Abstract
The amino acid sequence rules that specify β-sheet structure in proteins remain obscure. A subclass of β-sheet proteins, parallel β-helices, represent a processive folding of the chain into an elongated topologically simpler fold than globular β-sheets. In this paper, we present a computational approach that predicts the right-handed parallel β-helix supersecondary structural motif in primary amino acid sequences by using β-strand interactions learned from non-β-helix structures. A program called BETAWRAP (http://theory.lcs.mit.edu/betawrap) implements this method and recognizes each of the seven known parallel β-helix families, when trained on the known parallel β-helices from outside that family. BETAWRAP identifies 2,448 sequences among 595,890 screened from the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/) nonredundant protein database as likely parallel β-helices. It identifies surprisingly many bacterial and fungal protein sequences that play a role in human infectious disease; these include toxins, virulence factors, adhesins, and surface proteins of Chlamydia, Helicobacteria, Bordetella, Leishmania, Borrelia, Rickettsia, Neisseria, and Bacillus anthracis. Also unexpected was the rarity of the parallel β-helix fold and its predicted sequences among higher eukaryotes. The computational method introduced here can be called a three-dimensional dynamic profile method because it generates interstrand pairwise correlations from a processive sequence wrap. Such methods may be applicable to recognizing other beta structures for which strand topology and profiles of residue accessibility are well conserved.
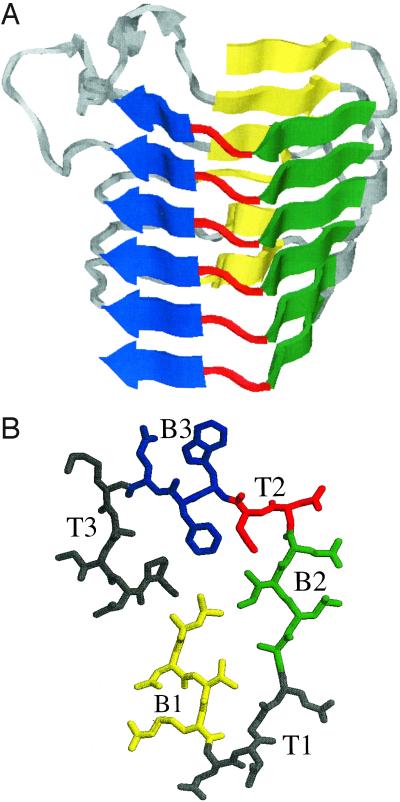
The right-handed parallel β-helix motif, first reported by Jurnak et al. (1), is characterized by a series of processive coils, each of which contributes to the three long β-sheets that come together in a triangular prism shape to comprise the fold (Fig. 1A). The cross-section, or rung, of a parallel β-helix consists of three β-strands connected by variable-length turn regions (Fig. 1B); the backbone folds up in a helical fashion with β-strands from adjacent rungs stacking on top of each other in a parallel orientation. The buried cylindrical core is predominantly composed of hydrophobic amino acids, as in globular β-sheets. However a distinct feature is the presence of stacks of hydrophobic side chains, as well as ladders of hydrogen bonding side chains such as asparagines in some structures in the superfamily (2). Although the known parallel β-helices vary in the number of complete rungs and in the lengths of the turn regions, the β-strand portions of the rungs have patterns of pleating and hydrogen bonding that are well conserved across the superfamily (3). The right-handed parallel β-helix motif (henceforth called β-helix) is not common, with 12 known three-dimensional structures in the Protein Data Bank (PDB; http://www.rcsb.org/pdb) (4). The β-helix structures include pectate lyases, important for bacterial infection of plants, the phage P22 tailspike adhesin, which binds the O-antigen of Salmonella typhimurium, and the P.69 pertactin toxin from Bordetella pertussis, the cause of Whooping Cough.
Figure 1.
(A) Side view of x-ray crystal structure (1) of Pectate lyase C from Erwinia chrysanthemi, residue 102–258, generated using the molecular graphics program rasmol (29). (B) Top view of a single rung of a β-helix (residues 242–263 of A), with β-strands B1 in yellow, B2 in green, and B3 in blue, and the intervening turns T1, T2 (in red), and T3. The alternating pattern of the strands before and after T2 and the T2 turn itself are conserved across the superfamily (3, 8).
The simple, repeating units of structure in the β-helices make them easy to classify from inspection of their three-dimensional structure; however, there is no regular repeat at the sequence level. Furthermore, the known β-helix proteins in different families according to the Structural Classification of Proteins (SCOP) database (5) mostly exhibit very low sequence homology with one another, making them unamenable to multiple sequence alignment methods such as psi-blast (6) and hmmer (7). Many of the new proteins predicted in this paper to form β-helices from their sequence information are not found when general sequence alignment methods (6, 7) are applied to the known β-helix structures.
Computational methods to recognize the β-helix fold must thus find a way to use more structural information. Heffron et al. (8) proposed a method to recognize β-helices based on a sequence-based profile of a pectate lyase template. Based on their template, they predicted potential β-helix folds in several sequences of unsolved structure, including additional pectate lyases. However, this method did not recognize known β-helices from outside the pectate and pectin lyase families. Likewise, threading methods (9) primarily find with reasonable confidence levels sequences from the same family as the query sequence, except in the case of the pectate and pectin lyases, which are also found to be similar by sequence-based methods. Examination of the solved β-helix structures (2, 10), together with analysis of mutants defective in the folding of β-helices (11), suggested that the interactions of the strand side chains in the buried core were critical determinants of the fold. We incorporated this interior packing emphasis in the development of betawrap.
It has been known for some time that in β-structural motifs amino acid residues that are close in space in the folded protein can exhibit marked statistical preferences, but using these correlations for prediction seemed difficult (12–16). betawrap successfully predicts the β-helix by dynamically parsing an amino acid sequence into stacking β-strands separated by variable and fixed length turns. It identifies possible parses by exploiting statistical preferences based on pairwise correlations between aligned residues in adjacent rungs. As such, it is a spatial generalization of window-based methods for recognizing secondary and supersecondary α-helical motifs (17–19). These pairwise correlations were learned from a database of β-strand interactions obtained from amphipathic β-sheets in non-β-helix structures. It was assumed that the core packing interactions within globular β-sheets would have the same general character as the core packing interactions within β-helices so that the alignment correlations could be learned from non-β-helix proteins. This avoided over-training on the small set of known solved β-helix structures.
The betawrap program, which implements the three-dimensional dynamic profile method presented here, does not produce any false positives or false negatives when tested on the PDB (4). Moreover, when run on the NCBI nonredundant protein sequence database, the program's top 200 scoring protein sequences contained over 60 diverse bacterial and fungal proteins that play a role in human infectious disease. In addition, high-scoring sequences from higher eukaryotes were significantly underrepresented, with only two sequences total from among human, mouse, fly, and worm sequences.
Methods
betawrap assigns a score and a corresponding Z-score to an amino acid sequence as outlined in this paragraph. First it identifies likely locations for the well conserved B2-T2-B3 rung segment (Fig. 1B) by using a simple hydrophobic-residue sequence pattern. From each such segment it searches forward and backward in the sequence for potential neighboring rungs that align well, using a rung–rung alignment score as explained below. This score incorporates the β-sheet pairwise correlations and additional information on turn lengths and stacking preferences from the known β-helices. Repeating this search process with the five best-scoring candidates in each direction, betawrap constructs a tree, with the candidate B2-T2-B3 segment as the root, of potential wraps of the sequence into a β-helical structure. After filtering the wraps to interleave the B1-strands and avoid α-helical and transmembrane regions, betawrap assigns a score to the sequence that is the average of the top ten scores over all generated wraps from all likely candidate initial segments.
PDB-minus, a nonredundant version of the PDB (4), with β-helices removed, was constructed from the PDB-select 25% list of June 2000 (20). (PDB-select is a subset of the PDB in which no two proteins have sequence similarity greater than a cutoff; in this case, 25%.) The database contained 1,346 sequences. The SCOP database (5) was used for classification of protein structures, with the exception of the pectin methylesterase protein from E. chrysanthemi (PDB ID code 1qjv), which was only recently solved and has not yet been placed in the SCOP database. Because of its low sequence and structural homology to other known β-helices, we placed it by itself as one of the seven families in the β-helix superfamily. In analyzing sequences identified by betawrap, hidden Markov models from the Pfam database (21) were used to assign protein families to sequences of unsolved structure.
The β-structure database of aligned residue pairs in amphipathic β-sheets was constructed from PDB-minus (with membrane proteins removed) by using the program stride (22). Amphipathic β-sheets were detected by using the residue surface accessibility values as reported by stride, and the hydrogen bonding patterns were used to determine residue alignment in the sheets. The method incorporated all sheets in PDB-minus whose residue surface accessibility values alternated between <0.05 and >0.15. There were 650 chains, all from non-β-helix protein structures, that contributed β-sheets or portions of sheets to this database. Mixed and antiparallel sheets were included in the database because there were not sufficiently many amphipathic parallel β-sheets to generate robust alignment statistics. Although alignment preferences differ somewhat between parallel and antiparallel β-sheets (14), there is some indication that the parallel β-sheets of the β-helices may have some features of antiparallel β-sheets (2).
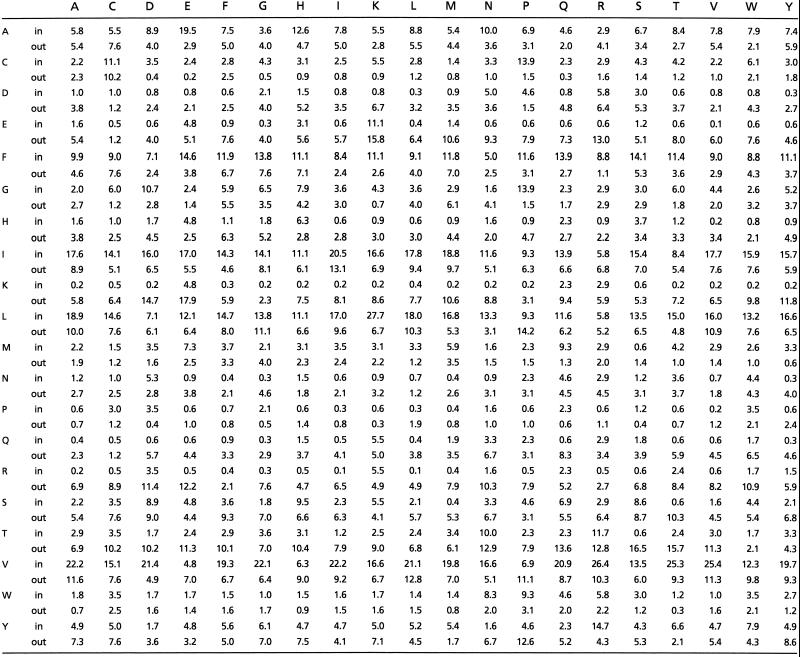
A rung–rung alignment score is calculated as follows. Residue pairs in the β-structure database are grouped into two classes depending on whether they are buried or exposed, and their pairwise frequencies are tabulated in each class. The conditional probability that a residue of type X will align with residue Y, given their orientation relative to the core, is estimated from the database by using standard methods (17). These conditional probabilities are shown in Table 1; for each of the 20 amino acids, the corresponding pair of rows (one for the inward orientation relative to the core and one for the outward) give the conditional probabilities of seeing that residue in alignment with the residues indexing the columns (for pairs that were not seen in the database, such as a buried arginine–arginine pair, the single-residue probabilities, conditioned on residue orientation relative to the core, were used for scoring). The natural logarithm of this conditional probability gives the pair score of a vertical alignment of two residues. The raw score of a rung–rung alignment is then calculated as the weighted sum of the seven alignment scores for the aligned pairs in the β-strands B2 and B3 (a weight of 1 is given to the scores for inward pairs and ½ for the scores of the outward pairs, to reflect the fact that the environment of the inward residues is better conserved between β-helices than that of the outer pairs).
Table 1.
β-sheet alignment probabilities (*100) used by betawrap
The rung–rung alignment score is computed from the raw rung–rung alignment score by incorporating bonuses and penalties learned from the known β-helices in the training set. These adjustments capture turn length distributions (a penalty of −1 for each standard deviation away from the mean rung–rung sequence separation); the avoidance of large hydrophobic residues at the turn positions that bound the β-strands (a penalty equal to the additional number of such residues as compared with sequences of the training set); and preferences for stacks of aliphatic, aromatic, and polar residues (3) in the core of the β-helix domain (a bonus of +1 for each stacked pair). The score attached to a wrap, or collection of stacked rungs, is the average of the rung–rung alignment scores over the stacked pairs in the wrap.
Wraps of an amino acid sequence into the β-helical structure are generated by a branched search starting from likely initial rungs and guided by the rung–rung alignment score. Initial B2-T2-B3 segments are detected by using a hydrophobic sequence pattern that reflects the conserved pattern of pleating in this segment. From each such segment, the algorithm searches forward and backward in sequence for sequence segments that align well according to the rung–rung alignment score (these segments need not match the hydrophobic sequence pattern). This search is repeated from each of the top-ranking aligned segments, and the process is iterated to generate a tree of wraps of the sequence into stacked rungs, all of which contain the initial rung segment. Experimentation led to an optimal wrap size of five rungs together with a branching factor of five for the search tree. The search is optimized by using dynamic programming and pruning of low-scoring branches.
Once complete wraps have been generated the algorithm searches for the strands of the B1 sheet (Fig. 1B) in the sequence gaps between consecutive B2-T2-B3 segments. Potential placements of the B1 strands are scored by using a rung–rung alignment score for pairs of β-strands; if a B1-sheet cannot be found that scores above a threshold score (set by using the sequences in the training set), the wrap is rejected. In addition, wraps intersecting transmembrane regions or regions of high α-helical content are discarded. Transmembrane regions are predicted by using the GES hydrophobicity scale (23), a window of size 21, and a threshold of −2 kcal/mol. Regions of excessive α content are identified by using a filter based on the gor iv program (19), which we found to be a reliable, straightforward, and efficient predictor of overall α-helical content.
The final score assigned to an amino acid sequence is the average of its top ten wrap scores; sequences with fewer than ten wraps remaining after the search and filtering processes are rejected. Note that although some of the known β-helices themselves have fewer than ten distinct, completely correct wraps, the search process applied to these proteins generates a large collection of potential wraps, among which are found the correct wraps but also many partially correct wraps with alternative placement of one or more rungs. For sequences that receive a final score, a Z-score is obtained from the final score by taking the mean and standard deviation of the set of scores of the non-β-helices in PDB-minus that pass the filtering stage, and calculating the number of standard deviations the score is from the mean. Note that this Z-score understates the significance of the raw score, as the majority of the sequences in PDB-minus are rejected before scoring.
Results
There is no overlap in the Z-scores computed by BETAWRAP when the histogram scores for the β-helix database are plotted against those for PDB-minus, a nonredundant version of the PDB (4, 20) with β-helices removed (Fig. 2). The scores reported for the β-helix proteins in Table 2 and in Fig. 2 are the scores from a leave-family-out cross experiment for that β-helix's protein family. In particular, a 7-fold cross-validation was performed on the seven β-helix families of closely related proteins in the SCOP database (5). For each cross, proteins in one β-helix family together with 40% of PDB-minus (531 structures chosen randomly) were placed in the test set, whereas the remainder of the β-helices and PDB-minus (815 structures) were placed in the training set. This was used to set those parameters of the algorithm that were learned from the known β-helices.
Figure 2.
Histogram of the protein Z-scores as computed by betawrap. The β-helix scores (12 proteins) were superimposed on the scores of the PDB-minus database (1,346 proteins), with the 1,091 proteins that could not be successfully wrapped given the arbitrary score −3.5. The β-helix histogram is in blue, and PDB-minus is in green. The following lists the three top-scoring non-β-helix proteins (PDB name, betawrap score, SCOP superfamily): 4SBV:A, 1.87, eight-stranded β-sandwich; 3TDT, 1.84, left-handed parallel β-helix; 1B35C, 1.83, eight-stranded β-sandwich.
Table 2.
Known β-helices, and their betawrap scores and Z-scores
| SCOP Family | Name | Source | PDB | Score | Z-score |
|---|---|---|---|---|---|
| Pectate lyase | Pectate lyase E | E. chrysanthemi | 1PCL | −16.02 | 4.66 |
| Pectate lyase | Pectate lyase C | E. chrysanthemi | 1PLU | −16.44 | 4.41 |
| Pectate lyase | Pectate lyase | Bacillus subtilis | 1BN8 | −18.42 | 3.26 |
| Pectin lyase | Pectin lyase B | Aspergillus niger | 1QCX | −17.09 | 4.03 |
| Pectin lyase | Pectin lyase A | A. niger | 1IDK | −17.99 | 3.51 |
| Galacturonase | Polygalacturonase | Erwinia carotovora | 1BHE | −18.80 | 3.03 |
| Galacturonase | Polygalacturonase II | A. niger | 1CZF | −19.32 | 2.72 |
| Galacturonase | Rhamnogalacturonase A | Aspergillus aculeatus | 1RMG | −20.12 | 2.26 |
| P22 tailspike | P22 tailspike | Salmonella typhimurium | 1TSP | −20.46 | 2.05 |
| P.69 pertactin | P.69 pertactin | B. pertussis | 1DAB | −17.84 | 3.60 |
| Chondroitinase | Chondroitinase B | Flavobacterium heparinium | 1DBO | −19.55 | 2.59 |
| Unclassified | Pectin methylesterase | E. chrysanthemi | 1QJV | −20.74 | 1.90 |
BETAWRAP scores 2,448 proteins higher than the lowest scoring β-helix when searching the 595,890 sequences in the NCBI nonredundant protein database. Table 4 (which is published as supporting information on the PNAS web site, www.pnas.org) lists the top 200 scoring proteins, each with its rank by score, accession number, name, source organism, Z-score, and the sequence positions of its highest scoring wrap. Because of space constraints Table 4 could not be printed in its entirety; Table 3 is a subset of the top 200 scoring proteins selected for their potential biomedical interest (on the basis of functional annotation and/or source organism). The top 200 scoring proteins in Table 4 include proteins that are functionally similar to the known β-helices, as well as some proteins that are similar at the sequence level. The pectate lyases and galacturonases are well represented, as are the pollen allergens, which are members of the pectate lyase superfamily and have been predicted to have a β-helical structure (1, 24). Also included in the list are seven members (ranks 9, 12, 17, 22, 168, 185, and 186) of the hexapeptide repeat family, which are likely to fold into a left-handed parallel β-helix (25). A significant fraction of the proteins found are characterized as outer membrane or cell-surface proteins; these include a large family of related membrane proteins from several Chlamydia species, as well as cell-surface glycoproteins from a number of bacterial and archaeal species.
Table 3.
Selected proteins from the top 200 scoring proteins in NCBI protein sequence database
| Rank | Accession no. | Protein name | Source organism | Z-score | Best wrap positions |
|---|---|---|---|---|---|
| 5 | 11355115 | Hypothetical protein | Vibrio cholerae | 6.23 | 206–332 |
| 19 | 541803 | Major allergen Cry j I precursor | Cryptomeria japonica | 5.19 | 147–267 |
| 23 | 7468997 | Probable outer membrane protein H | Chlamydia trachomatis | 5.11 | 232–382 |
| 24 | 8978911 | Polymorphic membrane protein B Family | Chlamydophila pneumoniae J138 | 5.11 | 769–904 |
| 32 | 1085822 | Coat protein gp1 | Ectocarpus siliculosus virus | 5.03 | 276–386 |
| 35 | 3255935 | Outer membrane protein 5 | C. pneumoniae | 5.00 | 238–375 |
| 42 | 7465392 | Toxin-like outer membrane protein HP0922 | Helicobacter pylori | 4.94 | 137–263 |
| 45 | 7468853 | Hypothetical protein pmpA | C. trachomatis | 4.92 | 240–368 |
| 49 | 7465395 | Toxin-like outer membrane protein jhp0856 | H. pylori | 4.88 | 194–301 |
| 57 | 1657778 | Putative 98 kDa outer membrane protein | Chlamydophila abortus | 4.79 | 148–278 |
| 61 | 11362555 | Polymorphic membrane protein G family | C. pneumoniae | 4.77 | 465–615 |
| 71 | 5566260 | RNA-editing-associated protein 1 | Trypanosoma brucei | 4.67 | 165–265 |
| 76 | 9964588 | AMVITR08 | Amsacta moorei entomopoxvirus | 4.64 | 55–188 |
| 81 | 7463535 | Hypothetical protein | Borrelia burgdorferi | 4.61 | 630–753 |
| 82 | 1078697 | Amastigote-specific protein A2 precursor | Leishmania donovani infantum | 4.61 | 93–231 |
| 83 | 9955515 | Major pollen allergen-like protein | Arabidopsis thaliana | 4.61 | 80–216 |
| 89 | 7327850 | Pertactin | Bordetella bronchiseptica | 4.57 | 163–296 |
| 95 | 7468995 | Probable outer membrane protein F | C. trachomatis | 4.55 | 242–382 |
| 101 | 79327 | 190K surface antigen precursor | Rickettsia rickettsii | 4.51 | 1,022–1,162 |
| 102 | 7468505 | Polymorphic membrane protein G family | C. pneumoniae | 4.50 | 203–347 |
| 108 | 9631583 | Asn/Thr/Ser/Val rich protein | Paramecium bursaria Chlorella virus 1 | 4.48 | 1,056–1,198 |
| 109 | 9087163 | Major pollen allergen cha o 1 precursor | Chamaecyparis obtusa | 4.48 | 141–267 |
| 112 | 283435 | Probable cell-surface protein | Trypanosoma cruzl | 4.48 | 1,496–1,631 |
| 127 | 10956404 | pXO2-14 | Bacillus anthracis | 4.41 | 793–919 |
| 128 | 95503 | Pertactin | B. parapertussis | 4.41 | 163–301 |
| 137 | 7468993 | Probable outer membrane protein D | C. trachomatis | 4.37 | 574–718 |
| 151 | 452482 | Pectate lyase | Aspergillus nidulans | 4.32 | 135–259 |
| 155 | 8978389 | Pmp-3 | C. pneumoniae J138 | 4.31 | 49–198 |
| 156 | 7468854 | Hypothetical protein pmpB | C. trachomatis | 4.31 | 1,010–1,133 |
| 158 | 74141 | Early E1B 44K protein I | Tupaia adenovirus 1 | 4.30 | 71–202 |
| 160 | 7465394 | Toxin-like outer membrane protein jhp0556 | H. pylori | 4.30 | 222–335 |
| 162 | 465388 | 47K protein | Bovine adenovirus 3 | 4.29 | 150–273 |
| 163 | 7464312 | Hypothetical protein | H. pylori | 4.28 | 638–792 |
| 166 | 9049492 | Pertactin | B. bronchiseptica | 4.27 | 43–159 |
| 167 | 2144235 | Outer membrane protein B precursor | Rickettsia japonica | 4.27 | 1,010–1,143 |
| 173 | 7493918 | Pectin lyase B precursor | A. niger | 4.26 | 165–297 |
| 175 | 7468500 | Polymorphic membrane protein g family | C. pneumoniae | 4.26 | 149–290 |
| 178 | 7468855 | Hypothetical protein pmpC | C. trachomatis | 4.26 | 876–1,021 |
| 180 | 7707789 | Protopectinase-AS | A. niger var. awamorii | 4.24 | 156–284 |
| 193 | 8101719 | Cup s 1 pollen allergen precursor | Cupressus sempervirens | 4.21 | 142–267 |
| 199 | 6688592 | Hypothetical protein | Legionella pneumophila | 4.19 | 59–203 |
A striking feature of the sequences identified is their association with known human pathogens: V. cholerae (cholera); H. pylori (ulcers); Plasmodium falciparum (malaria); C. trachomatis (venereal infection); C. pneumoniae (respiratory infection); Listeria monocytogenes (listeriosis); C. abortus (genital infection); T. brucei (sleeping sickness); B. burgdorferi (Lyme disease); L. donovani (Leishmaniasis); B. bronchiseptica (respiratory infection); R. rickettsii (Rocky Mountain spotted fever); T. cruzi (sleeping sickness); B. parapertussis (whooping cough); B. anthracis (anthrax); R. japonica (Oriental spotted fever); Neisseria meningitides (meningitis); and L. pneumophilia (Legionnaire's disease). Although a full phylogenetic analysis remains to done, high scores for sequences derived from soil and other environmental microorganisms are relatively rare, suggesting that the association with pathogens represents functional aspects of the β-helix fold.
There is an additional bias in the source organisms for the high-scoring proteins. Although proteins from humans, mice, nematode worms, and fruit flies account for over 20% of total sequences in the NCBI nonredundant protein database, only two proteins in the top 200 BETAWRAP scores come from these species. The likelihood of this occurring had the sequences been chosen randomly is less than 10−18. This bias agrees with the observed species distribution of the known β-helices, which are found primarily in bacteria, plants, and fungi.
A few of the 200 top-scoring putative proteins are likely to represent false positives from independent evidence. Two are among the very rare sequences from vertebrates: the dynein heavy chain from trout (Oncorhyncus) and the SON DNA binding protein from Humans. Characterization of the dynein chains and DNA binding proteins from other vertebrates makes it very unlikely that these proteins are β-helices. A third protein family, the leucine-rich repeat (LRR) family, is represented in Table 4. The structures of several proteins in this family have been determined and are characterized by a coiled fold in which α-helices and β-strands alternate along the chain (26). Thus the LRR sequences found by BETAWRAP most likely represent false positives; at the same time they have striking structural similarities to the β-helices, including stacks of aliphatic residues in the β-sheets and internal polar-residue stacks in the turns. Indeed, several LRR proteins were found to match a sequence profile developed from the pectate lyases (8). The 37 sequences identified as potential LRR family members (21) have been segregated at the bottom of Table 4.
Discussion
Our results indicate that there are spatial pairwise correlations in β-helices that can be recovered from sequence data and used to distinguish β-helical from non-β-helical domains. Pairwise alignment preferences learned from general amphipathic β-sheet proteins, together with family-specific stacking preferences, contain sufficient information to allow reasonably accurate reconstruction of the tertiary structure of these proteins. Our approach is in contrast to other attempts to predict β-strand pairings (e.g., ref. 16) in that we incorporate the amphipathic nature of β-helices, partitioning aligned pairs of residues according to their orientation relative to the core in the assumed structural template. It seems likely that these methods can enhance other structure prediction methods, and lead to better supersecondary structure predictors for β-structural motifs.
All of the β-helices whose structures are known are elongated proteins. The function of the majority involves recognition or interaction with long polysaccharides or lipopolysaccharides. The structure of the P22 tailspike complexed with its specific Salmonella lipopolysaccharide (LPS) substrate has been solved, and the LPS lies extended along the external face of the β-helix (27). Thus the active site of this protein is not in a crevice, but along a ribbon-like surface. These proteins may represent a group of proteins that have evolved to interact with elongated polysaccharides (3). These preferential substrates of the β-helices may explain their involvement in cell surface recognition, penetration, and pathogenesis. One of the exceptions, insect antifreeze protein, which interacts with ice crystal surfaces, may be a later functional divergence (28).
Jenkins et al. (3) have proposed that the right-handed parallel β-helices represent a single superfamily of folds evolved from a common ancestor, probably by duplication of rung sequences. The absence of this fold among higher eukaryotes suggests either that the fold evolved after the divergence of animals from prokaryotes, or that this fold has been lost or suppressed in higher eukaryotes.
The high frequency of occurrence of bacterial toxins, adhesins, and allergens in the list implies that the β-helix fold may have a more extensive role in human disease than was previously recognized. Identification of gene sequences as putative β-helices may serve as an early warning signal for uncharacterized proteins that contribute to bacterial virulence.
Supplementary Material
Acknowledgments
P.B. was supported in part by a Massachusetts Institute of Technology/Merck Graduate fellowship, L.C. by an Emmaline Bigelow Conland fellowship at the Radcliffe Institute for Advanced Study, J.K. by National Institutes of Health Grant GM 17980, and B.B. by a Charles E. Reed Faculty Initiatives Award.
Abbreviations
- PDB
Protein Data Bank
- NCBI
National Center for Biotechnology Information
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Yoder M D, Keen N T, Jurnak F. Science. 1993;260:1503–1507. doi: 10.1126/science.8502994. [DOI] [PubMed] [Google Scholar]
- 2.Yoder M D, Lietzke S E, Jurnak F. Structure. 1993;1:241–251. doi: 10.1016/0969-2126(93)90013-7. [DOI] [PubMed] [Google Scholar]
- 3.Jenkins J, Mayans O, Pickersgill R. J Struct Biol. 1998;122:236–246. doi: 10.1006/jsbi.1998.3985. [DOI] [PubMed] [Google Scholar]
- 4.Berman H M, Westbrook J, Feng Z, Gilliland G, Bhat T N, Weissig H, Shindyalov I N, Bourne P E. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murzin A G, Brenner S F, Hubbard T, Chothia C. J Mol Biol. 1995;297:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 6.Altschul S, Madden T, Schaffer A, Zhang J, Zhang Z, Miller W, Lipman L. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eddy S. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 8.Heffron S, Moe G, Sieber V, Mengaund J, Cossart P, Vitali J, Jurnak F. J Struct Biol. 1998;122:232–235. doi: 10.1006/jsbi.1998.3978. [DOI] [PubMed] [Google Scholar]
- 9.Jones D, Taylor W, Thornton J. Nature (London) 1992;358:86–89. doi: 10.1038/358086a0. [DOI] [PubMed] [Google Scholar]
- 10.Kreisberg J F, Betts S D, King J. Protein Sci. 2000;9:2338–2343. doi: 10.1110/ps.9.12.2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haase-Pettingel C, King J. J Mol Biol. 1997;267:88–102. doi: 10.1006/jmbi.1996.0841. [DOI] [PubMed] [Google Scholar]
- 12.Simons K T, Strauss C, Baker D. J Mol Biol. 2001;306:1191–1199. doi: 10.1006/jmbi.2000.4459. [DOI] [PubMed] [Google Scholar]
- 13.Koehl P, Levitt M. Nat Struct Biol. 1999;6:108–111. doi: 10.1038/5794. [DOI] [PubMed] [Google Scholar]
- 14.Lifson S, Sander C. J Mol Biol. 1980;139:627–629. doi: 10.1016/0022-2836(80)90052-2. [DOI] [PubMed] [Google Scholar]
- 15.Hubbard T, Park J. Proteins. 1996;3:398–402. doi: 10.1002/prot.340230313. [DOI] [PubMed] [Google Scholar]
- 16.Zhu H, Braun W. Protein Sci. 1999;8:326–342. doi: 10.1110/ps.8.2.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berger B. J Comput Biol. 1995;2:125–138. doi: 10.1089/cmb.1995.2.125. [DOI] [PubMed] [Google Scholar]
- 18.Berger B, Wilson D B, Wolf E, Tonchev T, Milla M, Kim P S. Proc Natl Acad Sci USA. 1995;92:8259–8263. doi: 10.1073/pnas.92.18.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garnier J, Gibrat J F, Robson B. Methods Enzymol. 1996;266:540–553. doi: 10.1016/s0076-6879(96)66034-0. [DOI] [PubMed] [Google Scholar]
- 20.Hobohm U, Scharf M, Schneider R, Sander C. Protein Sci. 1992;1:409–417. doi: 10.1002/pro.5560010313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bateman A, Birney E, Durbin R, Eddy S R, Howe K L, Sonnhammer E L L. Nucleic Acids Res. 2000;28:263–266. doi: 10.1093/nar/28.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frishman D, Argos P. Proteins. 1995;23:566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- 23.Engelman D, Steitz T, Goldman A. Annu Rev Biophys Biophys Chem. 1999;15:321–353. doi: 10.1146/annurev.bb.15.060186.001541. [DOI] [PubMed] [Google Scholar]
- 24.Henrissat B, Heffron S E, Yoder M D, Lietzke S D, Jurnak F. Plant Physiol. 1995;107:963–976. doi: 10.1104/pp.107.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vuorio R, Harkonen T, Tolvanen M, Vaara M. FEBS Lett. 1994;337:289–292. doi: 10.1016/0014-5793(94)80211-4. [DOI] [PubMed] [Google Scholar]
- 26.Kobe B, Deisenhofer D. Nature (London) 1995;374:183–186. doi: 10.1038/374183a0. [DOI] [PubMed] [Google Scholar]
- 27.Steinbacher S, Miller S, Baxa U, Weintraub A, Seckler R, Huber R. Proc Nat Acad Sci USA. 1996;93:10584–10588. doi: 10.1073/pnas.93.20.10584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Graether S P, Kuiper M J, Gagne S M, Walker V K, Jia Z, Sykes B D, Davies P L. Nature (London) 2000;406:325–328. doi: 10.1038/35018610. [DOI] [PubMed] [Google Scholar]
- 29.Sayle R A, Milner-White E J. Trends Biochem Sci. 1995;20:374. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.