curcumin may be used as a therapeutic agent in multimodal treatment of patients with glioma.
Conflict of Interest
The authors declare no conflict of interest.
Supporting information
Figure S1. Histological change by H & E analysis between the curcumin‐treated group and control group.
Acknowledgments
This work was supported by the National High Technology Research and Development Program 863 (2012AA02A508).
The first two authors contributed equally to this work.
References
- 1. Furnari FB, Fenton T, Bachoo RM, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev 2007;21:2683–2710. [DOI] [PubMed] [Google Scholar]
- 2. Soni D, King JA, Kaye AH, Hovens CM. Genetics of glioblastoma multiforme: mitogenic signaling and cell cycle pathways converge. J Clin Neurosci 2005;12:1–5. [DOI] [PubMed] [Google Scholar]
- 3. Rossi M, Magnoni L, Miracco C, et al. beta‐catenin and Gli1 are prognostic markers in glioblastoma. Cancer Biol Ther 2011;11:753–761. [DOI] [PubMed] [Google Scholar]
- 4. Clement V, Sanchez P, de Tribolet N, Radovanovic I, Ruiz i Altaba A. HEDGEHOG‐GLI1 signaling regulates human glioma growth, cancer stem cell self‐renewal, and tumorigenicity. Curr Biol 2007;17:165–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kinzler KW, Bigner SH, Bigner DD, et al. Identification of an amplified, highly expressed gene in a human glioma. Science 1987;236:70–73. [DOI] [PubMed] [Google Scholar]
- 6. Dahmane N, Sanchez P, Gitton Y, et al. The Sonic Hedgehog‐Gli pathway regulates dorsal brain growth and tumorigenesis. Development 2001;128:5201–5212. [DOI] [PubMed] [Google Scholar]
- 7. Katoh Y, Katoh M. Hedgehog target genes: mechanisms of carcinogenesis induced by aberrant hedgehog signaling activation. Curr Mol Med 2009;9:873–886. [DOI] [PubMed] [Google Scholar]
- 8. Liao S, Xia J, Chen Z, et al. Inhibitory effect of curcumin on oral carcinoma CAL‐27 cells via suppression of Notch‐1 and NF‐kappaB signaling pathways. J Cell Biochem 2011;112:1055–1065. [DOI] [PubMed] [Google Scholar]
- 9. Yang CL, Liu YY, Ma YG, et al. Curcumin blocks small cell lung cancer cells migration, invasion, angiogenesis, cell cycle and neoplasia through Janus kinase‐STAT3 signalling pathway. PLoS ONE 2012;7:e37960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zanotto‐Filho A, Braganhol E, Edelweiss MI, et al. The curry spice curcumin selectively inhibits cancer cells growth in vitro and in preclinical model of glioblastoma. J Nutr Biochem 2012;23:591–601. [DOI] [PubMed] [Google Scholar]
- 11. Weissenberger J, Priester M, Bernreuther C, et al. Dietary curcumin attenuates glioma growth in a syngeneic mouse model by inhibition of the JAK1,2/STAT3 signaling pathway. Clin Cancer Res 2010;16:5781–5795. [DOI] [PubMed] [Google Scholar]
- 12. Zhuang W, Long L, Zheng B, et al. Curcumin promotes differentiation of glioma‐initiating cells by inducing autophagy. Cancer Sci 2012;103:684–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Perry MC, Demeule M, Regina A, Moumdjian R, Beliveau R. Curcumin inhibits tumor growth and angiogenesis in glioblastoma xenografts. Mol Nutr Food Res 2010;54:1192–1201. [DOI] [PubMed] [Google Scholar]
- 14. Zhang J, Han L, Ge Y, et al. miR‐221/222 promote malignant progression of glioma through activation of the Akt pathway. Int J Oncol 2010;36:913–920. [DOI] [PubMed] [Google Scholar]
- 15. Ehtesham M, Sarangi A, Valadez JG, et al. Ligand‐dependent activation of the hedgehog pathway in glioma progenitor cells. Oncogene 2007;26:5752–5761. [DOI] [PubMed] [Google Scholar]
- 16. Kinzler KW, Vogelstein B. The GLI gene encodes a nuclear protein which binds specific sequences in the human genome. Mol Cell Biol 1990;10:634–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yoo YA, Kang MH, Kim JS, Oh SC. Sonic hedgehog signaling promotes motility and invasiveness of gastric cancer cells through TGF‐beta‐mediated activation of the ALK5‐Smad 3 pathway. Carcinogenesis 2008;29:480–490. [DOI] [PubMed] [Google Scholar]
- 18. Wang K, Pan L, Che X, Cui D, Li C. Gli1 inhibition induces cell‐cycle arrest and enhanced apoptosis in brain glioma cell lines. J Neurooncol 2010;98:319–327. [DOI] [PubMed] [Google Scholar]
- 19. Martin S, Toquet C, Oliver L, et al. Expression of bcl‐2, bax and bcl‐xl in human gliomas: a re‐appraisal. J Neurooncol 2001;52:129–139. [DOI] [PubMed] [Google Scholar]
- 20. Li Y, Zhang S, Huang S. FoxM1: a potential drug target for glioma. Future Oncol 2012;8:223–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rampling R, Sanson M, Gorlia T, et al. A phase I study of LY317615 (enzastaurin) and temozolomide in patients with gliomas (EORTC trial 26054). Neuro Oncol 2012;14:344–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen L, Han L, Shi Z, et al. LY294002 enhances cytotoxicity of temozolomide in glioma by down‐regulation of the PI3K/Akt pathway. Mol Med Report 2012;5:575–579. [DOI] [PubMed] [Google Scholar]
- 23. Darefsky AS, King JT Jr, Dubrow R. Adult glioblastoma multiforme survival in the temozolomide era: a population‐based analysis of Surveillance, Epidemiology, and End Results registries. Cancer 2012;118:2163–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang K, Pan L, Che X, Cui D, Li C. Sonic Hedgehog/GLI(1) signaling pathway inhibition restricts cell migration and invasion in human gliomas. Neurol Res 2010;32:975–980. [DOI] [PubMed] [Google Scholar]
- 25. Hsieh A, Ellsworth R, Hsieh D. Hedgehog/GLI1 regulates IGF dependent malignant behaviors in glioma stem cells. J Cell Physiol 2011;226:1118–1127. [DOI] [PubMed] [Google Scholar]
- 26. Bar EE, Chaudhry A, Lin A, et al. Cyclopamine‐mediated hedgehog pathway inhibition depletes stem‐like cancer cells in glioblastoma. Stem Cells 2007;25:2524–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Aggarwal BB, Sundaram C, Malani N, Ichikawa H. Curcumin: the Indian solid gold. Adv Exp Med Biol 2007;595:1–75. [DOI] [PubMed] [Google Scholar]
- 28. Parasramka MA, Gupta SV. Synergistic effect of garcinol and curcumin on antiproliferative and apoptotic activity in pancreatic cancer cells. J Oncol 2012;2012:709739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sundram V, Chauhan SC, Ebeling M, Jaggi M. Curcumin attenuates beta‐catenin signaling in prostate cancer cells through activation of protein kinase D1. PLoS ONE 2012;7:e35368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Milacic V, Banerjee S, Landis‐Piwowar KR, Sarkar FH, Majumdar AP, Dou QP. Curcumin inhibits the proteasome activity in human colon cancer cells in vitro and in vivo. Cancer Res 2008;68:7283–7292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ng AP, Chng WJ, Khan M. Curcumin sensitizes acute promyelocytic leukemia cells to unfolded protein response‐induced apoptosis by blocking the loss of misfolded N‐CoR protein. Mol Cancer Res 2011;9:878–888. [DOI] [PubMed] [Google Scholar]
- 32. Senft C, Polacin M, Priester M, Seifert V, Kogel D, Weissenberger J. The nontoxic natural compound Curcumin exerts anti‐proliferative, anti‐migratory, and anti‐invasive properties against malignant gliomas. BMC Cancer 2010;10:491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Aoki H, Takada Y, Kondo S, Sawaya R, Aggarwal BB, Kondo Y. Evidence that curcumin suppresses the growth of malignant gliomas in vitro and in vivo through induction of autophagy: role of Akt and extracellular signal‐regulated kinase signaling pathways. Mol Pharmacol 2007;72:29–39. [DOI] [PubMed] [Google Scholar]
- 34. Dhandapani KM, Mahesh VB, Brann DW. Curcumin suppresses growth and chemoresistance of human glioblastoma cells via AP‐1 and NFkappaB transcription factors. J Neurochem 2007;102:522–538. [DOI] [PubMed] [Google Scholar]
- 35. Kenney AM, Cole MD, Rowitch DH. Nmyc upregulation by sonic hedgehog signaling promotes proliferation in developing cerebellar granule neuron precursors. Development 2003;130:15–28. [DOI] [PubMed] [Google Scholar]
- 36. Kenney AM, Rowitch DH. Sonic hedgehog promotes G(1) cyclin expression and sustained cell cycle progression in mammalian neuronal precursors. Mol Cell Biol 2000;20:9055–9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Teh MT, Wong ST, Neill GW, Ghali LR, Philpott MP, Quinn AG. FOXM1 is a downstream target of Gli1 in basal cell carcinomas. Cancer Res 2002;62:4773–4780. [PubMed] [Google Scholar]
- 38. Schuller U, Zhao Q, Godinho SA, et al. Forkhead transcription factor FoxM1 regulates mitotic entry and prevents spindle defects in cerebellar granule neuron precursors. Mol Cell Biol 2007;27:8259–8270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kundu P, Mohanty C, Sahoo SK. Antiglioma activity of curcumin‐loaded lipid nanoparticles and its enhanced bioavailability in brain tissue for effective glioblastoma therapy. Acta Biomater 2012;8:2670–2687. [DOI] [PubMed] [Google Scholar]
- 40. Choi BH, Kim CG, Bae YS, Lim Y, Lee YH, Shin SY. p21 Waf1/Cip1 expression by curcumin in U‐87MG human glioma cells: role of early growth response‐1 expression. Cancer Res 2008;68:1369–1377. [DOI] [PubMed] [Google Scholar]
- 41. Karmakar S, Banik NL, Ray SK. Curcumin suppressed anti‐apoptotic signals and activated cysteine proteases for apoptosis in human malignant glioblastoma U87MG cells. Neurochem Res 2007;32:2103–2113. [DOI] [PubMed] [Google Scholar]
- 42. Luthra PM, Kumar R, Prakash A. Demethoxycurcumin induces Bcl‐2 mediated G2/M arrest and apoptosis in human glioma U87 cells. Biochem Biophys Res Commun 2009;384:420–425. [DOI] [PubMed] [Google Scholar]
- 43. Purkayastha S, Berliner A, Fernando SS, et al. Curcumin blocks brain tumor formation. Brain Res 2009;1266:130–138. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Histological change by H & E analysis between the curcumin‐treated group and control group.


