Abstract
Schizophrenia and bipolar disorder are complex mental disorders with risks contributed by multiple genes. Dysregulation of gene expression has been implicated, but little is known about such regulation systems in the human brain. We analyzed three transcriptome datasets using 394 brain tissue samples from patients with schizophrenia or bipolar disorder and healthy control individuals without known history of psychiatric disorders. We built genome wide co-expression networks that included microRNAs (miRNAs). We identified a co-expression network module that was differentially expressed between patients and control individuals. This module contained genes that were principally involved in glial and neural cell genesis and glial cell differentiation, and included schizophrenia risk genes carrying rare variants. This module included five miRNAs and 545 mRNAs, with six transcription factors serving as hub genes in this module. We found that the most connected transcription factor POU3F2, a gene also identified on a GWAS for bipolar disorder, could regulate hsa-miR-320e and other putative target mRNAs. These regulatory relationships were replicated by PsychENCODE/BrainGVEX data and validated by knockdown and overexpression experiments in the SH-SY5Y and neural progenitor cell lines in vitro. We identified a psychosis-associated brain gene expression module that was enriched for rare coding variants in genes associated with schizophrenia and contained the putative bipolar disorder risk gene POU3F2 as a key regulator of gene expression in this module.
Overline: Neuropsychiatric Disease
Single Sentence Summary: POU3F2 regulates expression of genes in brains of schizophrenia and bipolar disorder.
Accessible Summary: To reveal the dysregulated expression of genes and their regulators in brains of schizophrenia and bipolar disorder, we analyzed postmortem brain transcriptome data and discovered that POU3F2 was one of the core regulators of the gene coexpression network underlying schizophrenia and bipolar disorder risk, and further validated the regulatory relationships, and investigated related functions in cellular models.
Introduction
Schizophrenia (SCZ) and bipolar disorder (BD) are severe psychiatric diseases that each affects millions of people worldwide (1). Despite a century of evidence establishing their genetic basis, only recently have specific genetic risk factors been identified (2). However, there is not a simple Mendelian model between the risk alleles and these psychiatric disorders. Instead, these psychiatric disorders are very complex and are polygenic in nature involving hundreds of genes with small effect sizes (3, 4). With the polygenic nature of these disorders, many studies have focused on converging individual genes into functional networks in order to reveal the underlying disease etiology. For example, Fromer et al. have reported a brain co-expression network captured SCZ association and impact of polygenic risk for SCZ (5). Gandal et al. identified several disease-shared and disorder-specific co-expression modules in parallel with polygenic overlap among five major psychiatric disorders (6). However, it is still unclear how disease-associated module genes interact and contribute to the pathophysiology of SCZ and BD.
The most studied regulators of gene expression are transcription factors and microRNAs (miRNAs). Increasing evidence implicates connections between transcription factors and miRNAs with SCZ and BD. Transcription factors and miRNAs are known to impact brain development, are differentially expressed in postmortem brain tissue from patients with SCZ and BD, and their targets genes are enriched in SCZ and BD risk loci (7–9).
Based on the assumption that co-expression implies coregulation (10), and hub genes in co-expression modules are likely to be the regulators of gene co-expression, we integrated genotype, mRNA, miRNA data from brain tissue samples from patients with schizophrenia or bipolar disease to search for transcription factors and miRNAs at the hub of disease-associated modules, and to experimentally validate their putative regulatory relationships. We identified one hub gene, POU class 3 homeobox 2 (POU3F2), as the master regulator in the disease-associated module for schizophrenia and bipolar disease.
Results
Because SCZ and BD share genetic components and similar gene expression patterns (6, 11), and with the goal to explore the biology underlying potential shared disease mechanisms between SCZ and BD, we used postmortem brain transcriptome data from 95 patients with schizophrenia, 74 patients with bipolar disease, and 225 control individuals from multiple datasets; we combined SCZ and BD as major psychiatric disorders in all case-control studies. Both mRNA and miRNA data from microarray and RNA sequencing (RNA-seq) were incorporated. Detailed demographic information and quality control steps for the postmortem brain tissue data are provided in the Materials and Methods and tables S1, S2.
Disease-associated miRNA and mRNA co-expression network module
To detect disease-associated co-expression modules, we first analyzed the mRNA and miRNA expression in parietal cortex tissue samples from the Stanley Medical Research Institute (SMRI) using weighted gene co-expression network analysis (WGCNA) (12). To capture expression of a group of genes exhibiting case-control differences, we combined case-control data to construct networks and identified 46 co-expression modules. We found one disease-associated module (daM) after removing the effects of sex, age, brain pH, RNA integrity number (RIN), and post-mortem interval (PMI) (control vs. SCZ+BD, P = 4.3e-5, FDR q = 7.0e-3). Five miRNAs and 545 genes were included in the daM. Functional enrichment test by DAVID v6.8 (13) analysis showed that these genes were enriched for gliogenesis (P = 1.5e-11, FDR q = 2.8e-8), glial cell differentiation (P = 1.5e-8, FDR q = 2.8e-5) and neurogenesis (P = 3.8e-8, FDR q = 7.2e-5).
To investigate whether gene networks were conserved among patients with SCZ or BD and controls, we also constructed gene co-expression networks in other ways. We separated SCZ, BD and control samples to run WGCNA analysis independently, and compared their module differences. We used Zsummary to assess whether the connectivity level and pattern of a module in one dataset were preserved in another, where Zsummary > 2 implies moderate preservation and Zsummary > 10 indicates high preservation (14). We found that all modules detected in control samples were well preserved in SCZ or BD patients, indicating no significant module differences among those groups (Zsummary > 2, see Materials and Methods). We also applied WGCNA analysis on SCZ+control and BP+control brain tissue samples to determine if any disease specific modules existed. We detected one module showing significant association with SCZ in SCZ+control samples, but this did not survive multiple testing corrections (P < 0.05, FDR q value > 0.05). Genes in this module significantly overlapped with ones in the daM of the combined dataset (P < 0.01). We did not detect any disease-associated module in BP+control samples.
Module preservation in independent datasets
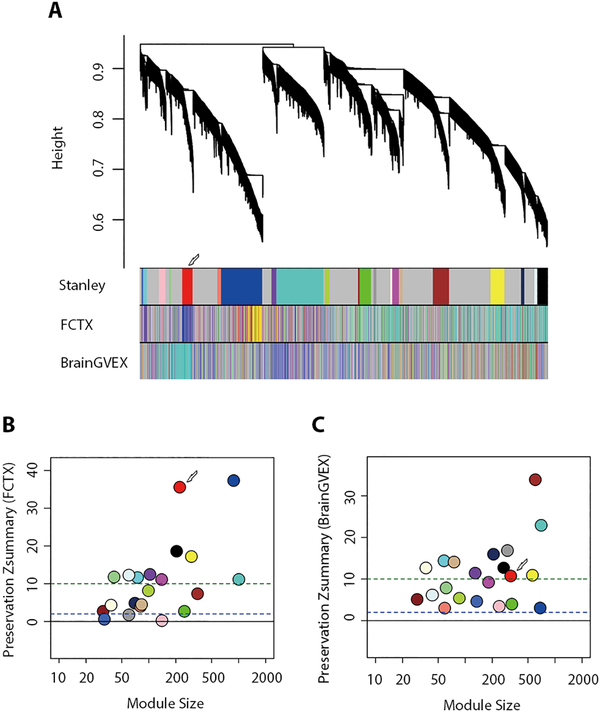
To test preservation of the daM, two independent datasets were used as replicates: one group comprised control samples from Andrew Singleton’s group, which also included miRNA and mRNA microarray expression data from 138 frontal cortex tissue samples (FCTX; GSE15745) (15); the other group comprised RNA-seq data for prefrontal cortex tissue samples from 63 controls, 70 SCZ, and 48 BD patients from the PsychENCODE/BrainGVEX project (16) (Fig. 1A). The daM in the SMRI parietal cortex tissue data had Zsummary = 36.8 in frontal cortex data from GSE15745 (Fig. 1B) and Zsummary = 10.9 in prefrontal cortex data from BrainGVEX (Fig. 1C), indicating well-preserved membership and connectivity of the daM. It is worth noting that the corresponding module in BrainGVEX also exhibited significant disease association in the same direction (P < 0.01).
Fig. 1. Conservation of disease-associated module genes in postmortem brain tissue from different sources.
(A) Genes detected in the disease-associated module (daM; red) for the Stanley postmortem brain samples were also clustered in the modules for FCTX postmortem brain samples (blue) and BrainGVEX postmortem brain samples (turquoise). The preservation Zsummary of the daM was 36.8 for FCTX samples (B) and 10.9 for BrainGVEX samples (C) (Zsummary > 10 indicates high preservation). MiRNA and mRNA microarray expression data were obtained for 138 postmortem frontal cortex samples from healthy control individuals in the FCTX data set. RNA-seq data was obtained for postmortem prefrontal cortex samples from the BrainGVEX data set for 63 healthy control individuals, 70 patients with SCZ and 48 patients with BD.
Enrichment of genetic variants associated with SCZ or BD
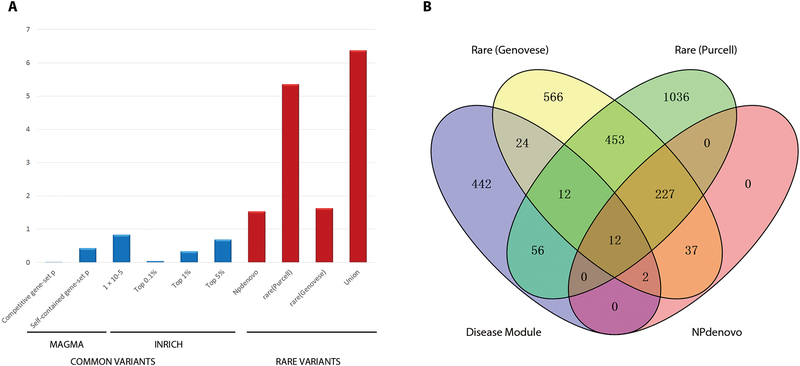
We tested whether genes in the daM were related to genetic association with SCZ or BD. For the genetic variants, we focused on common or rare single nucleotide variants and copy number variations (CNVs). For common variants, we applied MAGMA (17) and INRICH (18) to test the enrichment of daM genes with signals from SCZ or BD GWAS but did not detect any significant enrichment (Fig. 2A). For rare variants, we examined the rare variants burden of module genes associated with disease. Genes in the daM were significantly enriched for genes implicated in rare and de novo variants from three selected independent sources (19–21) (enriched P < 0.05) (Fig. 2A, B). The enrichment was more significant when we combined the three gene sets (overlapped gene number = 106, enriched P = 4.31e-7, Fig. 2A and table S3). As a negative control, we used type 2 diabetes associated genes from GWAS to test the enrichment with genes containing rare variants (22), and found no significant overlap with any of the three sources, nor the combined set. Other brain co-expression modules from our data were also used as controls to test the enrichment. We observed another nine out of 46 modules significantly enriched for genes with rare coding variants, and the daM was the second significant module after multiple testing correction (adjusted P < 0.0001). For CNVs, we tested the enrichment of genes implicated in CNV regions but did not observe any significant enrichment.
Fig. 2. Enrichment of common and rare genetic variants in the daM for SCZ or BD postmortem brain tissue.
(A) Enrichment of genes in the disease-associated module (daM) with common and rare variants. For common variants, we applied MAGMA and INRICH to test the enrichment. Self-contained gene-set analysis tested whether genes in a gene set showed joint association with SCZ, and competitive gene-set analysis tested whether those genes showed differential association with SCZ compared with other genes in the rest of the genome. Thresholds of 1 × 10−5, top 0.1%, top 1%, and top 5% of all significant SNPs were used as index SNPs in INRICH, and none of them detected significant enrichment. For rare variants, data from two exome sequencing studies and one database were used to test the enrichment. We applied a hypergeometric method to test the overlap with genes collected from Purcell’s study (20), NPdenovo database (19), and the combined gene sets from these two studies. We applied logistic regression to test the rare variant burden using disruptive and damaging ultra-rare variant (dURV) counts for each gene from Genovese’s study (21). (B) The number of genes containing de novo or rare mutations from three sources (19–21) overlapping with genes in daM.
Potential key regulators and their roles in the daM
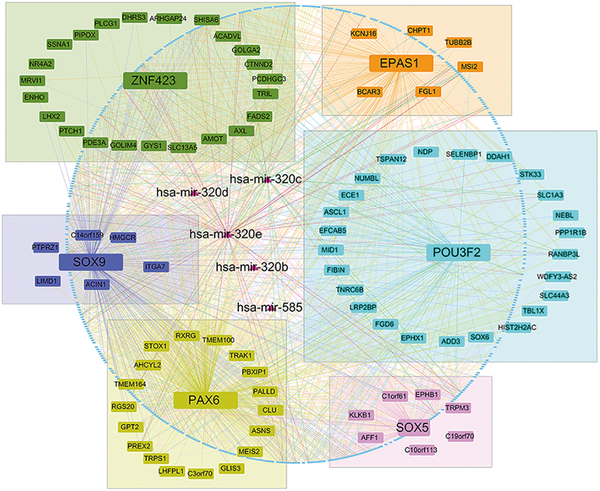
In the daM, we focused on identifying transcription factors and miRNAs as key regulators and explored their functional roles. Five miRNAs were in the module: hsa-miR-585, and four hsa-miR-320 family members including hsa-miR-320b, hsa-miR-320c, hsa-miR-320d, and hsa-miR-320e. Six transcription factors were also included in this module: POU3F2, endothelial PAS domain protein 1 (EPAS1), paired box 6 (PAX6), zinc finger protein 423 (ZNF423), SRY-box 5 (SOX5), and SRY-box 9 (SOX9).
We first explored different aspects of regulatory roles for miRNAs in the daM: module membership, pairwise weighted correlation with mRNA, and the enrichment of target genes with predicted binding. By calculating the module membership, we found these five miRNAs had significant correlations with the module eigengene (P < 0.001; Table 1). By extracting pairwise weighted correlation coefficient (rw) between the miRNA and mRNA, we detected strong correlations between miRNAs and mRNAs (rw > 0.05, original correlation r > 0.607). Numbers of correlated mRNAs for each miRNA are provided in Table 1. We observed that expression of most of the mRNAs was inversely correlated with expression of miRNAs (476 out of 545, 87.3%), suggesting that miRNAs may play a role in regulating the transcriptome through direct downregulation of their mRNA targets. Using mirWalk2.0, we examined the predicted binding targets of each miRNA and calculated the overlapped genes in the daM (23). We found that mRNA genes predicted to be targets of the five miRNAs were significantly enriched in this module (hypergeometric probability test P < 0.05, Table 1). This suggested that the predicted regulation relationships from mirWalk2.0 were consistent with the expression correlation results from WGCNA.
Table 1.
Characteristics of miRNAs in daM and their predicted binding targets.
| miRNA | Chr | Correlated with ME* (p-value) | miRWalk2.0 | WGCNA | |
|---|---|---|---|---|---|
| Genes predicted as binding targets in daM (# of total target genes) | Significance of binding targets in module (Enrichment p-value) | # of correlated genes in module (rw#>0.05) | |||
| Chr1 | 3.70E-05 | 333 (10126) | <1e-10 | 14 | |
| hsa-miR-320c | Chr18 | 1.31E-05 | 303 (9480) | <1e-10 | 34 |
| hsa-miR-320d | Chr13 | 1.09E-05 | 297 (8904) | <1e-10 | 44 |
| hsa-miR-320e | Chr19 | 4.79E-06 | 263 (7887) | <1e-10 | 68 |
| hsa-miR-585 | Chr5 | 1.30E-05 | 139 (7742) | 0.046 | 0 |
ME, module eigengene.
rw is the weighted correlation coefficient from the transformed pairwise correlation matrix, where rw>0.05 is equivalent to the original r > 0.607.
We next explored the regulatory roles of transcription factors in the daM. 101 putative targets of the six transcription factors were included using transcription factor binding information obtained from Fuxman et al. and Kheradpour et al. (24, 25). Among the six transcription factors detected in the module, POU3F2 had the most regulated putative targets (N = 26) in this module, including transcription factors PAX6 and SOX9. The other transcription factors EPAS1, PAX6, ZNF423, SOX5, and SOX9, have 9, 21, 24, 10, and 11 putative targets, respectively. We also provided those transcription factor pairwise correlation and disease association P values in replicated BrainGVEX data (tables S4, S5).
We visualized the relationships between transcription factors and their miRNA targets in this module with the six transcription factors and five miRNAs as hub genes (Fig. 3). We found the transcription factor-miRNA target binding relationship was consistent with the relationship extracted from module correlation testing. Hsa-miR-320e was the most connected node in this module according to the correlation data (68 nodes with rw > 0.05, Table 1).
Fig. 3. Transcription factor-target binding information in the daM.
All mRNAs, miRNAs, and their relationships in the daM were plotted. The colored lines indicate the pairwise correlation extracted from module testing with rw > 0.05. Six transcription factors (POU3F2, PAX6, EPAS1, ZNF423, SOX5, SOX9), their binding targets, and names of five miRNAs were labeled in this figure, and other genes in the module were plotted as dots in this network. Transcription factors and their targets were framed in six separate colored boxes. Rw is the weighted correlation coefficient from the transformed pairwise correlation matrix, where rw > 0.05 is equivalent to the original r > 0.607.
Causal relationships among key regulators in the daM
We tested whether transcription factors were upstream or downstream regulators of miRNAs or were regulated by other transcription factors through integration of genetic markers. Causal inference of correlated nodes (miRNAs, transcription factors, and their target protein-coding genes in this study) were inferred by the network edge orienting (NEO) method (26). Because we were interested in the causal relationship between five miRNAs and six transcription factors, only miRNA quantitative trait locus (miQTL) signals associated with the five miRNAs and expression QTL (eQTL) signals associated with the six transcription factors were included in this analysis. Among the five miRNAs, only hsa-miR-320e had significant miQTL signals (P < 0.05, FDR q < 0.05), so this miRNA and six transcription factors were selected for regulation direction tests.
We used modified NEO (see Materials and Methods) to build a local structure equation model and to obtain edge-oriented scores. The orthogonal causal anchors (LEO.NB.OCA) (A → B) > 0.3 and candidate pleiotropic anchor (LEO.NB.CPA) (A → B) > 0.8 indicated the regulation direction was A to B. We observed that LEO.NB.OCA (POU3F2 → hsa-miR-320e) = 0.526, LEO.NB.CPA (POU3F2 → hsa-miR-320e) = 1.55, which suggested that POU3F2 may be an upstream regulator that affected hsa-miR-320e’s expression (fig. S1; table S6). Meanwhile, NEO results indicated that POU3F2 was the upstream regulator of other transcription factors (PAX6, ZNF423 and SOX9) (fig. S1; table S6). These results suggested that POU3F2 was a key regulator in the daM.
Experimental validation of the putative causal regulatory relationship
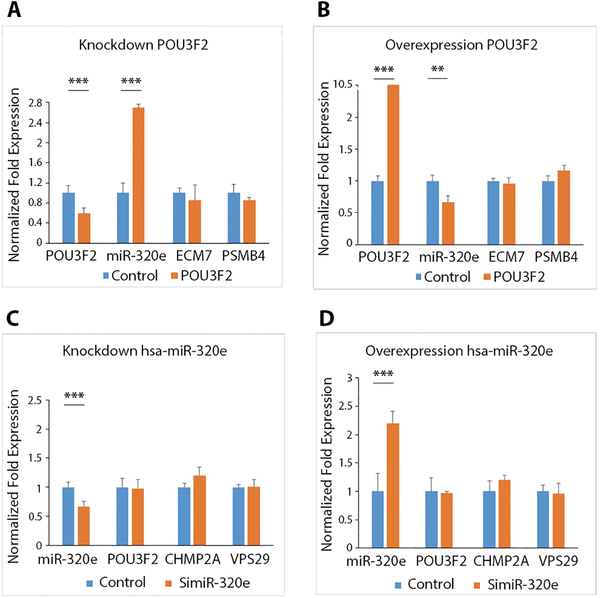
With the hub positions of POU3F2 and hsa-miR-320e in the regulation network, we tried to confirm their predicted relationships through in vitro experiments. We used RNA interference (RNAi) and a gene overexpression assay to induce expression alterations of POU3F2 and hsa-miR-320e in SH-SY5Y neuroblastoma cells and examined the expression changes of their predicted targets.
The expression of POU3F2 decreased by 41% after RNAi in SH-SY5Y cells (P < 0.001). As a result, hsa-miR-320e expression increased by 170% (P < 0.001); expression of two negative controls (ECM7 and PSMB4) that were not predicted targets of POU3F2 did not change (Fig. 4A). In overexpression experiments, POU3F2’s expression increased by nearly ten fold (P < 0.001). As a result, hsa-miR-320e’s expression significantly decreased by 33% (P < 0.01), and the expression of two negative controls was not significantly changed (Fig. 4B).
Fig. 4. The causal relationship between POU3F2 and hsa-miR-320e.
(A, B) Shown are the qPCR results after knocking down POU3F2 (A) and after overexpression of POU3F2 (B) in SH-SY5Y neuroblastoma cells. (C, D) The qPCR results after knocking down hsa-miR-320e (C) and after overexpression of hsa-miR-320e (D) in SH-SY5Y cells. Orange bars indicate genes’ expression of POU3F2, hsa-miR-320e and negative controls after knocking down or overexpressing POU3F2 or hsa-miR-320e, and the blue bars indicate gene expression in control groups before knocking down or overexpression of POU3F2 or hsa-miR-320e. ECM7 and PSMB4 were negative controls for POU3F2, and CHMP2A and VPS29 were negative controls for the miRNA hsa-miR-320e. Three biological replicates were used, and for each biological replicate we designed three technical replicates. * P < 0.05, ** P < 0.01, ***P < 0.001. Data are represented as mean ± standard error of the mean (SEM).
In the case of knocking down hsa-miR-320e, hsa-miR-320e’s expression decreased by 33% (P < 0.001) but had no effect on expression of POU3F2 (Fig. 4C). Overexpression of hsa-miR-320e (increased by 120%, P < 0.001) did not change expression of POU3F2 (Fig. 4D). Two negative controls (CHMP2A and VPS29), which were not potential targets of hsa-miR-320e, were not significantly changed in both knockdown and overexpression experiments (Fig. 4C, D). These in vitro results confirmed POU3F2 was the upstream regulator of hsa-miR-320e experimentally.
POU3F2 regulates proliferation and differentiation of neural progenitor cells
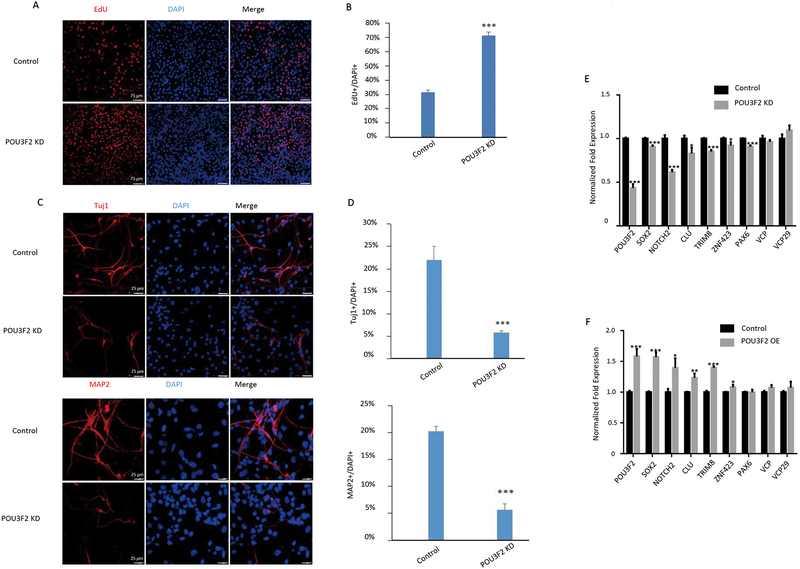
Accumulating evidence suggests that POU3F2 is primarily expressed in the central nervous system and plays an important role in brain development and cell differentiation (27, 28). To further characterize its functional roles, we decided to knockdown POU3F2 in human neural progenitor cells (NPCs). NPCs were transfected with small hairpin RNAs (shRNAs) against POU3F2, and their proliferation and differentiation were evaluated using an immunofluorescence assay. After knocking down POU3F2 (expression decreased by 51%, P < 0.001), we found that the proliferation ratio of EdU+ (5-ethynyl-2’-deoxyuridine, a marker for proliferating cells) to DAPI+ (4′,6-diamidino-2-phenylindole, a marker of live cells) was significantly increased compared to control groups (P < 0.001; Fig. 5A, B). We next analyzed the differentiation of NPCs to neurons and found the proportions of Tuj1+ (a marker of immature neurons) and MAP2+ (a marker of mature neurons) were significantly decreased compared to control groups (P < 0.001; Fig. 5C, D). These results indicated that POU3F2 knockdown could promote NPCs’ proliferation ability and inhibit NPCs’ differentiation to neurons.
Fig. 5. POU3F2 regulates proliferation and differentiation of NPCs.
(A, B) Immunofluorescence staining for EdU (a marker of proliferating cells) after POU3F2 knockdown in human NPCs (A); quantification of proliferation is shown in panel B. (C, D) Immunofluorescence staining for Tuj1 (a marker of immature neurons) and MAP2 (a marker of mature neurons) after POU3F2 knockdown in NPCs (C); quantification of differentiation of NPCs into neurons is shown in D. (E, F) qPCR data showing POU3F2 putative targets after knocking down (E) or overexpressing POU3F2 (F). Three biological replicates were used, and for each biological replicate we designed three technical replicates. * P < 0.05, ** P < 0.01, ***P < 0.001. Data are represented as mean ± SEM.
To investigate how POU3F2 affected cell proliferation and differentiation capabilities, we examined expression changes of six putative targets of POU3F2 in the daM in NPCs after POU3F2 knockdown or overexpression. These putative targets included SOX9, PAX6, ZNF423, NOTCH2, CLU, and TRIM8. The three transcription factors (SOX9, PAX6 and ZNF423) and the most connected gene in the daM (NOTCH2) have been reported to regulate brain development and neural differentiation in many studies (29–32). NOTCH2 was also reported to be associated with SCZ (33). TRIM8 and CLU were also hub genes in the daM. These two genes are located in the 108 significant SCZ GWAS loci (3) and are involved in tumor cell proliferation and differentiation (34, 35). Expression of SOX9, ZNF423, NOTCH2, CLU and TRIM8 significantly decreased by 10%, 7.9%, 39%, 17% and 15% after POU3F2 knockdown, and significantly increased by 57%, 8.0%, 39%, 23% and 40% after POU3F2 overexpression (P < 0.05; Fig. 5E, F). Expression of PAX6 was not significantly increased after POU3F2 overexpression, suggesting it may not be regulated by POU3F2 in our NPC model. Expression of negative controls (VPS29 and VCP) was not significantly changed (Fig. 5E, F).
Discussion
Prior work has documented abnormalities in coordinated gene expression networks in postmortem brain tissue from patients with SCZ or BD. However, these studies have involved either a single-dimension correlated network that cannot resolve the driver node within the module or have not done a regulatory relationship analysis. Most findings have been presented as correlations instead of causal relationships. In this study, we integrated multiple dimensional data sets and revealed a role for POU3F2 as a regulator of the network. POU3F2 is clearly only one regulator and there are many pathways that may potentially be involved in the etiology of SCZ or BD. Our study provides a framework to capture such pathways and to tease out their regulatory relationships.
POU3F2 has been reported to be associated with SCZ using brain activation level from functional magnetic resonance imaging as a quantitative phenotype (36). POU3F2 has been shown to lie close to the BD risk loci in the most recent Psychiatric Genomics Consortium BD GWAS data (37). Expression changes in POU3F2 have been observed in neurons derived from SCZ patient-specific induced pluripotent stem cells (38). POU3F2 was discovered based on the sequence similarity of the POU domain and is also known as Brain-2 (Brn-2) since it is expressed in the central nervous system (39). The function of POU3F2 was initially studied in melanocytic cells. In our study, we observed that POU3F2 knockdown could promote NPC proliferation and inhibit neuronal differentiation (Fig. 5A-5D). The aberrant expression of POU3F2 could lead to alterations in cell number and may be one possible explanation for anatomical changes in the brain tissue of patients with BD (40).
The genes in daM significantly overlapped with ones from the PsychENCODE Capstone One study, which detected one module associated with disease and was functionally enriched for genes involved in glia differentiation (geneM3/isoM1, Overlap P < 0.01). Genes in the daM also overlapped with our previous findings using datasets from multiple brain banks (33), and with results from the Torkamani et al. study (41) (overlapping P < 0.01).
In our study, we observed that the daM harbored risk genes carrying rare variants but not the common variants identified by GWAS studies. However, the genes in daM could still be related to common risk variants. For example, rs11191359, rs4146429, rs4146428 are eQTL signals of TRIM8, located in the promoter region of TRIM8. They are located in schizophrenia-associated region and have linkage disequilibrium with SCZ GWAS SNP rs7907645 (GWAS association P = 1.27e-11) (3). TRIM8 expression is regulated by the transcription factors POU3F2 and PAX6 (24). Variants in the TRIM8 promoter region may disrupt the binding efficiency of POU3F2 and PAX6 and reduce expression of TRIM8. Integrating genetic risk variants with regulatory networks may provide new insights about the transcriptional regulatory architecture that could underlie certain psychiatric disorders.
Our study does have several limitations. First, although hundreds of brain samples were used, the sample size was still relatively small. Possibly due to the small sample size, analyses of the co-expression modules in the uncombined SCZ and BD samples did not yield robust findings. Second, limited data were generated containing both miRNA and mRNA expression for building networks involving miRNAs. This could lead to missing some important miRNA regulatory relationships. Third, gene expression changes may be affected by altered cellular population in patients (42). In our study, we estimated the proportions of each cell type using a deconvolution method and found no significant difference (fig. S2). Larger sample sizes and new methods may be required to better estimate cell numbers. Fourth, we only validated a few regulatory pathways in the daM. Large-scale validation should be completed in the future. Lastly, we observed the daM genes involved in gliogenesis, glial cell differentiation and neurogenesis, but we only validated the regulatory loop in neuroblastoma and neural progenitor cell lines. Further investigation is needed to clarify whether these regulatory networks also occur in non-neural cells.
By integrating genotype, miRNA, and mRNA expression data, and transcription factor and miRNA binding information, we found cascade regulation relationships from SNP variants to transcription factors to miRNAs or other target genes. Our results suggest that complex diseases such as SCZ and BD require a systems biology approach, with an integration of multi-dimensional data sets to elucidate a better understanding of disease risk.
Materials and Methods
Study design
This study was designed to investigate the dysfunctional regulatory network and key regulators in postmortem brain tissue from patients with SCZ or BP. We analyzed genome-wide mRNA, miRNA, or genotyping data from postmortem brain tissue from 169 patients with SCZ or BD and 225 healthy control individuals who did not have a known history of psychiatric disorders. We first applied WGCNA and QTL analysis to reveal mRNA-miRNA and genotype-mRNA/miRNA relationships. DaM was detected exhibiting case control expression difference after removing confounding factors. We next used web resources to explore transcriptional regulators (transcription factors and miRNA binding information) and identified POU3F2 as a master regulator of other transcription factors and miRNAs in the daM. The function of POU3F2 was examined in cell differentiation and proliferation experiments and its regulatory activities were validated using RNAi, gene overexpression, and luciferase reporter experiments.
Samples
Discovery data:
Parietal cortex (PC) tissue specimens from the SMRI Neuropathology Consortium and Array collections included SCZ, BD, and control samples (table S1) (43). We removed non-Europeans, duplicates, and samples missing any of the mRNA, miRNA, or genotyping data. After filtering, we retained 75 samples (51 patients and 24 controls), yielding data for 19,984 mRNAs, 470 miRNAs, and 1,452,078 SNPs for subsequent analyses. The detailed demographic, clinic information and their distribution differences among groups are provided in table S1.
Replicate data:
The two replication datasets were microarray data from Andrew Singleton’s group (GEO Accession Number: GSE15745) and RNA-seq data of BrainGVEX from PsychENCODE. GSE15745 contains frozen frontal cortex tissue samples from 138 neurologically normal Caucasian subjects after quality control (15). The BrainGVEX project includes samples from SMRI, so we excluded those overlapping samples when using PsychENCODE as replicates. Seventy SCZ, 48 BD, and 63 controls from PsychENCODE were used for validation. The detailed demographic, clinic information and their distribution differences among groups are provided in table S2.
Module construction and preservation statistics
We identified mRNA and miRNA with correlated expression patterns using WGCNA (12). We calculated a correlation matrix for all possible pairwise nodes (mRNA and miRNA) and chose the power = 6 for weighting the correlation matrix following an approximate scale-free topology. We set minimum block size as 30 and biweight midcorrelation (bicor) to build the network. We detected network modules using the dynamic tree cut algorithm with the mergeCutHeight as 0.05 and deepSplit as 2. WGCNA and the dynamic tree cut algorithm were implemented in R (version 3.1.3) (12). The unsigned network type was used to keep the negative relationships between miRNAs and mRNAs. We plotted the pairwise connection network using Cytoscape v3.6.1 (44).
Because our sample size is relatively small, we used two additional datasets to assess the module preservation. The validation data sets include samples from BrainGVEX and GSE15745. We applied Zsummary test to assess module preservation between expression datasets (14). The recommended thresholds are Zsummary < 2 implies no evidence for module preservation, 2 < Zsummary < 10 implies weak to moderate evidence, and Zsummary > 10 implies strong evidence for module preservation.
Network Edge Orienting (NEO) analysis of transcription factor-miRNA interactions
In addition to binding information, we used modified NEO to investigate the causal relationship between transcription factors and miRNAs (26). The imported data was the expression data of Transcription factors and miRNAs, and genotype data. The outputs are local-structure edge orienting (LEO) scores, which use the likelihoods of local structural equation models to integrate selected traits and markers to assess the causal relationship between correlated quantitative variables. We selected the genotype data that included eQTLs of Transcription factors and miRNAs from our analysis of SMRI samples and from results of the GTEx portal (www.gtexportal.org), CommonMind Consortium (commonmind.org/), and UK Brain Expression Consortium (http://www.braineac.org/). Totally 901 SNPs were included in NEO analysis. The candidate pleiotropic anchor (CPA) model was used to test single marker edge orienting and the orthogonal causal anchor (OCA) model was used to test multiple genetic markers. The likelihood-based CPA score assessed whether the chosen model would yield a higher likelihood than did the alternative models. We used a threshold of 0.8, as the software suggested, which implies that the model likelihood score of the causal model was 100.8 = 6.3 fold higher than that of the next best model. For the OCA score, we used a threshold of 0.3, as suggested, which implies that the model likelihood score of the causal model was 100.3 = 2 fold higher than that of the next best model.
NPC proliferation and differentiation assay
We investigated the function of POU3F2 in neuronal cell proliferation using BeyoClick™ EdU Cell Proliferation Kit with Alexa Fluor 555 (Beyotime, C0075S). Briefly, after POU3F2 knockdown, NPCs were incubated with 10 μM EdU solution for 5 hours to label proliferating cells. And then Cells were fixed with 4% paraformaldehyde and permeabilized with 0.3% Triton X-100/PBS. The prepared Click Additive Solution was used to detect the EdU-incorporated cells. Finally, cells were labeled with Hoechst 33342 in order to count the proportion of EdU positive cells. For neuron differentiation assay, we used a STEMdiff Neuron Differentiation Kit (Stemcell, 08500) and a STEMdiff Neuron Maturation Kit (Stemcell, 08510) to generate neurons according to manufacturer’s instructions. For immunofluorescence staining assay, cells were fixed with 4% paraformaldehyde and permeabilized with 0.5% Triton X-100/PBS. Then cells were blocked using 5% BSA/PBS for 30 min at room temperature followed by incubation with indicated primary antibodies for one hour at room temperature: Tuj1 (1:300, CST, 5568), MAP2 (1:300, CST, 8707). After three washes with PBS, cells were incubated with fluorescently labeled secondary antibodies for one hour at room temperature, followed by staining with the fluorescent nuclear dye DAPI (beyotime, C1002). The proportions of EdU+, Tuj1+ and MAP2+ cells were quantified with Image J software.
Statistical analysis
Descriptive statistics are reported as mean and standard deviation (SD) or minimum to maximum values. Biological or technical replicates from experiments are reported as mean ± SEM. We applied Student’s t-test to compare the mean difference between two independent groups if the data were normally distributed in each group. Nonparametric Wilcoxon signed-rank test was used if the data were not normally distributed. For single testing, a two-tailed P value less than 0.05 was consided as statistical significance, *P < 0.05, **P < 0.01, ***P < 0.001. For multiple testing, the FDR q value was calculated based on the nominal distribution of P values. We performed all statistical analysis using the program R (version 3.1.3).
Supplementary Material
Materials and Methods
Fig. S1. Causal relationship between transcription factors and miRNAs.
Fig. S2. Differences in cell type proportions between cases (BP and SCZ) and controls.
Fig. S3. Immunofluorescence staining of NPCs.
Fig. S4. RT-qPCR analysis of POU3F2 knockdown efficiency.
Fig. S5. Distribution of eigengene values in daM.
Table S1. Demographic and clinical information for discovery SMRI data.
Table S2. Demographic and clinical information for replicated BrainGVEX data.
Table S3. Overlap of genes in the daM and genes with rare variants.
Table S4. Transcription factor expression associated with disease in SMRI and BrainGVEX data
Table S5. Transcription factor expression correlations in BrainGVEX data.
Table S6. Orthogonal causal anchor (LEO.NB.OCA) and candidate pleiotropic anchor (LEO.NB.CPA) scores for six transcription factors and one miRNA.
Table S7. Sequences of RT-qPCR primers and siRNA.
Table S8. Top 50 connected genes in the daM, miRNAs, transcription factors and their WGCNA parameters.
Table S9. POU3F2 and its putative targets in daM.
Acknowledgements:
The authors thank the tissue donors and their families. We thank all consortium members for discussions and feedback on this manuscript. We thank Jieqiong Tan for help with the cellular model experiments. We thank the Chicago Biomedical Consortium with support from the Searle Fund at The Chicago Community Trust. The PsychENCODE consortium projects are funded by the US National Institute of Mental Health.
Funding: This work was supported by NIH grant # 1 U01 MH103340–01, 1R01ES024988 (to C. Liu), National Natural Science Foundation of China grant # 81401114 and 31571312, the National Key Plan for Scientific Research and Development of China (# 2016YFC1306000 to C. Chen). This work was partially supported by the Cancer Prevention and Research Institute of Texas (# RP170005 to Cristian Coarfa), and the Research Council of Norway through the FRIPRO Mobility grant scheme (# 251134 to Yunpeng Wang). The FRIPRO Mobility grant scheme (FRICON) is co-funded by the European Union’s Seventh Framework Programme for research, technological development and demonstration under Marie Curie grant agreement # 608695. The University of Houston Sequencing Core Facility was made possible through a gift from the Cullen Foundation. Data were generated as part of the PsychENCODE Consortium, supported by NIH grant #: U01MH103392, U01MH103365, U01MH103346, U01MH103340, U01MH103339, R21MH109956, R21MH105881, R21MH105853, R21MH103877, R21MH102791, R01MH111721, R01MH110928, R01MH110927, R01MH110926, R01MH110921, R01MH110920, R01MH110905, R01MH109715, R01MH109677, R01MH105898, R01MH105898, R01MH094714, P50MH106934 awarded to: Schahram Akbarian (Icahn School of Medicine at Mount Sinai), Gregory Crawford (Duke University), Stella Dracheva (Icahn School of Medicine at Mount Sinai), Peggy Farnham (University of Southern California), Mark Gerstein (Yale University), Daniel Geschwind (University of California, Los Angeles), Fernando Goes (Johns Hopkins University), Thomas M. Hyde (Lieber Institute for Brain Development), Andrew Jaffe (Lieber Institute for Brain Development), James A. Knowles (University of Southern California), Chunyu Liu (SUNY Upstate Medical University), Dalila Pinto (Icahn School of Medicine at Mount Sinai), Panos Roussos (Icahn School of Medicine at Mount Sinai), Stephan Sanders (University of California, San Francisco), Nenad Sestan (Yale University), Pamela Sklar (Icahn School of Medicine at Mount Sinai), Matthew State (University of California, San Francisco), Patrick Sullivan (University of North Carolina), Flora Vaccarino (Yale University), Daniel Weinberger (Lieber Institute for Brain Development), Sherman Weissman (Yale University), Kevin White (University of Chicago), Jeremy Willsey (University of California, San Francisco), and Peter Zandi (Johns Hopkins University).
Footnotes
Competing interests: KPW is President and a shareholder of Tempus Labs, Inc. The other authors declare no competing interests.
Data and materials availability: All data associated with this study are in the paper or supplementary materials. The Stanley brain expression data has been submitted to GEO with accession # GSE35977. To apply for PsychENCODE data, researchers can submit an application form via the website: https://www.synapse.org//#!Synapse:syn4921369/wiki/235539
References and Notes
- 1.Perala J, Suvisaari J, Saarni SI, Kuoppasalmi K, Isometsa E, Pirkola S, Partonen T, Tuulio-Henriksson A, Hintikka J, Kieseppa T, Harkanen T, Koskinen S, Lonnqvist J, Lifetime prevalence of psychotic and bipolar I disorders in a general population. Archives of general psychiatry 64, 19–28 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Neale BM, Sklar P, Genetic analysis of schizophrenia and bipolar disorder reveals polygenicity but also suggests new directions for molecular interrogation. Current opinion in neurobiology 30, 131–138 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Schizophrenia Working Group of the Psychiatric Genomics Consortium, Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Psychiatric GWAS Consortium Bipolar Disorder Working Group, Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nature genetics 43, 977–983 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fromer M, Roussos P, Sieberts SK, Johnson JS, Kavanagh DH, Perumal TM, Ruderfer DM, Oh EC, Topol A, Shah HR, Klei LL, Kramer R, Pinto D, Gumus ZH, Cicek AE, Dang KK, Browne A, Lu C, Xie L, Readhead B, Stahl EA, Xiao J, Parvizi M, Hamamsy T, Fullard JF, Wang YC, Mahajan MC, Derry JM, Dudley JT, Hemby SE, Logsdon BA, Talbot K, Raj T, Bennett DA, De Jager PL, Zhu J, Zhang B, Sullivan PF, Chess A, Purcell SM, Shinobu LA, Mangravite LM, Toyoshiba H, Gur RE, Hahn CG, Lewis DA, Haroutunian V, Peters MA, Lipska BK, Buxbaum JD, Schadt EE, Hirai K, Roeder K, Brennand KJ, Katsanis N, Domenici E, Devlin B, Sklar P, Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nature neuroscience 19, 1442–1453 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gandal MJ, Haney JR, Parikshak NN, Leppa V, Ramaswami G, Hartl C, Schork AJ, Appadurai V, Buil A, Werge TM, Liu C, White KP, CommonMind Consortium, PsychENCODE Consortium, iPSYCH-BROAD Working Group, S. Horvath, D. H. Geschwind, Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science 359, 693–697 (2018).29439242 [Google Scholar]
- 7.Bavamian S, Mellios N, Lalonde J, Fass DM, Wang J, Sheridan SD, Madison JM, Zhou F, Rueckert EH, Barker D, Perlis RH, Sur M, Haggarty SJ, Dysregulation of miR-34a links neuronal development to genetic risk factors for bipolar disorder. Molecular psychiatry 20, 573–584 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wright C, Turner JA, Calhoun VD, Perrone-Bizzozero N, Potential Impact of miR-137 and Its Targets in Schizophrenia. Frontiers in genetics 4, 58 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ubhi K, Price J, Expression of POU-domain transcription factor, Oct-6, in schizophrenia, bipolar disorder and major depression. BMC psychiatry 5, 38 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quackenbush J, Genomics. Microarrays--guilt by association. Science 302, 240–241 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Cross-Disorder Group of the Psychiatric Genomics Consortium, Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nature genetics 45, 984–994 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Langfelder P, Horvath S, WGCNA: an R package for weighted correlation network analysis. BMC bioinformatics 9, 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang da W, Sherman BT, Lempicki RA, Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Langfelder P, Luo R, Oldham MC, Horvath S, Is my network module preserved and reproducible? PLoS computational biology 7, e1001057 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibbs JR, van der Brug MP, Hernandez DG, Traynor BJ, Nalls MA, Lai SL, Arepalli S, Dillman A, Rafferty IP, Troncoso J, Johnson R, Zielke HR, Ferrucci L, Longo DL, Cookson MR, Singleton AB, Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS genetics 6, e1000952 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.PsychENCODE Consortium Akbarian S, Liu C, Knowles JA, Vaccarino FM, Farnham PJ, Crawford GE, Jaffe AE, Pinto D, Dracheva S, Geschwind DH, Mill J, Nairn AC, Abyzov A, Pochareddy S, Prabhakar S, Weissman S, Sullivan PF, State MW, Weng Z, Peters MA, White KP, Gerstein MB, Amiri A, Armoskus C, Ashley-Koch AE, Bae T, Beckel-Mitchener A, Berman BP, Coetzee GA, Coppola G, Francoeur N, Fromer M, Gao R, Grennan K, Herstein J, Kavanagh DH, Ivanov NA, Jiang Y, Kitchen RR, Kozlenkov A, Kundakovic M, Li M, Li Z, Liu S, Mangravite LM, Mattei E, Markenscoff-Papadimitriou E, Navarro FC, North N, Omberg L, Panchision D, Parikshak N, Poschmann J, Price AJ, Purcaro M, Reddy TE, Roussos P, Schreiner S, Scuderi S, Sebra R, Shibata M, Shieh AW, Skarica M, Sun W, Swarup V, Thomas A, Tsuji J, van Bakel H, Wang D, Wang Y, Wang K, Werling DM, Willsey AJ, Witt H, Won H, Wong CC, Wray GA, Wu EY, Xu X, Yao L, Senthil G, Lehner T, Sklar P, Sestan N, The PsychENCODE project. Nature neuroscience 18, 1707–1712 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Leeuw CA, Mooij JM, Heskes T, Posthuma D, MAGMA: generalized gene-set analysis of GWAS data. PLoS computational biology 11, e1004219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee PH, O’Dushlaine C, Thomas B, Purcell SM, INRICH: interval-based enrichment analysis for genome-wide association studies. Bioinformatics 28, 1797–1799 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J, Cai T, Jiang Y, Chen H, He X, Chen C, Li X, Shao Q, Ran X, Li Z, Xia K, Liu C, Sun ZS, Wu J, Genes with de novo mutations are shared by four neuropsychiatric disorders discovered from NPdenovo database. Molecular psychiatry 21, 298 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P, O’Dushlaine C, Chambert K, Bergen SE, Kahler A, Duncan L, Stahl E, Genovese G, Fernandez E, Collins MO, Komiyama NH, Choudhary JS, Magnusson PK, Banks E, Shakir K, Garimella K, Fennell T, DePristo M, Grant SG, Haggarty SJ, Gabriel S, Scolnick EM, Lander ES, Hultman CM, Sullivan PF, McCarroll SA, Sklar P, A polygenic burden of rare disruptive mutations in schizophrenia. Nature 506, 185–190 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Genovese G, Fromer M, Stahl EA, Ruderfer DM, Chambert K, Landen M, Moran JL, Purcell SM, Sklar P, Sullivan PF, Hultman CM, McCarroll SA, Increased burden of ultra-rare protein-altering variants among 4,877 individuals with schizophrenia. Nature neuroscience 19, 1433–1441 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.the DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium, Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nature genetics 44, 981–990 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dweep H, Gretz N, miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nature methods 12, 697 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Fuxman Bass JI, Sahni N, Shrestha S, Garcia-Gonzalez A, Mori A, Bhat N, Yi S, Hill DE, Vidal M, Walhout AJM, Human gene-centered transcription factor networks for enhancers and disease variants. Cell 161, 661–673 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kheradpour P, Kellis M, Systematic discovery and characterization of regulatory motifs in ENCODE TF binding experiments. Nucleic acids research 42, 2976–2987 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aten JE, Fuller TF, Lusis AJ, Horvath S, Using genetic markers to orient the edges in quantitative trait networks: the NEO software. BMC systems biology 2, 34 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang TQ, Citri A, Sebastiano V, Marro S, Sudhof TC, Wernig M, Induction of human neuronal cells by defined transcription factors. Nature 476, 220–223 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McEvilly RJ, de Diaz MO, Schonemann MD, Hooshmand F, Rosenfeld MG, Transcriptional regulation of cortical neuron migration by POU domain factors. Science 295, 1528–1532 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Casoni F, Croci L, Bosone C, D’Ambrosio R, Badaloni A, Gaudesi D, Barili V, Sarna JR, Tessarollo L, Cremona O, Hawkes R, Warming S, Consalez GG, Zfp423/ZNF423 regulates cell cycle progression, the mode of cell division and the DNA-damage response in Purkinje neuron progenitors. Development 144, 3686–3697 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Georgala PA, Carr CB, Price DJ, The role of Pax6 in forebrain development. Developmental neurobiology 71, 690–709 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Scott CE, Wynn SL, Sesay A, Cruz C, Cheung M, Gomez Gaviro MV, Booth S, Gao B, Cheah KS, Lovell-Badge R, Briscoe J, SOX9 induces and maintains neural stem cells. Nature neuroscience 13, 1181–1189 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Ables JL, Breunig JJ, Eisch AJ, Rakic P, Not(ch) just development: Notch signalling in the adult brain. Nature reviews. Neuroscience 12, 269–283 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen C, Cheng L, Grennan K, Pibiri F, Zhang C, Badner JA, C. Members of the Bipolar Disorder Genome Study, Gershon ES, Liu C, Two gene co-expression modules differentiate psychotics and controls. Molecular psychiatry 18, 1308–1314 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang C, Mukherjee S, Tucker-Burden C, Ross JL, Chau MJ, Kong J, Brat DJ, TRIM8 regulates stemness in glioblastoma through PIAS3-STAT3. Molecular oncology 11, 280–294 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sala A, Bettuzzi S, Pucci S, Chayka O, Dews M, Thomas-Tikhonenko A, Regulation of CLU gene expression by oncogenes and epigenetic factors implications for tumorigenesis. Advances in cancer research 105, 115–132 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Potkin SG, Turner JA, Guffanti G, Lakatos A, Fallon JH, Nguyen DD, Mathalon D, Ford J, Lauriello J, Macciardi F, Fbirn A genome-wide association study of schizophrenia using brain activation as a quantitative phenotype. Schizophrenia bulletin 35, 96–108 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stahl E, Gerome Breen A Forstner A McQuillin S Ripke, Bipolar Disorder Working Group of the Psychiatric Genomics Consortium, Cichon S, Scott L, Ophoff R, Andreassen OA, Kelsoe J, Sklar P, Genomewide association study identifies 30 loci associated with bipolar disorder. bioRxiv, 173062 (2018). [Google Scholar]
- 38.Pedrosa E, Sandler V, Shah A, Carroll R, Chang C, Rockowitz S, Guo X, Zheng D, Lachman HM, Development of patient-specific neurons in schizophrenia using induced pluripotent stem cells. J Neurogenet 25, 88–103 (2011). [DOI] [PubMed] [Google Scholar]
- 39.He X, Treacy MN, Simmons DM, Ingraham HA, Swanson LW, Rosenfeld MG, Expression of a large family of POU-domain regulatory genes in mammalian brain development. Nature 340, 35–41 (1989). [DOI] [PubMed] [Google Scholar]
- 40.Morch-Johnsen L, Nerland S, Jorgensen KN, Osnes K, Hartberg CB, Andreassen OA, Melle I, Nesvag R, Agartz I, Cortical thickness abnormalities in bipolar disorder patients with a lifetime history of auditory hallucinations. Bipolar disorders 20, 647–657 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Torkamani A, Dean B, Schork NJ, Thomas EA, Coexpression network analysis of neural tissue reveals perturbations in developmental processes in schizophrenia. Genome research 20, 403–412 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmalbach B, Lepsveridze E, Djogo N, Papashvili G, Kuang F, Leshchyns’ka I, Sytnyk V, Nikonenko AG, Dityatev A, Jakovcevski I, Schachner M, Age-dependent loss of parvalbumin-expressing hippocampal interneurons in mice deficient in CHL1, a mental retardation and schizophrenia susceptibility gene. Journal of neurochemistry 135, 830–844 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Torrey EF, Webster M, Knable M, Johnston N, Yolken RH, The stanley foundation brain collection and neuropathology consortium. Schizophr Res 44, 151–155 (2000). [DOI] [PubMed] [Google Scholar]
- 44.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T, Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benovoy D, Kwan T, Majewski J, Effect of polymorphisms within probe-target sequences on olignonucleotide microarray experiments. Nucleic acids research 36, 4417–4423 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP, Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Willenbrock H, Salomon J, Sokilde R, Barken KB, Hansen TN, Nielsen FC, Moller S, Litman T, Quantitative miRNA expression analysis: comparing microarrays with next-generation sequencing. Rna 15, 2028–2034 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Langmead B, Salzberg SL, Fast gapped-read alignment with Bowtie 2. Nature methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tam S, Tsao MS, McPherson JD, Optimization of miRNA-seq data preprocessing. Briefings in bioinformatics 16, 950–963 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen C, Grennan K, Badner J, Zhang D, Gershon E, Jin L, Liu C, Removing batch effects in analysis of expression microarray data: an evaluation of six batch adjustment methods. PloS one 6, e17238 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martin M, Cutadapt Removes Adapter Sequences From High-Throughput Sequencing Reads. EMBnet.journal 17, 10–12 (2011). [Google Scholar]
- 52.Dobin A, Gingeras TR, Mapping RNA-seq Reads with STAR. Current protocols in bioinformatics 51, 11–14 11–19 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome S Project Data Processing, The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thompson JA, Tan J, Greene CS, Cross-platform normalization of microarray and RNA-seq data for machine learning applications. PeerJ 4, e1621 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu Z, Chang YC, Wang Y, Huang CL, Liu Y, Tian F, Granger B, Delisi C, VisANT 4.0: Integrative network platform to connect genes, drugs, diseases and therapies. Nucleic acids research 41, W225–231 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC, PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu C, Cheng L, Badner JA, Zhang D, Craig DW, Redman M, Gershon ES, Whole-genome association mapping of gene expression in the human prefrontal cortex. Mol Psychiatry 15, 779–784 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hauberg ME, Roussos P, Grove J, Borglum AD, Mattheisen M, Schizophrenia C Working Group of the Psychiatric Genomics, Analyzing the Role of MicroRNAs in Schizophrenia in the Context of Common Genetic Risk Variants. JAMA psychiatry 73, 369–377 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Uddin M, Tammimies K, Pellecchia G, Alipanahi B, Hu P, Wang Z, Pinto D, Lau L, Nalpathamkalam T, Marshall CR, Blencowe BJ, Frey BJ, Merico D, Yuen RK, Scherer SW, Brain-expressed exons under purifying selection are enriched for de novo mutations in autism spectrum disorder. Nature genetics 46, 742–747 (2014). [DOI] [PubMed] [Google Scholar]
- 60.Georgieva L, Rees E, Moran JL, Chambert KD, Milanova V, Craddock N, Purcell S, Sklar P, McCarroll S, Holmans P, O’Donovan MC, Owen MJ, Kirov G, De novo CNVs in bipolar affective disorder and schizophrenia. Human molecular genetics 23, 6677–6683 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gershon ES, Alliey-Rodriguez N, New ethical issues for genetic counseling in common mental disorders. The American journal of psychiatry 170, 968–976 (2013). [DOI] [PubMed] [Google Scholar]
- 62.Gaujoux R, Seoighe C, CellMix: a comprehensive toolbox for gene expression deconvolution. Bioinformatics 29, 2211–2212 (2013). [DOI] [PubMed] [Google Scholar]
- 63.Zhang Y, Sloan SA, Clarke LE, Caneda C, Plaza CA, Blumenthal PD, Vogel H, Steinberg GK, Edwards MS, Li G, Duncan JA 3rd, Cheshier SH, Shuer LM, Chang EF, Grant GA, Gephart MG, Barres BA, Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron 89, 37–53 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Materials and Methods
Fig. S1. Causal relationship between transcription factors and miRNAs.
Fig. S2. Differences in cell type proportions between cases (BP and SCZ) and controls.
Fig. S3. Immunofluorescence staining of NPCs.
Fig. S4. RT-qPCR analysis of POU3F2 knockdown efficiency.
Fig. S5. Distribution of eigengene values in daM.
Table S1. Demographic and clinical information for discovery SMRI data.
Table S2. Demographic and clinical information for replicated BrainGVEX data.
Table S3. Overlap of genes in the daM and genes with rare variants.
Table S4. Transcription factor expression associated with disease in SMRI and BrainGVEX data
Table S5. Transcription factor expression correlations in BrainGVEX data.
Table S6. Orthogonal causal anchor (LEO.NB.OCA) and candidate pleiotropic anchor (LEO.NB.CPA) scores for six transcription factors and one miRNA.
Table S7. Sequences of RT-qPCR primers and siRNA.
Table S8. Top 50 connected genes in the daM, miRNAs, transcription factors and their WGCNA parameters.
Table S9. POU3F2 and its putative targets in daM.