Abstract.
The mosquito Aedes aegypti is a transmission vector for dangerous epidemic diseases in humans. Insecticides have been used as the most general vector control method in the world. However, Ae. aegypti have developed many resistant mechanisms such as reduced neuronal sensitivity to insecticides (target-site resistance), enhanced insecticide metabolism (metabolic resistance), altered transport, sequestration, and other mechanisms. It has become a major problem for vector control programs. Transcriptome sequencing and bioinformatic analysis were used to compare transcription levels between a susceptible strain (Bora7) and a resistant strain (KhanhHoa7) collected from the field. A total of 161 million Illumina reads, including 66,076,678 reads from the Bora7 strain and 69,606,654 reads from the KhanhHoa7 strain, were generated and assembled into 11,174 genes. A comparison of the KhanhHoa7 transcriptome to that of Bora7 showed 672 upregulated genes and 488 downregulated genes. We identified the highly upregulated genes: cytochrome P450 4C1, 4C3, 4C21, 4D1, 4D1 isoform X2, 4D2, 4D2 isoform X2, 4G15, 6A2, 6A8, 6D3, and 9E2; Glutathione S transferase (GST1), UGT1-3, 1-7, 2B15, and 2B37; binding cassette transporter (ABC) transporter F family member 4 and ABC transporter G family member 20. Interestingly, there was a significant increase in the expression of the genes such as CYP9E2 (8.3-fold), CYP6A8 (5.9-fold), CYP6D3 (5.4-fold), CYP4C21 (5.4-fold), CYP4G15 (5.2-fold), GST1 (3.5-fold), and ABC transporter 4 (2.1-fold). Our results suggested a potential relationship between the expression of the genes in metabolic processes and insecticide resistance in the studied strain. These results may contribute to the understanding of the mechanisms of insecticide resistance in Ae. aegypti.
INTRODUCTION
The mosquito Aedes aegypti is a transmission vector for dangerous epidemic diseases in humans, such as dengue, yellow fever, and chikungunya.1 In particular, dengue is the most dangerous disease, affecting 50 million people per year, and the number of affected people has increased 30-fold in the last 50 years.1,2 Insecticides have been used as the most general vector control method in the world. However, excessive insecticide use has led to the development of resistance in mosquitoes and has become a major problem for vector control programs.
Two major mechanisms of resistance are reduced neuronal sensitivity to insecticides (target-site resistance) and enhanced insecticide metabolism (metabolic resistance). Target-site resistance caused by mutation in voltage-gated sodium channels (VGSC) is known as knockdown resistance.3 Metabolic resistance involves detoxification enzymes such as cytochrome P450 monooxygenases (P450s or CYPs for genes), carboxy/cholinesterases, glutathione S-transferases (GSTs), and UDP-glucosyltransferases.4–7 Many studies showed that resistance of mosquitoes to pyrethroids is linked to the induction of detoxification genes.8–10
Monooxygenase-mediated metabolism (by activity of cytochrome P450 monooxygenases—P450s) is the most common mechanism in insects to resist insecticides.11 Cytochrome P450s are known to play an important role in detoxifying exogenous compounds such as insecticides.11,12 In insects, increased metabolic resistance is a result of enhanced levels of P450 proteins and P450 activity because of the overexpression of P450 genes.5,13–16
There are many reports that have demonstrated a relationship between resistance and elevated P450 activity in Ae. aegypti. Diversity of resistance is conferred by the existence of multiple isoforms and differential expression of cytochrome P450s.17 To date, more than 600 P450 genes have been identified in insects. Some genes belonging to the families CYP4, CYP6, CYP9, and CYP12 are associated with insecticide resistance.6,18 Whole-genome sequencing has revealed that Ae. aegypti has 160 P450-coding genes, and 44 of these belong to the CYP6 family.19 CYP6Z9 was found to be 4-fold overexpressed in a permethrin-resistant strain collected in northern Thailand.19 CYP6M6, CYP6Z8, and CYP6M11 have also been identified as inducible by permethrin and pollutants.20 The overexpression of CYP6M6 and CYP6Z6 was found in the Vauclin strain in Martinique, indicating the involvement of CYP6 in pyrethroid resistance in Ae. aegypti.20 Bariami et al.21 detected differences in the expression of cytochrome genes such as CYP6BB2, CYP9J9, CYP9J10, CYP9J26, CYP9J27, and CYP9J28 in Ae. aegypti pyrethroid-resistant populations from the Cayman Islands and Cuba.
High GST activity related to insecticide resistance has also been reported in many species of insects.19,22–28 For pyrethroid detoxification, GSTs have no direct role in the metabolism of pyrethroids, but they have the capacity to reduce the peroxidative damage caused by pyrethroids by detoxifying lipid peroxidation products.29 Glutathione S-transferases can also bind pyrethroid insecticides in a sequestering mechanism as a passive way of detoxification.30 In mosquitoes, approximately 30 GST genes have been identified within different subfamilies.19,31 Members of the epsilon GST subfamily (GSTe7 and GSTe2) have been found in the dehydrochlorination of dichlorodiphenyltrichloroethane (DDT) and pyrethroid deltamethrin in Ae. aegypti.24,32,33 However, there are a large number of genes encoding detoxification enzymes,34–36 so determining the exact enzymes that play the main role in resistance in each mosquito species is a great challenge.37
In addition, many studies have indicated that there are potential polymorphisms affecting the function of detoxification enzymes and provided evidence for the roles of these enzymes in insecticide resistance in mosquitoes.38 Recently, polymorphisms of the P450 gene CYP9M10 have been demonstrated to be involved in pyrethroid resistance in Culex pipiens.39 This suggested that a deep analysis of the polymorphisms associated with resistance can improve our knowledge of the mechanisms developed by mosquitoes to resist insecticides. Today, thanks to the development of next-generation sequencing technologies such as mRNA sequencing (RNA-seq), the expression and polymorphism of genes involved in insecticide resistance have been evaluated.40–42
Vietnam is one of the countries that have the most dengue cases in Southeast Asia with 50,000 cases reported in 2016.43 To control transmission vectors, Vietnam used a large amount of insecticide with 24 tons of DDT (in 1993 and 1994) and 21,000 L of photo-stable pyrethroid formulations.44 Every year, this figure is estimated to be 4,000 tons of chemicals belonging to the pyrethroid group, leading to the development of resistance in many mosquito species, including the Ae. aegypti mosquito. In the past few years, many reports on the resistance in Ae. aegypti have showed that there was a reduction in susceptibility, and many populations were found to develop high resistance to pyrethroid and DDT.44–50 The aim of this study was to use RNA-seq to investigate transcription levels and analysis of the changes associated with resistance in Ae. aegypti in Vietnam.
METHODS
Sample collection and resistance testing.
Larvae were collected from natural habitats in Khanh Hoa Province of Vietnam between August and September 2016 and reared into adults under rearing conditions in the insectary. The samples were collected in accordance with the standard procedure of the Ministry of Health, Vietnam. The larvae were collected from clean water containers (rain water or domestic water) of households, 100 households for each sampling location and two locations for each province. Specimens were morphologically identified to species and used in the study.51 Adult Ae. aegypti mosquitoes were reared in standard insectary conditions (27°C ± 2°C, 12 hours/12 hours light/dark, and 70% ± 10% relative humidity) in net cages and fed with sugar water and then starved for 3–4 hours before insecticide tests. Laboratory strain, Bora Bora 52 (originating from Laboratori se Lutte contre les Insectes nuisibles, Montpallier, France) (denoted by Bora7 in this study) was used as a control strain to compare with a resistant insecticide strain (KhanhHoa7).
To determine the resistance level, bioassays were carried out according to the WHO procedure52 using 1,500 adult females of 2–5-day-old F1 generation with four replicates of 25 mosquitoes per tube. The insecticides tested were 0.75% permethrin (type I of pyrethroid), 0.05% deltamethrin (type II of pyrethroid), 0.05% lambda-cyhalothrin (pyrethroid), 4% DDT (organochlorine), 1% propoxur (carbamate), and 5% malathion (organophosphate). Resistance status was classified according to WHO52 criteria: resistance for < 90% mortality, probable resistance for 90–98% mortality, and susceptible for > 98% mortality. The resistant mosquitoes were used for transcriptome analysis.
RNA extraction and preparation of cDNA libraries.
For each sample, total RNA was extracted from 10 adult mosquitoes using the RNeasy Mini Kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. RNA quality and quantity were assessed with a NanoDrop instrument (NanoPhotometer – IMPLEN P3000, Munchen, Germany). Ten samples from each strain, representing a total of 100 mosquitoes, were pooled and then used for preparing cDNA libraries using the NEBNext Poly(A) mRNA Magnetic Isolation Module (NEB#E7490, Illumina, Ipswich, MA) and the NEBNext Ultra DNA Library Prep Kit (NEB#E7370, Illumina, Ipswich, MA). Two replicates of cDNA libraries were prepared for each strain.
Sequencing, read mapping, and gene expression analysis.
Sequence reads (100-bp paired end) from each cDNA library were generated in the Illumina HiSeq 2000 instrument. The reads were then mapped to the Ae. aegypti genome sequence (AaegL5.0 gene set, http://vectorbase.org) using the TopHat algorithm with default parameters (http://tophat.cbcb.umd.edu, v1.0.14),53 leading to an average mapping rate of 80%. The reads were first filtered based on the sequence quality (mean read quality ≥ 30 and with Ns < 10%) and mapping quality (alignment score ≥ 98). For the gene ontology (GO) annotation of the unigenes, the BLAST2GO program (https://www.blast2go.com/) was used.54
Transcript abundance of the unigenes in each sample was calculated and normalized using the fragments per kilobase million (FPKM) method.55 Differentially expressed gene (DEG) analysis was performed using the DEGseq R package with a threshold of |log2 (fold change)| > 1 and corrected P-value < 0.05.56 Gene ontology enrichment analysis was performed by mapping the DEGs to the GO database, and the gene numbers from each GO term were calculated as compared with the genomic background.57 Visualization of the clusters of the DEG expression pattern was performed using the MultiExperiment Viewer (MEV; ver. 4.9).
Single nucleotide polymorphism/indel analysis.
The mpileup function in SAMtools (v0.1.18)58 was used to compare each sample with the reference genome for single nucleotide polymorphism (SNP) detection. The correlation between mutation and gene information can be derived based on the annotated gene information in the database, enabling annotation of the mutation site. Single nucleotide polymorphism alleles were computed between the insecticide-resistant and susceptible strains.
RESULTS
Insecticide resistance levels.
The insecticide resistance levels of the Bora strain and the mosquito strain collected from Khanh Hoa are shown in Table 1. The results indicated that the Bora strain was sensitive to all types of insecticide and the KhanhHoa strain was sensitive to organophosphate but strongly resistant to organochlorine (DDT), carbamates, and pyrethroids. Twenty-four–hour mortalities were 37% for deltamethrin, 21% for lambda-cyhalothrin, and 7% for permethrin. These results suggested that insecticide resistance was present in the KhanhHoa population at high levels, and the KhanhHoa7 strain was hence used for transcriptome analysis.
Table 1.
Percentage mortality in field samples of Aedes aegypti 24 hours after exposure to insecticide-impregnated papers and resistance status using the WHO test (WHO, 2016)
| Insecticides (diagnostic dose) | Bora strain | KhanhHoa strain | ||
|---|---|---|---|---|
| Mortality (%) | Resistance status | Mortality (%) | Resistance status | |
| Dichlorodiphenyltrichloroethane (4%) | 100 | S | 0 | R |
| Propoxur (1%) | 100 | S | 42 | R |
| Malathion (5%) | 100 | S | 100 | S |
| Deltamethrin (0.05%) | 100 | S | 37 | R |
| Lambda-cyhalothrin (0.05%) | 100 | S | 21 | R |
| Permethrin (0.75%) | 100 | S | 7 | R |
R = resistant; S = susceptible.
Identification of DEGs in strains.
More than 161 million 150-bp cDNA reads were sequenced across two samples (77,385,050 in Bora7 and 83,870,558 in KhanhHoa7). More than 80% of the reads were successfully mapped to the Ae. aegypti genome, including 11,444,233,590 bases from Bora7 and 12,384,682,936 bases from KhanhHoa7. Filtering for sequence quality and mapping score (Table 2) revealed the percentage of bases with quality scores (Qphred) higher than 20 or 30. Mapped reads were assigned to genomic features—exons, introns, and intergenic regions. For the KhanhHoa7 strain, the percentages of reads mapped to exonic, intergenic, and intronic regions were 52.08%, 14.84%, and 33.08%, respectively. These percentages were 41.17% (exonic region), 24.77% (intergenic region), and 34.06% (intronic region) in the Bora7 strain. These reads were generated and assembled into 11,174 genes. We also identified 3,440 novel transcripts, which may indicate that novel genes are specifically expressed in resistant strains. RNA-seq sequence data have been deposited at NCBI under the accession number PRJNA429544.
Table 2.
Filtered data statistics in studied strains
| Sample | Read length | Number of reads | Total read mapped (%) | Number of bases | Q20 (%) | Q30 (%) | GC (%) | N (ppm) |
|---|---|---|---|---|---|---|---|---|
| Bora | 147.89 | 77,385,050 | 66,076,678 (85.38) | 11,444,233,590 | 95.55 | 89.13 | 34.57 | 4.64 |
| KhanhHoa | 147.66 | 83,870,558 | 69,606,654 (82.99) | 12,384,682,936 | 95.14 | 88.21 | 40.02 | 4.44 |
GC% = percentage of G + C in the read; N (ppm) = number of base “N” per million bases; Q20, Q30 = percentage of bases with quality scores (Qphred) higher than 20 or 30, respectively.
Analysis of differences in transcription was performed on 11,528 and 12,158 transcripts showing a transcription signal higher than 0.1 Reads per kilobase of exon model per million reads (RPKM) in Bora7 and KhanhHoa7 strains, respectively (Table 3). A total of 1,762 (14.49%) transcripts had RPKM > 1, and approximately 4,644 (38.20%) transcripts (which had RPKM values from 3 to 15), 2,257 (18.56%) transcripts (RPKM values from 15 to 60), and 890 (7.32%) transcripts (RPKM values > 60) were identified in the KhanhHoa7 strain.
Table 3.
Gene expression level in studied strains
| Sample | The number of gene expression in different levels (reads per kilobase of exon model per million reads) | |||||
|---|---|---|---|---|---|---|
| 0–0.1 | 0.1–1 | 1–3 | 3–15 | 15–60 | > 60 | |
| Bora7 | 561 (4.78%) | 2,603 (22.58%) | 2,403 (20.84%) | 3,814 (33,08%) | 1,486 (12.89%) | 661 (5.73%) |
| KhanhHoa7 | 574 (4.72%) | 2,031 (16.71%) | 1,762 (14.49%) | 4,644 (38.20%) | 2,257 (18.56%) | 890 (7.32%) |
A transcriptome comparison between KhanhHoa7 and Bora7 showed 672 upregulated genes and 488 downregulated genes. We identified a large number of genes involved in resistance, including carboxylesterase for three, cuticle protein for 100, esterase for 22, endocuticle structural glycoprotein for 14, cytochrome P450 for 72, GST for 35, larval/pupal cuticle protein for four, larval cuticle protein for 42, probable cytochrome P450 for 69, and UDP-glucuronosyltransferase for 30. These genes were carboxylesterase 1F with an FPKM value of 2.32; esterase B1 with an FPKM value of 28.20; cytochrome P450 4C1, 4C3, 4C21, 4D1, 4D1 isoform X2, 4D2, 4D2 isoform X2, 4G15, 6A2, 6A8, 6D3, and 9E2 with FPKM values ranging from 2.39 to 147.66; GST 1 with an FPKM value of 70.93; and UDP-glucuronosyltransferase 1-3, 1-7, 2B15, and 2B37 with FPKM values of 15.35, 57.27, 11.92, and 23.88 (P-value < 0.001), respectively (Table 4). In addition, there was increased expression of genes, including ABC transporter F family member 4 and ABC transporter F family member 20, with FPKM values of 31.28 and 6.13 (P-value < 0.001), respectively. However, the cuticle protein genes (including cuticle protein, endocuticle structural glycoprotein, and larval cuticle protein genes) were downregulated in KhanhHoa7 as compared with the susceptible strain Bora7.
Table 4.
Annotation for the genes involved in insecticide resistance
| Gene name | AAEL annotation | Physical positions of the genes | Fragments per kilobase million (P < 0.001) |
|---|---|---|---|
| Cytochrome P450 4C1 (CYP4C38) | AAEL012266 | 2.384053732–384080949 | 18.85 |
| Cytochrome P450 4C3 | AAEL008017 | 1.74332275–74367682 | 2.39 |
| Cytochrome P450 4C21 (CYP325V1) | AAEL017136 | 3.112513177–112534429 | 15.60 |
| Cytochrome P450 4D1 (CYP4K3) | AAEL007798 | 3.152871519–152883632 | 8.64 |
| Cytochrome P450 4D1 isoform X2 | AAEL007807 | 3.152793074–152802023 | 5.22 |
| Cytochrome P450 4D2 (CYP4AR2) | AAEL010154 | 2.380179249–380180990 | 5.11 |
| Cytochrome P450 4D2 isoform X2 | AAEL007816 | 3.152716768–152729259 | 15.79 |
| Cytochrome P450 4G15 | AAEL006824 | 1.82804948–82813910 | 91.34 |
| Cytochrome P450 6A2 (CYP6BY1) | AAEL017539 | 3.399253843–399255488 | 4.59 |
| Cytochrome P450 6A8 | AAEL017061 | 1.271375696–271377569 | 23.83 |
| Cytochrome P450 6D3 (CYP6Z6) | AAEL009123 | 1.2418888478–418890194 | 27.20 |
| Cytochrome P450 9E2 (CYP9J28) | AAEL014617 | 3.368627096–368628939 | 83.21 |
| GST1 (GST3) | AAEL007947 | 2.351708207–351709186 | 70.93 |
| UGT1-3 | AAEL000687 | 2.458603992–458605906 | 15.35 |
| UGT1-7 | AAEL010366 | 2.182796211–182798339 | 57.27 |
| UGT2B15 | AAEL003091 | 2.213572075–213585955 | 11.92 |
| UGT2B37 | AAEL002688 | 2.464868089–464912877 | 23.88 |
| ABC transporter F family member 4 | AAEL016973 | 2.294820170–294833814 | 31.28 |
| ABC transporter G family member 20 | AAEL014428 | 1.159149149–159331370 | 6.13 |
AAEL = Aedes aegypti Liverpool strain; GST = Glutathione S-transferase; UGT = UDP-glucosyltransferases.
These genes exhibited an increase in expression from 1.5- to 8.3-fold (P-value < 0.01) in the resistant strain compared with the control strain. Interestingly, there were significant increase in the expression of the genes such as CYP9E2 (CYP9J28) (8.3-fold), CYP6A8 (5.9-fold), CYP6D3 (CYP6Z6) (5.4-fold), CYP4C21 (5.4-fold), CYP4G15 (5.2-fold), GST1 (3.5-fold), and ABC transporter 4 (2.1-fold). These results suggested that these genes may be related to insecticide resistance in the studied strain. To validate our transcriptome data, three genes, including CYP4C1, CYP6Z6, CYP9J28, were selected randomly for quantitative real time-PCR (qRT-PCR) analysis, using the same RNA samples as for the transcriptome sequencing. The qRT-PCR results showed similar upregulated results based on transcriptome sequencing analysis. The results were 6.60-, 6.72-, and 18.00-fold, respectively, in qRT-PCR.59
Functional annotation and classification of the DEGs.
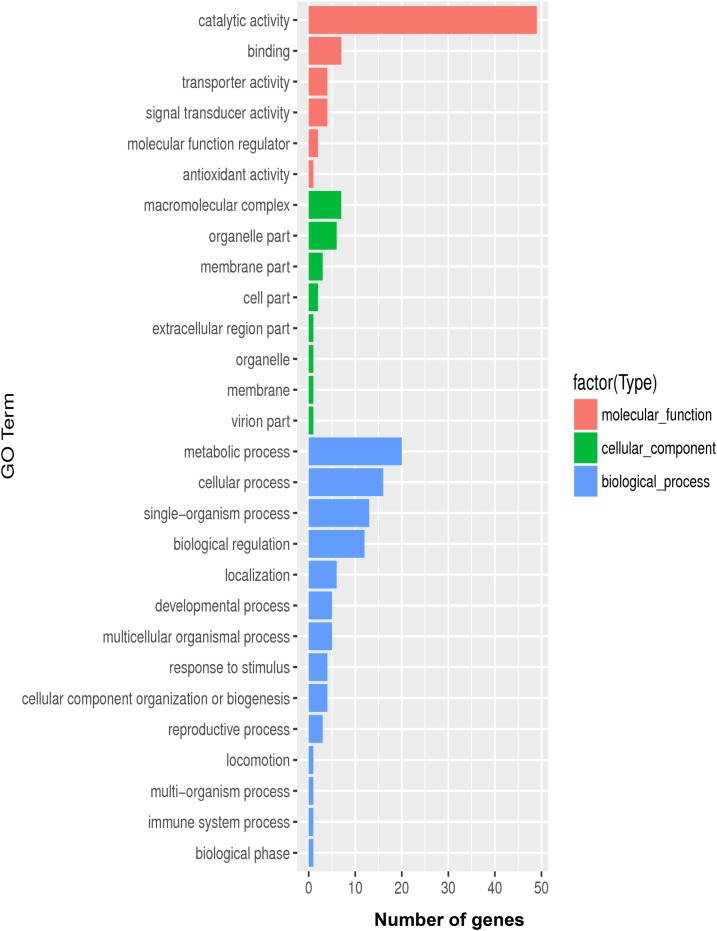
Gene ontology functional enrichment analyses can provide information about how DEGs are related to certain biological functions and hence were used to classify genes into the predicted functional groups. In this analysis, ∼50,000 transcripts were categorized into 28 functional groups, including biological processes, cellular components, and molecular functions (Figure 1). Among the biological processes, the most abundant groups included metabolic process, cellular process, single-organism process, biological regulation, localization, and development process; within this category, the dominant subcategories were metabolic process, cellular process, single-organism process, and biological regulation. The next most enriched category was molecular function, with the following subcategories: catalytic activity, binding, transporter, signal transducer, molecular function regulator, and antioxidant activity. Last was the group of cellular component functions.
Figure 1.
Histogram presentation of gene ontology (GO) classification. The results are summarized as three main categories: biological processes, cellular components, and molecular functions. The x axis shows the number of the matched genes and the y axis shows subgroups of molecular functions from GO classification. This figure appears in color at www.ajtmh.org.
Identification of SNPs/indels and alternative splicing in strains.
A total of 117,968 SNPs/indels loci were identified between the KhanhHoa7 strain and the Ae. aegypti reference genome. Distribution of these polymorphisms in the genome is shown in Table 5. These variations were distributed with ∼70% of them located in exonic, ∼13% in intergenic (within 1 kb of gene boundaries), ∼13% in intronic, ∼0.32% in upstream, and ∼2% in downstream regions of the insecticide-resistant strain. More than 164,328 SNPs and 4,517 indels were detected within the exonic region of 7,341 genes; another 4,824 SNPs and 399 indels were detected within the downstream regions of 1,685 genes; 802 SNPs and 29 indels were found in the upstream regions of 412 genes; and 126 SNPs/indels were found in alternative spliced sequences in these strains.
Table 5.
Single nucleotide polymorphism/indel genomic distribution in the studied strains
| Region | Number of single nucleotide polymorphisms/indels | Number of genes | |
|---|---|---|---|
| Bora7 strain (%) | KhanhHoa7 strain (%) | ||
| Exonic | 93,079 (70.00%) | 126,381 (71.01%) | 10,781 |
| Intergenic | 19,065 (14.34%) | 24,044 (13.51%) | – |
| Intronic | 17,609 (13.24%) | 23,247 (13.06%) | – |
| Splicing | 41 | 100 (0.06%) | 92 |
| Exonic splicing | 17 | 46 | – |
| Upstream | 325 (0.24%) | 575 (0.32%) | 412 |
| Downstream | 2,807 (2.11%) | 3,539 (1.98%) | 1,685 |
| Up/downstream | 22 | 36 | – |
| Total | 132,965 | 177,968 | – |
Among them, 1,010 SNPs were found in genes (113 genes) related to resistance, including ABC transporter, acetylcholinesterase, carboxylesterase, esterase, GST, cytochrome P450, and cuticle protein. These data will be useful in understanding the effect of SNPs on resistance in Ae. aegypti. However, the role of these SNPs in the genes has not been studied in this study. These results were the same as in the report of David et al.12 with 220,449 SNPs that were detected in Ae. aegypti pyrethroid-resistant strains. In this study, 60% of SNPs were located in coding regions, 20% in gene boundaries, 2.5% in 5′UTR (5′ untranslated region), and 9% in 3′UTR (3′ untranslated region).12
DISCUSSION
Studying resistance levels and mechanisms related to resistance in vectors is interesting. In this study, the test results showed that the KhanhHoa7 strain had strong resistance to DDT, carbamates, and pyrethroids. These results were perfectly consistent with those of the previous studies.46,48,50 In addition, studies have also shown that A. aegypti mosquitoes from Khanh Hoa carried mutations (Ile1011Val, Val1016Ile, Val1016Gly, Ala1007Gly, and Phe1558Cys) in the VGSC gene. To better understand the mechanisms that lead to the development of resistance in A. aegypti mosquitoes, we conducted transcriptome sequencing of susceptible and resistant strains.
High-throughput sequencing approaches such as RNAseq have provided information on gene expression and nucleotide variations over the whole transcriptome of a given sample.60,61 Transcriptome analysis allows the simultaneous study of the effects of multiple genes and can explain the mechanisms of gene action in different processes, including the enhancement of resistance in mosquitoes. Thus, the metabolic resistance mechanisms are known to enhance the expression levels and activities of enzymes to detoxify and decompose insecticide molecules.62 Resistance can result in the overexpression of a single enzyme or an active combination of different enzymes.
In mosquitoes, the CYP6Z subfamily was previously associated with the response to pyrethroids, carbamates, and organochlorines. Enhanced expression of Ae. aegypti CYP6Z9 was found to be 4-fold in a permethrin-resistant strain collected in northern Thailand.19 CYP9J23 is also identified as having an increase in expression of 5.3-fold in Ae. aegypti larvae due to inducible by permethrin and pollutants.20 Bariami et al.21 detected differences in the expression of cytochrome genes such as CYP6BB2, CYP9J9, CYP9J10, CYP9J19, CYP9J26, CYP9J27, and CYP9J28 (with 2.5 to 14.21-fold change) in Ae. aegypti pyrethroid-resistant populations from the Cayman Islands and Cuba. Cytochrome P450 CYP6Z9 has also been reported to show an increase in expression of more than 20% in Anophenles gambiae by Stevenson et al.63 In addition, elevated levels of GSTs have been found to be involved in insecticide resistance, as they are overexpressed in pyrethroid-resistant populations.12,19,22,23,25–28 Lima et al.64 reported the expression of GSTE7 and CYP6N12 in resistant Ae. aegypti from central Amazonia. The roles of the Ae. aegypti GSTE7 and GSTE2 genes in increased susceptibility to the pyrethroid deltamethrin were studied recently using RNA interference–mediated knockdown of the genes.33 Overexpression of ABC transporter 4 in Ae. aegypti was reported by Bariami et al.21 In our study, we also obtained the same results as those in the previous studies with an increase in the expression of the genes involved in the metabolisms.
Remarkably, in our study, the results indicated that there was downregulation of genes encoding cuticle protein in resistant compared with susceptible strains, although cuticular resistance was characterized recently by thickening of the cuticle layer, leading to slower penetration of the insecticide and, thus, reduction in the amount of insecticide within the insect. A similar resistance mechanism has been demonstrated in the cotton bollworm Helicoverpa armigera for pyrethroids.65 Cuticle thickening linked to pyrethroid resistance has also been identified in the oriental fruit fly Bactrocera dorsalis.66 Cuticular resistance has been mentioned often recently in Anopheles mosquitoes.67–71 In Ae. aegypti, differential expression of genes encoding cuticle proteins has been reported in mosquito larvae.60,72 The overexpression of multiple genes encoding cuticle proteins has been found in mosquito larvae that were selected with imidacloprid for several generations in the laboratory.72 However, Kasai et al.71 showed that there was no significant difference in the rate of permethrin reduction by the cuticle between resistant and susceptible strains. However, more detailed studies on the role of genes and resistance mechanisms in Ae. aegypti mosquitoes are needed.
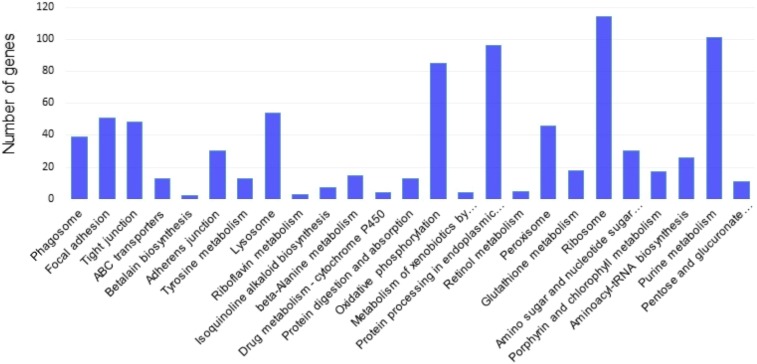
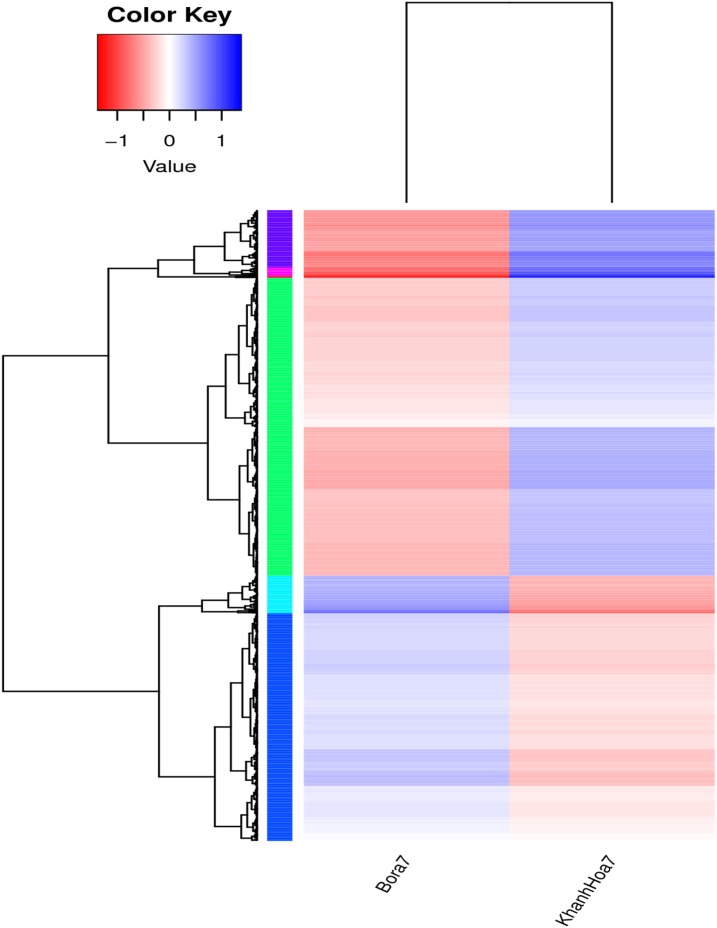
To analyze the interplay between the up- and downregulated genes, all DEGs were mapped to the referential canonical pathways in the Kyoto Encyclopedia of Genes and Genomes (KEGG) database using a cutoff E value of 10−5. The 25 pathways are presented in Figure 2. Distribution analysis of genes defined by KEGG revealed clustering of the DEGs in protein synthesis pathways and metabolism pathways. The six largest pathways were ribosome (ko03010), purine metabolism (ko00230), protein processing in endoplastic reticulum (ko04141), oxidative phosphorylation (ko00190), lysosome (ko04142), and peroxisome (ko04146). These were the main pathways involved in metabolism and detoxification processes. Significant differences in expression patterns between Bora7 and KhanhHoa7 strains were clearly observed in the heatmaps (Figure 3). The cluster analysis of these DEGs between the two strains revealed that the gene expression pattern in the resistant strain was distinctly different from that in the susceptible strain, suggesting that the transcriptional regulations of DEGs for their functions were unique in the resistant strain.
Figure 2.
Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of the top 25 pathways is shown. The x axis shows pathways from KEGG classification and the y axis shows the number of genes. This figure appears in color at www.ajtmh.org.
Figure 3.
Heatmap analysis showing the expression pattern of differentially expressed genes in different strains. Log10 (reads per kilobase of exon model per million reads + 1) values are used for clustering. Intensity of color indicates expression levels, with genes of high expression in blue and low expression in red.This figure appears in color at www.ajtmh.org.
CONCLUSION
In this study, a sample collected from the field was used to compare transcriptional differences with the susceptible strain (Bora7). Using RNA-Seq and DEG analysis, we identified the molecular diversity of genes involved in biological pathways or different expression of genes involved in metabolism and other biological processes. These changes likely resulted from SNPs/indels or differential expression of metabolic pathways. Some candidate genes were revealed, such as cytochrome P450, GST, UDP-glucuronosyltransferase, and ABC transporters. These results suggested a potential relationship between these genes and metabolic processes. However, in our study, there was a downregulation of genes that encoded cuticle proteins.
Financial support:
This study was supported by the National Foundation for Science and Technology Development, Vietnam (NAFOSTED, grant no. 106-NN.02-2015.17).
REFERENCES
- 1.World Health Organization , 2009. Dengue: Guidelines for Diagnosis, Treatment, Prevention and Control. Geneva Switzerland: WHO, 1–160. [PubMed] [Google Scholar]
- 2.Phillips ML, 2008. Dengue reborn: widespread resurgence of a resilient vector. Environ Health Perspect 116: A382–A388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soderlund DM, Bloomquist JR, 1990. Molecular mechanisms of insecticide resistance. Roush RT, Tabashnik BE, eds. Pesticide Resistance in Arthropods. London, United Kingdom: Chapman and Hall, 58–96. [Google Scholar]
- 4.Hemingway J, Hawkes NJ, McCarroll L, Ranson H, 2004. The molecular basis of insecticide resistance in mosquitoes. Insect Biochem Mol Biol 34: 653–655. [DOI] [PubMed] [Google Scholar]
- 5.Feyereisen R, 2005. Insect cytochrome P450. Gilbert LI, Iatrou K, Gill SS, eds. Comprehensive Molecular Insect Science, Vol. 4. Oxford, United Kingdom: Elsevier, 1–77. [Google Scholar]
- 6.Li X, Schuler MA, Berenbaum MR, 2007. Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu Rev Entomol 52: 231–253. [DOI] [PubMed] [Google Scholar]
- 7.Nkya TE, Akhouayri I, Kisinza W, David JP, 2013. Impact of environment on mosquito response to pyrethroid insecticides: facts, evidences and prospects. Insect Biochem Mol Biol 43: 407–416. [DOI] [PubMed] [Google Scholar]
- 8.Poupardin R, Reynaud S, Strode C, Ranson H, Vontas J, David JP, 2008. Cross-induction of detoxification genes by environmental xenobiotics and insecticides in the mosquito Aedes aegypti: impact on larval tolerance to chemical insecticides. Insect Biochem Mol Biol 38: 540–551. [DOI] [PubMed] [Google Scholar]
- 9.Poupardin R, Riaz MA, Vontas J, David JP, Reynaud S, 2010. Transcription profiling of eleven cytochrome P450s potentially involved in xenobiotic metabolism in the mosquito Aedes aegypti. Insect Mol Biol 19: 185–193. [DOI] [PubMed] [Google Scholar]
- 10.Riaz MA, Poupardin R, Reynaud S, Strode C, Ranson H, David JP, 2009. Impact of glyphosate and benzo[a]pyrene on the tolerance of mosquito larvae to chemical insecticides. Role of detoxification genes in response to xenobiotics. Aquat Toxicol 93: 61–69. [DOI] [PubMed] [Google Scholar]
- 11.Scott JG, 1999. Cytochromes P450 and insecticide resistance. Insect Biochem Mol Biol 29: 757–777. [DOI] [PubMed] [Google Scholar]
- 12.David JP, Faucon F, Chandor-Proust A, Poupardin R, Riaz MA, Bonin A, Navratil V, Reynaud S, 2014. Comparative analysis of response to selection with three insecticides in the dengue mosquito Aedes aegypti using mRNA sequencing. BMC Genomics 15: 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carino FA, Koener JF, Plapp FW, Jr., Feyereisen R, 1994. Constitutive overexpression of the cytochrome P450 gene CYP6A1 in a house fly strain with metabolic resistance to insecticides. Insect Biochem Mol Biol 24: 411–418. [DOI] [PubMed] [Google Scholar]
- 14.Liu N, Scott JG, 1997. Phenobarbital induction of CYP6D1 is due to a trans acting factor on autosome 2 in house flies, Musca domestica. Insect Mol Biol 6: 77–81. [DOI] [PubMed] [Google Scholar]
- 15.Liu N, Scott JG, 1998. Increased transcription of CYP6D1 causes cytochrome P450-mediated insecticide resistance in house fly. Insect Biochem Mol Biol 28: 531–535. [DOI] [PubMed] [Google Scholar]
- 16.Kasai S, Weerashinghe IS, Shono T, Yamakawa M, 2000. Molecular cloning, nucleotide sequence, and gene expression of a cytochrome P450 (CYP6F1) from the pyrethroid-resistant mosquito, Culex quinquefasciatus say. Insect Biochem Mol Biol 30: 163–171. [DOI] [PubMed] [Google Scholar]
- 17.Scott JG, Wen Z, 2001. Cytochromes P450 of insects: the tip of the iceberg. Pest Manag Sci 57: 958–967. [DOI] [PubMed] [Google Scholar]
- 18.Feyereisen R, 2015. Insect P450 inhibitors and insecticides: challenges and opportunities. Pest Manag Sci 71: 793–800. [DOI] [PubMed] [Google Scholar]
- 19.Strode C, et al. 2008. Genomic analysis of detoxification genes in the mosquito Aedes aegypti. Insect Biochem Mol Biol 38: 113–123. [DOI] [PubMed] [Google Scholar]
- 20.Marcombe S, et al. 2009. Exploring the molecular basis of insecticide resistance in the dengue vector Aedes aegypti: a case study in Martinique island (French West Indies). BMC Genomics 10: 494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bariami V, Jones CM, Poupardin R, Vontas J, Ranson H, 2012. Gene amplification, ABC transporters and cytochrome P450s: unraveling the molecular basis of pyrethroid resistance in the dengue vector, Aedes aegypti. PLoS Negl Trop Dis 6: e1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.David JP, Strode C, Vontas J, Nikou D, Vaughan A, Pignatelli PM, Louis C, Hemingway J, Ranson H, 2005. The Anopheles gambiae detoxification chip: a highly specific microarray to study metabolic-based insecticide resistance in malaria vectors. Proc Nat Acad Sci USA 102: 4080–4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Enayati AA, Ranson H, Hemingway J, 2005. Insect glutathione transferases and insecticide resistance. Insect Mol Biol 14: 3–8. [DOI] [PubMed] [Google Scholar]
- 24.Lumjuan N, McCarroll L, Prapanthadara LA, Hemingway J, Ranson H, 2005. Elevated activity of an epsilon class glutathione transferase confers DDT resistance in the dengue vector, Aedes aegypti. Insect Biochem Mol Biol 35: 861–871. [DOI] [PubMed] [Google Scholar]
- 25.Ranson H, Hemingway J, 2005. Mosquito glutathione transferases. Methods Enzymol 401: 226–241. [DOI] [PubMed] [Google Scholar]
- 26.Muller P, Donnelly MJ, Ranson H, 2007. Transcription profiling of a recently colonised pyrethroid resistant Anopheles gambiae strain from Ghana. BMC Genomics 8: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muller P, Chouaïbou M, Pignatelli P, Etang J, Walker ED, Donnelly MJ, Simard F, Ranson H, 2008. Pyrethroid tolerance is associated with elevated expression of antioxidants and agricultural practice in Anopheles arabiensis sampled from an area of cotton fields in northern Cameroon. Mol Ecol 17: 1145–1155. [DOI] [PubMed] [Google Scholar]
- 28.Marcombe S, et al. 2012. Insecticide resistance in the dengue vector Aedes aegypti from Martinique: distribution, mechanisms and relations with environmental factors. PLoS One 7: e30989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vontas JG, Small GJ, Hemingway J, 2001. Glutathione S-transferases as antioxidant defence agents confer pyrethroid resistance in Nilaparvata lugens. Biochem J 357: 65–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kostaropoulos I, Papadopoulos AI, Metaxakis A, Boukouvala E, Papadopoulou-Mourkidou E, 2001. Glutathione S-transferase in the defence against pyrethroids in insects. Insect Biochem Mol Biol 31: 313–319. [DOI] [PubMed] [Google Scholar]
- 31.Ranson H, Claudianos C, Ortelli F, Abgrall C, Hemingway J, Sharakhova MV, Unger MF, Collins FH, Feyereisen R, 2002. Evolution of supergene families associated with insecticide resistance. Science 298: 179–181. [DOI] [PubMed] [Google Scholar]
- 32.Ortelli F, Rossiter LC, Vontas J, Ranson H, Hemingway J, 2003. Heterologous expression of four glutathione transferase genes genetically linked to a major insecticide-resistance locus from the malaria vector Anopheles gambiae. Biochem J 373: 957–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lumjuan N, Rajatileka S, Changsom D, Wicheer J, Leelapat P, Prapanthadara LA, Somboon P, Lycett G, Ranson H, 2011. The role of the Aedes aegypti epsilon glutathione transferases in conferring resistance to DDT and pyrethroid insecticides. Insect Biochem Mol Biol 41: 203–209. [DOI] [PubMed] [Google Scholar]
- 34.Holt RA, et al. 2002. The genome sequence of the malaria mosquito Anopheles gambiae. Science 298: 129–149. [DOI] [PubMed] [Google Scholar]
- 35.Nene V, et al. 2007. Genome sequence of Aedes aegypti, a major arbovirus vector. Science 316: 1718–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arensburger P, et al. 2010. Sequencing of Culex quinquefasciatus establishes a platform for mosquito comparative genomics. Science 330: 86–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.David JP, Ismail HM, Chandor-Proust A, Paine MJ, 2013. Role of cytochrome P450s in insecticide resistance: impact on the control of mosquito-borne diseases and use of insecticides on earth. Philos Trans R Soc Lond B Biol Sci 368: 20120429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chiu TL, Wen Z, Rupasinghe SG, Schuler MA, 2008. Comparative molecular modeling of Anopheles gambiae CYP6Z1, a mosquito P450 capable of metabolizing DDT. Proc Natl Acad Sci USA 105: 8855–8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hardstone MC, Komagata O, Kasai S, Tomita T, Scott JG, 2010. Use of isogenic strains indicates CYP9M10 is linked to permethrin resistance in Culex pipiens quinquefasciatus. Insect Mol Biol 19: 717–726. [DOI] [PubMed] [Google Scholar]
- 40.Wang Z, Gerstein M, Snyder M, 2009. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilhelm BT, Landry JR, 2009. RNA-seq-quantitative measurement of expression through massively parallel RNA-sequencing. Methods 48: 249–257. [DOI] [PubMed] [Google Scholar]
- 42.Ozsolak F, Milos PM, 2011. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet 12: 87–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ministry of Health , 2016. Ministry of Health Annual Report 2016. Hanoi, Vietnam: MOH Vietnam. [Google Scholar]
- 44.Kawada H, Higa Y, Nguyen YT, Tran SH, Nguyen HT, Takagi M, 2009. Nationwide investigation of the pyrethroid susceptibility of mosquito larvae collected from used tires in Vietnam. PLoS Negl Trop Dis 3: e391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huber K, Le Loan L, Hoang TH, Tien TK, Rodhain F, Failloux AB, 2003. Aedes aegypti in south Vietnam: ecology, genetic structure, vectorial competence and resistance to insecticides. South East Asian J Trop Med Public Health 34: 81–86. [PubMed] [Google Scholar]
- 46.Bingham G, Strode C, Tran L, Khoa PT, Jamet HP, 2011. Can piperonyl butoxide enhance the efficacy of pyrethroids against pyrethroid-resistant Aedes aegypti? Trop Med Int Health 16: 492–500. [DOI] [PubMed] [Google Scholar]
- 47.Vu DH, Nguyen TBN, Do TH, Nguyen TBL, 2004. Susceptibility of Aedes aegypti to insecticides in Viet Nam. Dengue Bull 28: 179–183. [Google Scholar]
- 48.Khoa PT, Hieu HV, Hung MN, 2016. Major resistant mechanism to insecticides of Aedes aegypti mosquito: a vector of dengue and Zika virus in Vietnam. SM Trop Med J 1: 1010. [Google Scholar]
- 49.Tran TD, Nguyen VD, Vu DC, Ho DT, 2016. Mapping insecticide resistance in dengue vectors in the northern Viet Nam, 2010–2013. Vector Biol J 1: 1. [Google Scholar]
- 50.Lien NTK, Ngoc NTH, Hien NT, Hoang NH, Binh NTH, 2018. Two novel mutations in the voltage-gated sodium channel associated with knockdown resistance (kdr) in the dengue vector Aedes aegypti in Vietnam. J Vector Ecol 43: 184–189. [DOI] [PubMed] [Google Scholar]
- 51.Rueda LM, 2004. Pictorial keys for the identification of mosquitoes (Diptera: Culicidae) associated with dengue virus transmission. Zootaxa 589: 1. [Google Scholar]
- 52.World Health Organization , 2016. Test Procedures for Insecticide Resistance Monitoring in Malaria Vector Mosquitoes. Geneva, Switzerland: WHO. [Google Scholar]
- 53.Trapnell C, Pachter L, Salzberg SL, 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M, 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676. [DOI] [PubMed] [Google Scholar]
- 55.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B, 2008. Mapping and quantifying mammalian transcriptomes by RNAseq. Nat Methods 5: 621–628. [DOI] [PubMed] [Google Scholar]
- 56.Wang K, Li M, Hakonarson H, 2010. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M, 2012. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res 40: 109–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup , 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Duong TT, Ngoc NTH, Binh NTH, Kien LT, Khiet DM, Lien NTK, 2018. Assessment responsibility of Aedes aegypti with insecticide in some study sides by real-time PCR. J Malar Parasite Dis Control 3: 74–81. [Google Scholar]
- 60.David JP, Coissac E, Melodelima C, Poupardin R, Riaz MA, Chandor-Proust A, Reynaud S, 2010. Transcriptome response to pollutants and insecticides in the dengue vector Aedes aegypti using next-generation sequencing technology. BMC Genomics 11: 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vontas J, Ranson H, Alphey L, 2010. Transcriptomics and disease vector control. BMC Biol 8: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brooke BD, Koekemoer LL, 2010. Major effect genes or loose confederations? The development of insecticide resistance in the malaria vector Anopheles gambiae. Parasit Vectors 3: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stevenson BJ, et al. 2011. Cytochrome P450 6M2 from the malaria vector Anopheles gambiae metabolizes pyrethroids: sequential metabolism of deltamethrin revealed. Insect Biochem Mol Biol 41: 492–502. [DOI] [PubMed] [Google Scholar]
- 64.Lima VS, Pinto AC, Rafael MS, 2015. Effect of isodillapiole on the expression of the insecticide resistance genes GSTE7 and CYP6N12 in Aedes aegypti from central Amazonia. Genet Mol Res 14: 16728–16735. [DOI] [PubMed] [Google Scholar]
- 65.Ahmad M, Denholm I, Bromilow RH, 2006. Delayed cuticular penetration and enhanced metabolism of deltamethrin in pyrethroid-resistant strains of Helicoverpa armigera from China and Pakistan. Pest Manag Sci 62: 805–810. [DOI] [PubMed] [Google Scholar]
- 66.Lin Y, Jin T, Zeng L, Lu Y, 2012. Cuticular penetration of b-cypermethrin in insecticide-susceptible and resistant strains of Bactrocera dorsalis. Pestic Biochem Physiol 103: 189–193. [Google Scholar]
- 67.Wood OR, Hanrahan S, Coetzee M, Koekemoer LL, Brooke BD, 2010. Cuticle thickening associated with pyrethroid resistance in the major malaria vector Anopheles funestus. Parasit Vectors 3: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gregory R, et al. 2011. A de novo expression profiling of Anopheles funestus, malaria vector in Africa, using 454 pyrosequencing. PLoS One 6: e17418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Balabanidou V, et al. 2016. Cytochrome P450 associated with insecticide resistance catalyzes cuticular hydrocarbon production in Anopheles gambiae. Proc Natl Acad Sci USA 113: 9268–9273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yahouédo GA, Chandre F, Rossignol M, Ginibre C, Balabanidou V, Mendez NGA, Pigeon O, Vontas J, Cornelie S, 2017. Contributions of cuticle permeability and enzyme detoxification to pyrethroid resistance in the major malaria vector Anopheles gambiae. Sci Rep 7: 11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kasai S, Komagata O, Itokawa K, Shono T, Ng LC, Kobayashi M, Tomita T, 2014. Mechanisms of pyrethroid resistance in the dengue mosquito vector, Aedes aegypti: target site insensitivity, penetration, and metabolism. PLoS Negl Trop Dis 8: e2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Riaz MA, Chandor-Proust A, Dauphin-Villemant C, Poupardin R, Jones CM, Strode C, Régent-Kloeckner M, David JP, Reynaud S, 2013. Molecular mechanisms associated with increased tolerance to the neonicotinoid insecticide imidacloprid in the dengue vector Aedes aegypti. Aquat Toxicol 126: 326–337. [DOI] [PubMed] [Google Scholar]