Abstract.
Plasmodium malariae is a neglected malaria parasite. It has wide geographic distribution and, although often associated with mild malaria, is linked to a high burden of anemia and nephrotic syndromes. Here, we report a cohort study conducted in the Kanchanaburi Province of Thailand during May 2013–June 2014 in which P. malariae infection was detected. Of the 812 study participants, two were found to be infected with P. malariae. One had an infection that led to acute malaria, but the other was positive for P. malariae at multiple visits during the study and apparently had chronic asymptomatic infection. Such persistent infection may explain how P. malariae has been able to thrive at very low prevalence and represents a challenge for malaria elimination.
INTRODUCTION
Plasmodium malariae is one of the five Plasmodium species causing human malaria. It has broad geographic distribution, being present in many parts of Asia, the western Pacific, South America, and Africa. Although commonly believed to cause mild malaria, P. malariae infection has been linked to a high burden of anemia and a chronic nephrotic syndrome.1,2 Plasmodium malariae infection can last a very long time, sometimes several decades.3–5
Compared with Plasmodium falciparum and Plasmodium vivax, the two major malaria parasite species, P. malariae has received much less attention from the research community. Much of the current knowledge about the parasitology and clinical illness caused by this parasite was obtained through records of malaria therapy, a treatment of neurosyphilis by induced malaria in the first half of the 20th century.6,7 There is only a dearth of information about the parasite in its natural settings. In Africa, P. malariae is mostly found as mixed infection with P. falciparum.8–11 In Papua New Guinea (PNG), P. malariae contributed to approximately 4% of malaria infection.12 In Brazil, asymptomatic infection of P. malariae was detected by nested polymerase chain reaction (PCR) in a few individuals at multiple time points during a 21-month follow-up, consistent with long-lasting low-density infection.13 The recent use of sensitive molecular diagnosis suggested that the prevalence of P. malariae in many early studies may have been significantly underestimated because of the limited sensitivity of light microscopy (LM).14–16 By quantitative PCR (qPCR), 15% P. malariae prevalence was observed in 5- to 9-year-old children from PNG.17
In this study, we report P. malariae infection in a hypoendemic area of Thailand near the Thailand–Myanmar border. Two unrelated individuals were found to be infected with the parasite during a cohort study of 812 participants between May 2013 and June 2014.
METHODS
Characteristics of the study area.
The study was carried out in Ban Bong Ti Bon village of Bong Ti subdistrict, Sai Yok district, Kanchanaburi Province. This village had a population of approximately 800, which was composed mostly of Thai and Karen ethnic origin. Agriculture was the main occupation. Malaria is seasonal with the peak season during May–August. According to a report from the Thai Ministry of Public Health, the number of malaria patients in the country has significantly declined during the years 2012–2016. The annual malaria case numbers were 16,196 (P. malariae: 48) in 2012, 14,740 (P. malariae: 80) in 2013, 11,352 (no P. malariae infection reported) in 2014, 12,637 (P. malariae: 26) in 2015, and 15,451 (P. malariae: 26) in 2016.18 In Bong Ti subdistrict, P. vivax and P. falciparum were the predominant species contributing to more than 99% of all infections. Four clinical cases of P. malariae detected by LM and none for Plasmodium ovale and Plasmodium knowlesi were reported by the ministry during these 4 years.18
Sample collection.
Blood samples were collected from each of 812 individuals every 4 weeks during May 2013–June 2014. In total, 14 active case detection (ACD) visits per individual were made during the study. In addition, samples were obtained by passive case detection (PCD) when participants presented to the local malaria clinic. Demographic and behavioral factors often associated with malaria infection were collected by interview based on questionnaire. Finger-prick blood samples (250 μL) were collected into BD microtainers™ with K2 ethylene diamine tetraacetic acid (EDTA) as anticoagulant, 50 µL of which was transferred immediately to another tube with 250 µL of RNAprotect cell reagent (Qiagen, Hilden, Germany) for RNA preservation. Both the original blood and blood in RNAprotect were stored on dry ice until arrival at the laboratory in Bangkok where they were transferred to −20°C until DNA/RNA extraction. Plasmodium detection was completed approximately 1 month after each visit. At the time of the study (2013–2014), the Thai national guideline for malaria treatment required confirmation by positive blood smears within 7 days. As a result, no antimalarial treatment was given to PCR-positive individuals of our study.
Informed consent was obtained from each individual or a legal guardian before participation in the study. The study was approved by the Ethics Committee of the Faculty of Tropical Medicine, Mahidol University (Ethics Committee approval number TMEC 13-020).
DNA extraction and purification.
Genomic DNA (gDNA) was extracted from 200 µL blood pellet samples with FavorPrep 96-well genomic DNA extraction kit (Favorgen, Ping-Tung, Taiwan) and stored in 100 µL elution buffer. Genomic DNA samples were kept at −20°C before analyses.
Plasmodium detection.
All DNA samples were screened by genus-specific qPCR assay (QMAL) targeting 18S rRNA genes as previously described.19 Four microliters of purified DNA, corresponding to 8 µL whole blood, was used as the template for QMAL. Six hundred and nine samples were identified as Plasmodium positive. Analyses of P. falciparum and P. vivax infections are being performed for separate publication.
Detection of P. malariae, P. ovale, and P. knowlesi.
Because of the limited quantity of available DNA, detection of P. malariae, P. ovale, and P. knowlesi was by quantitative reverse transcription PCR (qRT-PCR). Only QMAL-positive blood samples were examined. RNA was extracted from the 50-µL portion of blood stored in RNAprotect using the RNeasy Plus Mini Kit (Qiagen). Purified RNA was eluted in 50 µL water and stored at −80°C. Quantitative reverse transcription PCR was performed using 2 µL of RNA as the template, using the SuperScript III Platinum one-step qRT-PCR kit. Most primer and probe sequences have been previously published (Table 1).19,20
Table 1.
Primers and probes for quantitative reverse transcription PCR (qRT-PCR)
| Species | Primer | Sequence (5′ > 3′) |
|---|---|---|
| Plasmodium malariae20 | Mal_fw | TGC CGA CTA GGT GTT GGA TGA T |
| Mal_rev | CTA GTG AGT TTC CCC GTG TTG AGT | |
| Mal_probe | HEX – TGT TTC TTT TAG ATA GCT TCC TTC AG – BFQ | |
| Plasmodium ovale20 | Ova_fw | CCA GCT CCA ATA GCG TAT ATT AAA |
| Ova_rev* | ACA CAT TTT GSA TAA GGA ATG CAA AG | |
| Ova_probe* | HEX – TAT AAG ATG CTT AGR CAA TAC AAC GTA TCT G – BFQ | |
| Plasmodium knowlesi | Kno_fw | GTT AGC GAG AGC CAC AAA AAA GCG AAT |
| Kno_rev* | ACT CAA AGT AAC AAA ATC TTC CAT A | |
| Kno_probe* | HEX – TGC TTT ATG TGC GCA TCC TCT ACC TA – BFQ |
Primers and probes validated for qRT-PCR targeting 18S rRNA.
* Oligo includes a wobble: S = G/C; R = A/G.
Nested PCR to confirm Plasmodium species in PCD samples.
Nested PCR for malaria diagnosis was modified from the original method by Kimura et al.,21 using purified gDNA as the template. The sequences of primers used in nested PCR are shown in Table 2 with modified oligonucleotides (MR_W2 and KR_W1). The first PCR uses primers P1F and P2R (genus-specific primers) to amplify Plasmodium 18S rRNA genes. The PCR product for this first reaction was then used as the template for species-specific semi-nested amplification with FR, MR_W2, OR, VR, or KR_W1 as the reverse primer and PF1 as the forward primer. The thermocycle of both the first and the nested reaction was as follows: 1) 94.0°C for 10:00 minutes; 2) 35 rounds of denaturation at 95.0°C for 0:30 minutes, annealing at 60.0°C for 1:30 minutes, and extension at 72.0°C for 1:00 minutes; and 3) 72.0°C for 5:00 minutes. The PCR product was then analyzed by agarose gel electrophoresis.
Table 2.
Primers and probes for nested PCR21
| Primer name | Sequence |
|---|---|
| P1F | 5′-ACG ATC AGA TAC CGT CGT AAT CTT-3′ |
| P2R | 5′-ACG ATC AGA TAC CGT CGT AAT CTT-3′ |
| FR | 5′-CAA TCT AAA AGT CAC CTC GAA AGA TG-3′ |
| MR_W2 | 5′-AAG GAA GCT ATC TAA AAG AAA CAC TCA T-3′ |
| OR | 5′-ACT GAA GGA AGC AAT CTA AGA AAT TT-3′ |
| VR | 5′-CAA TCT AAG AAT AAA CTC CGA GAG GAA A-3′ |
| KR_W1 | 5′-CTG AAG GAA GCA ATC TAA GAG TTC-3′ |
Primers used in nested PCR to identify the species of Plasmodium parasite.
Plasmodium malariae merozoite surface protein-1 gene (Pmmsp-1) sequence analysis.
Nested PCR was used to amplify region 1, a highly polymorphic region, of Pmmsp-1 as described previously.22 The PCR amplicon was subjected to dye-terminator sequencing (Bioneer, Daejeon, Republic of Korea). DNA sequence chromatograms were examined visually to confirm good quality before sequence alignment.
RESULTS
Plasmodium malariae infection.
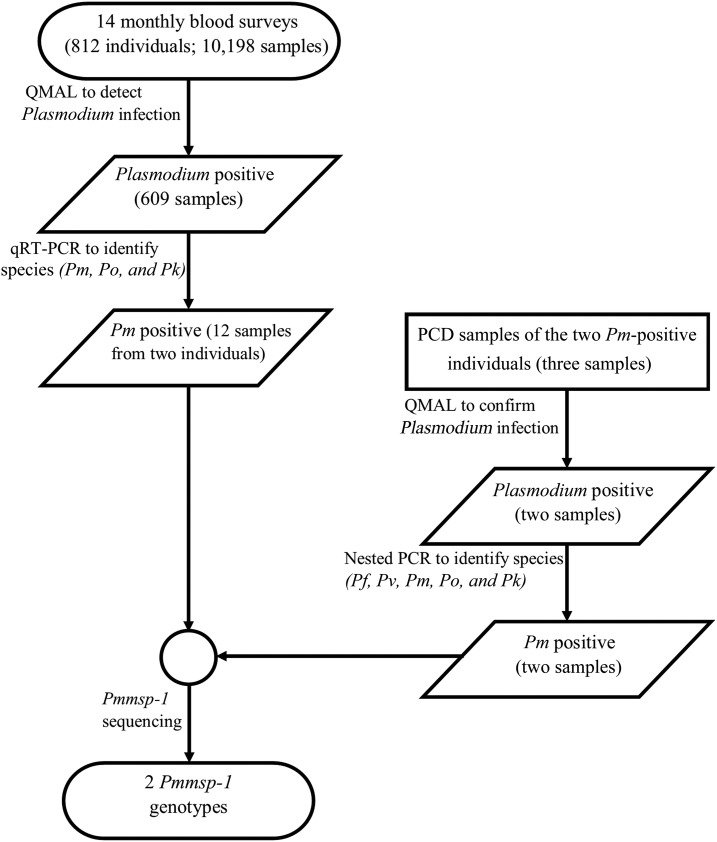
Parasite detection workflow is summarized in Figure 1. Finger-prick blood samples were collected from each of the 812 participants at monthly intervals. In total, 14 ACD visits were made, resulting in 10,198 blood samples. These samples were examined for Plasmodium infection by a genus-specific qPCR assay QMAL,19 609 of which were positive for Plasmodium sp. Quantitative reverse transcription PCR was performed to identify infection of P. ovale, P. knowlesi, and P. malariae. Of the 609 Plasmodium-positive samples, 12 were positive for P. malariae by qRT-PCR and none for P. ovale and P. knowlesi. In addition to these 12 P. malariae–positive samples from ACD, two additional P. malariae–positive samples were identified by nested PCR; these two samples were obtained from PCD visits when the participants presented to the local malaria clinic. These 14 P. malariae–positive samples were derived from only two study participants, 11 from the first and three from the second.
Figure 1.
Study outline. Pf = Plasmodium falciparum; Pk = Plasmodium knowlesi; Pm = Plasmodium malariae; Pmmsp-1 = Plasmodium malariae merozoite surface protein-1 gene; Po = Plasmodium ovale; Pv = Plasmodium vivax; PCD = passive case detection; QMAL = genus-specific quantitative PCR assay; qRT-PCR = quantitative reverse transcription PCR.
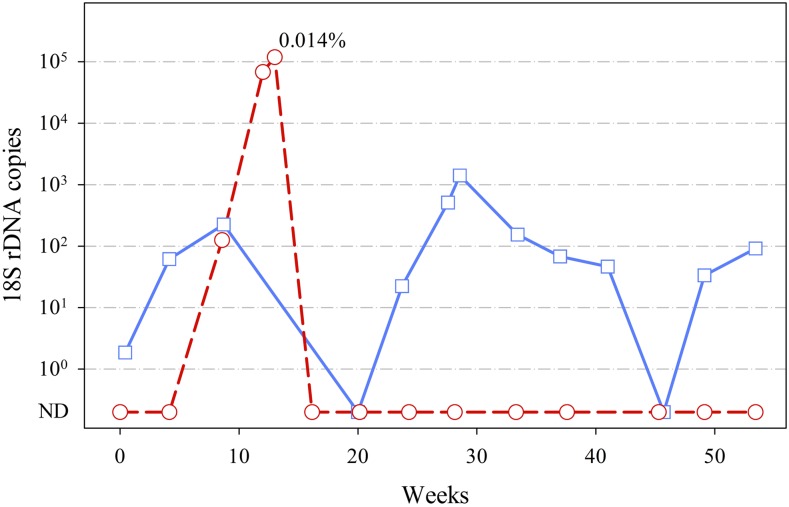
The first infected participant was a 41-year-old man. Despite repeated detections of P. malariae infection, he had no malaria-like symptoms at all ACD visits. The participant presented once to the Bong Ti malaria clinic on November 29, 2013 (week 27), with fever, but his blood sample was negative by LM, and he did not receive antimalaria treatment. Parasite densities of these P. malariae–positive samples were always very low. Their 18S rDNA copy numbers from QMAL were at least 100-fold lower than that of the second participant who suffered a clinical episode of malaria (Figure 2). Samples were not collected at weeks 12 and 16 because the participant temporarily left the study site. The samples from weeks 20 and 45 were negative by QMAL, suggesting that the parasite was either absent or present below to the limit of detection.
Figure 2.
Time course of Plasmodium malariae infections. Shown are the copy numbers of Plasmodium 18S rDNA detected in each QMAL reaction (equivalent to 8 μL of blood). Blue solid line with squares, 1st participant; red dashed line with circles, 2nd participant. 18S rDNA copy below 100 considered as QMAL-negative samples in which parasite was ND; 0.014% indicates the parasitemia of participant 2 at the clinical passive case detection visit on August 20, 2013, as determined by light microscopy. ND = not detected; QMAL = genus-specific quantitative PCR assay. This figure appears in color at www.ajtmh.org.
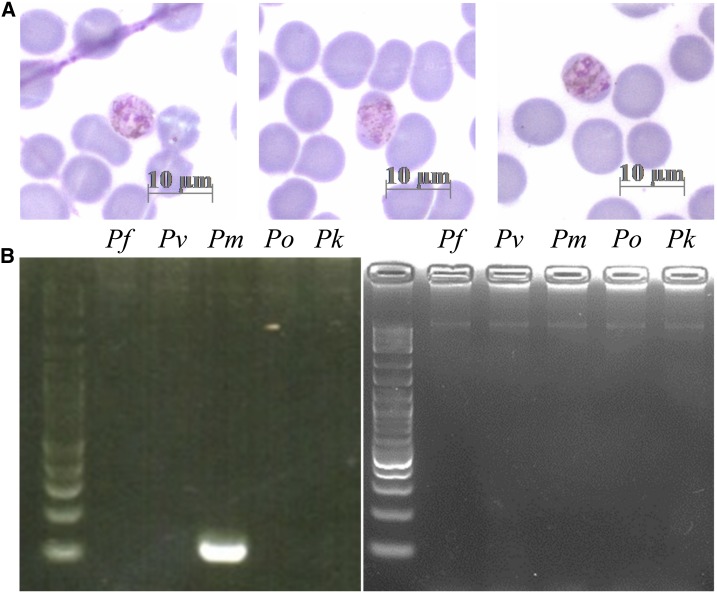
The second infected participant contributed three infected blood samples. This participant was a 54-year-old woman who lived in the same village as the first participant. She reported never to have had malaria before the study. The first two samples were from two consecutive ACD visits on July 19, 2013 (week 9), and August 13, 2013 (week 12). At both times, this individual appeared healthy and had none of the usual malaria symptoms. The third sample was obtained from PCD on August 20, 2013 (week 13), when the participant presented to the Bong Ti malaria clinic with fever. This time, the participant’s blood was positive for malaria parasites by LM with 0.014% parasitemia. The parasite showed characteristics of P. malariae trophozoites, including lack of erythrocyte enlargement and lack of Schüffner’s dots (Figure 3A). Nested PCR of this infection confirmed that the infection was due to P. malariae (Figure 3B). The parasite was apparently cleared by the P. vivax radical cure prescribed by the malaria clinic staff. None of the following nine blood samples collected from this individual over the next months was PCR positive for any Plasmodium species. Taken the results together, it appears that a new infection which was first detected on July 19, 2013 (week 9), developed into acute illness on August 20, 2013 (week 13).
Figure 3.
Blood sample diagnosis of clinical Plasmodium malariae. (A) A thin smear of clinical P. malariae infection from the second participant on August 20, 2013. (B) Nested PCR to identify Plasmodium species. Left panel: gel electrophoresis of PCR products using blood DNA of the second participant from August 20, 2013, as the template. Right panel: negative controls of PCR without DNA template. Left lane of each panel: 100-bp molecular weight marker. Pf = Plasmodium falciparum; Pk = Plasmodium knowlesi; Pm = Plasmodium malariae; Po = Plasmodium ovale; Pv = Plasmodium vivax. This figure appears in color at www.ajtmh.org.
Plasmodium malariae infections in the two participants were genetically distinct.
Because the two participants infected with P. malariae lived only 1.2 km from each other, it was possible that the infection was transmitted from one to the other through local Anopheline mosquitoes. To determine whether the parasites carried by the participants shared the same genotype, we sequenced segments of the highly polymorphic gene Pmmsp-1 from all P. malariae samples. All samples from the first participant had the same Pmmsp-1 sequence, but this sequence was different from that of the second participant (Table 3). Thus, the parasites from the two participants were clearly distinct.
Table 3.
Nucleotide sequences of the polymorphic regions (base 140–146, 175–220, and 240–265) of Plasmodium malariae merozoite surface protein-1 gene (Pmmsp-1) from 14 P. malariae–positive blood samples
| Samples | Nucleotide position of Pmmsp-1 | ||
|---|---|---|---|
| 140–146 | 175–220 | 240–265 | |
| MSP1-MM1A* | TAATAT | ACATCGCTGATGAGAATAAAAAATTAGAGGCTCCTAGTGAATCAGG | AAGAATTGTAATGAAAAACAGAAAAT |
| 1st participant | |||
| Week 0 | TAAAAT | ACATCGCTACTAAGAATAAAGAATTAGAGGCTCATAGTGGATCAGG | AAGAATTGTGCTAAAAAACAGGAAAT |
| Week 4 | TAAAAT | ACATCGCTACTAAGAATAAAGAATTAGAGGCTCATAGTGGATCAGG | AAGAATTGTGCTAAAAAACAGGAAAT |
| Week 9 | TAAAAT | ACATCGCTACTAAGAATAAAGAATTAGAGGCTCATAGTGGATCAGG | AAGAATTGTGCTAAAAAACAGGAAAT |
| Week 24 | TAAAAT | ACATCGCTACTAAGAATAAAGAATTAGAGGCTCATAGTGGATCAGG | AAGAATTGTGCTAAAAAACAGGAAAT |
| Week 27† | TAAAAT | ACATCGCTACTAAGAATAAAGAATTAGAGGCTCATAGTGGATCAGG | AAGAATTGTGCTAAAAAACAGGAAAT |
| Week 28 | TAAAAT | ACATCGCTACTAAGAATAAAGAATTAGAGGCTCATAGTGGATCAGG | AAGAATTGTGCTAAAAAACAGGAAAT |
| Week 33 | TAAAAT | ACATCGCTACTAAGAATAAAGAATTAGAGGCTCATAGTGGATCAGG | AAGAATTGTGCTAAAAAACAGGAAAT |
| Week 37 | TAAAAT | ACATCGCTACTAAGAATAAAGAATTAGAGGCTCATAGTGGATCAGG | AAGAATTGTGCTAAAAAACAGGAAAT |
| Week 41 | TAAAAT | ACATCGCTACTAAGAATAAAGAATTAGAGGCTCATAGTGGATCAGG | AAGAATTGTGCTAAAAAACAGGAAAT |
| Week 49 | TAAAAT | ACATCGCTACTAAGAATAAAGAATTAGAGGCTCATAGTGGATCAGG | AAGAATTGTGCTAAAAAACAGGAAAT |
| Week 53 | TAAAAT | ACATCGCTACTAAGAATAAAGAATTAGAGGCTCATAGTGGATCAGG | AAGAATTGTGCTAAAAAACAGGAAAT |
| 2nd participant | |||
| Week 9 | TAATAT | ACATCACTACTACGAATAACGAATTAGTGACGTCTAATGTACCAAG | ACGAATTGTGATAAAAAACAGAAAAT |
| Week 12 | TAATAT | ACATCACTACTACGAATAACGAATTAGTGACGTCTAATGTACCAAG | ACGAATTGTGATAAAAAACAGAAAAT |
| Week 13† | TAATAT | ACATCACTACTACGAATAACGAATTAGTGACGTCTAATGTACCAAG | ACGAATTGTGATAAAAAACAGAAAAT |
Bases with bold characters represent single-nucleotide polymorphism found. Only highly polymorphic segments of the gene are shown for clarity. All other sequenced gene segments display the same clustering within each individual.
* Reference sequence (GenBank: FJ824669.1).
† Samples obtained through passive case detection.
DISCUSSION
Only a handful of studies have described P. malariae infections in the malaria-endemic areas of mainland Southeast Asia.23–26 Plasmodium malariae malaria is sporadically reported and caused only 0.2% of total malaria cases in Thailand during 2012–2016.18 Our study, conducted between May 2013 and June 2014, identified two individuals out of 812 residents infected with P. malariae. The finding was consistent with the recent report by the Thai Ministry of Public Health, in which P. malariae was present in the study area.16
Our first infected participant was a 41-year-old male agriculturist. Throughout the study, this participant had no symptoms at all ACD visits. However, he was positive for P. malariae at multiple visits. Because this participant reported to have had malaria several times in the past, it is likely that he had acquired protective immunity from previous exposure to the parasite. DNA sequencing revealed a single Pmmsp-1 genotype, suggesting persistent infection of a single parasite line. The absence of parasites at ACD visit 6 (week 20) and visit 12 (week 46) was likely due to fluctuation of parasitemia to a level lower than the detection limit of qPCR. The participant presented to the local malaria clinic with fever once on November 29, 2013 (week 27), but was diagnosed negative for malaria by LM. He was not treated with antimalarial drug. Given that the parasitemia at this time was low (on the order of 0.0001%) and that he did not return to the clinic again during the study period, the fever was likely due to another cause. The repeated detections of a clonal line of the parasite at multiple time points from this participant provide direct evidence for chronic low-density asymptomatic infection of P. malariae in its natural setting.
In contrast to the first participant, the second participant developed clinical malaria after two consecutive positive detections by qPCR. The timing of the first detection suggests that the parasite could be detected as early as 5 weeks before the onset of symptoms. This incubation time sits well within the range of the reported pre-patent period (time from mosquito transmission to first appearance of parasite on thick film) of 16–59 days.11 Although Giemsa smear showed features of P. malariae, the parasite was initially misidentified as P. vivax by the malaria clinic personnel, and the patient was treated with P. vivax radical cure according to Thailand’s malaria treatment guideline, that is, 3 days of chloroquine (25 mg/kg total dose) + 14 days of primaquine (15 mg/day). Although the treatment was successful, this misdiagnosis exemplifies the frequent incorrect species identification of P. malariae27,28 and microscopist bias toward the more prevalent P. vivax.
In summary, two cases of P. malariae infection were identified in a cohort of 812 participants living in an endemic site of Thailand. The characteristics of infection suggest that one infection was chronic and asymptomatic, whereas the other culminated in acute illness. The observed persistence of infection without symptoms may provide a source of transmission that helps sustain P. malariae at very low prevalence. To accelerate malaria elimination in Thailand and other endemic countries, intervention such as mass screening and treatment aiming to remove such a residual source of transmission may be necessary.
Acknowledgments:
This project was supported by the Office of Higher Education Commission and Mahidol University under the National Research Universities Initiative, the TransEPI consortium funded by the Bill & Melinda Gates Foundation (www.gatesfoundation.org), and grants (D43TW006571 and U19AI089672) from the National Institutes of Health (NIH). W. N. was supported by a Wellcome Trust Intermediate Fellowship in Public Health and Tropical Medicine (101073/Z/13Z) and I. M. by an NHMRC Senior Research Fellowship (#1043345).
REFERENCES
- 1.Langford S, Douglas NM, Lampah DA, Simpson JA, Kenangalem E, Sugiarto P, Anstey NM, Poespoprodjo JR, Price RN, 2015. Plasmodium malariae infection associated with a high burden of anemia: a hospital-based surveillance study. PLoS Negl Trop Dis 9: e0004195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eiam-Ong S, 2003. Malarial nephropathy. Semin Nephrol 23: 21–33. [DOI] [PubMed] [Google Scholar]
- 3.Hommel B, Galloula A, Simon A, Buffet P, 2013. Hyposplenism revealed by Plasmodium malariae infection. Malar J 12: 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morovic M, Poljak I, Miletic B, Troselj-Vukic B, Seili-Bekafigo I, Milotic I, 2003. Late symptomatic Plasmodium malariae relapse in the territory of the former Yugoslavia. J Travel Med 10: 301–302. [DOI] [PubMed] [Google Scholar]
- 5.Vinetz JM, Li J, McCutchan TF, Kaslow DC, 1998. Plasmodium malariae infection in an asymptomatic 74-year-old Greek woman with splenomegaly. N Engl J Med 338: 367–371. [DOI] [PubMed] [Google Scholar]
- 6.Lupascu G, Constantinescu P, Negulici E, Shute PG, Maryon ME, 1968. Parasitological and clinical investigations on infections with the VS Romanian strain of Plasmodium malariae transmitted by Anopheles labranchiae atroparvus. Bull World Health Organ 38: 61–67. [PMC free article] [PubMed] [Google Scholar]
- 7.McKenzie FE, Jeffery GM, Collins WE, 2001. Plasmodium malariae blood-stage dynamics. J Parasitol 87: 626–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Browne EN, Frimpong E, Sievertsen J, Hagen J, Hamelmann C, Dietz K, Horstmann RD, Burchard GD, 2000. Malariometric update for the rainforest and savanna of Ashanti region, Ghana. Ann Trop Med Parasitol 94: 15–22. [PubMed] [Google Scholar]
- 9.Molineaux L, Storey J, Cohen JE, Thomas A, 1980. A longitudinal study of human malaria in the west African Savanna in the absence of control measures: relationships between different Plasmodium species, in particular P. falciparum and P. malariae. Am J Trop Med Hyg 29: 725–737. [DOI] [PubMed] [Google Scholar]
- 10.McKenzie FE, Bossert WH, 1999. Multispecies Plasmodium infections of humans. J Parasitol 85: 12–18. [PMC free article] [PubMed] [Google Scholar]
- 11.Collins WE, Jeffery GM, 2007. Plasmodium malariae: parasite and disease. Clin Microbiol Rev 20: 579–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Genton B, D’Acremont V, Rare L, Baea K, Reeder JC, Alpers MP, Muller I, 2008. Plasmodium vivax and mixed infections are associated with severe malaria in children: a prospective cohort study from Papua New Guinea. PLoS Med 5: e127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Alencar FEC, Malafronte RDS, Cerutti Junior C, Natal Fernandes L, Buery JC, Fux B, Rezende HR, Duarte AMRC, Medeiros-Sousa AR, Miranda AE, 2018. Assessment of asymptomatic Plasmodium spp. infection by detection of parasite DNA in residents of an extra-Amazonian region of Brazil. Malar J 17: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Camargo-Ayala PA, Cubides JR, Nino CH, Camargo M, Rodriguez-Celis CA, Quinones T, Sanchez-Suarez L, Patarroyo ME, Patarroyo MA, 2016. High Plasmodium malariae prevalence in an endemic area of the Colombian Amazon region. PLoS One 11: e0159968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roh ME, Oyet C, Orikiriza P, Wade M, Kiwanuka GN, Mwanga-Amumpaire J, Parikh S, Boum Y, 2nd, 2016. Asymptomatic Plasmodium infections in children in low malaria transmission setting, southwestern Uganda. Emerg Infect Dis 22: 1494–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Alencar FEC, Malafronte RDS, Cerutti C, Jr., Natal Fernandes L, Buery JC, Fux B, Rezende HR, Miranda AE, 2017. Reassessment of asymptomatic carriers of Plasmodium spp. in an endemic area with a very low incidence of malaria in extra-Amazonian Brazil. Malar J 16: 452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hofmann NE, et al. 2017. The complex relationship of exposure to new Plasmodium infections and incidence of clinical malaria in Papua New Guinea. Elife 6: e23708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Department of Disease Control MoPH, Thailand , 2018. Thailand Malaria Elimination Program. Available at: http://203.157.41.215/malariar10/report/malaria_home_main.php. Accessed March 15, 2018.
- 19.Wampfler R, Mwingira F, Javati S, Robinson L, Betuela I, Siba P, Beck HP, Mueller I, Felger I, 2013. Strategies for detection of Plasmodium species gametocytes. PLoS One 8: e76316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosanas-Urgell A, Mueller D, Betuela I, Barnadas C, Iga J, Zimmerman PA, del Portillo HA, Siba P, Mueller I, Felger I, 2010. Comparison of diagnostic methods for the detection and quantification of the four sympatric Plasmodium species in field samples from Papua New Guinea. Malar J 9: 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimura M, Kaneko O, Qing L, Mian Z, Kawamoto F, Wataya Y, Otani S, Yamaguchi Y, Tanabe K, 1997. Identification of the Four Species of Human PCR Diagnosis for Four Human Malaria Parasites. Bangkok, Thailand: The Toyota Foundation Mini-Symposium on Malaria, Faculty of Tropical Medicine, Mahidol University. [Google Scholar]
- 22.Guimaraes LO, et al. 2015. Merozoite surface protein-1 genetic diversity in Plasmodium malariae and Plasmodium brasilianum from Brazil. BMC Infect Dis 15: 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanomsing N, Mayxay M, Newton PN, Nosten F, Dolecek C, Hien TT, White NJ, Day NP, Dondorp AM, Imwong M, 2014. Genetic variability of Plasmodium malariae dihydropteroate synthase (dhps) in four Asian countries. PLoS One 9: e93942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakeesathit S, Saralamba N, Pukrittayakamee S, Dondorp A, Nosten F, White NJ, Imwong M, 2016. Limited polymorphism of the Kelch propeller domain in Plasmodium malariae and P. ovale isolates from Thailand. Antimicrob Agents Chemother 60: 4055–4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khim N, et al. 2012. Reduced impact of pyrimethamine drug pressure on Plasmodium malariae dihydrofolate reductase gene. Antimicrob Agents Chemother 56: 863–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pongponratn E, Prommano O, Chaisri U, Viriyavejakul P, Wilairatana P, 2012. Plasmodium malariae-infected erythrocytes in the peripheral blood, liver, stomach and duodenum: an ultrastructural study. Southeast Asian J Trop Med Public Health 43: 1080–1086. [PubMed] [Google Scholar]
- 27.Savargaonkar D, Shah N, Das MK, Srivastava B, Valecha N, 2014. Plasmodium malariae infection: a case of missed diagnosis. J Vector Borne Dis 51: 149–151. [PubMed] [Google Scholar]
- 28.Cao Y, et al. 2016. The increasing importance of Plasmodium ovale and Plasmodium malariae in a malaria elimination setting: an observational study of imported cases in Jiangsu province, China, 2011–2014. Malar J 15: 459. [DOI] [PMC free article] [PubMed] [Google Scholar]