Summary
Background
EXOC3L2 gene rs597668 polymorphism was identified to be significantly associated with Alzheimer's disease (AD) in Caucasian population. However, recent studies reported consistent and inconsistent results in Caucasian and Asian populations.
Aims
In order to assess this association, we performed a meta‐analysis of rs597668 polymorphism using RevMan (v.5.1) software.
Methods
We searched PubMed and Google scholar databases and selected 4 independent publications, which included 16 independent studies. We conducted sensitivity analysis and evaluated the publication bias. In the end, we calculated the odds ratio (OR) using fixed effect model (Mantel‐Haenszel).
Results
We observed significant association between rs597668 polymorphism and AD using allele model (P = 0.006, OR = 1.09, 95% CI 1.03–1.16) and the dominant model (P = 0.008, OR = 1.11, 95% CI 1.03–1.21).
Discussion and Conclusions
To our knowledge, this is the first study that assesses the association between rs597668 polymorphism and AD by meta‐analysis. We believe that our findings will be very useful for future genetic studies in AD.
Keywords: Alzheimer's disease, Caucasian, East Asian, EXOC3L2 rs597668 polymorphism, Genome‐wide association studies
Introduction
Alzheimer's disease (AD) is a complex and neurodegenerative disease in the elderly 1. It is estimated that there are 35.6 million dementia people in the world in 2010 2. This number will be 65.7 million in 2030 2. In order to identify common AD variants, large‐scale genome‐wide association studies (GWAS) of AD have been conducted 3, 4, 5, 6, 7. Some common AD variants in new AD susceptibility genes (CR1, BIN1, CLU, PICALM, MS4A4/MS4A6E, CD2AP, CD33, EPHA1, EXOC3L2 and ABCA7) have been reported 3, 4, 5, 6, 7.
A single‐nucleotide polymorphism (SNP) rs597668 near EXOC3L2 gene was reported to be significantly associated with AD in Caucasian population (P = 6.450E−09) 5. The following studies investigated the association and reported consistent and inconsistent results in Caucasian population and Asian population. Carrasquillo et al. investigated the rs597668 polymorphism in a large dataset from the USA and Europe (3287 AD cases and 4396 controls). The result showed that rs597668 variant was not significantly associated with AD (P = 0.090) 8. Lambert et al. analyzed rs597668 polymorphism in three contrasting European populations (Finland, Italy, and Spain) with 2816 AD cases and 2706 controls. They successfully replicated this association with P = 2.000E−03 9.
In addition to the Caucasian population, this polymorphism was also investigated in Chinese and Japanese populations. Liu et al. investigated rs597668 polymorphism in 1205 unrelated Northern Han Chinese subjects (598 AD patients and 607 healthy controls). However, they did not report significant association between rs597668 and AD using genotype test (P = 0.653) and allele test (P = 0.603) 10. Ohara et al. investigated rs597668 polymorphism using 3758 Japanese subjects (825 AD cases and 2933 controls). However, they still did not report any significant association (P = 0.440) 11. Considering the inconsistent results, in this research, we investigated the association between rs597668 and AD by a meta‐analysis.
Materials and Methods
Literature Search and Inclusion Criteria
We searched PubMed and Google scholar databases to select all possible studies using the key words “Alzheimer's disease”, “EXOC3L2”. The literature search was updated on 3/20/2013. We included the studies meeting the following criteria: (1) the study evaluated the association between rs597668 polymorphism and AD by a case‐control design and (2) the study provided the numbers of rs597668 genotype. In the end, we selected four independent publications, which included 16 independent studies (Figure 1).
Figure 1.
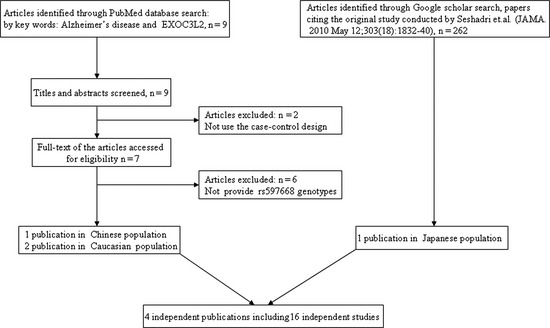
Flow chart of meta‐analysis for exclusion or inclusion of individual articles.
Data Extraction
For the case‐control genetics studies, the following information was extracted from each study: (1) the name of the first author, (2) the year of publication, (3) the ethnicity of the studied population, (4) the number of cases and controls, (5) the genotype numbers of rs597668 polymorphism in cases and controls, and (6) the type of AD (Table 1).
Table 1.
Characteristics of 16 studies selected for meta‐analysis
| Study | Population | Type of AD | Case # | Case genotype # | Control # | Control genotype # | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| TT | TC | CC | TT | TC | CC | |||||
| Carrasquillo 2011 8 | Autopsy | LOAD | 304 | 205 | 92 | 7 | 100 | 73 | 24 | 3 |
| Carrasquillo 2011 8 | Jacksonville | LOAD | 500 | 332 | 150 | 18 | 959 | 684 | 246 | 29 |
| Carrasquillo 2011 8 | Norway | LOAD | 338 | 224 | 102 | 12 | 544 | 351 | 175 | 18 |
| Carrasquillo 2011 8 | Rochester | LOAD | 311 | 215 | 81 | 15 | 1617 | 1142 | 436 | 39 |
| Carrasquillo 2011 8 | Southampton | LOAD | 36 | 24 | 11 | 1 | 128 | 79 | 44 | 5 |
| Carrasquillo 2011 8 | Bristol | LOAD | 200 | 141 | 56 | 3 | 37 | 23 | 13 | 1 |
| Carrasquillo 2011 8 | Leeds | LOAD | 113 | 72 | 36 | 5 | 275 | 190 | 81 | 4 |
| Carrasquillo 2011 8 | Man/Notts | LOAD | 177 | 111 | 60 | 6 | 87 | 57 | 29 | 1 |
| Carrasquillo 2011 8 | NCRAD | LOAD | 698 | 460 | 209 | 29 | 209 | 138 | 64 | 7 |
| Carrasquillo 2011 8 | Oxford | LOAD | 97 | 64 | 28 | 5 | 204 | 149 | 50 | 5 |
| Carrasquillo 2011 8 | Poland | LOAD | 470 | 273 | 169 | 28 | 182 | 128 | 51 | 3 |
| Lambert 2011 9 | Finland | LOAD | 562 | 265 | 241 | 56 | 529 | 302 | 184 | 43 |
| Lambert 2011 9 | Italy | LOAD | 1541 | 1115 | 336 | 90 | 1268 | 980 | 266 | 22 |
| Lambert 2011 9 | Spain | LOAD | 627 | 559 | 54 | 14 | 832 | 669 | 150 | 13 |
| Liu 2012 10 | China | LOAD | 571 | 261 | 236 | 74 | 607 | 267 | 275 | 65 |
| Ohara 2012 11 | Japan | LOAD | 825 | 265 | 418 | 142 | 2933 | 937 | 1449 | 547 |
| All | – | – | 7370 | 4586 | 2279 | 505 | 10511 | 6169 | 3537 | 805 |
LOAD, Late‐onset Alzheimer's disease.
Genetic Model
In order to ensure that interesting findings were not missed because of the different methods, we used three genetic models, which included the allele model (C vs. T), the dominant model (CC + CT vs. TT), and the recessive model (CC vs. CT + TT).
Heterogeneity Test
The heterogeneity among the selected studies was evaluated using Cochran's Q test. It approximately follows a χ2 distribution with k‐1 degrees of freedom (k is the number of studies for analysis) 12. The threshold of significant difference is 0.01. was also used to evaluate the heterogeneity. Low, moderate, large and extreme heterogeneity corresponded to 0–25%, 25–50%, 50–75% and 75–100%, respectively 12.
Meta‐Analysis
The fixed effect model (Mantel‐Haenszel) or random‐effect model (DerSimonian‐Laird) was used to calculate the odds ratio (OR). If there is no significant heterogeneity among the studies included (P > 0.01 and I 2 < 50%), odds ratio (OR) is calculated by the fixed effect model (Mantel‐Haenszel). Otherwise, the OR is calculated by random‐effect model (DerSimonian‐Laird). The significance level of OR was determined using the Z‐test. We used the RevMan (v.5.1) software to calculate all the tests above (http://ims.cochrane.org/revman).
Sensitivity Analysis
We performed a sensitivity analysis to illustrate the influence of specific studies. In sensitivity analysis, relative influence of each study was assessed by omitting one study at a time.
Publication Bias Analysis
The results of meta‐analysis may be influenced by potential publication bias 13. The likely presence or absence of bias should be routinely examined 13. Here, we evaluated the potential publication bias using funnel plots. If there is no bias, the plot will be a symmetrical inverted funnel. Otherwise, it will be an asymmetrical inverted funnel 12.
Results
The Results from Meta‐Analysis
We observed significant heterogeneity among these 16 studies using the allele (P < 1.000E−05 and I 2 = 71%), the recessive (P = 7.00E−04 and I 2 = 61%), and the dominant (P < 1.00E−05 and I 2 = 71%) models. We then calculated the overall OR using random‐effect model. Significant result was identified using recessive model (P = 6.000E−03, OR = 1.48, 95% CI 1.12–1.95). However, we did not identify significant association between rs597668 polymorphism and AD using allele model (P = 0.100, OR = 1.11, 95% CI 0.98–1.26) and dominant model (P = 0.280, OR = 1.08, 95% CI 0.94–1.25).
The Results from Sensitivity Analysis
To verify the findings from meta‐analysis, we performed a sensitivity analysis and the relative influence of each study was assessed by omitting one study at a time. We found that the study from Lambert et al. (Italy and Spain) 9 played an important role in causing the different association between rs597668 polymorphism and AD (Table 2).
Table 2.
Sensitivity analysis with each study omitted in meta‐analysis
| Study omitted | C vs. T | CC vs. CT + TT | CC vs. CT + TT | ||||||
|---|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | P | OR | 95% CI | P | OR | 95% CI | P | |
| None | 1.11 | 0.98–1.26 | 0.10 | 1.48 | 1.12–1.95 | 0.006 | 1.08 | 0.94–1.25 | 0.28 |
| Carrasquillo 2011 Autopsy | 1.11 | 0.97–1.26 | 0.13 | 1.51 | 1.14–2.01 | 0.004 | 1.07 | 0.92–1.25 | 0.36 |
| Carrasquillo 2011 Jacksonville | 1.10 | 0.98–1.26 | 0.16 | 1.51 | 1.12–2.05 | 0.007 | 1.07 | 0.91–1.25 | 0.41 |
| Carrasquillo 2011 Norway | 1.12 | 0.98–1.28 | 0.09 | 1.52 | 1.13–2.04 | 0.006 | 1.10 | 0.94–1.28 | 0.24 |
| Carrasquillo 2011 Rochester | 1.11 | 0.97–1.26 | 0.14 | 1.44 | 1.07–1.92 | 0.01 | 1.08 | 0.93–1.27 | 0.32 |
| Carrasquillo 2011 Southampton | 1.12 | 0.99–1.27 | 0.08 | 1.50 | 1.13–1.99 | 0.005 | 1.09 | 0.94–1.26 | 0.25 |
| Carrasquillo 2011 Bristol | 1.12 | 0.99–1.27 | 0.07 | 1.50 | 1.13–1.99 | 0.005 | 1.10 | 0.95–1.27 | 0.22 |
| Carrasquillo 2011 Leeds | 1.10 | 0.97–1.25 | 0.15 | 1.44 | 1.09–1.91 | 0.01 | 1.07 | 0.92–1.25 | 0.36 |
| Carrasquillo 2011 Man/Notts | 1.11 | 0.97–1.26 | 0.12 | 1.46 | 1.10–1.94 | 0.008 | 1.08 | 0.93–1.26 | 0.31 |
| Carrasquillo 2011 NCRAD | 1.12 | 0.98–1.27 | 0.11 | 1.50 | 1.12–2.01 | 0.007 | 1.09 | 0.93–1.27 | 0.28 |
| Carrasquillo 2011 Oxford | 1.10 | 0.97–1.25 | 0.15 | 1.46 | 1.10–1.94 | 0.009 | 1.07 | 0.92–1.24 | 0.37 |
| Carrasquillo 2011 Poland | 1.08 | 0.96–1.22 | 0.22 | 1.44 | 1.08–1.87 | 0.01 | 1.05 | 0.91–1.21 | 0.50 |
| Lambert 2011 Finland | 1.09 | 0.96–1.24 | 0.19 | 1.52 | 1.11–2.07 | 0.009 | 1.05 | 0.91–1.20 | 0.49 |
| Lambert 2011 Italy | 1.08 | 0.96–1.22 | 0.21 | 1.13 | 0.99–1.30 | 0.08 | 1.06 | 0.91–1.24 | 0.44 |
| Lambert 2011 Spain | 1.16 | 1.05–1.29 | 0.006 | 1.49 | 1.11–2.00 | 0.009 | 1.15 | 1.07–1.23 | 3.00E−04 |
| Liu 2012 China | 1.12 | 0.98–1.28 | 0.11 | 1.52 | 1.11–2.10 | 0.01 | 1.10 | 0.94–1.28 | 0.24 |
| Ohara 2012 Japan | 1.12 | 0.98–1.29 | 0.09 | 1.62 | 1.36–1.93 | 5.96E−08 | 1.09 | 0.93–1.28 | 0.29 |
The bold indicates the significance level is 0.05.
After both studies were excluded, we did not identify significant heterogeneity among the remaining 14 studies for allele model (P = 0.030 and I 2 = 47%), the recessive model (P = 0.170 and I 2 = 26%), and the dominant model (P = 0.050 and I 2 = 41%). We calculated the OR using fixed effect model. The robust association between rs597668 polymorphism and AD was detected using allele model (P = 0.006, OR = 1.09, 95% CI 1.03–1.16) and the dominant model (P = 0.008, OR = 1.11, 95% CI 1.03–1.21; Figure 2).
Figure 2.
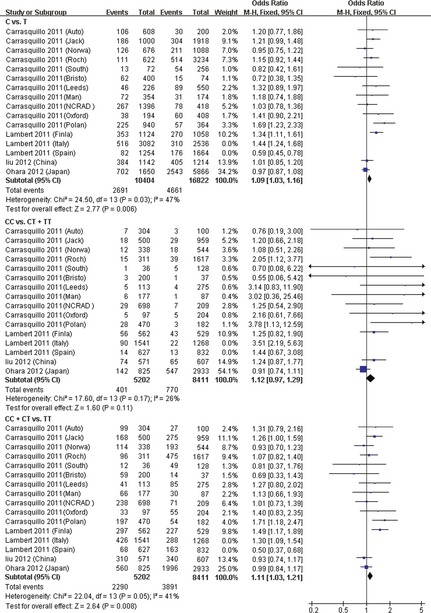
Forest plot for the meta‐analysis of rs597668 polymorphism in AD.
The Result from Publication Bias Analysis
We evaluated the potential publication bias among the 14 studies using funnel plots. Interestingly, all the three plots were symmetrical inverted funnels for allele model, the recessive model, and the dominant model, which indicated no publication bias (Figure 3).
Figure 3.
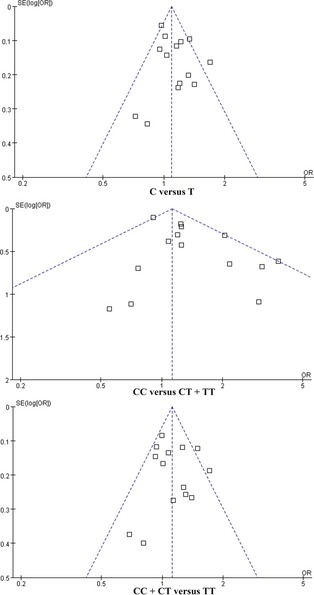
The publication bias analysis of rs597668 polymorphism in AD.
Discussion
Previous studies investigated the association between rs597668 and AD in Caucasian and East Asian populations and reported consistent and inconsistent results. In this research, we performed a meta‐analysis. We first evaluated the genetic heterogeneity among the selected studies and found significant heterogeneity. After the sensitivity analysis, we found that the study from Lambert et al. (Italy and Spain) contributed to the large heterogeneity. After removing both studies, no heterogeneity was observed. We calculated the OR using fixed effect model and found that rs597668 polymorphism was significantly associated with AD. Using the funnel plots, we identified no publication bias.
Recent study investigated the mechanisms of rs597668 polymorphism in AD pathogenesis 14. Schmidt et al. analyzed rs597668 polymorphism of 40 AD patients by a longitudinal study. They identified that EXOC3L2 rs597668 polymorphism was significantly associated with more aggressive disease courses 14. The C allele was associated with the risk of faster decline 14. For rs597668 polymorphism, in addition to being a risk factor for disease development, it also might act as prognostic disease marker 14. Considering the findings from our study and previous studies, rs597668 polymorphism may be a new therapeutic target to treat or prevent AD.
In our research, considering only two studies in Asia population and the results were consistent, we did not separately perform a meta‐analysis in Caucasian and East Asian populations. The reason can be seen from Li et al.'s study 15. In order to collect all possible studies with various types of study design from all over the world, we searched PubMed and Google scholar databases. In the end, we selected 17 independent studies, among which 16 studies evaluated the association between rs597668 polymorphism and AD by a case‐control design. Considering that only one study analyzed rs597668 polymorphism of 40 AD patients by a longitudinal study 14, we only performed a meta‐analysis of rs597668 polymorphism using the results from case‐control design.
Before our submission (3/22/2013), we accessed the AlzGene (www.alzgene.org) 16 and PubMed (www.ncbi.nlm.nih.gov/pubmed) databases. We found no study investigating the association between rs597668 polymorphism and AD by a meta‐analysis method. To our knowledge, this is the first study that assesses the association between rs597668 polymorphism and AD by meta‐analysis. We believe that our findings will be very useful for future genetic studies in AD.
Conflict of Interest
The authors declare no conflict of interest.
Acknowledgments
This work was supported by General Program (No. 81171120) and Youth Program (No. 81100940) of National Natural Science Foundation of China. We appreciate the useful comments made by anonymous reviewers. We also want to thank our colleagues, who play an important role in our revised manuscript.
References
- 1. Pedersen NL. Reaching the limits of genome‐wide significance in Alzheimer disease: Back to the environment. JAMA 2010;303:1864–1865. [DOI] [PubMed] [Google Scholar]
- 2. Bettens K, Sleegers K, Van Broeckhoven C. Current status on Alzheimer disease molecular genetics: From past, to present, to future. Hum Mol Genet 2010;19:R4–R11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hollingworth P, Harold D, Sims R, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat Genet 2011;43:429–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Naj AC, Jun G, Beecham GW, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late‐onset Alzheimer's disease. Nat Genet 2011;43:436–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Seshadri S, Fitzpatrick AL, Ikram MA, et al. Genome‐wide analysis of genetic loci associated with Alzheimer disease. JAMA 2010;303:1832–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Harold D, Abraham R, Hollingworth P, et al. Genome‐wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet 2009;41:1088–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lambert JC, Heath S, Even G, et al. Genome‐wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet 2009;41:1094–1099. [DOI] [PubMed] [Google Scholar]
- 8. Carrasquillo MM, Belbin O, Hunter TA, et al. Replication of BIN1 association with Alzheimer's disease and evaluation of genetic interactions. J Alzheimers Dis 2011;24:751–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lambert JC, Zelenika D, Hiltunen M, et al. Evidence of the association of BIN1 and PICALM with the AD risk in contrasting European populations. Neurobiol Aging 2011;32:756–e711. [DOI] [PubMed] [Google Scholar]
- 10. Liu QY, Miao D, Yu JT, et al. Lack of association between rs597668 polymorphism near EXOC3L2 and late‐onset Alzheimer's disease in Han Chinese. Neurosci Lett 2012;513:174–177. [DOI] [PubMed] [Google Scholar]
- 11. Ohara T, Ninomiya T, Hirakawa Y, et al. Association study of susceptibility genes for late‐onset Alzheimer's disease in the Japanese population. Psychiatr Genet 2012;22:290–293. [DOI] [PubMed] [Google Scholar]
- 12. Ioannidis JP, Patsopoulos NA, Evangelou E. Heterogeneity in meta‐analyses of genome‐wide association investigations. PLoS One 2007;2:e841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Egger M, Smith GD. Bias in location and selection of studies. BMJ 1998;316:61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schmidt C, Wolff M, von Ahsen N, Zerr I. Alzheimer's disease: Genetic polymorphisms and rate of decline. Dement Geriatr Cogn Disord 2012;33:84–89. [DOI] [PubMed] [Google Scholar]
- 15. Li XY, Cai XL, Bian PD, Hu LR. High salt intake and stroke: Meta‐analysis of the epidemiologic evidence. CNS Neurosci Ther 2012;18:691–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta‐analyses of Alzheimer disease genetic association studies: The AlzGene database. Nat Genet 2007;39:17–23. [DOI] [PubMed] [Google Scholar]


