Abstract
PURPOSE
CheckMate 568 is an open-label phase II trial that evaluated the efficacy and safety of nivolumab plus low-dose ipilimumab as first-line treatment of advanced/metastatic non–small-cell lung cancer (NSCLC). We assessed the association of efficacy with programmed death ligand 1 (PD-L1) expression and tumor mutational burden (TMB).
PATIENTS AND METHODS
Two hundred eighty-eight patients with previously untreated, recurrent stage IIIB/IV NSCLC received nivolumab 3 mg/kg every 2 weeks plus ipilimumab 1 mg/kg every 6 weeks. The primary end point was objective response rate (ORR) in patients with 1% or more and less than 1% tumor PD-L1 expression. Efficacy on the basis of TMB (FoundationOne CDx assay) was a secondary end point.
RESULTS
Of treated patients with tumor available for testing, 252 patients (88%) of 288 were evaluable for PD-L1 expression and 98 patients (82%) of 120 for TMB. ORR was 30% overall and 41% and 15% in patients with 1% or greater and less than 1% tumor PD-L1 expression, respectively. ORR increased with higher TMB, plateauing at 10 or more mutations/megabase (mut/Mb). Regardless of PD-L1 expression, ORRs were higher in patients with TMB of 10 or more mut/Mb (n = 48: PD-L1, ≥ 1%, 48%; PD-L1, < 1%, 47%) versus TMB of fewer than 10 mut/Mb (n = 50: PD-L1, ≥ 1%, 18%; PD-L1, < 1%, 5%), and progression-free survival was longer in patients with TMB of 10 or more mut/Mb versus TMB of fewer than 10 mut/Mb (median, 7.1 v 2.6 months). Grade 3 to 4 treatment-related adverse events occurred in 29% of patients.
CONCLUSION
Nivolumab plus low-dose ipilimumab was effective and tolerable as a first-line treatment of advanced/metastatic NSCLC. TMB of 10 or more mut/Mb was associated with improved response and prolonged progression-free survival in both tumor PD-L1 expression 1% or greater and less than 1% subgroups and was thus identified as a potentially relevant cutoff in the assessment of TMB as a biomarker for first-line nivolumab plus ipilimumab.
INTRODUCTION
Current first-line treatments for metastatic non–small-cell lung cancer (NSCLC) without mutations that are sensitive to targeted therapies include platinum-based chemotherapy,1,2 pembrolizumab—an anti–programmed death (PD) -1 immune checkpoint inhibitor—in patients with 50% or greater tumor PD ligand 1 (PD-L1) expression,1-3 and pembrolizumab combined with chemotherapy.1-3 The introduction of first-line immunotherapy has improved outcomes for patients, but an unmet need remains for biomarkers to identify patients who benefit most from this treatment approach. Combination immunotherapy and the identification of clinically meaningful biomarkers beyond PD-L1 have the potential to allow more patients to experience durable clinical benefit while deferring chemotherapy to the second line.
Nivolumab, a fully human immunoglobulin G4 PD-1 antibody, currently approved for previously treated advanced or metastatic NSCLC,4 and ipilimumab, an anti–cytotoxic T lymphocyte antigen-4 antibody, are immune checkpoint inhibitors with complementary mechanisms of action.5 The combination of these two agents has shown promising results in multiple cancers6-8 and is currently approved for the treatment of unresectable or metastatic melanoma and renal cell carcinoma.4 In the phase I CheckMate 012 trial, nivolumab plus low-dose ipilimumab demonstrated tolerable safety and durable clinical activity as a first-line treatment of advanced NSCLC.7,9,10
Tumor PD-L1 expression has been identified as a predictive biomarker for response to immune checkpoint inhibitors in several studies in both newly diagnosed and previously treated NSCLC.11-13 Tumor mutational burden (TMB) is an emerging biomarker of response to immune checkpoint inhibitors in various cancers, including NSCLC, small-cell lung cancer, bladder cancer, and melanoma.14-20 In NSCLC, PD-L1 and TMB are independent biomarkers.14,17-19,21 The phase III CheckMate 026 trial suggested the potential utility of TMB in identifying patients in whom response and prolonged progression-free survival (PFS) are more likely with first-line treatment with nivolumab in NSCLC.14 In addition, a retrospective analysis of CheckMate 012 found that high TMB—at or above the median—was associated with improved objective response rate (ORR) and PFS with first-line nivolumab plus ipilimumab.21
CheckMate 568 is a two-part, open-label, single-arm, phase II trial that evaluated nivolumab plus low-dose ipilimumab for first-line treatment of advanced or metastatic NSCLC. We report efficacy by PD-L1 expression and safety from part 1, and the association of TMB with response and PFS with this regimen. We also report the identification of a potentially clinically meaningful TMB cutoff, which was subsequently validated through a preplanned coprimary end point analysis in CheckMate 227 (ClinicalTrials.gov identifier: NCT02477826). CheckMate 227 met its coprimary end point, with significantly longer PFS (hazard ratio, 0.58 [97.5% CI, 0.41 to 0.81]; P = .0002) with nivolumab plus ipilimumab versus platinum-based chemotherapy in patients with TMB of 10 or more mutations per megabase (mut/Mb).22
PATIENTS AND METHODS
Patients and Treatment
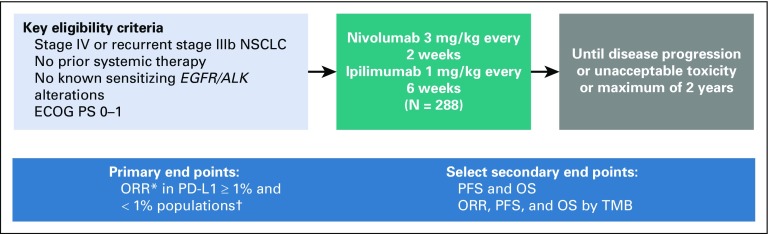
Patients age 18 years or older from the United States and Canada with histologically confirmed stage IV or recurrent stage IIIB NSCLC per 7th International Association for the Study of Lung Cancer classification who met the following criteria were eligible: measurable disease per RECIST version 1.1, Eastern Cooperative Oncology Group performance status of 0 or 1, and no prior systemic therapy for stage IV disease (Appendix Fig A1, online only). Prior definitive chemoradiation for locally advanced disease was permitted if the last chemotherapy or radiotherapy was 6 months or more before enrollment. Prior adjuvant or neoadjuvant chemotherapy for early-stage disease was permitted if completed 6 months or more before initiating study treatment. Tumor tissue was required to be available for central PD-L1 testing.
Key exclusion criteria included known EGFR mutations or ALK alterations that are sensitive to targeted therapy, autoimmune disease, or untreated CNS metastases. Patients with CNS metastases were eligible if those were treated adequately and the patients received no corticosteroids or a dose of prednisone 10 mg or less per day (or equivalent) for 2 or more weeks before first treatment. For additional details, see the Appendix (online only).
Patients received nivolumab 3 mg/kg intravenously over 30 minutes every 2 weeks plus ipilimumab 1 mg/kg intravenously over 30 minutes every 6 weeks for 2 or more years or until disease progression or unacceptable toxicity. Treatment beyond initial investigator-assessed RECIST v1.1–defined progression was permitted if the patient had investigator-assessed clinical benefit and tolerated treatment.
End Points and Assessments
The primary end point was ORR by blinded independent central review (BICR) per RECIST v1.1, assessed in patients with 1% or greater and less than 1% tumor PD-L1 expression. ORR was defined as the proportion of patients who achieved a best response of complete response (CR) or partial response between the dates of first treatment and documented disease progression or subsequent anticancer therapy, whichever occurred first. ORR required confirmation at a subsequent time point of 4 or more weeks later.
Secondary end points included ORR and PFS by BICR per RECIST v1.1, overall survival, and efficacy by TMB and PD-L1 expression. Exploratory end points included safety and tolerability.
After the first dose, we conducted tumor assessments every 6 weeks (±7 days) until week 48, then every 12 weeks (±7 days) thereafter until BICR-assessed progression. Adverse events (AEs) were graded using the US National Cancer Institute’s Common Terminology Criteria for Adverse Events, version 4.0.
PD-L1 Biomarker Analysis
Fresh or archival tumor biopsy specimens were tested for PD-L1 by a centralized laboratory (Agilent Technologies/Dako PD-L1 immunohistochemistry 28-8 pharmDx test) before study treatment.
TMB Analysis
As part of a protocol amendment that was implemented after enrollment was completed, efficacy by TMB was added as a secondary end point to assess the role of TMB in first-line treatment with nivolumab plus ipilimumab and identify a TMB cutoff for additional validation in the phase III CheckMate 227 trial. We conducted TMB and secondary end point analyses in patients with available tissue at the time of the amendment. The FoundationOne CDx (Foundation Medicine, Cambridge, MA) platform is an analytically and clinically validated companion diagnostic for targeted therapy in solid tumors.23,24 The assay used next-generation sequencing (NGS) to detect substitutions, insertions and deletions (indels), and copy number alterations in 324 genes and selected gene rearrangements on archival or fresh formalin-fixed, paraffin-embedded tumor samples.24 A minimum of 25 mm2 of tumor tissue, or 10 slides cut at 4 to 5 μm, were required.25
TMB was defined as the number of somatic, coding, base substitutions, and short indels per Mb of genome examined. All base substitutions and indels in the coding region of targeted genes, including synonymous mutations, were filtered for both oncogenic driver events according to the Catalogue of Somatic Mutations in Cancer (COSMIC) and germline status per the Single Nucleotide Polymorphism Database (dbSNP) and the Exome Aggregation Consortium (ExAC) database, as well as with a private database of rare germline events compiled in the Foundation Medicine clinical cohort. Additional filtering on the basis of a computational assessment of germline status using the somatic-germline-zygosity tool was also performed.26 TMB was calculated according to previously defined methods.27 Independent reports have demonstrated concordance between TMB estimated from whole-exome sequencing and TMB estimated from targeted NGS in NSCLC and across multiple solid tumors.18,28,29
Trial Oversight
The study protocol was approved by an institutional review board or independent ethics committee at each participating center. The study was conducted in accordance with the Declaration of Helsinki and Good Clinical Practice guidelines as defined by the International Conference on Harmonization. All patients provided written informed consent before enrollment. The Bristol-Myers Squibb policy on data sharing may be found online.30 The database lock for this analysis was August 24, 2017.
Statistical Analysis
For the primary end point of ORR, a sample size of approximately 300 treated patients (60 patients with ≥ 50% tumor PD-L1 expression and 100 patients with < 1% tumor PD-L1 expression) provided 95% confidence that the observed ORR was estimated to within 5.8% of estimates for all treated patients, 13.5% of estimates for patients with 50% or greater tumor PD-L1 expression, and 10.5% of estimates for patients with less than 1% tumor PD-L1 expression. ORR was summarized by a binomial response rate and its corresponding two-sided 95% exact CIs using the Clopper-Pearson method. Best overall response was summarized by response category—CR, partial response, stable disease, or progressive disease. We estimated PFS time-to-event distributions using the Kaplan-Meier method.
We used nonparametric receiver operating characteristic (ROC) curve estimates to evaluate the performance of TMB as a classifier of ORR per BICR. Area under the ROC curve was estimated to summarize TMB classification accuracy across all possible TMB cutoffs. For additional details, see the Appendix.
Safety was summarized for all treated patients and assessed through summaries and by subject listings of deaths, serious AEs, AEs leading to discontinuation or modification, overall AEs, select AEs, and laboratory abnormalities.
RESULTS
Patients and Treatment
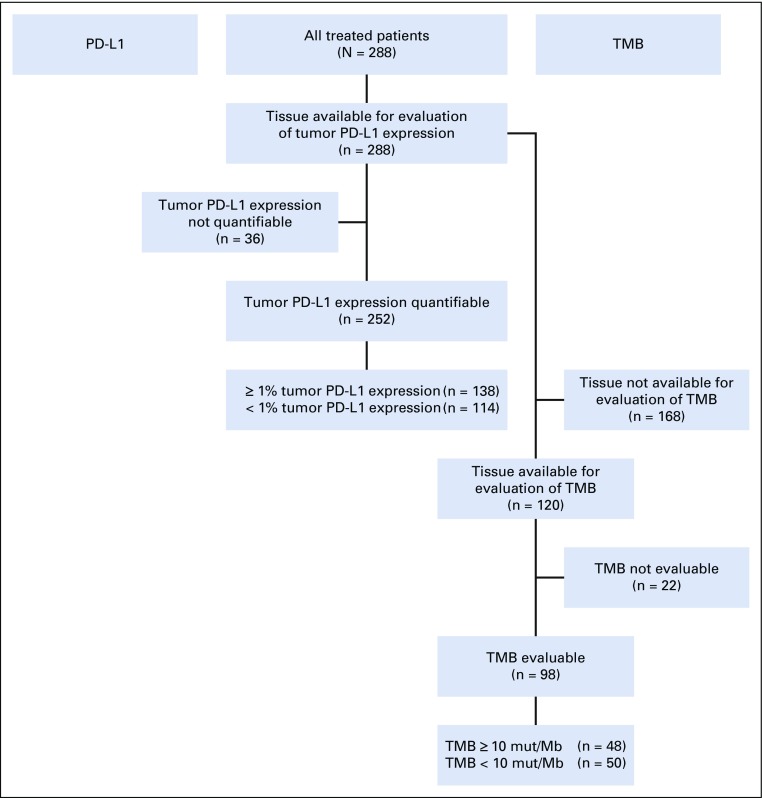
Overall, 427 patients were enrolled from February 2016 through November 2016, and 288 patients (67%) were treated. Of 139 enrolled patients who did not enter the treatment period, 106 no longer met the study criteria, 15 withdrew consent, seven died, one developed an AE during screening and was not able to proceed with enrollment, one was lost to follow-up, and nine did not receive treatment for other reasons. Tumor samples for PD-L1 testing were available from all treated patients, and 252 patients (88%) were evaluable for tumor PD-L1 expression (testing success rate, 88%). In patients with evaluable PD-L1, 138 (55%) had 1% or greater tumor PD-L1 expression and 114 (45%) had less than 1% tumor PD-L1 expression (Appendix Fig A2, online only).
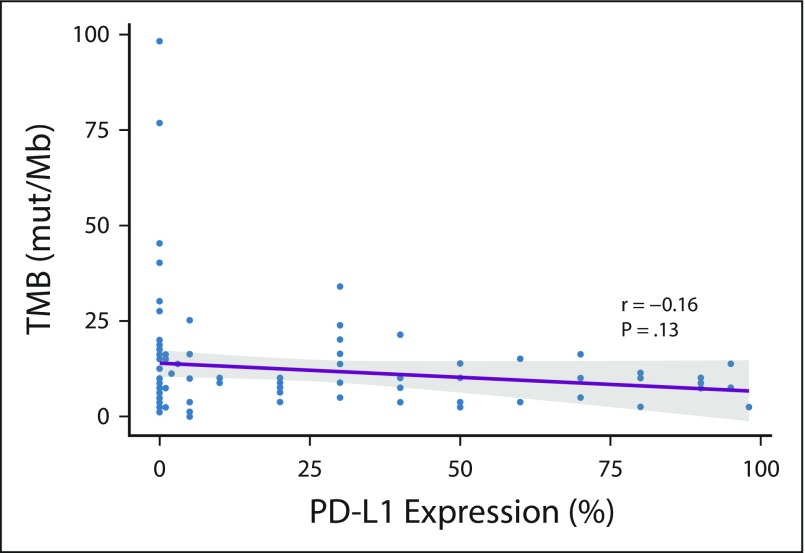
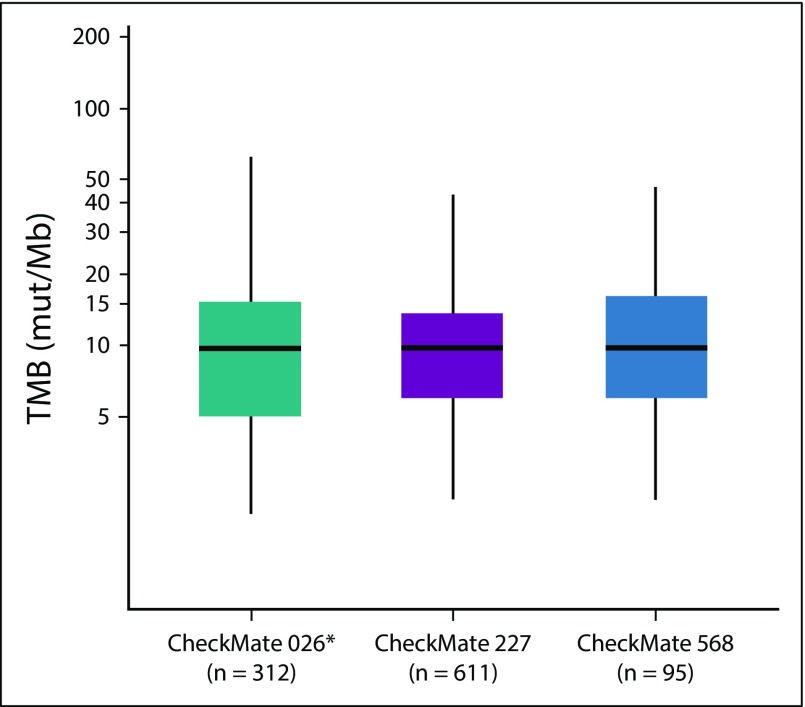
At the time of protocol amendment, 120 treated patients (42%) of 288 had sufficient tumor tissue available after PD-L1 testing for TMB analysis, and 98 (34%) had a successful TMB result (testing success rate, 82%). Within the TMB-evaluable population (n = 98), 50 patients (51%) had TMB of fewer than 10 mut/Mb and 48 (49%) had TMB of 10 or more mut/Mb (Appendix Fig A2). Ninety-five patients (33% of all treated patients) were evaluable for both tumor PD-L1 expression and TMB. A regression analysis demonstrated no association between TMB and PD-L1 expression (Appendix Fig A3, online only). Distribution and prevalence of TMB were similar to CheckMate 026 and CheckMate 227 (Appendix Fig A4, online only).
Baseline characteristics were generally consistent among the all-treated, PD-L1–evaluable, and TMB-evaluable populations. There was a slightly higher rate of squamous histology in TMB-evaluable patients versus all-treated and PD-L1–evaluable patients (35% v 26% v 25%, respectively; Table 1).
TABLE 1.
Baseline Characteristics

With a minimum follow-up of 6 months, 88 all-treated patients (31%) remained on treatment (Appendix Table A1, online only). Median duration of nivolumab plus ipilimumab was 4.0 months (range, 0.03 to ≥ 17.5 months). The median number of doses received was 8.5 (range, 1 to 38) for nivolumab and 3.0 (range, 1 to 13) for ipilimumab. Patients with 1% or greater tumor PD-L1 expression had a longer median duration of therapy versus those with less than 1% tumor PD-L1 expression (5.7 [range, 0.03 to ≥ 17.5] v 2.8 [range, 0.03 to ≥ 15.7] months).
Efficacy
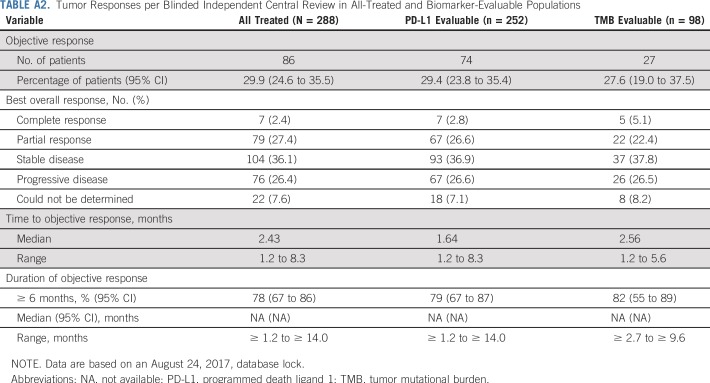
ORR was 30% in all treated patients (Table 2 and Appendix Table A2, online only) and similar in PD-L1–evaluable (29%) and TMB-evaluable (28%) patients (Appendix Table A2). CRs were observed in 2% (n = 7) of all treated patients. Responses were durable across populations. Median duration of response was not reached at the time of this analysis (median follow-up, 8.8 months). In all treated patients, median PFS was 4.2 months (95% CI, 3.0 to 5.6 months) and the 6-month PFS rate was 43% (Appendix Fig A5, online only).
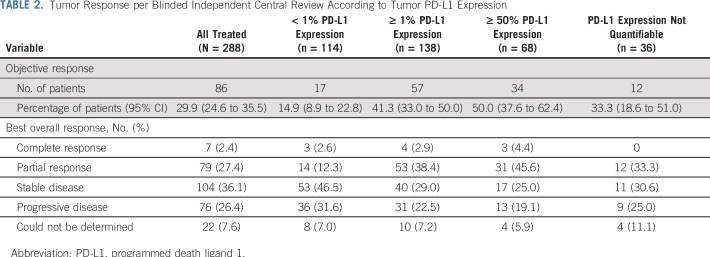
TABLE 2.
Tumor Response per Blinded Independent Central Review According to Tumor PD-L1 Expression
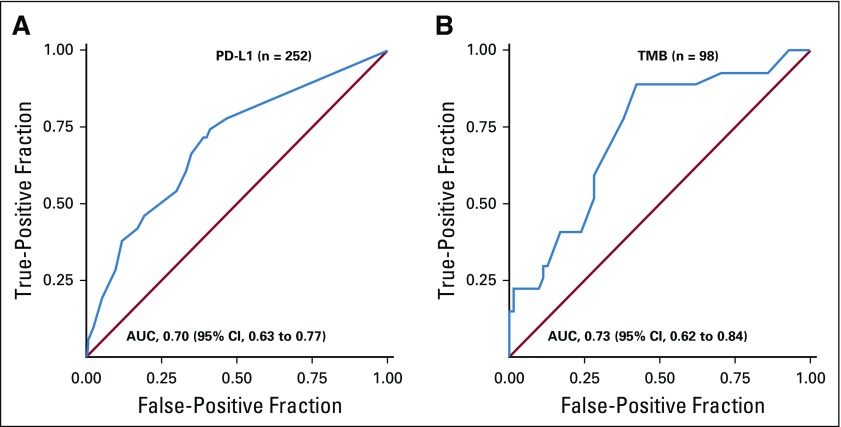
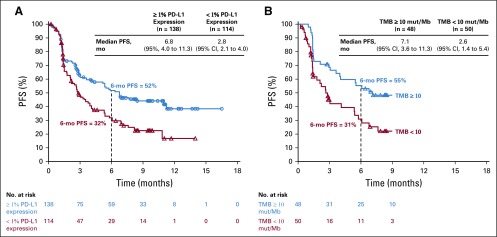
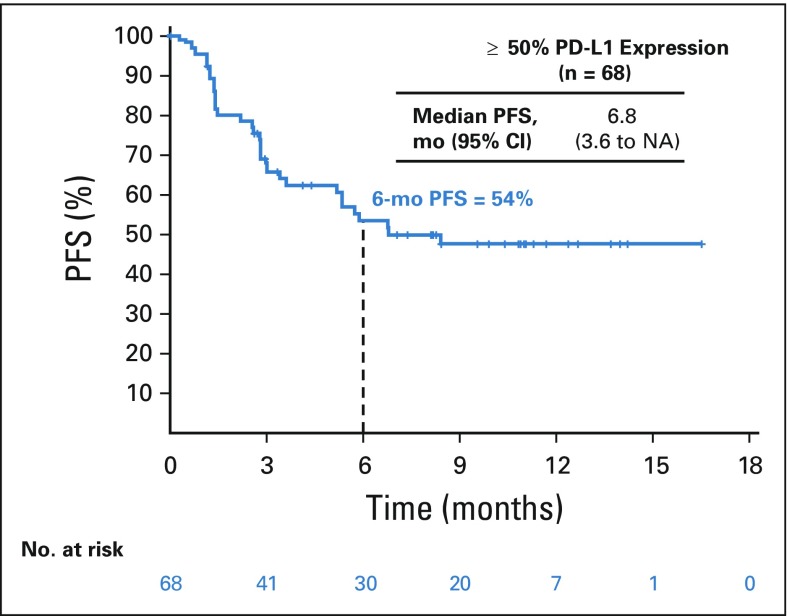
In patients with 1% or greater tumor PD-L1 expression (n = 138), ORR was 41%, including CRs in four patients (3%; Table 2). In patients with less than 1% tumor PD-L1 expression (n = 114), ORR was 15%, with CRs in three patients (3%). PD-L1 was an informative classifier for ORR (Fig 1A; area under the curve [AUC], 0.70; 95% CI, 0.63 to 0.77). True positive fraction (TPF) and false positive fraction (FPF) at the PD-L1 1% cutoff on the basis of 2,000 bootstrap replicates was 0.77 (95% CI, 0.68 to 0.87) and 0.55 (95% CI, 0.47 to 0.62), respectively. Median PFS was longer (6.8 [95% CI, 4.0 to 11.3] months v 2.8 [95% CI, 2.1 to 4.0] months) and the 6-month PFS rate was higher (52% v 32%) in patients with 1% or greater versus less than 1% tumor PD-L1 expression (Fig 2A). In patients with 50% or greater tumor PD-L1 expression (n = 68), ORR was 50%, including CRs in three patients (4%; Table 2), median PFS was 6.8 months, and the 6-month PFS rate was 54% (Appendix Fig A6, online only).
FIG 1.
Receiver operating characteristic curves of objective response rate by (A) tumor programmed death ligand 1 (PD-L1) expression and (B) tumor mutational burden (TMB). AUC, area under the curve.
FIG 2.
Progression-free survival (PFS) by (A) tumor programmed death ligand 1 (PD-L1) expression and (B) tumor mutational burden (TMB) cutoff. mut/Mb, mutations per megabase.
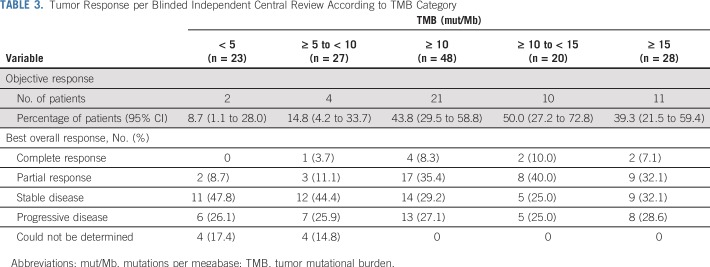
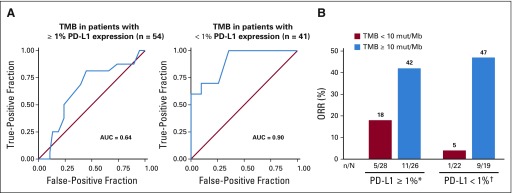
In the TMB-evaluable population, ORR increased in subgroups of patients with higher TMB, plateauing at 10 or more mut/Mb (Table 3). ORR was 44% versus 12% in patients with TMB of 10 or more mut/Mb versus TMB of fewer than 10 mut/Mb. No additional ORR benefit was observed in patients with TMB of 15 or more mut/Mb (Table 3). The association of efficacy with TMB did not depend on tumor PD-L1 expression. Regardless of PD-L1 expression, ORRs were greater in patients with TMB of 10 or more mut/Mb (42% PD-L1 ≥ 1% and 47% PD-L1 < 1%) versus TMB of fewer than 10 mut/Mb (18% PD-L1 ≥ 1% and 5% PD-L1 < 1%; Appendix Fig A7B, online only). ROC analysis demonstrated that TMB may be an informative classifier for ORR with nivolumab plus ipilimumab (Fig 1B; AUC, 0.73; 95% CI, 0.62 to 0.84), including for patients with 1% or greater and less than 1% tumor PD-L1 expression (Appendix Fig A7A). AUC (95% CIs) for PD-L1 and TMB ROC curves overlapped. TPF and FPF at the TMB 10 mut/Mb cutoff on the basis of 2,000 bootstrap replicates was 0.78 (95% CI, 0.63 to 0.93) and 0.62 (95% CI, 0.49 to 0.73), respectively.
TABLE 3.
Tumor Response per Blinded Independent Central Review According to TMB Category
Median PFS was longer in patients with TMB of 10 or more mut/Mb (7.1 months [95% CI, 3.6 to 11.3 months]) versus TMB of fewer than 10 mut/Mb (2.6 months [95% CI, 1.4 to 5.4 months]; Fig 2B). The percentage of patients who remained progression free at 6 months was 55% and 31% for the TMB of 10 or more mut/Mb and TMB of fewer than 10 mut/Mb subgroups, respectively.
Safety
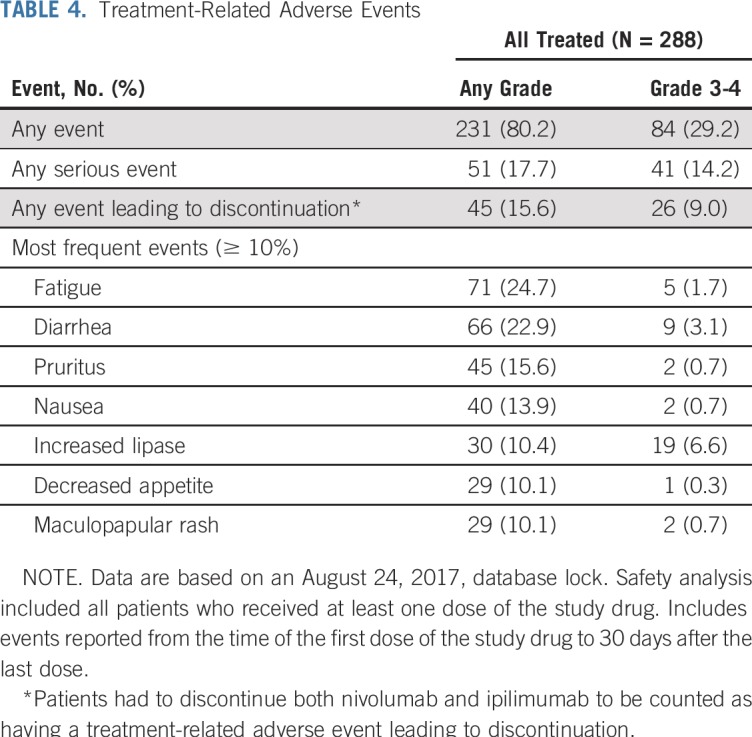
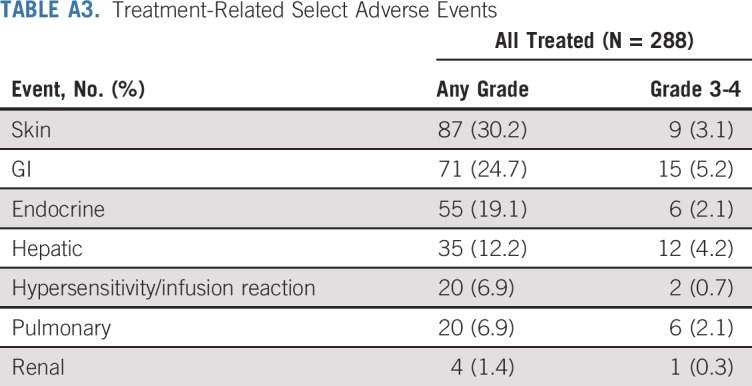
Treatment-related AEs (TRAEs) of any grade were reported in 80% of patients, and grade 3 to 4 TRAEs occurred in 29% of patients (Table 4). Any grade and grade 3 to 4 TRAEs led to discontinuation in 16% and 9% of patients, respectively. Most treatment-related select AEs—those with a potential immunologic cause—were grade 1 to 2 (Appendix Table A3, online only). Most common treatment-related select AEs of any grade were skin reactions (30%), and the most common grade 3 to 4 treatment-related select AEs were GI toxicities (5%).
TABLE 4.
Treatment-Related Adverse Events
Four treatment-related deaths were reported within 100 days of the last dose of the study drug, attributed (one each) to cardiac arrhythmia (known prior history of cardiac disease), pneumonitis, hypoxia, and other. Per autopsy, patients with pneumonitis and hypoxia both had documented extensive underlying lung cancer with lymphangitic spread and alveolar damage. Primary reasons per investigator for cause of death classified as other were toxicity related to immune-mediated hepatitis in the presence of worsening liver metastasis, grade 4 elevated bilirubin, grade 1 aspartate aminotransferase elevation, and grade 2 alanine aminotransferase elevation.
DISCUSSION
Nivolumab plus low-dose ipilimumab was effective in the first-line treatment of patients with advanced or metastatic NSCLC in this phase II study. ORR was 30% in all treated patients and responses were durable—median duration of response not reached. Enhanced benefit was observed in patients with 1% or greater versus less than 1% tumor PD-L1 expression. Higher response rates and improved PFS were also observed in patients with TMB of 10 or more mut/Mb versus TMB of fewer than 10 mut/Mb, irrespective of PD-L1 expression. Results of CheckMate 568 were similar to those reported for the phase I study CheckMate 012.7,21 Treatment was tolerable, as evidenced by a relatively low discontinuation rate as a result of TRAEs, which is consistent with previous findings for this dosing regimen.7
Several factors were considered in identifying the TMB of 10 or more mut/Mb cutoff for selecting patients who are most likely to benefit from first-line nivolumab plus ipilimumab in NSCLC: ORR was greater than 40% at this cutoff, regardless of tumor PD-L1 expression, and higher than historically observed responses with chemotherapy in this population31; a higher TMB cutoff did not result in additional improvement; and classification performance (TPF and FPF) on the basis of ROC analysis indicated that TMB may be an informative classifier at the 10 mut/Mb cutoff. The identified TMB cutoff with first-line nivolumab plus ipilimumab required subsequent prospective validation in a larger randomized study in this patient population.
TMB assessment in Checkmate 568 required 10 unstained slides typical of standard-of-care NGS testing and was introduced during the course of the study on the basis of emerging data with the intention of predefining a cutoff for additional assessment and validation in the phase III CheckMate 227 study. However, as biospecimens were used initially for assessment of PD-L1 expression, only one third of the overall patient population had sufficient tissue remaining to assess TMB. Despite this limitation, 98 patients (82%) of 120 with available tumor were successfully tested for TMB. Of note, 49% of TMB-evaluable patients had TMB of 10 or more mut/Mb, which is similar to the prevalence observed in CheckMate 026 and CheckMate 227, with more than 1,000 tumors successfully analyzed for TMB across the three studies.14,22 Baseline characteristics and outcomes in the TMB-evaluable population were representative of all treated patients, and an association between high TMB and efficacy was demonstrated with the combination regimen. These results informed the statistical plan of CheckMate 227 and the coprimary end point of PFS in patients with TMB of 10 or more mut/Mb. Results of CheckMate 227 subsequently validated the TMB cutoff by demonstrating that patients with TMB of 10 or more mut/Mb had significantly prolonged PFS with first-line nivolumab plus ipilimumab versus chemotherapy.22 No significant difference in PFS between the two treatments was observed in patients with TMB of fewer than 10 mut/Mb.22
CheckMate 568 supported PD-L1 and TMB as biomarkers associated with tumor response to combination immune checkpoint inhibitor therapy, as previously demonstrated in several studies.14,17-19,21 Tumor PD-L1 expression and TMB were each associated with ORR, with no meaningful correlation between the two biomarkers observed. Improved ORR in patients with TMB of 10 or more mut/Mb was observed irrespective of tumor PD-L1 expression.
TMB assessment was conducted by the NGS-based FoundationOne CDx assay, which has since been approved by the US Food and Drug Administration as a companion diagnostic for targeted therapies, with indications in five solid tumors.23 The cost of NGS has dramatically decreased as a result of emerging technical advances; thus, increased accessibility for the assessment of multiple markers within a single tissue sample increases the feasibility of including TMB as part of the routine diagnostic work-up of patients.32
The CheckMate 568 single-arm, phase II trial demonstrated that nivolumab plus low-dose ipilimumab was effective and well tolerated as a first-line treatment of advanced or metastatic NSCLC. TMB and PD-L1 are each associated with enhanced ORR and PFS. This analysis supported TMB of 10 or more mut/Mb as a clinically meaningful cutoff for response and PFS with this regimen in this patient population, and informed the preplanned coprimary end point analysis in CheckMate 227.
ACKNOWLEDGMENT
The authors thank the patients and their families, as well as the participating study teams, for making this study possible (see the Appendix for a full list of investigators); Jessica Proszynski and Katleen Van Hoefs of Bristol-Myers Squibb for their contributions as protocol managers of this study; Foundation Medicine for collaborative development of the FoundationOne CDx tumor gene profiling assay; and the staff of Dako for collaborative development of the PD-L1 IHC 28-8 pharmDx assay. Medical writing and editorial assistance, including writing of the first draft, was provided by Stefanie Puglielli, PhD, of Evidence Scientific Solutions Inc., with funding from Bristol-Myers Squibb.
APPENDIX
Supplemental Methods
Patients with the need for systemic treatment with corticosteroids (prednisone greater than 10 mg per day or equivalent) or other immunosuppressive medications within 2 weeks of study drug administration were also excluded from this study. However, inhaled or topical steroids and adrenal replacement corticosteroid doses (greater than 10 mg per day equivalent) were permitted in the absence of autoimmune disease.
Patients with known EGFR mutations and those with nonsquamous histology and unknown/indeterminate EGFR status were excluded. Patients with sensitizing ALK mutations were excluded, although testing was not mandatory, and those with unknown/indeterminate ALK status could be enrolled.
For the evaluation of the performance of tumor mutational burden (TMB) as a classifier of objective response rate per blinded independent central review, we generated receiver operating characteristic curves of the estimated true positive fraction versus false positive fraction for each possible observed TMB cutoff. Bootstrapping from the programmed death ligand 1–evaluable (n = 252) and TMB-evaluable samples (n = 98) was used to estimate 95% CIs for true positive fraction and false positive fraction at the 1% programmed death ligand 1 expression and 10 mut/Mb TMB cutoffs, respectively. Each of these confidence interval estimates was determined using 2,000 bootstrap replicate samples as implemented in the R statistical package pROC (Robin X, et al: BMC Bioinformatics 12:77, 2011). The 95% CI for the area under the curve was estimated using the method by Delong et al (Biometrics 44:837-845, 1988) and used to summarize TMB classification accuracy across all possible TMB cutoffs.
List of CheckMate 568 Part 1 Investigators
Canada: Jacques Jolivet (St. Jerome Medical Research Inc.), Silvana Spadafora (Sault Area Hospital), Anna Tomiak (Kingston Regional Cancer Centre); United States: Eric Avery (Nebraska Hematology Oncology), Bruno Bastos (Cleveland Clinic Florida), Hossein Borghaei (Fox Chase Cancer Center), Julie Brahmer (Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins University), Brian Byrne (Cancer Center of Central Connecticut), Franklin Chen (Novant Health Oncology Specialists), Jason Chesney (James Graham Brown Cancer Center), Shaker Dakhil (Cancer Center of Kansas), Gene Finley (Allegheny Health Network), Justin Gainor (Harvard Cancer Hospital), Leena Gandhi (Massachusetts General Hospital), Martin Gutierrez (John Theurer Cancer Center at Hackensack University Medical Center), Matthew Hellmann (Memorial Sloan Kettering Cancer Center), Leora Horn (Vanderbilt-Ingram Cancer Center), James Orcutt (Charleston Hematology Oncology Associates), Gregory Otterson (The Ohio State University), Ian Rabinowitz (University of New Mexico Comprehensive Cancer Center), Suresh Ramalingam (Winship Cancer Institute, Emory University), Neal Ready (Duke University Medical Center), Igor Rybkin (Henry Ford Health System), Jeffrey Schneider (Winthrop University Hospital), David Spigel (Sarah Cannon Research Institute), Mark Taylor (Summit Cancer Care), Kai Zu (Sharp Memorial Hospital).
FIG A1.
Study design. (*) Efficacy analyses by blinded independent central review. (†) Programmed death ligand 1 (PD-L1) status determined by Dako PD-L1 IHC 28-8 pharmDx immunohistochemical test. Database lock: August 24, 2017; minimum follow-up: 6 months; median follow-up: 8.8 months. ECOG PS, Eastern Cooperative Oncology Group performance status; NSCLC, non–small-cell lung cancer; ORR, objective response rate; OS, overall survival; PFS, progression-free survival; TMB, tumor mutational burden.
FIG A2.
Biomarker assessment flow diagram. PD-L1, programmed death ligand 1; TMB, tumor mutational burden.
FIG A3.
Regression analysis of tumor mutational burden (TMB) and tumor programmed death ligand 1 (PD-L1) expression. mut/Mb, mutations per megabase.
FIG A4.
Tumor mutational burden (TMB) distribution across first-line non–small-cell lung cancer data sets. (*) In CheckMate 026, TMB was evaluated by whole-exome sequencing and values were converted to mutations per megabase (mut/Mb) using TMB bridging analysis.14
FIG A5.
Progression-free survival (PFS) in all treated patients.
FIG A6.
Progression-free survival (PFS) in patients with 50% or greater tumor programmed death ligand 1 (PD-L1) expression. NA, not available.
FIG A7.
Tumor responses by tumor programmed death ligand 1 (PD-L1) expression and tumor mutational burden (TMB) category. (A) Receiver operating characteristic (ROC) curve for TMB and objective response rate (ORR) by tumor PD-L1 expression. (B) ORRs per blinded independent central review. (*)Complete responses were 4% for both TMB less than 10 mutations per megabase (mut/Mb) and TMB 10 or more mut/Mb. (†) Complete responses were 0% for TMB less than 10 mut/Mb and 16% for TMB 10 or more mut/Mb. AUC, area under the curve.
TABLE A1.
End-of-Treatment Summary
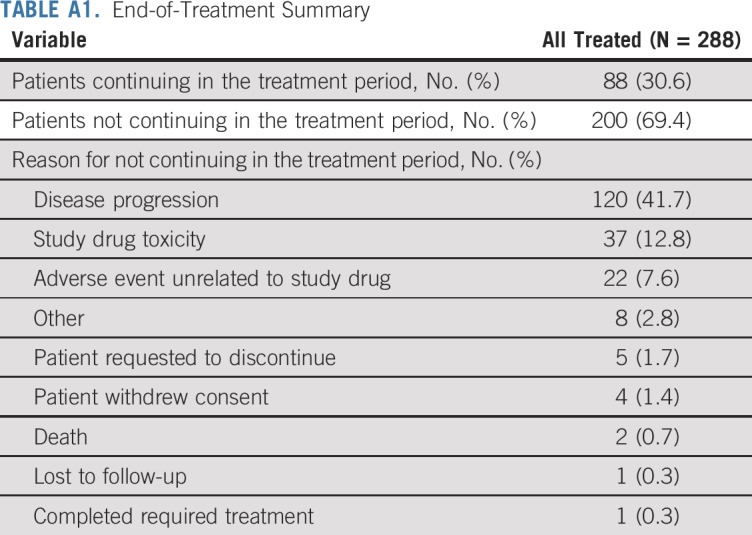
TABLE A2.
Tumor Responses per Blinded Independent Central Review in All-Treated and Biomarker-Evaluable Populations
TABLE A3.
Treatment-Related Select Adverse Events
Footnotes
Presented at the 2018 American Association for Cancer Research Annual Meeting, Chicago, IL, April 14-18, 2018.
Funded by Bristol-Myers Squibb, (Princeton, NJ) and ONO Pharmaceutical (Osaka, Japan).
Clinical trial information: NCT02659059.
AUTHOR CONTRIBUTIONS
Conception and design: Neal Ready, Matthew D. Hellmann, Julie Brahmer, Xuemei Li, William J. Geese, Joseph D. Szustakowski, Han Chang, Suresh S. Ramalingam
Provision of study materials or patients: Matthew D. Hellmann, Mark M. Awad, Martin Gutierrez, Justin F. Gainor, Hossein Borghaei, Leora Horn, Mihaela Mates, Julie Brahmer, Ian Rabinowitz, Pavan S. Reddy, James Orcutt, David R. Spigel, Luis Paz-Ares, Suresh S. Ramalingam
Collection and assembly of data: Neal Ready, Matthew D. Hellmann, Mark M. Awad, Gregory A. Otterson, Martin Gutierrez, Justin F. Gainor, Hossein Borghaei, Jacques Jolivet, Leora Horn, Pavan S. Reddy, Jason Chesney, James Orcutt, Martin Reck, Xuemei Li, George Green, Han Chang
Data analysis and interpretation: Neal Ready, Matthew D. Hellmann, Mark M. Awad, Gregory A. Otterson, Martin Gutierrez, Justin F. Gainor, Hossein Borghaei, Jacques Jolivet, Leora Horn, Mihaela Mates, Julie Brahmer, Ian Rabinowitz, Pavan S. Reddy, Jason Chesney, David R. Spigel, Martin Reck, Kenneth John O’Byrne, Luis Paz-Ares, Wenhua Hu, Kim Zerba, Xuemei Li, Brian Lestini, William J. Geese, Joseph D. Szustakowski, George Green, Han Chang, Suresh S. Ramalingam
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
First-Line Nivolumab Plus Ipilimumab in Advanced Non–Small-Cell Lung Cancer (CheckMate 568): Outcomes by Programmed Death Ligand 1 and Tumor Mutational Burden as Biomarkers
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/site/ifc.
Neal Ready
Consulting or Advisory Role: Bristol-Myers Squibb, Novartis, Merck, AbbVie, Celgene, Merck Serono, AstraZeneca
Research Funding: Bristol-Myers Squibb, Merck
Matthew D. Hellmann
Stock and Other Ownership Interests: Shattuck Labs
Honoraria: AstraZeneca, Bristol-Myers Squibb
Consulting or Advisory Role: Bristol-Myers Squibb, Merck, Genentech, AstraZeneca, MedImmune, Novartis, Janssen Pharmaceuticals, Nektar, Syndax, Mirati Therapeutics, Shattuck Labs
Research Funding: Bristol-Myers Squibb (Inst)
Patents, Royalties, Other Intellectual Property: Patent filed by Memorial Sloan-Kettering (PCT/US2015/062208) for the use of tumor mutation burden for prediction of immunotherapy efficacy, licensed to Personal Genome Diagnostics (Inst)
Travel, Accommodations, Expenses: AstraZeneca, Bristol-Myers Squibb
Mark M. Awad
Consulting or Advisory Role: Genentech, Merck, Pfizer, Boehringer Ingelheim, AbbVie, AstraZeneca, MedImmune, Clovis Oncology, Nektar, Bristol-Myers Squibb, ARIAD Pharmaceuticals, Foundation Medicine, Syndax, Novartis, Blueprint Medicines
Research Funding: Bristol-Myers Squibb
Gregory A. Otterson
Consulting or Advisory Role: Novartis (Inst), Takeda (Inst), Novocure (Inst)
Research Funding: Genentech (Inst), Pfizer (Inst), Bristol-Myers Squibb (Inst), Clovis Oncology (Inst), Novartis (Inst), Acerta Pharma (Inst), Ignyta (Inst), Merck (Inst), AstraZeneca (Inst)
Martin Gutierrez
Stock and Other Ownership Interests: COTA
Consulting or Advisory Role: Bayer, Eli Lilly, Karyopharm Therapeutics
Speakers' Bureau: Bristol-Myers Squibb, Merck, Foundation One, Eli Lilly
Research Funding: Novartis (Inst), AbbVie (Inst), MedImmune (Inst), Bristol-Myers Squibb (Inst), Rexahn Pharmaceuticals (Inst), Esanex (Inst), Karyopharm Therapeutics (Inst), TG Therapeutics (Inst), Daiichi Sankyo (Inst), EMD Serono (Inst), Eli Lilly (Inst), Acceleron Pharma (Inst), Gilead Sciences (Inst), Incyte (Inst), Merck (Inst), Loxo (Inst), Bayer (Inst), Celgene (Inst), Eisai (Inst), Genentech (Inst), Mirati Therapeutics (Inst), Moderna Therapeutics (Inst), Regeneron (Inst), Sanofi (Inst), Seattle Genetics (Inst), Tesaro (Inst)
Justin F. Gainor
Honoraria: Merck, Incyte, ARIAD Pharmaceuticals, Novartis, Pfizer
Consulting or Advisory Role: Genentech, Bristol-Myers Squibb, Theravance, Loxo, Takeda, Array BioPharma, Amgen, Merck, Agios, Regeneron, Oncorus, Jounce Therapeutics
Research Funding: Merck (Inst), Novartis (Inst), Genentech (Inst), Bristol-Myers Squibb (Inst), Adaptimmune (Inst), AstraZeneca (Inst), ARIAD Pharmaceuticals (Inst), Jounce Therapeutics (Inst), Blueprint Medicines (Inst), Moderna Therapeutics (Inst), Tesaro (Inst), Alexo Therapeutics (Inst), Array BioPharma (Inst)
Hossein Borghaei
Honoraria: Bristol-Myers Squibb, Celgene, Axiom Biotechnologies
Consulting or Advisory Role: Bristol-Myers Squibb, Eli Lilly, Celgene, Genentech, Pfizer, Boehringer Ingelheim, EMD Serono, Trovagene, Novartis, Merck, AstraZeneca, Genmab, Regeneron, Cantargia AB, BioNTech AG, AbbVie
Research Funding: Millennium Pharmaceuticals (Inst), Merck (Inst), Celgene (Inst), Bristol-Myers Squibb (Inst), Eli Lilly (Inst)
Travel, Accommodations, Expenses: Bristol-Myers Squibb, Eli Lilly, Clovis Oncology, Celgene, Genentech, Novartis, Merck
Other Relationship: University of Pennsylvania, Takeda
Jacques Jolivet
Honoraria: Alethia Biotherapeutique
Research Funding: Bristol-Myers Squibb (Inst), Merck (Inst)
Leora Horn
Consulting or Advisory Role: Merck, Xcovery, Genentech, Eli Lilly, AbbVie, AstraZeneca, Janssen Pharmaceuticals, Bristol-Myers Squibb, Incyte, EMD Serono, Tesaro
Research Funding: Boehringer Ingelheim (Inst), Xcovery (Inst)
Other Relationship: Bristol-Myers Squibb, Boehringer Ingelheim
Mihaela Mates
Consulting or Advisory Role: Roche
Travel, Accommodations, Expenses: AstraZeneca
Julie Brahmer
Consulting or Advisory Role: Bristol-Myers Squibb, Eli Lilly, Celgene, Syndax, Janssen Oncology, Merck, Amgen, Genentech
Research Funding: Bristol-Myers Squibb (Inst), Merck (Inst), AstraZeneca (Inst), Incyte (Inst), Janssen Oncology (Inst)
Travel, Accommodations, Expenses: Bristol-Myers Squibb, Merck
Other Relationship: Bristol-Myers Squibb, Merck
Pavan S. Reddy
Consulting or Advisory Role: Merck
Speakers' Bureau: Clovis Oncology
Jason Chesney
Consulting or Advisory Role: Amgen, Replimune
Research Funding: Amgen, Replimune, Iovance Biotherapeutics, Bristol-Myers Squibb, MedImmune
Patents, Royalties, Other Intellectual Property: University of Louisville US patents
Travel, Accommodations, Expenses: Amgen, Replimune
David R. Spigel
Leadership: Centennial Medical Center
Consulting or Advisory Role: Genentech (Inst), Novartis (Inst), Celgene (Inst), Bristol-Myers Squibb (Inst), AstraZeneca (Inst), Pfizer (Inst), Boehringer Ingelheim (Inst), AbbVie (Inst), Foundation Medicine (Inst), GlaxoSmithKline (Inst), Eli Lilly (Inst), Merck (Inst), Moderna Therapeutics (Inst), Nektar (Inst), Takeda (Inst), Amgen (Inst), TRM Oncology (Inst), Precision Oncology (Inst), Evelo Therapeutics (Inst), Illumina (Inst), PharmaMar (Inst)
Research Funding: Genentech (Inst), Novartis (Inst), Celgene (Inst), Bristol-Myers Squibb (Inst), Eli Lilly (Inst), AstraZeneca (Inst), Pfizer (Inst), Boehringer Ingelheim (Inst), University of Texas Southwestern Medical Center, Simmons Cancer Center (Inst), Merck (Inst), AbbVie (Inst), GlaxoSmithKline (Inst), G1 Therapeutics (Inst), Neon Therapeutics (Inst), Takeda (Inst), Foundation Medicine (Inst), Nektar (Inst), Celldex (Inst), Clovis Oncology (Inst), Daiichi Sankyo (Inst), EMD Serono (Inst), Acerta Pharma (Inst), Oncogenex (Inst), Astellas Pharma (Inst), GRAIL (Inst), Transgene (Inst), Aeglea Biotherapeutics (Inst), Tesaro (Inst), Ipsen (Inst), ARMO BioSciences (Inst), Amgen (Inst), Millennium Pharmaceuticals (Inst)
Travel, Accommodations, Expenses: AstraZeneca, Boehringer Ingelheim, Celgene, Eli Lilly, EMD Serono, Bristol-Myers Squibb, Genentech, Genzyme, Intuitive Surgical, Merck, Pfizer, Purdue Pharma, Spectrum Pharmaceuticals, Sysmex
Martin Reck
Consulting or Advisory Role: Eli Lilly, MSD Oncology, Merck Serono, Bristol-Myers Squibb, AstraZeneca, Boehringer Ingelheim, Celgene, Pfizer, Novartis, Genentech, AbbVie, Amgen
Speakers' Bureau: Genentech, Eli Lilly, MSD Oncology, Merck Serono, AstraZeneca, Bristol-Myers Squibb, Boehringer Ingelheim, Celgene, Pfizer, Novartis, Amgen
Kenneth John O’Byrne
Stock and Other Ownership Interests: CARP Pharmaceuticals
Honoraria: Merck Sharp & Dohme, Bristol-Myers Squibb, Roche, Teva Pharmaceuticals, Boehringer Ingelheim, AstraZeneca, Pfizer, EMD Serono, Novartis, Janssen-Cilag, Mundipharma
Consulting or Advisory Role: Merck Sharp & Dohme, Boehringer Ingelheim, Genentech, Janssen-Cilag, Pfizer, AstraZeneca, MedImmune, Bristol-Myers Squibb, Novartis, Teva Pharmaceuticals, Natera
Speakers' Bureau: Merck Sharp & Dohme, Boehringer Ingelheim, Bristol-Myers Squibb, Roche, Janssen-Cilag, Pfizer, Mundipharma, Novartis, Foundation Medicine
Research Funding: AstraZeneca, MedImmune, Bristol-Myers Squibb
Patents, Royalties, Other Intellectual Property: Four patents/provisional patents held by the University of Queensland (Inst)
Travel, Accommodations, Expenses: Boehringer Ingelheim, Bristol-Myers Squibb, Roche
Luis Paz-Ares
Leadership: Genomica, EMA SAG (I)
Honoraria: Genentech, Eli Lilly, Pfizer, Boehringer Ingelheim, Bristol-Myers Squibb, MSD Oncology, AstraZeneca, Merck Serono, PharmaMar, Novartis, Celgene, Sysmex, Amgen, Incyte
Travel, Accommodations, Expenses: Roche, AstraZeneca, MSD Oncology, Bristol-Myers Squibb, Eli Lilly, Pfizer
Other Relationship: Novartis (I), Ipsen (I), Pfizer (I), Servier (I), Sanofi (I), Roche (I), Amgen (I), Merck (I)
Wenhua Hu
Employment: Bristol-Myers Squibb
Stock and Other Ownership Interests: Bristol-Myers Squibb
Kim Zerba
Employment: Bristol-Myers Squibb
Stock and Other Ownership Interests: Bristol-Myers Squibb
Xuemei Li
Employment: Bristol-Myers Squibb
Stock and Other Ownership Interests: Bristol-Myers Squibb
Research Funding: Bristol-Myers Squibb
Travel, Accommodations, Expenses: Bristol-Myers Squibb
Brian Lestini
Employment: Bristol-Myers Squibb
Stock and Other Ownership Interests: Bristol-Myers Squibb
William J. Geese
Employment: Bristol-Myers Squibb
Stock and Other Ownership Interests: Bristol-Myers Squibb
Patents, Royalties, Other Intellectual Property: Inventor on one or more pending patent applications for tumor mutational burden as a predictive biomarker for immunotherapy (Inst)
Joseph D. Szustakowski
Employment: Bristol-Myers Squibb
Stock and Other Ownership Interests: Bristol-Myers Squibb
Research Funding: Bristol-Myers Squibb
Patents, Royalties, Other Intellectual Property: Inventor on one or more pending patent applications, including applications for tumor mutational burden as a predictive biomarker of immunotherapy
Travel, Accommodations, Expenses: Bristol-Myers Squibb
George Green
Employment: Bristol-Myers Squibb
Stock and Other Ownership Interests: Bristol-Myers Squibb
Han Chang
Employment: Bristol-Myers Squibb
Stock and Other Ownership Interests: Bristol-Myers Squibb
Research Funding: Bristol-Myers Squibb
Patents, Royalties, Other Intellectual Property: Inventor on one or more pending patent applications, including applications for tumor mutational burden as a predictive biomarker of immunotherapy
Travel, Accommodations, Expenses: Bristol-Myers Squibb
Suresh S. Ramalingam
Consulting or Advisory Role: Amgen, Boehringer Ingelheim, Celgene, Genentech, Eli Lilly, ImClone, Bristol-Myers Squibb, AstraZeneca, AbbVie, Merck, Takeda, Tesaro, Nektar, Loxo
Research Funding: AbbVie (Inst), Bristol-Myers Squibb (Inst), Pfizer (Inst), Merck (Inst), AstraZeneca (Inst), MedImmune (Inst), Vertex (Inst), Takeda (Inst), EMD Serono (Inst)
Travel, Accommodations, Expenses: EMD Serono, Pfizer, AstraZeneca
No other potential conflicts of interest were reported.
REFERENCES
- 1.National Comprehensive Cancer Network NCCN guidelines: Non-small cell lung cancer. https://www.nccn.org/professionals/physician_gls/pdf/nscl.pdf
- 2.Hanna N, Johnson D, Temin S, et al. Systemic therapy for stage IV non–small-cell lung cancer: American Society of Clinical Oncology Clinical Practice Guideline update. J Clin Oncol. 2017;35:3484–3515. doi: 10.1200/JCO.2017.74.6065. [DOI] [PubMed] [Google Scholar]
- 3.Keytruda (pembrolizumab) [package insert] Whitehouse Station, NJ: Merck & Co.; 2018. [Google Scholar]
- 4.Opdivo (nivolumab) [package insert] Princeton, NJ: Bristol-Myers Squibb Company; 2018. [Google Scholar]
- 5.Curran MA, Montalvo W, Yagita H, et al. PD-1 and CTLA-4 combination blockade expands infiltrating T cells and reduces regulatory T and myeloid cells within B16 melanoma tumors. Proc Natl Acad Sci USA. 2010;107:4275–4280. doi: 10.1073/pnas.0915174107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Antonia SJ, López-Martin JA, Bendell J, et al. Nivolumab alone and nivolumab plus ipilimumab in recurrent small-cell lung cancer (CheckMate 032): A multicentre, open-label, phase 1/2 trial. Lancet Oncol. 2016;17:883–895. doi: 10.1016/S1470-2045(16)30098-5. [DOI] [PubMed] [Google Scholar]
- 7.Hellmann MD, Rizvi NA, Goldman JW, et al. Nivolumab plus ipilimumab as first-line treatment for advanced non-small-cell lung cancer (CheckMate 012): Results of an open-label, phase 1, multicohort study. Lancet Oncol. 2017;18:31–41. doi: 10.1016/S1470-2045(16)30624-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Larkin J, Chiarion-Sileni V, Gonzalez R, et al. Combined nivolumab and ipilimumab or monotherapy in untreated melanoma. N Engl J Med. 2015;373:23–34. doi: 10.1056/NEJMoa1504030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldman JW, Antonia SJ, Gettinger SN, et al. Nivolumab (N) plus ipilimumab (I) as first-line (1L) treatment for advanced (adv) NSCLC: 2-yr OS and long-term outcomes from CheckMate 012. J Clin Oncol. 2017;35(suppl; abstr 9093) [Google Scholar]
- 10.Antonia S, Gettinger S, Borghaei H, et al. Long-term outcomes with first-line nivolumab plus ipilimumab in advanced NSCLC: 3-year follow-up from CheckMate 012; 19th World Conference on Lung Cancer; Toronto, ON, Canada. September 23-26, 2018. [Google Scholar]
- 11.Borghaei H, Paz-Ares L, Horn L, et al. Nivolumab versus docetaxel in advanced nonsquamous non–small-cell lung cancer. N Engl J Med. 2015;373:1627–1639. doi: 10.1056/NEJMoa1507643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reck M, Rodríguez-Abreu D, Robinson AG, et al. Pembrolizumab versus chemotherapy for PD-L1–positive non–small-cell lung cancer. N Engl J Med. 2016;375:1823–1833. doi: 10.1056/NEJMoa1606774. [DOI] [PubMed] [Google Scholar]
- 13.Rittmeyer A, Barlesi F, Waterkamp D, et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): A phase 3, open-label, multicentre randomised controlled trial. Lancet. 2017;389:255–265. doi: 10.1016/S0140-6736(16)32517-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carbone DP, Reck M, Paz-Ares L, et al. First-line nivolumab in stage IV or recurrent non–small-cell lung cancer. N Engl J Med. 2017;376:2415–2426. doi: 10.1056/NEJMoa1613493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodman AM, Kato S, Bazhenova L, et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther. 2017;16:2598–2608. doi: 10.1158/1535-7163.MCT-17-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hellmann MD, Callahan MK, Awad MM, et al. Tumor mutational burden and efficacy of nivolumab monotherapy and in combination with ipilimumab in small-cell lung cancer. Cancer Cell. 2018;33:853–861.e4. doi: 10.1016/j.ccell.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kowanetz M, Zou W, Shames D, et al. Tumor mutation burden (TMB) is associated with improved efficacy of atezolizumab in 1L and 2L+ NSCLC patients. J Thorac Oncol. 2017;12(suppl; abstr OA20.01):S321–S322. [Google Scholar]
- 18.Rizvi H, Sanchez-Vega F, La K, et al. Molecular determinants of response to anti–programmed cell death (PD)-1 and anti–programmed death-ligand 1 (PD-L1) blockade in patients with non–small-cell lung cancer profiled with targeted next-generation sequencing. J Clin Oncol. 2018;36:633–641. doi: 10.1200/JCO.2017.75.3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med. 2017;377:2500–2501. doi: 10.1056/NEJMc1713444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hellmann MD, Nathanson T, Rizvi H, et al. Genomic features of response to combination immunotherapy in patients with advanced non-small-cell lung cancer. Cancer Cell. 2018;33:843–852.e4. doi: 10.1016/j.ccell.2018.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hellmann MD, Ciuleanu TE, Pluzanski A, et al. Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N Engl J Med. 2018;378:2093–2104. doi: 10.1056/NEJMoa1801946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Foundation Medicine FoundationOne CDx technical information. https://www.foundationmedicine.com/genomic-testing/foundation-one-cdx#overview
- 24.Foundation Medicine FoundationOne CDx. https://www.foundationmedicine.com/genomic-testing/foundation-one-cdx
- 25.Foundation Medicine FoundationOne CDx specimen instructions. https://assets.ctfassets.net/vhribv12lmne/6ms7OiT5PaQgGiMWue2MAM/52d91048be64b72e73ffa0c1cab043c0/F1CDx_Specimen_Instructions.pdf
- 26.Sun JX, He Y, Sanford E, et al. A computational approach to distinguish somatic vs. germline origin of genomic alterations from deep sequencing of cancer specimens without a matched normal. PLOS Comput Biol. 2018;14:e1005965. doi: 10.1371/journal.pcbi.1005965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9:34. doi: 10.1186/s13073-017-0424-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szustakowski J, Green G, Geese WJ, et al. Evaluation of tumor mutational burden as a biomarker for immune checkpoint inhibitor efficacy: A calibration study of whole exome sequencing with FoundationOne. Cancer Res. 2018;78(suppl; abstr 5528) [Google Scholar]
- 29.Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703–713. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bristol-Myers Squibb Data sharing request process. https://www.bms.com/researchers-and-partners/independent-research/data-sharing-request-process.html
- 31.Capelletto E, Novello S, Scagliotti GV. First-line therapeutic options for advanced non-small-cell lung cancer in the molecular medicine era. Future Oncol. 2014;10:1081–1093. doi: 10.2217/fon.13.247. [DOI] [PubMed] [Google Scholar]
- 32.Kruglyak KM, Lin E, Ong FS. Next-generation sequencing and applications to the diagnosis and treatment of lung cancer. Adv Exp Med Biol. 2016;890:123–136. doi: 10.1007/978-3-319-24932-2_7. [DOI] [PubMed] [Google Scholar]