Abstract
Purpose:
Inflammation is linked to prostate cancer (PrCa) progression and is mediated by Nuclear Factor Kappa B (NFκB). Tristetraprolin is a key node of NFκB activation and we investigated its biological and prognostic role in lethal PrCa.
Methods:
In vitro assays assessed the function of tristetraprolin and the association between low mRNA tristetraprolin levels and lethal PrCa (metastatic disease or death) was assessed across independent prostatectomy cohorts: (i) nested case-control studies from Health Professionals Follow-up Study and Physicians’ Health Study, and (ii) prostatectomy samples from Cleveland Clinic, Mayo Clinic, Johns Hopkins and MSKCC. Tristetraprolin expression levels in prostatectomy samples from patients with localized disease and biopsies of metastatic castration resistant PrCa (mCRPC) were assessed in a Cornell University cohort.
Results:
In vitro tristetraprolin expression was inversely associated with NFκB-controlled genes, proliferation and enzalutamide sensitivity. Men with localized PrCa and lower quartile of tumor tristetraprolin expression had a significant, nearly 2-fold higher risk of lethal PrCa after adjusting for known clinical and histological prognostic features (age, RP Gleason score, T-stage). Tristetraprolin expression was also significantly lower in mCRPC compared with localized PrCa.
Conclusions:
Lower levels of tristetraprolin in human PrCa prostatectomy tissue are associated with more aggressive PrCa and may serve as an actionable prognostic and predictive biomarker.
Impact:
There is a clear need for improved biomarkers to identify patients with localized prostate cancer in need of treatment intensification, such as adjuvant testosterone suppression, or treatment de-intensification, such as active surveillance. Tristetraprolin levels may serve as informative biomarkers in localized PrCa.
Introduction:
Prostate cancer has a varied clinical course, with some patients having localized disease which does not require intervention while others present with metastatic disease that responds poorly to therapy1,2. Clinicopathological staging such as higher Gleason score, larger primary, and findings of extension beyond the prostate at prostatectomy increase chances of developing metastatic and lethal disease3,4. In addition, commercially available assays of RNA transcript profiles representing biological processes such as cell cycle and apoptosis provide further prognostic information on risk of relapse after prostatectomy or radiation for localized disease5.
Biological drivers for the development of prostate cancer and its lethal subtypes are diverse. Epidemiological and biological studies have implicated aberrant metabolism and inflammation6. In the case of metabolism, obesity and markers of insulin resistance have been linked a greater chance of dying of prostate cancer.7 With respect to inflammation, elevation of serum cytokines such as IL6 have been associated with lethal prostate cancer and inflammatory changes have been associated with prostate cancer lesions in the prostate6,8. Nuclear Factor Kappa B (NFκB) activation is a transcription factor that controls inflammation and can either promote cancer progression or cancer cell death depending on its associated regulators9. Despite numerous preclinical studies demonstrating a central role of NFκB activation in promoting proliferation, development of metastases and evasion of apoptosis in prostate cancer10–12, biomarkers of NFκB activation, such as presence of nuclear staining of the active subunit p65 in prostate cancer cells in localized disease have not been found to reliably identify patients with a higher risk of metastatic disease after a prostatectomy or radiation therapy given with curative intent13–15. Identification of patients with loss of negative regulators of NFκB activity which, in turn, promote aggressive or lethal prostate cancer, may therefore identify patients who need intensification of therapy such as adjuvant androgen deprivation therapy (ADT) with radiation or prostatectomy. Moreover, such patients may also benefit from adding drugs that block cancer-promoting NFκB activity9,16.
Using a bioinformatic analytical approach with Bayesian data integration of publicly available genomic datasets, we defined an NFκB pathway that was enriched in patients with prostate cancer who relapsed with metastatic disease after a prostatectomy17,18. After reviewing the literature of potential candidates for known functions and biological data supporting a role in prostate cancer, tristetraprolin was considered the lead candidate and was taken forward for further evaluation. Other candidates included ATF3, CXCL2, DUSP5, JUNB, NEDD9, SELE, and TRIB118 but tristetraprolin was considered to be the most promising and novel based on it known function and limited data in prostate cancer. Tristetraprolin homolog was found to be one of the key nodes in this pathway, and we postulated is a regulator of NFκB activity. We hypothesized that low tristetraprolin levels in prostatectomy specimens would be associated with a higher likelihood of metastasis or fatal disease.
Tristetraprolin (TTP), also known as ZFP36, is member of the TIS11 (TPA-induced sequence) family which binds specific mRNA sequences and leads to transcript deadenylation and degradation19. The physiological effects of tristetraprolin loss include inflammatory conditions such as arthritis and dermatitis20 with increased levels of pro-inflammatory cytokines21. Tristetraprolin control of NFκB activity has been reported to occur via a number of mechanisms, including mediation of TNF-alpha mRNA (an NFκB activator) and IL-1β22 degradation, as well as blocking the nuclear translocation of the p65 subunit of NF-κ B and physically interacting with histone deacetylases (HDACs), HDAC-1, HDAC-3, and HDAC-7 and these can also serve as co-repressors of NF-κ B-dependent transcription23. Tristetraprolin also controls other critical genes overexpressed in inflammation and cancer including c-myc, c-JUN and p5319. Previous work adds further plausibility that loss of tristetraprolin function is a driver of cancer in general and prostate cancer specifically using in vitro modeling experiments as well as gene expression of human tissues showing association with biochemical relapse and decreased levels in metastatic deposits versus localized disease24,25.
We therefore, sought to further define the biological activity of tristetraprolin and control of NFκB-related genes within prostate cancer cells in vitro, and whether loss of this tumor suppressor in human tumors is prognostic for the more clinically relevant endpoints of metastatic or lethal prostate cancer.
Patients and Methods:
In vitro
Human cell lines
LNCaP cell line was purchased from the American Type Culture Collection. RWPE-1 cell line was obtained from Dr. Myles Brown lab in Dana-Farber Cancer Institute. LNCaP cells were maintained in RPMI 1640 and supplemented with 10% FBS. RWPE-1 cells were cultured in Keratinocyte-SFM with EGF and BPE from life technologies. All culture mediums include 100 IU of penicillin, streptomycin (100 μg/ml). All cells were maintained at 37°C, 5% CO2, and 100% relative humidity and regularly screened for mycoplasma using a Venor GeM Mycoplasma Detection Kit (Sigma). RWPE-1 and LNCaP cell lines were originally obtained from ATCC. LNCaP was authenticated by whole genome sequencing. RWPE-1 was authenticated by determining the expression of a set of known genes and isoenzymes, specified by ATCC. These cell lines were routinely screened for mycoplasma contamination using the mycoplasma detection kit from Sigma. Freshly defrosted cell lines were used for experiments and experiments were completed within two or three passaging period. All our stocks are less than 10 passages.
siRNA and overexpression
Cell lines were cultured until ~80% confluence and then transfected with siRNAs or overexpression construct for tristetraprolin (Origene) using lipofectamine 2000. Knockdown or overexpression efficiency of tristetraprolin was detected by RT-PCR (forward 5’- GCTATGTCGGACCTTCTCAGAG −3’, reverse 5’- CCTGGAGGTAGAACTTGTGACAG −3’) at 3 days after transfection. All RT-PCR experiments were performed in triplicate.
Cell proliferation
Cells were split into 96-well plate with a confluence of ~ 40% after transfection for 24 hrs. Cell proliferation assay was carried out at different days after splitting using the WST-1 assay (Roche) with the detection of the absorption at a wavelength of 450 nm following the manufacture instruction. Each experiment was performed in triplicate.
Human NFκB Signaling Pathway PCR Array
Genes involved in the NFκB signaling pathway were detected by the Human NFκB Signaling Pathway PCR Array kit from Sabioscience following the manufacture instruction. Briefly, 1 µg total RNA was extracted to synthesis cDNA for 96-well plate formats after cells were transfected by siRNA or overexpression construct of tristetraprolin for 3 days. Gene change was treated as significant when the fold change > 1.5 and P value < 0.05.
Statistical analysis of in vitro studies
Statistical analysis for all in vitro studies was performed with Prism 6.0 software (GraphPad Software Inc.). All values in figures are presented as means ± SD. Statistical significance was calculated on the basis of Student’s t test (two-tailed) and the level of significance was set at P < 0.05.
Patho-clinical outcome studies in newly diagnosed prostate cancer
To assess the utility of tristetraprolin mRNA levels as a prognostic marker, we first leveraged data from a study nested in the Health Professionals Follow-up Study (HPFS) and Physicians’ Health Study (PHS). Gene expression of tristetraprolin was quantified in archival surgical tumor tissue using Human Gene 1.0 ST microarrays (Affymetrix, Santa Clara, CA; GSE79021). Low and high tristetraprolin were defined as expression below the lower quartile and greater than or equal to the lower quartile, respectively, where the lower quartile cutoff was selected to discriminate tumors with very low tristetraprolin expression from those with normal levels. In this “extreme” case-control design26 cases (n=113) were men who died of prostate cancer or developed metastatic disease at any time over follow-up, and controls (n = 291) were men who lived at least 8 years after diagnosis and remained metastasis free. Further study details have been reported previously27. Odds ratios and 95% confidence intervals for lethal disease were estimated through logistic regression in R version 3.4.0.
Using the same definition of lethal cases and prostate cancer controls, the prognostic utility of tristetraprolin was further assessed in 788 NCCN intermediate and high-risk patients pooled from 5 cohorts in GenomeDx Bioscience’s Genomics Resource Information Database (GRID). Tristetraprolin expression was quantified from radical prostatectomy specimens using the Affymetrix Human Exon 1.0 ST GeneChip28 (GSE79957), and low and high tristetraprolin was again defined as below versus greater than or equal to the lower expression quartile. Because of differences among the 5 study cohorts, particularly in baseline tristetraprolin expression, analyses were stratified by cohort using conditional logistic regression in SAS v9.429.
Tristetraprolin levels in benign prostate and primary prostate cancer versus metastatic CRPC samples
RNA-seq profiling data from the Weill Cornell Cohort have been previously published30,31 (dbGAPphs000909.v.p1). They include 117 samples: 26 benign prostate, 39 localized prostate cancer (PCa), and 52 CRPC cases, processed according to the protocols described in those papers. For the purpose of this analysis, FPKM (fragment per kilobase of exonic regions per million mapped reads) values quantified tristetraprolin expression. Batch normalization with COMBAT32 was used to account for differences in library preparation, specifically poly-A selection or ribosomal depletion. Oncomine data was accessed to recapitulate the findings.
Results:
In vitro silencing and overexpressing of tristetraprolin was undertaken in the RWPE1 and LNCaP cell lines, respectively (Figure 1A). Silencing tristetraprolin in the RWPE-1 cell line increased the proliferative potential whereas restoring the tumor suppressor in LNCaP resulted in decreased proliferation (Figure 1B). Silencing tristetraprolin resulted in increased expression of many NFκB related genes and overexpression of tristetraprolin decreased levels of NFκB related genes (Figure 1C). Notably, increasing tristetraprolin led to decreased BCL2A1, CSF2 and CSF3, and decreasing tristetraprolin led to increases in these same NFκB regulated genes (Supplementary table 1). Decrease of the NFκB inhibitor, NFKBIA, with overexpression of tristetraprolin in LNCaP presumably relates to decreased NFκB activity, and silencing tristetraprolin results in increased NFκB activity and compensatory increase in the inhibitor, NFKBIA as part of tightly controlled feedback loops9. It was also noted that over-expression of tristetraprolin augmented the sensitivity of LNCaP cells to enzalutamide whereas silencing tristetraprolin again increased proliferation of the RWPE1 cells and notably the enzalutamide had only a mild effect on decreasing the proliferation (Figure 1D).
Figure 1.
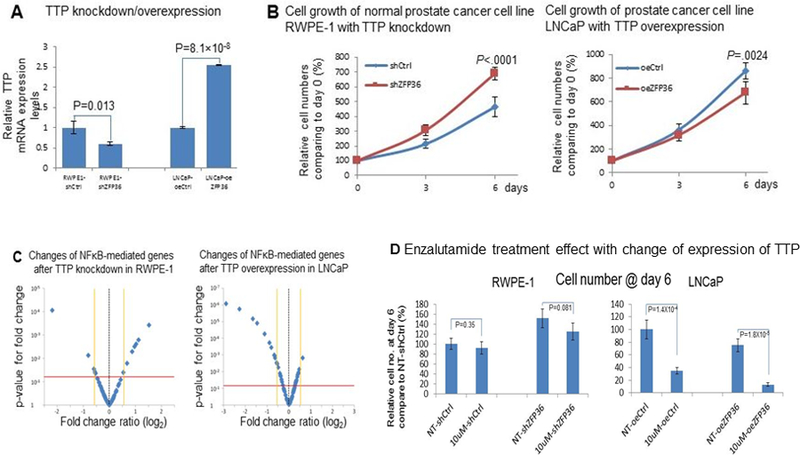
Knockdown of tristetraprolin in non-cancer prostate cell line (RWPE-1) and overexpression in prostate cancer cell line (LNCaP) (A) resulted in significantly increased and decreased proliferation respectively (B) and made LNCaP more sensitive to enzalutamide (E) and also impacted NFκB related genes (C, D)
Patient characteristics for the HPFS and PHS case-control design are summarized in Table 1. Mean tristetraprolin expression was significantly lower in the lethal cases compared to the non-lethal controls (p<0.001). The crude odds ratio for lethal disease comparing those with low versus high tristetraprolin expression was 3.52 (95% CI: 2.18–5.69; p<0.001). This association persisted with adjustment for Gleason category (<7, 3+4, 4+3, 8, 9–10), age at diagnosis, and clinical stage (T1/T2 N0/Nx M0/Mx, T3 N0/Nx M0/Mx, T4/N1/M1), with an adjusted odds ratio of 1.83 (95% CI: 0.99–3.33; p=0.05).
Table 1:
Clinical characteristics of the Harvard Prostate Tumor Cohorts (HPFS and PHS) stratified by lowest Tristetraprolin quartile
| TristetraprolinLow (n=101) | TristetraprolinHigh (n=303) | p-value | |
|---|---|---|---|
| Cohort – n (%) | 0.55 | ||
| HPFS | 66 (26) | 188 (74) | |
| PHS | 35 (23) | 115 (77) | |
| Age at diagnosis – median (SD; range) | 67.9 (7.0; 47−81) |
65.0 (6.2; 49−79) |
0.003 |
| Pathologic RP Gleason score – n (%) | <0.001 | ||
| 2−6 | 10 (18) | 47 (82) | |
| 7 | 49 (20) | 192 (80) | |
| 8 | 13 (30) | 30 (70) | |
| 9−10 | 29 (46) | 34 (54) | |
| Clinical stage – n (%) | 0.001 | ||
| T1/T2 | 78 (22) | 272 (78) | |
| T3 | 7 (26) | 20 (74) | |
| T4 | 12 (60) | 8 (40) | |
| Total PSA at diagnosis, ng/mL – n (%) | 0.11 | ||
| 0−4 | 4 (10) | 37 (90) | |
| 4−10 | 37 (19) | 159 (81) | |
| 10−20 | 16 (25) | 47 (75) | |
| >20 | 12 (29) | 29 (71) | |
| Not available | 32 (51) | 31 (49) | |
| Case status – (%) | <0.001 | ||
| Lethal | 49 (43) | 64 (57) | |
| Indolent | 52 (18) | 239 (82) | |
| RP: radical Prostatectomy | |||
HPFS refers to Health Professionals Follow-up Study and PHS refers to Physicians’ Health Study
Similar associations were observed in the pooled GenomeDX cohorts (Table 2). The crude odds ratio for metastatic disease comparing those with low to those with high tristetraprolin expression was 1.93 (95% CI: 1.26–2.44; p<0.001). The odds ratio adjusted for Gleason score, diagnostic age, and clinical stage was 1.84 (1.24–2.75; p=0.003).
Table 2:
Patient Characteristics of the GenomeDX Cohorts stratified by lowest Tristetraprolin quartile
| TristetraprolinLow (n=197) | TristetraprolinHigh (n=591) | p-value | |
|---|---|---|---|
| Study Cohort − n (%) | <0.001 | ||
| CCF | 6 (6) | 101 (94) | |
| JHU | 96 (36) | 170 (64) | |
| MSK | 2 (13) | 14 (88) | |
| Mayo I* | 78 (30) | 186 (70) | |
| Mayo II | 15 (11) | 120 (89) | |
| Age at diagnosis, median (SD; range) | 63.0 (6.8, 38–77) | 62.5 (6.9, 42–79) | 0.22 |
| Pathologic RP Gleason Score – n (%) | 0.043 | ||
| 2 − 6 | 12 (14) | 72 (86) | |
| 7 | 68 (24) | 217 (76) | |
| 8 | 33 (25) | 99 (75) | |
| 9−10 | 84 (29) | 203 (71) | |
| Clinical Stage – n (%) | 0.31 | ||
| T1/T2 | 173 (24) | 534 (76) | |
| T3 | 24 (30) | 57 (70) | |
| Adjuvant ADT – n (%)** | 0.91 | ||
| No | 62 (23) | 202 (77) | |
| Yes | 31 (23) | 104 (77) | |
| Adjuvant RT – n (%)** | 0.63 | ||
| No | 80 (23) | 269 (77) | |
| Yes | 13 (26) | 37 (74) | |
| Total PSA at diagnosis, ng/mL – n (%) | 0.036 | ||
| 0−4 | 20 (26) | 57 (74) | |
| 4−10 | 84 (24) | 271 (76) | |
| 10−20 | 47 (23) | 155 (77) | |
| >20 | 44 (34) | 85 (66) | |
| Not available | 2 (8) | 23 (92) | |
| Case status – n (%) | <0.001 | ||
| Lethal | 120 (30) | 278 (70) | |
| Indolent | 77 (20) | 313 (80) | |
| RP: radical Prostatectomy | |||
Mayo I and II refer to distinct patient cohorts that were used in the original discovery and validation, respectively, of GenomeDx Decipher genomic classifier (Erho,2013).
Based on total n=399 (93 TristetrapolinLow, 309 TristetrapolinHigh.
In the further independent cohort from Weill Cornell Medicine, tristetraprolin expression levels from RNA-Seq were approximately the same in benign and localized disease specimens, but significantly decreased in the metastatic castration-resistant prostate cancer tissues compared to each (p=7.1e-10 and p=9.0e-14, respectively; Figure 2). These findings were confirmed by analysis of seven publicly available datasets in Oncomine (Figure 3).
Figure 2.
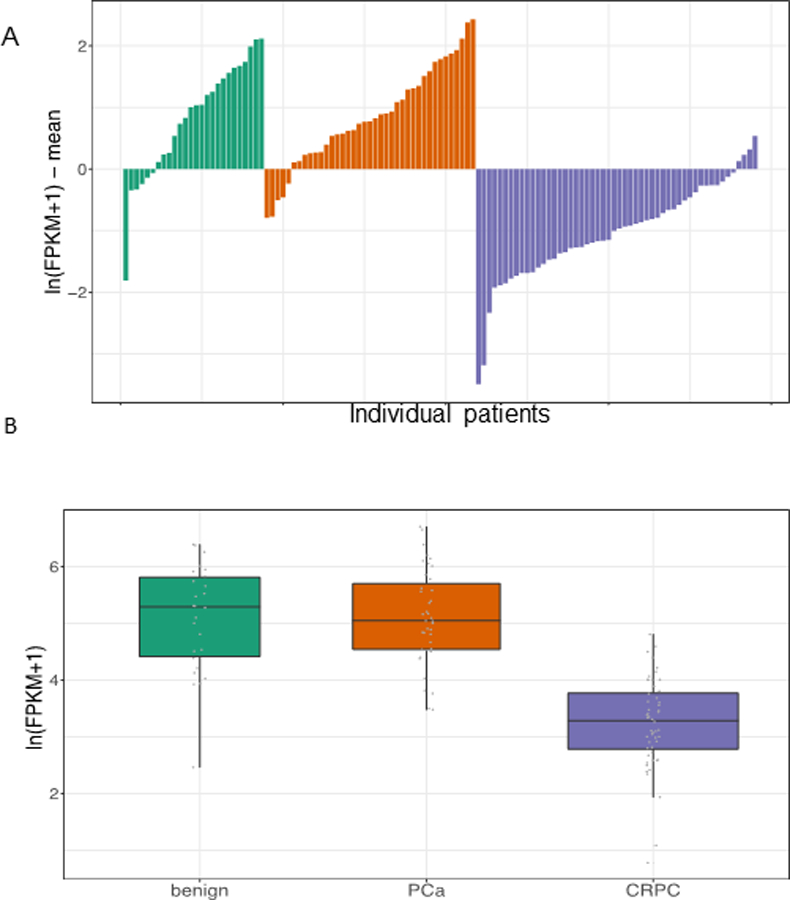
Tristetraprolin expression in WCM prostate cancer cohorts. The top panel (A) depicts the expression of tristetraprolin for all samples in the cohort where each bar corresponds to a sample. The bottom panel (B) summarizes the data in whisker plots. Tristetraprolin expression is significantly downregulated in CRPC compared to benign prostate (Wilcoxon test p=7.1e-10) and localized prostate cancer (PCa) (p=9.0e-14).
Figure 3.
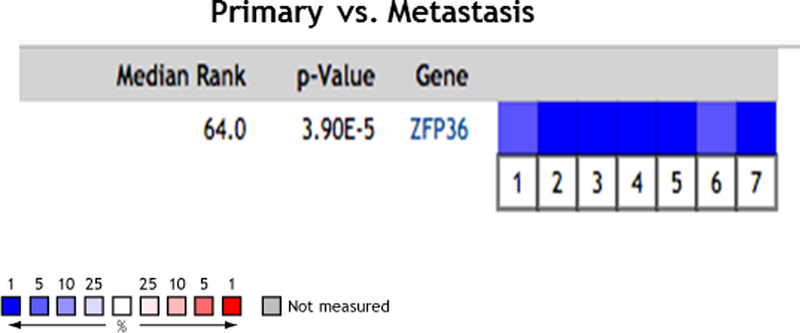
Figure 3: Oncomine analysis - https://www.oncomine.com/resource/main.html. The expression of tristetraprolin in several Oncomine prostate cancer datasets show significant downregulation in metastatic samples compared to primary tissues. 1. Prostate Cancer – Metastasis; Grasso Prostate, Nature, 201239; 2. Prostate Cancer – Metastasis; Holzbeierlein Prostate, Am J Pathol, 200440 3. Prostate Cancer - Metastasis LaTulippe Prostate, Cancer Res, 200241; 4. Prostate Cancer- Metastasis; Ramaswamy Multi-cancer, Proc Natl Acad Sci U S A, 200142; 5. Prostate Cancer - Metastasis Ramaswamy Multi-cancer 2, Nat Genet, 200342; 6. Prostate Cancer - Metastasis Taylor Prostate 3, Cancer Cell, 201043; 7. Prostate Cancer - Metastasis Yu Prostate, J Clin Oncol, 200444
Discussion
We previously applied an integrated Bayesian and systems biology approach to interrogate 18 the network signaling of inflammatory processes implicated in prostate cancer development. This identified tristetraprolin as a potential regulator of NFκB signaling. Tristetraprolin was chosen as the lead candidate based on prior literature indicating it may have a role in prostate cancer biology. In the present work, we first confirmed using in vitro studies that tristetraprolin was biologically relevant in prostate cancer by at least partially controlling NFκB-related genes and proliferation in prostate non-cancerous and cancerous cell lines and partially modulating resistance to androgen receptor inhibition. The latter observation is consistent with previous work reporting that overexpression of tristetraprolin represses multiple steroid nuclear receptors including androgen receptor (AR) transactivation in breast cancer cell lines33. As such, LNCaP with overexpressed tristetraprolin may be made more sensitive to enzalutamide treatment by dual mechanisms of AR signaling inhibition both pharmaceutically with enzalutamide and by the effects of tristetraprolin suppression on NFκB regulated AR transcription as well as blocking NFκB mediated resistance mechanisms.
Having confirmed tristetraprolin has a biological role in prostate cancer, we then assessed the hypothesis that mRNA levels of tristetraprolin would be low in human prostate tissue that is associated with lethal or metastatic disease after a prostatectomy. The results of the present gene expression profiling across several prostatectomy cohorts reproducibly and convincingly showed that tristetraprolin gene expression is lower in patients who develop metastatic disease. Analyses of an additional cohort and seven publicly available cohorts confirmed that tristetraprolin levels are lower in metastatic deposits. The relationship has important prognostic features, in particular that the association of low tristetraprolin gene levels with a significant increased risk of lethal prostate cancer after treatment with curative intent is independent of Gleason score. It is also noted that beyond its prognostic role for lethal disease, tristetraprolin may additionally serve as a predictive biomarker for patients who might benefit from NFκB inhibition.
Strengths of this study include use of the clinically relevant endpoint of metastatic and lethal prostate cancer from prostate cancer cohorts with long-term prospective follow-up and consistent effect estimates across a variety of patient populations. These findings further the findings from prior work which used less clinically relevant endpoints such as biochemical recurrence and are bolstered by in vitro work both detailed in this manuscript and prior publications24,34–36. Importantly, addition of tristetraprolin expression improves the prognostic accuracy of known clinical factors, as evidenced by the magnitude of the tristetraprolin effect on metastatic progression in the multivariable models across the HPFS/PHS and GenomeDX cohorts. Additional area under the curve (AUC) analyses were not possible in the GenomeDX data, due to the need to stratify by cohort. When modeled as a continuous variable in a logistic regression for prognosis in HPFS/PHS, tristetraprolin improved the AUC of the clinical factor model from 0.84 (95% CI: 0.80–0.88) to 0.85 (95% CI: 0.81–0.89). Although this improvement was not statistically significant (p=0.23), tests for AUC improvement are known to have low power, and it has been shown that the ability of a biomarker such as low tristetraprolin to improve the clinical model can more powerfully be inferred from its effect in the multivariable logistic regression presented in Results37.
Limitations of this work are the preliminary nature of the preclinical findings which are correlative at this time. Nonetheless, the data completed to date is consistent with and adds to the prior but limited preclinical work and provides guidance for further studies. These include in vivo tumorigenesis and more detailed mechanistic and functional studies dissecting the role of tristetraprolin in controlling NFκB versus other pathways such as myc and JUN and whether it also impacts inflammatory factors in the stroma. This could be achieved with silencing RNA of tristetrpolin RNA with and without inhibition of NFκB via IκB super-repressor38 or pharmacologically with dimethylaminoparthenolide16. Other limitations include use of retrospective cohorts and limited prognostic variables for the clinical association studies. This data supports use of samples from prospective trials to address these limitations.
In conclusion, this work provides robust evidence that tristetraprolin is a potentially actionable prognostic and predictive biomarker that was chosen following an integrated Bayesian-systems biology approach to interrogating gene expression data. The next tranche of work is to further explore the biology of tristetraprolin in prostate cancer and assess whether patients with low tristetraprolin benefit from adjuvant androgen deprivation and/or specific NFκB inhibition to define its clinical utility.
Supplementary Material
Acknowledgements
Funding: DOD W81XWH-11–1-0379 (Sweeney). The Health Professionals Follow-Up Study was supported by U01CA167552 (to Mucci and Willett). This study was additionally supported by CA136578 (Mucci), CA141298 (Stampfer), CA131945 (Loda), and P50CA090381 (Kantoff). L.A. Mucci was supported by a Prostate Cancer Foundation Young Investigator Award.
Footnotes
Disclosure of Potential Conflicts of Interest: E. Davicioni holds ownership interest (including patents) in GenomeDx Biosciences. A.E. Ross holds ownership interest (including patents) in and is a consultant/advisory board member for GenomeDx Biosciences. E.A. Klein holds consulting or advisory roles for GenomeDx Biosciences, Berg, and Genomic Health. R.J. Karnes reports receiving other commercial research support from GenomeDx Biosciences. Drs Sweeney, Mucci, Gerke, Lee, Börnigen, Wang and Huttenhower hold a patent for TTP as a biomarker in prostate cancer. No other potential conflicts were disclosed by the other authors.
References
- 1.Hamdy FC, Donovan JL, Lane JA, et al. : 10-Year Outcomes after Monitoring, Surgery, or Radiotherapy for Localized Prostate Cancer. N Engl J Med 375:1415–1424, 2016 [DOI] [PubMed] [Google Scholar]
- 2.Sweeney CJ, Chen YH, Carducci M, et al. : Chemohormonal Therapy in Metastatic Hormone-Sensitive Prostate Cancer. N Engl J Med 373:737–46, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolla M, Van Tienhoven G, Warde P, et al. : External irradiation with or without long-term androgen suppression for prostate cancer with high metastatic risk: 10-year results of an EORTC randomised study. Lancet Oncol 11:1066–73, 2010 [DOI] [PubMed] [Google Scholar]
- 4.Thompson IM Jr., Tangen CM, Paradelo J, et al. : Adjuvant radiotherapy for pathologically advanced prostate cancer: a randomized clinical trial. JAMA 296:2329–35, 2006 [DOI] [PubMed] [Google Scholar]
- 5.Bostrom PJ, Bjartell AS, Catto JW, et al. : Genomic Predictors of Outcome in Prostate Cancer. Eur Urol 68:1033–44, 2015 [DOI] [PubMed] [Google Scholar]
- 6.Nelson WG, De Marzo AM, Isaacs WB: Prostate cancer. N Engl J Med 349:366–81, 2003 [DOI] [PubMed] [Google Scholar]
- 7.Ma J, Li H, Giovannucci E, et al. : Prediagnostic body-mass index, plasma C-peptide concentration, and prostate cancer-specific mortality in men with prostate cancer: a long-term survival analysis. Lancet Oncol 9:1039–47, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stark JR, Li H, Kraft P, et al. : Circulating prediagnostic interleukin-6 and C-reactive protein and prostate cancer incidence and mortality. Int J Cancer 124:2683–9, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perkins ND: The diverse and complex roles of NF-kappaB subunits in cancer. Nat Rev Cancer 12:121–32, 2012 [DOI] [PubMed] [Google Scholar]
- 10.Min J, Zaslavsky A, Fedele G, et al. : An oncogene-tumor suppressor cascade drives metastatic prostate cancer by coordinately activating Ras and nuclear factor-kappaB. Nat Med 16:286–294, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sweeney C, Li L, Shanmugam R, et al. : Nuclear factor-kappaB is constitutively activated in prostate cancer in vitro and is overexpressed in prostatic intraepithelial neoplasia and adenocarcinoma of the prostate. Clin Cancer Res 10:5501–7, 2004 [DOI] [PubMed] [Google Scholar]
- 12.Zhang L, Altuwaijri S, Deng F, et al. : NF-kappaB regulates androgen receptor expression and prostate cancer growth. Am J Pathol 175:489–99, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lessard L, Karakiewicz PI, Bellon-Gagnon P, et al. : Nuclear localization of nuclear factor-kappaB p65 in primary prostate tumors is highly predictive of pelvic lymph node metastases. Clin Cancer Res 12:5741–5, 2006 [DOI] [PubMed] [Google Scholar]
- 14.Okera M, Bae K, Bernstein E, Cheng L, Lawton C, Wolkov H, Pollack A, Dicker A, Sandler H, Sweeney CJ. Evaluation of nuclear factor κB and chemokine receptor CXCR4 co-expression in patients with prostate cancer in the Radiation Therapy Oncology Group (RTOG) 8610. BJU Int, 2010 [DOI] [PMC free article] [PubMed]
- 15.Gannon PO, Lessard L, Stevens LM, et al. : Large-scale independent validation of the nuclear factor-kappa B p65 prognostic biomarker in prostate cancer. Eur J Cancer 49:2441–8, 2013 [DOI] [PubMed] [Google Scholar]
- 16.Shanmugam R, Kusumanchi P, Cheng L, et al. : A water-soluble parthenolide analogue suppresses in vivo prostate cancer growth by targeting NFkappaB and generating reactive oxygen species. Prostate 70:1074–86, 2010 [DOI] [PubMed] [Google Scholar]
- 17.Bornigen D, Moon YS, Rahnavard G, et al. : A reproducible approach to high-throughput biological data acquisition and integration. PeerJ 3:e791, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bornigen D, Tyekucheva S, Wang X, et al. : Computational Reconstruction of NFkappaB Pathway Interaction Mechanisms during Prostate Cancer. PLoS Comput Biol 12:e1004820, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanduja S, Blanco FF, Young LE, et al. : The role of tristetraprolin in cancer and inflammation. Front Biosci (Landmark Ed) 17:174–88, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor GA, Carballo E, Lee DM, et al. : A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity 4:445–54, 1996 [DOI] [PubMed] [Google Scholar]
- 21.Cappuzzo F, Ciuleanu T, Stelmakh L, et al. : Erlotinib as maintenance treatment in advanced non-small-cell lung cancer: a multicentre, randomised, placebo-controlled phase 3 study. Lancet Oncol 11:521–9, 2010 [DOI] [PubMed] [Google Scholar]
- 22.King EM, Kaur M, Gong W, et al. : Regulation of tristetraprolin expression by interleukin-1 beta and dexamethasone in human pulmonary epithelial cells: roles for nuclear factor-kappa B and p38 mitogen-activated protein kinase. J Pharmacol Exp Ther 330:575–85, 2009 [DOI] [PubMed] [Google Scholar]
- 23.Liang J, Lei T, Song Y, et al. : RNA-destabilizing factor tristetraprolin negatively regulates NF-kappaB signaling. J Biol Chem 284:29383–90, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berglund AE, Scott KE, Li W, et al. : Tristetraprolin disables prostate cancer maintenance by impairing proliferation and metabolic function. Oncotarget 7:83462–83475, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu JG, Yuan DB, Chen WH, et al. : Prognostic value of ZFP36 and SOCS3 expressions in human prostate cancer. Clin Transl Oncol 18:782–91, 2016 [DOI] [PubMed] [Google Scholar]
- 26.Sinnott JA, Rider JR, Carlsson J, et al. : Molecular differences in transition zone and peripheral zone prostate tumors. Carcinogenesis 36:632–8, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sinnott JA, Peisch SF, Tyekucheva S, et al. : Prognostic Utility of a New mRNA Expression Signature of Gleason Score. Clin Cancer Res 23:81–87, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Erho N, Crisan A, Vergara IA, et al. : Discovery and validation of a prostate cancer genomic classifier that predicts early metastasis following radical prostatectomy. PLoS One 8:e66855, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karnes RJ, Choeurng V, Ross AE, et al. : Validation of a Genomic Risk Classifier to Predict Prostate Cancer-specific Mortality in Men with Adverse Pathologic Features. Eur Urol, 2017 [DOI] [PMC free article] [PubMed]
- 30.Chakravarti A, Winter K, Wu CL, et al. : Expression of the epidermal growth factor receptor and Her-2 are predictors of favorable outcome and reduced complete response rates, respectively, in patients with muscle-invading bladder cancers treated by concurrent radiation and cisplatin-based chemotherapy: a report from the Radiation Therapy Oncology Group. Int J Radiat Oncol Biol Phys 62:309–17, 2005 [DOI] [PubMed] [Google Scholar]
- 31.Beltran H, Prandi D, Mosquera JM, et al. : Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med 22:298–305, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson WE, Li C, Rabinovic A: Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8:118–27, 2007 [DOI] [PubMed] [Google Scholar]
- 33.Barrios-Garcia T, Gomez-Romero V, Tecalco-Cruz A, et al. : Nuclear tristetraprolin acts as a corepressor of multiple steroid nuclear receptors in breast cancer cells. Mol Genet Metab Rep 7:20–6, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fallahi M, Amelio AL, Cleveland JL, et al. : CREB targets define the gene expression signature of malignancies having reduced levels of the tumor suppressor tristetraprolin. PLoS One 9:e115517, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haaland CM, Heaphy CM, Butler KS, et al. : Differential gene expression in tumor adjacent histologically normal prostatic tissue indicates field cancerization. Int J Oncol 35:537–46, 2009 [DOI] [PubMed] [Google Scholar]
- 36.Lee HH, Lee SR, Leem SH: Tristetraprolin regulates prostate cancer cell growth through suppression of E2F1. J Microbiol Biotechnol 24:287–94, 2014 [DOI] [PubMed] [Google Scholar]
- 37.Pepe MS, Kerr KF, Longton G, et al. : Testing for improvement in prediction model performance. Stat Med 32:1467–82, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deraska PV, O’Leary C, Reavis HD, et al. : NF-kappaB inhibition by dimethylaminoparthenolide radiosensitizes non-small-cell lung carcinoma by blocking DNA double-strand break repair. Cell Death Discov 4:10, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grasso CS, Wu YM, Robinson DR, et al. : The mutational landscape of lethal castration-resistant prostate cancer. Nature 487:239–43, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holzbeierlein J, Lal P, LaTulippe E, et al. : Gene expression analysis of human prostate carcinoma during hormonal therapy identifies androgen-responsive genes and mechanisms of therapy resistance. Am J Pathol 164:217–27, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.LaTulippe E, Satagopan J, Smith A, et al. : Comprehensive gene expression analysis of prostate cancer reveals distinct transcriptional programs associated with metastatic disease. Cancer Res 62:4499–506, 2002 [PubMed] [Google Scholar]
- 42.Ramaswamy S, Tamayo P, Rifkin R, et al. : Multiclass cancer diagnosis using tumor gene expression signatures. Proc Natl Acad Sci U S A 98:15149–54, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taylor BS, Schultz N, Hieronymus H, et al. : Integrative genomic profiling of human prostate cancer. Cancer Cell 18:11–22, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu YP, Landsittel D, Jing L, et al. : Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol 22:2790–9, 2004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


