Abstract.
Rotavirus has been one of the major etiological agents causing severe diarrhea in infants and young children worldwide. In Thailand, rotavirus contributes to one-third of reported pediatric diarrheal cases. We studied stool samples from 1,709 children with acute gastroenteritis and 1,761 children with no reported gastroenteritis whose age ranged from 3 months to 5 years from four different regions in Thailand between March 2008 and August 2010. The samples were tested for the presence of rotavirus by real-time reverse transcription–polymerase chain reaction (RT-PCR) amplification of vp6 gene and enzyme-linked immunosorbent assay. The positive samples were further characterized for their G and P genotypes (vp7 and vp4 genes) by conventional RT-PCR. From all four regions, 26.8% of cases and 1.6% of controls were positive for rotavirus, and G1P[8] was the most predominant genotype, followed by G2P[4], G3P[8], and G9P[8]. In addition, the uncommon genotypes including G1P[4], G1P[6], G2P[6], G2P[8], G4P[6], G9P[4], G9P[6], G12P[6], and G12P[8] were also detected at approximately 14% of all samples tested. Interestingly, G5P[19], a recombinant genotype between human and animal strains, and G1P7[5], a reassortant vaccine strain which is closely related to four human-bovine reassortant strains of RotaTeq™ vaccine, were detected in control samples. Data reported in this study will provide additional information on molecular epidemiology of rotavirus infection in Thailand before the impending national implementation of rotavirus vaccination program.
INTRODUCTION
Rotavirus is one of the major causes of diarrhea in infants and young children worldwide. The WHO estimated that rotavirus was responsible for more than 200,000 deaths among children younger than 5 years each year in 2010–2013.1,2 Most of the mortality from rotavirus occurred in developing countries, especially in Asia and sub-Saharan Africa.2 In Asia, the proportion of all diarrheal deaths due to rotavirus by region between 2000 and 2013 was 50.7–54.6% in Southeast Asia, 44.9–49.2% in Western Asia, 35.2–39.2% in Eastern Asia, and 34.1–38.6% in Southern Asia.2
The detection of rotavirus via real-time polymerase chain reaction (PCR) is often analyzed further for its G and P genotypes to yield molecular epidemiological information. Sequences of the two outer capsid proteins, VP4 and VP7, are used to classify rotavirus P and G genotypes, respectively.3 Presently, 36 G genotypes and 51 P genotypes have been identified in humans and animals with various G- and P-genotype combinations.4 The most common global human rotavirus genotypes associated with diarrhea are G1P[8], G2P[4], G3P[8], G4P[8], and G9[8].5 These rotavirus genotype combinations are the major cause of human rotavirus infection in developed countries, causing greater than 90% of rotavirus gastroenteritis cases in children.6 Uncommon G- and P-genotype combinations, which are the result of genetic reassortment between human and animal rotavirus strains, are prevalent in infected children in developing countries.7,8 The WHO reported that 28.8–58% of rotavirus detected in 2013 were of uncommon genotypes in the Southeast Asia region, the Africa region, the eastern Mediterranean region, and the Region of the Americas.9 The presently available rotavirus vaccines, RotaTeq® and Rotarix®, were developed from rotavirus genotypes G1–G4 in combination with P[5] and G1P[8], respectively.10 The prevalence of uncommon rotavirus genotypes detected in different geographical regions of the world may warrant a vaccine that provides cross-protection or coverage against a broader group of genotype combinations.
In Thailand, rotavirus is a major cause of pediatric diarrheal cases contributing to one-third of reported diarrheal cases in children aged 6–11 months, with a peak incidence during the dry and cool season (October–February).11 Genotype G9 was the most predominant genotype detected in Thailand in 2000–2002,12–14 G2 in 2003, G1 in 2004–2009,13–17 and G3 in 2009–2011.17 The P genotype, P[8], predominated in 2000–2011, followed by P[4].13–18 The most common rotavirus genotype combinations detected in Thailand are G1P[8], G9P[8], G2P[4], and G3P[8]. In addition, the uncommon genotypes G2P[8], G3P[3], G3P[9], G3P[10], G3P[19], G12P[6], and G12P[8] were also detected in 2000–2011.19
This study reports the prevalence and molecular epidemiology of rotavirus infections among children with and without acute gastroenteritis from a hospital-based surveillance in four different regions in Thailand in 2008–2010. This study provides useful epidemiological data regarding the circulating rotavirus genotypes before the implementation of a rotavirus vaccine in Thailand.
MATERIALS AND METHODS
Source of specimens.
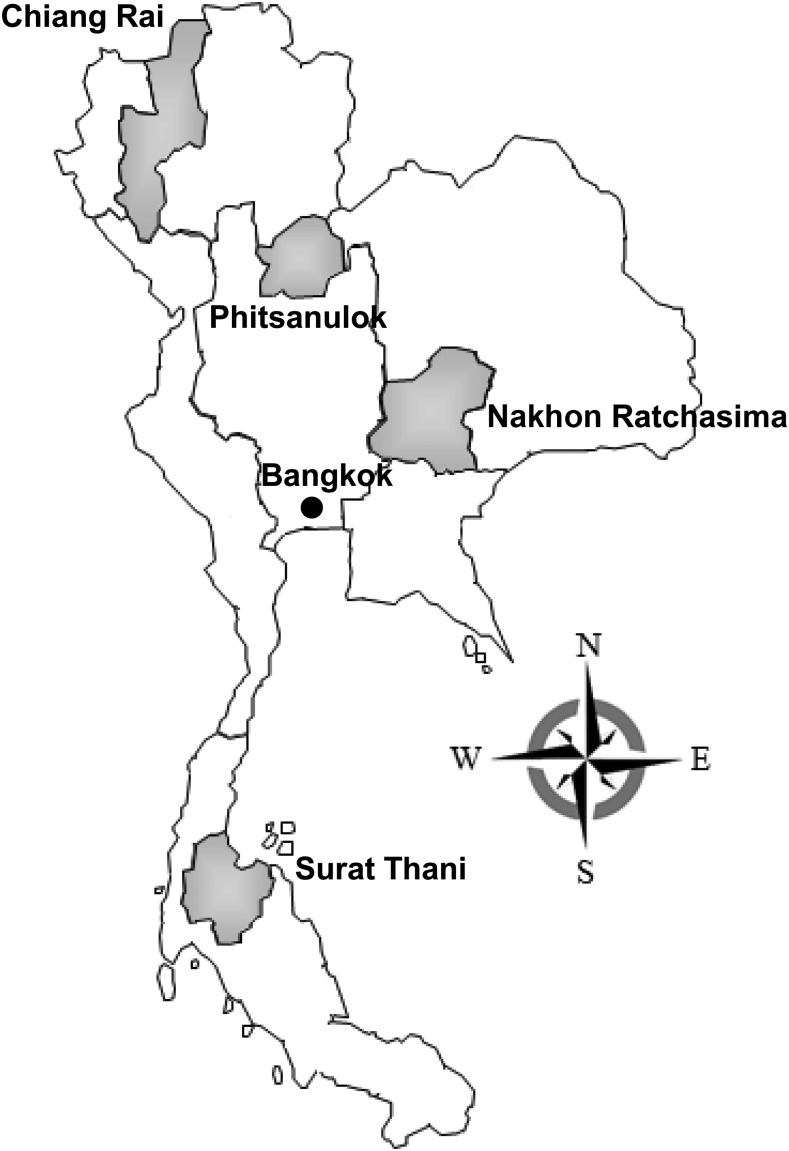
Stool samples were collected as part of regional surveillance for diarrhea etiology in Thai children aged 3–60 months. The surveillance was set up at major hospitals in four regions of Thailand: Chiang Rai (CR) in the northern region, Nakhon Ratchasima (Korat) (KR) in the northeastern region, Phitsanulok (PN) in the central region, and Surat Thani (SR) in the southern region: from March 2008 to August 2010 (Figure 1). Cases were inpatient or outpatient children who had three or more unformed stools per 24 hours with at least one additional symptom (nausea, vomiting, abdominal pain, fatigue/lethargy, or fever). Controls were defined as children of the same age range who were seen or admitted at the same hospital with no history of diarrhea within the past 2 weeks. Samples were frozen at −70°C until processed. The study was approved by the ethical review committees of the Ministry of Public Health, Thailand, and the Walter Reed Army Institute of Research, Silver Spring, MD.
Figure 1.
Location of the four regional sites used in this study: Chiang Rai Province in the northern region, Nakorn Ratchasima Province in the northeastern region, Phitsanulok Province in the central region, and Surat Thani Province in the southern region.
Detection of group a rotavirus.
RNA were extracted from stool samples using NucliSens® Magnetic Extraction kit according to the manufacturer’s instruction (bioMérieux, Lyon, France). The extracted RNA was detected by real-time reverse transcription–polymerase chain reaction (RT-PCR) to amplify the vp6 gene as described previously.20 Antigen detection of rotavirus by enzyme-linked immunosorbent assay (ELISA) using RIDASCREEN® Rotavirus EIA kit (R-Biopharm AG, Darmstadt, Germany) was also performed in parallel.
Genotyping of rotavirus by conventional RT-PCR.
Samples that were positive by real-time RT-PCR and/or ELISA were further characterized for their G and P genotypes by conventional RT-PCR using a method described previously by Silapong et al.20 Briefly, RNA was treated with DNase and was used as a template to generate complementary DNA fragments of vp4 and vp7 genes, which were then used as a template to amplify a smaller fragment for each gene. A size-specific band indicates each G and P genotype. Samples that did not generate a usable template by RT-PCR were characterized further by nested multiple PCR using various primer combinations.20 The presence of a size-specific band indicates each G and P genotype. For samples that did not have identifiable band, vp4 and vp7 genes were amplified and cloned into pSC-A-amp/kan PCR Cloning Vector (StrataClone PCR Cloning Kit; Agilent Technologies, Santa Clara, CA) and sent to a commercial company for sequencing (Macrogen, Seoul, South Korea).
Phylogenetic analyses.
Nucleotide sequences of vp4 (P genotype) and vp7 (G genotype) genes were verified for consensus sequences using Sequencher software version 4.1.2 (Gene Codes Corporation, Ann Arbor, MI). A phylogenetic tree for each genotype was generated using neighbor joining with Kimura’s two-parameter model with 1,000 bootstrap replicates in MEGA version 6.21 Sequences of prototypes for each G and P genotype from GenBank were used as references.
Statistical analysis.
The differences among proportions were analyzed by χ2 test in IBM SPSS® Statistics version 22 (IBM Corporation, Armonk, NY).
RESULTS
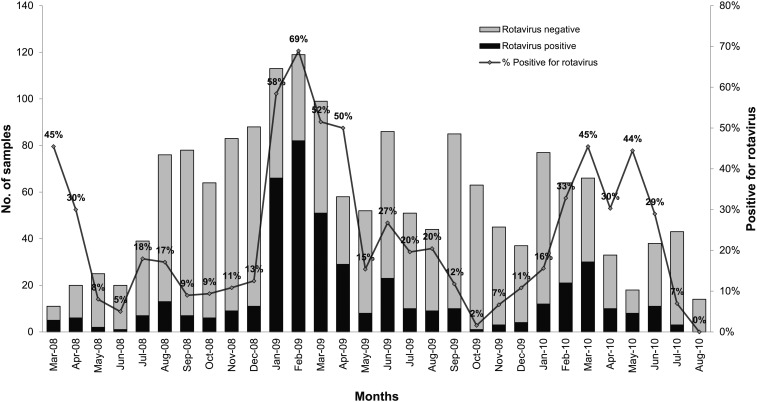
A total of 3,470 stool samples (1,709 cases and 1,761 controls) were tested for rotavirus by real-time RT-PCR and/or ELISA. The results showed that rotavirus was positive in 486 samples (458 of cases, 26.8% and 28 of controls, 1.6%). Greater than 70% of children with rotavirus infection were younger than 2 years. Cases were most prevalent during the cooler months, specifically from January 2009 to March 2009, with a 52.0–69.0% detection rate (Figure 2). Rotavirus was detected more among hospitalized cases (34%) than outpatient cases (18%) (P < 0.001). Of the 458 cases in four regions, rotavirus was detected at the highest percentage in CR (37.1%; 198/533), followed by SR at 30.8% (111/360), KR at 24.1% (86/357), and PN at 13.7% (63/459). Rotavirus was detected every month in CR, located in the northern part of Thailand, where the weather is relatively cooler than that in the rest of the country.
Figure 2.
Distribution of rotavirus group A in young Thai children with acute diarrhea cases from four regions of Thailand by month from March 2008 to April 2010.
All the rotavirus positive samples by real-time RT-PCR or ELISA were analyzed further for their G and P genotypes. G1 predominated in all regions (36.4–50.0%), followed by G2 (13.2–34.4%), except in the central region (PN), where G12 was the second most common genotype. Overall, G3, G9, and G12 were detected at 8.2%, 9.1%, and 5.8 %, respectively, whereas G4 was detected at 1%. There was no G4 detected in KR and SR sites, and G12 was not detected in SR. Eleven samples were non-typeable for G genotype (Table 1). Three common P genotypes—P[4], P[6], and P[8]—were detected at all four sites. P[8] was the most abundant P genotype detected at 67.5%, followed by P[4] and P[6] at 25.7% and 4.1%, respectively. There were 11 non-typeable P genotypes detected during the study (Table 1).
Table 1.
Distribution of G and P genotypes from rotavirus-positive stool samples in cases and controls by real-time reverse transcription–polymerase chain reaction and/or enzyme-linked immunosorbent assay in Thai children from March 2008 to April 2010
| Genotype | No. of rotavirus strains (%) | Total (%) (N = 486) | |||
|---|---|---|---|---|---|
| CR site (N = 208) | KR site (N = 88) | PN site (N = 68) | SR site (N = 122) | ||
| G genotype | |||||
| G1 | 104 (50.0) | 32 (36.4) | 30 (44.1) | 61 (50.0) | 227 (46.7) |
| G2 | 56 (26.9) | 23 (26.2) | 9 (13.2) | 42 (34.4) | 130 (26.7) |
| G3 | 21 (10.1) | 9 (10.2) | 8 (11.8) | 2 (1.6) | 40 (8.2) |
| G4 | 4 (1.9) | 0 (0.0) | 1 (1.5) | 0 (0.0) | 5 (1.0) |
| G5 | 0 (0.0) | 1 (1.1) | 0 (0.0) | 0 (0.0) | 1 (0.2) |
| G9 | 9 (4.3) | 14 (15.9) | 7 (10.3) | 14 (11.5) | 44 (9.1) |
| G12 | 7 (3.4) | 9 (10.2) | 12 (17.6) | 0 (0.0) | 28 (5.8) |
| G-non-typeable | 7 (3.4) | 0 (0.0) | 1 (1.5) | 3 (2.5) | 11 (2.3) |
| P genotype | |||||
| P[4] | 55 (26.4) | 20 (22.7) | 7 (10.3) | 43 (35.3) | 125 (25.7) |
| P7[5] | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (0.8) | 1 (0.2) |
| P[6] | 4 (1.9) | 14 (15.9) | 2 (2.9) | 0 (0.0) | 20 (4.1) |
| P[8] | 147 (70.7) | 53 (60.2) | 56 (82.4) | 72 (59.0) | 328 (67.5) |
| P[19] | 0 (0.0) | 1 (1.1) | 0 (0.0) | 0 (0.0) | 1 (0.2) |
| P-non-typeable | 2 (1.0) | 0 (0.0) | 3 (4.4) | 6 (4.9) | 11 (2.3) |
CR = Chiang Rai; KR = Nakorn Rachasrima (Korat); PN = Phitsanulok; SR = Surat Thani. Bold values indicate genotypes identified with the highest percentage at each site.
The distribution of rotavirus genotype combinations detected in the four regions of Thailand is shown in Table 2. The most prominent genotype combinations from all four sites were G1P[8] and G2P[4]. The uncommon genotype combinations—G1P[4], G1P7[5], G1P[6], G2P[6], G2P[8], G4P[6], G5P[19], G9P[4], G9P[6], G12P[6], and G12P[8]—were also detected at a combined rate of 13.4%. Sixteen samples (3.3%) were not-typeable for either or both genotypes.
Table 2.
Distribution of detected rotavirus strains in Thai children from four regions of Thailand, 2008–2010
| Genotypes | CR (N = 208) | KR (N = 88) | PN (N = 68) | SR (N = 122) | Total (%) N = 486 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2008 (%) (N = 40) | 2009 (%) (N = 128) | 2010 (%) (N = 40) | 2008 (%) (N = 14) | 2009 (%) (N = 40) | 2010 (%) (N = 34) | 2008 (%) (N = 3) | 2009 (%) (N = 37) | 2010 (%) (N = 28) | 2008 (%) (N = 11) | 2009 (%) (N = 108) | 2010 (%) (N = 3) | ||
| Common genotypes | |||||||||||||
| G1P[8] | 1 (2.5) | 70 (54.7) | 28 (70.0) | 2 (14.3) | 5 (12.5) | 21 (61.8) | 2 (66.7) | 16 (43.2) | 12 (42.9) | 9 (81.8) | 48 (44.4) | – | 214 (44.0) |
| G2P[4] | 34 (85.0) | 15 (11.7) | – | 2 (14.3) | 16 (40.0) | – | 1 (33.3) | 6 (16.2) | – | 1 (9.1) | 38 (35.2) | – | 113 (23.3) |
| G3P[8] | – | 17 (13.3) | 4 (10.0) | – | 3 (7.5) | 6 (17.6) | – | 1 (2.7) | 7 (25.0) | 1 (9.1) | – | 1 (33.3) | 40 (8.2) |
| G4P[8] | – | 2 (1.6) | – | – | – | – | – | – | – | – | – | – | 2 (0.4) |
| G9P[8] | 2 (5.0) | 4 (3.1) | 3 (7.5) | 5 (35.7) | 8 (20.0) | – | – | 1 (2.7) | 3 (10.7) | – | 10 (9.3) | – | 36 (7.4) |
| Uncommon genotypes* | |||||||||||||
| G1P[4] | – | 4 (3.1) | – | – | 2 (5.0) | – | – | – | – | – | 2 (1.9) | – | 8 (1.7) |
| G1P7[5] | – | – | – | – | – | – | – | – | – | – | – | 1 (33.3) | 1 (0.2) |
| G1P[6] | – | 1 (0.8) | – | – | – | 2 (5.9) | – | – | – | – | – | – | 3 (0.6) |
| G2P[6] | – | – | – | 1 (7.1) | 1 (2.5) | 1 (2.9) | – | – | – | – | – | – | 3 (0.6) |
| G2P[8] | 1 (2.5) | 4 (3.1) | 2 (5.0) | – | 2 (5.0) | – | – | 2 (5.4) | – | – | 3 (2.8) | – | 14 (2.9) |
| G4P[6] | 1 (2.5) | 1 (0.8) | – | – | – | – | – | – | 1 (3.6) | – | – | – | 3 (0.6) |
| G5P[19] | – | – | – | – | 1 (2.5) | – | – | – | – | – | – | – | 1 (0.2) |
| G9P[4] | – | – | – | – | – | – | – | – | – | – | 2 (1.9) | – | 2 (0.4) |
| G9P[6] | – | – | – | – | – | 1 (2.9) | – | 1 (2.7) | – | – | – | – | 2 (0.4) |
| G12P[6] | 1 (2.5) | – | – | 4 (28.6) | 1 (2.5) | 3 (8.8) | – | – | – | – | – | – | 9 (1.9) |
| G12P[8] | – | 3 (2.3) | 3 (7.5) | – | 1 (2.5) | – | – | 10 (27.0) | 2 (7.1) | – | – | – | 19 (3.9) |
| Non-typeables | |||||||||||||
| G1 PNT | – | – | – | – | – | – | – | – | – | – | 1 (0.9) | – | 1 (0.2) |
| G9 PNT | – | – | – | – | – | – | – | – | 2 (7.1) | – | 1 (0.9) | 1 (33.3) | 4 (0.8) |
| GNT P[4] | – | 2 (1.6) | – | – | – | – | – | – | – | – | – | – | 2 (0.4) |
| GNT P[8] | – | 3 (2.3) | – | – | – | – | – | – | – | – | – | – | 3 (0.6) |
| GNT PNT | – | 2 (1.6) | – | – | – | – | – | – | 1 (3.6) | – | 3 (2.8) | – | 6 (1.2) |
CR = Chiang Rai; GNT = non-typeable G-genotype; KR = Nakorn Rachasrima (Korat); PN = Phitsanulok; SR = Surat Thani; PNT = on-typeable P-genotype. Bold values indicate genotypes identified with the highest percentage in a given year.
* Uncommon genotypes were classified based on the definition provided by the WHO.9
An annual analysis showed that G2P[4] was the most common genotype combination detected in 2008 in the north (CR), but G1P[8] became the most common genotype combination in the subsequent surveillance year (2009), except KR, where G2P[4] predominated. G1P[8] remained the major strain in the third surveillance year (2010), except at SR. An uncommon genotype, G12P[8], was the second most predominant genotype at PN in 2009 (Table 2).
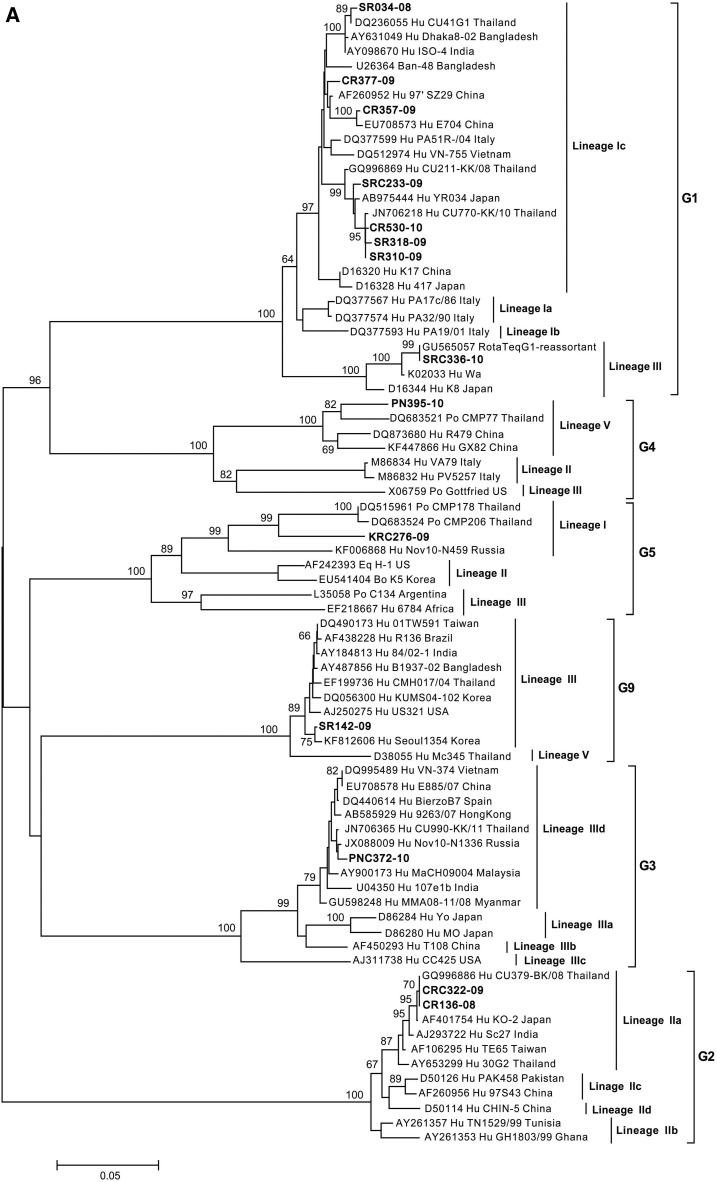
Nucleotide sequences of the vp7 gene (829 bp) showed 14 samples that clustered with the G genotypes: G1, G2, G3, G4, G5, and G9 (Figure 3A). Seven of the sequenced samples were clustered into sub-lineage Ic of lineage I of rotavirus G1, which includes strains from Asian countries such as Bangladesh, India, China, and Vietnam, with nucleotide identities ranging from 96.1% to 99.4%. In addition, one sample (SRC336-10) clustered into lineage III of rotavirus G1 which associated with P7[5], showing 100% identity to RotaTeq G1-reassortant vaccine strain containing G1, G2, G3, and G4 with P7[5].
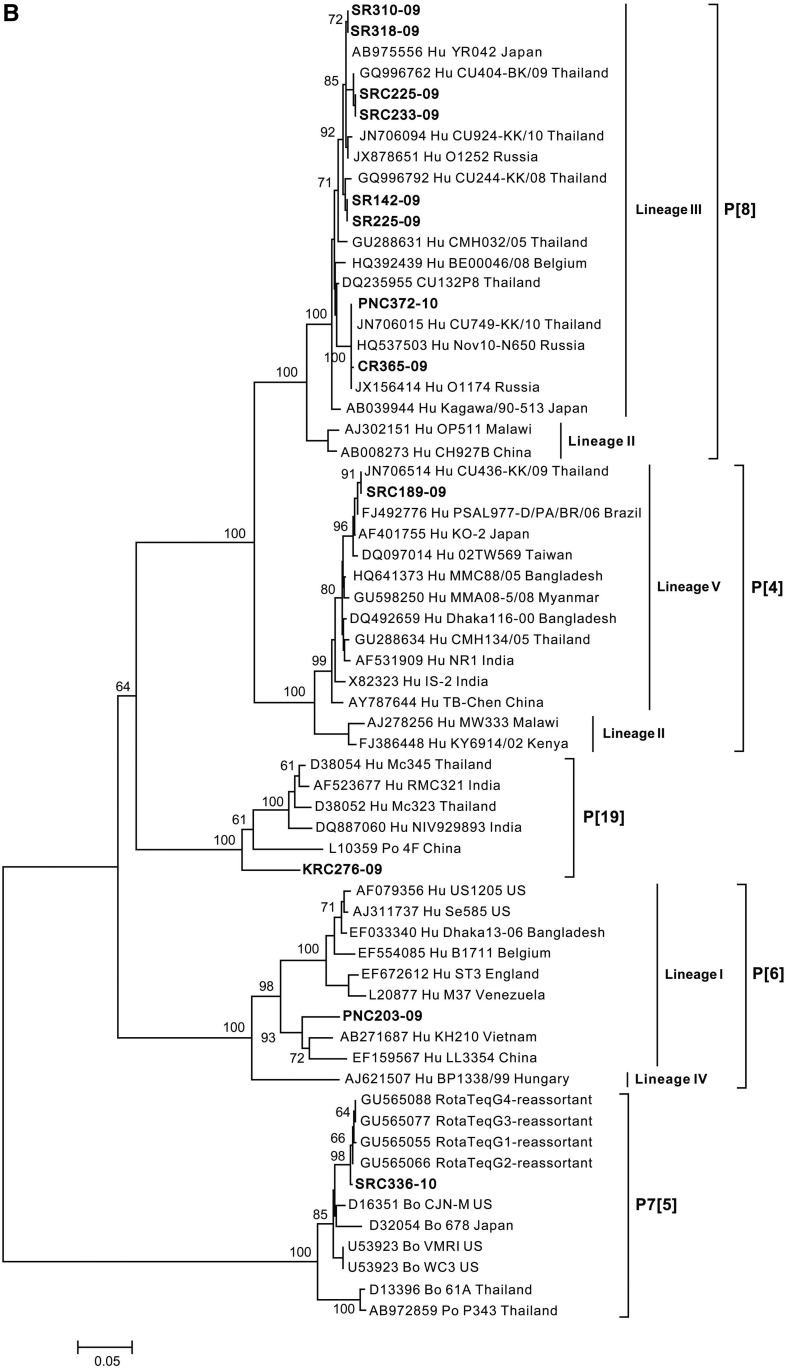
Figure 3.
Neighbor-joining phylogenetic tree generated from partially sequenced (A) vp7 gene (G genotype) and (B) vp4 gene (P genotype). Representative sequence prototypes for each genotype were used in the analysis, and accession numbers are listed next to the name. Numbers at nodes indicate bootstrap values.
Two samples from the north clustered into sub-lineage IIa of rotavirus G2. PNC372-10 and PN395-10 from the central part clustered into sub-lineage IIId of rotavirus G3 lineage III, and lineage V of rotavirus G4, respectively (Figure 3A). KRC276-09 from the northeastern region was clustered into lineage I of rotavirus G5 which was associated with P[19] and showed 92–92.4% sequence similarity to strains CMP178 and 206 which were detected in piglets in Chiang Mai.22 Last, SR142-09 from the south was clustered into lineage III of rotavirus G9, which is similar to strains isolated from Asia, Europe, and the United States (Figure 3A).
Phylogenetic analysis of the vp4 gene (640 bp) clustered 12 samples into different P genotypes: P[4], P7[5], P[6], P[8] and P[19] (Figure 3B). Eight samples were clustered into lineage III of rotavirus P[8], which is the most common P genotype. These samples had sequence similarity ranging from 96.9% to 100% to strains isolated from Belgium, Japan, Russia, and Thailand. Rotavirus P[4] (SRC189-09) and P[6] (PNC203-09) samples were clustered into lineage V and lineage I of their respective genotypes. Both samples had sequence similarity (93–97.8%) to strains isolated from Asian countries.
DISCUSSION
This study reports the molecular epidemiology of rotavirus from 2008 to 2010 in young Thai children from four study sites located in northern, northeastern, central, and southern parts of Thailand, representing the major geographic regions (Figure 1). Rotavirus was responsible for a quarter of children with acute gastroenteritis, whereas only a small percentage of rotavirus was detected among children without gastroenteritis symptoms.
Epidemiological surveillance of rotavirus by several groups in Thailand has shown a shift in the predominant genotype combinations over the years, starting with G9P[8] during 2000–2002,13,14 G2P[4] in 2003,14 G1P[8] during 2004–2009,14–16,18,23 and G3P[8] during 2009–2011.17 However, most of the studies only focused on one geographical region which might not be completely representative of the shift in strains and circulating strains in other regions of Thailand during the same period. Moreover, predominant genotypes of rotavirus may differ by population studied, such as hospitalized cases versus outpatient diarrhea cases. Certain genotypes, for example, G9, have been reported to be associated with more severe diseases.20 In this study, G1P[8] was the most common genotype detected between 2009 and 2010. However, analysis based on geography revealed a slightly different trend of the predominating rotavirus strains as G2P[4] predominated during the first and second years at CR and KR sites (Table 2), which further indicates the cycling of the predominating genotype. In addition, G4 was detected at 1.0%, which is lower than 5.3% reported by Jiraphongsa et al.,12 whereas other similar studies have no reported G4 genotypes in Thailand between 2002 and 2011.13–18,23 The phylogenetic analysis of rotavirus strains into lineages/sublineages of G1, G2, G3, G9, P[4], P[6], and P[8] (Figure 3A and B) shows that rotavirus genotypes in Thailand are similar to genotypes found in other geographical regions,15,17,24–27 including Europe, America, and Asia
The detection of 1.6% of rotavirus in control samples is similar to data reported by Mullick et al. (2014), where 2% of asymptomatic Indian children were rotavirus positive.28 Previous studies have demonstrated that the fecal excretion by asymptomatic persons may be a source of infection for susceptible person.29,30 However, information on asymptomatic rotavirus infection is rarely reported. Interestingly, G5P[19] from non-diarrhea controls may have derived from a close association between human and piglets (Figure 3), as there have been reports of G3P[19] and G5P[13] from piglet stool in Chiang Mai in 2000–2001 and 2008, respectively.31,32 These results indicate a possible recombination between human and animal rotavirus strains, demonstrating the diversity of circulating strains in Thailand. This study is the first to report rotavirus G5P[19] in Thailand from an asymptomatic patient. This study was a passive surveillance and was limited by the small sample size; however, the surveillance was conducted at multiple regional sites representing all of the four main regions in Thailand, which provides a wider range of rotavirus epidemiological pattern.
The WHO reported that five common genotypes (G1P[8], G2P[4], G3P[8], G4P[8], and G9P[8]) cause nearly 100% of rotavirus infection in developed countries, whereas uncommon genotypes cause 10–35% of rotavirus infection in developing countries.9 The percentage of uncommon rotavirus genotypes (13.4%) reported in this study is consistent with the reported trend.9 Uncommon genotypes found in this study—G1P7[5], G5P[19], G9P[4], and G9P[6]—add to the existing list of uncommon genotypes reported to circulate in Thailand.19,33,34 The presence of uncommon genotypes and the regular shifting of predominant genotypes may contribute to the likelihood of uncommon genotypes to become common global genotypes.5,7 This will have an important implication for the development of an effective vaccine with genotype cross protection against rotavirus in the future, specifically in Thailand.
Acknowledgments:
We would like to thank the children and parents; staff at the participating Regional Medical Science Centers and hospitals; staff, Orntipa Sethabutr, at the Department of Enteric Diseases, AFRIMS, for significant contributions during the conduct and laboratory testing of the study; and Krongkaew Supawat at the Department of Medical Sciences, Ministry of Public Health, MoPH, for her significant contributions during the study.
Disclaimer: The manuscript has been reviewed by the Walter Reed Army Institute of Research. There is no objection to this publication. This opinion or assertions contained herein are the private views of the author(s), and are not to be construed as official or as reflecting true views of the Department of the Army or the Department of Defense. The investigators have adhered to the policies for protection of human subjects as prescribed in AR 70-25.
REFERENCES
- 1.World Health Organization , 2016. Estimated Rotavirus Deaths for Children under 5 Years of Age: 2013, 215 000. Available at: http://www.who.int/immunization/monitoring_surveillance/burden/estimates/rotavirus/en/. Accessed January 26, 2018. [Google Scholar]
- 2.Tate JE, Burton AH, Boschi-Pinto C, Parashar UD; World Health Organization-Coordinated Global Rotavirus Surveillance Network , 2016. Global, regional, and national estimates of rotavirus mortality in children <5 years of age, 2000–2013. Clin Infect Dis 62 (Suppl 2): S96–S105. [DOI] [PubMed] [Google Scholar]
- 3.Matthijnssens J, et al. 2008. Recommendations for the classification of group A rotaviruses using all 11 genomic RNA segments. Arch Virol 153: 1621–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.KU LEUVEN, Laboratory of Viral Metagenomics , 2018. Rotavirus Classification Working Group. Available at: http://www.rega.kuleuven.be/cev/viralmetagenomics/virus-classification/rcwg. Accessed July 23, 2018. [Google Scholar]
- 5.Santos N, Hoshino Y, 2005. Global distribution of rotavirus serotypes/genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Rev Med Virol 15: 29–56. [DOI] [PubMed] [Google Scholar]
- 6.Iturriza-Gomara M, 2014. EuroRotaNet: Annual Report 2014. Available at: http://www.thl.fi/documents/533963/1527557/EuroRotaNetAnnualReport6th_2014.pdf/7e4386c0-0e09-4e18-8576-95031eaf2680. Accessed September 5, 2018. [Google Scholar]
- 7.Patton JT, 2012. Rotavirus diversity and evolution in the post-vaccine world. Discov Med 13: 85–97. [PMC free article] [PubMed] [Google Scholar]
- 8.Armah GE, Steele AD, Esona MD, Akran VA, Nimzing L, Pennap G, 2010. Diversity of rotavirus strains circulating in West Africa from 1996 to 2000. J Infect Dis 202: S64–S71. [DOI] [PubMed] [Google Scholar]
- 9.World Health Organization , 2015. Vaccine Preventable Diseases Surveillance: Global Rotavirus Surveillance and Information Bulletin. Data period: January–December 2013. Available at: http://www.who.int/immunization/monitoring_surveillance/resources/WHO_Global_RV_Surv_Bulletin_Jan_2015_Final.pdf?ua=1. Accessed November 21, 2018. [Google Scholar]
- 10.Dennehy PH, 2012. Rotavirus infection: an update on management and prevention. Adv Pediatr 59: 47–74. [DOI] [PubMed] [Google Scholar]
- 11.Maneekarn N, Ushijima H, 2000. Epidemiology of rotavirus infection in Thailand. Pediatr Int 42: 415–421. [DOI] [PubMed] [Google Scholar]
- 12.Jiraphongsa C, Bresee JS, Pongsuwanna Y, Kluabwang P, Poonawagul U, Arporntip P, Kanoksil M, Premsri N, Intusoma U; Rotavirus Surveillance Project Thailand Study Group , 2005. Epidemiology and burden of rotavirus diarrhea in Thailand: results of sentinel surveillance. J Infect Dis 192 (Suppl 1): S87–S93. [DOI] [PubMed] [Google Scholar]
- 13.Khamrin P, et al. 2006. Emergence of human G9 rotavirus with an exceptionally high frequency in children admitted to hospital with diarrhea in Chiang Mai, Thailand. J Med Virol 78: 273–280. [DOI] [PubMed] [Google Scholar]
- 14.Khamrin P, Peerakome S, Tonusin S, Malasao R, Okitsu S, Mizuguchi M, Ushijima H, Maneekarn N, 2007. Changing pattern of rotavirus G genotype distribution in Chiang Mai, Thailand from 2002 to 2004: decline of G9 and reemergence of G1 and G2. J Med Virol 79: 1775–1782. [DOI] [PubMed] [Google Scholar]
- 15.Khananurak K, Vutithanachot V, Simakachorn N, Theamboonlers A, Chongsrisawat V, Poovorawan Y, 2010. Prevalence and phylogenetic analysis of rotavirus genotypes in Thailand between 2007 and 2009. Infect Genet Evol 10: 537–545. [DOI] [PubMed] [Google Scholar]
- 16.Theamboonlers A, Bhattarakosol P, Chongsrisawat V, Sungkapalee T, Wutthirattanakowit N, Poovorawan Y, 2008. Molecular characterization of group A human rotaviruses in Bangkok and Buriram, Thailand during 2004–2006 reveals the predominance of G1P[8], G9P[8] and a rare G3P[19] strain. Virus Genes 36: 289–298. [DOI] [PubMed] [Google Scholar]
- 17.Maiklang O, Vutithanachot V, Vutithanachot C, Hacharoen P, Chieochansin T, Poovorawan Y, 2012. Prevalence of group A genotype human rotavirus among children with dirarrhea in Thailand, 2009–2011. Southeast Asian J Trop Med Public Health 43: 904–916. [PubMed] [Google Scholar]
- 18.Khamrin P, Maneekarn N, Malasao R, Nguyen TA, Ishida S, Okitsu S, Ushijima H, 2010. Genotypic linkages of VP4, VP6, VP7, NSP4, NSP5 genes of rotaviruses circulating among children with acute gastroenteritis in Thailand. Infect Genet Evol 10: 467–472. [DOI] [PubMed] [Google Scholar]
- 19.Maneekarn N, Khamrin P, 2014. Rotavirus associated gastroenteritis in Thailand. Virus Dis 25: 201–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silapong S, Sakpaisal P, Bodhidatta L, Lertsethtakarn P, Sethabutr O, Vansith K, Meng CY, Swierczewski BE, Mason CJ, 2017. Genotypic distribution of rotavirus in Phnom Penh, Cambodia: an association of G9 with more severe diseases. Am J Trop Med Hyg 96: 947–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S, 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan-It W, et al. 2011. Detection and genetic characterization of norovirus infections in children with acute gastroenteritis in Japan, 2007–2009. Clin Lab 57: 213–220. [PubMed] [Google Scholar]
- 23.Chaimongkol N, Khamrin P, Malasao R, Thongprachum A, Ushijima H, Maneekarn N, 2012. Genotypic linkages of gene segments of rotaviruses circulating in pediatric patients with acute gastroenteritis in Thailand. Infect Genet Evol 12: 1381–1391. [DOI] [PubMed] [Google Scholar]
- 24.Arista S, Giammanco GM, De Grazia S, Ramirez S, Lo Biundo C, Colomba C, Cascio A, Martella V, 2006. Heterogeneity and temporal dynamics of evolution of G1 human rotaviruses in a settled population. J Virol 80: 10724–10733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abdel-Haq N, Amjad M, McGrath E, Chearskul P, Amer A, Salimnia H, Asmar BI, 2011. Emergence of human rotavirus genotype G9 in metropolitan Detroit between 2007 and 2009. J Med Microbiol 60: 761–767. [DOI] [PubMed] [Google Scholar]
- 26.Oh HK, Hong SH, Ahn BY, Min HK, 2012. Phylogenetic analysis of the rotavirus genotypes originated from children < 5 years of age in 16 cities in South Korea, between 2000 and 2004. Osong Public Health Res Perspect 3: 36–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong HJ, Qian Y, Huang T, Zhu RN, Zhao LQ, Zhang Y, Li RC, Li YP, 2013. Identification of circulating porcine-human reassortant G4P[6] rotavirus from children with acute diarrhea in China by whole genome analyses. Infect Genet Evol 20: 155–162. [DOI] [PubMed] [Google Scholar]
- 28.Mullick S, Mukherjee A, Ghosh S, Pazhani GP, Sur D, Manna B, Nataro JP, Levine MM, Ramamurthy T, Chawla-Sarkar M, 2014. Community based case-control study of rotavirus gastroenteritis among young children during 2008--2010 reveals vast genetic diversity and increased prevalence of G9 strains in Kolkata. Plos One 9: e112970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barnes GL, Callaghan SL, Kirkwood CD, Bogdanovic-Sakran N, Johnston LJ, Bishop RF, 2003. Excretion of serotype G1 rotavirus strains by asymptomatic staff: a possible source of nosocomial infection. J Pediatr 142: 722–725. [DOI] [PubMed] [Google Scholar]
- 30.Phillips G, Lopman B, Rodrigues LC, Tam CC, 2010. Asymptomatic rotavirus infections in England: prevalence, characteristics, and risk factors. Am J Epidemiol 171: 1023–1030. [DOI] [PubMed] [Google Scholar]
- 31.Maneekarn N, Khamrin P, Chan-it W, Peerakome S, Sukchai S, Pringprao K, Ushijima H, 2006. Detection of rare G3P[19] porcine rotavirus strains in Chiang Mai, Thailand, provides evidence for origin of the VP4 genes of Mc323 and Mc345 human rotaviruses. J Clin Microbiol 44: 4113–4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan-It W, Khamrin P, Saekhow P, Pantip C, Thongprachum A, Peerakorne S, Ushijima H, Maneekarn N, 2008. Multiple combinations of P[13]-like genotype with G3, G4, and G5 in porcine rotaviruses. J Clin Microbiol 46: 1169–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kittigul L, Swangsri T, Pombubpa K, Howteerakul N, Diraphat P, Hirunpetcharat C, 2014. Rotavirus infection in children and adults with acute gastroenteritis in Thailand. Southeast Asian J Trop Med Public Health 45: 816–824. [PubMed] [Google Scholar]
- 34.Yodmeeklin A, Khamrin P, Kumthip K, Malasao R, Ukarapol N, Ushijima H, Maneekarn N, 2018. Increasing predominance of G8P[8] species A rotaviruses in children admitted to hospital with acute gastroenteritis in Thailand, 2010–2013. Arch Virol 163: 2165–2178. [DOI] [PubMed] [Google Scholar]