Abstract
Background
The time-consuming and user-dependent postprocessing of biochemical cartilage MRI has limited the use of delayed gadolinium-enhanced MRI of cartilage (dGEMRIC). An automated analysis of biochemical three-dimensional (3-D) images could deliver a more time-efficient and objective evaluation of cartilage composition, and provide comprehensive information about cartilage thickness, surface area, and volume compared with manual two-dimensional (2-D) analysis.
Questions/purposes
(1) How does the 3-D analysis of cartilage thickness and dGEMRIC index using both a manual and a new automated method compare with the manual 2-D analysis (gold standard)? (2) How does the manual 3-D analysis of regional patterns of dGEMRIC index, cartilage thickness, surface area and volume compare with a new automatic method? (3) What is the interobserver reliability and intraobserver reproducibility of software-assisted manual 3-D and automated 3-D analysis of dGEMRIC indices, thickness, surface, and volume for two readers on two time points?
Methods
In this IRB-approved, retrospective, diagnostic study, we identified the first 25 symptomatic hips (23 patients) who underwent a contrast-enhanced MRI at 3T including a 3-D dGEMRIC sequence for intraarticular pathology assessment due to structural hip deformities. Of the 23 patients, 10 (43%) were male, 16 (64%) hips had a cam deformity and 16 (64%) hips had either a pincer deformity or acetabular dysplasia. The development of an automated deep-learning-based approach for 3-D segmentation of hip cartilage models was based on two steps: First, one reader (FS) provided a manual 3-D segmentation of hip cartilage, which served as training data for the neural network and was used as input data for the manual 3-D analysis. Next, we developed the deep convolutional neural network to obtain an automated 3-D cartilage segmentation that we used as input data for the automated 3-D analysis. For actual analysis of the manually and automatically generated 3-D cartilage models, a dedicated software was developed. Manual 2-D analysis of dGEMRIC indices and cartilage thickness was performed at each “full-hour” position on radial images and served as the gold standard for comparison with the corresponding measurements of the manual and the automated 3-D analysis. We measured dGEMRIC index, cartilage thickness, surface area, and volume for each of the four joint quadrants and compared the manual and the automated 3-D analyses using mean differences. Agreement between the techniques was assessed using intraclass correlation coefficients (ICC). The overlap between 3-D cartilage volumes was assessed using dice coefficients and means of all distances between surface points of the models were calculated as average surface distance. The interobserver reliability and intraobserver reproducibility of the software-assisted manual 3-D and the automated 3-D analysis of dGEMRIC indices, thickness, surface and volume was assessed for two readers on two different time points using ICCs.
Results
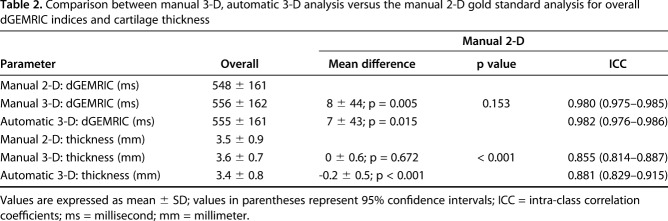
Comparable mean overall difference and almost-perfect agreement in dGEMRIC indices was found between the manual 3-D analysis (8 ± 44 ms, p = 0.005; ICC = 0.980), the automated 3-D analysis (7 ± 43 ms, p = 0.015; ICC = 0.982), and the manual 2-D analysis.
Agreement for measuring overall cartilage thickness was almost perfect for both 3-D methods (ICC = 0.855 and 0.881) versus the manual 2-D analysis. A mean difference of -0.2 ± 0.5 mm (p < 0.001) was observed for overall cartilage thickness between the automated 3-D analysis and the manual 2-D analysis; no such difference was observed between the manual 3-D and the manual 2-D analysis.
Regional patterns were comparable for both 3-D methods. The highest dGEMRIC indices were found posterosuperiorly (manual: 602 ± 158 ms; p = 0.013, automated: 602 ± 158 ms; p = 0.012). The thickest cartilage was found anteroinferiorly (manual: 5.3 ± 0.8 mm, p < 0.001; automated: 4.3 ± 0.6 mm; p < 0.001). The smallest surface area was found anteroinferiorly (manual: 134 ± 60 mm2; p < 0.001, automated: 155 ± 60 mm2; p < 0.001). The largest volume was found anterosuperiorly (manual: 2343 ± 492 mm3; p < 0.001, automated: 2294 ± 467 mm3; p < 0.001). Mean average surface distance was 0.26 ± 0.13 mm and mean Dice coefficient was 86% ± 3%. Intraobserver reproducibility and interobserver reliability was near perfect for overall analysis of dGEMRIC indices, thickness, surface area, and volume (ICC range, 0.962–1).
Conclusions
The presented deep learning approach for a fully automatic segmentation of hip cartilage enables an accurate, reliable and reproducible analysis of dGEMRIC indices, thickness, surface area, and volume. This time-efficient and objective analysis of biochemical cartilage composition and morphology yields the potential to improve patient selection in femoroacetabular impingement (FAI) surgery and to aid surgeons with planning of acetabuloplasty and periacetabular osteotomies in pincer FAI and hip dysplasia. In addition, this validation paves way to the large-scale use of this method for prospective trials which longitudinally monitor the effect of reconstructive hip surgery and the natural course of osteoarthritis.
Level of Evidence
Level III, diagnostic study.
Introduction
Cartilage damage at the time of surgery is a major predictor of clinical outcome after the surgical correction of femoroacetabular impingement (FAI) [16, 17, 31] and hip dysplasia [3, 28, 61]. Assessment of classic radiographic signs of osteoarthritis is important for surgical decision-making, but changes on plain x-rays become visible only late in the natural history of the disease, and it might be helpful both prognostically and therapeutically if more sensitive tests were available. Although more accurate than radiographs, morphologic MRI with intraarticular contrast cannot visualize early biochemical damage to the cartilage matrix [17]. By contrast, the use of biochemical MRI sequences such as delayed gadolinium-enhanced MRI of cartilage (dGEMRIC) [12], T2 [21], T2* [8], or T1rho [4] are sensitive to early degenerative changes in cartilage composition. Among these techniques, dGEMRIC is perhaps the one used most commonly in the hip [11, 12, 23, 24, 26, 43, 44]. dGEMRIC is based on the depletion of negatively-charged glycosaminoglycan molecules that are replaced by the negatively charged gadolinium. A low dGEMRIC index indicates advanced biochemical cartilage damage and is an accurate predictor for mid-term failure after periacetabular osteotomy (PAO) for hip dysplasia [12, 26].
Although rapid three-dimensional (3-D) high-resolution MRI techniques for biochemical assessment of cartilage quality are available, they are not widely used in clinical practice [66]. Instead, an expert reader assesses biochemical MRIs manually on reformatted two-dimensional (2-D) images. Despite being prone to inter- and intraobserver variability and quite labor intensive, manual assessment of reformatted 2-D images is the current gold standard for assessing biochemical cartilage MRI [66]. In current practice, most of the 3-D data are not used, in favor of a more time-efficient analysis [49]. To overcome this limitation, researchers have developed techniques for a more automated [10, 14, 20, 39, 49] computer-assisted 3-D assessment of biochemical cartilage damage. These techniques are based on the manual labelling or segmentation of cartilage to generate a 3-D cartilage model. These data then are used to train an algorithm to automatically generate 3-D models of hip cartilage. However, some of the available approaches require multiple manual steps [14, 20, 49] and are based on asymptomatic volunteers only [10], while others used MRI sequences, which are difficult to implement in clinical practice because of relatively long image-acquisition times [14, 20, 39]. In addition to assessing cartilage quality, the use of 3-D MRI offers the possibility to simultaneously provide information on cartilage thickness, cartilage surface area, and cartilage volume, thus allowing for a potentially improved comprehensive hip assessment. However, to our knowledge, there are no studies validating a fully automatic MRI-based 3-D segmentation of cartilage models for the assessment of dGEMRIC, thickness, surface, and volume of hip cartilage in symptomatic patients with structural hip deformities using a deep learning approach. Briefly, deep learning is a class of newer machine-learning methods that can produce a mapping from raw inputs to desired outputs, such as image segmentation. Unlike traditional machine-learning methods, which require hand-crafted feature extraction from input data, deep-learning methods can learn features directly from data. Deep learning-based methods, particularly those based on deep convolutional neural networks, can directly map whole volumetric data to its volume-wise labels. To date, deep learning approaches have been applied successfully in medical image analysis including musculoskeletal applications such as knee segmentation [35, 40, 65].
Therefore, we asked: (1) How does the 3-D analysis of cartilage thickness and dGEMRIC index using both a manual and a new automated method compare with the manual 2-D analysis (gold standard)? (2) How does the manual 3-D analysis of regional patterns of dGEMRIC index, cartilage thickness, surface area and volume compare with a new automatic method? (3) What is the interobserver reliability and intraobserver reproducibility of software-assisted manual 3-D and automated 3-D analysis of dGEMRIC indices, thickness, surface, and volume for two readers on two time points?
Patients and Methods
Study Design and Patient Population
Following institutional review board approval, we performed a retrospective, diagnostic study on the first 25 symptomatic hips undergoing a contrast-enhanced MRI at 3T including a 3-D dGEMRIC sequence to assess intraarticular pathology due to structural hip deformities at a tertiary care center.
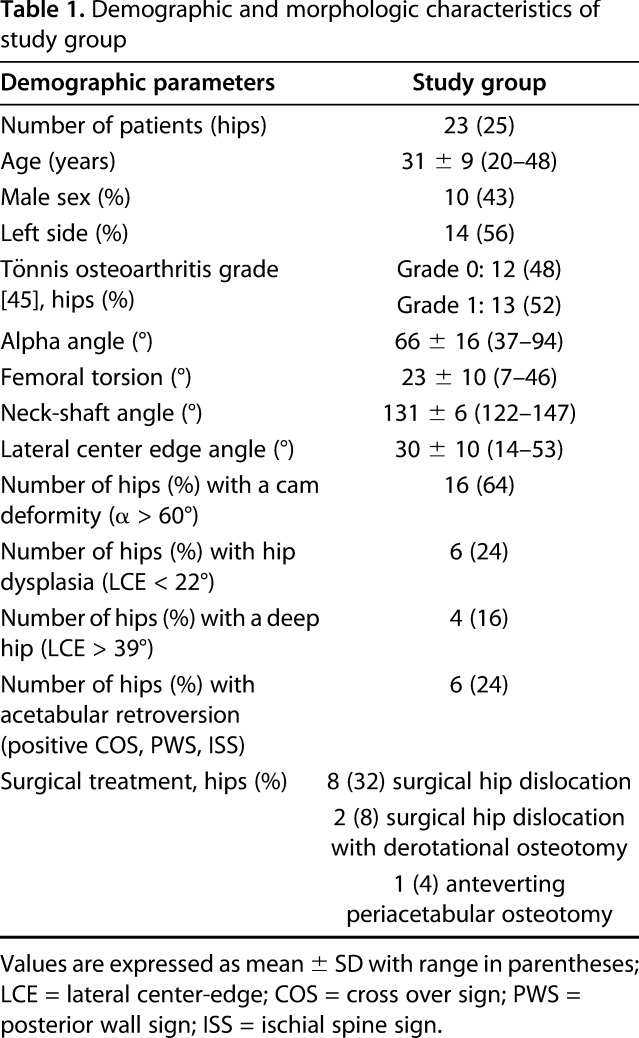
We included the first 25 hips (23 patients) who had not undergone previous hip surgery and had a complete dGEMRIC scan. This group of patients was identified within a sample of 105 patients (116 hips) who underwent a dGEMRIC scan between September 2012 and April 2016. Eleven hips (11 patients) underwent subsequent joint-preserving hip surgery. Ten hips (10 patients) underwent a surgical hip dislocation for FAI correction, and one hip (one patient) underwent an anteverting periacetabular osteotomy to correct an acetabular retroversion. Mean age was 31 ± 9 years (range, 20–48 years) and 10 males (43%) and 13 females were included. A cam deformity was present in 16 hips (64%). Six hips (24%) had acetabular dysplasia, four hips (16%) had a deep acetabular socket, and six hips (24%) presented with acetabular retroversion (Table 1).
Table 1.
Demographic and morphologic characteristics of study group
Radiographic Evaluation
All patients underwent biplanar radiographic hip imaging, including a supine AP pelvis view and a cross-table view [60]. One author (FS) who has 5 years of experience in diagnostic imaging of degenerative hip disease performed an image analysis and assessed the presence of osseous deformities on the AP pelvic view. A validated computerized method was used (software Hip2Norm, University of Bern, Bern, Switzerland) for measuring radiographic parameters [55, 59, 64]. The alpha angle was measured on radial MRIs [36] and angles > 60° served as cut-off for a cam deformity [54]. Femoral torsion was measured according to Murphy et al. [33]. Hip dysplasia was defined as a lateral center-edge (LCE) < 22° [57], a deep hip as a LCE > 39° [57] and acetabular retroversion as a positive cross-over sign, posterior wall sign and ischial spine sign [48], respectively.
MR Imaging Protocol
All patients underwent an indirect MR hip arthrography according to a standardized technique for dGEMRIC on a 3T scanner (Trio; Siemens, Erlangen, Germany) [44] using a 6-channel flexible body-matrix phased-array coil. In the two patients who had dGEMRIC scans of both hips, the image acquisition was performed on two different days, one day for each hip. After intravenous injection of gadolinium contrast agent (0.2 mmol/kg, Gd-DTPA2-, Magnevist, Bayer, Leverkusen, Germany) patients were instructed to walk for 15 minutes. The dGEMRIC sequence was acquired 45 minutes to 70 minutes after injection. A standard dual-flip angle, isovoxel 3-D gradient-echo technique was acquired in the axial-oblique plane [44]: Repetition time of 15 milliseconds (ms), echo time of 3.3 ms, flip angles of 4°, and 24°, 160 x 160 mm field of view, matrix size of 192 x 192. Original voxel size was 0.83 x 0.83 x 0.8 mm, the integrated interpolation was used and final voxel size of the axially reformatted images was 0.24 x 0.24 x 1 mm. Acquisition time was 8:46 minutes for 128 slices. No additional prescan for B1 correction was performed.
Post-imaging Processing and Development of the Neural Network
We have developed a deep-learning-based approach for 3-D segmentation of hip cartilage models. Deep learning is a class of machine-learning methods. One of the big advantages of deep-learning methods over traditional machine learning methods is that deep learning methods can produce a mapping from raw inputs to desired outputs, such as image classes, segmentation, etc, by learning features directly from data while traditional machine learning methods require hand-crafted feature extraction from inputs. With the advent of large datasets and increased computing power offered by the graphical processing units, deep-learning-based methods, especially those based on convolutional neural networks, have become more widely available. In these methods, artificial neurons are arranged in multiple layers and there are weighted connections between neurons in different layers. The exact weights are obtained via a process called training where example pairs of inputs and target outputs are used to iteratively adjust the weights by back-propagating a corrective error signal through the network. As soon as a deep network is trained, it can be used to obtain the desired output from a given testing data.
Development of the fully automatic deep-learning-based approach for 3-D segmentation of hip cartilage models was based on two steps (Fig. 1A-C). At first, 3-D training data of hip cartilage was provided, to develop a deep neural network capable of performing fully automatic 3-D segmentation of hip cartilage. To obtain the training 3-D cartilage models, one reader (RH) who was specifically trained performed a manual threshold-assisted segmentation of combined hip cartilage layers using Amira software (FEI, Hillsboro, Oregon, USA) (Fig. 1B). This was performed on 1-mm thick, axial reformatted gradient echo images obtained at a flip angle of 24°, which yield a better morphologic visualization of cartilage than the actual dGEMRIC map. True axial reformats were obtained since this orientation is preferred when performing manual cartilage segmentation in our institution. The gradient echo images obtained at a flip angle of 24° were used for training the deep neural network and later on for testing. Since flip angle images represent the raw data for calculating dGEMRIC values, both image sets are perfectly aligned. After completion, all images were reviewed for accuracy in consensus with a reader (FS), leading to the final manual 3-D segmentation. Automatic segmentation was performed using a deep-learning-based method.
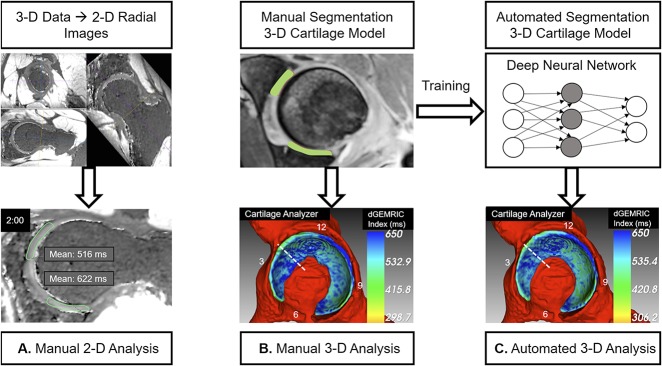
Fig. 1 A-C.
The three different methods for assessment of morphologic and biochemical cartilage damage are shown. (A) For the manual two-dimension (2-D) analysis, which served as the gold-standard for measuring dGEMRIC indices and cartilage thickness, radial 2-D images were reformatted from the three-dimension (3-D) dataset. Measurement was performed at each full-hour position of the clock face. (B) For the manual 3-D analysis, we performed a manual segmentation of the cartilage layers. These manual 3-D models were used as input data into the cartilage analyzer software (University of Bern, Bern, Switzerland) to calculate dGEMRIC indices, thickness, surface area, and volume. (C) The manually segmented 3-D cartilage models were used to train a deep neural network to be a fully automatically segmented 3-D cartilage model. This automatically segmented 3-D model was used as input data into analysis software to calculate dGEMRIC indices, thickness, surface area, and volume (automated 3-D analysis). Study design: (1) dGEMRIC indices and cartilage thickness as measured with the manual 3-D- and the automated 3-D analysis were compared against the manual 2D analysis. (2) Measurements of dGEMRIC indices, thickness, surface area and volume of each quadrant were compared between the 3-D manual analysis and the 3-D automated analysis.
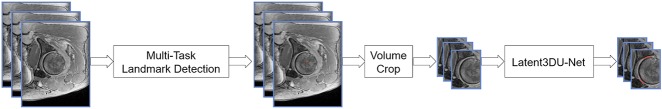
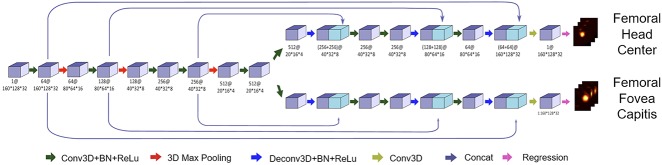
The development of the actual neural network was based on a two-stage method (Fig. 2). In the first stage, two landmarks, such as the femoral head center and femoral fovea capitis, were detected by using a multitask landmark detection network (Fig. 3). The landmark detection network follows an encoder-decoder architecture with long and short skip connections. The encoder part focuses on analysis and feature representation learning from the input data while the decoder part generates landmark heatmaps. It was designed to regress heatmaps of all landmarks. Each landmark detector had its own decoder part but shared the same encoder part to encourage effective feature learning. The detected landmarks allowed us to crop the original data around the femoral head region such that at the second stage, we could train another network to segment cartilage from the cropped volume. Our cartilage segmentation network was based on the Latent3DU-net that we introduced in our previous work [63], where prior shape knowledge were incorporated into a voxelwise semantic segmentation. Additionally, based on the manual 3-D segmentation, we also defined the locations for two anatomical landmarks for each patient, such as the femoral head center and femoral fovea capitis, which were then used to generate the ground truth heatmaps for training landmark detectors. Our method was implemented with Python using TensorFlow framework on a workstation with a 3.6GHz Intel® i7 CPU and a GTX 1080 Ti graphics card with 11GB GPU memory.
Fig. 2.
A schematic illustration of the two-stage deep learning based method for cartilage segmentation is shown. At first, the femoral head center and the femoral fovea capitis were identified with the use of a multitask landmark detection network (Fig. 3). After that the original data was cropped to include the hip only. Then the Latent3DU-net cartilage segmentation network was applied.
Fig. 3.
A schematic view of the multitask landmark detection network that was used for detection of the femoral head center and the fovea capitis is shown. This is based on an encoder-decoder architecture with long and short skip connections. The encoder part was used for feature learning from the input data, the decoder part was used to generate the landmark heatmaps. The numbers below each block follow the pattern of “number of feature stack@data size.”
Given the manual segmentation of 25 hip MRIs, we developed a standard five-fold cross-validation study to evaluate the efficacy of the developed method. More specifically, we evenly split the 25 hip MRIs and the associated manual segmentation into five groups. Each time, one group was taken out as the testing data while the remaining four groups were taken as the training data. This procedure was repeated five times such that each group was used once as the testing data. To visualize the dGEMRIC indices in 3-D, we developed an in-house program (Cartilage Analyzer, University of Bern, Bern, Switzerland). This program first uses the generated segmentation results (either manually or automatically) to mask out the cartilage from the dGEMRIC volume and further to create surface meshes of the cartilage. The 3-D visualization of the color-coded maps correspond to the voxels located at the respective surfaces (Fig. 4A-C). A dGEMRIC index-to-color map is then displayed in a 3-D rendering of the corresponding cartilage meshes, taking the 3-D rendering of the surrounding osseous structures as background. The morphologic (thickness, surface area, and volume) and biochemical measurements can be allocated to four quadrants in the region specified by the user.
Fig. 4 A-C.
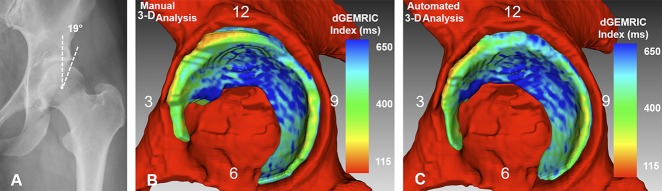
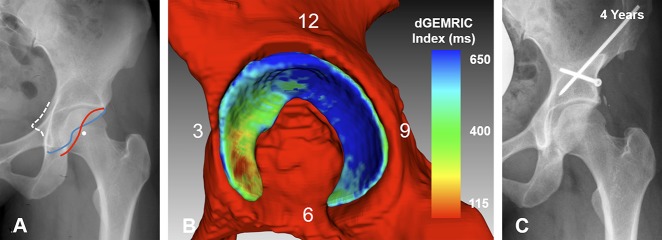
(A) An AP pelvis x-ray shows a 48-year-old woman with mild hip dysplasia (LCE angle = 19°). (B) The Manual 3-D analysis is shown. (C) The automated 3-D analysis is shown with dGEMRIC indices color-coded. The red indicates regions of low dGEMRIC indices demonstrating low glycosaminoglycan content corresponding to biochemical cartilage damage and vice-versa for blue. (B) Using the manual 3D analysis, the mean dGEMRIC index was 513 ms and cartilage surface area was 1394 mm2. This was comparable to (C) the automated 3-D analysis which yielded a mean dGEMRIC index of 514 ms and a cartilage surface area of 1257 mm2. (B) Overestimation of the manual 3-D analysis (Table 3) for measuring thickness in the anterioinferior quadrant resulted in a difference of 1.6 mm compared to the automated 3-D analysis.
Variables, Outcome Measures
Manual placement of regions of interest (ROI) was performed by one reader (FS) (Fig. 1A). Twelve radial dGEMRIC- and radial morphologic flip-angle images were reformatted and allocated to the 12 clock-face positions using a previously described method [27, 47]. For anatomic allocation, the acetabular teardrop, which reflects the midpoint of the acetabular notch, was used as a reference for the 6 o’clock position with the 12 o’clock position located directly opposed to it. Based on that, each “full hour” position on the clock-face position was defined at every two slice gaps of the 12 reformats [27, 47]. For assessment of dGEMRIC indices, ROIs included both directly opposed femoral and acetabular cartilage layers (Fig. 1A). We assessed cartilage thickness by measuring the peripheral and central joint space widths and calculating the mean. This was performed for 10 of 12 possible clock-face positions per hip. No measurements were performed at the 5 and 6 o’clock positions, which are usually devoid of cartilage. This manual 2-D analysis was performed with Osirix software (Version 6.0, Geneva, Switzerland [42]). For comparison against the manual 2-D assessment, the manually segmented 3-D cartilage models (Fig. 1B) and automatically segmented 3-D cartilage models (Fig. 1C) were used as input data for the Cartilage Analyzer software for 3-D analysis. Analogously to the manual 2-D analysis the mean dGEMRIC indices and mean cartilage thickness was calculated with ROIs at the same 10 clock-face positions except for the 5- and 6 o’clock positions. For anatomic allocation, we inspected the 3-D models and identified the midpoint of the inferior acetabular notch as the 6 o’clock position with the 12 o’clock position located directly opposite to it. Based on that each full-hour position was defined at every 30° of the 360° circumference. For comparison between the manual 2-D analysis and the two 3-D analyses the ROIs of each clock-face position were clustered into the four quadrants for each of the 25 hips [44]: 100 (25 hips each 4 clock-face positions, 12–3 o’clock) ROIs for the anterosuperior-, 100 (25 hips each 4 clock-face positions, 9–12 o’clock) ROIs for the posterosuperior-, 50 (25 hips each 2 clock-face positions, 3–4 o’clock) ROIs for the anteroinferior- and 75 (25 hips each 3 clock-face positions, 7–9 o’clock) ROIs for the posteroinferior quadrant. dGEMRIC indices and cartilage thickness of the manually and automatic segmentation 3-D cartilage models were compared against the manual 2-D analysis, which served as gold standard. Differences in accuracy of the 3-D methods relative to the manual 2-D method were compared.
The manually and automatically segmented 3-D models were used for calculation of mean dGEMRIC indices, mean thickness, surface, and volume of hip cartilage using the Cartilage Analyzer software. The four parameters were compared among the four quadrants for both 3-D techniques. To assess the accuracy of the fully automatic cartilage segmentation method, we used the corresponding manual segmentation as the reference to calculate dice coefficients [13] and average surface distance. The dice coefficient quantifies the overlap of two binary volumetric data by normalizing the sizes of their intersection over the average of their sizes. The average surface distance is the average of all the distances from points on the boundary of an automatically segmented cartilage to the boundary of a manually segmented cartilage.
Two independent readers (FS, TDL) performed a repeated analysis for all 25 hips. They analyzed each quadrant using the Cartilage Analyzer to assess the intraobserver reproducibility and the interobserver reliability of this software to calculate dGEMRIC, cartilage thickness, cartilage surface, and cartilage volume. The repeated analysis was performed 4 weeks after the original analysis. The manually and automatically segmentation 3-D models were used as input data for the Cartilage Analyzer software.
Statistical Analysis
Descriptive statistics (mean, SD, range) were calculated. Normal distribution was evaluated using the Kolmogorov-Smirnov test. Mean differences were calculated and tested with paired t-tests or Wilcoxon rank-sum tests. Of note, inherent to the way differences in means are calculated, results can be smaller than the nominal image resolution. Agreement between the segmentation techniques was assessed with intraclass correlation coefficients (ICC) including 95% confidence intervals (CIs). To assess the accuracy of the fully automatic cartilage segmentation method, we calculated the average surface distance and dice coefficients between the automatically segmented cartilages, and the manually segmented cartilages. Analysis of variance was performed to assess regional patterns using both 3-D methods. If a significant difference was observed pairwise comparisons were performed and p values (0.05/4 = 0.013 adjusted for four groups) were adjusted with the Bonferroni correction for the four quadrants. Reproducibility and reliability were assessed with the ICC including 95% CIs. The ICC was graded as ICC < 0.20 for slight; 0.21 – 0.40 for fair; 0.41 – 0.60 for moderate; 0.61 – 0.80 for substantial; and > 0.80 for almost-perfect agreement [32]. We used SPSS version 25.0 (IBM, Armonk, New York, USA) for statistical analysis.
Results
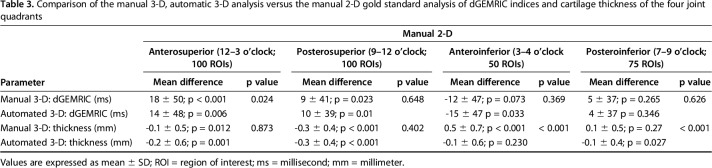
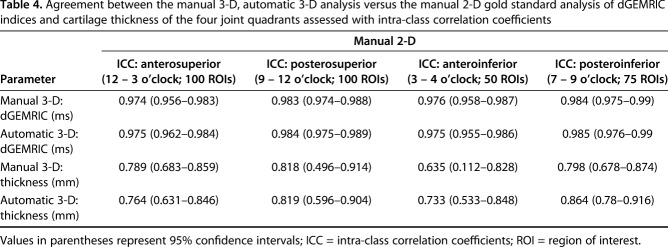
Comparable mean overall difference and almost perfect agreement in dGEMRIC indices were found between the manual 3-D analysis (8 ± 44 ms, p = 0.005), the automated 3-D analysis (7 ± 43 ms, p = 0.015), and the manual 2-D analysis (Table 2). The regional dGEMRIC comparison showed comparable differences (range, -15 ms to 18 ms; Table 3) and almost perfect agreement (range of ICCs: 0.974 – 0.985; Table 4). Agreement for measuring overall cartilage thickness was almost perfect for both 3-D methods (ICC = 0.855 and 0.881) versus the manual 2-D analysis (Table 2). A mean difference of -0.2 ± 0.5 mm (p < 0.001) was observed for overall cartilage thickness between the automated 3-D analysis and the manual 2-D analysis; no such difference was observed between the manual 3-D and the manual 2-D analysis (Table 2). In the anteroinferior quadrant, cartilage thickness assessed with the manual 3-D analysis differed by 0.5 ± 0.7 mm (p < 0.001) compared with the manual 2-D analysis (Table 3); no such difference was observed between the automated 3-D analysis and the manual 2-D analysis (Table 3). Agreement for measuring regional cartilage thickness ranged from substantial to almost perfect (ICC range, 0.635–0.864) for both 3-D methods versus the manual 2-D analysis (Table 4).
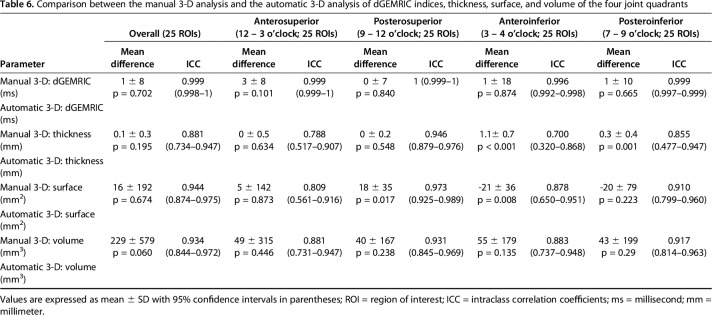
Table 2.
Comparison between manual 3-D, automatic 3-D analysis versus the manual 2-D gold standard analysis for overall dGEMRIC indices and cartilage thickness
Table 3.
Comparison of the manual 3-D, automatic 3-D analysis versus the manual 2-D gold standard analysis of dGEMRIC indices and cartilage thickness of the four joint quadrants
Table 4.
Agreement between the manual 3-D, automatic 3-D analysis versus the manual 2-D gold standard analysis of dGEMRIC indices and cartilage thickness of the four joint quadrants assessed with intra-class correlation coefficients
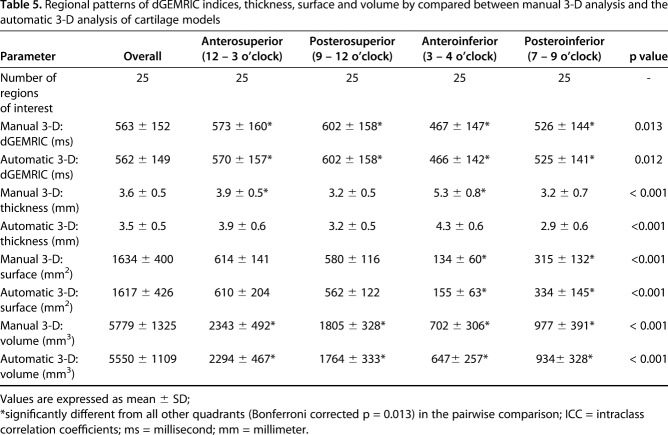
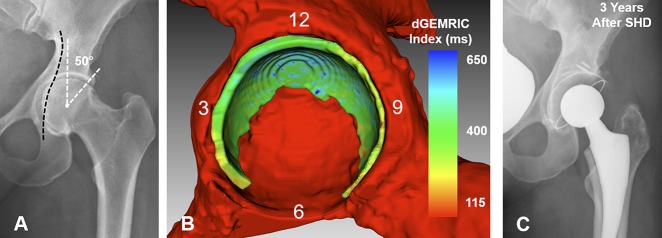
Regional morphologic and biochemical cartilage patterns were not different with the numbers available for both 3-D methods (Table 5). We found the highest dGEMRIC indices posterosuperiorly (manual: 602 ± 158 ms, p = 0.013; automated: 602 ± 158 ms, p = 0.012). We found the thickest cartilage anteroinferiorly (manual: 5.3 ± 0.8 mm, p < 0.001; automated: 4.3 ± 0.6 mm, p < 0.001). The smallest surface area was found anteroinferiorly (manual: 134 ± 60 mm2, p < 0.001; automated: 155 ± 60 mm2, p <0.001). Overall surface area ranged from 1088 mm2 to 2378 mm2 for the manual 3-D and from 1089 mm2 to 2812 mm2 for the automated 3-D analysis. The largest volume was found anterosuperiorly (manual: 2343 ± 492 mm3, p < 0.001; automated: 2294 ± 467 mm3, p < 0.001). Overall volume ranged from 3871 mm3 to 8284 mm3 for the manual 3-D and from 3868 mm3 to 7237 mm3 for the automated 3-D analysis. The greatest mean difference was found in cartilage thickness (1.1 ± 0.7 mm; p < 0.001) and cartilage surface area (−21 ± 36 mm2; p = 0.008) of the anteroinferior quadrant (Table 6). Comparison of both 3-D cartilages models showed and a mean dice coefficient of 86 ± 3% (range, 79%–90%) and a mean average surface distance of 0.26 ± 0.13 mm (range, 0.11–0.62 mm).
Table 5.
Regional patterns of dGEMRIC indices, thickness, surface and volume by compared between manual 3-D analysis and the automatic 3-D analysis of cartilage models
Table 6.
Comparison between the manual 3-D analysis and the automatic 3-D analysis of dGEMRIC indices, thickness, surface, and volume of the four joint quadrants
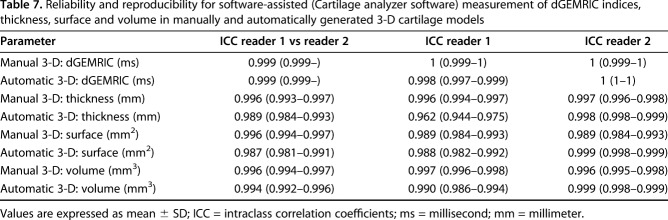
Intraobserver reproducibility and interobserver reliability were almost perfect for analysis of dGEMRIC indices, thickness, surface area, and volume (ICC range, 0.962–1) using either 3-D method (Table 7) for two readers and two different timepoints.
Table 7.
Reliability and reproducibility for software-assisted (Cartilage analyzer software) measurement of dGEMRIC indices, thickness, surface and volume in manually and automatically generated 3-D cartilage models
Discussion
Compared with radiographic- and 2-D morphologic MR imaging, 3-D biochemical imaging MR imaging techniques are more sensitive to the prearthritic cartilage degeneration that precedes morphological damage, and they offer the theoretical possibility to perform a volumetric analysis. However, the time-consuming and user-dependent postprocessing of biochemical cartilage MR images has limited the wide use of techniques like dGEMRIC [66]. To overcome this limitation, we developed a fully-automated method for 3-D cartilage segmentation using a deep-learning-based approach and validated this approach in patients with structural hip deformities, taking manual segmentation as the reference. This approach, after training, could directly map a volumetric 3-D cartilage to its volume-wise labels thereby providing an accurate, reliable, and reproducible automated 3-D analysis of cartilage composition, thickness, surface area, and volume. This time-efficient and objective analysis of biochemical cartilage composition and morphology yields the potential to improve patient selection in FAI surgery and to aid surgeons with planning of acetabuloplasty and periacetabular osteotomies in pincer FAI and hip dysplasia. In addition, this validation paves way to the large-scale use of this method for prospective trials that longitudinally monitor the effect of reconstructive hip surgery and the natural course of osteoarthritis.
This study has a number of limitations. First, the method used for automated 3-D cartilage segmentation enables assessment of combined femoral and acetabular cartilage layers only. Acetabular cartilage degeneration typically precedes femoral cartilage damage in FAI and hip dysplasia [6, 62]. Therefore, a separate analysis of acetabular and femoral cartilage would theoretically allow a better estimation of actual damage to the joint early in the degeneration process. Clear differentiation between the thin cartilage layers on MRI is a substantial challenge due to the tight coaptation of the hip. In contrast to other joints such as the knee, the femoral and acetabular cartilage layers are directly opposed to each other in the hip and together with the sealing effect of the labrum, this prevents contrast agent or synovial fluid from accumulating in the joint space [45]. To overcome this limitation, some authors used leg traction to achieve a distinct visualization of femoral and acetabular cartilage layers [1, 34, 46], while others used special algorithms [10]. However, a combined assessment of femoral and acetabular cartilage damage has proven to be highly accurate in predicting failure after PAO for correction of hip dysplasia [12, 26]. Furthermore the use of planar dGEMRIC maps generated from 3-D cartilage models enabled a moderate-to-strong correlation with the joint damage observed at surgery [9]. Thus, we believe the chosen approach for automated cartilage segmentation is reasonable. Second, it was not possible to validate the calculated 3-D cartilage surface area against a reference gold standard. This was related to the fact that the current study is based on symptomatic patients, which precludes a cadaveric comparison. However, our findings (surface area: 1088.8 mm2–2812.1 mm2) (Table 6) are comparable to the reported size of the lunate surface ranging from 1328 mm2–3610 mm2 in symptomatic patients [53]. Third, the results of this approach for automatic cartilage segmentation may differ when being applied to older patients with more severe radiographic cartilage degeneration. We selected a typical cohort of young and active patients without severe radiographic osteoarthritis (Tönnis 0 or 1) since these are the patients who are most likely to benefit from a comprehensive 3-D cartilage imaging analysis for an improved planning of joint-preserving hip surgery. Fourth, at the time of image acquisition our imaging protocol did not include a prescan to correct for B1 inhomogeneity, which reportedly can alter the resultant dGEMRIC values [50]. However, this does not affect the accuracy of the segmentation itself but leads to a greater variation in dGEMRIC indices among patients. Since it was not our aim to define thresholds to predict the presence of cartilage lesions or to define a prognostic threshold, this may be negligible.
Due to its thick cartilage layers, its superficial location, and the availability of dedicated coils, advanced compositional cartilage imaging techniques are usually first validated in the knee [15, 30]. To accelerate and standardize the postprocessing of morphologic- and biochemical 3-D images via automated segmentation, deep-learning-based solutions using deep convolutional neural networks have been introduced [35, 65]. In these studies a high accuracy for automatic segmentation of knee cartilage compartments with dice coefficients ranging from 0.770–0.878 [35] and 0.806–0.807 [65] has been reported. To the best of our knowledge, this is the first study to apply a deep-learning approach for fully automatic segmentation of hip cartilage. Although the cartilage layers are naturally thinner and more difficult to identify in the hip, we found a comparable accuracy with a strong mean dice coefficient of 86% and a range of 79%–90%. This was higher than previous studies for automated T2 mapping (mean dice coefficient of 73%) using a 3-D deformable model method [10] or a multitemplate based label fusion technique for automated dGEMRIC analysis (range of dice coefficients between 76% and 82%) [49]. Of note, previous studies presenting different techniques for automated cartilage segmentation did not describe the osseous acetabular morphology in their patient population [20, 39, 49], excluded hips with pincer FAI or acetabular dysplasia [14], or only included asymptomatic volunteers [10]. Acetabular morphology reportedly affects the size and shape of the lunate surface in patients eligible for joint-preserving hip surgery [53]. Consequently, an automated segmentation approach will be less accurate if applied to a hip morphology largely different from the used for initial training and testing. To ensure a wide applicability of our segmentation approach, we chose a representative patient cohort that included hips with acetabular dysplasia, normal acetabular coverage, acetabular retroversion, and acetabular overcoverage (Table 1). Furthermore, our approach yielded the advantage of providing a more comprehensive assessment of hip cartilage, including thickness, surface area, and volume while previous studies only focused on validating different approaches for extracting biochemical cartilage composition using dGEMRIC [14, 20, 49], T2 [10, 39], and T1rho [39]. In addition, it is important to mention that the methodology for automated cartilage segmentation can be applied to any other high-resolution 3-D cartilage MR technique such as T2* mapping [22]. Imaging parameters and image characteristics between different biochemical cartilage techniques differ from each other. Therefore, a segmentation approach that has been validated with one sequence technique would require adjustments and a new validation study before it can be applied to another type of sequence.
In contrast to previous studies [10, 14, 49], we compared dGEMRIC indices and cartilage thickness of the automated- and manual 3-D segmentation against the manual ROI placement on reformatted 2-D images, which is still considered the standard method (Fig. 1) [66]. We found minor differences in mean dGEMRIC indices (range, -15 to 18 ms; Table 3) for the manual 3-D and the automated 3-D analysis compared to the manual 2-D analysis. This deviation is acceptable as it is below the clinically relevant range of 50 ms to 100 ms that corresponds to different stages of the macroscopic cartilage damage [9]. Reference values of combined femoroacetabular cartilage thickness on MRI for symptomatic patients are sparse. However, a range of 2.8 mm to 4.3 mm has been reported for superolateral cartilage thickness using manual measurements, and a difference of 1.1 mm between dysplastic hips and normal hips has been shown [5]. In our study, the mean differences in cartilage thickness for the manual 3-D analysis ranged from -0.3 mm to 0.5 mm compared to the manual 2-D gold standard (Table 3). The mean differences in cartilage thickness for the automated 3-D analysis ranged from -0.3 mm to -0.1 mm compared to the manual 2-D gold standard (Table 3). Notably, the automated 3-D analysis was more accurate than the manual 3-D analysis anteroinferiorly, where the largest deviation compared with the manual 2-D analysis was observed (Table 3). This supports the use of more objective methods, such as our deep learning approach, for segmentation.
Distinct patterns of cartilage damage have been linked with different pathomechanisms and osseous deformities of the hip. This includes hip instability secondary to acetabular dysplasia, inclusion, and impaction impingement due to cam and pincer deformities [6, 56]. However, a reliable preoperative assessment of the damage pattern is challenging using standard, morphologic imaging [51]. To overcome this limitation, we validated a new automated 3-D analysis for regional assessment of hip cartilage and showed dGEMRIC patterns comparable to the manual 3-D reference (Table 6). Using the 3-D visualization, a more global pattern of biochemical damage was observed in a dysplastic hip (Fig. 4) and in a deep hip (Fig. 5A-B). By contrast, we observed an even distribution of high dGEMRIC values in a young patient with normal acetabular coverage (Fig. 6A-C). We observed an anterior decline in dGEMRIC indices in a hip with acetabular retroversion (Fig. 7A-C). The most pronounced uniform decrease in dGEMRIC indices was observed in a hip with severe global overcoverage secondary to protrusio acetabuli (Fig. 8A-C). A 3-D visualization of these patterns may have prognostic implications as shown in dysplasia in which anterior dGEMRIC indices were more accurate predictors of failure after PAO than global dGEMRIC indices [12, 26]. By contrast, the definition of reliable thresholds to predict the success of FAI surgery based on preoperative dGEMRIC indices is more challenging [11, 25]. This may be related to the more focal damage pattern of FAI [51], which makes detection and manual measurement of actual cartilage damage difficult on reformatted 2-D dGEMRIC images. In comparison, a fast 3-D visualization of dGEMRIC indices at a glance enables a volumetric dGEMRIC analysis of the most severely affected cartilage area, including regions with healthy cartilage as internal reference. Thus, this proposed more time-efficient and objective 3-D analysis can improve patient selection with the goal to identify those patients who benefit most from FAI surgery (Fig. 7) as opposed to others with too advanced cartilage degeneration (Fig. 8). In addition, such a tool yields promise to facilitate the standardized longitudinal analysis of cartilage composition in large prospective trials that aim to monitor the effect of reconstructive hip surgery and the natural course of osteoarthritis. We could further show that the new automated 3-D analysis enabled an accurate detection of regional patterns of cartilage thickness and volume (Table 6). A 3-D analysis of cartilage thickness and volume can provide complementary information or may be used as alternative biomarker, if no 3-D biochemical cartilage sequences are acquired.
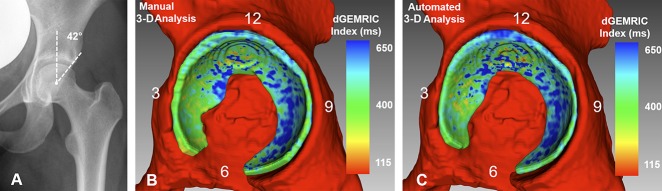
Fig. 5 A-C.
(A) AP pelvis of a 21-year-old woman with a deep hip (LCE angle = 42°) and a cam deformity is shown. (B) Manual 3-D analysis is shown with dGEMRIC indices color-coded. (C) Automated 3-D analysis is shown with dGEMRIC indices color-coded. The red indicates regions of low dGEMRIC indices demonstrating low glycosaminoglycan content corresponding to biochemical cartilage damage and vice-versa for blue color. (B) Using the manual 3D analysis, the mean dGEMRIC index was 482 ms and cartilage surface area was 2128 mm2. This was comparable to (C) the automated 3-D analysis which yielded a mean dGEMRIC index of 489 ms and a cartilage surface of 1912 mm2. (B) Overestimation of the manual 3-D analysis (Table 3) for measuring thickness in the anterioinferior quadrant resulted in a difference of 0.8 mm compared to the automated 3-D analysis.
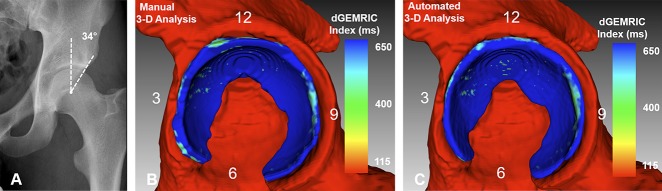
Fig. 6 A-C.
(A) AP pelvis of a 22-year-old man with normal acetabular coverage (LCE angle = 34°) and a cam deformity is shown. (B) The manual 3-D analysis is shown with dGEMRIC indices color-coded. (C) An automated 3-D analysis is shown with the red indicating regions of low dGEMRIC indices demonstrating low glycosaminoglycan content corresponding to biochemical cartilage damage and vice-versa for blue. (B) Using the manual 3-D analysis the mean dGEMRIC index was 767 ms, mean thickness was 3.8 mm, surface area was 1972 mm2 and volume was 7569 mm3. This was comparable to (C) the automated 3-D analysis which yielded a mean dGEMRIC index of 765 ms, a mean thickness of 3.7 mm, a surface area of 1981 mm2 and a volume of 7236 mm3.
Fig. 7 A-C.
(A) An AP pelvis image shows a 27-year-old woman with severe acetabular retroversion (positive cross-over sign, posterior wall sign, and ischial spine sign). (B) Fully-automated 3-D cartilage model is shown. The dGEMRIC indices are color-coded. Biochemical cartilage damage as shown with lower (green) dGEMRIC indices is present anteriorly compared to the high dGEMRIC indices posteriorly (blue). The overall (1250 mm2) and the anterosuperior cartilage surface area were smaller (457 mm2) compared with the respective means of the entire study group (overall: 1617 ± 426 mm2; anterosuperior: 610 ± 204 mm2; Table 7). The patient underwent anteverting periacetabular osteotomy for correction of malrotation of the hemipelvis. (C) AP pelvis view at the last clinical followup 4 years after surgery shows no progression of osteoarthritis and the patient was asymptomatic.
Fig. 8 A-C.
(A) The AP pelvis x-ray shows a 40-year-old woman with protrusio acetabuli and an LCE angle of 50°. (B) Fully-automated 3-D cartilage model is shown. dGEMRIC indices are color-coded. Biochemical cartilage damage as shown with lower dGEMRIC indices (green) is uniformly present. Cartilage surface area is slightly increased with 1817 mm2 compared with the mean of 1617 ± 426 mm2 of the entire study group. Although the LCE was higher compared with the deep hip shown in Fig. 5 (LCE 42°), the cartilage surface area was smaller compared with that hip (1912 mm2). (C) The patient underwent a surgical hip dislocation for circumferential rim trimming. After 3 years, the patient experienced a worsening of pain and underwent total hip replacement.
The most appropriate surgical treatment for different acetabular deformities is still subject of debate. More specifically, there is no consensus how to best surgically correct borderline dysplastic hips [2, 37] and hips with pincer FAI secondary to acetabular retroversion [19, 53, 67] and overcoverage [7, 18, 29]. This may be related to the fact that surgical planning relies on the radiographic anatomy of the acetabulum which is affected by the acquisition technique and can only estimate the size of the weightbearing cartilage surface area [55, 57]. Therefore, we performed an automated regional and global 3-D analysis of the cartilage surface area which was comparable to the manual 3-D analysis (Table 5). Calculation of total cartilage surface area and visualization of regional acetabular deficiency based on an automatic 3-D cartilage segmentation may aid in surgical decision making between PAO to restore normal coverage or hip arthroscopy for soft tissue repair in patients with mild or borderline dysplasia (Fig. 4). Acetabular retroversion is considered focal anterior overcoverage in hips with a cranial crossover sign or a rotational deformity in global cases in hips with severe radiographic retroversion [48]. In addition, there are hips with a continuous spectrum of global acetabular overcoverage [58]. Anteverting PAO has been performed to correct malrotation of the acetabulum with durable mid- to long-term results [38, 48, 67] while arthroscopic rim trimming has been proposed as a less invasive alternative [19]. In patients with radiographic signs of a too-deep socket, rim trimming is routinely performed [18]. However, it has been shown that more severe radiographic signs of acetabular overcoverage (Fig. 8) do not necessarily reflect a larger lunate surface compared with deep hips with less severe signs of overcoverage (Fig. 5), and that the acetabular cartilage surface area is smaller in hips with severe retroversion (Fig. 7) compared with normal hips (Fig.6) [41, 53]. Consequently, such hips are at risk of an iatrogenic increase in joint contact stresses as a result of a too aggressive acetabuloplasty [7, 53, 67], which is associated with long-term failure of FAI surgery [52]. In the future, the proposed method for automated 3-D cartilage segmentation may be used to define reference values for cartilage surface area for an individual case-by case comparison. Thereby, the used methodology can potentially improve surgical planning of acetabuloplasty and periacetabular osteotomies.
While a quick inspection of the 3-D cartilage models may be helpful to get an overview of biochemical cartilage damage and cartilage morphology [9], we developed a software for calculation of the actual cartilage parameters. Similar to previous studies [10, 14, 39] that used MR-based cartilage segmentation, we showed that the developed software for analyzing the 3-D cartilage models enabled assessment with almost perfect intraobserver reproducibility and interobserver reliability (ICC range, 0.962–1; Table 7).
To conclude, we developed a fully-automated method for 3-D cartilage segmentation using a deep-learning-based approach and validated this approach in patients with structural hip deformities, taking manual segmentation as the reference. This approach, after training, could directly map a volumetric 3-D cartilage to its volume-wise labels, thereby providing an accurate, reliable, and reproducible automated 3-D analysis of cartilage composition, thickness, surface area, and volume. The time-efficient and objective analysis of biochemical cartilage composition and cartilage thickness using this automatic approach for segmentation yields potential to improve patient selection and identify those patients who benefit most from FAI surgery. In addition, this validation may lead to the large-scale use of this methodology for prospective trials that longitudinally monitor the effect of reconstructive hip surgery and the natural course of osteoarthritis. Furthermore, the calculation of size of the weightbearing cartilage surface area may improve surgical planning of acetabuloplasty and periacetabular osteotomies for treatment of hip dysplasia and pincer FAI.
Footnotes
All ICMJE Conflict of Interest Forms for authors and Clinical Orthopaedics and Related Research® editors and board members are on file with the publication and can be viewed on request.
One of the authors certifies that he (FS) has received or may receive payments or benefits, during the study period, in an amount of USD 10,000 to USD 100,000, from the Swiss National Science Foundation (Bern, Switzerland, Grant No. P1BEP3_181643). Two of the authors certifies that they (GZ, GZ) have received or may receive payments or benefits, during the study period, in an amount of USD 10,000 to USD 100,000, from the Swiss National Science Foundation (Bern, Switzerland, Grant No.205321 163224/1). The institution of one or more of the authors (MT, KAS) has received funding from the Swiss National Science Foundation, outside the submitted work. The remaining authors (RH, TDL, ENN, JDW, YJK) certify that neither he or she, nor any member of his or her immediate family, have funding or commercial associations (consultancies, stock ownership, equity interest, patent/licensing arrangements, etc) that might pose a conflict of interest in connection with the submitted article.
Each author certifies that his or her institution approved the reporting of this investigation and that all investigations were conducted in conformity with ethical principles of research.
This work was performed at the Department of Orthopaedic Surgery, Inselspital Bern, University of Bern, Bern, Switzerland.
References
- 1.Abraham CL, Bangerter NK, McGavin LS, Peters CL, Drew AJ, Hanrahan CJ, Anderson AE. Accuracy of 3D dual echo steady state (DESS) MR arthrography to quantify acetabular cartilage thickness. J Magn Reson Imaging. 2015;42:1329–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adler KL, Giordano BD. The utility of hip arthroscopy in the setting of acetabular dysplasia: a systematic review. Arthroscopy. 2019;35:237–248. [DOI] [PubMed] [Google Scholar]
- 3.Albers CE, Steppacher SD, Ganz R, Tannast M, Siebenrock KA. Impingement adversely affects 10-year survivorship after periacetabular osteotomy for DDH. Clin Orthop Relat Res. 2013;471:1602–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anwander H, Melkus G, Rakhra KS, Beaulé PE. T1ρ MRI detects cartilage damage in asymptomatic individuals with a cam deformity. J Orthop Res. 2016;34:1004–1009. [DOI] [PubMed] [Google Scholar]
- 5.Ashwell ZR, Flug J, Chadayammuri V, Pascual-Garrido C, Garabekyan T, Mei-Dan O. Lateral acetabular coverage as a predictor of femoroacetabular cartilage thickness. J Hip Preserv Surg. 2016;3:262–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beck M, Kalhor M, Leunig M, Ganz R. Hip morphology influences the pattern of damage to the acetabular cartilage: femoroacetabular impingement as a cause of early osteoarthritis of the hip. J Bone Joint Surg Br. 2005;87:1012–1018. [DOI] [PubMed] [Google Scholar]
- 7.Bhatia S, Lee S, Shewman E, Mather RC, Salata MJ, Bush-Joseph CA, Nho SJ. Effects of acetabular rim trimming on hip joint contact pressures: how much is too much? Am J Sports Med. 2015;43:2138–2145. [DOI] [PubMed] [Google Scholar]
- 8.Bittersohl B, Miese FR, Hosalkar HS, Herten M, Antoch G, Krauspe R, Zilkens C. T2* mapping of hip joint cartilage in various histological grades of degeneration. Osteoarthritis Cartilage. 2012;20:653–660. [DOI] [PubMed] [Google Scholar]
- 9.Bulat E, Bixby SD, Siversson C, Kalish LA, Warfield SK, Kim Y-J. Planar dGEMRIC maps may aid imaging assessment of cartilage damage in femoroacetabular impingement. Clin Orthop Relat Res. 2016;474:467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chandra SS, Surowiec R, Ho C, Xia Y, Engstrom C, Crozier S, Fripp J. Automated analysis of hip joint cartilage combining MR T2 and three-dimensional fast-spin-echo images. Magn Reson Med. 2016;75:403–413. [DOI] [PubMed] [Google Scholar]
- 11.Chandrasekaran S, Vemula SP, Lindner D, Lodhia P, Suarez-Ahedo C, Domb BG. Preoperative delayed gadolinium-enhanced magnetic resonance imaging of cartilage (dGEMRIC) for patients undergoing hip arthroscopy: Indices are predictive of magnitude of improvement in two-year patient-reported outcomes. J Bone Joint Surg Am. 2015;97:1305–1315. [DOI] [PubMed] [Google Scholar]
- 12.Cunningham T, Jessel R, Zurakowski D, Millis MB, Kim Y-J. Delayed gadolinium-enhanced magnetic resonance imaging of cartilage to predict early failure of Bernese periacetabular osteotomy for hip dysplasia. J Bone Joint Surg Am. 2006;88:1540–1548. [DOI] [PubMed] [Google Scholar]
- 13.Dice L. Measures of the amount of ecologic association between species. Ecology. 1945;26:297–302. [Google Scholar]
- 14.Fernquest S, Park D, Marcan M, Palmer A, Voiculescu I, Glyn-Jones S. Segmentation of hip cartilage in compositional magnetic resonance imaging: A fast, accurate, reproducible, and clinically viable semi-automated methodology. J Orthop Res. 2018. [DOI] [PubMed] [Google Scholar]
- 15.Guermazi A, Roemer FW, Alizai H, Winalski CS, Welsch G, Brittberg M, Trattnig S. State of the art: MR imaging after knee cartilage repair surgery. Radiology. 2015;277:23–43. [DOI] [PubMed] [Google Scholar]
- 16.Haefeli P, Albers C, Steppacher S, Tannast M, Büchler L. What are the risk factors for revision surgery after hip arthroscopy for femoroacetabular impingement at 7-year followup? Clin Orthop Relat Res. 2017;475:1169–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanke MS, Steppacher SD, Anwander H, Werlen S, Siebenrock KA, Tannast M. What MRI findings predict failure 10 years after surgery for femoroacetabular impingement? Clin Orthop Relat Res. 2017;475:1192–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanke MS, Steppacher SD, Zurmühle CA, Siebenrock KA, Tannast M. Hips with protrusio acetabuli Are at increased risk for failure after femoroacetabular impingement surgery: A 10-year followup. Clin Orthop Relat Res. 2016;474:2168-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartigan DE, Perets I, Walsh JP, Close MR, Domb BG. Clinical outcomes of hip arthroscopy in radiographically diagnosed retroverted acetabula. Am J Sports Med. 2016;44:2531–2536. [DOI] [PubMed] [Google Scholar]
- 20.Hesper T, Bittersohl B, Schleich C, Hosalkar H, Krauspe R, Krekel P, Zilkens C. Automatic cartilage segmentation for delayed gadolinium-enhanced magnetic resonance imaging of hip joint cartilage: a feasibility study. Cartilage. [Published online ahead of print June 1, 2018]. DOI: 10.1177/1947603518783481. [DOI] [PMC free article] [PubMed]
- 21.Hesper T, Bulat E, Bixby S, Akhondi-Asl A, Afacan O, Miller P, Bowen G, Warfield S, Kim Y-J. Both 3-T dGEMRIC and acetabular-femoral T2 difference may detect cartilage damage at the chondrolabral junction. Clin Orthop Relat Res. 2017;475:1058–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hesper T, Neugroda C, Schleich C, Antoch G, Hosalkar H, Krauspe R, Zilkens C, Bittersohl B. T2*-mapping of acetabular cartilage in patients with femoroacetabular impingement at 3 Tesla: comparative analysis with arthroscopic findings. Cartilage. 2018;9:118–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hingsammer AM, Kalish LA, Stelzeneder D, Bixby S, Mamisch TC, Connell P, Millis MB, Kim Y-J. Does periacetabular osteotomy for hip dysplasia modulate cartilage biochemistry? J Bone Joint Surg Am. 2015;97:544–550. [DOI] [PubMed] [Google Scholar]
- 24.Hingsammer AM, Miller PE, Millis MB, Kim Y-J. Does periacetabular osteotomy have depth-related effects on the articular cartilage of the hip? Clin Orthop Relat Res. 2015;473:3735–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hingsammer AM, Stelzeneder D, Kalish LA, Millis MB, Kim Y-J. Prognostic factors for mid-term symptom relief after open surgical correction for femoroacetabular impingement. Hip Int. 2015;25:406–412. [DOI] [PubMed] [Google Scholar]
- 26.Kim SD, Jessel R, Zurakowski D, Millis MB, Kim Y-J. Anterior delayed gadolinium-enhanced MRI of cartilage values predict joint failure after periacetabular osteotomy. Clin Orthop Relat Res. 2012;470:3332–3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klenke F, Hoffmann D, Cross B, Siebenrock K. Validation of a standardized mapping system of the hip joint for radial MRA sequencing. Skeletal Radiol. 2015;44:339–343. [DOI] [PubMed] [Google Scholar]
- 28.Lerch TD, Steppacher SD, Liechti EF, Tannast M, Siebenrock KA. One-third of hips after periacetabular osteotomy survive 30 years with good clinical results, no progression of arthritis, or conversion to THA. Clin Orthop Relat Res. 2017;475:1154–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liechti EF, Ferguson SJ, Tannast M. Protrusio acetabuli: joint loading with severe pincer impingement and its theoretical implications for surgical therapy. J. Orthop. Res. 2015;33:106–113. [DOI] [PubMed] [Google Scholar]
- 30.Link TM, Neumann J, Li X. Prestructural cartilage assessment using MRI. J Magn Reson Imaging. 2017;45:949–965. [DOI] [PubMed] [Google Scholar]
- 31.Menge TJ, Briggs KK, Dornan GJ, McNamara SC, Philippon MJ. Survivorship and outcomes 10 years following hip arthroscopy for femoroacetabular impingement: Labral debridement compared with labral repair. J Bone Joint Surg Am. 2017;99:997–1004. [DOI] [PubMed] [Google Scholar]
- 32.Montgomery AA, Graham A, Evans PH, Fahey T. Inter-rater agreement in the scoring of abstracts submitted to a primary care research conference. BMC Health Serv Res. 2002;2:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murphy SB, Simon SR, Kijewski PK, Wilkinson RH, Griscom NT. Femoral anteversion. J Bone Joint Surg Am. 1987;69:1169–1176. [PubMed] [Google Scholar]
- 34.Nishii T, Sugano N, Sato Y, Tanaka H, Miki H, Yoshikawa H. Three-dimensional distribution of acetabular cartilage thickness in patients with hip dysplasia: a fully automated computational analysis of MR imaging. Osteoarthritis Cartilage . 2004;12:650–657. [DOI] [PubMed] [Google Scholar]
- 35.Norman B, Pedoia V, Majumdar S. Use of 2D U-Net convolutional neural networks for automated cartilage and meniscus segmentation of knee MR imaging data to determine relaxometry and morphometry. Radiology. 2018;288:177–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nötzli HP, Wyss TF, Stoecklin CH, Schmid MR, Treiber K, Hodler J. The contour of the femoral head-neck junction as a predictor for the risk of anterior impingement. J Bone Joint Surg Br. 2002;84:556–560. [DOI] [PubMed] [Google Scholar]
- 37.Novais EN, Coobs BR, Nepple JJ, Clohisy JC, ANCHOR Study Group. Previous failed hip arthroscopy negatively impacts early patient-reported outcomes of the periacetabular osteotomy: an ANCHOR Matched Cohort Study. J Hip Preserv Surg. 2018;5:370–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parry JA, Swann RP, Erickson JA, Peters CL, Trousdale RT, Sierra RJ. Midterm outcomes of reverse (anteverting) periacetabular osteotomy in patients with hip impingement secondary to acetabular retroversion. Am J Sports Med. 2016;44:672–676. [DOI] [PubMed] [Google Scholar]
- 39.Pedoia V, Gallo MC, Souza RB, Majumdar S. Longitudinal study using voxel-based relaxometry: Association between cartilage T1ρ and T2 and patient reported outcome changes in hip osteoarthritis. J Magn Reson Imaging. 2017;45:1523–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pedoia V, Norman B, Mehany SN, Bucknor MD, Link TM, Majumdar S. 3D convolutional neural networks for detection and severity staging of meniscus and PFJ cartilage morphological degenerative changes in osteoarthritis and anterior cruciate ligament subjects. J Magn Reson Imaging. 2019;49:400-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pun SY, Hingsammer A, Millis MB, Kim Y-J. Is increased acetabular cartilage or fossa size associated with pincer femoroacetabular impingement? Clin Orthop Relat Res. 2017;475:1013–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosset A, Spadola L, Ratib O. OsiriX: an open-source software for navigating in multidimensional DICOM images. J Digit Imaging. 2004;17:205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmaranzer F, Arendt L, Liechti EF, Nuss K, von Rechenberg B, Kircher PR, Tannast M. Do dGEMRIC and T2 imaging correlate with histologic cartilage degeneration in an experimental ovine FAI model? Clin Orthop Relat Res. [Published online ahead of print November 29, 2018]. DOI: 10.1097/CORR.0000000000000593. [DOI] [PMC free article] [PubMed]
- 44.Schmaranzer F, Haefeli P, Hanke M, Liechti E, Werlen S, Siebenrock K, Tannast M. How does the dGEMRIC index change after surgical treatment for FAI? A prospective controlled study: Preliminary results. Clin Orthop Relat Res. 2017;475:1080–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmaranzer F, Klauser A, Kogler M, Henninger B, Forstner T, Reichkendler M, Schmaranzer E. Improving visualization of the central compartment of the hip with direct MR arthrography under axial leg traction: a feasibility study. Acad Radiol. 2014;21:1240–1247. [DOI] [PubMed] [Google Scholar]
- 46.Schmaranzer F, Klauser A, Kogler M, Henninger B, Forstner T, Reichkendler M, Schmaranzer E. Diagnostic performance of direct traction MR arthrography of the hip: detection of chondral and labral lesions with arthroscopic comparison. Eur Radiol. 2015;25:1721–1730. [DOI] [PubMed] [Google Scholar]
- 47.Schmaranzer F, Todorski IAS, Lerch TD, Schwab J, Cullmann-Bastian J, Tannast M. Intra-articular lesions: Imaging and surgical correlation. Semin Musculoskelet Radiol. 2017;21:487–506. [DOI] [PubMed] [Google Scholar]
- 48.Siebenrock KA, Schaller C, Tannast M, Keel M, Büchler L. Anteverting periacetabular osteotomy for symptomatic acetabular retroversion: results at ten years. J Bone Joint Surg Am. 2014;96:1785–1792. [DOI] [PubMed] [Google Scholar]
- 49.Siversson C, Akhondi-Asl A, Bixby S, Kim Y-J, Warfield SK. Three-dimensional hip cartilage quality assessment of morphology and dGEMRIC by planar maps and automated segmentation. Osteoarthritis Cartilage. 2014;22:1511–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Siversson C, Chan J, Tiderius C-J, Mamisch TC, Jellus V, Svensson J, Kim Y-J. Effects of B1 inhomogeneity correction for three-dimensional variable flip angle T1 measurements in hip dGEMRIC at 3 T and 1.5 T. Magn Reson Med. 2012;67:1776–1781. [DOI] [PubMed] [Google Scholar]
- 51.Stelzeneder D, Mamisch TC, Kress I, Domayer SE, Werlen S, Bixby SD, Millis MB, Kim Y-J. Patterns of joint damage seen on MRI in early hip osteoarthritis due to structural hip deformities. Osteoarthritis Cartilage. 2012;20:661–669. [DOI] [PubMed] [Google Scholar]
- 52.Steppacher S, Anwander H, Zurmühle C, Tannast M, Siebenrock K. Eighty percent of patients with surgical hip dislocation for femoroacetabular impingement have a good clinical result without osteoarthritis progression at 10 years. Clin Orthop Relat Res. 2015;473:1333–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Steppacher SD, Lerch TD, Gharanizadeh K, Liechti EF, Werlen SF, Puls M, Tannast M, Siebenrock KA. Size and shape of the lunate surface in different types of pincer impingement: theoretical implications for surgical therapy. Osteoarthritis Cartilage. 2014;22:951–958. [DOI] [PubMed] [Google Scholar]
- 54.Sutter R, Dietrich TJ, Zingg PO, Pfirrmann CWA. How useful is the alpha angle for discriminating between symptomatic patients with cam-type femoroacetabular impingement and asymptomatic volunteers? Radiology. 2012;264:514–521. [DOI] [PubMed] [Google Scholar]
- 55.Tannast M, Fritsch S, Zheng G, Siebenrock KA, Steppacher SD. Which radiographic hip parameters do not have to be corrected for pelvic rotation and tilt? Clin Orthop Relat Res. 2015;473:1255–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tannast M, Goricki D, Beck M, Murphy SB, Siebenrock KA. Hip damage occurs at the zone of femoroacetabular impingement. Clin Orthop Relat Res. 2008;466:273–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tannast M, Hanke MS, Zheng G, Steppacher SD, Siebenrock KA. What are the radiographic reference values for acetabular under- and overcoverage? Clin Orthop Relat Res. 2015;473:1234–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tannast M, Leunig M, Session participants. Report of breakout session: Coxa profunda/protrusio management. Clin Orthop Relat Res. 2012;470:3459–3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tannast M, Mistry S, Steppacher SD, Reichenbach S, Langlotz F, Siebenrock KA, Zheng G. Radiographic analysis of femoroacetabular impingement with Hip2Norm-reliable and validated. J Orthop Res. 2008;26:1199–1205. [DOI] [PubMed] [Google Scholar]
- 60.Tannast M, Siebenrock KA, Anderson SE. Femoroacetabular impingement: radiographic diagnosis--what the radiologist should know. AJR Am J Roentgenol. 2007;188:1540–1552. [DOI] [PubMed] [Google Scholar]
- 61.Wells J, Millis M, Kim Y-J, Bulat E, Miller P, Matheney T. Survivorship of the Bernese periacetabular osteotomy: What factors are associated with long-term failure? Clin Orthop Relat Res. 2017;475:396–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zaltz I, Leunig M. Parafoveal chondral defects associated with femoroacetabular impingement. Clin Orthop Relat Res. 2012;470:3383–3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zeng G, Wang Q, Lerch T, Schmaranzer F, Tannast M, Siebenrock K, Zheng G. Latent3DU-net: Multi-level latent shape space constrained 3D U-net for automatic segmentation of the proximal femur from radial MRI of the hip. In: Shi Y, Suk H-I, Liu M, eds. Machine Learning in Medical Imaging.Vol 11046 Cham: Springer International Publishing; 2018:188–196. Available at: http://link.springer.com/10.1007/978-3-030-00919-9_22 [Accessed February 1, 2019]. [Google Scholar]
- 64.Zheng G, Tannast M, Anderegg C, Siebenrock KA, Langlotz F. Hip2Norm: an object-oriented cross-platform program for 3D analysis of hip joint morphology using 2D pelvic radiographs. Comput Methods Programs Biomed. 2007;87:36–45. [DOI] [PubMed] [Google Scholar]
- 65.Zhou Z, Zhao G, Kijowski R, Liu F. Deep convolutional neural network for segmentation of knee joint anatomy. Magn Reson Med. 2018;80:2759–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zilkens C, Tiderius CJ, Krauspe R, Bittersohl B. Current knowledge and importance of dGEMRIC techniques in diagnosis of hip joint diseases. Skeletal Radiol. 2015;44:1073–1083. [DOI] [PubMed] [Google Scholar]
- 67.Zurmühle CA, Anwander H, Albers CE, Hanke MS, Steppacher SD, Siebenrock KA, Tannast M. Periacetabular osteotomy provides higher survivorship than rim trimming for acetabular retroversion. Clin Orthop Relat Res. 2017;475:1138–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]