Abstract
PURPOSE
The current diagnostic testing algorithm for Lynch syndrome (LS) is complex and often involves multiple follow-up germline and somatic tests. We aimed to describe the results of paired tumor/germline testing performed on a large cohort of patients with colorectal cancer (CRC) and endometrial cancer (EC) to better determine the utility of this novel testing methodology.
MATERIALS AND METHODS
We retrospectively reviewed a consecutive series of patients with CRC and EC undergoing paired tumor/germline analysis of the LS genes at a clinical diagnostic laboratory (N = 702). Microsatellite instability, MLH1 promoter hypermethylation, and germline testing of additional genes were performed if ordered. Patients were assigned to one of five groups on the basis of prior tumor screening and germline testing outcomes. Results for each group are described.
RESULTS
Overall results were informative regarding an LS diagnosis for 76.1% and 60.8% of patients with mismatch-repair–deficient (MMRd) CRC and EC without and with prior germline testing, respectively. LS germline mutations were identified in 24.8% of patients in the group without prior germline testing, and interestingly, in 9.5% of patients with previous germline testing; four of these were discordant with prior tumor screening. Upon excluding patients with MLH1 promoter hypermethylation and germline mutations, biallelic somatic inactivation was seen in approximately 50% of patients with MMRd tumors across groups.
CONCLUSION
Paired testing identified a cause for MMRd tumors in 76% and 61% of patients without and with prior LS germline testing, respectively. Findings support inclusion of tumor sequencing as well as comprehensive LS germline testing in the LS testing algorithm. Paired testing offers a complete, convenient evaluation for LS with high diagnostic resolution.
INTRODUCTION
Lynch syndrome (LS) is the most common inherited cancer predisposition syndrome, accounting for 2% to 3% of patients with colorectal cancer (CRC)1,2 and 2% to 3% of patients with endometrial cancer (EC).3,4 LS is caused primarily by pathogenic germline variants in the mismatch repair (MMR) genes (MLH1, MSH2, MSH6, PMS2) and 3′ deletions of EPCAM, which result in silencing of MSH2. Herein, these five are referred to as LS genes, and prevalence estimates range from 1 in 440 to 1 in 279 in the US population.5,6 Without intervention, individuals with LS face lifetime cancer risks of up to 82% for CRC and 60% for EC, along with increased risks for other cancer types.7-12
Universal screening for LS among newly diagnosed patients with CRC and EC has been widely adopted across the United States because it is supported by a number of national organizations13-18 and has been proven cost effective.19,20 Screening approaches involve performing immunohistochemical (IHC) staining for MMR proteins and/or microsatellite instability (MSI) analysis on tumor specimens to identify MMR deficiency, followed by MLH1 promoter hypermethylation (MPH) or BRAF V600E analysis for tumors lacking MLH1 protein expression to identify likely sporadic tumors.
Molecular germline testing confirms a diagnosis of LS in 24% to 67% of patients with MMR-deficient (MMRd) CRC1 and in 16% to 80% of those with MMRd EC,4 depending on the pattern of IHC protein expression and excluding patients with MPH. In the absence of a formal LS diagnosis, screening recommendations for the patient and other family members can remain unclear; recent National Comprehensive Cancer Network guidelines advise that these patients be observed based on family history.13 In recent years, somatic inactivation has been identified in 52% to 69% of unexplained MMRd tumors,21-23 and follow-up to abnormal MSI/IHC now includes consideration of tumor sequencing for patients with no germline pathogenic variant detected.13 A recent study suggests that tumor sequencing with follow-up germline testing is superior to current screening algorithms in regard to sensitivity and ability to inform treatment decisisons.24 We describe the use of a paired tumor/germline assay25 to aid in the diagnostic evaluation of LS in more than 700 patients with CRC and EC with MMRd tumors and/or clinical histories suggestive of LS and discuss the benefits and limitations of this concurrent testing approach.
MATERIALS AND METHODS
Patients
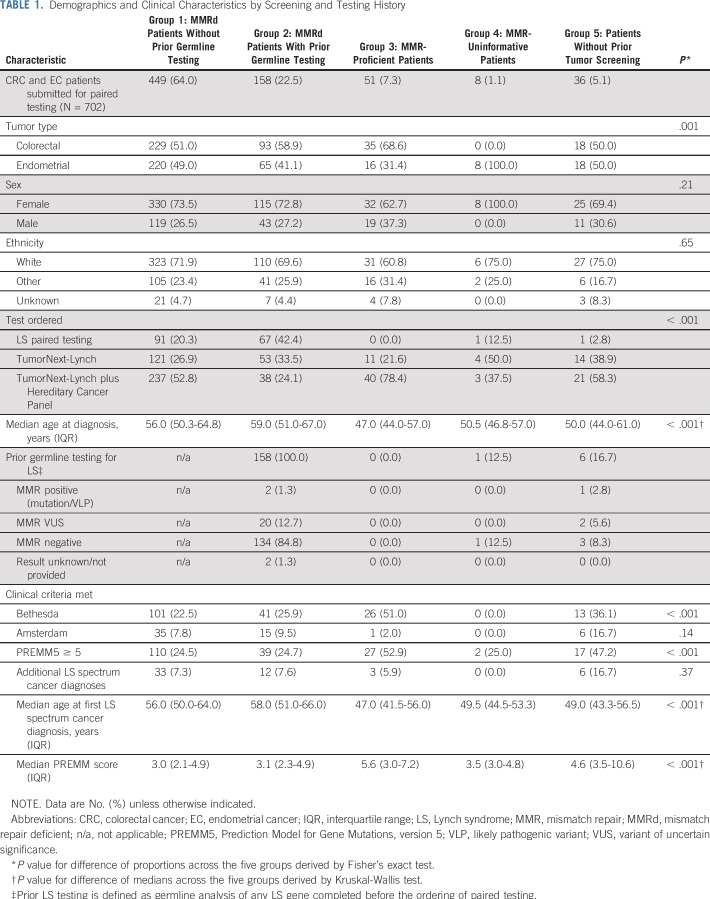
Consecutive patients with CRC and EC undergoing one of five paired tumor/germline tests for LS between 2016 and 2018 at a single clinical diagnostic laboratory (Ambry Genetics, Aliso Viejo, CA) were retrospectively reviewed. Patient demographic information and clinical, family, and genetic testing histories were obtained from test requisition forms and other clinical documentation, such as pedigrees, clinic notes, and pathology reports, when available. Personal and family histories were reviewed to determine whether patients met revised Bethesda guidelines26 or Amsterdam criteria II,27 and mutation probabilities were calculated using the published PREMM5 (Prediction Model for Gene Mutations, version 5) prediction model28 (Table 1). This study was determined to be exempt from review by Western Institutional Review Board.
TABLE 1.
Demographics and Clinical Characteristics by Screening and Testing History
Paired Tumor/Germline Testing
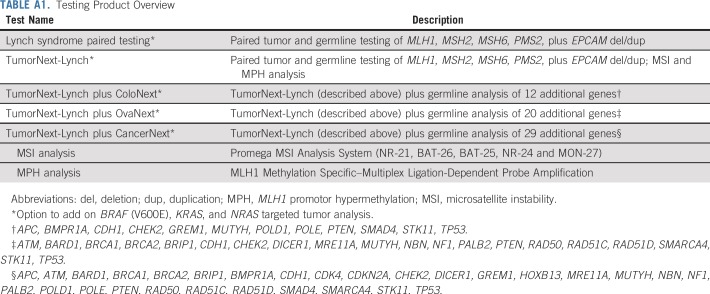
The paired tumor/germline assay analyzed DNA isolated from both blood and tumor samples in parallel.25 Briefly, paired tumor/germline analysis was performed on the five LS genes using a customized next-generation sequencing panel. Somatic variants, copy number variants, and loss of heterozygosity were determined by comparing the data between tumor and blood samples. MSI analysis, MPH analysis, and germline testing of additional cancer susceptibility genes were performed if indicated by the specific test ordered (Table A1). Detailed methods for all components of the paired tumor/germline assay are included in the Appendix.
Interpretation of Results
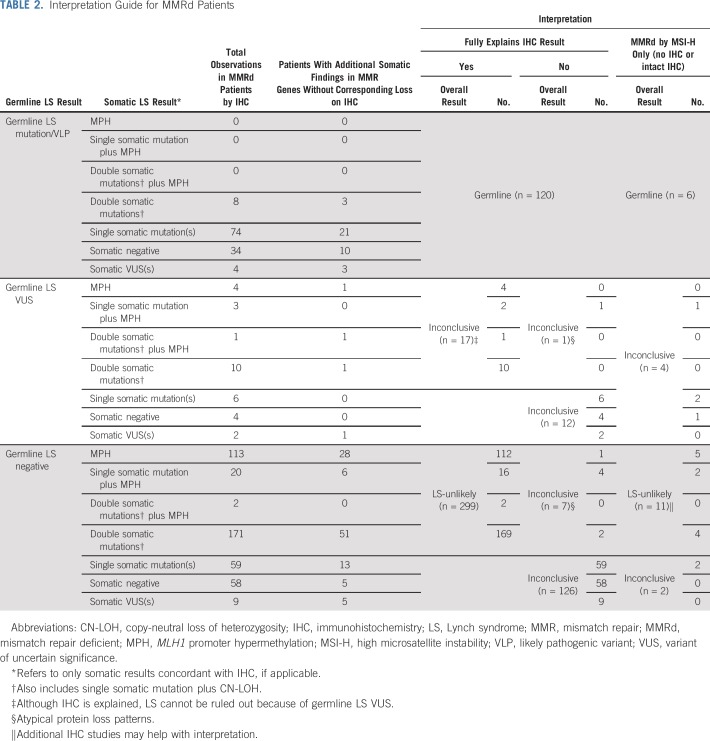
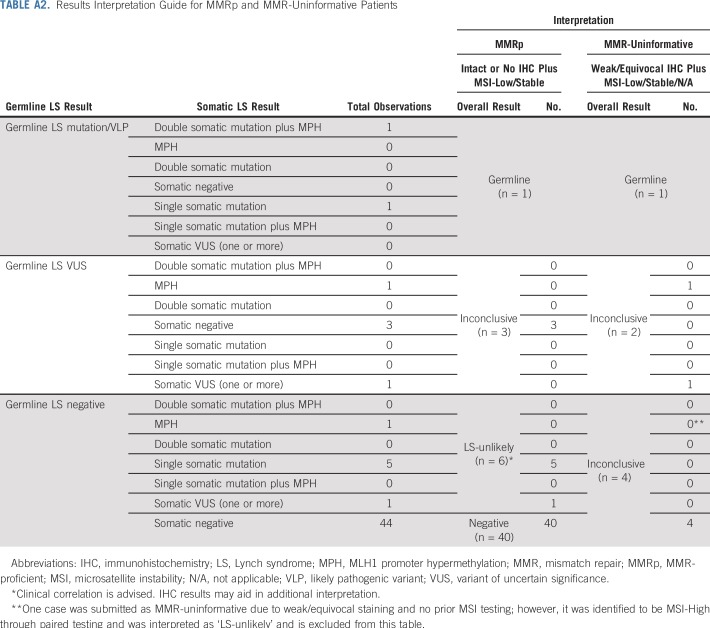
Overall results of paired tumor/germline testing were interpreted in the context of available IHC and/or MSI results (Tables 2 and A2). Results were interpreted as germline if a pathogenic mutation or likely pathogenic variant (VLP) of germline origin was identified in an LS gene, regardless of somatic findings or prior tumor screening results. For patients with MMRd tumors with abnormal IHC, overall results were interpreted as LS-unlikely if no germline mutation or variant of uncertain significance (VUS) was identified and somatic findings could explain protein loss by IHC (MLH1-, PMS2-, MSH2-, MSH6-). Explanatory somatic findings included identifying (1) two mutations/VLPs of somatic origin or (2) copy-neutral loss of heterozygosity (CN-LOH) and a somatic mutation/VLP in the same LS gene that were concordant with abnormal IHC. Results were also interpreted as LS-unlikely if these findings were seen in an MSI-high (MSI-H) tumor with no IHC results (Table 2). Identifying somatic MPH in the context of MLH1 protein loss (MLH1-) and/or PMS2 protein loss (PMS2-) or MSI-H with no IHC results was also considered explanatory. Of note, for patients in whom MSI results from prior screening were discordant with the MSI results obtained through paired testing, the latter were used in the overall results interpretation. Results were interpreted as inconclusive if MMRd remained unexplained or if IHC was explained by somatic findings, but LS could not be as confidently ruled out because of the presence of a germline LS VUS (Table 2).
TABLE 2.
Interpretation Guide for MMRd Patients
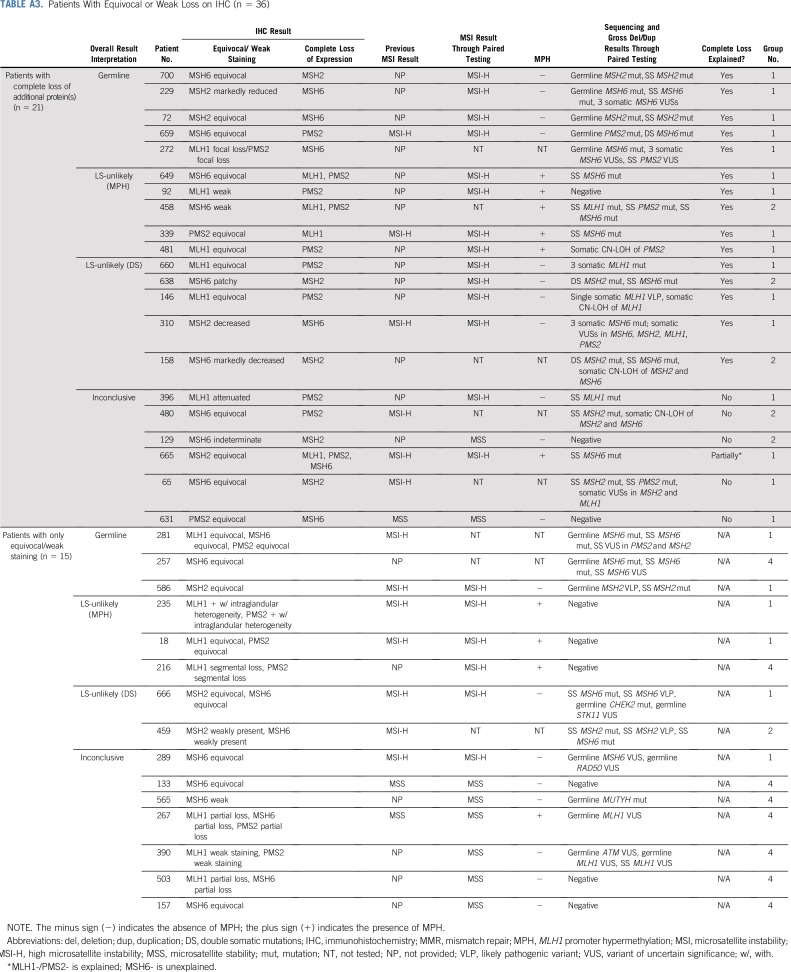
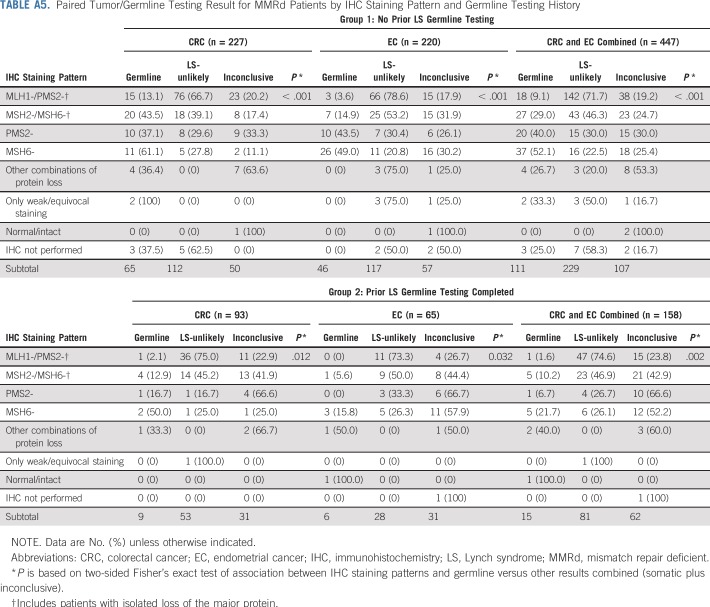
Tumors demonstrating microsatellite stability (MSS), MSI-low (MSI-L), and/or intact MMR protein expression by IHC were considered MMR-proficient (MMRp). MSS/MSI-L tumors with IHC results not reported as loss/absent (eg, equivocal, weak) without complete loss of any other protein on the pathology report were considered MMR-uninformative. Interpretation of results for patients with MMRp and MMR-uninformative tumors are listed in Table A2. Fifteen patients in this series were found to be MMR-uninformative, with results listed in Table A3. These patients were excluded from the patients described as MMRd.
Statistical Methods
Descriptive characteristics were summarized with medians for continuous variables and proportions for categorical variables. Differences in medians and proportions for patients with CRC versus EC were assessed by Wilcoxon rank sum test and Fisher’s exact test, respectively. Fisher’s exact test was also used to assess test result association with prior LS testing and IHC staining pattern, respectively.
RESULTS
Demographic and Clinical Characteristics
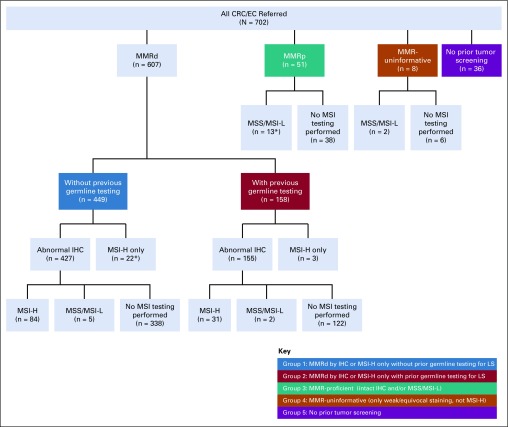
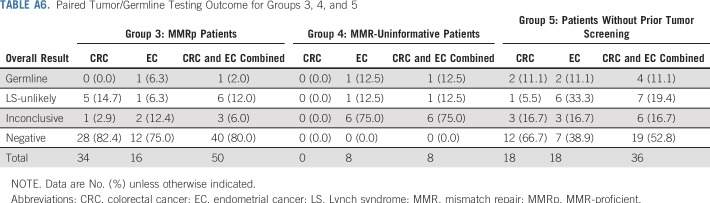
In total, 702 paired tumor/germline tests were performed in patients with CRC (n = 375) and/or EC (n = 327) from more than 200 institutions. Overall, 94.9% (n = 666) of specimens had prior IHC and/or MSI analysis, and 23.5% (n = 165) had prior germline testing for LS, defined as germline analysis of any LS gene completed before the ordering of paired testing. Patients were placed into one of five groups on the basis of prior tumor screening results and germline testing history (Table 1; Fig 1). Patients with MMRd tumors were stratified into two groups: group 1, which had no prior LS germline testing (n = 449), and group 2, which did have testing (n = 158). Groups 3, 4, and 5 included patients with MMRp tumors (n = 51), patients with uninformative tumor screening (n = 8), and patients without prior tumor screening (n = 36), respectively. Personal and family cancer histories of each group were analyzed (Table 1).
FIG 1.
Paired tumor/germline patients by prior testing and tumor screening history. Abnormal immunohistochemistry (IHC): abnormal IHC screening because of loss of expression of at least one mismatch repair (MMR) protein as reported on external final IHC result; weak/equivocal staining: MMR protein expression reported as weak, equivocal, attenuated, nearly absent, segmental, partial, focal, decreased, or indeterminate without complete loss of any other MMR proteins on external final IHC result. (*) Three patients submitted for paired testing were removed from additional analysis; two from group 1 because microsatellite instability (MSI) results from paired testing were microsatellite stable (MSS) and one from group 3 because MSI results from paired testing were MSI high (MSI-H). CRC, colorectal cancer; EC, endometrial cancer; MSI-L, low microsatellite instability.
Patients Referred With MMRd Tumors (groups 1 and 2)
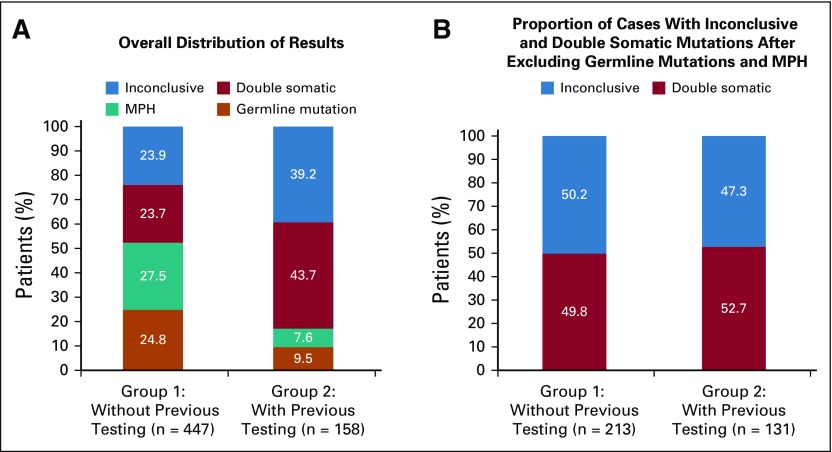
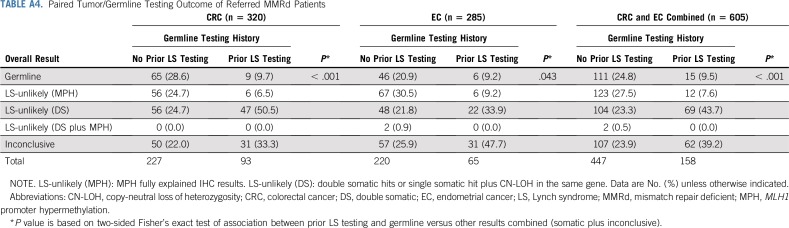
As illustrated in Figure 1, the majority of paired testing was performed on patients with CRC or EC with abnormal IHC, with or without MSI testing (82.9%; n = 582) or only MSI-H (3.6%; n = 25). Paired testing was able to resolve 76.1% (n = 340 of 447) of patients without prior germline testing and 60.8% (n = 96 of 158) of patients with prior germline testing. Patients were considered resolved when either an LS germline mutation/VLP was identified or IHC was fully explained by somatic findings making LS-unlikely (Table A4). When comparing groups 1 and 2, a higher proportion of patients without prior testing had overall results of either a germline mutation or MPH, whereas a higher proportion of patients with prior germline testing had double somatic mutations and inconclusive results (Fig 2).
FIG 2.
Comparison of paired tumor/germline testing outcome for patients with mismatch repair deficient (MMRd) tumors by germline testing history. (A) Overall distribution of paired testing results in patients with MMRd tumors. (B) Paired testing outcomes after removing patients with germline Lynch syndrome mutations and MLH1 promoter hypermethylation from each group. Inconclusive: patients in whom abnormal immunohistochemistry (IHC) result is unexplained by somatic or germline findings. Double somatic: patients with somatic inactivation of an MMR gene as a result of double somatic mutations (or single somatic mutation with copy-neutral loss of heterozygosity) in the same gene and concordant with IHC result. MLH1 promoter hypermethylation (MPH): patients with MLH1 promoter hypermethylation identified as explanatory for MMRd. Germline mutation: patients with a germline MMR mutation/likely pathogenic variant. Group 1 without previous testing: patients in whom germline testing of one or more MMR genes, either in whole or in part, was not previously performed. Group 2 with previous testing: patients in whom germline testing of one or more MMR genes was previously performed.
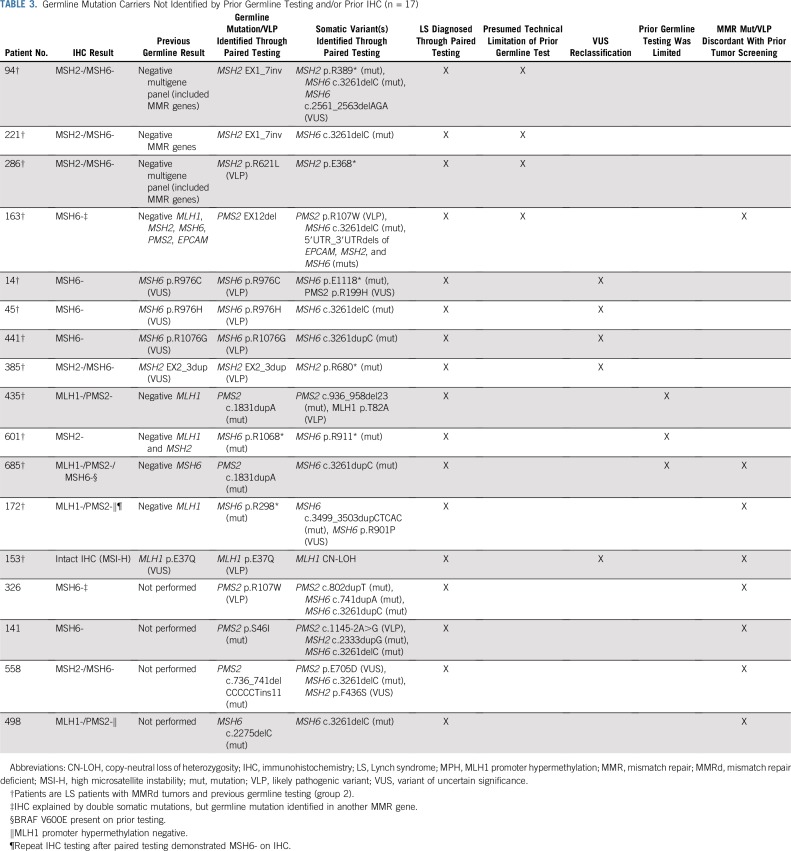
Of patients with no prior germline testing for LS (group 1), 25% (n = 111 of 447) were diagnosed with LS by paired tumor/germline testing, although results varied based on protein staining pattern (Table A5). Unexpectedly, 9.5% (n = 15 of 158) of group 2 patients were found to have germline mutations/VLPs by paired tumor/germline testing (Table A4). In two of these, the germline mutation had been previously identified, but protein loss on IHC remained unexplained by that variant. In the remaining 13 germline patients from group 2, the mutation/VLP was either not previously identified (n = 8) or was originally called a VUS and classified as a VLP on paired testing (n = 5; patients 14, 45, 441, 385, and 153; Table 3). Of the eight patients with previously negative germline testing, four had mutations/VLPs identified because of presumed technical limitations of prior testing (patients 94, 221, 286, and 163), three were missed because of clinicians ordering limited germline testing (patients 435, 601, and 685), and two had a germline mutation in an unexpected gene on the basis of prior tumor screening (patients 685 and 172; Table 3).
TABLE 3.
Germline Mutation Carriers Not Identified by Prior Germline Testing and/or Prior IHC (n = 17)
Tumor MMRd was also explained by somatic findings. In the group with no prior testing, LS-unlikely patients were resolved by either biallelic somatic inactivation (23.3%; n = 104 of 447), MPH (27.5%; n = 123 of 447), or both (0.5%; n = 2 of 447; Table A4). In the group with prior testing, LS-unlikely patients were resolved by either biallelic somatic inactivation (43.7%; n = 69 of 158) or MPH (7.6%; 12 of 158). When excluding patients with explanatory MPH and germline mutations, the proportions of patients with double somatic or inconclusive results did not significantly vary between groups (P = .65).
For the remaining patients with MMRd tumors, paired tumor/germline testing yielded inconclusive results (n = 169 of 605; 27.9% for groups 1 and 2 combined; Table A4 and A5). The most common inconclusive result was identification of only one mutation/VLP of somatic origin that was concordant with loss on IHC (n = 59 of 169; 34.9%; Table 2). Other inconclusive results included identification of an LS germline or somatic VUS (n = 43 of 169; 25.4%) or negative LS germline results in conjunction with negative somatic results (n = 58 of 169; 34.3%; Table 2).
Other Referrals
Results for the subset of patients with MMRp tumors (n = 51), uninformative tumor screening (n = 8), or no prior tumor screening (n = 36) are listed in Table A6. A higher proportion of these patients met Bethesda guidelines and had a greater than 5% likelihood of carrying an MMR mutation predicted by PREMM5 relative to patients with MMRd tumors, suggesting that patients with MMRp tumors and those without prior screening had paired testing ordered by clinicians when there remained indicators of LS, such as clinical or family history (Table 1).
DISCUSSION
In this study, we demonstrate a novel LS testing approach that pairs comprehensive germline testing with concurrent tumor analysis. This paired testing approach provided a molecular etiology for MMRd in 76.1% and 60.8% of patients without and with prior germline testing, respectively.
As expected, a higher percentage of germline mutations and promoter hypermethylation were identified in group 1 compared with group 2 (24.8% v 9.5% and 27.5% v 7.6%, respectively; Fig 2A). When excluding patients with MPH, the rate of germline mutation in group 1 (34.3%) is similar to published literature.1,4 In contrast, germline LS mutations were unexpectedly identified in 9.5% (n = 15) of patients with prior germline testing (group 2). Three of these patients had germline mutations detected in genes not initially selected for germline mutation analysis but still within the scope of plausible explanations for the IHC finding (Table 3; patients 435, 601, and 685). For example, one patient with an MLH1-/PMS2- IHC result was found to carry a PMS2 mutation but only had previous MLH1 testing. In another three of these patients (Table 3; patients 685, 172, and 153) and in four patients without prior germline testing (n = 4 of 447; 0.8%; Table 3; patients 326, 141, 558, and 498), germline mutations were detected in genes that were not considered as concordant with prior tumor screening. For example, one patient with an MLH1-/PMS2- IHC result was identified as carrying an MSH6 mutation. These findings highlight the ability of comprehensive germline testing of all five LS genes to overcome limitations of prior tumor screening and germline testing, including possible erroneous IHC, and support concurrent, comprehensive germline testing of all LS genes, regardless of the pattern of protein loss on IHC.
Results from this study also support adding tumor analysis to the diagnostic workup for LS. When excluding patients with a germline mutation or MPH, double somatic mutations were identified in the same proportion (approximately 50%) in both groups (Fig 2B), consistent with previous studies.21-23 Identifying double somatic mutations decreases our suspicion of LS as we currently understand it21-23; however, additional research and longitudinal studies examining clinical outcome in patients with double somatic mutations are warranted.
In this cohort, the rate of inconclusive results for patients with MMRd tumors was 27.9% overall. A subset of these had unexplained abnormal IHC and negative paired tumor/germline test results, suggesting the possibility of false-positive IHC results, which is supported by the fact that 25 of 58 (43.1%) of these patients were MSS on paired testing. Likewise, the inconclusive rate may be inflated compared with expected, because a larger percentage of patients in groups 1 and 2 likely could have been resolved if MPH studies had been ordered. For example, 26.9% (n = 21 of 78) of inconclusive patients were MLH1- and/or PMS2- with no MPH testing ordered by the clinician. In addition, 22.5% (n = 38 of 169) of inconclusive patients demonstrated unexplained MSH6-, suggesting the possibility of loss secondary to neoadjuvant chemoradiation29-31 or MLH1/PMS2 deficiency.32
A combination of germline and tumor testing inarguably leads to the highest resolution of patients with MMRd tumors; however, additional studies are needed to determine the optimal testing strategy with respect to cost effectiveness and to further explore the clinical utility of somatic and paired mutational analysis. When comparing test results from groups 1 and 2, we expect that if comprehensive germline testing was ordered and germline analysis had been previously performed in group 1, the proportion of resolved patients would be unchanged regardless of whether tumor sequencing was performed concurrently with or subsequent to comprehensive germline testing. Although clinic-based studies are needed to further inform an optimal testing strategy, we note several potential benefits of paired testing on the basis of our experience.
One major benefit is the ability to have a single unified interpretation of all genetic testing results in one laboratory. Also, based on recent literature about somatic mutations in MMRd tumors,33,34 paired testing has the ability to further inform variant classification. Five individuals from group 2 with previously identified germline VUSs were found to also have a mutation or CN-LOH of somatic origin in the same LS gene, providing an additional line of evidence to support reclassification to VLP (Table 3). Although somatic data were used as supporting evidence, future studies are needed to understand how heavily somatic data should be weighed.
Additionally, paired testing demonstrates the ability to simultaneously diagnose LS while also explaining atypical IHC patterns in patients where germline mutations were not concordant with the specific protein expression. For example, tumors in our cohort were identified to carry double somatic mutations/VLPs, which explained IHC staining in addition to a germline mutation in a different LS gene (Table 3; patients 326 and 163). Interestingly, in one patient, the germline mutation identified was a single exon deletion in the PMS2 pseudogene region. Testing strategies employing only tumor sequencing to screen for germline alterations may have missed this because of limitations in detecting exon-level somatic copy number variants, especially in this region with significant pseudogene homology. Thus, the paired testing approach reduces the risk of unidentified large rearrangements of germline origin.
Last, paired testing is a more convenient approach compared with sequential testing. Concurrent germline and tumor testing has the potential to reduce the risk of losing patients to follow-up and reduces clinician burden of ordering different tests and sending multiple specimens to various laboratories. Thus, clinicians and patients may prefer paired testing, especially given the current uncertainty regarding insurance coverage for multiple LS-related tests.
Because this study was performed at a single clinical diagnostic laboratory, complete information/records may not have been provided by the ordering clinician. Similarly, patients underwent different variations of paired testing depending on the product ordered by their clinical provider (Table A1), which affects interpretation of overall results. Study outcomes are also not generalizable to all CRC and EC patients, because the majority of this cohort comprised individuals with some indication for LS genetic testing. However, these results may be more widely applicable, given that only 25% of patients met revised Bethesda guidelines, and only 8% met Amsterdam criteria II.
In addition, interpretation of these data and placement of patients into overall result categories relied on external IHC results, without availability of internal pathology review. We also acknowledge that methods to differentiate between driver and passenger mutations would be helpful in refining the interpretation of results independent of external testing.37
Importantly, although double somatic mutations or a somatic mutation with CN-LOH may explain protein loss by IHC, they may not always rule out LS (Tables 2 and 3), because we found that germline mutations can be identified with double somatic mutations in the same or different LS gene. Similarly, it is often not possible to definitively determine phase (cis or trans) of sequencing alterations. Although these data argue against a diagnosis of LS in which somatic findings explain IHC in the absence of a germline mutation or VUS, one cannot entirely rule out that technical limitations of current methodologies may lead to undetected germline or somatic variants. Likewise, low tumor cellularity may lead to false-negative somatic results, as there are some cases in which MPH signal is detected but is below reporting threshold and therefore interpreted as negative. For these reasons, it is advised that clinicians interpret results in the context of clinical history. Additional cancer screening recommendations on the basis of personal or family history may still be warranted, even in patients where double somatic mutations are identified, or MMRd is completely explained by comprehensive paired testing. Future clinical correlation and outcome studies would aid in better understanding the relationship between molecular findings and clinical history.
In conclusion, paired tumor/germline testing offers a novel approach to the traditional step-wise testing algorithm for LS, in the setting of abnormal MSI/IHC testing. Paired testing resolved 76% of patients with MMRd tumors without prior LS germline testing and 61% of patients with MMRd tumors with prior LS germline testing. This supports the current recommendation of including tumor sequencing in the LS testing algorithm and confirms the importance of performing comprehensive germline LS testing regardless of IHC staining pattern. Given these two findings, paired testing provides an opportunity for clinicians and patients to obtain both germline and somatic results simultaneously to aid in making the most informed management recommendations for patients with suspected LS.
ACKNOWLEDGMENT
We acknowledge Lily Hoang for her assistance with PREMM5 score curation.
Appendix
Materials and Methods
Sequencing and copy number variant analyses.
The paired tumor/germline assay analyzed DNA isolated from both blood and tumor sample in parallel.25 The customized next-generation sequencing panel consisted of 508 exons, 81 introns (partial), and 13 untranslated regions in 39 genes. The panel was composed of biotinylated xGen Lockdown probes synthesized by Integrated DNA Technologies (IDT, Coralville, IA). Next-generation sequencing was performed on the Illumina (San Diego, CA) HiSEquation 2500. Paired tumor/germline analysis was performed on five genes associated with Lynch syndrome. Germline analysis was performed on additional cancer predisposition genes depending on ordering request. In addition, somatic variants associated with resistance to anti–epidermal growth factor receptor therapy were also analyzed. Sequencing data generated from tumor and blood paired samples was processed with an inhouse-developed bioinformatics pipeline.25 Sequencing reads were aligned to human genome (UCSC hg19) with Novoalign V3.02.07 (Novocraft Technologies Sdn Bhd, Selangor, Malaysia). Germline variants were called with GATK (v3.2.2; Broad Institute, Cambridge, MA) using data from blood samples. Common polymorphisms and false-positive variants generated as a result of sequencing artifact were filtered out by using population frequency data from multiple sources, including National Center for Biotechnology Information dbSNP; National Heart, Lung, and Blood Institute Exome Sequencing Project; 1000 Genomes; and internal data. Germline copy number variants were detected using either multiplex ligation-dependent probe amplification (MLPA) or targeted array comparative genomic hybridization. Somatic variants, copy number variants, and loss of heterozygosity were determined by comparing the data between tumor and blood samples. Microsatellite instability (MSI) and MLH1 promoter hypermethylation analyses were performed in accordance with the specific paired/tumor germline test ordered.
MSI analysis.
MSI analysis was performed on Promega MSI Analysis System (Promega, Madison, WI). The kit analyzes the mononucleotide repeat regions NR-21, BAT-26, BAT-25, NR-24, and MON-27. DNA from both tumor and matched normal blood was amplified by polymerase chain reaction to generate fluorescently labeled amplicons of the repeat regions. The amplicons were separated based on size using capillary gel electrophoresis on an ABI 3730xl DNA Analyzer (Applied Biosystems, Carlsbad, CA). Fragment analysis was performed using GeneMapper software (Thermo Fisher Scientific, Waltham, MA), which compares the allelic patterns of the matched normal and tumor samples to determine alterations in the length of each of the five repeat regions. Two or more repeat regions with altered amplicon length was considered MSI-high, one repeat region with altered amplicon length was considered MSI-low, and zero repeat region with altered amplicon was considered microsatellite stable.
MLH1 promoter hypermethylation analysis.
The MLH1 promoter methylation status was determined by using the MLH1 Methylation Specific–Multiplex Ligation-Dependent Probe Amplification kit (ME011-B3; MRC Holland, the Netherlands), which contains probes covering the MLH1 promoter at five CpG islands. Briefly, genomic DNA from tumor sample was denatured and incubated with MLPA probes for 16 hours to facilitate probe hybridization. The sample was subsequently split into two parts; one was treated with methylation-sensitive enzyme, HhaI, and one without the enzymatic reaction. Both parts were subsequently amplified. Fragment analysis was performed on ABI 3730xL. methylation-specific–MLPA data were analyzed using Coffalyser software (MRC-Holland, Amsterdam, Netherlands), which compares the amplicon signal intensity between the digested and undigested portions of the same sample and reports a ratio. The MLH1 promoter methylation status was determined by the ratios of two individual probes covering the critical region for gene silencing (Deng G, et al: Cancer Res 59, 2029-2033, 1999). Either both probes with a ratio greater than 0.15, or one of the two probes with a ratio greater than 0.30 were interpreted as positive.
TABLE A1.
Testing Product Overview
TABLE A2.
Results Interpretation Guide for MMRp and MMR-Uninformative Patients
TABLE A3.
Patients With Equivocal or Weak Loss on IHC (n = 36)
TABLE A4.
Paired Tumor/Germline Testing Outcome of Referred MMRd Patients
TABLE A5.
Paired Tumor/Germline Testing Result for MMRd Patients by IHC Staining Pattern and Germline Testing History
TABLE A6.
Paired Tumor/Germline Testing Outcome for Groups 3, 4, and 5
Footnotes
Presented at the Collaborative Group of the Americas on Inherited Colorectal Cancer 2017, October 20-21, Orlando, FL; National Society of Genetic Counselors 2017, September 13-16, Columbus, OH; Society of Gynecologic Oncology 2018, March 24-27, New Orleans, LA; American College of Medical Genetics and Genomics 2018, April 10-14, Charlotte, NC; and Collaborative Group of the Americas on Inherited Colorectal Cancer 2018, October 14-15, San Diego, CA.
AUTHOR CONTRIBUTIONS
Conception and design: Monalyn U. Salvador, Melissa R.F. Truelson, Carla Mason, Beth Souders, Kelly Fulk, Jessica Profato, Kory Jasperson, Brigette Tippin-Davis, Phillip Gray, Daniel Chen
Financial support: Brigette Tippin-Davis, Daniel Chen
Administrative support: Brigette Tippin-Davis, Daniel Chen
Provision of study materials or patients: Daniel Chen
Collection and assembly of data: Monalyn U. Salvador, Melissa R.F. Truelson, Carla Mason, Beth Souders, Holly LaDuca, Brittany Dougall, Stephanie Gutierrez, Kory Jasperson, Hsiao-Mei Lu, Swati Shah, Daniel Chen
Data analysis and interpretation: Monalyn U. Salvador, Melissa R.F. Truelson, Carla Mason, Beth Souders, Holly LaDuca, Mary Helen Black, Kelly Fulk, Kory Jasperson, Hsiao-Mei Lu, Elizabeth C. Chao, Negar Ghahramani, Megan Landsverk, Chia-Ling Gau, Daniel Chen, Melissa Pronold
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Comprehensive Paired Tumor/Germline Testing for Lynch Syndrome: Bringing Resolution to the Diagnostic Process
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/site/ifc.
Monalyn U. Salvador
Employment: Ambry Genetics, Ambry Genetics (I)
Stock and Other Ownership Interests: Ambry Genetics (I)
Travel, Accommodations, Expenses: Ambry Genetics, Ambry Genetics (I)
Melissa R.F. Truelson
Employment: Ambry Genetics
Travel, Accommodations, Expenses: Ambry Genetics
Carla Mason
Employment: Ambry Genetics
Beth Souders
Employment: Ambry Genetics
Travel, Accommodations, Expenses: Ambry Genetics
Holly LaDuca
Employment: Ambry Genetics
Stock and Other Ownership Interests: Ambry Genetics
Mary Helen Black
Employment: Ambry Genetics
Research Funding: Ambry Genetics
Travel, Accommodations, Expenses: Ambry Genetics
Kelly Fulk
Employment: Ambry Genetics
Jessica Profato
Employment: Ambry Genetics
Stephanie Gutierrez
Employment: Ambry Genetics
Kory Jasperson
Employment: Ambry Genetics
Brigette Tippin-Davis
Employment: Ambry Genetics
Stock and Other Ownership Interests: Ambry Genetics
Research Funding: Ambry Genetics (Inst)
Travel, Accommodations, Expenses: Ambry Genetics
Hsiao-Mei Lu
Employment: Ambry Genetics
Stock and Other Ownership Interests: Ambry Genetics
Phillip Gray
Employment: Ambry Genetics
Swati Shah
Employment: Ambry Genetics
Stock and Other Ownership Interests: Novartis
Travel, Accommodations, Expenses: Ambry Genetics
Elizabeth C. Chao
Employment: Ambry Genetics
Stock and Other Ownership Interests: Ambry Genetics
Consulting or Advisory Role: Premier Genomics
Negar Ghahramani
Employment: Ambry Genetics
Research Funding: Ambry Genetics
Travel, Accommodations, Expenses: Ambry Genetics
Megan Landsverk
Employment: Ambry Genetics
Chia-Ling Gau
Employment: Ambry Genetics
Stock and Other Ownership Interests: Ambry Genetics
No other potential conflicts of interest were reported.
REFERENCES
- 1.Hampel H, Frankel WL, Martin E, et al. : Feasibility of screening for Lynch syndrome among patients with colorectal cancer. J Clin Oncol 26:5783-5788, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hampel H, Frankel WL, Martin E, et al. : Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer). N Engl J Med 352:1851-1860, 2005 [DOI] [PubMed] [Google Scholar]
- 3.Hampel H, Frankel W, Panescu J, et al. : Screening for Lynch syndrome (hereditary nonpolyposis colorectal cancer) among endometrial cancer patients. Cancer Res 66:7810-7817, 2006 [DOI] [PubMed] [Google Scholar]
- 4.Buchanan DD, Rosty C, Clendenning M, et al. : Clinical problems of colorectal cancer and endometrial cancer cases with unknown cause of tumor mismatch repair deficiency (suspected Lynch syndrome). Appl Clin Genet 7:183-193, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Win AK, Jenkins MA, Dowty JG, et al. : Prevalence and penetrance of major genes and polygenes for colorectal cancer. Cancer Epidemiol Biomarkers Prev 26:404-412, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen S, Wang W, Lee S, et al. : Prediction of germline mutations and cancer risk in the Lynch syndrome. JAMA 296:1479-1487, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Aarnio M, Sankila R, Pukkala E, et al: Cancer risk in mutation carriers of DNA-mismatch-repair genes. Int J Cancer 81:214-218, 1999. [DOI] [PubMed]
- 8.Baglietto L, Lindor NM, Dowty JG, et al. : Risks of Lynch syndrome cancers for MSH6 mutation carriers. J Natl Cancer Inst 102:193-201, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Capelle LG, Van Grieken NC, Lingsma HF, et al. : Risk and epidemiological time trends of gastric cancer in Lynch syndrome carriers in the Netherlands. Gastroenterology 138:487-492, 2010 [DOI] [PubMed] [Google Scholar]
- 10.Hampel H, Stephens JA, Pukkala E, et al. : Cancer risk in hereditary nonpolyposis colorectal cancer syndrome: Later age of onset. Gastroenterology 129:415-421, 2005 [DOI] [PubMed] [Google Scholar]
- 11.Stoffel E, Mukherjee B, Raymond VM, et al. : Calculation of risk of colorectal and endometrial cancer among patients with Lynch syndrome. Gastroenterology 137:1621-1627, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Watson P, Vasen HF, Mecklin JP, et al: The risk of extra-colonic, extra-endometrial cancer in the Lynch syndrome. Int J Cancer 123:444-449, 2008. [DOI] [PMC free article] [PubMed]
- 13. National Comprehensive Cancer Network: Genetic/familial high-risk assessment: Colorectal (version 1.2018). https://www.nccn.org/professionals/physician_gls/pdf/genetics_colon.pdf.
- 14.Committee on Practice Bulletins-GynecologySociety of Gynecologic Oncology : ACOG practice bulletin No. 147: Lynch syndrome. Obstet Gynecol 124:1042-1054, 2014 [DOI] [PubMed] [Google Scholar]
- 15.Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Working Group : Recommendations from the EGAPP Working Group: Genetic testing strategies in newly diagnosed individuals with colorectal cancer aimed at reducing morbidity and mortality from Lynch syndrome in relatives. Genet Med 11:35-41, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giardiello FM, Allen JI, Axilbund JE, et al. : Guidelines on genetic evaluation and management of Lynch syndrome: A consensus statement by the US Multi-Society Task Force on colorectal cancer. Am J Gastroenterol 109:1159-1179, 2014 [DOI] [PubMed] [Google Scholar]
- 17.Syngal S, Brand RE, Church JM, et al. : ACG clinical guideline: Genetic testing and management of hereditary gastrointestinal cancer syndromes. Am J Gastroenterol 110:223-262, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lancaster JM, Powell CB, Kauff ND, et al. : Society of Gynecologic Oncologists Education Committee statement on risk assessment for inherited gynecologic cancer predispositions. Gynecol Oncol 107:159-162, 2007 [DOI] [PubMed] [Google Scholar]
- 19.Goverde A, Spaander MC, van Doorn HC, et al. : Cost-effectiveness of routine screening for Lynch syndrome in endometrial cancer patients up to 70 years of age. Gynecol Oncol 143:453-459, 2016 [DOI] [PubMed] [Google Scholar]
- 20.Leenen CH, Goverde A, de Bekker-Grob EW, et al. : Cost-effectiveness of routine screening for Lynch syndrome in colorectal cancer patients up to 70 years of age. Genet Med 18:966-973, 2016 [DOI] [PubMed] [Google Scholar]
- 21. Haraldsdottir S, Hampel H, Tomsic J, et al: Colon and endometrial cancers with mismatch repair deficiency can arise from somatic, rather than germline, mutations. Gastroenterology 147:1308-13016.e1, 2014. [DOI] [PMC free article] [PubMed]
- 22. Mensenkamp AR, Vogelaar IP, van Zelst-Stams WA, et al: Somatic mutations in MLH1 and MSH2 are a frequent cause of mismatch-repair deficiency in Lynch syndrome-like tumors. Gastroenterology 146:643-6436.e8, 2014. [DOI] [PubMed]
- 23.Sourrouille I, Coulet F, Lefevre JH, et al. : Somatic mosaicism and double somatic hits can lead to MSI colorectal tumors. Fam Cancer 12:27-33, 2013 [DOI] [PubMed] [Google Scholar]
- 24.Hampel H, Pearlman R, Beightol M, et al. : Assessment of tumor sequencing as a replacement for lynch syndrome screening and current molecular tests for patients with colorectal cancer. JAMA Oncol 4:806-813, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gray PN, Tsai P, Chen D, et al. : TumorNext-Lynch-MMR: A comprehensive next generation sequencing assay for the detection of germline and somatic mutations in genes associated with mismatch repair deficiency and Lynch syndrome. Oncotarget 9:20304-20322, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Umar A, Boland CR, Terdiman JP, et al. : Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst 96:261-268, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vasen HF, Watson P, Mecklin JP, et al. : New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative group on HNPCC. Gastroenterology 116:1453-1456, 1999 [DOI] [PubMed] [Google Scholar]
- 28.Kastrinos F, Uno H, Ukaegbu C, et al. : Development and validation of the PREMM5 model for comprehensive risk assessment of Lynch syndrome. J Clin Oncol 35:2165-2172, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bao F, Panarelli NC, Rennert H, et al. : Neoadjuvant therapy induces loss of MSH6 expression in colorectal carcinoma. Am J Surg Pathol 34:1798-1804, 2010 [DOI] [PubMed] [Google Scholar]
- 30.Graham RP, Kerr SE, Butz ML, et al. : Heterogenous MSH6 loss is a result of microsatellite instability within MSH6 and occurs in sporadic and hereditary colorectal and endometrial carcinomas. Am J Surg Pathol 39:1370-1376, 2015 [DOI] [PubMed] [Google Scholar]
- 31.Radu OM, Nikiforova MN, Farkas LM, et al. : Challenging cases encountered in colorectal cancer screening for Lynch syndrome reveal novel findings: nucleolar MSH6 staining and impact of prior chemoradiation therapy. Hum Pathol 42:1247-1258, 2011 [DOI] [PubMed] [Google Scholar]
- 32.Shia J, Zhang L, Shike M, et al. : Secondary mutation in a coding mononucleotide tract in MSH6 causes loss of immunoexpression of MSH6 in colorectal carcinomas with MLH1/PMS2 deficiency. Mod Pathol 26:131-138, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pearlman R, Frankel WL, Swanson B, et al. : Prevalence and spectrum of germline cancer susceptibility gene mutations among patients with early-onset colorectal cancer. JAMA Oncol 3:464-471, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shirts BH, Konnick EQ, Upham S, et al. : Using somatic mutations from tumors to classify variants in mismatch repair genes. Am J Hum Genet 103:19-29, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]