Abstract.
Eastern equine encephalitis virus (EEEV) infection results in high mortality in infected horses and humans. Florida has been identified as an important source of EEEV epidemics to other states in the United States. In this study, we further characterized the epidemiological and evolutionary dynamics of EEEV in Florida. Epidemiological analysis of sentinel chicken seroconversion rates to EEEV infections during 2005–2016 suggested significant seasonality of EEEV activity in Florida. We observed significant annual activity of EEEV in the North and North Central regions, with little significant seasonality in the Panhandle region. Phylogenetic analysis of complete EEEV genome sequences from different host sources and regions in Florida during 1986–2014 revealed extensive genetic diversity and spatial dispersal of the virus within Florida and relatively more clustering of the viruses in the Panhandle region. We found no significant association between EEEV genetic variation and host source. Overall, our study revealed a complex epidemiological dynamic of EEEV within Florida, implicating the Panhandle region as a possible source of the virus with sustained year-round transmission. These findings will help in implementing targeted control measures that can have the most impact in reducing or eliminating EEEV and other mosquito-borne viral infections within Florida and in the rest of the United States.
INTRODUCTION
Eastern equine encephalitis virus (EEEV), family Togaviridae, genus Alphavirus, is a single-stranded positive-sense RNA arbovirus (arthropod-borne virus) that can infect humans, resulting in an approximately 40% mortality rate, and, interestingly, is only found east of the Mississippi River.1 Survivors usually have lifelong neurological impairments that can result in eventual death.2,3 Generally, EEEV circulates between passerine birds and its primary enzootic mosquito vector, Culiseta melanura. During periods of increased circulation, additional competent mosquito vectors can transmit EEEV to other dead-end hosts such as humans, horses, and other mammals.4
Transmission to these other dead-end hosts results in outbreaks, such as those seen in Massachusetts and New Hampshire in 20055 and Massachusetts in 2012,6 and periodic cases elsewhere. The number of neuroinvasive cases reported to the CDC for the United States from 2007 to 2016 was 68 (ranging from 3 to 15 cases per year).1 Florida had the second highest number of neuroinvasive cases (10 cases) after Massachusetts for those years.1 Florida is bound by both the Atlantic and Gulf coasts, and coastal counties tend to have higher incidences of human cases of eastern equine encephalitis (EEE).7 Differing epidemiological dynamics have been found for EEEV, which is transmitted during July–October in most of the northeastern United States,3,8 but in Florida, it is transmitted year-round.9 In addition, the number of horse cases in Florida is high compared with that in other states.10 Because of this historically high activity of EEEV and other arboviruses (St. Louis encephalitis virus [SLEV] and West Nile virus) in Florida, there are surveillance systems in place for monitoring human and animal cases, including a sentinel chicken surveillance program. This program, established in 1978, provides early warning of human and animal outbreaks by detecting increased arbovirus transmission through monitoring antibody seroconversion in chicken flocks.11 In a previous study of Florida sentinel chicken seroconversion data, Day and Stark (1996) reported higher annual EEEV antibody seroconversion rates in the Panhandle and northern part of the state.12 These regions also have a high incidence of horse cases12,13 and the highest average annual incidence of human neuroinvasive disease,1 indicating the Panhandle and the northern part of the state have more active EEEV transmission when compared with the rest of the state.
In a recent study, using multistate whole genome sequencing and phylogenetic analysis, we revealed Florida is an important reservoir of EEEV for northern states because of high genetic diversity, multiyear persistence, and minimal genetic spatial structure indicative of more geographic mixing of EEEV than in northern states.14 This was coupled with a statistically significant pattern of movement out of Florida to northern states.14 In this study, we further investigated the seasonality of EEEV infection systematically and examined the extent of viral spatial diversity by performing an in-depth analysis by integrating long-term epidemiological data and genetic data of EEEV within Florida by region. These findings will have an impact on public health and improving EEEV control in Florida to prevent animal and human outbreaks and further spread to northern states of the United States.
METHODS
Surveillance data.
We obtained sentinel chicken surveillance data from the Florida Department of Health, Bureau of Public Health Laboratories surveillance program. This program involves weekly testing of chicken serum mostly during peak EEEV transmission months (May–August) from sites across the state. The Bureau of Public Health Laboratories performed a screening assay, hemagglutination inhibition (HAI), as previously described, to detect antibody to alphaviruses with EEEV suckling mouse brain sucrose–acetone-extracted antigen.11 The Bureau of Public Health Laboratories performed a confirmatory assay, IgM antibody capture ELISA (MAC-ELISA) with goat anti–chicken IgM capture antibody and a cutoff P/N value of 2.6,15 on HAI-positive, equivocal, and inconclusive sera. Negative, equivocal, or inconclusive MAC-ELISA results were followed with a second confirmatory assay, the plaque reduction neutralization test (PRNT), to detect virus-specific neutralizing antibody16 with a 95% plaque reduction level. The Bureau of Public Health Laboratories performed the PRNT for EEEV antibody and for Highlands J virus antibody, a HAI antibody cross-reactive alphavirus that maintains an enzootic cycle in passerine birds and Cs. melanura mosquitoes in Florida, but does not pose a major health risk to humans.17,18 Positive EEEV IgM MAC-ELISA or PRNT results were reported as confirmed positive seroconversions. Submitters removed confirmed chickens from flocks typically within 1–2 weeks of reporting. We calculated confirmed EEEV seroconversion rates (the number of confirmed chickens divided by the number of susceptible chickens bled) for the state and regions by week, month, and year from 2005 to 2016. Susceptible chickens were those that had not had a previous confirmed seroconversion to EEEV. We calculated mean monthly seroconversion rates (MMSRs) by dividing the number of EEEV-confirmed seroconversions by the total number of susceptible sentinel chickens in the state or by region for the entire study period. We divided the state into five regions (Panhandle, North, North Central, South Central, and South; Figure 1) similar to the previously reported divisions by Day and Stark (1996), with some modification to increase the number of regions, allowing for improved statistical analysis. EEE human case data were obtained from 2005 to 2016 from the Centers for Disease Control and Prevention ArboNet website,19 and EEE equine case data were obtained from 2005 to 2016 for the U.S. Department of Agriculture equine health encephalitis website.10
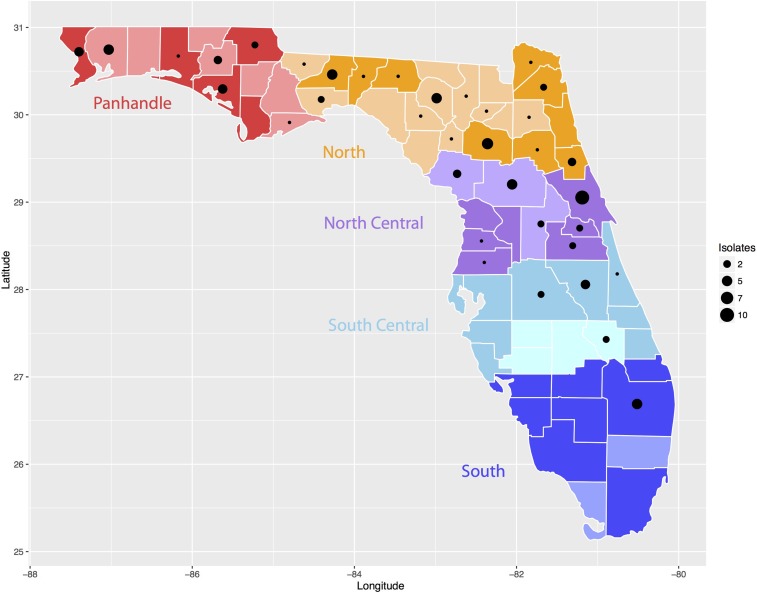
Figure 1.
Map of Florida with five regions from which the eastern equine encephalitis virus (EEEV) isolates and sentinel chickens were sampled. The x axis represents longitude and y axis represents latitude. The five regions in Florida are colored and marked in the figure. The darker shading in each region indicates counties in the region participating in the sentinel chicken surveillance program during the study period, 2005 to 2016, whereas the lighter shade indicates no chicken surveillance data were available from those counties. Black dots in each county represent EEEV isolates and size of the dot represents the total number of isolates. This figure appears in color at www.ajtmh.org.
Sentinel chicken EEEV seroconversion seasonality and periodicity analysis.
We created weekly and monthly time series from the weekly and monthly confirmed sentinel chicken EEEV seroconversion rates during 2005–2016 for the entire state and for the five regions. Because of the paucity of seroconversions in the South region (despite having a number of serum collections consistent with other regions), we combined the South Central and South regions in the analysis. To characterize the seasonality and periodicity of the seroconversion rates, we applied wavelet methods to the time series in R 3.4.1 with RStudio 1.1.383,20 as previously described.21–23 All time series were square root–transformed before analysis to control for variations in sentinel chicken serum collections over the study period.
We measured epidemic synchrony between regions with significant seasonality by estimating the monthly phase angle difference in wavelet-reconstructed time series, after extraction of the main annual cycle (0.8- to 1.2-year periods; Morlet continuous wavelet).24
Eastern equine encephalitis virus complete genome sequence data.
We used a total of 93 EEEV whole genome sequences (WGSs) from Florida previously sequenced by us that are available in GenBank (see Supplemental Table 1).14 The EEEV isolate sequences in this study represent a broad range and diversity based on the county of detection, year of collection, and host source (avian, equine, mosquito, or other mammals) in Florida. The collection dates of the original samples were from 1986 to 2014. The number of sequences per year ranged from 1 in 1986 to 15 in 2003, with most isolates dating from 2001 to 2010.
Phylogeographic analysis and the association between virus phylogeny and geographic information.
We studied a total of 93 complete genome sequences of Florida EEEV with available host and geographic information by comparative phylogenetic analysis with their sampling locations and host sources. We inferred phylogenetic trees using the Bayesian Markov chain Monte Carlo (BMCMC) method available in MrBayes version 3.2.5,25 run for 1 × 108 steps using a general time reversal (GTR) nucleotide substitution model with a gamma distribution of among-site rate variation (GTR + Γ) (selected as the best-fit model by Modeltest in MEGA 6.0). Trees were sampled every 1 × 104 steps, with the first 1,000 trees discarded as burn-in. The phylogeny was rooted with the oldest EEEV sequence sampled from Florida in 1986.
To determine whether the viral phylogeny displayed geographic clustering by regions, we grouped the viruses by location of where they were detected with the same five regions used for the sentinel chicken data (Figure 1). The grouping is similar to that designated in a previous small-scale (six WGSs and 15 partial sequences) phylogenetic study of Florida EEEV.26 In addition, we tested whether viral phylogenetic clustering is influenced by viral source (host) or sampling dates, we grouped viruses into four host sources (mosquito, avian, equine, and other mammals) and four sampling periods (before 2000, 2000–2004, 2005–2009, and 2010–2014) separately. We determined the overall statistical significance of association between the EEEV phylogenies and the viral traits described above (i.e., geography, host, and sampling date) using two phylogeny–trait association statistical tests: the parsimony score (PS) and the association index (AI). The null hypothesis was that clustering by host and geographic information is not less than that expected by chance. In addition, we used the maximum clade (MC) statistic to compare the strength of clustering at each group by calculating the expected and observed mean clade size from each group. We implemented all three statistics in the Bayesian tip-association significance testing (BaTS) program27 with a significance level of P < 0.05. A null distribution of these statistics was determined using the posterior distribution of BMCMC phylogenies.
RESULTS
Sentinel chicken surveillance data.
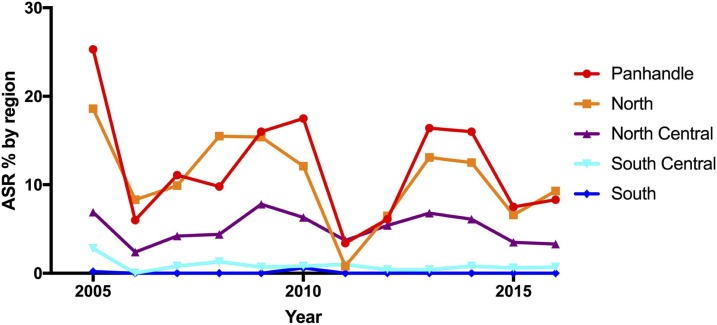
Thirty-eight of Florida’s 67 counties participated in the sentinel chicken surveillance program from 2005 to 2016 by placing sentinel flocks in the field for at least one season (Figure 1). Annual seroconversion rates (ASRs) to EEE and annual numbers of human and horse EEE cases are listed in Table 1. Over the 12-year study, ASR ranged from 1.6% in 2011 to 9.7% in 2005 with a mean ASR of 4.8% from 2005 to 2016. The annual seroconversion rate by region (Figure 2) indicated the Panhandle had the highest ASR most years (as high as 25%, never below 3%), followed by the North (as high as 19%) and the North Central (as high as 8%) regions. The South Central region ASR never exceeded 3% and the South region never exceeded 1% (most years 0%). The number of counties participating, number of sentinel chickens tested, and the MMSR by month and region are listed in Table 2. The statewide monthly number of counties participating ranged from 22 to 38, with most counties participating during the summer months (June–September). Most collections were from the South Central region. The statewide MMSR ranged from a low of 0.4% in February to a high of 2.3% in June with the three highest months being May–July. Regional MMSR ranged from a low of 0.0% (in the North, South Central, and South regions) to a high of 6.0% (in the Panhandle region). The Panhandle and North regions had the overall highest MMSR each month with both peaking in June, despite the mean monthly number of chickens tested being the lowest in these regions (1,713 and 2,138 respectfully) compared with the North Central and South Central regions. The lowest MMSR in the Panhandle region was 1.4% in March.
Table 1.
Annual eastern equine encephalitis (EEE) seroconversion rates in sentinel chickens in Florida and number of human EEE cases, 2005–2016
| Year | Annual seroconversion rate (%) | No. of counties | No. of human EEE FL cases | No. of human EEE U.S. cases | No. of equine EEE FL cases | No. of equine EEE U.S. cases |
|---|---|---|---|---|---|---|
| 2005 | 9.7 | 32 | 5 | 21 | 150 | 330 |
| 2006 | 2.7 | 34 | 0 | 8 | 17 | 111 |
| 2007 | 4.2 | 33 | 0 | 4 | 18 | 206 |
| 2008 | 5.1 | 31 | 1 | 4 | 89 | 185 |
| 2009 | 6.9 | 31 | 0 | 4 | 75 | 301 |
| 2010 | 5.7 | 29 | 4 | 10 | 92 | 247 |
| 2011 | 1.6 | 29 | 0 | 4 | 6 | 60 |
| 2012 | 2.9 | 28 | 2 | 15 | 34 | 209 |
| 2013 | 6.3 | 28 | 3 | 8 | 38 | 192 |
| 2014 | 5.7 | 28 | 0 | 8 | 59 | 136 |
| 2015 | 2.7 | 29 | 0 | 6 | 23 | 70 |
| 2016 | 3.4 | 28 | 0 | 7 | 24 | 116 |
| Overall | 4.8 | 38 | 15 | 99 | 625 | 2,163 |
FL = Florida.
Figure 2.
Annual seroconversion rate in percent of sentinel chickens seroconverting to eastern equine encephalitis virus in Florida by region, 2005 to 2016. Annual seroconversion rate in percent (the number of confirmed chickens divided by the number of susceptible chickens bled in each year and represented as percent) for the Panhandle region in red, North region in orange, North Central region in purple, South Central region in light blue, and the South region in dark blue. This figure appears in color at www.ajtmh.org.
Table 2.
Total number of monthly eastern equine encephalitis seroconversions in sentinel chickens and mean monthly seroconversion rates (MMSR) by region in Florida, 2005–2016
| Panhandle Region | North Region | |||||||
|---|---|---|---|---|---|---|---|---|
| Month | No. counties | Total no. chickens tested | No. chick. seropos. | MMSR, % | No. counties | Total no. chickens tested | No. chickens seropos. | MMSR, % |
| January | 4 | 1218 | 23 | 1.9 | 2 | 100 | 0 | 0.0 |
| Februray | 4 | 1406 | 22 | 1.6 | 2 | 162 | 4 | 2.5 |
| March | 4 | 1520 | 21 | 1.4 | 3 | 1006 | 22 | 2.2 |
| April | 5 | 1797 | 28 | 1.6 | 6 | 1972 | 37 | 1.9 |
| May | 5 | 1893 | 42 | 2.2 | 9 | 3159 | 71 | 2.2 |
| June | 5 | 1894 | 114 | 6.0 | 9 | 3405 | 164 | 4.8 |
| July | 5 | 1911 | 75 | 3.9 | 9 | 3474 | 161 | 4.6 |
| August | 5 | 1890 | 46 | 2.4 | 9 | 3482 | 108 | 3.1 |
| September | 5 | 1859 | 36 | 1.9 | 9 | 3432 | 61 | 1.8 |
| October | 4 | 1845 | 55 | 3.0 | 8 | 2915 | 34 | 1.2 |
| November | 4 | 1741 | 47 | 2.7 | 7 | 1943 | 10 | 0.5 |
| December | 4 | 1581 | 28 | 1.8 | 4 | 604 | 0 | 0.0 |
| North Central Region | South Central Region | |||||||
| January | 4 | 2582 | 20 | 0.8 | 5 | 3342 | 0 | 0.0 |
| Februray | 4 | 2653 | 11 | 0.4 | 6 | 3485 | 3 | 0.1 |
| March | 5 | 3104 | 16 | 0.5 | 6 | 3791 | 1 | 0.0 |
| April | 6 | 3612 | 17 | 0.5 | 7 | 4912 | 5 | 0.1 |
| May | 6 | 3744 | 51 | 1.4 | 8 | 5437 | 10 | 0.2 |
| June | 7 | 3904 | 87 | 2.2 | 9 | 5753 | 32 | 0.6 |
| July | 7 | 3923 | 61 | 1.6 | 9 | 5551 | 11 | 0.2 |
| August | 7 | 3836 | 27 | 0.7 | 9 | 5583 | 7 | 0.1 |
| September | 7 | 3864 | 13 | 0.3 | 9 | 5775 | 12 | 0.2 |
| October | 7 | 3876 | 31 | 0.8 | 9 | 5500 | 7 | 0.1 |
| November | 7 | 3667 | 41 | 1.1 | 9 | 5023 | 5 | 0.1 |
| December | 6 | 2938 | 28 | 1.0 | 8 | 3859 | 3 | 0.1 |
| South Region | Statewide | |||||||
| January | 7 | 1291 | 0 | 0.0 | 22 | 8533 | 43 | 0.5 |
| Februray | 6 | 1456 | 0 | 0.0 | 22 | 9162 | 40 | 0.4 |
| March | 5 | 1373 | 0 | 0.0 | 23 | 10794 | 60 | 0.6 |
| April | 6 | 2026 | 0 | 0.0 | 30 | 14319 | 87 | 0.6 |
| May | 7 | 2311 | 2 | 0.1 | 35 | 16544 | 176 | 1.1 |
| June | 8 | 2514 | 2 | 0.1 | 38 | 17470 | 399 | 2.3 |
| July | 8 | 2528 | 1 | 0.0 | 38 | 17387 | 309 | 1.8 |
| August | 8 | 2521 | 0 | 0.0 | 38 | 17312 | 188 | 1.1 |
| September | 8 | 2552 | 0 | 0.0 | 38 | 17482 | 122 | 0.7 |
| October | 8 | 2415 | 0 | 0.0 | 36 | 16551 | 127 | 0.8 |
| November | 8 | 2264 | 0 | 0.0 | 35 | 14638 | 103 | 0.7 |
| December | 7 | 1688 | 0 | 0.0 | 29 | 10670 | 59 | 0.6 |
Sentinel chicken seroconversion periodicity and seasonality.
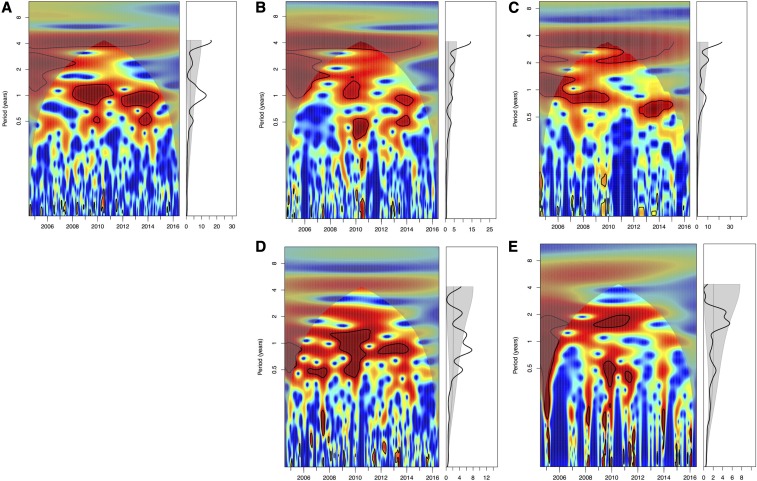
To determine if the peak seroconversion rates observed in the sentinel chickens for the state and regions showed statistically significant seasonality, we performed wavelet analysis on the weekly and monthly confirmed sentinel chicken seroconversion rates. The weekly analysis showed significant annual periodicity (P < 0.05) in Florida as a whole, in the North region, and in the North Central region, but not in either the Panhandle region or South Central/South combined region (Figure 3). The North Central region also showed significant semiannual periodicity (P < 0.05). The analysis of monthly data showed similar results, significant annual periodicity in Florida as a whole and in the North region. However, the North Central region did not show significant annual periodicity but semiannual periodicity only (Supplemental Figure 1).
Figure 3.
Seasonality of eastern equine encephalitis virus activities in Florida, 2005–2016. Wavelet power spectrum of weekly confirmed seroconversion rates for (A) entire Florida, (B) Panhandle region, (C) North region, (D) North Central region, (E) South and South Central regions. Wavelet power spectrums identify changes in periodicities over time (left) and average periodicity (right). Power increases from blue to red, and red indicates stronger periodicities. Black lines highlight statistically significant periodicities, the annual/1-year periodicities in this study. This figure appears in color at www.ajtmh.org.
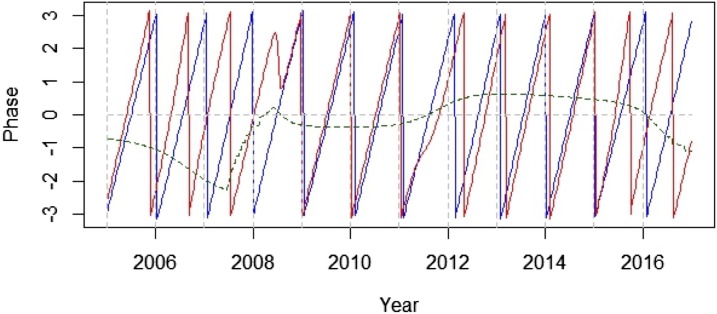
The North and North Central regions with significant periodicity based on the weekly data were further analyzed by wavelet phase coherence to determine if one region tended to peak in seasonality before the other, indicating potential virus movement in one direction or the other (Figure 4). The analysis suggested periodic fluctuations in leading and lagging between the two regions: the North Central region leading in seasonality for the first 4 years of the study period (2005–2008), followed by synchronization for several years (2009–2011), then shifting to the North region leading for a couple years (2012–2014), followed by another synchronization of 1 year (2015), and finally a shift back to the North Central region leading for the remainder of the study period (2016).
Figure 4.
Phase analysis of sentinel chicken seroconversion rates in North and North Central regions in Florida. The blue and red lines represent the phase of time series of eastern equine encephalitis virus weekly seroconversion rates in the North region and North Central region at a periodicity of 0.8–1.2 years separately. Green line represents phase differences between the North and North Central regions. This figure appears in color at www.ajtmh.org.
Analysis of phylogeographic clustering of EEEV within Florida.
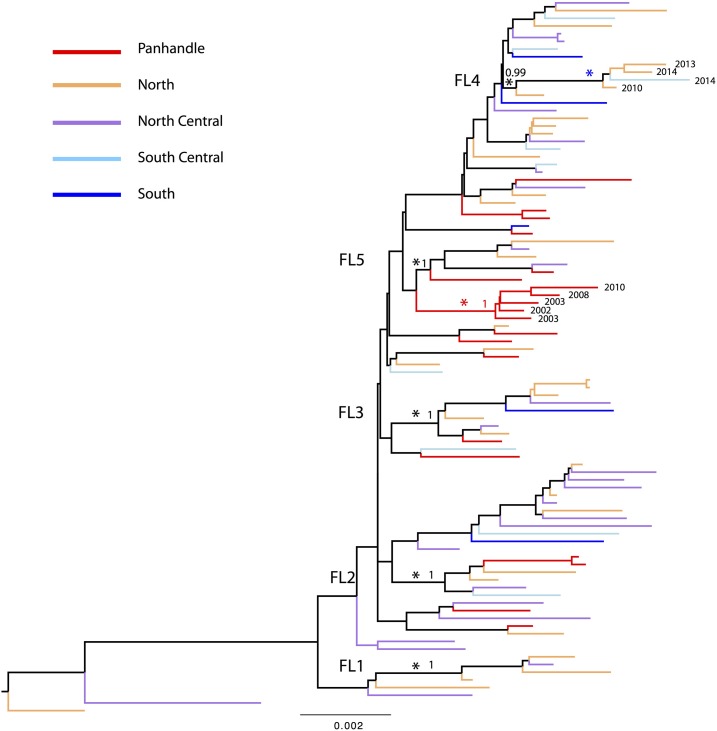
As previously found, there is notable genetic diversity of EEEV in Florida with five small monophyletic groups, FL1, FL2, FL3, FL4, and FL5.14 Further analysis by region had similar results (Figure 5). To determine the phylogeographic structure of EEEV within Florida, we performed phylogeny-trait association (BaTS) tests (Table 3) on the phylogenies of the 93 complete genome sequences of Florida strains. The results showed no significant phylogenetic clustering of the five Florida regions (P-values for AI and PS > 0.05), suggesting extensive spatial mixing of the virus among the regions, as also noted from a previous phylogenetic analysis of strains sampled from Florida and other states.26 However, the MC statistic for the Panhandle region was significant (P = 0.003), suggesting significant spatial structure and more localized evolution of EEEV in the Panhandle region, which is largely characterized by a Panhandle-only virus group in the BMCMC phylogenies (indicated by red asterisk in Figure 5). In addition, the results of BaTS analysis suggested the source of the virus (host) did not influence the phylogenetic clustering of Florida sequences, P > 0.05 in AI, PS, and MC values (Table 3). Although the phylogenetic tree showed significant temporal structure (P < 0.001 in both AI and PS), only the MC statistic for the sampling period 2010–2014 was significant (P = 0.019), which is characterized by a group of recent strains (2010–2014, blue asterisk in Figure 5), suggesting more localized evolution of EEEV during 2010–2014.
Figure 5.
Bayesian Markov chain Monte Carlo phylogenetic tree of eastern equine encephalitis virus (EEEV) sequences sampled from Florida. Sequences sampled from different regions in Florida are colored and described in the key. Five previously defined small monophyletic groups of Florida sequences are marked by black asterisk and indicated in the tree. Panhandle-only group is marked by red asterisk in the tree. The phylogenetic tree was rooted using the oldest EEEV sequence sampled from Florida in 1986, and scale bars represent the number of nucleotide substitutions per site. This figure appears in color at www.ajtmh.org.
Table 3.
Results of the phylogeny–trait association tests for different regions, host types, and sampling dates of eastern equine encephalitis virus in Florida
| Comparison | Statistic | Cases (n = 93) | Significance |
|---|---|---|---|
| Florida regions | AI | 93 | P = 0.102 |
| PS | 93 | P = 0.069 | |
| MC (Panhandle) | 20 | P = 0.003* | |
| MC (North) | 33 | P = 0.972 | |
| MC (North Central) | 26 | P = 0.833 | |
| MC (South Central) | 9 | P = 1 | |
| MC (South) | 5 | P = 1 | |
| Host | AI | 93 | P = 0.095 |
| PS | 93 | P = 0.155 | |
| MC (mosquito) | 15 | P = 0.362 | |
| MC (avian) | 32 | P = 0.947 | |
| MC (equine) | 44 | P = 0.689 | |
| MC (other mammals) | 2 | P = 1 | |
| Sampling dates | AI | 93 | P < 0.001* |
| PS | 93 | P < 0.001* | |
| MC (before 2000) | 10 | P = 0.423 | |
| MC (2000–2004) | 36 | P = 0.175 | |
| MC (2005–2009) | 25 | P = 0.149 | |
| MC (2010–2014) | 22 | P = 0.019* |
AI = association index; MC = maximum clade; PS = parsimony score.
* P-value less than 0.05 is considered statistically significant.
DISCUSSION
This is the first in-depth long-term study of the epidemiological dynamics of EEEV in Florida, which is proposed as one of the major source regions seeding EEEV epizootics in northeast regions of the United States. The sentinel chicken seroconversion data suggested significant seasonality of EEEV activity in Florida, especially in the North and North Central regions. However, there was no significant seasonality of EEEV activity observed in the Panhandle region. We also analyzed 93 complete genome sequences of EEEV strains collected from different hosts and Florida regions during 1986–2014 and found that there was little evidence for overall phylogeographic structure of the virus in Florida, except for the Panhandle region, which showed significant spatial structure and more localized evolution of EEEV in the region. This study indicates complex epidemiological dynamics of EEEV in Florida with year-round EEEV activity and significant viral spatial structure of EEEV in the Florida Panhandle, which suggests this region to be a potential source of EEEV for the rest of the state.
Our study used sentinel chicken seroconversion data from the Florida statewide surveillance program. Sentinel chicken annual confirmed seroconversion rates showed peaks coinciding with many peaks in human and equine cases in Florida and the United States, but the correspondence was not perfect. Years where the seroconversions were high, but human and/or equine cases were low may indicate successes in the surveillance program resulting in preventative activity. But years where the seroconversions were low, but human and/or equine cases were high may indicate program shortcomings. Other factors in the epizootic cycle could exist that are difficult to detect or predict that may influence transmission to humans and animals such as movement of EEEV into bridge vectors and abundance of these competent vectors (Aedes, Coquillettidia, and Culex species). Culiseta melanura may also play a role as an enzootic vector because it has been found to feed on humans and mammals in addition to birds.28–32 For this reason, mosquito control measures do not rely solely on sentinel chicken seroconversion rates but also mosquito data. The sentinel chicken annual confirmed seroconversion rates by region showed rates at their highest in the Panhandle and decreasing as one moved south, with very low to nonexistent activity in the southernmost part of the state. This is consistent with a previous analysis of sentinel chicken seroconversion rates,11 and matches human and horse cases for Florida.1,12,13 The May–August peak of the season for EEE sentinel chicken seroconversion rates was consistent with a previous analysis of Florida surveillance data from 1955 to 1974,9 but the peak of sentinel chicken collections, June–September for the entire state, July–August for the Panhandle and northern counties, and July–September for the central and southern counties, did not coincide. The timing and location of chicken placements in the field may be a result of historical precedence for SLEV peak transmission from late September to early October in the central to southern part of the state.33 Sentinel chicken surveillance for EEEV might be improved by placement of larger numbers of chickens in the Panhandle and northern counties earlier in the arbovirus season, such as in April, and maintaining a minimal level of chickens year-round. This could help prevent in-state transmission and movement of the virus out of Florida to other states by identifying an increase in virus transmission early, to allow implementation of interventions that can cost much less than the treatment and support of an infected person34 or the costs associated to losses in the horse industry in the state.
The most important epidemiological dynamic of EEEV in Florida found in this study is annual seasonality in Florida as a whole but different seasonality patterns in regions of the state. In particular, the North and North Central regions showed significant annual periodicity, whereas little seasonality was observed in the Panhandle region. Florida has been suggested as one of the major source regions seeding EEEV epidemics in Northeast regions of the United States because of its subtropical climate and year-round EEEV activities. Strikingly, in our study, for the first time we showed an annual seasonality pattern of EEEV activity in Florida despite the year-round mosquito activity in the state. However, the Panhandle region showed support for sustained year-round enzootic transmission from the sentinel chicken confirmed EEEV seroconversion data. Despite appearing to have seasonality in the Panhandle with the highest MMSR (6.0%) in June, the wavelet analysis revealed little significant seasonality in the region. This is because the rates never dropped below 1.4%, while other regions declined or showed no viral activity during the winter months. Although the North and North Central regions also had year-round transmission, they had statistically significant annual seasonality from the wavelet analysis. Furthermore, complex epidemiological dynamics of EEEV in the state were also supported by phase coherence analysis, which suggested no pattern of one region with a peak seasonality over the other but rather an oscillating coherence between the North and North Central regions every few years. This likely indicates extensive mixing of transmission between the two regions. In contrast to the Panhandle, these regions did not have transmission that continues at high seroconversion rates all year. There could be bias in the analysis because the location of the chicken flocks in the Panhandle region may be more appropriate for EEEV because of a long history of known transmission in this region, but this is unlikely because the locations of the flocks is generally based on convenience and has not been systematically determined.
In addition, the complexity of EEEV dynamics in Florida was also supported by extensive spatial dispersal of the virus within the five regions in Florida and relatively more clustering of the viruses in the Panhandle region, as suggested by our phylogeographic analysis. Some previous studies have divided Florida into four regions (Panhandle, North, Central, and South) based on the physiographic divisions of Florida that were created in 1981.12,35 We adopted a five-region grouping rather than a four-region grouping not only because these five regions are geographically distinct and represent different ecological biomes, but also because there are relatively balanced sample sizes in these five regions. A previous study with fewer viral strains and the four-region grouping showed no clustering by location.26 Our finding of significant clustering of the Panhandle viruses suggests some extent of localized transmission and evolution of EEEV in the Panhandle region and viruses can persist in the region for multiple years (2002–2010, Figure 5). Our BaTS analyses also suggest little influence of viral source (host) on viral phylogenetic clustering. In addition, we found the sampling date had significant influence on viral phylogeny (temporal structure); however, this is due to the strict “clock-like” evolution of EEEV.14 Furthermore, more localized evolution of EEEV recently (during 2010–2014) could also have an impact on the complexity of EEEV dynamics in the state.
A relatively different topography and ecology in the Panhandle region might be the reason why the Panhandle region is different from other regions in Florida. Spatial analysis of EEEV in Florida has found an association of epizootic and enzootic transmission associated with tree plantations, which are in greater abundance in the Panhandle region.12,35,36 The ecology of the Panhandle is also conducive to year-round Cs. melanura feeding on passerine birds,37 which in the winter months may be primarily northern cardinals that are in greater abundance as compared with southern Florida.28,38,39 A study comparing Cs. melanura blood meals in the winter to the spring in Florida, including one county in the Panhandle, revealed primary feeding from birds in the winter to a shift to reptiles in the spring with a county in the Panhandle having the highest percentage of blood meals from reptiles.28 It has been suggested overwintering of EEEV in ectotherms may play a significant role in sustaining transmission when the ectotherms emerge from brumation in the spring.40 The conditions in the Panhandle may be ideal for sustained enzootic transmission because the habitat for passerine birds, Cs. melanura mosquitoes, and potential overwintering ectotherms is optimal, creating a closed ecosystem-like environment for virus evolution. This may explain the significant clustering of the Panhandle sequences, the higher sentinel chicken seroconversion rates, and the lack of sentinel chicken seroconversion seasonality found in our study.
In addition to improved sentinel chicken surveillance in the future, additional genetic studies are needed. As next generation sequencing becomes a more common laboratory method, EEEV WGSs of additional historical and future isolates will be instrumental in further characterizing the virus in Florida. With additional sequences, a transmission analysis between the Panhandle and other states could be performed to definitively determine if the Panhandle is the region within the state that is the source of EEEV to other northern states. This analysis should be expanded to more of the southeastern United States, especially states bordering the Florida Panhandle (Alabama and Georgia) to better understand the extent of the sustained transmission and localized viral evolution found in the Panhandle.
In conclusion, this study has revealed the complex epidemiological dynamics of EEEV in Florida, characterized by different seasonality patterns and different spatial dispersal degrees of EEEV in different regions. Our study also showed the year-round viral activity and more localized evolution of EEEV in the Panhandle region, thus suggesting the Panhandle region may play an important role in seeding the virus for the rest of the state as well as be the possible source in the source-sink model in which Florida was identified as a location from which EEEV moves to northern states. The findings in this study will have an impact on targeting EEEV control in Florida to prevent animal and human outbreaks within the state and potentially out of state.
Supplementary Files
Acknowledgments:
The sequencing and sequence data analysis work was supported by the NIAID/NIH Genomic Centers for Infectious Diseases (GCID) program (U19-AI-110819 [to S. R. D.]). S. R. D. is also supported by NIH-funded Tennessee Center for AIDS Research (P30 AI110527). This study was also supported by Cooperative Agreement Number U01CK000510 to T. R. U., funded by the Centers for Disease Control and Prevention. The content is solely the responsibility of the authors and does not represent official views of the National Institutes of Health, Department of Health and Human Services, or Florida Department of Health.
Note: Supplemental figure and table appears at www.ajtmh.org.
REFERENCES
- 1.Centers for Disease Control and Prevention , 2017. Eastern Equine Encephalitis Epidemiology and Geographic Distribution. Available at: https://www.cdc.gov/easternequineencephalitis/tech/epi.html. Accessed November 12, 2018. [Google Scholar]
- 2.Calisher CH, 1994. Medically important arboviruses of the United States and Canada. Clin Microbiol Rev 7: 89–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morris CD, 1988. Eastern equine encephalomyelitis. Monath TP, ed. The Arboviruses: Epidemiology and Ecology. Boca Raton, FL: CRC Press, 1–20. [Google Scholar]
- 4.Scott TW, Weaver SC, 1989. Eastern equine encephalomyelitis virus: epidemiology and evolution of mosquito transmission. Adv Virus Res 37: 277–328. [DOI] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention (CDC) , 2006. Eastern equine encephalitis—New Hampshire and Massachusetts, August–September 2005. Morb Mortal Wkly Rep 55: 697–700. [PubMed] [Google Scholar]
- 6.Massachusetts Department of Public Health , 2012. Arbovirus Surveillance Summary, 2012. Available at: https://www.mass.gov/files/documents/2016/07/us/2012-summary.pdf. Accessed November 12, 2018. [Google Scholar]
- 7.Reimann CA, Hayes EB, DiGuiseppi C, Hoffman R, Lehman JA, Lindsey NP, Campbell GL, Fischer M, 2008. Epidemiology of neuroinvasive arboviral disease in the United States, 1999–2007. Am J Trop Med Hyg 79: 974–979. [PubMed] [Google Scholar]
- 8.Howard JJ, Morris CD, Emord DE, Grayson MA, 1988. Epizootiology of eastern equine encephalitis virus in upstate New York, USA. VII. Virus surveillance 1978–85, description of 1983 outbreak, and series conclusions. J Med Entomol 25: 501–514. [DOI] [PubMed] [Google Scholar]
- 9.Bigler WJ, Lassing EB, Buff EE, Prather EC, Beck EC, Hoff GL, 1976. Endemic eastern equine encephalomyelitis in Florida: a twenty-year analysis, 1955–1974. Am J Trop Med Hyg 25: 884–890. [DOI] [PubMed] [Google Scholar]
- 10.United States Department of Agriculture , 2018. Eastern and Western Equine Encephalitis Cases and Surveillance Information. Available at: https://www.aphis.usda.gov/aphis/ourfocus/animalhealth/animal-disease-information/horse-disease-information/sa_encephalitis/ct_ee_index. Accessed July 26, 2018. [Google Scholar]
- 11.Day JF, Stark LM, 1996. Transmission patterns of St. Louis encephalitis and eastern equine encephalitis viruses in Florida: 1978–1993. J Med Entomol 33: 132–139. [DOI] [PubMed] [Google Scholar]
- 12.Vander Kelen PT, Downs JA, Stark LM, Loraamm RW, Anderson JH, Unnasch TR, 2012. Spatial epidemiology of eastern equine encephalitis in Florida. Int J Health Geogr 11: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Downs J, Vaziri M, Jenkins A, Unnasch T, 2018. Validation of a risk index model for predicting eastern equine encephalitis virus transmission to horses in Florida. J Med Entomol 55: 1143–1149. [DOI] [PubMed] [Google Scholar]
- 14.Tan Y, et al. 2018. Large scale complete genome sequencing and phylodynamic analysis of eastern equine encephalitis virus reveal source-sink transmission dynamics in the United States. J Virol JVI.00074-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin DA, Muth DA, Brown T, Johnson AJ, Karabatsos N, Roehrig JT, 2000. Standardization of immunoglobulin M capture enzyme-linked immunosorbent assays for routine diagnosis of arboviral infections. J Clin Microbiol 38: 1823–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beaty BJ, Calisher CH, Shope RE, 1989. Arboviruses. Schmidt NJ, Emmons RW, eds. Diagnostic Procedures for Viral, Rickettsial and Chlamydial Infections, 6th edition Washington, DC: American Public Health Association, 797–856. [Google Scholar]
- 17.Hayes CG, Wallis RC, 1977. Ecology of Western equine encephalomyelitis in the eastern United States. Adv Virus Res 21: 37–83. [DOI] [PubMed] [Google Scholar]
- 18.Karabatsos N, Lewis AL, Calisher CH, Hunt AR, Roehrig JT, 1988. Identification of Highlands J virus from a Florida horse. Am J Trop Med Hyg 39: 603–606. [DOI] [PubMed] [Google Scholar]
- 19.Centers for Disease Control and Prevention , 2018. ArboNet Maps. Available at: https://wwwn.cdc.gov/arbonet/maps/ADB_Diseases_Map/index.html. Accessed November 12, 2018. [Google Scholar]
- 20.R Foundation for Statistical Computing , 2017. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 21.Johansson MA, Cummings DA, Glass GE, 2009. Multiyear climate variability and dengue–El Nino southern oscillation, weather, and dengue incidence in Puerto Rico, Mexico, and Thailand: a longitudinal data analysis. PLoS Med 6: e1000168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng X, et al. 2013. Epidemiological dynamics and phylogeography of influenza virus in southern China. J Infect Dis 207: 106–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang L, Wong CM, Lau EH, Chan KP, Ou CQ, Peiris JS, 2008. Synchrony of clinical and laboratory surveillance for influenza in Hong Kong. PLoS One 3: e1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grenfell BT, Bjornstad ON, Kappey J, 2001. Travelling waves and spatial hierarchies in measles epidemics. Nature 414: 716–723. [DOI] [PubMed] [Google Scholar]
- 25.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP, 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.White GS, Pickett BE, Lefkowitz EJ, Johnson AG, Ottendorfer C, Stark LM, Unnasch TR, 2011. Phylogenetic analysis of eastern equine encephalitis virus isolates from Florida. Am J Trop Med Hyg 84: 709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parker J, Rambaut A, Pybus OG, 2008. Correlating viral phenotypes with phylogeny: accounting for phylogenetic uncertainty. Infect Genet Evol 8: 239–246. [DOI] [PubMed] [Google Scholar]
- 28.Blosser EM, Lord CC, Stenn T, Acevedo C, Hassan HK, Reeves LE, Unnasch TR, Burkett-Cadena ND, 2017. Environmental drivers of seasonal patterns of host utilization by Culiseta melanura (Diptera: Culicidae) in Florida. J Med Entomol 54: 1365–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Molaei G, et al. 2013. Vector-host interactions and epizootiology of eastern equine encephalitis virus in Massachusetts. Vector Borne Zoonotic Dis 13: 312–323. [DOI] [PubMed] [Google Scholar]
- 30.Molaei G, Armstrong PM, Abadam CF, Akaratovic KI, Kiser JP, Andreadis TG, 2015. Vector-host interactions of Culiseta melanura in a focus of eastern equine encephalitis virus activity in southeastern Virginia. PLoS One 10: e0136743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Molaei G, Armstrong PM, Graham AC, Kramer LD, Andreadis TG, 2015. Insights into the recent emergence and expansion of eastern equine encephalitis virus in a new focus in the Northern New England USA. Parasit Vectors 8: 516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Molaei G, Oliver J, Andreadis TG, Armstrong PM, Howard JJ, 2006. Molecular identification of blood-meal sources in Culiseta melanura and Culiseta morsitans from an endemic focus of eastern equine encephalitis virus in New York. Am J Trop Med Hyg 75: 1140–1147. [PubMed] [Google Scholar]
- 33.Nelson DB, Kappus KD, Janowski HT, Buff E, Wellings FM, Schneider HJ, 1983. St. Louis encephalitis - Florida 1977. Patterns of widespread outbreaks. Am J Trop Med Hyg 32: 412–416. [PubMed] [Google Scholar]
- 34.Villari P, Spielman A, McDowell M, Komar N, Timperi RJ, 1995. The economic burden imposed by a residual case of eastern encephalitis. Am J Trop Med Hyg 52: 8–13. [DOI] [PubMed] [Google Scholar]
- 35.Vander Kelen PT, Downs JA, Unnasch T, Stark L, 2014. A risk index model for predicting eastern equine encephalitis virus transmission to horses in Florida. Appl Geogr 48: 79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vander Kelen PT, et al. 2012. Habitat associations of eastern equine encephalitis transmission in Walton County Florida. J Med Entomol 49: 746–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burkett-Cadena ND, Bingham AM, Hunt B, Morse G, Unnasch TR, 2015. Ecology of Culiseta melanura and other mosquitoes (Diptera: Culicidae) from Walton County, FL, during winter period 2013–2014. J Med Entomol 52: 1074–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Estep LK, et al. 2013. Risk of exposure to eastern equine encephalomyelitis virus increases with the density of northern cardinals. PLoS One 8: e57879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sauer JR, Niven DK, Hines JE, Ziolkowski DJ, Pardieck KL, Fallon JE, Link WA, 2017. The North American Breeding Bird Survey, Results and Analysis 1966–2015. Version 2.07.2017. Laurel, MD: USGS Patuxent Wildlife Research Center. [Google Scholar]
- 40.Bingham AM, Graham SP, Burkett-Cadena ND, White GS, Hassan HK, Unnasch TR, 2012. Detection of eastern equine encephalomyelitis virus RNA in North American snakes. Am J Trop Med Hyg 87: 1140–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.