Abstract
Scope:
We investigated and compared the effects of green tea polyphenols – Polyphenon E (PPE) and black tea polyphenols – theaflavins (TFs) on gut microbiota and development of diabetes in db/db mice.
Methods and results:
Supplementation of PPE (0.1%) in the diet to female db/db mice for 7 weeks decreased fasting blood glucose levels and mesenteric fat, while increasing the serum level of insulin, possibly through protection against β-cell damage. However, TFs were less or not effective. Microbiome analysis through 16S rRNA gene sequencing showed that PPE and TFs treatments significantly altered the bacterial community structure in the cecum and colon, but not in the ileum. The key bacterial phylotypes responding to the treatments were then clustered into 11 co-abundance groups (CAGs). CAGs 6 and 7, significantly increased by PPE but not by TFs, were negatively associated with blood glucose levels. The OTUs in these CAGs were from two different phyla, Firmicutes and Bacteroidetes. CAG 10, decreased by PPE and TFs, was positively associated with blood glucose levels.
Conclusion:
Gut microbiota respond to tea polyphenol treatments as CAGs, instead of taxa. Some of the CAGs associated with the blood glucose lowering effect were enriched by PPE, but not TFs.
Keywords: Tea polyphenols, db/db mice, intestinal microbiota, metabolism, diabetes
Graphical Abstract
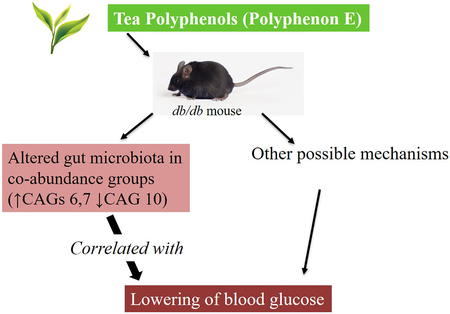
Dietary supplementation of green tea Polyphenon E (PPE) to db/db mice for seven weeks decreased fasting blood glucose levels. It also altered gut microbiota in co-abundance groups (CAGs). CAGs 6 and 7 (increased by PPE) were negatively correlated with blood glucose levels; while CAG10 (decreased by PPE) was positively correlated with blood glucose levels. Black tea theaflavins (TFs) also decreased CAG10, but had little effects on CAGs 6 and 7 as well as on blood glucose levels.
1. Introduction
Green tea extracts and tea polyphenol preparations have shown activities in mitigation or prevention of obesity, diabetes and related diseases [1, 2]. The polyphenols in green tea are known as catechins, which include the most abundant (−)-epigallocatechin-3-gallate (EGCG), as well as other epicatechins (Fig. 1a). Using a high-fat diet (HFD) induced obesity model, we found that supplementation with EGCG significantly reduced body weight gain, visceral fat weights and fasting blood glucose level [3]. In db/db mice, a diabetes model, dietary EGCG was found to prevent the progression of glucose intolerance [4, 5]. Many epidemiological studies and short-term randomized controlled trials (RCT) also showed an association between tea or catechin consumption and lower incidence of metabolic syndrome and diabetes, even though the results are not conclusive [1, 6, 7]. In most studies, the beneficial effects were observed in individuals consuming 3–4 cups of tea (or 600–900 mg of tea catechins) or more daily. Green tea was usually more effective than black tea [1, 2]. The possible mechanisms of action include reduction of fat absorption [1, 3] and promotion of catabolism by EGCG through metabolic regulators, such as AMP-activated protein kinase (AMPK) [1, 8]. However, the exact mechanisms remain elusive.
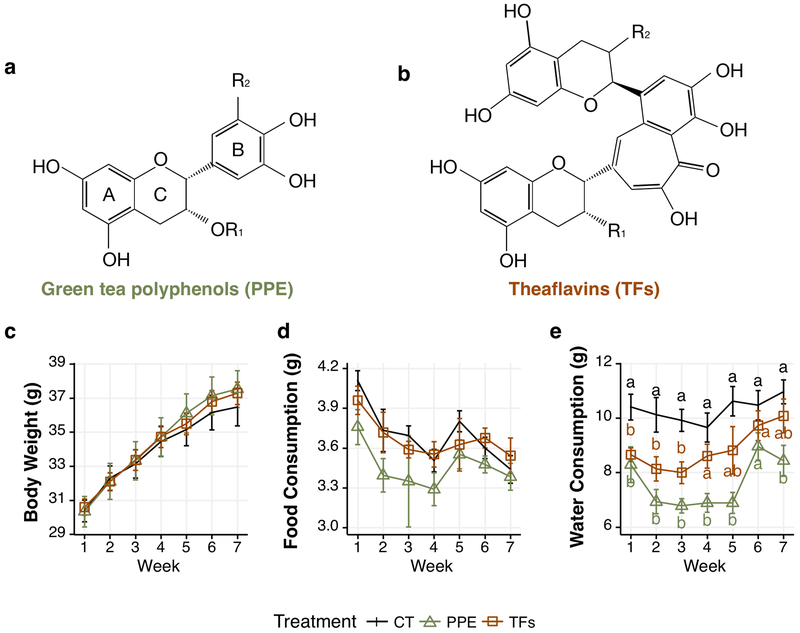
Figure 1.
Effects of PPE and TFs treatment on mouse body weights, food and water consumption. Structures of green tea polyphenols (a) and theaflavins (b); effects of PPE and TFs treatment on mouse body weights (c), food consumption (d) and water consumption (e). For green tea polyphenols – Epigallocatechin 3-gallate (EGCG): R1=galloyl, R2= OH; Epigallocatechin (EGC): R1 = H, R2 = OH; Epicatechin 3-gallate: R1 = galloyl, R2 = H; and Epicatechin: R1= R2= H. For theaflavins – Theaflavin (TF): R1 = R2 = H; TF-3-gallate: R1 = galloyl, R2 = H; TF-3‘-gallate: R1 = H, R2 = galloyl and TF-3,3‘digallate: R1 = R2 = galloyl. Body weight (n=12 mice) and food or water consumption (n=3 cages) were measured 3 times per week. For clarity, only the weekly data are shown as mean ± S.E. a,b indicate difference among groups by ANOVA (p < 0.05).
Many publications reported that dietary tea polyphenols, at different doses, attenuated body weight gain and decreased the ratio of Firmicutes to Bacteroidetes in mice fed a HFD [9–13]. It is unclear, however, whether the results were due to the prevention of HFD-induced changes in microbiota by tea polyphenol treatment, and the role of intestinal microbiota in mediating the beneficial effects of tea, remain unclear. There are few studies [10] dealing with the effects of black tea polyphenols on gut microbes.
In the present work, we investigated and compared the effects of polyphenols from green and black tea on the development of diabetes and on gut microbiome in db/db mice, which develop diabetes and obesity without using a high-fat diet. A standardized and well-studied [14, 15] green tea polyphenol preparation, Polyphenon E (PPE), was used at 0.1% in the diet. This level approximates human consumption of two cups of tea a day. As a comparison, a preparation of black tea theaflavins (TFs) (Fig. 1b), was also used at 0.1% in the diet in this study for seven weeks. Parameters related to diabetes were monitored. Gut microbiota were analyzed using 16S rRNA gene sequencing and the bacteria were characterized as species-level operational taxonomic units (OTUs, at 97% homology cutoff), and grouped in co-abundance groups (CAGs). Some CAGs were found to respond to tea polyphenol treatments and also correlated with the fasting glucose levels of mice.
2. Materials and methods
2.1. Chemicals and animal diet
PPE was a gift from Dr. Yukihiko Hara (Mitsui Norin Co., Shuzuoka, Japan). It contained 65.6% EGCG and other catechins, with a total catechin content of 91%. The sample of TFs was a gift from Dr. Yushun Gong of Hunan Agricultural University (Changsha, Hunan, China), prepared by treating a tea polyphenol solution with crushed tea leaves (to provide phenol oxidase as a catalyst). The theaflavins formed were purified with a column of nanoporous resin. The final product contained 73.7% tea polyphenols by weight. Analysis with HPLC in our laboratory showed that it contained 31.0% theaflavins, with theaflavin (TF), TF-3-gallate, TF-3`-gallate and TF-3,3`-digallate presented at ratios of 1 : 1.8 : 0.44 : 1.89, the remnants are uncharacterized polyphenols. The control diet (AIN93M) and diet supplemented at 1mg/g with PPE or TFs, respectively) were prepared by Research Diets Inc. (New Brunswick, NJ).
2.2. Animals, treatments and monitoring
All animal experiments were carried out in the animal facility in the Department of Chemical Biology under protocol 02–027 of Rutgers University. Female BKS (Cg-Dock7m +/+ Leprdb/J) db/db mice, obtained from Jackson Laboratory (Bar Harbor, ME) at 5 weeks of age, were acclimated on AIN93M diet for ten days. The mice then were randomized into three treatment groups (12 mice per group): Group 1 – control diet, Group 2 – PPE diet and Group 3 – TFs diet. Mice were maintained in plastic cages with corn cob bedding, four mice per cage, in a controlled room (temperature 24 to 25 °C, humidity 70–75%, and lighting regimen of 12-hour light-dark cycles). The location of the 9 animal cages in the rack was rotated on a weekly basis so that the environment of the animals would not be affected by a fixed location. Body weight, liquid consumption and food consumption were monitored three times a week.
Fasting blood glucose levels were measured at 1, 3 and 6 weeks after initiating the experimental diet using test strips on an Ascensia Contour Blood glucose meter (Bayer Healthcare LLC, Mishawaka, IN). On the day of the measurement, cage bedding was changed and the mice were fasted for 8 h (7:30AM to 3:30PM). After blood glucose measurement, a fasting blood sample (~100 µl) was collected from the tail vein. Serum was prepared, divided into small aliquots and stored frozen at −80°C. Serum insulin levels were measured using a Rat/Mouse Insulin ELISA kit (Millipore Corporation, Billerica, MA).
The mice were euthanized on week 7 by CO2 asphyxiation; blood was collected by cardiac puncture, serum was prepared and stored at −80°C. The liver, pancreas, intestine, adipose tissues, and spleen were collected; samples were stored frozen at −80°C or fixed in formalin for analysis.
2.3. Immunohistochemistry (IHC)
Sections of pancreas were deparaffinized, rehydrated and heated in a microwave oven in an antigen unmasking solution (Vector Laboratories, Burlingame, CA) for antigen retrieval. Endogenous peroxidase was quenched with 3% H2O2. The sections were incubated in PBS containing 5% normal serum to block nonspecific binding and incubated with antibodies against insulin (7.5 mg/ml, Cell Signaling, Danvers, MA) overnight at 4°C. The slides were incubated in biotin-conjugated secondary antibody (IgG, 1:200 dilution) and avidin-biotin peroxidase (Vector Laboratories) at room temperature for 1 h each. Negative controls were run in parallel without primary antibody in the incubation. The results of the IHC staining were analyzed with an Aperio ScanScope Scanner (Aperio Technologies, Vista, CA) and calculated using Images Scope software (Aperio Technologies, Inc.).
2.4. Sample collection and analysis of intestinal microbiome
For microbiota analysis, the contents of the ileum (4 cm in length, 3–4 cm from the ileocecal junction), cecum and middle colon (2 cm in length) were extruded into sterile, DNase/RNase-free tubes and flashed frozen in liquid nitrogen. The intestinal mucosa from each segment were rinsed, scraped into a tube and frozen in liquid nitrogen. Thus, six samples (ileal mucosa, ileal content, cecal mucosa, cecal content, colonic mucosa and colonic content) from each mouse were collected. They were stored at −80°C and shipped on dry ice to the Alkek Center for Metagenomics and Microbiome Research (Houston, TX) for DNA extractions and sequencing. DNA was extracted using the PowerSoil DNA Isolation Kit (MO BIO Laboratories). The 16S rRNA gene V4 region (515F-806R) was amplified by PCR and sequenced in the MiSeq platform (Illumina) using the 2 × 250 bp paired-end protocol [16].
The raw sequence was subject to USEARCH v10.0.240 [17] for quality assurance and OTU picking. In brief, the raw sequence was first demultiplexed. The demultiplexed reads then were merged into paired reads and the primer was stripped. For quality filtering, merged reads with expected error threshold larger than 1.0 or read length shorter than 160 was discarded. The quality filtered reads were dereplicated into unique sequences. Based on the abundances of the unique sequences, singletons were discarded. Then the sequences were subject to OTU clustering at 97% similarity. A Chimera filter was built in this OTU clustering implementation; therefore, no additional chimera filtering was needed. An OTU table was constructed by mapping all merged sequences to the OTUs picked by UPARSE. The OTU table were subject to QIIME 1.9.1 [18] for the following analyses: alpha diversity, including number of observed OTUs and Shannon diversity, jackknifed beta diversity at 8600 sequencing depth based on UniFrac distance. Mean value of the jackknifed UniFrac coordinates were plotted. One sample with less than 8600 reads was discarded. The phylogenetic information of the OTUs was obtained using the RDP classifier 11.5 with reference sequencing from 16S rRNA training set 16 of Ribosomal Database Project [19] using a bootstrap cutoff of 50%. The OTU table was then rarefied at the sequencing depth of 8600 for the following analyses.
2.5. Statistical Analysis
For metabolism data, one-way analysis of variance (ANOVA) followed by Tukey multiple range analysis was used to assess the differences among treatment groups. A significance level of p < 0.05 was set for all tests. Methods for statistical analysis of microbiome are shown in the next section. All statistical analyses were performed using software R 3.3.2.
The differences among treatment groups for α-diversity was assessed by Kruskal-Wallis multiple comparison followed by Wilcoxon pairwise comparison for significantly different groups. PERMANOVA test was used for β-diversity to determine microbial composition and structure differences between groups, followed by pairwise PERMANOVA. PERMANOVA test was conducted on the UniFrac distance to determine β-diversity and the difference of microbial composition and structure among groups. This was followed by pairwise PERMANOVA. The p-value was adjusted by False-Discovery Rate (FDR, threshold 0.05) for pairwise comparisons to avoid Type 1 errors. For samples from each location, Random Forest model [20] was applied to identify the OTUs that are different among the control, PPE and TFs groups. The important OTUs were selected by Boruta feature selection using Boruta Package in R [21]. Boruta is an algorithm wrap around the random forest algorithm, which captures all the important, interesting features in response to an outcome variable. The relative abundance of the selected OTUs were shown in heatmap. Each column represents one sample of each sampling site. The columns were arranged by Ward cluster [22] of the Euclidean distance among those samples. Each row of the heatmap corresponds to one OTU. The selected important OTUs were assigned to co-abundance groups (CAGs) based on Ward cluster on SparCC distances. Simply, the OTU table with the selected OTUs in the four sampling sites were subject to SparCC correlation analysis [23]. The SparCC distance was calculated by 1- SparCC correlation, and then used for Ward cluster analysis. A tree was built based on Ward clustering results. PERMANOVA test was conducted along the tree from top to bottom. The nodes with no significant difference (PERMANOVA, p>0.01) are considered as one CAG. The cutoff for PERMANOVA p-value was chosen based on previous publication and the obtained number of CAGs[24]. The relative abundance of each CAG was calculated as the sum of the OTUs belonging to that CAG. One-way ANOVA and Tukey multiple comparison was used to analyze the relative abundance of CAGs among treatment. The OTUs in each CAG were increased or decreased together corresponding to treatments. Therefore, spearman correlation with FDR adjustment for multiple comparison between the relative abundance of the CAGs and the fasting blood glucose levels and serum insulin levels was analyzed.
3. Results
3.1. Body weight gain, food consumption and water consumption
From daily observations, all the animals were in good health (Fig. 1c). The treatments did not significantly affect the body weight and food consumption (Fig. 1c, d). The water consumption of the db/db mice (approximately 10 mL/day) was very high and was significantly decreased in the PPE group and modestly in the TFs group (in weeks 1 to 3) (Fig. 1e).
3.2. Effects on weights of organs and fat pads and levels of blood glucose and insulin
After sacrifice, the weights of the liver, spleen, pancreas and fat pads were measured and expressed as percentages of body weight (Table 1). Liver and spleen weights were not significantly altered by treatments with PPE and TFs. However, pancreas weights were significantly increased (22%) by TFs. PPE treatment significantly decreased the weights of the mesenteric fat pads (by 19%), but did not affect the weights of retroperitoneal fat pads or the total fat. On the other hand, TFs treatment had no effect on the weights of all types of fat pads measured.
Table 1.
Weights of liver, spleen, pancreas, and fat pads (% body weight)
| Control |
PPE |
TFs |
|
|---|---|---|---|
| Liver | 6.92 ± 0.45 | 6.67 ± 0.96 | 7.14 ± 1.06 |
| Spleen | 0.24 ± 0.17 | 0.21 ± 0.08 | 0.23 ± 0.11 |
| Pancreas | 0.27 ± 0.05 a | 0.30 ± 0.11 ab | 0.33 ± 0.06 b |
| Mesenteric fat | 3.27 ± 0.40 a | 2.66 ± 0.38 b | 3.14 ± 0.49 a |
| Peritoneal fat | 7.14 ± 1.05 | 7.81 ± 0.34 | 7.68 ± 0.57 |
| Retroperitoneal fat | 2.73 ± 0.46 | 2.70 ± 0.61 | 2.91 ± 0.66 |
| Total fat | 13.13 ± 1.41 | 13.17 ± 0.85 | 13.73 ± 1.22 |
After sacrificing (at week 7) the mice, the weight of the organs and fat pads were weighted. The data are shown as mean ± SD (n=12).
indicate significant difference among group by ANOVA (p < 0.05).
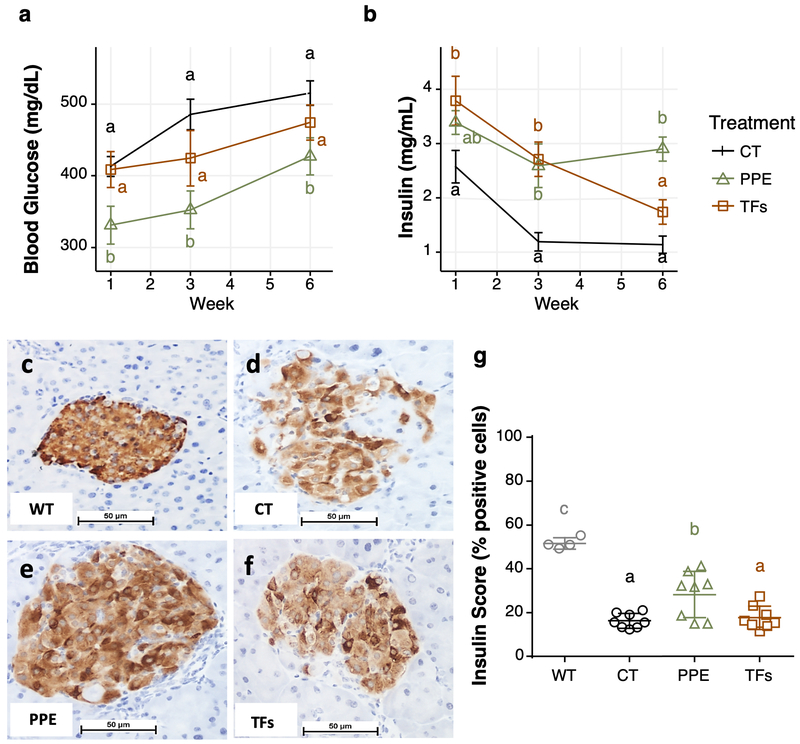
The fasting blood glucose levels were increased due to the deficiency in leptin receptor in the db/db mice model. The mice treated with PPE had significantly lower fasting blood glucose levels compared to the corresponding control groups on weeks 1, 3 and 6. TFs also appeared to lower blood glucose levels on weeks 3 and 6, but the difference was not statistically significant (Fig. 2a). Because of the progressive damage to the pancreatic β-cells with age in db/db mice, blood insulin levels decreased during the experimental period (Fig. 2b). Treatment with PPE significantly prevented the decrease of serum insulin levels on weeks 3 and 6. TFs, however, prevented the decrease in insulin levels on weeks 1 and 3, but not on week 6 (Fig. 2b).
Figure 2.
Effects of PPE and TFs treatments on blood glucose (a), insulin levels (b), insulin staining and insulin score of pancreas samples (c-g). The data for glucose and insulin levels are shown as mean ± S.E. (n=12). Panel C was from a female wild-type mouse at 7 weeks of age and fasted overnight before sacrifice; the size of the pancreas islet was smaller than the db/db mice at age 13 weeks (without fasting), shown in panels d-f. a,b,c indicate difference among groups by ANOVA (p < 0.05).
3.3. Effect on pancreatic pathology and β-cells
The above results suggest that PPE and TFs protected pancreatic β-cells from cellular damage during the development of diabetes in db/db mice. Histopathological examination of the pancreas of db/db mice on week 7 showed areas of confluent acute, chronic or septal inflammation and tissue or fat necrosis, and some islets showed focal hyperplasia and mitotic or apoptotic cells; PPE and TFs treatments did not produce significant changes (results not shown). For insulin IHC, pancreatic tissue slides from db/db mice at week 7 were analyzed together with an untreated female wild-type mouse from a previous experiment (Fig. 2). Islets of the normal wild-type mouse were round to ovoid masses of cells, demonstrating a regular interface with the surrounding exocrine cells, and β-cells were uniformly and densely stained by the anti-insulin antibody (Fig. 2c). Comparatively, islets of db/db mice (control group) were reduced in size and staining intensity; some heavily labeled cells occurred in clusters within the islet, with adjacent β-cells displaying very sparse labeling (Fig. 2d). In the PPE group, the decrease in islet overall size and staining intensity was less extensive than that of the control group; β-cells showed considerable variation of insulin labeling, with some cells intensely stained while others displayed a faint granular pattern, reflecting a reduction of stored insulin granules (Fig. 2e). In the TFs group, the number and size of stained islets were markedly reduced (Fig. 2f), displaying a striking reduction in the number of stained β-cells randomly distributed within the islet, similar to the db/db control group. Analysis of insulin staining intensity with a ScanScope (Fig. 2g) and followed by Tukey multiple comparison test showed that the pancreatic β-cells of db/db mice severely impaired its insulin secretion ability. PPE treatment significantly prevented this impairment, but TFs did not.
3.4. Effects of treatments with PPE and TFs on the intestinal microbiome
Because the gut microbiota change extensively during the development of diabetes in db/db mice [25], this study was not designed as a self-controlled study; rather we compared the microbiome among the control, PPE and TFs groups after 7 weeks of dietary treatment. Fresh samples were collected at six intestinal sites from each mouse for microbiome analysis.
Structural changes of the microbiota in different parts of the intestine.
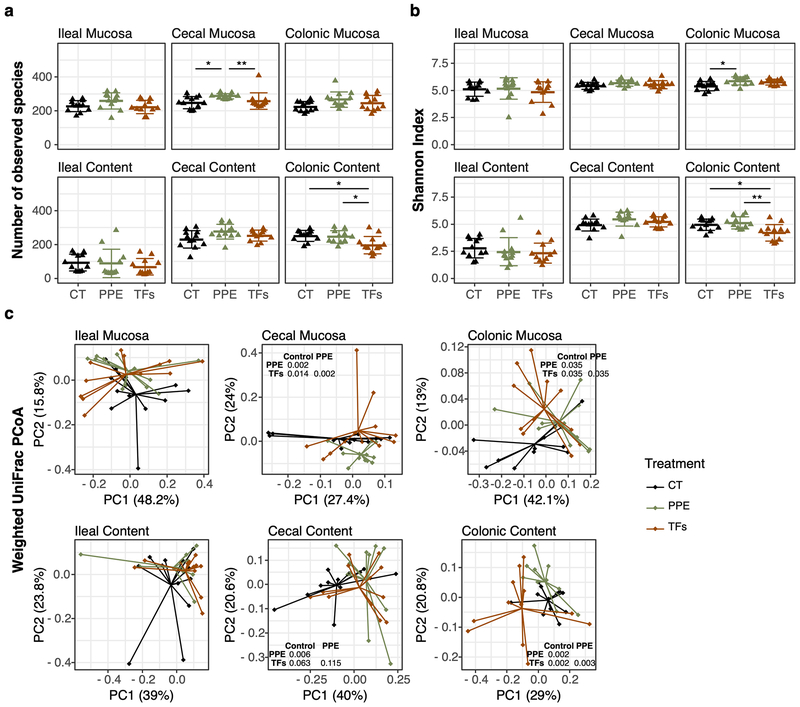
The global structures of gut microbiome were characterized among the samples collected at the six sampling sites. The number of observed OTUs (α-diversity), reflecting the richness of the bacterial community (Fig. 3a), was significantly increased by PPE only in cecal mucosa (p < 0.05). Shannon index, which indicates abundance and evenness of the bacterial community, however, was significantly increased by PPE only in the colonic mucosa (p<0.05) (Fig.3b). On the other hand, TFs treatment decreased α-diversity in colonic content in both α-diversity metrics shown in Fig. 3. β-Diversity was evaluated by the principal coordinate plot on weighted UniFrac distance. The microbiome in the six sampling sites significantly differed from one another (Supplementary Fig. S1). Permutational multivariate analysis of variance (PERMANOVA) [26] of the UniFrac distances [27] showed no significant difference among treatment groups in ileal mucosa and ileal content (Fig. 3c). Lactobacillus was the dominant genus in ileal content (Fig. S2). In the other four sampling sites, PERMANOVA indicated significant differences among treatment groups (Fig 3c). Pairwise PERMANOVA test showed significant differences among treatment groups (PPE vs. control groups, TFs vs control groups, or PPE vs. TFs groups) in three sampling sites: cecal mucosa, colonic mucosa and colonic content (p<0.05). In cecal content, PPE significantly changed the microbiota composition (p<0.05), but TFs did not (p=0.063). Because treatment with PPE and TFs did not significantly affect the microbial compositions of the ileal mucosa and content, these samples were not included in further microbiome analyses.
Figure 3.
α-Diversity and β-diversity of microbiota composition at each sampling site. The number of observed species in each sample (a), Shannon index (b), and weighted UniFrac PCoA (c) are shown. α-Diversities were compared using Wilcoxon test (* p < 0.05, ** p < 0.01, *** p < 0.001). β-Diversities were compared using PERMANOVA among three groups first. If p-value < 0.05, then β-diversities between treatment groups were analyzed by pairwise PERMANOVA, and p-value was listed in each panel.
OTUs responsible for the differences among treatment groups.
The overall microbiome composition from cecal mucosa, cecal content, colonic mucosa and colonic content were significantly different from each other and Random Forest classification models were successfully established [20]. The OTUs responsible for the compositional differences of each treatment group were selected based on the Random Forest models at each sampling site. The relative abundance of the selected OTUs from each sampling site is shown in a heatmap (Fig. S3). There were 32, 30, 30 and 23 OTUs identified that explain the microbial differences among treatment groups in samples of cecal mucosa, colonic content, colonic mucosa and cecal content, respectively. In total 61 unique OTUs were selected by Random Forest algorithm as responsible for the microbial difference among treatment groups. The relative abundance of the 61 OTUs in each treatment group and their taxonomic information are shown in Table S1. As these OTUs were picked due to changes induced by different polyphenol treatments, we hereafter refer them as responding key OTUs. The relative abundance of the OTUs in cecal content were further analyzed in Fig. S4. The results show that, for example, OTU5 Barnesiella was significantly decreased by both PPE and TFs, and OTU12 Desulfovibrio was decreased by TFs (Fig. S4).
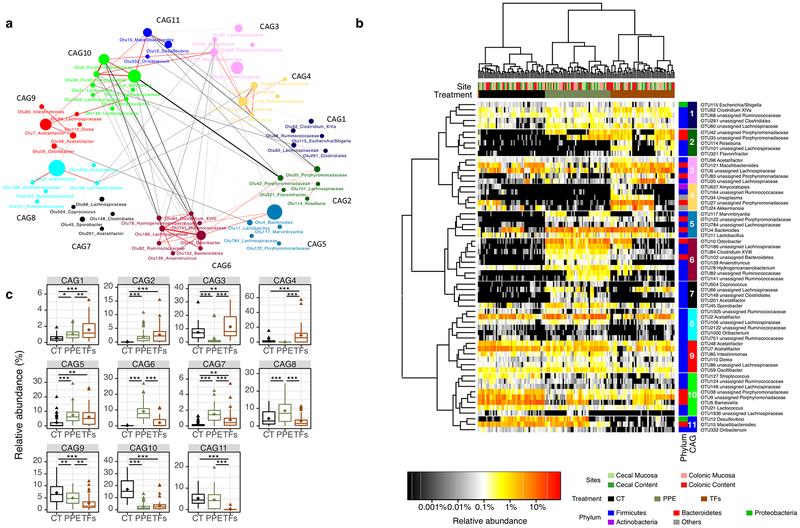
To study the correlation among the OTUs, SparCC analysis [23] was used to calculate the relationship among the responding OTUs. The OTUs that co-occurred were assigned to 11 CAGs. Fig. 4a depicts the interaction among the OTUs and their CAGs. Positive correlations were indicated in red, and negative correlations were shown in grey. CAGs 1, 2, 5 and 6 were negatively correlated with CAGs 9, 10, 11, 3 and 4. Among those, the negative correlation between CAG6 and CAG10 indicated a strong mutual exclusive relationship. The relative abundance of these 61 responding OTUs in the four sampling sites is shown in Fig. 4b. All samples were primarily clustered by treatment groups. The results indicate that the changes of key OTUs, caused by PPE and TFs, were similar throughout the intestinal tract, although the microbiota compositions at different sampling sites were significantly different from one another. Those assignments with less than 50% confidence were designated as unassigned and the higher taxonomy rank was used to identify the OTUs. In the heatmap, the relative abundance of those OTUs in cecum and colon were clustered by treatment (cluster on columns). The average abundance of the CAGs in the four sampling sites are shown in Fig. 4c.
Figure 4.
Co-abundance groups of OTUs that were significantly altered by PPE and/or TFs treatment. Microbial interaction network showed SparCC correlation among the OTUs (a). Each node represents a bacterial OTU and its size is proportional to the relative abundance. OTUs were grouped into different co-abundance groups (CAGs) by PERMANOVA on the SparCC distance tree (999 permutations, p < 0.01). Heatmap shows the relative abundance of the 61 OTUs (b). Colors from black to red indicates the increase of the relative abundance. OTUs (rows) were clustered based on SparCC correlation distance. Samples (columns) were clustered based on Euclidean distance. Both rows and columns were grouped by Ward cluster algorithm. The average abundance of each CAGs among the four sampling sites was shown in boxplot (c). * p < 0.05, ** p < 0.01, *** p < 0.001
Despite the differences in overall microbiome structure in different sampling sites, OTUs were influenced similarly by treatments. Fig. S5 shows the correlation network of the OTUs identified to be the key responding OTUs in each sampling sites. Each circle represents one OTU. The OTUs are colored based on the previously obtained CAG assignments. Although different OTUs existed at different sampling sites, the OTUs belonging to CAG6 were negative correlated with OTUs in CAG10 in all four sampling sites (connected by black lines).
3.5. Association between CAGs and levels of blood glucose and insulin
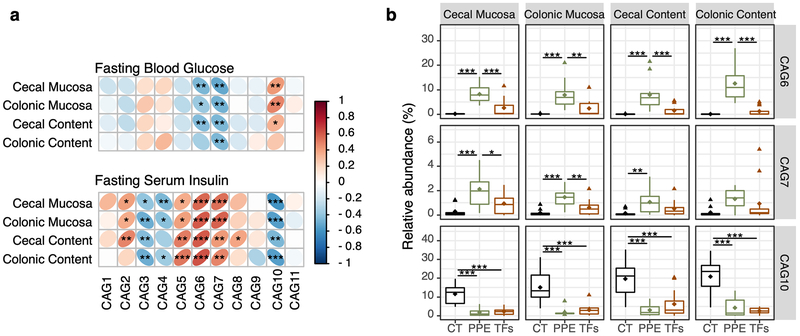
Because non-fasting mice were used for sacrifice at week 7, the fasting blood glucose and insulin levels of week 6 were used to study their association with the microbiome data. Spearman’s rank correlation between the relative abundance of CAGs in the four sampling sites and the fasting blood glucose level at week 6 in each mouse indicated that CAGs 6 and 7 were negatively correlated, while CAG10 was positively correlated, with the fasting blood glucose level (Fig. 5a). Inversely, CAGs 6 and 7 were positively correlated, while CAG10 was negatively correlated, with the fasting serum insulin level (Fig. 5a). In addition, CAGs 2 and 5 were positively correlated, while CAGs 3, 4 and 10 were negatively correlated with the insulin level. Since, the correlation between blood glucose and CAGs 6, 7 and 10 with PPE treatment were most significant (p < 0.01), the effect of PPE on the identified taxa were compared (Table S2). CAGs 6 and 7 were presented in low abundance (less than 1%) in control group, and significantly increased by PPE, but not or slightly increased by TFs. CAG10 were decreased by both PPE and TFs (Fig 5b). The taxonomic information of OTUs in CAGs 6, 7 and 10 are listed in Table S2 and the relative abundance of those OTUs in each treatment groups were listed in Fig S6.
Figure 5.
CAGs correlated with glucose and insulin levels at different sampling sites. Spearman correlation of the relative abundance of each CAG from cecal mucosa, cecal content, colonic mucosa, and colonic content with fasting blood glucose levels and serum insulin levels at week 6 (a). The relative abundance of CAGs 6, 7 and 10 in each of the four sampling sites are shown (b). * p < 0.05, ** p < 0.01, *** p < 0.001.
4. Discussion
In this study, we demonstrated that oral administration of a standardized green tea polyphenol preparation PPE (0.1% in the diet) to db/db mice significantly decreased fasting blood glucose levels. It also increased blood insulin levels and reduced the damage to pancreatic β-cells. These results suggest that PPE protected pancreatic β-cells from cellular damage during the progression of the diabetic state in db/db mice, possibly through its antioxidant activity [4]. PPE also reduced water consumption of db/db mice, an effect which has been reported with antidiabetic agents, such as rosiglitazone [28]. However, TFs was less or not effective.
The intestinal microbiota has been associated with the development of type 2 diabetes [29], and altering the microbiota structure has been shown to alleviate this disease [30]. Our results showed that OTUs from the same taxa could respond differently to the treatment by tea polyphenols and OTUs from different taxa could respond similarly in CAGs to the treatment (Supplementary Table 1). Such a response pattern has also been reported in studies with other agents, for example, fiber [30]. The CAG approach is an effective method to reduce the dimension of the dataset, especially for functional studies. We found that both PPE and TFs changed the gut microbiota but in different ways, commensurate with their health effects. OTUs in CAGs 6 and 7, which were increased by PPE but not by TFs, were negatively associated with fasting blood glucose levels, suggesting their glucose lowering effects. The most abundant member of these CAGs – OTU10 – belongs to the genus of Odoribacter, whose lowered abundance has been observed in Crohn’s disease [31]. Two OTUs belong to the Lachnospiraceae family, which are common butyrate producing commensal group in gut microbiota [32, 33], and their restoration has been associated with the remission of recurrent Clostridium difficile infection [34]. Despite these known beneficial effects, the contributions of these bacteria to the blood glucose lowering effect are still unclear. OTUs in CAG 10, which were decreased by PPE and TFs and positively associated with the blood glucose levels, are likely potential contributing factors to the development of diabetes in db/db mice. The most abundant OTU in CAG10 was in Barnesiella, which was decreased significantly by PPE and TFs. Barnesiella has been reported to be enriched in db/db mice compared to the wild-type counterpart [25], and its decrease may provide beneficial effects in regulating glucose metabolism. The microbiota altered by PPE may change the subsequent short chain fatty acid production. Upon binding to their receptor – free fatty acid receptors 2 and 3 (FFA2 and FFA3), these fatty acids have been suggested to increase insulin secretion and beta-cell survival and proliferation.[35].
The shift of microbiome by tea polyphenols may be caused by their selective anti-microbial activities[25, 36]. The bacteria that can degrade and utilize, or are resistant to, tea polyphenols could flourish and contribute to the overall microbiome shift. Some previous studies showed decreased Firmicutes to Bacteroidetes ratio by green tea polyphenols [9–13]. In our study, PPE significantly increased the ratio in the intestine, except in cecal mucosa and colonic content (Fig. S7). Nevertheless, presenting results on taxa may not be that useful in functional studies, because different bacterial species in the same phylum may have different functions. Bacteria from different phyla can cooperate through reciprocal feedback or cross feeding, and respond similarly to the experimental conditions in CAGs [37]. For the first time, our study identified CAGs of bacteria that were associated with the improved regulation of glucose metabolism in PPE treated db/db mice. Most of the OTUs in these CAGs can only be identified to the family level, indicating that they are novel and not well characterized before. OTUs in the same CAG can be identified to different phyla. For example, OTU10 Odoribacter in the phylum of Bacteroidetes and OTU186, an unidentified Lachnospiraceae in the phylum of Firmicutes, were both increased by PPE and were assigned to the same CAG. The present study forms the basis for a functional guild approach in studying the role of gut microbiome in mediating health effects by tea and other dietary factors.
In summary, green tea PPE is shown to have beneficial effects in lowering fasting blood glucose level in db/db mice. Such an effect is positively correlated with increased abundance of bacteria in CAGs 6 and 7, while negatively correlated with guild CAG10, in the cecum and colon. On the other hand, the black tea TFs only decreased the abundance of CAG10 and did not significantly decrease blood glucose levels. The presently observed correlations suggest that altering specific functional groups of the gut microbiota could significantly contribute to the beneficial effects of green tea polyphenols, in addition to other mechanisms as discussed above [1, 4]. Further research, including studying microbiome as functional guilds and their further characterizations, is needed to further understand the extent and entailed mechanisms of the contribution of gut microbiome in mediating the health effects of tea polyphenols.
Supplementary Material
Acknowledgements
We thank Dr. Y. Hara (Mitsui Norin Co.) and Dr. Y. Gong (Hunan Agricultural University) for providing the PPE and TFs for our study; Rutgers Discovery Informatics Institute (RDI2) and Advanced Computing and Data Cyberinfrastructure for providing computing system; Mr. Jing Wang and Dr. Guojun Wu of Jiao Tong University for technical support on the bioinformatics data analysis; and Ms. Vi Dan for assistance in the preparation of this manuscript. This work was supported by a pilot project award from CEED at Rutgers University (NIH grant ES005022), and NIH grant CA133024 (to CSY), and NIH grant ES023512 to the Center for Precision Environmental Health at the Baylor College of Medicine (NJA, MCR). The raw Illumina sequence data has been uploaded to the sequence read archive (SRA) at NCBI under Bioproject accession PRJNA480760.
Abbreviations:
- PPE
Polyphenone E
- TFs
Preparation of theaflavins
- EGCG
(−)-epigallocatechin-3-gallate
- AMPK
AMP-activated protein kinase
- T2D
Type 2 diabetes
- IHC
Immunohistochemistry
- ANOVA
Analysis of variance
- PERMANOVA
Permutational multivariate analysis of variance
- CAGs
Co-abundance groups
- OTUs
Operational taxonomic units
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- [1].Yang CS, Zhang J, Zhang L, Huang J, Wang Y, Mechanisms of body weight reduction and metabolic syndrome alleviation by tea. Mol Nutr Food Res 2016, 60, 160–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Yang CS, Hong J, Prevention of chronic diseases by tea: Possible mechanisms and human relevance. Annu Rev Nutr 2013, 33, 161–181. [DOI] [PubMed] [Google Scholar]
- [3].Bose M, Lambert JD, Ju J, Reuhl KR, Shapses SA, Yang CS, The major green tea polyphenol, (−)-epigallocatechin-3-gallate, inhibits obesity, metabolic syndrome, and fatty liver disease in high-fat-fed mice. The Journal of nutrition 2008, 138, 1677–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ortsater H, Grankvist N, Wolfram S, Kuehn N, Sjoholm A, Diet supplementation with green tea extract epigallocatechin gallate prevents progression to glucose intolerance in db/db mice. Nutr Metab (Lond) 2012, 9, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wein S, Schrader E, Rimbach G, Wolffram S, Oral green tea catechins transiently lower plasma glucose concentrations in female db/db mice. J Med Food 2013, 16, 312–317. [DOI] [PubMed] [Google Scholar]
- [6].Li Y, Wang C, Huai Q, Guo F, Liu L, Feng R, Sun C, Effects of tea or tea extract on metabolic profiles in patients with type 2 diabetes mellitus: A meta-analysis of ten randomized controlled trials. Diabetes Metab Res Rev 2016, 32, 2–10. [DOI] [PubMed] [Google Scholar]
- [7].Yu J, Song P, Perry R, Penfold C, Cooper AR, The effectiveness of green tea or green tea extract on insulin resistance and glycemic control in type 2 diabetes mellitus: A meta-analysis. Diabetes Metab J 2017, 41, 251–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hardie DG, Ampk: Positive and negative regulation, and its role in whole-body energy homeostasis. Current opinion in cell biology 2015, 33, 1–7. [DOI] [PubMed] [Google Scholar]
- [9].Guo XJ, Cheng M, Zhang X, Cao JX, Wu ZF, Weng PF, Green tea polyphenols reduce obesity in high-fat diet-induced mice by modulating intestinal microbiota composition. Int J Food Sci Tech 2017, 52, 1723–1730. [Google Scholar]
- [10].Henning SM, Yang J, Hsu M, Lee RP, Grojean EM, Ly A, Tseng CH, Heber D, Li Z, Decaffeinated green and black tea polyphenols decrease weight gain and alter microbiome populations and function in diet-induced obese mice. Eur J Nutr 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Remely M, Ferk F, Sterneder S, Setayesh T, Roth S, Kepcija T, Noorizadeh R, Rebhan I, Greunz M, Beckmann J, Wagner KH, Knasmuller S, Haslberger AG, Egcg prevents high fat diet-induced changes in gut microbiota, decreases of DNA strand breaks, and changes in expression and DNA methylation of dnmt1 and mlh1 in c57bl/6j male mice. Oxid Med Cell Longev 2017, 2017, 3079148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Cheng M, Zhang X, Miao Y, Cao J, Wu Z, Weng P, The modulatory effect of (−)-epigallocatechin 3-o-(3-o-methyl) gallate (egcg3''me) on intestinal microbiota of high fat diet-induced obesity mice model. Food Res Int 2017, 92, 9–16. [DOI] [PubMed] [Google Scholar]
- [13].Singh DP, Singh J, Boparai RK, Zhu J, Mantri S, Khare P, Khardori R, Kondepudi KK, Chopra K, Bishnoi M, Isomalto-oligosaccharides, a prebiotic, functionally augment green tea effects against high fat diet-induced metabolic alterations via preventing gut dysbacteriosis in mice. Pharmacol Res 2017, 123, 103–113. [DOI] [PubMed] [Google Scholar]
- [14].Chow HH, Hakim IA, Vining DR, Crowell JA, Ranger-Moore J, Chew WM, Celaya CA, Rodney SR, Hara Y, Alberts DS, Effects of dosing condition on the oral bioavailability of green tea catechins after single-dose administration of polyphenon e in healthy individuals. Clin Cancer Res 2005, 11, 4627–4633. [DOI] [PubMed] [Google Scholar]
- [15].Joe AK, Schnoll-Sussman F, Bresalier RS, Abrams JA, Hibshoosh H, Cheung K, Friedman RA, Yang CS, Milne GL, Liu DD, Lee JJ, Abdul K, Bigg M, Foreman J, Su T, Wang X, Ahmed A, Neugut AI, Akpa E, Lippman SM, Perloff M, Brown PH, Lightdale CJ, Phase ib randomized, double-blinded, placebo-controlled, dose escalation study of polyphenon e in patients with barrett's esophagus. Cancer Prev Res (Phila) 2015, 8, 1131–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R, Ultra-high-throughput microbial community analysis on the illumina hiseq and miseq platforms. ISME J 2012, 6, 1621–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Edgar RC, Uparse: Highly accurate otu sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [DOI] [PubMed] [Google Scholar]
- [18].Kuczynski J, Stombaugh J, Walters WA, Gonzalez A, Caporaso JG, Knight R, Using qiime to analyze 16s rrna gene sequences from microbial communities. Curr Protoc Bioinformatics 2011, Chapter 10, Unit 10 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wang Q, Garrity GM, Tiedje JM, Cole JR, Naive bayesian classifier for rapid assignment of rrna sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Liaw A, Wiener M, Classification and regression by randomforest. R News 2002, 2, 18–22. [Google Scholar]
- [21].Kursa MB, Rudnicki WR, Feature selection with the boruta package. Journal of Statistical Software 2010, 36, 1–13. [Google Scholar]
- [22].Murtagh F, Legendre P, Ward’s hierarchical agglomerative clustering method: Which algorithms implement ward’s criterion? Journal of Classification 2014, 31, 274–295. [Google Scholar]
- [23].Friedman J, Alm EJ, Inferring correlation networks from genomic survey data. PLoS Comput. Biol. 2012, 8, e1002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhang Q, Wu Y, Wang J, Wu G, Long W, Xue Z, Wang L, Zhang X, Pang X, Zhao Y, Zhao L, Zhang C, Accelerated dysbiosis of gut microbiota during aggravation of dss-induced colitis by a butyrate-producing bacterium. Sci. Rep. 2016, 6, 27572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Geurts L, Lazarevic V, Derrien M, Everard A, Van Roye M, Knauf C, Valet P, Girard M, Muccioli GG, Francois P, de Vos WM, Schrenzel J, Delzenne NM, Cani PD, Altered gut microbiota and endocannabinoid system tone in obese and diabetic leptin-resistant mice: Impact on apelin regulation in adipose tissue. Front Microbiol 2011, 2, 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Dixon P, Vegan, a package of r functions for community ecology. Journal of Vegetation Science 2003, 14, 927–930. [Google Scholar]
- [27].Lozupone C, Knight R, Unifrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Chodavarapu H, Grobe N, Somineni HK, Salem ES, Madhu M, Elased KM, Rosiglitazone treatment of type 2 diabetic db/db mice attenuates urinary albumin and angiotensin converting enzyme 2 excretion. PLoS One 2013, 8, e62833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, Peng Y, Zhang D, Jie Z, Wu W, Qin Y, Xue W, Li J, Han L, Lu D, Wu P, Dai Y, Sun X, Li Z, Tang A, Zhong S, Li X, Chen W, Xu R, Wang M, Feng Q, Gong M, Yu J, Zhang Y, Zhang M, Hansen T, Sanchez G, Raes J, Falony G, Okuda S, Almeida M, LeChatelier E, Renault P, Pons N, Batto JM, Zhang Z, Chen H, Yang R, Zheng W, Li S, Yang H, Wang J, Ehrlich SD, Nielsen R, Pedersen O, Kristiansen K, Wang J, A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012, 490, 55–60. [DOI] [PubMed] [Google Scholar]
- [30].Zhao L, Zhang F, Ding X, Wu G, Lam YY, Wang X, Fu H, Xue X, Lu C, Ma J, Yu L, Xu C, Ren Z, Xu Y, Xu S, Shen H, Zhu X, Shi Y, Shen Q, Dong W, Liu R, Ling Y, Zeng Y, Wang X, Zhang Q, Wang J, Wang L, Wu Y, Zeng B, Wei H, Zhang M, Peng Y, Zhang C, Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science 2018, 359, 1151–1156. [DOI] [PubMed] [Google Scholar]
- [31].Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, Bousvaros A, Korzenik J, Sands BE, Xavier RJ, Huttenhower C, Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012, 13, R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sagheddu V, Patrone V, Miragoli F, Puglisi E, Morelli L, Infant early gut colonization by lachnospiraceae: High frequency of ruminococcus gnavus. Front Pediatr 2016, 4, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Duncan SH, Barcenilla A, Stewart CS, Pryde SE, Flint HJ, Acetate utilization and butyryl coenzyme a (coa):Acetate-coa transferase in butyrate-producing bacteria from the human large intestine. Appl. Environ. Microbiol. 2002, 68, 5186–5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Song Y, Garg S, Girotra M, Maddox C, von Rosenvinge EC, Dutta A, Dutta S, Fricke WF, Microbiota dynamics in patients treated with fecal microbiota transplantation for recurrent clostridium difficile infection. PLoS One 2013, 8, e81330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Priyadarshini M, Navarro G, Layden BT, Gut microbiota: Ffar reaching effects on islets. Endocrinology 2018, 159, 2495–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hui X, Yue Q, Zhang DD, Li H, Yang SQ, Gao WY, Antimicrobial mechanism of theaflavins: They target 1-deoxy-d-xylulose 5-phosphate reductoisomerase, the key enzyme of the mep terpenoid biosynthetic pathway. Sci. Rep. 2016, 6, 38945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Rakoff-Nahoum S, Foster KR, Comstock LE, The evolution of cooperation within the gut microbiota. Nature 2016, 533, 255–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.