Abstract
Host genetic variation has a major impact on infectious disease susceptibility. The study of pathogen resistance genes, largely aided by mouse models, has significantly advanced our understanding of infectious disease pathogenesis. The Collaborative Cross (CC), a newly developed multi-parental mouse genetic reference population, serves as a tractable model system to study how pathogens interact with genetically diverse populations. In this review, we summarize progress utilizing the CC as a platform to develop improved models of pathogen-induced disease and to map polymorphic host response loci associated with variation in susceptibility to pathogens.
Keywords: genetics reference populations, Collaborative Cross, infection, immunity, immune genes
Host genetic variation plays an important role in shaping infectious disease susceptibility. Noll et al. review the application of a genetically diverse mouse reference population, the Collaborative Cross, to study variation in disease response across multiple pathogens, highlighting advances in model development and genetic mapping.
Main Text
Introduction
Pathogens often cause variable disease outcomes across infected individuals, ranging from asymptomatic infection to severe or fatal disease. Multiple factors influence an individual’s susceptibility to a pathogen, including variation in pathogen dose or virulence as well as host age, prior immune experience, microbiome/coinfection, and genetics. A role for host genetics in pathogen susceptibility is supported both by twin studies (Kallmann and Reisner, 1943) and by evidence of pathogen-driven selective pressure on the evolution of the human immune system (Cagliani and Sironi, 2013). Human genetic studies have demonstrated a role for host genetics in pathogen susceptibility and identified polymorphic genetic loci and genes associated with variation in susceptibility to specific pathogens (e.g., HIV) (McLaren et al., 2015), classes of pathogens (e.g., mycobacteria) (Bustamante et al., 2014), and more generalized primary immunodeficiencies (PIDs) that result in severe immune defects (Chapman and Hill, 2012). This information has enhanced our understanding of how host genetic variation affects pathogen susceptibility and how specific genes regulate human immunity and host-pathogen interactions.
While advances in human genetic analysis have led to the identification of several host genes that regulate pathogen susceptibility in humans (Newport and Finan, 2011), much of our understanding of how specific genes affect infectious disease pathogenesis in mammals has come from studies using mouse models (Masopust et al., 2017). This is in large part due to the existence of inbred mouse strains as well as the vast amount of immunological and molecular genetic tools available for the mouse, including the early generation of the mouse reference genome. Inbred strains, in which each mouse is essentially genetically identical, allow control of host genetics as well as other factors that can confound human studies, such as pathogen dose, nutrition, and prior immune exposure. Additionally, targeted knockout technology has been used in the mouse genome for almost 30 years (Bouabe and Okkenhaug, 2013). While advances in gene editing techniques (e.g., CRISPR) have recently extended these approaches to other species, genetically modified mice have been an unparalleled resource for studying the role of specific genes in the response to a variety of pathogens (Masopust et al., 2017). Furthermore, the use of classical genetic crossing approaches (e.g., intercrosses), as well as reproducible mouse genetic reference populations (GRPs), has facilitated the discovery of key regulators of host immunity and pathogen susceptibility such as the innate immune receptor Tlr4, oligoadenylate-synthetase Oas1b, and divalent metal transporter Nramp1 (Perelygin et al., 2002, Qureshi et al., 1999, Vidal et al., 1993). The recent development of multi-parental GRPs, such as the Collaborative Cross (CC), promises to further extend genetic mapping capabilities, while also leading to the development of mouse models that more accurately reproduce the genetic diversity and phenotypic range seen in human populations (Churchill et al., 2004). This review will summarize recent advances in the use of mouse GRPs, with an emphasis on the use of the CC for studying infectious diseases and immunity.
Human Genetic Variation and Infectious Disease
The impact of host genetics on pathogen susceptibility can be illustrated by genes exhibiting large effects such as the chemokine receptor CCR5 and fucosyltransferase 2 (FUT2), for which specific variants have a major impact on resistance to viral infection (HIV and norovirus, respectively) (Quillent et al., 1998, Thorven et al., 2005). Likewise, deleterious mutations in immune genes can result in PIDs, which cause enhanced susceptibility to specific or entire classes of pathogens (Carneiro-Sampaio and Coutinho, 2007). To date, over 350 genetically driven PIDs have been identified in humans (Picard et al., 2018). The identification of these monogenic resistance and susceptibility genes has enhanced our understanding of human immunity and host-pathogen interactions while contributing to the development of antiviral therapies, such as HIV entry inhibitors, as well as gene therapy and immune replacement therapies (Collins and Thrasher, 2015, Lenardo et al., 2016).
Though genes of large effect can impact human health, inheritance patterns suggest that susceptibility to infectious disease is largely polygenic (driven by tens to thousands of variants of smaller effect) (Hill, 2012). This is supported by results from genome-wide association studies (GWAS) designed to map loci associated with variation in pathogen susceptibility and vaccine response. For example, GWAS of HIV viral load have shown that mutations in the HIV co-receptor CCR5 and variants in human leukocyte antigen (HLA) explain approximately 15% of the total variance, with an additional 5% of heritable variance explained by additive effects of other genes (McLaren et al., 2015). GWAS on malaria susceptibility have identified variants in multiple genes including the calcium ATPase ATP2B4, the glucose phosphate dehydrogenase GP6D, and HLAs, as well as confirming previously identified variants in hemoglobin genes (sickle cell and thalassemia) (Hedrick, 2011, Timmann et al., 2012). Multiple loci have been associated with clearance of hepatitis C virus, including the cytokine IL-28b and HLA genes (Duggal et al., 2013), and over 20 loci have been associated with leprosy susceptibility (Wang et al., 2016).
While GWAS have been applied to and enhanced our understanding of infectious diseases, these studies explain only a fraction of the heritable variation in pathogen susceptibility (Hill, 2012). This is partly attributable to factors such as small cohort sizes, ethnic striations, and lack of replicates, which limit detection and interpretation of results (Du et al., 2012). Rare genetic variants are unlikely to be detected by GWAS, due to both lack of power and lack of tagging on conventional genotyping arrays (Auer and Lettre, 2015). A specific concern for GWAS in infectious diseases is that studies can be confounded by pathogen genetics, pathogen dose, and other environmental factors. Furthermore, validation and mechanistic analysis can be difficult in humans due to factors such as lack of replicates and inability to routinely access many affected tissue types. For these reasons, the use of appropriate animal models has been a critical resource for identifying and studying pathogen susceptibility genes.
Mouse Models of Disease
While a variety of model genetic systems have been used to analyze host-pathogen interactions (e.g., Caenorhabditis elegans, Danio rerio, and Drosophila melanogaster) (Allen and Neely, 2010, Dionne and Schneider, 2008, Marsh and May, 2012), the laboratory mouse (Mus musculus) provides the most robust mammalian system for dissecting genetic regulation of immune responses in human-relevant diseases. Mice are relatively inexpensive, can be easily controlled in diet and environment, and reproduce quickly. A wide variety of well-characterized mouse immunological reagents are available, as well as genetic tools such as reproducible inbred strains, gene-specific knockouts, genome-wide mutagenesis strategies (e.g., N-ethyl-N-nitrosourea [ENU] and CRISPR/Cas9), and transgenic technologies. These systems facilitate the identification and mechanistic characterization of specific host genes that affect disease pathogenesis and/or immunity (Pelletier et al., 2015, Takaki et al., 2017).
Though over 450 commercially available inbred mouse strains and outbred stocks are available (Beck et al., 2000), the majority of research in infectious disease and immunology is conducted in a limited set of strains. One of the major reasons for the focus on specific strains is the need to control for genetic background when analyzing gene-specific knockouts or performing large-scale mutagenesis screens. Large-scale initiatives such as the International Mouse Phenotyping Consortium (IMPC) depend upon the ability to control background strain to analyze the impact of specific gene deletions or mutations on a wide range of developmental and immune phenotypes (Muñoz-Fuentes et al., 2018), and have thus generated these mutants on common backgrounds using largely identical procedures. C57BL/6J is the most commonly used inbred mouse strain; it is the source of the mouse reference genome and the genetic background for the majority of knockout mice. Other strains, such as the BALB/c substrains, also have a rich history of use in immunology and infectious disease research (Watanabe et al., 2004). While the ongoing use of such strains is driven by the need to compare results across studies, it is important to note that every inbred strain has a unique set of genetic features, and thus no one strain is representative of all mice, let alone of a genetically complex population such as humans.
Although genetically modified mice have been an essential resource for gene discovery and mechanistic analysis, there has been a growing acknowledgment in the field that knockout approaches have limitations. Approximately 15% of genes cannot be knocked out because they are developmentally essential (NIH, 2015). Similarly, other knockouts have no observable phenotype, due to genetic redundancy and/or lack of robustness in the phenotyping pipeline (Barbaric et al., 2007). In other cases, knockouts have disparate phenotypes on different genetic backgrounds due to gene-gene interactions (Doetschman, 2009). Furthermore, gene knockouts result in null alleles (complete loss of gene function), whereas naturally occurring genetic variants are most likely to be hypomorphic or hypermorphic alleles (partial loss or gain of gene function; alterations in timing and magnitude of expression). Lastly, while single-gene approaches are straightforward, they are isolated systems that do not model the simultaneous contribution of variants in multiple pathways, as would most likely be observed in a natural system. Thus, to better model and understand the role of genes in complex phenotypes such as immunity and infectious disease, it is pertinent to consider complex genetic interactions across diverse genetic backgrounds.
Genetic Reference Populations
In contrast to the approaches discussed above, researchers have also studied the role of natural genetic variants by leveraging differential phenotypes across inbred strains, using classical genetic breeding strategies to identify pathogen susceptibility genes such as Oas1b, immune cell activating receptor Ly49H, and large interferon-induced GTPase Mx1 (Casanova et al., 2002). In contrast to mapping crosses between stocks (e.g., F2 crosses), where each mouse is unique and therefore ephemeral, GRPs serve as reproducible models to study the role of genetic diversity. GRPs consist of anywhere from a few to hundreds of inbred lines derived and split from a common ancestral population, and each line has a fixed and known genome that can be replicated indefinitely. The existence of reproducible yet genetically diverse individual mouse strains facilitates study designs that involve case versus control, genotype by environment interactions (GxE), and phenotypic penetrance or threshold traits. These populations further facilitate the integration of phenotypic, genotypic, and molecular data for systems-level data analysis while also allowing retrospective integration of new layers of data.
The first recombinant inbred panel, the CXB (BALB/cJ × C57BL/6J), was created by Donald Bailey to facilitate mapping of the MHC locus (Bailey, 1971). Subsequent biparental recombinant inbred panels include the AXB/BXA (A/J × C57BL/6J, C57BL/6J × A/J), the BXH (C57BL/6J × C3H/HeJ), and the BXD (C57BL/6J × DBA/2J), which is the largest and most extensively used of these panels (Peirce et al., 2004). Though originally developed to study monogenic traits, use of these panels has been extended to study genetically complex traits such as susceptibility to murine plasmodia (Hoffmann et al., 1984), murine leukemia virus (Panoutsakopoulou et al., 1998), group A streptococcus (Chella Krishnan et al., 2016), and influenza A virus (IAV) (Nedelko et al., 2012). As results from these panels accumulated, there was a community-wide recognition that larger and more genetically diverse GRPs would improve the ability to genetically dissect more complex traits (Threadgill et al., 2002).
The Collaborative Cross
Based on results from the early GRPs as well as genetic mapping studies, the Complex Trait Consortium proposed to create a second-generation, multi-parent advanced intercross GRP to promote complex trait genetic mapping and systems genetics research (Threadgill et al., 2002). In contrast to single-gene approaches, systems genetics considers phenotypes in the context of global genetic variation as well as intermediate molecular factors such as gene expression, protein abundance, and environmental interactions. The resource ultimately derived from this proposal was the CC, a large panel of recombinant inbred strains derived from eight genetically diverse founder strains. The CC was designed to facilitate systems genetics approaches with improved resolution and higher statistical power compared to traditional GRPs, to expand the pool of genetic variation segregating in the population, and to bridge the analysis of genetic and molecular networks across diverse phenotypes (Churchill et al., 2004).
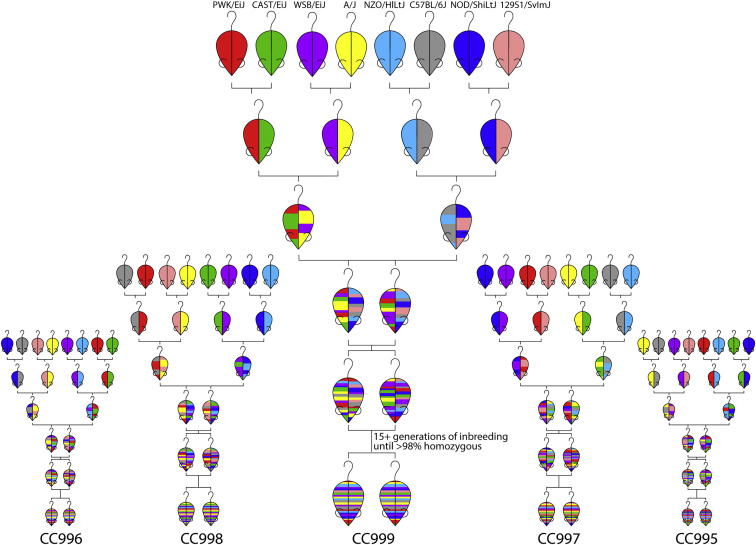
Of the eight CC founder strains, five are classical laboratory strains chosen for their rich history of use in mouse genetics (C57BL/6J and 129S1/SvImJ) or their relevance as models of common diseases including cancer, type 1 diabetes, metabolic syndrome, and obesity (A/J, NOD/ShiLtJ, and NZO/HiLtJ). The other three CC founders are wild-derived inbred strains, which represent the three major subspecies of Mus musculus: casteneus (CAST/EiJ), musculus (PWK/PhJ), and domesticus (WSB/EiJ), and introduce much of the genetic diversity into the CC. These eight founder strains were interbred in a funnel breeding scheme to produce recombinant mice with genomic contributions from each founder, which were then bred to homozygosity to produce recombinant inbred (RI) CC strains (Figure 1 ). Although the initial goal was to produce 1,000 CC strains, only around 100 CC strains survived the inbreeding process due to high rates of infertility and breeding issues, largely caused by genomic incompatibility introduced by the wild-derived strains. Extinction of CC strains during the inbreeding process inspired the creation of the Diversity Outbred (DO) panel, an outbred population derived from a set of incipient CC strains (Churchill et al., 2012). The DO now serves as its own important genetic resource that has proven useful for high-resolution genetic mapping as well as exploration of phenotypic diversity (Chick et al., 2016, Gatti et al., 2014, Smallwood et al., 2014, Shorter et al., 2018). While the DO has only been used in a limited number of infectious disease studies (McHugh et al., 2013, Niazi et al., 2015), the DO and CC should be viewed as complementary resources for studying how genetic variation affects pathogen susceptibility and immunity.
Figure 1.
Representative CC Funnels
CC founders were bred in funnel breeding schemes to produce progeny with genetic contributions from each of the eight founders, at which point they were inbred for generations until reaching near homozygosity. Many different funnels were set up, each to produce a unique CC RI line.
While classical mouse GRPs remain actively used, and in some ways provide a more straightforward analytical path than the CC due to reduced complexity, several aspects of the CC make it more attractive for discovery-based research. The CC captures approximately 90% of common genetic variants described within laboratory mice, with up to eight distinct haplotypes at any region of the genome (Roberts et al., 2007). Due to higher genetic diversity with novel allele combinations and epistatic interactions, the CC yields more phenotypic variation and extreme phenotypes than classical GRPs. The selection of founder strains and the breeding design, as well as higher levels of recombination, result in lower levels of long-range disequilibrium and population structure than classical GRPs, allowing for higher resolution quantitative trait locus (QTL) mapping (Iraqi et al., 2012, Threadgill et al., 2002). Genetic variation is also more uniformly distributed across the genome, removing several genomic “blind spots” that interfere with mapping in classical inbred strains and GRPs (Roberts et al., 2007).
Although the CC has only been available for a relatively short period of time, it has been utilized across a number of disciplines, including response to environmental toxins (Venkatratnam et al., 2017), nutrition (Schoenrock et al., 2018), body composition (Mathes et al., 2011), and immunity and pathogenesis (discussed further here). Importantly, these studies have highlighted previously uncharacterized phenotypic diversity, including expansion of the phenotypic range, disassociation of traits previously thought to be connected, and identification of completely novel phenotypes (Srivastava et al., 2017). The CC is an especially useful resource for studying the role of host genetics in host-pathogen interactions because it allows for control of many of the factors described above that confound infectious disease studies in humans. Over the past decade, a variety of studies utilizing the CC, including non-fully inbred incipient CC lines (commonly termed the pre-CC) and derived CC populations (such as CC-F1s), have been used to probe the role of host genetics in the response to infectious disease. Here we will discuss the current work in this field, including studies focusing on phenotypic characterization (e.g., model development and molecular signatures of disease response) and genetic mapping of disease trait-associated QTL.
Phenotypic Characterization in the CC
There is an ongoing debate over the validity of mouse models in recapitulating human disease (Seok et al., 2013, Takao and Miyakawa, 2015). While the utility of the mouse is undeniable in the infectious disease field, it is equally true that individual classical inbred strains do not fully recapitulate human disease and most models recapitulate only a portion of the disease phenotypes observed in humans. In an effort to improve model fidelity to human disease, a number of researchers have turned to the CC to develop new mouse models. Genetically diverse CC strains show a range of responses to pathogens, from variation in disease severity to completely novel disease phenotypes. This phenotypic variability better mimics the diversity of disease presentation across the human population, thus providing a more comprehensive platform to develop newer, more representative mouse models.
Ebola Virus
Ebola virus (EBOV) causes severe, and often lethal, hemorrhagic fever in humans. While classical inbred strains are susceptible to mouse-adapted EBOV (MA-EBOV), they do not display many of the characteristic symptoms observed in severe human cases such as coagulopathy, hemorrhagic manifestations, and rash (St Claire et al., 2017). Rasmussen et al. infected a panel of F1 crosses between CC strains (CC-F1s) following an initial assessment in founder strains (Rasmussen et al., 2014). They observed significant phenotypic variation following MA-EBOV infection, from high resistance to complete lethality, and covering a spectrum of different pathologies similar to the range of clinical disease observed in humans. Importantly, some strains presented with severe hemorrhagic disease and liver damage, consistent with human EBOV disease. Follow-up studies with representative susceptible and resistant strains found differential transcriptional responses, highlighting the central transcriptional regulatory gene Tek, for which haplotypes across these CC-F1s correlated with weight loss and mortality following MA-EBOV infection.
West Nile Virus
West Nile virus (WNV), a neuroinvasive flavivirus, induces a diverse spectrum of immune responses and clinical outcomes in humans that classical mouse models do not fully recapitulate (Graham et al., 2015). Graham et al. assessed WNV susceptibility in a panel of CC-F1 crosses that were heterozygous for the H-2b MHC haplotype, which allowed quantification of WNV-specific T cells with the same MHC tetramer. WNV outcome ranged from highly resistant to highly susceptible, including a novel outcome group in which mice were outwardly asymptomatic despite higher viral titers and immunopathology in the brain.
Graham et al. followed up on one unique CC-F1 ((CC032/GeniUnc × CC013/GeniUnc)-F1), in which half of the mice that survived infection displayed sustained weight loss and persistent viral loads in the CNS. They found that these mice exhibited a rapid early innate inflammatory response characterized by increased early expression of IFN-β and the interferon-stimulated gene IFN1, as well as high early viral titers and the ability to control, but not clear, viral replication in the CNS (Graham et al., 2016). Additionally, (CC032/GeniUnc × CC013/GeniUnc)-F1 mice had a unique immunoregulatory signature, with a high number of T regulatory cells in both infected and uninfected animals and a distinctive post-infection gene expression profile that indicated reduced cytolytic ability.
The initial Graham et al. study (Graham et al., 2015) found that WNV response largely tracked with the haplotype of Oas1b, a known flavivirus resistance gene, although there was still considerable disease variation within Oas1b haplotype groups. Green et al. continued to dissect the impact of Oas1b on the innate immune transcriptional landscape following WNV infection (Green et al., 2017) by performing genome-wide micro-array analysis across multiple time points post-infection in seven CC-F1s carrying different combinations of functional (wild-derived) or non-functional (classical inbred strain-derived) Oas1b haplotypes. Transcriptional correlation analysis of differentially expressed immune genes across CC-F1s yielded three gene clusters, and pathway analysis led to the construction of innate immune regulatory networks associated with WNV infection between Oas1b haplotype groups.
Mycobacterium tuberculosis
Mycobacterium tuberculosis (Mtb) infection varies broadly in disease presentation and pathology in humans. Likewise, the efficacy of the standard TB vaccine, the live-attenuated BCG, is variable across individuals (Mangtani et al., 2014). Smith et al. studied Mtb pathogenesis and BCG vaccine efficacy in the CC founders and three RI strains that were selected based on a pilot study assessing Mtb susceptibility (Smith et al., 2016). The authors observed a wide range of susceptibility to Mtb and dissociation of previously associated phenotypes such as bacterial burden, immune cell recruitment, and tissue damage. When the BCG vaccine was administered prior to Mtb challenge, vaccine efficacy was low overall and not correlated with susceptibility to primary infection. While BCG vaccination reduced Mtb burden in four strains, there was an increased bacterial burden in NZO/HlLtJ mice, a model for obesity and type 2 diabetes. In addition to highlighting the importance of host genetics on Mtb susceptibility and BCG-induced vaccine responses, the study illustrates the potential impact of comorbidities, such as type 2 diabetes in NZO/HlLtJ mice, on vaccine responses in genetically diverse populations.
Influenza A Virus and SARS-Coronavirus
Maurizio et al. analyzed the heritability of IAV-induced disease using CC-F1s as well as F1 crosses between founder strains (Maurizio et al., 2018). These studies determined that IAV-induced weight loss was 57% heritable at day 4 post-infection, and that this heritability was mostly composed of additive effects largely attributable to the haplotype of Mx1, a polymorphic large-effect anti-IAV gene. The genetic dominance of the protective Mx1 haplotype varied depending on subspecies origin, with the M. musculus musculus allele acting dominantly and the CAST/EiJ allele, identified by Ferris et al. (2013), acting additively. Consistent with Ferris et al. (discussed below), these authors determined that when controlling for Mx1, non-Mx1 heritable effects still accounted for 34% of the phenotypic variation, and this effect was consistent across founder diallel, pre-CC, and CC-F1 populations.
Human studies have identified blood transcriptomic and proteomic signatures associated with IAV infection that could be used to predict patient outcome. Elbahesh and Schughart observed that gene signatures from IAV-infected humans were reproduced in the peripheral blood of CC founders (Elbahesh and Schughart, 2016). In a follow-up study, Kollmus et al. characterized the transcriptional response to IAV in the peripheral blood of eleven CC strains (Kollmus et al., 2018). Though both human and mouse datasets were globally heterogeneous, the CC dataset showed that genetic background strongly influenced gene expression, highlighting the importance of genetics in driving the immune response. For the most differentially expressed genes, the transcriptional responses in mice and humans were largely similar, demonstrating the utility of the CC in recapitulating IAV-induced host response in humans.
Xiong et al. characterized transcriptomic variation in response to severe acute respiratory syndrome coronavirus (SARS-CoV) and IAV across the eight CC founder strains (Xiong et al., 2014). Differential gene expression analysis at days 2 and 4 post SARS-CoV or IAV infection showed significant differences driven by mouse strain, infection status, and time point. Genes with strain-specific differential expression patterns were largely enriched for immune pathways, while more generic differential expression patterns were enriched for basic biological functions. The study also noted the presence of strain-specific isoforms and novel transcripts not present in the C57BL/6J reference annotation.
Pseudomonas aeruginosa
Pseudomonas aeruginosa is an opportunistic bacterial pathogen with variable clinical outcomes across susceptible individuals. Human studies, including twin studies as well as association studies, have identified multiple genes associated with susceptibility to P. aeruginosa. Lorè et al. studied P. aeruginosa infection in 17 pre-CC lines, focusing on survival time and early body weight change, and observed a large amount of variation in these phenotypes (Lorè et al., 2015).
Genetic Mapping of Disease Response in the CC
As described above, the CC was initially envisioned as a genetic mapping population with approximately 1,000 strains (Threadgill et al., 2002). While the number of available CC RI strains is closer to 100, this population has proven to be sufficient for successful QTL mapping. Mapping studies have identified QTLs spanning the genome that are driven by variants from all eight CC founders and show very little overlap across pathogens (Figure 2 ; Table 1 ; discussed below). Overall, infectious disease phenotypes mapped in the CC show a broad range in overall heritability (the proportion of population-wide variation explained by differences between strains) as well as effect sizes (the proportion of population-wide variation explained by specific loci). Across the studies described in Table 1, reported estimates for heritability range from 12% to over 80%, while reported estimates for effect size range from 5% (for a QTL of small effect influencing SARS-CoV viral titer) to over 40% (for a large effect QTL containing the gene Mx1 on influenza-induced weight loss). Authors have narrowed many of these QTLs to lists of candidate genes, and a small number of these candidates have been subsequently validated.
Figure 2.
Summary of Infectious Disease and Immunity QTLs Mapped in the CC
QTLs are shown mapped onto MGSCv37/mm9; genome positions for QTLs that were mapped on GSCv38/mm10 were converted to corresponding MGSCv37/mm9 coordinates.
Table 1.
Summary of Infectious Disease and Immunity QTL Mapped in the CC
| Chr | Start (Mb) | End (Mb) | Pathogen | Phenotype | Name | Sig | Population | Reference |
|---|---|---|---|---|---|---|---|---|
| 1 | 6.3 | 12.7 | A. fumigatus | survival day | Asprl7 | ∗ | pre-CC | Durrant et al., 2011 |
| 3 | 109.6 | 123.6 | A. fumigatus | survival day | Asprl6 | ∗ | pre-CC | Durrant et al., 2011 |
| 8 | 44.2 | 55.5 | A. fumigatus | survival day | Asprl1 | ∗∗∗ | pre-CC | Durrant et al., 2011 |
| 10 | 12 | 23 | A. fumigatus | survival day | Asprl4 | ∗∗ | pre-CC | Durrant et al., 2011 |
| 10 | 94.3 | 99 | A. fumigatus | survival day | Asprl2 | ∗∗∗ | pre-CC | Durrant et al., 2011 |
| 15 | 31 | 37.5 | A. fumigatus | survival day | Asprl3 | ∗∗∗ | pre-CC | Durrant et al., 2011 |
| 18 | 5.2 | 13.6 | A. fumigatus | survival day | Asprl5 | ∗ | pre-CC | Durrant et al., 2011 |
| 4 | 54.3 | 58.3 | K. pneuomonia | day 2 survival | Kprl1 | ∗∗∗ | pre-CC | Vered et al., 2014 |
| 8 | 29.7 | 33.6 | K. pneuomonia | day 8 survival | Kprl2 | ∗∗∗ | pre-CC | Vered et al., 2014 |
| 18 | 19.9 | 36.4 | K. pneuomonia | day 8 survival | Kprl3 | ∗ | pre-CC | Vered et al., 2014 |
| 1 | 74.1 | 81.8 | S. Typhimurium | spleen bacterial load | StSl3b | ∗ | CC | Zhang et al., 2018 |
| 1 | 77.5 | 95.6 | S. Typhimurium | spleen bacterial load | StSl3a | ∗ | CC | Zhang et al., 2018 |
| 6 | 77.1 | 90 | S. Typhimurium | liver bacterial load | StSl4 | ∗ | CC | Zhang et al., 2018 |
| 8 | 11.3 | 17 | S. Typhimurium | spleen bacterial load | StSl1 | ∗∗∗ | CC | Zhang et al., 2018 |
| 10 | 46.4 | 54 | S. Typhimurium | spleen bacterial load | StSl2 | ∗∗ | CC | Zhang et al., 2018 |
| 17 | 80.5 | 91.1 | S. Typhimurium | liver bacterial load | StSl5 | ∗ | CC | Zhang et al., 2018 |
| 1 | 21.7 | 29 | influenza A virus | pulmonary edema (conditioned on Mx1) | HrI3 | ∗∗∗ | pre-CC | Ferris et al., 2013 |
| 7 | 89.1 | 96.7 | influenza A virus | weight loss | HrI2 | ∗∗∗ | pre-CC | Ferris et al., 2013 |
| 15 | 77.4 | 86.6 | influenza A virus | airway neutrophils | HrI4 | ∗∗∗ | pre-CC | Ferris et al., 2013 |
| 16 | 97.5 | 98.2 | influenza A virus | weight loss, viral titer, lung pathology score, clinical score, inflammation, airway damage, and expression modules | HrI1 | ∗∗∗ | pre-CC | Ferris et al., 2013 |
| 3 | 18.3 | 26.7 | SARS-coronavirus | vascular cuffing | HrS1 | ∗∗∗ | pre-CC | Gralinski et al., 2015 |
| 13 | 52.8 | 54.9 | SARS-coronavirus | vascular cuffing (conditioned on HrS1) | HrS4 | ∗∗∗ | pre-CC | Gralinski et al., 2015 |
| 15 | 72.1 | 75.8 | SARS-coronavirus | eosinophilia | HrS3 | ∗∗ | pre-CC | Gralinski et al., 2015 |
| 16 | 31.6 | 36.7 | SARS-coronavirus | viral titer | HrS2 | ∗∗ | pre-CC | Gralinski et al., 2015 |
| 7 | 55.2′ | 117.2′ | SARS-coronavirus | viral titer | HrS7 | ∗∗∗ | CC-F2 | Gralinski et al., 2015 |
| 9 | 116.5′ | 124.6′ | SARS-coronavirus | day 3 weight loss | HrS6 | ∗∗∗ | CC-F2 | Gralinski et al., 2015 |
| 12 | 81.6′ | 108.5′ | SARS-coronavirus | viral titer | HrS8 | ∗∗∗ | CC-F2 | Gralinski et al., 2015 |
| 15 | 0′ | 64.4′ | SARS-coronavirus | hemorrhage | HrS9 | ∗∗∗ | CC-F2 | Gralinski et al., 2015 |
| 18 | 27.1′ | 58.7′ | SARS-coronavirus | day 3 and 4 weight loss, viral titer, and hemorrhage | HrS5 | ∗∗∗ | CC-F2 | Gralinski et al., 2015 |
| 3 | 129 | 130 | none | transitional B cells | ∗∗∗ | pre-CC | Phillippi et al., 2014 | |
| 4 | 148.8 | 151.1 | none | H57+ (total T cells) | ∗∗ | pre-CC | Phillippi et al., 2014 | |
| 6 | 23.2 | 23.8 | none | B cell to T cell ratio, H57+ (total T cells) CD19+ (total B cells) | ∗∗∗ | pre-CC | Phillippi et al., 2014 | |
| 6 | 88.1 | 92.7 | none | B cell to T cell ratio | ∗∗∗ | pre-CC | Phillippi et al., 2014 | |
| 7 | 136.5 | 138.6 | none | CD11c mean fluorescence intensity (MFI) | ∗∗∗ | pre-CC | Phillippi et al., 2014 | |
| 7 | 141.1 | 142.2 | none | CD4+/CD8+ ratio, CD4+ T cells, CD8+ T cells, and CD11c MFI | ∗∗∗ | pre-CC | Phillippi et al., 2014 | |
| 8 | 3.1 | 16.8 | none | CD23 MFI | ∗∗∗ | pre-CC | Phillippi et al., 2014 | |
| X | 100′ | 106′ | none | CXCR3+ T regulatory cells | HI2 | ∗∗∗ | CC-F1 | Graham et al., 2017 |
| X | 140′ | 145′ | none | ICOS+ T regulatory cells | HI3 | ∗∗∗ | CC-F1 | Graham et al., 2017 |
| X | 160′ | 171′ | none | CD73+ T regulatory cells | HI1 | ∗∗∗ | CC-F1 | Graham et al., 2017 |
| 2 | 26 | 31.1 | none | GP23 | ∗∗ | CC | Krištić et al., 2018 | |
| 4 | 51.7 | 63.3 | none | GP15 | ∗∗ | CC | Krištić et al., 2018 | |
| 7 | 149.6 | 151.4 | none | GP25 | ∗∗∗ | CC | Krištić et al., 2018 | |
| 10 | 93 | 94.1 | none | GB17b | ∗∗∗ | CC | Krištić et al., 2018 | |
| 12 | 114.5 | 117.9 | none | GP1, GP14, GP16, GP17a, and GP20 | ∗∗∗ | CC | Krištić et al., 2018 | |
| 15 | 77.9 | 82.8 | none | GP6 and GP10 | ∗∗∗ | CC | Krištić et al., 2018 | |
| 16 | 15 | 20.3 | none | GP17b | ∗∗∗ | CC | Krištić et al., 2018 |
Significance threshold levels: ∗<0.1, ∗∗0.1, ∗∗∗0.05. Genome coordinates marked with ′ refer to genome assembly GSCv38/mm10; otherwise, coordinates refer to MGSCv37/mm9.
To date, most publications have highlighted QTLs mapped by phenotyping large panels of CC strains; however, this is not the only successful strategy. As mentioned above, some CC strains present with unique disease phenotypes, which may be driven by complex genetic regulatory networks involving multiple loci with epistatic interactions. When one or a few CC strains possess the combination of genetic variants needed to manifest the novel phenotype, population-wide studies may not possess sufficient mapping power. In these cases, researchers have successfully used more traditional F2 or backcross approaches to identify causal genes (Rogala et al., 2014). In this section, we will highlight results of both population-wide and intercross-mediated CC studies of relevance to infectious disease research.
Aspergillus fumigatus
The first host-pathogen study in the CC examined susceptibility to the fungus Aspergillus fumigatus, which is pathogenic in immunocompromised individuals. Durrant et al. found that survival time varied between 4 days and over 28 days post-infection in 66 pre-CC lines (Durrant et al., 2011). The authors mapped multiple genome-wide significant QTLs for survival time that were largely driven by wild-derived founder haplotypes. Candidate genes were identified using merge analysis, a procedure that compares the pattern of sequence variants (e.g., SNPs or indels) to the pattern of haplotype effects (impact of each founder strain haplotype on the phenotype) observed under the QTL (Yalcin et al., 2005). The application of merge analysis to the CC enables more effective gene refinement compared to studies in classic biparental GRPs, where the presence of only two haplotypes means that every genetic variant in a locus is a putative candidate causal variant. Merge analysis supported the candidate gene Irf2, an interferon regulatory transcriptional factor gene, under the most significant QTL; however, for the remaining QTLs there was no strong concordance between a priori and merge-supported candidates.
Klebsiella pneumoniae
Vered et al. challenged 73 pre-CC lines with K. pneuomonia, which causes severe pneumonia and sepsis in immunocompromised individuals (Vered et al., 2014). The pre-CC mice displayed broad and heritable variation in survival time exceeding that observed in classic inbred strains, as well as dissociation of survival time from temperature or body weight changes. The study identified one QTL at day 2 post-infection and two at day 8 post-infection. The authors used merge analysis to refine candidate genes; however, they found that the best merge candidates showed simpler allele effect patterns than the more complex haplotype effects observed under the QTL. The strongest candidate genes identified were Ikbkap, a transcriptional elongation factor complex component, and Actl7a, Actl7b, and Ctnnal1, which are involved in cell adhesion and cytoskeletal structure.
Influenza A Virus
Bottomly et al. analyzed a set of 99 pre-CC lines infected with mouse-adapted influenza A/PR/8/34 and assessed weight loss, clinical score, and mortality through 4 days post-infection (Bottomly et al., 2012). A subset of pre-CC lines, classified as high and low responders based on a composite metric of weight loss and histopathological scoring, were selected for transcriptomic profiling. Over 2,000 transcripts were differentially expressed between susceptible and resistant classes, and mapping identified 21 significant expression QTLs (eQTLs). Twelve of the eQTLs were validated in CC founder strains, and structural equation modeling was applied to these candidates to infer reactive expression networks underlying the transcriptional differences.
In a companion study, Ferris et al. further characterized variation in response to influenza A/PR/8/34 in 155 pre-CC lines (Ferris et al., 2013). While disease-associated phenotypes (weight loss, viral titer, and inflammation) were largely correlated, subsets of these pre-CC mice showed breakdowns in these relationships, resulting in novel phenotypic combinations not observed in classical mouse strains (e.g., high weight loss with minimal inflammation). This study identified four significant QTLs, including one strong QTL that explained approximately 42% of IAV-induced weight loss, over the IAV resistance gene Mx1. Importantly, while Mx1 is well studied, the authors identified a novel allelic variant derived from CAST/EiJ that protects from weight loss but less efficiently inhibits viral replication compared to the functional M. musculus musculus-derived allele carried by NZO/HiLtJ and PWK/PhJ. Consistent with Ferris et al.’s finding of a uniquely functional CAST/EiJ Mx1 variant, Leist et al. found that CAST/EiJ mice have a unique response to an H3N2 strain of IAV (Leist et al., 2016). While the Mx1 locus had a dominating effect in the Ferris et al. study, large phenotypic variation occurred within Mx1 classes, suggesting the presence of modifier alleles. Three additional QTLs were mapped when controlling for Mx1 haplotype, corresponding to variation in weight loss, pulmonary edema, and airway neutrophils. Notably, aside from the Mx1 locus, none of the QTLs identified by Ferris et al. matched influenza susceptibility loci found in mapping studies conducted in the BXD population. (Boon et al., 2009, Nedelko et al., 2012). These differences may reflect the more complex genetics of the CC compared to the BXD and/or differences in the strain of influenza or other experimental variables across studies.
Salmonella enterica
Salmonella enterica serovar Typhimurium (S. Typhimurium) causes typhoid fever, and previous mouse mapping studies have identified multiple loci associated with susceptibility (Roy et al., 2006, Sebastiani et al., 1998). To identify novel loci associated with S. Typhimurium susceptibility, Zhang et al. infected 35 CC RI strains and measured bacterial load in the liver and spleen at 4 days post-infection (Zhang et al., 2018). QTL scans for bacterial load mapped multiple significant or suggestive QTL (Figure 2; Table 1), which differed from those identified in studies conducted in other mapping populations (Roy et al., 2006, Sebastiani et al., 1998). Zhang et al. used merge analysis in combination with immune cell gene expression, gene ontology, and analysis of known protein functions to narrow the list of candidate genes under the two significant loci. They identified candidate genes Cul4a (ubiquitin ligase), Lamp1 (lysosomal membrane protein), Mcf2l (guanine nucleotide exchange factor), Pcid2 (TREX-2 complex component involved in mRNA nuclear export), and a high-priority candidate Slc35f1, which has lactate dehydrogenase activity that may be important in the S. Typhimurium pyruvate metabolism pathway. The study also noted that one strain, CC042/GeniUnc, had extremely high bacterial loads, suggesting that it may be uniquely susceptible to S. Typhimurium.
SARS-Coronavirus
SARS-CoV causes severe acute respiratory syndrome in humans, but the genes regulating these processes are poorly understood. Gralinski et al. infected the CC founder strains and 147 pre-CC lines with mouse-adapted SARS-CoV and studied disease through day 4 post-infection (Gralinski et al., 2015). Variation in weight loss and viral titer was significant in the CC founders and even broader in the pre-CC mice. Similar to IAV, phenotypic dissociation was observed in some pre-CC lines, with viral titer not correlated with weight loss. QTL mapping identified a significant main effect QTL, as well as a modifier QTL for vascular cuffing in the lungs, explaining 26% and 21% of the variation, respectively. Suggestive QTLs were identified for eosinophilia (26% of variation) and viral titer (22% of variation) (Figure 2; Table 1). The candidate region for the main effect vascular cuffing QTL was narrowed to a small 450 kb region of shared ancestry between the high-response haplotypes. This region contained only one functional gene, Trim55, a RING zinc-finger-containing protein expressed in smooth muscle, which had not previously been implicated in any immune phenotypes. Follow-up validation studies using Trim55 knockout mice, which showed altered chemokine and tight junction gene expression as well as altered inflammatory cell recruitment, confirmed a role for this gene in SARS-CoV-induced vascular cuffing. The authors also noted a high-priority candidate under the modifier QTL, the cadherin family member Cdhr2, which may be involved in migration of inflammatory cell from the blood into the lung.
In a subsequent study, Gralinksi et al. used an F2 cross between two CC-RI strains with highly divergent SARS-CoV susceptibilities to map five significant loci affecting weight loss, viral titer, and other disease phenotypes, including one main effect QTL that explained between 6% and 12% of the variation in every phenotype (Gralinski et al., 2017). Ticam2, a TLR4 adaptor protein, was identified as a high-priority candidate gene under the main effect QTL given the known importance of TLR4 in SARS-CoV pathogenesis (Totura et al., 2015). Ticam2 knockout mice had increased SARS-CoV-induced weight loss, early viral titers, and pulmonary hemorrhage.
Homeostatic Immunity
In addition to exhibiting high levels of variation in pathogen-induced responses, the CC also exhibits variation in baseline (homeostatic) immunity. Variation in immune homeostasis has been associated with variation in vaccine responses in humans (Tsang et al., 2014), and the CC provides a novel resource to study how variation in pre-existing immunity affects the host response to infection or vaccination. Phillippi et al. analyzed subsets of lymphocytes and antigen-presenting cells in the CC founders, CC founder F1s, and 66 pre-CC strains (Phillippi et al., 2014). They observed variation in the pre-CC exceeding that in the founders, as well as novel extreme phenotypes such as lymphopenia and inverted CD4/CD8 ratios. While some immune phenotypes were highly correlated, others showed little or no correlation to other immune populations. Mapping identified 10 significant QTLs across 8 phenotypes, including B/T cell ratio and mean fluorescence intensity for CD23, also known as Fc epsilon RII. The CD23 QTL contains the gene Fcer2a, which codes for CD23 itself, and a combination of merge analysis, conditional association, and residue conservation was used to identify specific coding polymorphisms within Fcer2a driving the QTL.
Graham et al. also performed an extensive examination of homeostatic immunity in over 100 CC-F1 crosses, with a focus on T cell populations, expression of activation markers, and production of inflammatory cytokines (Graham et al., 2017). CC strains exhibited high levels of phenotypic variation that extended well beyond the variation observed between C57BL/6J and BALB/c, the most common laboratory strains in immunological studies. Mapping identified two highly significant QTLs driving the frequency of specific T regulatory subsets, and candidates were chosen based on concordance of haplotype effects with founder polymorphisms that were coding or splice variants. Similar to the cis-QTL for CD23 expression identified by Phillippi et al., the QTL mapped by Graham et al. for CXCR3+ T regulatory cells contains the Cxcr3 gene itself.
Krištić et al. studied variation in IgG glycosylation, which is important for antibody structure and function, in 95 CC strains and observed nearly double the variation observed in humans (Krištić et al., 2018). Glycosylation patterns were up to 80% heritable, and multiple QTLs associated with variation in different glycans were identified. Variation in 5 different glycans all mapped to the immunoglobulin heavy chain locus. Variation in 2 other glycans mapped to a locus containing the glycosyltransferase-encoding gene Mgat3, which was identified as a strong candidate.
Commensals and the Microbiome in the CC
While the focus of this review is on genetic control of immune populations and response to infectious agents, there is a growing appreciation for the role commensals play in shaping health and modulating immune responses. Two studies have addressed these responses in the CC (Snijders et al., 2016) and DO (Carmody et al., 2015) populations. Both found that there were significant contributions of host genetic backgrounds. Further, the CC study found strong genomic signals for many bacterial groups (OTUs) around the genome. Given the increasing appreciation and awareness of the microbiome in modulating a variety of biomedically relevant traits, as well as the impact of commensals and the virome (Virgin, 2014) on directly modulating immune responses (Masopust et al., 2017), future work within the CC will allow for the disentangling of direct roles on host genetic variants on disease responses, as well as indirect roles mediated through effects on commensals or prior immune exposure.
Utilization of the CC
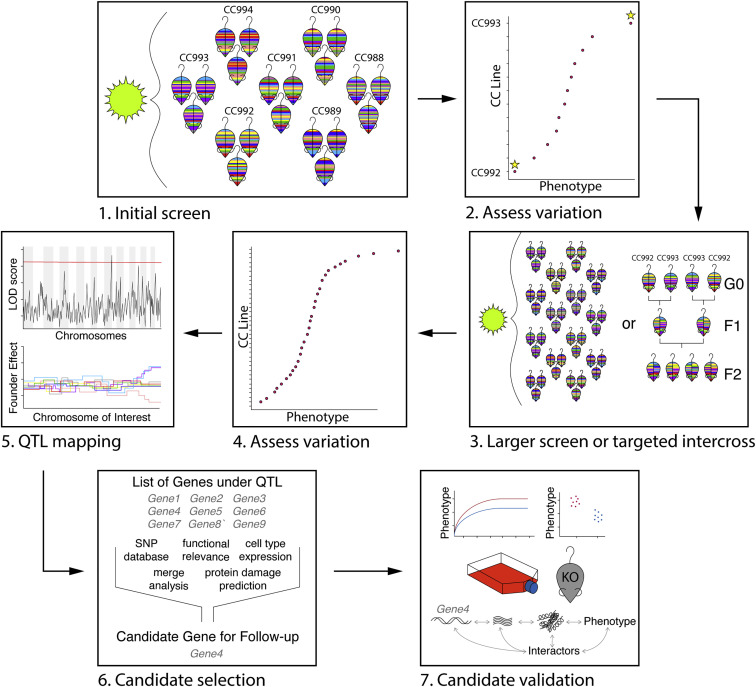
The CC is a powerful resource for model development, extreme phenotype discovery, population-based QTL mapping, intercross mapping, validation of candidate genes across genetic backgrounds, and a variety of other approaches. Here, we provide an overview of different study designs that can be implemented to study the impact of host genetic variation on pathogen outcomes or immunity using the CC (Figure 3 ).
Figure 3.
CC Workflow Schematic
(1) Small screen in a subset of CC strains (12–16), selected based on availability or a genotype of interest (e.g., MHC haplotype).
(2) Assess variation across strains, identify outliers, measure heritability, perform power calculations, and identify confounding variables and reassess experimental design if necessary.
(3) Perform a larger screen either in the full CC population or a targeted intercross. Following mapping, this can be repeated to follow-up on modifier alleles or other unexplained loci.
(4) Reassess variation, outliers, and heritability across mapping population.
(5) Map QTLs driving phenotype of interest; analyze founder haplotype effects if mapping is done in the CC.
(6) Rationally select candidate genes using different tools as applicable.
(7) Perform validation studies in vitro and/or in vivo to confirm effect of candidate gene on phenotype.
Many of the genetic mapping studies described above utilized large panels of CC strains and were able to identify loci of moderate to large effects on pathogen outcomes. However, these types of studies are extremely resource intensive and may not always be the optimal approach. Rather, we suggest a tiered approach to study design within the CC, starting with a small-scale analysis in a set of strains (e.g., 12–16) that sample most of the haplotypes present across the CC genomes, and expanding as necessary. The efficacy of this type of approach is demonstrated by the success of small CC screens that have identified strains with unique phenotypes and proceeded with further targeted studies such as F2 crosses to reveal complex polygenic networks driving the outlier phenotype (Gralinski et al., 2017, Rogala et al., 2014). Furthermore, even when investigators opt for large mapping studies, they may still identify strains with unique outlier phenotypes that cannot be explained by QTLs identified in the initial screen. This missing heritability may be driven by complex genetic interactions such as epistasis, which are difficult to study in complex populations and are more effectively studied in targeted crosses (Rogala et al., 2014).
Initially screening small sets of CC strains (e.g., 12–16) allows the investigator to obtain insight into the distribution of trait variation (e.g., continuous, bimodal), while testing whether any strains show unique or strong outlier phenotypes (Gralinski et al., 2015, Rasmussen et al., 2014, Rogala et al., 2014). These small screens also facilitate estimation of the proportion of phenotypic variation explained by genetics within the test population (heritability), identification of potential confounding factors (e.g., mouse size and activity levels), and performance of power calculations before embarking on a large-scale screen. In contrast to conducting the initial screen in the CC founders, CC strains exhibit more phenotypic diversity due to re-assortment of allelic variants throughout the genome. Furthermore, a specific set of CC strains can be chosen to avoid the presence of large effect resistance genes such as Mx1 or Oas1b that may dominate the response to some pathogens. Likewise, strains can be selected that carry specific gene haplotypes to facilitate specific assays (e.g., MHC haplotypes for antigen-specific T cell analysis) (Graham et al., 2015, Graham et al., 2017).
Following an initial screen, researchers interested in developing new models can pursue the strains with the most relevant phenotypes, while those interested in genetic mapping may either perform a more extensive population-wide study (e.g., across the entire CC) or conduct a focused analysis of one or more strains with extreme or novel phenotypes via classical intercrosses. Unlike CC mice, which are fully inbred and genotyped, each mouse in an F2 or other intercross mapping population will need to be genotyped. In both cases, genetic mapping should identify a subset of the variants impacting the phenotype of interest (e.g., disease). While an in-depth discussion of mapping methods is beyond the scope of this review, several robust mapping strategies have been successfully applied to the CC, including DOQTL, BAGPIPE, and R/qtl (Arends et al., 2010, Gatti et al., 2014, Valdar et al., 2009). Once QTLs have been mapped, investigators can confirm the underlying haplotype effects by screening additional CC strains with high or low responder haplotypes under the QTLs that were not included in the initial screen, or by screening a set of mice from the related DO population. While not essential, this step confirms the initial mapping studies and provides confidence in the QTL effects prior to embarking on candidate gene investigation.
The identification and validation of candidate genes can be a challenging and intensive process. QTLs are often large, containing tens to hundreds of genes. In contrast to mapping in biparental GRPs or intercross populations, where there are only two haplotypes and every variant is a putative candidate, in the CC, merge analysis (Yalcin et al., 2005) can be used to narrow candidates based on how the pattern of variants (e.g., SNPs or indels) for each founder matches the distribution of phenotypic responses associated with each founder haplotype. As described above, merge analysis has been a useful tool in the CC; however, a drawback of merge analysis is that it assumes a single SNP variant driving the locus. In multi-parental populations, different SNPs or other mutations within the same gene may phenocopy one another (e.g., different Mx1 null alleles; Ferris et al., 2013). Furthermore, other genetic variants (e.g., copy number variants and transposable elements) are often missed by traditional merge databases. Other in silico tools, including baseline tissue and/or haplotype-specific gene expression (e.g., ImmGen and GECCO) (Shay and Kang, 2013; http://csbio.unc.edu/gecco) and protein functional consequence prediction (e.g., SIFT; Sim et al., 2012), can help narrow candidates to a single high-priority candidate gene or small set of candidates (Ferris et al., 2013, Gralinski et al., 2015). Alternatively, if causal variants are suspected of altering transcript levels, de novo gene expression analysis can be used to further refine candidate genes under the locus.
Following candidate gene identification, there are several options for testing whether the candidate is actually causal. When the phenotype is driven by an individual cellular factor, such as an antiviral molecule or receptor, gene function can be validated in vitro using techniques such as CRISPR or ectopic expression of different variants. When the phenotype has a more systemic cause, in vivo validation is generally necessary. This can be facilitated by the wide availability of knockout mouse lines (Gralinski et al., 2015, Gralinski et al., 2017). However, in vivo validation studies should take into account factors such as genetic background of the knockout, the effect of the variant, and the distribution of haplotypes (e.g., a knockout on a C57BL/6J background may not provide an interpretable phenotype if, in the CC, the C57BL/6J haplotype is an extreme response haplotype, or if the C57BL/6J allele is hypomorphic or amorphic). More recent advances in CRISPR technology open up the possibility of knocking out or performing allele swaps in genes directly in CC founders or RI strains, enhancing the utility of the CC as a resource for both identifying and studying polymorphic genes affecting pathogen susceptibility or other phenotypes of interest.
Challenges and Future Directions
In less than two decades since the CC was conceptualized (Churchill et al., 2004), the CC has proven to be a fruitful resource across many fields, including infectious disease and immunology. Expanding use of the CC has highlighted certain challenges and considerations that should be recognized to drive future development in both experimental design and resource expansion.
Genetic mapping studies in the CC have successfully identified loci underlying infectious disease and immune phenotypes; however, relatively few have subsequently validated the initially identified candidate genes. Even more challenging than candidate gene validation is the identification of specific causal variants within a gene. While strategies such as merge analysis can help refine candidates and variants, these tools are not always informative. There is a need for more refinement methodologies that can be applied in combination with experimental validation to narrow and identify causal variants.
The wide array of genomic, molecular, and immunological reagents available is a valuable asset for mouse research; however, these do not necessarily uniformly work across the CC and should be validated before use. Existing CC-specific resources, such as GECCO, are not necessarily informative for infectious disease research since lymphoid organs, such as the spleen, thymus, and bone marrow, are not profiled. Furthermore, the development of new CC-specific tools, such as CC-derived cell lines (e.g., MEFs and IPSCs), will expand the utility of the CC across fields, including the infectious diseases and immunology fields.
One of the largest challenges across all of model organism research is connecting back to human relevancy. The CC has already proven useful in providing improved models of human-like disease, as highlighted above. A number of genes that have been highlighted in the CC, such as Mx1 and Oas1b, have human paralogs relevant in infectious disease susceptibility (Lin and Brass, 2013, Simon-Loriere et al., 2015), demonstrating the overlap between important genetically diverse infectious disease pathways in mouse and humans. However, the majority of studies have not yet made direct connections back to human datasets. As CC research develops, it will be increasingly important to continue to extend analysis from the CC to human genetics and vice versa. This will require more crosstalk between mouse and human geneticists to bridge gaps in research and communication.
Conclusions
Host immunity and susceptibility to infectious disease is a complex response driven by a multitude of factors, from environment and immune experience to the genetics of both the pathogen and the host. Studying the role of host genetics in infectious disease directly in humans is challenging because of this complexity, and therefore much of the research in the field leans heavily on the mouse as a model organism. While the role of traditional laboratory strains and genetically modified mice in driving advances in the field cannot be understated, these models do not capture the genetic diversity and phenotypic complexity observed in the natural population. Building on the success of classical intercrosses and biparental GRPs, the CC is a highly diverse and reproducible resource for studying and mapping complex traits, including infectious disease susceptibility. The reproducibility of CC strains will allow the community to cross-compare genes and genetic networks that regulate response across pathogens (e.g., IAV and SARS-CoV; Xiong et al., 2014), while also exploring the impact of factors such as the microbiome, co-infections, and genetic variation of the pathogen in the context of a controlled, genetically diverse model population. The CC also holds promise for the development of new models and testing platforms that will better reproduce specific human disease outcomes or host responses to pathogens and vaccines.
Acknowledgments
We would like to thank Sharon Taft-Benz, Sanjay Sarkar, Emily Madden, and Brea Hampton for critical reading of the manuscript. Chromosome ideograms shown in Figure 2 were generated using the karyoploteR package for R (Gel and Serra, 2017). This work was supported by U19 AI100625 and P01AI132130. K.E.N. received support from T32AI007419.
Contributor Information
Martin T. Ferris, Email: mtferris@email.unc.edu.
Mark T. Heise, Email: mark_heisem@med.unc.edu.
References
- Allen J.P., Neely M.N. Trolling for the ideal model host: zebrafish take the bait. Future Microbiol. 2010;5:563–569. doi: 10.2217/fmb.10.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arends D., Prins P., Jansen R.C., Broman K.W. R/qtl: high-throughput multiple QTL mapping. Bioinformatics. 2010;26:2990–2992. doi: 10.1093/bioinformatics/btq565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auer P.L., Lettre G. Rare variant association studies: considerations, challenges and opportunities. Genome Med. 2015;7:16. doi: 10.1186/s13073-015-0138-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey D.W. Recombinant-inbred strains. An aid to finding identity, linkage, and function of histocompatibility and other genes. Transplantation. 1971;11:325–327. doi: 10.1097/00007890-197103000-00013. [DOI] [PubMed] [Google Scholar]
- Barbaric I., Miller G., Dear T.N. Appearances can be deceiving: phenotypes of knockout mice. Brief. Funct. Genomics Proteomics. 2007;6:91–103. doi: 10.1093/bfgp/elm008. [DOI] [PubMed] [Google Scholar]
- Beck J.A., Lloyd S., Hafezparast M., Lennon-Pierce M., Eppig J.T., Festing M.F.W., Fisher E.M.C. Genealogies of mouse inbred strains. Nat. Genet. 2000;24:23–25. doi: 10.1038/71641. [DOI] [PubMed] [Google Scholar]
- Boon A.C.M., deBeauchamp J., Hollmann A., Luke J., Kotb M., Rowe S., Finkelstein D., Neale G., Lu L., Williams R.W., Webby R.J. Host genetic variation affects resistance to infection with a highly pathogenic H5N1 influenza A virus in mice. J. Virol. 2009;83:10417–10426. doi: 10.1128/JVI.00514-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottomly D., Ferris M.T., Aicher L.D., Rosenzweig E., Whitmore A., Aylor D.L., Haagmans B.L., Gralinski L.E., Bradel-Tretheway B.G., Bryan J.T. Expression quantitative trait Loci for extreme host response to influenza a in pre-collaborative cross mice. G3 (Bethesda) 2012;2:213–221. doi: 10.1534/g3.111.001800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouabe H., Okkenhaug K. Gene targeting in mice: a review. Methods Mol. Biol. 2013;1064:315–336. doi: 10.1007/978-1-62703-601-6_23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante J., Boisson-Dupuis S., Abel L., Casanova J.-L. Mendelian susceptibility to mycobacterial disease: genetic, immunological, and clinical features of inborn errors of IFN-γ immunity. Semin. Immunol. 2014;26:454–470. doi: 10.1016/j.smim.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagliani R., Sironi M. Pathogen-driven selection in the human genome. Int. J. Evol. Biol. 2013;2013:204240. doi: 10.1155/2013/204240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmody R.N., Gerber G.K., Luevano J.M., Jr., Gatti D.M., Somes L., Svenson K.L., Turnbaugh P.J. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe. 2015;17:72–84. doi: 10.1016/j.chom.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro-Sampaio M., Coutinho A. Immunity to microbes: lessons from primary immunodeficiencies. Infect. Immun. 2007;75:1545–1555. doi: 10.1128/IAI.00787-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova J.L., Schurr E., Abel L., Skamene E. Forward genetics of infectious diseases: immunological impact. Trends Immunol. 2002;23:469–472. doi: 10.1016/s1471-4906(02)02289-5. [DOI] [PubMed] [Google Scholar]
- Chapman S.J., Hill A.V.S. Human genetic susceptibility to infectious disease. Nat. Rev. Genet. 2012;13:175–188. doi: 10.1038/nrg3114. [DOI] [PubMed] [Google Scholar]
- Chella Krishnan K., Mukundan S., Alagarsamy J., Hur J., Nookala S., Siemens N., Svensson M., Hyldegaard O., Norrby-Teglund A., Kotb M. Genetic architecture of group A Streptococcal necrotizing soft tissue infections in the mouse. PLoS Pathog. 2016;12:e1005732. doi: 10.1371/journal.ppat.1005732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chick J.M., Munger S.C., Simecek P., Huttlin E.L., Choi K., Gatti D.M., Raghupathy N., Svenson K.L., Churchill G.A., Gygi S.P. Defining the consequences of genetic variation on a proteome-wide scale. Nature. 2016;534:500–505. doi: 10.1038/nature18270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill G.A., Airey D.C., Allayee H., Angel J.M., Attie A.D., Beatty J., Beavis W.D., Belknap J.K., Bennett B., Berrettini W., Complex Trait Consortium The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat. Genet. 2004;36:1133–1137. doi: 10.1038/ng1104-1133. [DOI] [PubMed] [Google Scholar]
- Churchill G.A., Gatti D.M., Munger S.C., Svenson K.L. The Diversity Outbred mouse population. Mamm. Genome. 2012;23:713–718. doi: 10.1007/s00335-012-9414-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins M., Thrasher A. Gene therapy: progress and predictions. Proc. Biol. Sci. 2015;282:20143003. doi: 10.1098/rspb.2014.3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionne M.S., Schneider D.S. Models of infectious diseases in the fruit fly Drosophila melanogaster. Dis. Model. Mech. 2008;1:43–49. doi: 10.1242/dmm.000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetschman T. Influence of genetic background on genetically engineered mouse phenotypes. Methods Mol. Biol. 2009;530:423–433. doi: 10.1007/978-1-59745-471-1_23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Xie J., Chang W., Han Y., Cao G. Genome-wide association studies: inherent limitations and future challenges. Front. Med. 2012;6:444–450. doi: 10.1007/s11684-012-0225-3. [DOI] [PubMed] [Google Scholar]
- Duggal P., Thio C.L., Wojcik G.L., Goedert J.J., Mangia A., Latanich R., Kim A.Y., Lauer G.M., Chung R.T., Peters M.G. Genome-wide association study of spontaneous resolution of hepatitis C virus infection: data from multiple cohorts. Ann. Intern. Med. 2013;158:235–245. doi: 10.7326/0003-4819-158-4-201302190-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant C., Tayem H., Yalcin B., Cleak J., Goodstadt L., de Villena F.P., Mott R., Iraqi F.A. Collaborative Cross mice and their power to map host susceptibility to Aspergillus fumigatus infection. Genome Res. 2011;21:1239–1248. doi: 10.1101/gr.118786.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbahesh H., Schughart K. Genetically diverse CC-founder mouse strains replicate the human influenza gene expression signature. Sci. Rep. 2016;6:26437. doi: 10.1038/srep26437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris M.T., Aylor D.L., Bottomly D., Whitmore A.C., Aicher L.D., Bell T.A., Bradel-Tretheway B., Bryan J.T., Buus R.J., Gralinski L.E. Modeling host genetic regulation of influenza pathogenesis in the collaborative cross. PLoS Pathog. 2013;9:e1003196. doi: 10.1371/journal.ppat.1003196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti D.M., Svenson K.L., Shabalin A., Wu L.-Y., Valdar W., Simecek P., Goodwin N., Cheng R., Pomp D., Palmer A. Quantitative trait locus mapping methods for diversity outbred mice. G3 (Bethesda) 2014;4:1623–1633. doi: 10.1534/g3.114.013748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gel B., Serra E. karyoploteR: an R/Bioconductor package to plot customizable genomes displaying arbitrary data. Bioinformatics. 2017;33:3088–3090. doi: 10.1093/bioinformatics/btx346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham J.B., Thomas S., Swarts J., McMillan A.A., Ferris M.T., Suthar M.S., Treuting P.M., Ireton R., Gale M., Jr., Lund J.M. Genetic diversity in the collaborative cross model recapitulates human West Nile virus disease outcomes. MBio. 2015;6 doi: 10.1128/mBio.00493-15. e00493–e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham J.B., Swarts J.L., Wilkins C., Thomas S., Green R., Sekine A., Voss K.M., Ireton R.C., Mooney M., Choonoo G. A mouse model of chronic West Nile virus disease. PLoS Pathog. 2016;12:e1005996. doi: 10.1371/journal.ppat.1005996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham J.B., Swarts J.L., Mooney M., Choonoo G., Jeng S., Miller D.R., Ferris M.T., McWeeney S., Lund J.M. Extensive homeostatic T cell phenotypic variation within the Collaborative Cross. Cell Rep. 2017;21:2313–2325. doi: 10.1016/j.celrep.2017.10.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralinski L.E., Ferris M.T., Aylor D.L., Whitmore A.C., Green R., Frieman M.B., Deming D., Menachery V.D., Miller D.R., Buus R.J. Genome wide identification of SARS-CoV susceptibility loci using the Collaborative Cross. PLoS Genet. 2015;11:e1005504. doi: 10.1371/journal.pgen.1005504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralinski L.E., Menachery V.D., Morgan A.P., Totura A.L., Beall A., Kocher J., Plante J., Harrison-Shostak D.C., Schäfer A., Pardo-Manuel de Villena F. Allelic variation in the Toll-like receptor adaptor protein Ticam2 contributes to SARS-coronavirus pathogenesis in mice. G3 (Bethesda) 2017;7:1653–1663. doi: 10.1534/g3.117.041434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R., Wilkins C., Thomas S., Sekine A., Hendrick D.M., Voss K., Ireton R.C., Mooney M., Go J.T., Choonoo G. Oas1b-dependent immune transcriptional profiles of West Nile virus infection in the Collaborative Cross. G3 (Bethesda) 2017;7:1665–1682. doi: 10.1534/g3.117.041624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick P.W. Population genetics of malaria resistance in humans. Heredity (Edinb) 2011;107:283–304. doi: 10.1038/hdy.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill A.V.S. Evolution, revolution and heresy in the genetics of infectious disease susceptibility. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2012;367:840–849. doi: 10.1098/rstb.2011.0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann E.J., Weidanz W.P., Long C.A. Susceptibility of CXB recombinant inbred mice to murine plasmodia. Infect. Immun. 1984;43:981–985. doi: 10.1128/iai.43.3.981-985.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iraqi F.A., Mahajne M., Salaymah Y., Sandovski H., Tayem H., Vered K., Balmer L., Hall M., Manship G., Morahan G., Collaborative Cross Consortium The genome architecture of the Collaborative Cross mouse genetic reference population. Genetics. 2012;190:389–401. doi: 10.1534/genetics.111.132639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallmann F.J., Reisner D. Twin studies on genetic variations in resistance to tuberculosis. J. Hered. 1943;34:293–301. [Google Scholar]
- Kollmus H., Pilzner C., Leist S.R., Heise M., Geffers R., Schughart K. Of mice and men: the host response to influenza virus infection. Mamm. Genome. 2018;29:446–470. doi: 10.1007/s00335-018-9750-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krištić J., Zaytseva O.O., Ram R., Nguyen Q., Novokmet M., Vučković F., Vilaj M., Trbojević-Akmačić I., Pezer M., Davern K.M. Profiling and genetic control of the murine immunoglobulin G glycome. Nat. Chem. Biol. 2018;14:516–524. doi: 10.1038/s41589-018-0034-3. [DOI] [PubMed] [Google Scholar]
- Leist S.R., Pilzner C., van den Brand J.M.A., Dengler L., Geffers R., Kuiken T., Balling R., Kollmus H., Schughart K. Influenza H3N2 infection of the collaborative cross founder strains reveals highly divergent host responses and identifies a unique phenotype in CAST/EiJ mice. BMC Genomics. 2016;17:143. doi: 10.1186/s12864-016-2483-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenardo M., Lo B., Lucas C.L. Genomics of immune diseases and new therapies. Annu. Rev. Immunol. 2016;34:121–149. doi: 10.1146/annurev-immunol-041015-055620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T.-Y., Brass A.L. Host genetic determinants of influenza pathogenicity. Curr. Opin. Virol. 2013;3:531–536. doi: 10.1016/j.coviro.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorè N.I., Iraqi F.A., Bragonzi A. Host genetic diversity influences the severity of Pseudomonas aeruginosa pneumonia in the Collaborative Cross mice. BMC Genet. 2015;16:106. doi: 10.1186/s12863-015-0260-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangtani P., Abubakar I., Ariti C., Beynon R., Pimpin L., Fine P.E.M., Rodrigues L.C., Smith P.G., Lipman M., Whiting P.F., Sterne J.A. Protection by BCG vaccine against tuberculosis: a systematic review of randomized controlled trials. Clin. Infect. Dis. 2014;58:470–480. doi: 10.1093/cid/cit790. [DOI] [PubMed] [Google Scholar]
- Marsh E.K., May R.C. Caenorhabditis elegans, a model organism for investigating immunity. Appl. Environ. Microbiol. 2012;78:2075–2081. doi: 10.1128/AEM.07486-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masopust D., Sivula C.P., Jameson S.C. Of mice, dirty mice, and men: using mice to understand human immunology. J. Immunol. 2017;199:383–388. doi: 10.4049/jimmunol.1700453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathes W.F., Aylor D.L., Miller D.R., Churchill G.A., Chesler E.J., de Villena F.P., Threadgill D.W., Pomp D. Architecture of energy balance traits in emerging lines of the Collaborative Cross. Am. J. Physiol. Endocrinol. Metab. 2011;300:E1124–E1134. doi: 10.1152/ajpendo.00707.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurizio P.L., Ferris M.T., Keele G.R., Miller D.R., Shaw G.D., Whitmore A.C., West A., Morrison C.R., Noll K.E., Plante K.S. Bayesian diallel analysis reveals Mx1-dependent and Mx1-independent effects on response to influenza A virus in mice. G3 (Bethesda) 2018;8:427–445. doi: 10.1534/g3.117.300438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh K.J., Mandalapu S., Kolls J.K., Ross T.M., Alcorn J.F. A novel outbred mouse model of 2009 pandemic influenza and bacterial co-infection severity. PLoS ONE. 2013;8:e82865. doi: 10.1371/journal.pone.0082865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren P.J., Coulonges C., Bartha I., Lenz T.L., Deutsch A.J., Bashirova A., Buchbinder S., Carrington M.N., Cossarizza A., Dalmau J. Polymorphisms of large effect explain the majority of the host genetic contribution to variation of HIV-1 virus load. Proc. Natl. Acad. Sci. USA. 2015;112:14658–14663. doi: 10.1073/pnas.1514867112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Fuentes V., Cacheiro P., Meehan T.F., Aguilar-Pimentel J.A., Brown S.D.M., Flenniken A.M., Flicek P., Galli A., Mashhadi H.H., Hrabě de Angelis M., IMPC consortium The International Mouse Phenotyping Consortium (IMPC): a functional catalogue of the mammalian genome that informs conservation. Conserv. Genet. 2018;19:995–1005. doi: 10.1007/s10592-018-1072-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedelko T., Kollmus H., Klawonn F., Spijker S., Lu L., Heßman M., Alberts R., Williams R.W., Schughart K. Distinct gene loci control the host response to influenza H1N1 virus infection in a time-dependent manner. BMC Genomics. 2012;13:411. doi: 10.1186/1471-2164-13-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newport M.J., Finan C. Genome-wide association studies and susceptibility to infectious diseases. Brief. Funct. Genomics. 2011;10:98–107. doi: 10.1093/bfgp/elq037. [DOI] [PubMed] [Google Scholar]
- Niazi M.K.K., Dhulekar N., Schmidt D., Major S., Cooper R., Abeijon C., Gatti D.M., Kramnik I., Yener B., Gurcan M., Beamer G. Lung necrosis and neutrophils reflect common pathways of susceptibility to Mycobacterium tuberculosis in genetically diverse, immune-competent mice. Dis. Model. Mech. 2015;8:1141–1153. doi: 10.1242/dmm.020867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIH Knockout mouse fact sheet. 2015. https://www.genome.gov/12514551/knockout-mice-fact-sheet/
- Panoutsakopoulou V., Little C.S., Sieck T.G., Blankenhorn E.P., Blank K.J. Differences in the immune response during the acute phase of E-55+ murine leukemia virus infection in progressor BALB and long term nonprogressor C57BL mice. J. Immunol. 1998;161:17–26. [PubMed] [Google Scholar]
- Peirce J.L., Lu L., Gu J., Silver L.M., Williams R.W. A new set of BXD recombinant inbred lines from advanced intercross populations in mice. BMC Genet. 2004;5:7. doi: 10.1186/1471-2156-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier S., Gingras S., Green D.R. Mouse genome engineering via CRISPR-Cas9 for study of immune function. Immunity. 2015;42:18–27. doi: 10.1016/j.immuni.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelygin A.A., Scherbik S.V., Zhulin I.B., Stockman B.M., Li Y., Brinton M.A. Positional cloning of the murine flavivirus resistance gene. Proc. Natl. Acad. Sci. USA. 2002;99:9322–9327. doi: 10.1073/pnas.142287799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillippi J., Xie Y., Miller D.R., Bell T.A., Zhang Z., Lenarcic A.B., Aylor D.L., Krovi S.H., Threadgill D.W., de Villena F.P. Using the emerging Collaborative Cross to probe the immune system. Genes Immun. 2014;15:38–46. doi: 10.1038/gene.2013.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard C., Bobby Gaspar H., Al-Herz W., Bousfiha A., Casanova J.-L., Chatila T., Crow Y.J., Cunningham-Rundles C., Etzioni A., Franco J.L. International Union of Immunological Societies: 2017 Primary Immunodeficiency Diseases Committee Report on Inborn Errors of Immunity. J. Clin. Immunol. 2018;38:96–128. doi: 10.1007/s10875-017-0464-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quillent C., Oberlin E., Braun J., Rousset D., Gonzalez-Canali G., Métais P., Montagnier L., Virelizier J.L., Arenzana-Seisdedos F., Beretta A. HIV-1-resistance phenotype conferred by combination of two separate inherited mutations of CCR5 gene. Lancet. 1998;351:14–18. doi: 10.1016/S0140-6736(97)09185-X. [DOI] [PubMed] [Google Scholar]
- Qureshi S.T., Larivière L., Leveque G., Clermont S., Moore K.J., Gros P., Malo D. Endotoxin-tolerant mice have mutations in Toll-like receptor 4 (Tlr4) J. Exp. Med. 1999;189:615–625. doi: 10.1084/jem.189.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen A.L., Okumura A., Ferris M.T., Green R., Feldmann F., Kelly S.M., Scott D.P., Safronetz D., Haddock E., LaCasse R. Host genetic diversity enables Ebola hemorrhagic fever pathogenesis and resistance. Science. 2014;346:987–991. doi: 10.1126/science.1259595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A., Pardo-Manuel de Villena F., Wang W., McMillan L., Threadgill D.W. The polymorphism architecture of mouse genetic resources elucidated using genome-wide resequencing data: implications for QTL discovery and systems genetics. Mamm. Genome. 2007;18:473–481. doi: 10.1007/s00335-007-9045-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogala A.R., Morgan A.P., Christensen A.M., Gooch T.J., Bell T.A., Miller D.R., Godfrey V.L., de Villena F.P.M. The Collaborative Cross as a resource for modeling human disease: CC011/Unc, a new mouse model for spontaneous colitis. Mamm. Genome. 2014;25:95–108. doi: 10.1007/s00335-013-9499-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy M.F., Riendeau N., Loredo-Osti J.C., Malo D. Complexity in the host response to Salmonella Typhimurium infection in AcB and BcA recombinant congenic strains. Genes Immun. 2006;7:655–666. doi: 10.1038/sj.gene.6364344. [DOI] [PubMed] [Google Scholar]
- Schoenrock S.A., Oreper D., Farrington J., McMullan R.C., Ervin R., Miller D.R., Pardo-Manuel de Villena F., Valdar W., Tarantino L.M. Perinatal nutrition interacts with genetic background to alter behavior in a parent-of-origin-dependent manner in adult Collaborative Cross mice. Genes Brain Behav. 2018;17:e12438. doi: 10.1111/gbb.12438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastiani G., Olien L., Gauthier S., Skamene E., Morgan K., Gros P., Malo D. Mapping of genetic modulators of natural resistance to infection with Salmonella typhimurium in wild-derived mice. Genomics. 1998;47:180–186. doi: 10.1006/geno.1997.5116. [DOI] [PubMed] [Google Scholar]
- Seok J., Warren H.S., Cuenca A.G., Mindrinos M.N., Baker H.V., Xu W., Richards D.R., McDonald-Smith G.P., Gao H., Hennessy L., Inflammation and Host Response to Injury, Large Scale Collaborative Research Program Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc. Natl. Acad. Sci. USA. 2013;110:3507–3512. doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay T., Kang J. Immunological Genome Project and systems immunology. Trends Immunol. 2013;34:602–609. doi: 10.1016/j.it.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J.R., Huang W., Beak J.Y., Hua K., Gatti D.M., de Villena F.P.-M., Pomp D., Jensen B.C. Quantitative trait mapping in Diversity Outbred mice identifies two genomic regions associated with heart size. Mamm. Genome. 2018;29:80–89. doi: 10.1007/s00335-017-9730-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim N.-L., Kumar P., Hu J., Henikoff S., Schneider G., Ng P.C. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40:W452-7. doi: 10.1093/nar/gks539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon-Loriere E., Lin R.-J., Kalayanarooj S.M., Chuansumrit A., Casademont I., Lin S.-Y., Yu H.-P., Lert-Itthiporn W., Chaiyaratana W., Tangthawornchaikul N. High anti-Dengue virus activity of the OAS gene family is associated with increased severity of Dengue. J. Infect. Dis. 2015;212:2011–2020. doi: 10.1093/infdis/jiv321. [DOI] [PubMed] [Google Scholar]
- Smallwood T.L., Gatti D.M., Quizon P., Weinstock G.M., Jung K.C., Zhao L., Hua K., Pomp D., Bennett B.J. High-resolution genetic mapping in the diversity outbred mouse population identifies Apobec1 as a candidate gene for atherosclerosis. G3 (Bethesda) 2014;4:2353–2363. doi: 10.1534/g3.114.014704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C.M., Proulx M.K., Olive A.J., Laddy D., Mishra B.B., Moss C., Gutierrez N.M., Bellerose M.M., Barreira-Silva P., Phuah J.Y. Tuberculosis susceptibility and vaccine protection are independently controlled by host genotype. MBio. 2016;7:1–13. doi: 10.1128/mBio.01516-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijders A.M., Langley S.A., Kim Y.M., Brislawn C.J., Noecker C., Zink E.M., Fansler S.J., Casey C.P., Miller D.R., Huang Y. Influence of early life exposure, host genetics and diet on the mouse gut microbiome and metabolome. Nat. Microbiol. 2016;2:16221. doi: 10.1038/nmicrobiol.2016.221. [DOI] [PubMed] [Google Scholar]
- Srivastava A., Morgan A.P., Najarian M.L., Sarsani V.K., Sigmon J.S., Shorter J.R., Kashfeen A., McMullan R.C., Williams L.H., Giusti-Rodríguez P. Genomes of the mouse Collaborative Cross. Genetics. 2017;206:537–556. doi: 10.1534/genetics.116.198838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Claire M.C., Ragland D.R., Bollinger L., Jahrling P.B. Animal models of Ebolavirus infection. Comp. Med. 2017;67:253–262. [PMC free article] [PubMed] [Google Scholar]
- Takaki H., Oshiumi H., Shingai M., Matsumoto M., Seya T. Development of mouse models for analysis of human virus infections. Microbiol. Immunol. 2017;61:107–113. doi: 10.1111/1348-0421.12477. [DOI] [PubMed] [Google Scholar]
- Takao K., Miyakawa T. Genomic responses in mouse models greatly mimic human inflammatory diseases. Proc. Natl. Acad. Sci. USA. 2015;112:1167–1172. doi: 10.1073/pnas.1401965111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorven M., Grahn A., Hedlund K.-O., Johansson H., Wahlfrid C., Larson G., Svensson L. A homozygous nonsense mutation (428G-->A) in the human secretor (FUT2) gene provides resistance to symptomatic norovirus (GGII) infections. J. Virol. 2005;79:15351–15355. doi: 10.1128/JVI.79.24.15351-15355.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threadgill D.W., Hunter K.W., Williams R.W. Genetic dissection of complex and quantitative traits: from fantasy to reality via a community effort. Mamm. Genome. 2002;13:175–178. doi: 10.1007/s00335-001-4001-Y. [DOI] [PubMed] [Google Scholar]
- Timmann C., Thye T., Vens M., Evans J., May J., Ehmen C., Sievertsen J., Muntau B., Ruge G., Loag W. Genome-wide association study indicates two novel resistance loci for severe malaria. Nature. 2012;489:443–446. doi: 10.1038/nature11334. [DOI] [PubMed] [Google Scholar]
- Totura A.L., Whitmore A., Agnihothram S., Schäfer A., Katze M.G., Heise M.T., Baric R.S. Toll-like receptor 3 signaling via TRIF contributes to a protective innate immune response to severe acute respiratory syndrome coronavirus infection. MBio. 2015;6 doi: 10.1128/mBio.00638-15. e00638–e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang J.S., Schwartzberg P.L., Kotliarov Y., Biancotto A., Xie Z., Germain R.N., Wang E., Olnes M.J., Narayanan M., Golding H., Baylor HIPC Center. CHI Consortium Global analyses of human immune variation reveal baseline predictors of postvaccination responses. Cell. 2014;157:499–513. doi: 10.1016/j.cell.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdar W., Holmes C.C., Mott R., Flint J. Mapping in structured populations by resample model averaging. Genetics. 2009;182:1263–1277. doi: 10.1534/genetics.109.100727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatratnam A., Furuya S., Kosyk O., Gold A., Bodnar W., Konganti K., Threadgill D.W., Gillespie K.M., Aylor D.L., Wright F.A. Editor’s highlight: Collaborative Cross mouse population enables refinements to characterization of the variability in toxicokinetics of trichloroethylene and provides genetic evidence for the role of PPAR pathway in its oxidative metabolism. Toxicol. Sci. 2017;158:48–62. doi: 10.1093/toxsci/kfx065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vered K., Durrant C., Mott R., Iraqi F.A. Susceptibility to Klebsiella pneumonaie infection in collaborative cross mice is a complex trait controlled by at least three loci acting at different time points. BMC Genomics. 2014;15:865. doi: 10.1186/1471-2164-15-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal S.M., Malo D., Vogan K., Skamene E., Gros P. Natural resistance to infection with intracellular parasites: isolation of a candidate for Bcg. Cell. 1993;73:469–485. doi: 10.1016/0092-8674(93)90135-d. [DOI] [PubMed] [Google Scholar]
- Virgin H.W. The virome in mammalian physiology and disease. Cell. 2014;157:142–150. doi: 10.1016/j.cell.2014.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Sun Y., Fu X., Yu G., Wang C., Bao F., Yue Z., Li J., Sun L., Irwanto A. A large-scale genome-wide association and meta-analysis identified four novel susceptibility loci for leprosy. Nat. Commun. 2016;7:13760. doi: 10.1038/ncomms13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe H., Numata K., Ito T., Takagi K., Matsukawa A. Innate immune response in Th1- and Th2-dominant mouse strains. Shock. 2004;22:460–466. doi: 10.1097/01.shk.0000142249.08135.e9. [DOI] [PubMed] [Google Scholar]
- Xiong H., Morrison J., Ferris M.T., Gralinski L.E., Whitmore A.C., Green R., Thomas M.J., Tisoncik-Go J., Schroth G.P., Pardo-Manuel de Villena F. Genomic profiling of collaborative cross founder mice infected with respiratory viruses reveals novel transcripts and infection-related strain-specific gene and isoform expression. G3 (Bethesda) 2014;4:1429–1444. doi: 10.1534/g3.114.011759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin B., Flint J., Mott R. Using progenitor strain information to identify quantitative trait nucleotides in outbred mice. Genetics. 2005;171:673–681. doi: 10.1534/genetics.104.028902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Malo D., Mott R., Panthier J.J., Montagutelli X., Jaubert J. Identification of new loci involved in the host susceptibility to Salmonella Typhimurium in collaborative cross mice. BMC Genomics. 2018;19:303. doi: 10.1186/s12864-018-4667-0. [DOI] [PMC free article] [PubMed] [Google Scholar]