Abstract.
In 2016, a chikungunya virus (CHIKV) outbreak was reported in Mandera, Kenya. This was the first major CHIKV outbreak in the country since the global reemergence of this virus in Kenya in 2004. We collected samples and sequenced viral genomes from this outbreak. All Kenyan genomes contained two mutations, E1:K211E and E2:V264A, recently reported to have an association with increased infectivity, dissemination, and transmission in the Aedes aegypti vector. Phylogeographic inference of temporal and spatial virus relationships showed that this variant emerged within the East, Central, and South African lineage between 2005 and 2008, most probably in India. It was also in India where the first large outbreak caused by this virus appeared, in New Delhi, 2010. More importantly, our results also showed that this variant is no longer contained to India. We found it present in several major outbreaks, including the 2016 outbreaks in Pakistan and Kenya, and the 2017 outbreak in Bangladesh. Thus, this variant may have a capability of driving large CHIKV outbreaks in different regions of the world. Our results point to the importance of continued genomic-based surveillance and prompt urgent vector competence studies to assess the level of vector susceptibility and virus transmission, and the impact this might have on this variant’s epidemic potential and global spread.
INTRODUCTION
In May 2016, the Kenyan Ministry of Health (KMoH) reported an outbreak of chikungunya virus (CHIKV) in Mandera County on the border with Somalia. During this time in Somalia, outbreaks of CHIKV were occurring in the neighboring Bula Hawa region, originating from Mogadishu. In Mandera town, 1,792 cases were detected, and an estimated 50% of the health work force was affected by this virus. A cross-border joint response was coordinated between Kenya and Somalia to control the outbreak.1 This was the first reported outbreak of CHIKV in Kenya since 2004.
The previous large CHIKV outbreak in Kenya occurred in Lamu Island in 2004, with an estimated 75% of the population infected.2 The disease also spread to the coastal city of Mombasa by the end of 2004, and further to the Comoros and La Réunion islands, causing large outbreaks in 2005–2006. On La Réunion island, unusual clinical complications were reported in association with CHIKV infection, and viral isolate sequences revealed the presence of an alanine-to-valine mutation in the E1 glycoprotein at position 226 (E1:A226V).3 This specific mutation was shown to confer significant increase in CHIKV infectivity for the Aedes albopictus vector, which was also the dominant mosquito species suspected to be responsible for the transmission of CHIKV on the island of La Réunion.4,5 Since then, the E1:A226V mutation has been observed in many of the genomes in the CHIKV lineage, spreading in the East, Central, and South African region (ECSA lineage), and has been shown to have emerged through convergent evolution in at least four different occasions.6
The remarkable emergence and spread of CHIKV adaptation to the Ae. albopictus vector prompted additional studies on the genetic plasticity of this virus, looking for additional biomarkers associated with virus transmission capacity, fitness, and pathogenicity.6–8 Mutations with the ability to enhance infection in this vector are of increased importance, as Ae. albopictus is rapidly expanding throughout the world.9 The Ae. albopictus mosquito is believed to have originated in Asia and is today most commonly found in east Asia. Aedes albopictus is also common in some parts of South and Southeast Asia, India, and Africa, and it has shown increased spread to regions with lower temperatures, such as southern Europe, southern Brazil, northern China, and the northern United States.9 In Europe, this vector has been associated with autochthonous transmission of CHIKV.10
Although some of the more recent CHIKV outbreaks have been transmitted by Ae. albopictus, most of the CHIKV transmissions in the world are associated with Aedes aegypti. Aedes aegypti is a container-breeding, domesticated mosquito mainly found in urban areas and feeding largely on humans during early and late daytime hours. Aedes aegypti originated from the ancestral zoophilic Ae. aegypti formosus native to Africa. Aedes aegypti now is most common in tropical and subtropical regions, such as Africa, India, Southeast Asia, and Australia.9 Recently, two mutations, E1:K211E and E2:V264A, have been reported in the CHIKV to be associated with increased fitness to Ae. aegypti vectors.11 These two mutations, in the background of the wild-type E1:226A, are believed to increase virus infectivity, dissemination, and transmission in Ae. aegypti, with no impact on virus fitness for the Ae. albopictus vector.6,11 E1:K211E was first observed in genomes sampled in 2006 from the Kerala and Puducherry regions, India, and simultaneous presence of both mutations was first observed in 2009–2010 in Tamil Nadu and Andhra Pradesh, India.12,13
In Kenya, the predominant CHIKV vector is Ae. aegypti, and vector competence studies using local vector populations have shown it is capable of transmitting the virus in this region.14 Given the recent large outbreak of CHIKV in the rural setting of Mandera County, Kenya, we analyzed CHIKV genomes sequenced from this outbreak for the presence of adaptive mutations associated with both Ae. albopictus and Ae. aegypti. Along with estimating origins and the time of emergence of a variant that carried two of these previously reported mutations, we also estimated time and origins of the Mandera CHIKV outbreak. Our results indicate the spread of a CHIKV that carries mutations previously associated with increased fitness for Ae. aegypti in Kenya. We also show that this variant is now connected with new large outbreaks in other regions of the world.
MATERIALS AND METHODS
Ethics statement.
The study was carried out on a protocol approved by the Walter Reed Army Institute of Research (#2189) and Kenya Medical Research Institute’s (KEMRI) Scientific and Ethics Review Unit (#3035) as an overarching protocol guiding investigation and reporting of arbovirus/hemorrhagic fever outbreaks in Kenya. Because the blood samples were collected from an outbreak and no human data were collected, this was deemed to be a nonhuman research study and no consent was required. The protocol was approved for additional analysis of outbreak samples and for publication of results.
Samples and sequencing.
From May 2016, following reports of widespread incidence of febrile illness with severe joint pains in Mandera (a city at the border with Somalia) and its environs, samples were collected from suspected cases of all ages and gender, using standard practices by the KMoH staff. The case definition used was “any patient presenting with sudden onset of fever > 38.5°C, with severe joint/muscle pains and headache within the last 3–5 days within Mandera County, the person should either be a resident of or visiting Mandera.” Chikungunya infection was confirmed by CHIKV-specific reverse transcription polymerase chain reaction (RT-PCR) and partial genome Sanger sequencing at the KEMRI laboratories. Vero cell culture inoculations were performed on the samples to obtain isolates for in-depth studies. For method details on infection confirmation, virus isolation, and Sanger sequencing, see Supplemental Appendix 1. A subset of chikungunya-confirmed positive samples from acutely ill patients were further subjected to high throughput full-genome sequencing at the KEMRI laboratories. The prepared complementary DNA (cDNA) was quantified using Qubit 3.0 fluorimeter and the dsDNA HS Assay Kit (Life Technologies–Thermo Fisher Scientific, Saint Aubin, France). The cDNA was fragmented enzymatically and tagged using the Nextera XT DNA Library kit (Illumina, San Diego, CA). Each sample was assigned a unique barcode sequence using the sequence libraries prepared with the Nextera-XT kit (Illumina), following the protocols and reagents supplied by Illumina, and sequenced on the MiSeq platform according to the manufacturer’s instructions (Illumina). Ten full genomes and five partial genomes were assembled by combining both de novo and reference mapping assemblies. De novo assemblies were performed using Abyss and Trinity, and reference mapping was performed using ngs_mapper.15–17 Sequences have been submitted to the GenBank under accession numbers MH423797–MH423811.
Phylogenetic analyses.
All full-genome CHIKV genomes were downloaded from the GenBank and curated in TempEst.18 The curation consisted of inference of a neighbor joining tree followed by linear regression of root to tip distances, given the sampling time. Genomes with too much or too little divergence as would be expected, given their root to tip distance and collection date, were considered as outliers and were removed from the dataset. In addition, all genomes with long stretches of Ns, genomes without collection date, and genomes without sampling location were also removed from the reference dataset. To determine the CHIKV lineage of the assembled genomes from Mandera, Kenya, these sequences were aligned to the constructed reference dataset (n = 466), using MEGAv7.19 This large dataset was also scanned for the presence of recombination using Recombination Detection Program version 4, with a minimum of three methods with significant (P < 0.05) recombination signal required to call a genome recombinant.20 Maximum likelihood (ML) trees were inferred using PhyML and RaxML, with general time reversible + gamma + invariant sites (GTR+G+I) (GTR+G for RaxML) model of evolution, as determined by jModelTest2.21–23 Node confidence values were derived by approximate likelihood-ratio test (aLRT) (PhyML) and bootstrap of 500 (RaxML). After evaluating the temporal structure using TempEst, the trees were used for an informed down-sampling of the dataset. For this analysis, only ECSA lineage genomes were kept as reference, identical genomes from the same location and sampling year were removed, as were genomes from a phylogenetically distant sub-clade only found to spread in Asia. The down-sampled dataset (ECSA lineage only, n = 115) was analyzed using the BEAST package v1.8.3, with GTR+G+I model of substitution, bayesian skyline plot, relaxed lognormal molecular clock, and asymmetrical trait (location) distribution.24 BEAST was run for 800 million generations, with subsampling every 80,000 and 10% burn-in. The maximum clade credibility (MCC) tree was summarized using TreeAnnotator. Ancestral states were reconstructed using ML, and all amino acid mutations at each node for the trees were mapped using TreeTime.25 Because viral genomes from several additional recent CHIKV outbreaks form India and Bangladesh were only sequenced in their E1 gene, we also inferred the ML tree of the partial E1 region including these sequences, using PhyML and TN93+G+I model of evolution, as determined by jModelTest2.
Selection and protein structure analyses.
Selection analyses were performed using the HyPhy package.26 The presence of site-specific selection was determined by a likelihood approach using FEL and by Bayesian method FUBAR with a probability level threshold of 0.95.27,28 Episodic selection was investigated using MEME, and any selection acting on tree branches was determined by aBSREL.29,30 Selection analyses were performed on the large full-genome dataset used for the ML trees (ECSA only) and the small dataset used for BEAST analyses. FUBAR was performed on separated structural and nonstructural genes from both datasets because of their large sizes. Positive selection on a site was determined present if the site was found to be under selection by all three methods that determine site-specific selective pressures (FEL, FUBAR, and MEME). Structure figures of CHIKV E1E2 (Protein Data Bank ID:3J2W31) were visualized and prepared using PyMOL (The PyMOL Molecular Graphics System; DeLano Scientific, Palo Alto, CA).
RESULTS
Chikungunya virus causing the 2016 outbreak in Mandera, Kenya, was introduced in 2015.
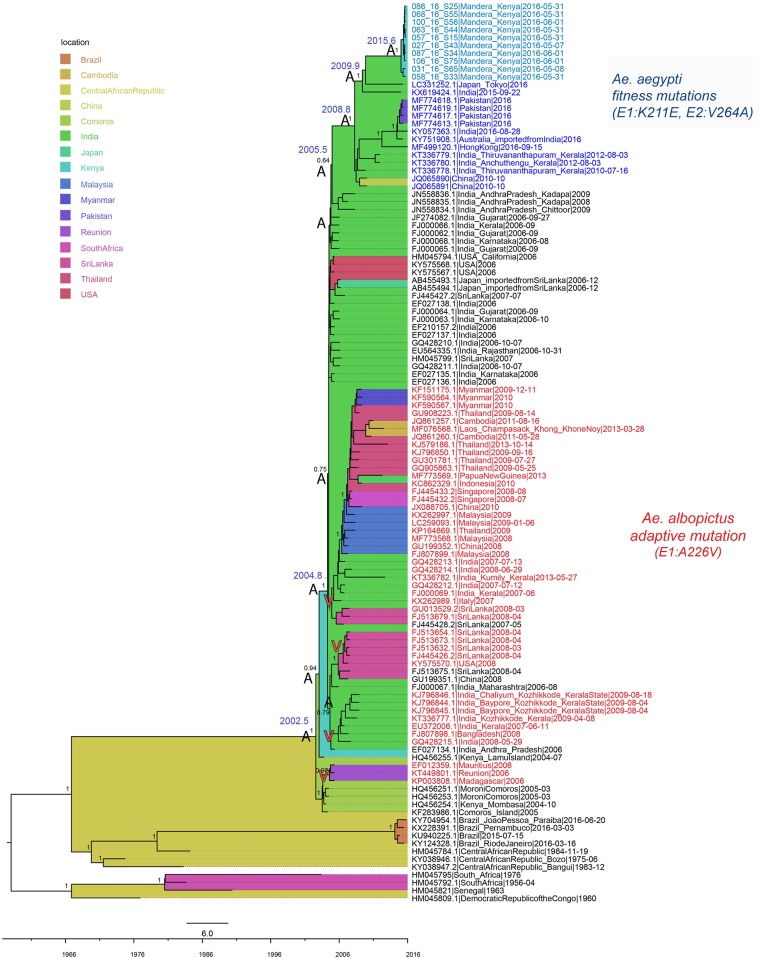
A total of 15 samples from the CHIKV outbreak in Mandera, Kenya, collected in May and June 2016, were sequenced. Ten of the sequenced samples produced full CHIKV genomes and five of the samples had partial genomes. No recombination was detected in the genomes. Maximum likelihood trees inferred by PhyML and RaxML were concordant and showed that all Kenyan genomes belonged to the ECSA lineage of CHIKV. Consistently with their outbreak origins, the genomes from Mandera clustered in a monophyletic clade defined by very short branches, indicating limited genetic diversity (Figure 1). The Kenyan cluster was most closely related to genomes from Japan and India; however, the long branch leading to this cluster indicated a probable lack of sampling of viruses more related to the Kenyan outbreak. Two genomes from the CHIKV outbreak in Kenya in 2004 were located more basally in the tree and did not cluster together with the viruses causing the Kenyan outbreak of 2016. The most recent common ancestor (MRCA) of the Kenyan 2016 viruses existed in mid-2015 (2015.6; highest posterior density (HPD) 95% = 2015.1–2015.9), making this the latest possible time point of introduction of this virus into Mandera, Kenya. The ancestor shared with the most closely related genomes from Japan and India existed in late 2009 (2009.9; 95% HPD = 2008.6–2011.5) and was estimated to have originated in India. These results indicate that the CHIKV causing the 2016 outbreak in Kenya was introduced into this region by 2015. Although the results suggest that the Mandera virus originated from a virus that existed in India in 2009, the long branch leading to the Kenyan cluster indicates missing data. Thus, India might not have been the direct source of the CHIKV introduction into Kenya, and more sampling is needed to, with greater precision, determine the exact origin of this introduction.
Figure 1.
Full-genome maximum clade credibility tree of the chikungunya virus East, Central, and South African lineage. Estimated location origin is marked in colored tree background, according to the legend. Taxa in red text represent genomes containing the Aedes albopictus–adaptive E1:A226V mutation, whereas all other taxa contain the wild type. Taxa in blue (light blue for Kenya) represent genomes with the E1:K211E and E2:V264A mutations, previously associated with increased fitness in Aedes aegypti. Most important node supports are shown, as well as the estimated times of the most recent common ancestors (TMRCAs) for nodes of interest. Ancestral amino acid states (A and V) at position E1:226 are plotted on the nodes to illustrate evolutionary paths of the Aedes albopictus and Ae. aegypti mutation variants. This figure appears in color at www.ajtmh.org.
Mutations associated with possible increased fitness to Ae. aegypti emerged by 2008.
Careful analyses of amino acid mutations associated with vector adaptation revealed that all Kenyan viruses contained two mutations, E1:K211E and E2:V264A, as well as the E1:226A background amino acid (Table 1). In the background of E1:226A, E1:K211E and E2:V264A have recently been correlated with enhancement of CHIKV fitness for the Ae. aegypti vectors from India. Further investigation of amino acids from the genomes surrounding the Kenyan samples in both the ML and MCC phylogenetic trees revealed a cluster of genomes, sampled from various regions of the world (Asia, Africa, and Australia), also containing the two Ae. aegypti fitness-associated mutations in the background of E1:226A (Figure 1, Table 1). These two amino acid changes were the only ones that characterized this cluster.
Table 1.
Changes in positions previously associated with vector competence and pathology of chikungunya virus
| Protein | Amino acid substitution | Phenotype | Asian lineage | ECSA lineage without Aedes aegypti–adapted cluster | ECSA Ae. aegypti–adapted cluster | Kenya |
|---|---|---|---|---|---|---|
| E1 | A98T6,7 | Epistatic covariant on E1:A226V | T | A | A | A |
| E1 | A226V4 | Enhanced infection of Aedes albopictus | A | A, V | A | A |
| E1 | K211E11 | Enhanced fitness in Ae. aegypti in background of E1:226A | E | K, T, N | E | E |
| E2 | V264A11 | V | V | A | A | |
| E2 | R198Q6 | Enhanced infection of Ae. albopictus, synergistic with E3:18F in background of E1:226V | R | R, Q | R | R |
| E2 | L210Q6,8 | Enhanced infection of Ae. albopictus, secondary to E2:A226V | L | L, Q | L | L |
| E2 | K233E/Q6 | Enhanced infection of Ae. albopictus | K | K, E | K | K |
| E2 | K234E6 | Enhanced infection of Ae. albopictus | K | K | K | K |
| E2 | K252Q6 | Enhanced infection of Ae. albopictus—secondary mutation | K | K, Q | K | K |
| E3 | S18F6 | Enhanced infection of Ae. albopictus, synergistic with E2:R198Q | S, F | S, F | S | S |
| nsP1 | G230R32 | Increase replication in Ae. albopictus in combination with nsP3:524* | G, R | G, R | G | G |
| V326M32 | V, M | V | V | V | ||
| nsP3 | *524R33 | Attenuation of arthritis and pathology | *, L, C, R | *, C, R | * | * |
ECSA = East, Central, and South African. Amino acids associated with each phenotype are shown in bold underlined font. Only available full genomes from each lineage were compared. Amino acids associated with each phenotype are highlighted in the table in bold and underscored font.
* Opal STOP codon.
The cluster containing the genomes with E1:K211E and E2:V264A mutations consisted of viruses from two different outbreaks, Kenya in 2016 and Pakistan in 2016, and additional genomes from India sampled between 2010 and 2016, and Japan, Hong Kong, and Australia, all sampled in 2016. The MRCA of this cluster was estimated to have existed in India since late 2008 (2008.8; 95% HPD = 2007.9–2009.5), indicating that the virus with dual E1:K211E and E2:V264A mutations emerged by the end of 2008 in this area of the world. The cluster shared a common ancestor with genomes from India, which did not contain the two suggested Ae. aegypti fitness mutations, in mid-2005 (2005.5; 95% HPD = 2005.0–2006.1). Although the node support for the ancestor of viruses with and without the two mutations was low, probably because of lack of sampling that would reveal the exact evolutionary relationships, it was estimated to have existed in India because of well-supported ancestral nodes preceding this one. Thus, these results indicated that the variant with possible increased fitness to Ae. aegypti most probably arose in India sometime between 2005 and 2008. The background E1:226A amino acid (non-red taxa, Figure 1) predominated in this part of the tree, whereas the Ae. albopictus–adaptive E1:226V amino acid (red taxa, Figure 1) was mainly found in the sub-lineage containing genomes from Southeast Asia. Genomes from India sampled in years 2007–2009 were found containing E1:226A and E1:226V mutations, meaning that this country experienced simultaneous spread of both the ancestral wild-type strain and the Ae. albopictus–adapted strain. None of the Ae. albopictus–adapted viruses contained the E1:K211E and E2:V264A changes, which were found occurring only in the presence of the E1:226A wild-type residue. Ancestral state reconstruction further revealed that the variant with the dual E1:K211E and E2:V264A Ae. aegypti fitness mutations did not arise from the Ae. albopictus–adapted strain (Figure 1). Rather, the Ae. aegypti variant evolved directly from the E1:226A wild type that was circulating in India at the same time as the E1:226V Ae. albopictus–adapted virus.
Maximum likelihood trees of the partial E1 region, including additional sequences from the CHIKV outbreaks in 2010 and 2016 from New Delhi, India, and from 2017 in Bangladesh, showed that genomes from these outbreaks also belonged to the cluster with the Ae. aegypti fitness-associated mutations (Figure 2). Despite low phylogenetic signal due to the shorter E1 segment, the cluster was supported by a high confidence value, 0.93. These results indicated that, in addition to the outbreaks of Pakistan and Kenya, the more recent outbreaks in India and Bangladesh were also most probably caused by the variant carrying mutations with possible increased fitness to Ae. aegypti.
Figure 2.
Partial E1 gene maximum likelihood tree of the chikungunya virus East, Central, and South African lineage. Taxa in blue (light blue for Kenya) represent genomes with the Aedes aegypti fitness-associated E1:K211E mutation. This figure appears in color at www.ajtmh.org.
Positive selection was acting on the E1:K211 but not the E2:264 position.
To investigate whether the E1:K211E and E2:V264A mutations appeared because of selective pressure and adaptation of the virus, we performed several tests for the presence of positive selection, on both the large CHIKV dataset used for the ML trees and on the smaller dataset used for BEAST analyses. Significant presence of positive selection in position E1:K211 was detected by FUBAR (probability > 0.99), FEL (P ≤ 0.05), and MEME (P < 0.05) in all tested datasets. Position E2:264 did not show any evidence of positive selection. No branch-specific selection was detected by aBSREL. Positions found to be under positive selection by all methods are listed in Table 2.
Table 2.
Positively selected positions by method
| FUBAR | FEL | MEME | |
|---|---|---|---|
| Maximum likelihood dataset | E1:211 | nsP1:171, nsP3:117, E1:211 | nsP1:4, nsP1:82, nsP1:157, nsP1:171, nsP1:301, nsP1:407, nsP2:349, nsP2:457, nsP2:604, nsP3:117, nsP3:303, nsP4:81, nsP4:467, nsP4:605, E2:57, E2:178, 6K:47, E1:146, E1:211, E1:382 |
| BEAST dataset | E1:211 | nsP1:171, nsP3:117, E1:211 | nsP1:4, nsP1:101, nsP1:171, nsP2:349, nsP3:117, nsP4:81, nsP4:467, E2:57, 6K:47, E1:146, E1:211 |
E1:K211E and E2:V264A mutations introduce charge and hydrophobicity changes to the CHIKV E protein.
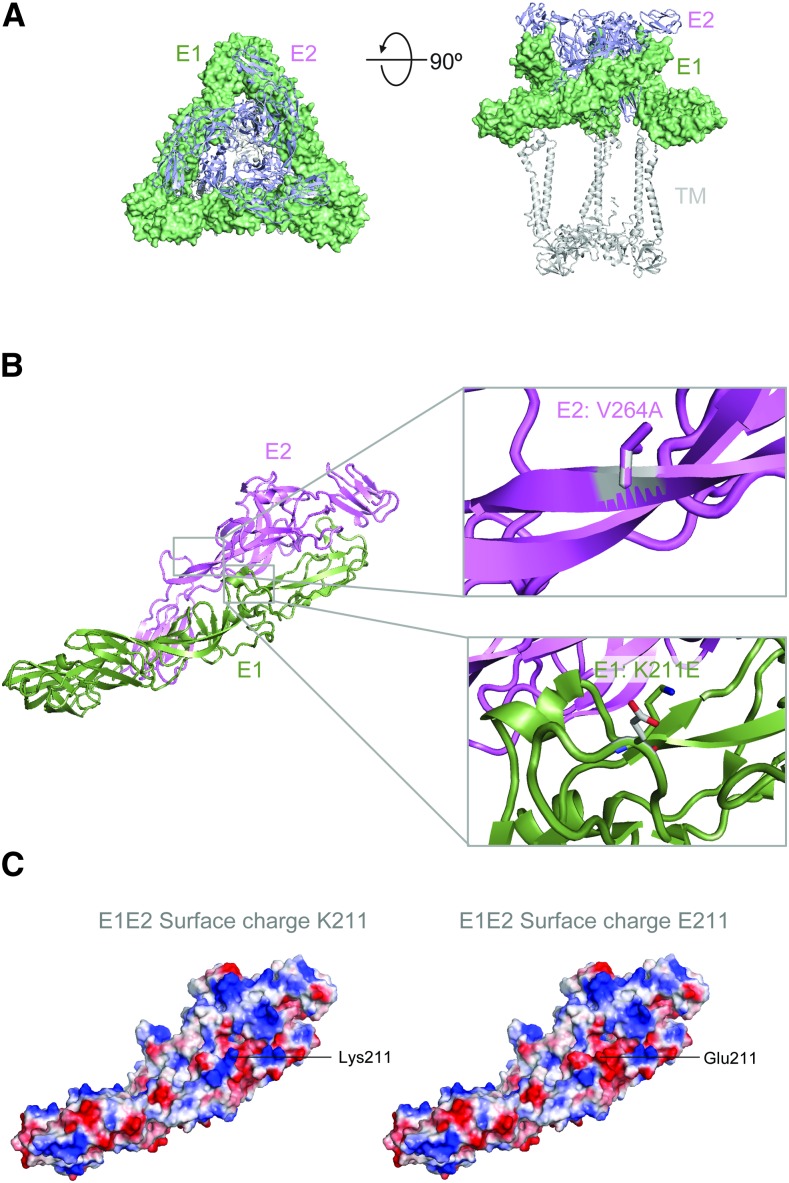
To investigate the potential impact of the E1:K211E and E2:V264A mutations on the structure of the CHIKV envelope protein, we modeled these mutations on a 3D structure of the E1E2 viral surface glycoprotein trimer molecule (Figure 3A).31 Our results show that both E1:211 and E2:264 residues are located centrally within the E1E2 heterodimer (Figure 3B). Our results also indicate that the K211E mutation causes a local change in the surface charge of the molecule (Figure 3C) from positive to negative. The V264A mutation reduces the hydrophobicity of this surface-exposed region.34,35
Figure 3.
Structure analysis of chikungunya virus (CHIKV) mutations. (A) Structure of the CHIKV E1E2 viral surface glycoprotein trimer molecule31 (PDB ID: 3J2W) is shown in two orientations with E1 in surface representation (green) and E2 in ribbon representation (blue). (B) A single E1E2 heterodimer is shown in ribbon representation with close-up windows showing the V264A and K211E mutations in stick representation. (C) The E1E2 heterodimer is shown in surface representation with the surface colored by charge (blue: positive, red: negative, and white: neutral). The Lys211 (left) and mutant Glu211 (right) variants are modeled. This figure appears in color at www.ajtmh.org.
DISCUSSION
Recent reemergence and global spread of CHIKV, coupled with its high morbidity and economic burden, has made it one of the more medically important arboviral diseases with major public health implications. Chikungunya virus originated in Africa, and the first known outbreak was recorded in today’s Tanzania in 1952–1953. Subsequently, sporadic outbreaks in Africa and larger epidemics in Asia were observed until the 1980s, followed by a period of decreased activity. The virus reemerged in the early 2000s in Africa, resulting in extensive and rapid spread throughout the world. In this study, we analyzed 10 CHIKV genomes sampled from the 2016 outbreak in Mandera, Kenya, and compared them with the genomes from the Kenya outbreak in 2004. We estimated the time of the 2016 outbreak emergence, as well as traced the emergence and movement of the previously reported Ae. aegypti fitness mutations that characterized these genomes. Variants with increased vector fitness may have the potential for more efficient transmission, resulting in large outbreaks, and may outcompete wild-type viral variants in the regions with abundant vector populations.
Our results confirm previous findings that reemerging CHIKV reached Kenya in mid-2002.3,36 Here, the virus splits into two variants. One variant caused disease in Mombasa in 2004 and spread further to Comoros, Madagascar, and La Réunion, where it obtained the Ae. albopictus–adaptive mutation E1:226V. The other variant, still carrying the wild-type E1:226A, spread north, causing the Lamu Island outbreak. Although the Lamu Island outbreak was observed first and was the largest in Kenya in 2004, present results suggest it did not seed the virus in Mombasa. Rather, the two shared a common ancestor that split into two different directions. Given that only one genome from each region is available, more data would be needed to completely resolve virus movement within Kenya during this period of time. Exact estimations of intercontinental routes of geographic spread are also limited by sampling skew; however, the wild-type variant from Lamu Island was next observed causing outbreaks in 2005–2006 in India, mainly on the west and south of the country (Rajasthan, Gujarat, Maharashtra, Andhra Pradesh, and Karnataka).37–39 Shortly thereafter, in 2007, a variant carrying the Ae. albopictus–adaptive E1:226V mutation was recorded in the province of Kerala.40 Both variants were found circulating in the following years in the country, between 2007 and 2013.41,42
In India, both Ae. albopictus and Ae. aegypti are prevalent vectors, and the presence of the recently described Ae. aegypti fitness-enhancing mutation E1:K211E was first observed in the Kerala–Puducherry CHIKV outbreak of 2006.12 Shortly thereafter, E1:K211E was accompanied by the second Ae. aegypti fitness-associated mutation, E2:V264A. This novel dual mutation variant was first observed in Kerala in 2009; however, the viruses from the Kerala–Puducherry CHIKV outbreak of 2006 were not sequenced in the E2 region.13 These results support our estimate that the Ae. aegypti variant carrying the E1:K211E and E2:V264A mutations probably arose in India sometime between 2005 and 2008. We show that this variant evolved directly from the E1:226A wild-type virus, and not from the E1:A226V virus with increased transmission efficiency by Ae. albopictus. In fact, the E1:K211E and E2:V264A mutations have not yet been detected in the naturally occurring Ae. albopictus–adapted virus. However, it is worth investigating whether the whole reemerging ECSA genotype virus has other amino acid residues that make it generally more fit for spread by Ae. albopictus.43 In that case, the two reported Ae. aegypti fitness mutations might represent compensatory adaptation of ECSA CHIKV, selected by reintroduction of an Ae. albopictus–adapted virus into regions infested with Ae. aegypti.
Our analyses suggest that the first indication of the E1:K211E and E2:V264A variant’s association with major outbreaks comes from India, where it caused an outbreak in New Delhi in 2010.44 Following this, CHIKV continued to circulate in discrete regions throughout India, and in 2016, the dual mutation variant appeared in another large outbreak in New Delhi.45 Simultaneously, it was also driving the outbreaks in Pakistan and Kenya.46 The Kenyan outbreak was estimated to have originated from a virus imported by 2015 and was most closely related to genomes from India. It is important to note that the implications of spread from India to Kenya are limited by sampling gaps, and India might not have been the direct source of the Kenyan outbreak. Indeed, the connection between the Somalian and Kenyan outbreaks indicates that the virus was most probably introduced from Somalia and, thus, that this variant is most likely spreading in African countries other than Kenya. Further sampling and sequencing in this region will reveal whether this is the case, or if the strong social, cultural, and human movement ties between Kenya and India aided in the dispersion of the virus between the two continents. Importantly, however, our results indicate that the variant carrying the two mutations with possible increased fitness to Ae. aegypti is no longer confined to India, but is spreading and associating with large outbreaks in other countries and other continents of the world. Interestingly, all recent CHIKV ECSA lineage genomes from the Ae. aegypti prevalent regions of the Indian Ocean sub-lineage contain the two Ae. aegypti fitness mutations. More sampling and surveillance are needed to determine whether the emergence of this new variant can thus lead to the replacement of the endemic wild-type virus.
After the 2016 Kenyan outbreak resolved, a new CHIKV outbreak started in the coastal city of Mombasa in 2017–2018, with a large proportion of cases (70%) reporting severe joint pain and high fever.47 No publicly available genomes exist from this event as of yet, but given the proximity of the Ae. aegypti–associated dual mutation virus outbreak of 2016, and given this variant’s association with large outbreaks, it is very possible that it is also responsible for this recent occurrence in Mombasa. Furthermore, other simultaneous CHIKV outbreaks have been reported in proximate regions to both Kenya and India, the 2016 outbreak in Mozambique and the 2017 outbreak in Dhaka, Bangladesh.48,49 The virus from the Dhaka outbreak contained the K211E mutation, and despite the lack of E2 sequencing, our analyses of the E1 region placed it within the Ae. aegypti dual mutation cluster. Thus, these results suggested that this outbreak was also caused by the variant with mutations previously associated with increased CHIKV fitness for Ae. aegypti.49 Sequencing and analyzing complete viral genomes from these and other countries will aid in the tracking of this variant’s spread, and it will provide insight into its possible replacement of the wild type in the Ae. aegypti–prevalent regions of the world.
The epidemic potential of a vector-borne agent depends on several interconnected intrinsic and extrinsic factors, such as viral and vector genetics, vector and host competence, vector abundance, temperature, and rainfall.50 Viral genetics, and the appearance of adaptive vector mutations, has previously been shown to play a role for the vector competence and spread of CHIKV by Ae. albopictus.4 The genetic impact of the recently described E1:K211E and E2:V264A mutations for the spread of CHIKV by Ae. aegypti is still not very well known. Our structural modeling suggests that these two mutations introduce changes in charge and hydrophobicity of the CHIKV envelope surface proteins. Previous studies in chikungunya and other alphaviruses indicate that changes in the E1/E2 proteins affect pH sensitivity and can have dramatic effects on structure and virus production levels.51–53 This phenomenon is seen with surface viral glycoproteins from other viruses as well, including HIV-1 and respiratory syncytial virus, where variants with reduced conformational flexibility have improved expression.54,55 Vector competence has been shown to vary greatly between different populations of Aedes mosquitos, implying importance of vector genetics, in addition to changes in important viral surface protein residues, in driving the spread of CHIKV.56,57 Thus, the possible impact of E1/E2 structural changes by the E1:K211E and E2:V264A mutations on virus infectivity, dissemination, and transmission by Ae. aegypti in different parts of the world could differ, and should be examined further. However, the presence of positive selection on E1:K211E, coupled with the increased spread of E1:K211E and E2:V264A CHIKV variant throughout the world, highlights the importance and the urgency of such studies. The possible level of increased susceptibility of these vectors to CHIKV infection may provide information for implementation of additional or alternate vector-control strategies.
In conclusion, we show that the CHIKV variant, carrying the E1:K211E and E2:A264V mutations previously associated with increased fitness to Ae. aegypti, is capable of rapid spread in the regions of high Ae. aegypti presence. It has been associated with several recent large CHIKV outbreaks in Africa and Asia, showing capacity for global spread. Our study highlights the importance of further sampling, genomic surveillance, and vector competence studies to promptly assess the epidemic potential of this CHIKV variant.
Supplementary Files
Note: Supplemental appendix appears at www.ajtmh.org.
Disclaimer: Material has been reviewed by the Walter Reed Army Institute of Research. There is no objection to its presentation and/or publication. The opinions or assertions contained herein are the private views of the author, and are not to be construed as official, or as reflecting true views of the Department of the Army or the Department of Defense. The investigators have adhered to the policies for protection of human subjects as prescribed in AR 70–25.
REFERENCES
- 1.World Health Organization , 2016. Chikungunya-Kenya. Available at: http://www.who.int/csr/don/09-august-2016-chikungunya-kenya/en/. Accessed July 12, 2018.
- 2.Sergon K, et al. 2008. Seroprevalence of chikungunya virus (CHIKV) infection on Lamu Island, Kenya, October 2004. Am J Trop Med Hyg 78: 333–337. [PubMed] [Google Scholar]
- 3.Schuffenecker I, et al. 2006. Genome microevolution of chikungunya viruses causing the Indian Ocean outbreak. PLoS Med 3: e263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsetsarkin KA, Vanlandingham DL, McGee CE, Higgs S, 2007. A single mutation in chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog 3: e201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weaver SC, 2014. Arrival of chikungunya virus in the new world: prospects for spread and impact on public health. PLoS Negl Trop Dis 8: e2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsetsarkin KA, et al. 2014. Multi-peaked adaptive landscape for chikungunya virus evolution predicts continued fitness optimization in Aedes albopictus mosquitoes. Nat Commun 5: 4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsetsarkin KA, Chen R, Leal G, Forrester N, Higgs S, Huang J, Weaver SC, 2011. Chikungunya virus emergence is constrained in Asia by lineage-specific adaptive landscapes. Proc Natl Acad Sci USA 108: 7872–7877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsetsarkin KA, Weaver SC, 2011. Sequential adaptive mutations enhance efficient vector switching by chikungunya virus and its epidemic emergence. PLoS Pathog 7: e1002412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kraemer MU, et al. 2015. The global distribution of the arbovirus vectors Aedes aegypti and Ae. albopictus. Elife 4: e08347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonilauri P, et al. 2008. Chikungunya virus in Aedes albopictus, Italy. Emerg Infect Dis 14: 852–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Agarwal A, Sharma AK, Sukumaran D, Parida M, Dash PK, 2016. Two novel epistatic mutations (E1:K211E and E2:V264A) in structural proteins of chikungunya virus enhance fitness in Aedes aegypti. Virology 497: 59–68. [DOI] [PubMed] [Google Scholar]
- 12.Kumar NP, Mitha MM, Krishnamoorthy N, Kamaraj T, Joseph R, Jambulingam P, 2007. Genotyping of virus involved in the 2006 chikungunya outbreak in South India (Kerala and Puducherry). Curr Sci 93: 1412–1416. [Google Scholar]
- 13.Sumathy K, Ella KM, 2012. Genetic diversity of chikungunya virus, India 2006–2010: evolutionary dynamics and serotype analyses. J Med Virol 84:462–470. [DOI] [PubMed] [Google Scholar]
- 14.Agha SB, et al. 2017. Vector competence of populations of Aedes aegypti from three distinct cities in Kenya for chikungunya virus. PLoS Negl Trop Dis 11: e0005860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ngs_Mapper. Available at: https://github.com/VDBWRAIR/ngs_mapper.
- 16.Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I. 2009. ABySS: a parallel assembler for short read sequence data. Genome Res 19: 1117–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29: 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rambaut A, Lam TT, Max Carvalho L, Pybus OG, 2016. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol 2: vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar S, Stecher G, Tamura K, 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin DP, Murrell B, Golden M, Khoosal A, Muhire B, 2015. RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol 1: vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guindon S, Gascuel O, 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52: 696–704. [DOI] [PubMed] [Google Scholar]
- 22.Stamatakis A, 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Posada D, 2008. jModelTest: phylogenetic model averaging. Mol Biol Evol 25: 1253–1256. [DOI] [PubMed] [Google Scholar]
- 24.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29: 1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sagulenko P, Puller V, Neher RA, 2018. TreeTime: maximum-likelihood phylodynamic analysis. Virus Evol 4: vex042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pond SL, Frost SD, Muse SV, 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21: 676–679. [DOI] [PubMed] [Google Scholar]
- 27.Murrell B, et al. 2013. FUBAR: a fast, unconstrained bayesian approximation for inferring selection. Mol Biol Evol 30: 1196–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kosakovsky Pond SL, Frost SD, 2005. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol 22: 1208–1222. [DOI] [PubMed] [Google Scholar]
- 29.Murrell B, Wertheim JO, Moola S, Weighill T, Scheffler K, Kosakovsky Pond SL, 2012. Detecting individual sites subject to episodic diversifying selection. PLoS Genet 8: e1002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith MD, Wertheim JO, Weaver S, Murrell B, Scheffler K, Kosakovsky Pond SL, 2015. Less is more: an adaptive branch-site random effects model for efficient detection of episodic diversifying selection. Mol Biol Evol 32: 1342–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun S, Xiang Y, Akahata W, Holdaway H, Pal P, Zhang X, Diamond MS, Nabel GJ, Rossmann MG, 2013. Structural analyses at pseudo atomic resolution of chikungunya virus and antibodies show mechanisms of neutralization. Elife 2: e00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mounce BC, Cesaro T, Vlajnic L, Vidina A, Vallet T, Weger-Lucarelli J, Passoni G, Stapleford KA, Levraud JP, Vignuzzi M, 2017. Chikungunya virus overcomes polyamine depletion by mutation of nsP1 and the opal stop codon to confer enhanced replication and fitness. J Virol 91: e00344-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones JE, et al. 2017. Disruption of the opal stop codon attenuates chikungunya virus-induced arthritis and pathology. MBio 8: e01456-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kyte J, Doolittle RF, 1982. A simple method for displaying the hydropathic character of a protein. J Mol Biol 157: 105–132. [DOI] [PubMed] [Google Scholar]
- 35.Rose GD, Wolfenden R, 1993. Hydrogen bonding, hydrophobicity, packing, and protein folding. Annu Rev Biophys Biomol Struct 22: 381–415. [DOI] [PubMed] [Google Scholar]
- 36.Lo Presti A, Cella E, Angeletti S, Ciccozzi M, 2016. Molecular epidemiology, evolution and phylogeny of chikungunya virus: an updating review. Infect Genet Evol 41: 270–278. [DOI] [PubMed] [Google Scholar]
- 37.Cherian SS, Walimbe AM, Jadhav SM, Gandhe SS, Hundekar SL, Mishra AC, Arankalle VA, 2009. Evolutionary rates and timescale comparison of chikungunya viruses inferred from the whole genome/E1 gene with special reference to the 2005–07 outbreak in the Indian subcontinent. Infect Genet Evol 9:16–23. [DOI] [PubMed] [Google Scholar]
- 38.Pfeffer M, Zoller G, Essbauer S, Tomaso H, Behrens-Riha N, Loscher T, Dobler G, 2008. Clinical and virological characterization of imported cases of chikungunya fever. Wien Klin Wochenschr 120: 95–100. [DOI] [PubMed] [Google Scholar]
- 39.Arankalle VA, Shrivastava S, Cherian S, Gunjikar RS, Walimbe AM, Jadhav SM, Sudeep AB, Mishra AC, 2007. Genetic divergence of chikungunya viruses in India (1963–2006) with special reference to the 2005–2006 explosive epidemic. J Gen Virol 88: 1967–1976. [DOI] [PubMed] [Google Scholar]
- 40.Kumar NP, Joseph R, Kamaraj T, Jambulingam P, 2008. A226V mutation in virus during the 2007 chikungunya outbreak in Kerala, India. J Gen Virol 89: 1945–1948. [DOI] [PubMed] [Google Scholar]
- 41.Naresh Kumar CV, Sivaprasad Y, Sai Gopal DV, 2016. Genetic diversity of 2006–2009 chikungunya virus outbreaks in Andhra Pradesh, India, reveals complete absence of E1:A226V mutation. Acta Virol 60: 114–117. [PubMed] [Google Scholar]
- 42.Abraham R, Manakkadan A, Mudaliar P, Joseph I, Sivakumar KC, Nair RR, Sreekumar E, 2016. Correlation of phylogenetic clade diversification and in vitro infectivity differences among Cosmopolitan genotype strains of chikungunya virus. Infect Genet Evol 37: 174–184. [DOI] [PubMed] [Google Scholar]
- 43.Tsetsarkin KA, McGee CE, Volk SM, Vanlandingham DL, Weaver SC, Higgs S, 2009. Epistatic roles of E2 glycoprotein mutations in adaption of chikungunya virus to Aedes albopictus and Ae. aegypti mosquitoes. PLoS One 4: e6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shrinet J, et al. 2012. Genetic characterization of chikungunya virus from New Delhi reveal emergence of a new molecular signature in Indian isolates. Virol J 9: 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaur N, Jain J, Kumar A, Narang M, Zakaria MK, Marcello A, Kumar D, Gaind R, Sunil S, 2017. Chikungunya outbreak in Delhi, India, 2016: report on coinfection status and comorbid conditions in patients. New Microbes New Infect 20: 39–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu SQ, et al. 2017. Detection, isolation, and characterization of chikungunya viruses associated with the Pakistan outbreak of 2016–2017. Virol Sin 32: 511–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.World Health Organization , 2018. Chikungunya-Mombasa, Kenya. Available at: http://www.who.int/csr/don/27-february-2018-chikungunya-kenya/en/. Accessed July 12, 2018.
- 48.Mugabe VA, et al. 2018. Evidence for chikungunya and dengue transmission in Quelimane, Mozambique: results from an investigation of a potential outbreak of chikungunya virus. PLoS One 13: e0192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Melan A, Aung MS, Khanam F, Paul SK, Riaz BK, Tahmina S, Kabir MI, Hossain MA, Kobayashi N, 2018. Molecular characterization of chikungunya virus causing the 2017 outbreak in Dhaka, Bangladesh. New Microbes New Infect 24: 14–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lounibos LP, Kramer LD, 2016. Invasiveness of Aedes aegypti and Aedes albopictus and vectorial capacity for chikungunya virus. J Infect Dis 214: S453–S458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Akahata W, Nabel GJ, 2012. A specific domain of the chikungunya virus E2 protein regulates particle formation in human cells: implications for alphavirus vaccine design. J Virol 86: 8879–8883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu SR, Haag L, Hammar L, Wu B, Garoff H, Xing L, Murata K, Cheng RH, 2007. The dynamic envelope of a fusion class II virus. Prefusion stages of semliki forest virus revealed by electron cryomicroscopy. J Biol Chem 282: 6752–6762. [DOI] [PubMed] [Google Scholar]
- 53.Lu YE, Eng CH, Shome SG, Kielian M, 2001. In vivo generation and characterization of a soluble form of the Semliki forest virus fusion protein. J Virol 75: 8329–8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanders RW, et al. 2002. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol 76: 8875–8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krarup A, et al. 2015. A highly stable prefusion RSV F vaccine derived from structural analysis of the fusion mechanism. Nat Commun 6: 8143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Honorio NA, Wiggins K, Camara DCP, Eastmond B, Alto BW, 2018. Chikungunya virus vector competency of Brazilian and Florida mosquito vectors. PLoS Negl Trop Dis 12: e0006521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vega-Rua A, Zouache K, Girod R, Failloux AB, Lourenco-de-Oliveira R, 2014. High level of vector competence of Aedes aegypti and Aedes albopictus from ten American countries as a crucial factor in the spread of chikungunya virus. J Virol 88: 6294–6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.