Abstract
Studies involving transcriptomics have revealed multiple molecular subtypes of hepatocellular carcinoma (HCC). PET/CT has also identified distinct molecular imaging subtypes, including those with increased and decreased choline metabolism as measured by tissue uptake of the radiopharmaceutical 18F-fluorocholine. Gene signatures reflecting the molecular heterogeneity of HCC may identify the biological and clinical significance of these imaging subtypes. In this study, 41 patients underwent 18F-fluorocholine PET/CT followed by tumor resection and gene expression profiling. Over- and under-expressed components of previously published gene signatures were evaluated for enrichment between tumors with high and low 18F-fluorocholine uptake using gene set analysis. Significant gene sets were enumerated by false discovery rate (FDR) based on phenotype permutation. Associations with overall survival were analyzed by univariate and multivariate proportional hazards regression. Ten gene sets related to HCC were significantly associated with high tumor 18F-fluorocholine uptake at FDR q < 0.05, including those from 3 different clinical molecular classification systems and 2 prognostic signatures for HCC that showed predictive value in the study cohort. Tumor avidity for 18F-fluorocholine was associated with favorable characteristics based on these signatures, with lower mortality based on survival analysis (hazard ratio 0.36, 95% confidence interval 0.14 to 0.95). Tumors demonstrating high 18F-fluorocholine uptake were also enriched for genes involved in oxidative phosphorylation, fatty acid metabolism, peroxisome, bile acid metabolism, xenobiotic metabolism, and adipogenesis. These results provide a pathobiological framework to further evaluate 18F-fluorocholine PET/CT as a molecular and prognostic classifier in HCC.
INTRODUCTION
Hepatocellular carcinoma (HCC), the third leading cause of cancer death worldwide(1), is rising sharply in incidence and mortality in the US(2). Efforts aimed at molecularly characterizing this disease, particularly by transcriptomic analysis, have revealed significant heterogeneity across tumors(3–7). This heterogeneity has the potential to confound clinical trials by diluting molecular therapeutic targets amongst otherwise uniformly selected patients(8, 9), and contributes to variability in clinical outcomes across all stages of this disease, including early stage (10–12). Accordingly, there is a constant search for biomarkers to provide better molecular target identification and risk-stratification for patients with HCC. However, increasing use of radiologic criteria as the basis for HCC diagnosis in the clinical setting has inadvertently limited the availability of tumor tissue for molecular profiling(13, 14). Although liver biopsies can be pursued to profile tumors, this invasive procedure causes significant morbidity and carries the risks of sampling error and tumor seeding(15). As a potential alternative, molecular imaging techniques such as positron emission tomography/ computed tomography (PET/CT) might provide a less invasive means to gain insight about tumor heterogeneity and pathobiology.
Fluorine-18 fluorocholine (FCh) is a radiopharmaceutical analog of choline designed to allow PET to trace the initial steps of phosphatidylcholine biosynthesis(16). It is approved by the European Medicines Agency as an imaging agent for HCC(17, 18). Anomalies in choline metabolism have been found in many different cancers(19), including HCC(20, 21), providing the premise for tumor detection using FCh PET/CT. When performed in tandem with 18F-fluorodeoxy-D-glucose (FDG) PET/CT, FCh PET/CT is associated with a diagnostic sensitivity of 90% or greater and results in improved tumor staging and treatment allocation for HCC(17, 18, 22). In comparison, FDG PET/CT and FCh PET/CT individually are associated with site-based sensitivities of 67% and 84 % respectively (p = 0.01)(17). Failures to detect HCC with FDG are predominantly the result of poor tumor contrast in the liver(17, 23), which was something anticipated by pre-clinical studies(24). In comparison, HCC can be detected with FCh based on either its increased metabolism relative to surrounding liver, as found in approximately 75% of patients, or its decreased metabolism, which occurs in another 10%−15%(17, 18). As a diagnostic imaging modality, FCh PET/CT is unique in this capacity to display two divergent “imaging phenotypes” for detecting HCC (examples shown in Figures 1A and1B).
Figure 1:
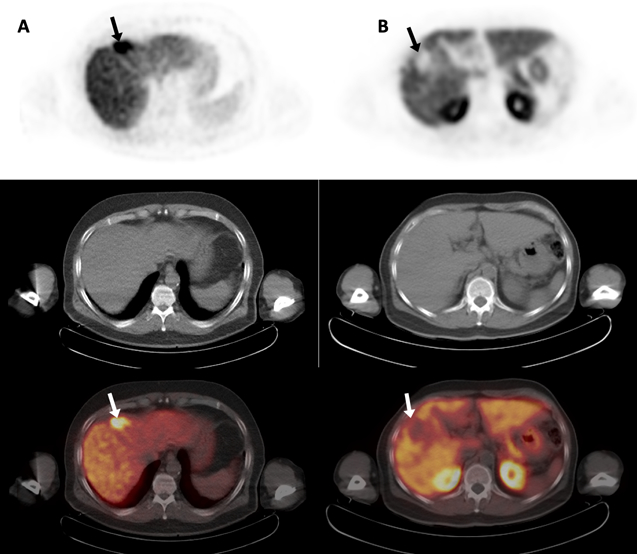
Two examples of hepatocellular carcinoma imaged by 18F-fluorocholine PET/CT. Transaxial PET (top row), non-contrast CT (middle row), and PET/CT images (bottom row) are shown. Column A: An HCC tumor demonstrating increased 18F-fluorocholine uptake (arrows). Column B: An HCC tumor demonstrating decreased 18F-fluorocholine uptake (arrows). In both cases, no corresponding structural abnormality (ie. necrosis or hemorrhage) is evident on CT. Assuming the underlying tissue is intact as in these cases, both imaging phenotypes can facilitate HCC detection. Based on the Hoshida classification system, these tumors were of different molecular sub-types, with the tumor in column A classified as a S3 sub-type and the tumor in column B classified as a S2 sub-type.
We hypothesized that gene expression differences are associated with these FCh PET/CT imaging phenotypes. Given that a large number of gene signatures have already been developed to characterize HCC, we further hypothesized that a subset of these existing gene signatures will contain components that correspond to an imaging phenotype. To test this hypothesis, we conducted a series of tumor enrichment analyses using collections of gene sets from a public repository to identify published gene signatures associated with tumor FCh uptake. In following this approach, we also sought to examine the clinical potential of FCh PET/CT as a non-invasive image-based molecular marker that could have value for patient risk-stratification and clinical trial enrichment in HCC.
METHODS
Patients
Between February 2012 and March 2016, 51 consecutive patients gave written informed consent to undergo FCh PET/CT prior to liver tumor resection as participants in an IRB-approved, federally-registered (NCT01395030), diagnostic trial. This trial was listed prospectively on clinicaltrials.gov in accordance with ICMJE. Briefly, patients were enrolled if they had HCC diagnosed histologically or suspected radiographically (ie. a Liver Imaging Reporting and Data System (LI-RADS) category 3, 4, or 5 lesion detected on a contrast enhanced CT or MRI examination(25)) or had a liver mass with imaging features of another primary malignancy; and were surgical candidates agreeable to liver tumor resection with Child-Pugh score < 10. Patients were excluded if they weighed greater than 350 pounds, were pregnant or lactating, had a serious underlying medical condition that would make FCh PET/CT intolerable, or previously received chemotherapeutic, molecularly-targeted, biological, or radiotherapeutic treatment for HCC. Based on these criteria, the study enrolled patients with suspected HCC and other primary liver cancers. Because the present study pertains to HCC, 8 patients with final diagnoses of other liver tumors (7 intrahepatic cholangiocarcinoma and 1 primary sarcoma) were not included. Two HCC patients with inadequate tissue for gene expression analysis were also not included, leaving 41 patients for analysis.
18F-fluorocholine PET/CT
Liver imaging was performed pre-operatively within two weeks of surgery using a Gemini TF-64 PET/CT scanner (Philips, Cambridge, MA). An in-line X-ray CT transmission scan of the torso was performed without intravenous contrast and completed in the supine position using these parameters: 64-channels, 120 kV, 50 mA/slice, rotation time 0.75 seconds, slice thickness/interval 5.0mm. For positron emission scanning, a dose of 2.2 to 3.0 MBq/kg of FCh was administered intravenously followed by emission scans of the liver at 30 to 40 minutes post-injection. PET image reconstruction was completed using the manufacturer’s protocol, with corrections for radioactive decay, dead-time, random coincidences, and scatter. Emission data was corrected for non-uniform attenuation using CT data. FCh doses were synthesized on the day of scanning using an automated chemical process control unit (CPCU, CTI/Siemens, Knoxville, TN) in accordance to an FDA Investigational New Drug Application. All doses passed assays for radiochemical purity, radionuclide identity, chemical purity, and non-pyrogenicity before use.
Tissues and Arrays
Tumor tissue samples were collected upon resection of the affected hepatic segment. Sampling was performed only if it did not interfere with surgical pathology evaluation, and limited to areas distant from the resection margin, avoiding any capsular, necrotic, hemorrhagic, or adjacent liver tissue. For radiopathologic correspondence, gross pathologic tumor size and sampling locations relative to the tumor-liver interface and surgical margin were recorded. Samples were preserved in RNALater (ThermoFisher, Waltham, MA) and stored at −80 C. RNA was extracted using the AllPrep DNA/RNA kit (Qiagen, Germantown, MD), checked for integrity using an Agilent Bioanalyzer with RNA 6000 Nano Chips (Agilent Technologies, Inc., Santa Clara, CA), processed using whole-genome direct annealing, selection, extension, and ligation (WG-DASL, Illumina, San Diego, CA), and hybridized onto Human HT-12 v4 Expression BeadChips (Illumina, San Diego, CA) covering 24,000 transcripts of genes, gene candidates, and splice variants. Final data arrays consisted of 20818 gene expression values for each patient. Formalin-fixed sections from each specimen also underwent histopathological review by a board-certified pathologist to confirm tumor cellularity and exclude the presence of benign liver tissue, hemorrhage, necrosis, and fibrosis.
Image Analysis
FCh PET/CT images were analyzed by an experienced reader (SAK, 15 years) in an unblinded fashion alongside clinically-available contrast-enhanced CT or MRI examinations. Areas of tumor sampling were then visually classified as demonstrating increased (FChHIGH), decreased (FChLOW), or iso-intense (FChISO) tracer activity relative to the adjacent liver on FCh PET/CT images. Tracer uptake in these areas was also quantified using a 1.2 cm diameter region of interest (ROI) to measure the mean and maximum standardized uptake value (SUVmean and SUVmax)(26). SUV was defined as the ratio of voxel to injected radioactivity normalized to body weight. The SUVmean of the liver was also measured using an ROI placed over tumor-adjacent liver. All ROIs excluded areas of hemorrhage or necrosis found on any of the imaging examinations. Tumor to adjacent liver ratios of SUVmean (TLRSUV) were calculated by dividing tumor SUVmean by liver SUVmean.
Gene Set Enrichment Analysis (GSEA)
GSEA (v3.0, Broad Institute, Cambridge, MA) was used to determine whether a priori defined sets of genes showed statistically significant concordant differences between tumors demonstrating the FChHIGH and FChLOW PET phenotypes(27). All gene sets used for analysis were downloaded from the Broad Institute Molecular Signature Database (mSigDB v6.2, obtained September 7, 2018 from software.broadinstitute.org/gsea/msigdb).
To test the study hypothesis that previously-developed gene sets related to HCC were associated with different FCh PET/CT imaging phenotypes, a custom collection of HCC-related gene sets was created using mSigDB. Using search operations, a list of gene sets whose descriptions contained the keyword “hepatocellular” was generated and limited by species to Homo sapien and by sample type to human tumor tissue, resulting in a user-defined collection of 70 HCC-related gene sets. GSEA was then performed with 1000 phenotype permutations using a FDR q-value of 0.05 as the significance threshold based on the number of gene sets tested(27).
Additional analyses using entire gene set collections from mSigDB were also conducted. To identify biological themes and pathways associated with imaging phenotype, GSEA was performed using a canonical collection containing 50 gene sets (mSigDB Hallmarks Collection v6.2). An exploratory analysis for gene set enrichment across a variety of clinical diseases and biological conditions was also performed using the entire mSigDB Chemical and Genetic Perturbations collection (CGP v6.2), a curated collection of 2701 gene sets encompassing a broad range of molecular signatures published in biomedical journals. This latter analysis was performed with an exploratory FDR q threshold of 0.25(27).
Molecular Tumor Classification
Molecular signatures corresponding to the highest scoring HCC-related gene sets on GSEA were used to classify tumors into sub-groups according to the referenced literature. Classes were assigned based on correlation with class genes(28). A FDR q < 0.05 was used to define confidence in class-based sub-group assignments.
Differential Gene Expression
Genes differentially expressed between FChHIGH and FChLOW tumors were identified at FDR q < 0.05 (GenePattern 3.9.10, genepattern.broadinstitute.org, Broad Institute, Cambridge, MA). Functional profiling of these results was performed using the Gene Functional Classification and Annotation Clustering Tools of the Database for Annotation, Visualization, and Integrated Discovery (DAVID v6.8, david.ncifcrf.gov)(29, 30).
Statistical Analysis
Other statistical analyses were performed using JMP Pro 14 (SAS Institute, Cary, NC) and MedCalc (v18.9, MedCalc bvba, Ostend, Belgium). Differences between groups were tested using Wilcoxon-Mann-Whitney, Fisher’s Exact, or chi-square test as appropriate. Survival was analyzed using the Kaplan-Meier method and log-rank test, followed by calculation of mortality hazard ratios with 95% confidence intervals (CI) by Cox univariate and multivariate proportional hazards regression. Multivariate analysis was adjusted for age and gender. Deaths occurring within 30 days of surgery were censored. Unless noted, significance was based on a two-tailed p-value < 0.05.
RESULTS
Patients and Imaging
Patient characteristics are summarized in Table 1. Regions of abnormal FCh uptake corresponding to the LIRADS category 3, 4, or 5 liver lesions found on diagnostic CT or MRI examinations were identified on PET/CT images in all 41 patients. In 31 (76%) patients, sampled tumor regions were assigned the FChHIGH phenotype based on visually increased FCh uptake relative to liver background. In the remaining 10 (24%) patients, tumor regions demonstrated decreased FCh uptake and were assigned the FChLOW phenotype. No tumor regions were assigned the FChISO phenotype. No significant association was noted between FCh PET/CT imaging phenotype and LI-RADS category (p = 0.54).
Table 1.
Patient characteristics and comparison between patients with FCHHIGH vs. FCHLOW tumors.
| Characteristic | Overall | FCHHIGH | FCHLOW | p-value |
|---|---|---|---|---|
| Mean Age (years) | 63.8 (±10.6) | 64.2 | 62.7 | 0.93 |
| Gender, Male/Female | 30 / 11 | 27/4 | 3/7 | <0.01 |
| Body Mass Index (kg/m2) | 26.2 (±4.8) | 26.3 | 26.1 | 0.80 |
| MELD Score | 8.7 (±2.4) | 8.05 | 8.96 | 0.90 |
| AFP level (ng/mL) | 1359 (±3676.7) | 948.5 | 2632.3 | <0.01 |
| Etiology/Risk Factors: | ||||
| HBV infected/non-infected | 11/30 | 8/23 | 3/7 | 1.00 |
| HCV infected/non-infected | 17/24 | 15/16 | 2/8 | 0.15 |
| >2 alcoholic beverages/day ( yes/no) | 16/25 | 13/18 | 3/7 | 0.71 |
| Tumor Characteristics: | ||||
| Tumor Size (cm) | 5.3 (±4.1) | 4.8 | 6.9 | 0.15 |
| Edmondson-Steiner grade (1–2 / 3–4) | 22/19 | 20/11 | 2/8 | 0.03 |
| Focality (multi-focal / solitary) | 8/33 | 5/26 | 3/7 | 0.38 |
| Vascular invasion (present/ absent) | 13/28 | 9/22 | 4/6 | 0.70 |
| Imaging Characteristics: | ||||
| LI-RADS category 3/4/5 | 8/18/15 | 6/15/10 | 2/3/5 | 0.54 |
| Tumor mean SUV | 9.5 (±3.9) | 11.2 | 4.3 | <0.001 |
| Liver mean SUV | 8.1(±2.0) | 8.1 | 8.2 | 0.90 |
| Classifications by Gene SIgnatures: | ||||
| Hoshida classification1 (S3/ S1–2) | 24/17 | 24/7 | 0/10 | <0.001 |
| Lee survival2 (good/poor) | 22/13 | 22/5 | 0/8 | <0.001 |
| Villanueva survival3 (good/poor) | 21/14 | 21/5 | 0/9 | <0.001 |
| Chiang classification4 (group1/group2) | 23/18 | 22/9 | 1/9 | <0.01 |
| Boyault classification5 (G1–3/G5–6) | 30/8 | 20/8 | 10/0 | 0.06 |
based on classification signature developed by Hoshida et al (2009).
based on survival signature developed by Lee et al (2004). Note, only 35 tumors successfully classified.
based on survival signature developed by Villanueva et al (2011). Note, only 35 tumors successfully classified.
based on classification signature developed by Chiang et al (2008).
based on classification signature developed by Boyault et al (2007). Note, only 38 tumors successfully classified.
Count or mean values are shown. Tumor size based on gross pathology geometric mean. Standard deviations parenthesized after overall values. MELD=Model for End-stage Liver Disease, AFP= alpha-fetoprotein, HBV=hepatitis B virus, HCV=hepatitis C virus, LI-RAD=Liver Imaging Reporting and Data System, SUV=standardized uptake value.
TLRSUV ranged from 1.21 to 2.00 (mean 1.47) for FChHIGH tumors and from 0.12 to 0.78 (mean 0.52) for FChLOW tumors. There were no TLRSUV values between 0.78 and 1.21. Therefore, visual classification and phenotype assignment based on a TLRSUV threshold of 1.0 classified the tumors identically (Supplementary Figure S1). Well and moderately-differentiated tumors (ie. Edmondson-Steiner grade 1 and 2), lower AFP values, and male gender were significantly associated with the FChHIGH phenotype (p = 0.027, p = 0.002, and p = 0.001, respectively). There were no significant associations with patient age, body mass index, Model for End-stage Liver Disease (MELD) score, tumor size, presence of microvascular invasion, or HCC risk factor.
Gene Set Analysis and Tumor Molecular Classification
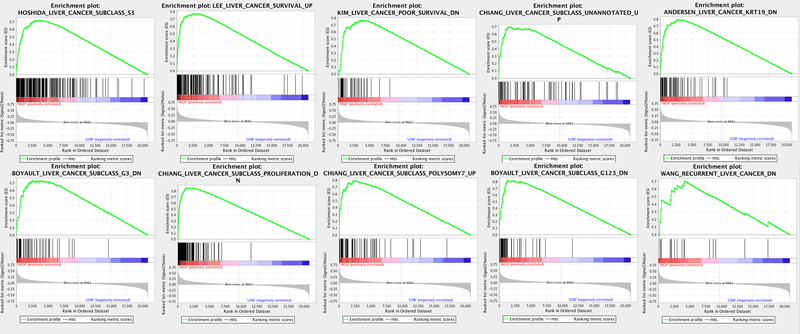
From among the 70 tumor-derived HCC-related gene sets available from mSigDB, 10 demonstrated enrichment by FChHIGH tumors relative to FChLOW tumors at a FDR < 0.05 (Table 2). The list of significant gene sets included those from previously developed gene signatures for predicting overall survival or recurrence in early-stage HCC (italicized in Table 2)(31–33). The list also included gene sets corresponding to 3 different molecular classification systems for HCC. Enrichment score plots for these gene sets are shown in Figure 2. The top scoring gene set, HOSHIDA_LIVER_CANCER_SUBCLASS_S3, defines an S3 sub-type of HCC based on a molecular classification system developed by Hoshida et al(5). Applying the complete Hoshida gene signature for classification, all tumor samples were successfully categorized into the different subtypes at FDR q < 0.05. Mirroring previous cohorts(5), the S3 sub-type comprised the largest sub-group with 24 tumors. Consistent with the GSEA results, all tumors classified as S3 displayed the FChHIGH phenotype. The S2 sub-type comprised the smallest sub-group with 4 tumors, with all displaying the FChLOW phenotype, while the S1 sub-type corresponded to 6 FChLOW and 7 FChHIGH tumors. S3 was significantly associated with the FChHIGH phenotype (p < 0.001) and lower AFP levels (435.8 vs. 2662.7 for non-S3, p < 0.005).
Table 2.
Among 70 gene sets from previously published HCC-related gene signatures derived from human tumor tissue, 10 were significantly enriched (FDR < 0.05) by tumors with FCHHIGH tumor phenotype.
| NAME | # genes | ES | NES | Nominal p-value | FDR q-value | PMID |
|---|---|---|---|---|---|---|
| HOSHIDA_LIVER_CANCER_SUBCLASS_S3 | 263 | 0.72 | 1.96 | <0.001 | 0.008 | 19723656 |
| LEE_LIVER_CANCER_SURVIVAL_UP | 170 | 0.78 | 1.83 | <0.001 | 0.028 | 15349906 |
| KIM_LIVER_CANCER_POOR_SURVIVAL_DN | 41 | 0.77 | 1.71 | 0.006 | 0.046 | 21320499 |
| CHIANG_LIVER_CANCER_SUBCLASS_UNANNOTATED_UP | 76 | 0.70 | 1.70 | 0.010 | 0.046 | 18701503 |
| ANDERSEN_LIVER_CANCER_KRT19_DN | 75 | 0.80 | 1.70 | 0.004 | 0.040 | 21320499 |
| BOYAULT_LIVER_CANCER_SUBCLASS_G3_DN | 51 | 0.73 | 1.69 | 0.012 | 0.040 | 17187432 |
| CHIANG_LIVER_CANCER_SUBCLASS_PROLIFERATION_DN | 174 | 0.86 | 1.69 | <0.001 | 0.037 | 18701503 |
| CHIANG_LIVER_CANCER_SUBCLASS_POLYSOMY7_UP | 72 | 0.79 | 1.65 | 0.006 | 0.049 | 18701503 |
| BOYAULT_LIVER_CANCER_SUBCLASS_G123_DN | 50 | 0.82 | 1.65 | <0.001 | 0.046 | 17187432 |
| WANG_RECURRENT_LIVER_CANCER_DN | 16 | 0.70 | 1.64 | 0.010 | 0.047 | 17975138 |
ES=enrichment score, NES=nominal ES, FDR=false discovery rate, PMID=PubMed ID of literature reference. The italicized gene sets are from prognostic signatures predictive of clinical outcomes after hepatic resection for HCC.
Figure 2:
Enrichment score plots of the significant HCC-related gene sets identified by gene set enrichment analysis. The running enrichment score profiles (green curves) indicate significant enrichment of member genes (vertical black bars). The corresponding rank ordered plots (lower graphs) show significant and positive correlation with the FChHIGH imaging phenotype.
There were also 3 significant gene sets that belonged to an HCC molecular classification system developed by Chiang et al(6). The gene sets identified as significant contained the over-expressed genes corresponding to a “polysomy 7” class and an “unannotated” class, and the under-expressed genes corresponding to a “proliferation” class (Table 2). The complete classification signature successfully classified all 41 tumor samples at FDR q < 0.05. Because 32 of the under-expressed genes of the “proliferation” class were also over-expressed genes of a CTNNB1 (catenin beta-1) class, two “Chiang groups” were created for the purpose of making study comparisons: Chiang group 1, containing the CTNNB1, polysomy 7, and unannotated classes, and Chiang group 2, containing the “interferon” and “proliferation” classes described in the original publication(6). Chiang group 1 was significantly associated with FChHIGH phenotype (p = 0.001) as well as with lower AFP levels (807.1 ng/mL vs. 2064.6 ng/mL, p < 0.05).
GSEA also identified 2 significant gene sets belonging to a molecular classification system developed by Boyault et al(4). The significant gene sets identified by GSEA comprised the under-expressed genes corresponding to groups termed G3 and G123 (Table 2). Applying the corresponding molecular classification signature, 38/41 tumor samples were confidently classified at FDR q < 0.05. There were no significant associations between imaging phenotype and groups defined by this signature.
Gene sets belonging to 2 prognostic signatures for HCC were also significantly associated with the FChHIGH phenotype. The first, LEE_LIVER_CANCER_SURVIVAL_UP, comprises the over-expressed genes of a gene signature developed by Lee et al. to predict survival after hepatic resection(31). This signature successfully classified 35/41 tumor samples at FDR q < 0.05, assigning 22 to a good survival group and 13 to a poor survival group. AFP levels were significantly lower for the good survival group compared to the poor survival group (471.0 ng/mL vs. 2763.7 ng/mL, p < 0.01). The other significant survival-related gene set was KIM_LIVER_CANCER_POOR_SURVIVAL_DN. It is comprised of under-expressed genes from a survival signature developed by Villanueva et al.(32). This signature successfully classified 35/41 tumors, assigning 21 into good survival and 14 into poor survival groups. AFP levels also differed significantly between good and poor survival groups defined by this signature (435.8 ng/mL vs. 2662.7 ng/mL, p < 0.005). Good survival groups defined by both signatures were significantly associated with the FChHIGH phenotype (Table 1). Gene signatures corresponding to the remaining gene sets shown in Table 2 were unable to classify enough samples at FDR < 0.05 for statistical comparisons.
Survival Results
A total of 20 patients died at follow-up. Excluding 3 patients that died within 30 days of surgery, the median duration of censored follow-up was 1463 days (range 814 to 2200 days). Significantly longer overall survival was observed in patients whose tumors demonstrated the FChHIGH phenotype compared to the FChLOW phenotype (Figure 3). Although significantly more males had FChHIGH tumors, gender was not significantly associated with overall survival. Age, HCC risk factor, AFP level, and Edmondson-Steiner grade were also not significantly associated with overall survival (all p > 0.05). Tumor size > 4cm was significantly associated with shorter overall survival and was a significant univariate predictor of increased mortality based on Cox regression analysis (Table 3). FChHIGH imaging phenotype, Hoshida sub-type S3 (vs. S1/S2), and the good survival groups of both Lee and Villanueva survival signatures were significant univariate predictors of lower mortality based on Cox regression analysis. FChHIGH imaging phenotype and the Villanueva signature remained significant on multivariate regression analysis (Table 3). No subgroups based on the Chiang and Boyault signatures were associated with significantly lower mortality, consistent with these signatures not being intended as prognostic classifiers.
Figure 3:
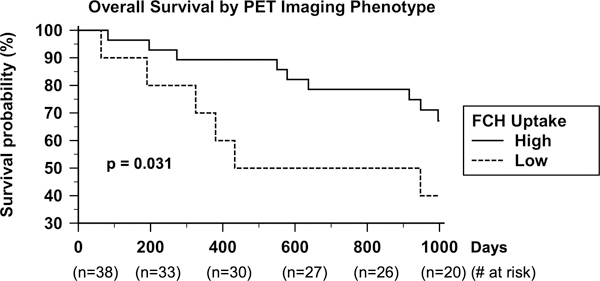
Kaplan-Meier survival plots show significantly worse survival in patients with tumors exhibiting low 18F-fluorocholine (FCH) uptake versus patients with tumors exhibiting high FCH uptake (median survival > 1000 days vs. 690 days, p = 0.031).
Table 3:
Univariate and multivariate proportional hazards regression analysis.
| Univariate analysis | Multivariate analysis1 | |||||
|---|---|---|---|---|---|---|
| Hazard Ratio | 95% CI | p-value | Hazard Ratio | 95% CI | p-value | |
| Age > 65 years | 1.39 | 0.53–3.70 | 0.50 | |||
| Gender (male) | 1.02 | 0.38–3.21 | 0.97 | |||
| Hepatitis B + | 0.98 | 0.28–2.77 | 0.98 | |||
| Hepatitis C + | 0.65 | 0.22–1.71 | 0.39 | |||
| Edmondson-Steiner Grade (1,2 vs. 3,4) | 1.10 | 0.42–2.93 | 0.85 | |||
| Serum alpha-fetoprotein > 400 ng/mL | 2.43 | 0.83–6.45 | 0.10 | |||
| Tumor Size > 4cm | 3.8 | 1.34–13.5 | 0.01 | 4.7 | 1.50–18.4 | <0.01 |
| PET Phenotype (FChHIGH vs. FChLOW) | 0.36 | 0.14–0.95 | 0.04 | 0.13 | 0.03–0.58 | <0.01 |
| Hoshida signature2 (S3 vs. S1-S2) | 0.33 | 0.12–0.88 | 0.03 | 0.43 | 0.15–1.26 | 0.13 |
| Lee signature3 (good vs. poor survival) | 0.36 | 0.11–0.93 | 0.04 | 0.46 | 0.15–1.35 | 0.16 |
| Villanueva signature4 (good vs. poor survival) | 0.23 | 0.07–0.64 | 0.01 | 0.30 | 0.09–0.87 | 0.03 |
adjusted for age, gender, and tumor size.
From Hoshida et al. (2009).
From Lee et al. (2004). Limited to 35 patients confidently classified by this signature.
From Villanueva et al (2011). Limited to 35 patients confidently classified by this signature.
Additional Transcriptomic Analyses
GSEA performed using 50 Hallmark gene sets identified 6 as significantly comprised of genes enriched in FChHIGH tumors relative to FChLOW tumors, including the gene sets representing oxidative phosphorylation (normalized enrichment score (NES) 2.00, FDR 0.018), fatty acid metabolism (NES 1.96, FDR 0.011), peroxisome (NES 1.90, FDR 0.013), bile acid metabolism (NES 1.89, FDR 0.011), xenobiotic metabolism (NES 1.85, FDR 0.013), and adipogenesis (NES 1.84, FDR 0.013). Correspondingly, 540 differentially expressed genes were identified between FChHIGH and FChLOW tumors at FDR q< 0.05. The top DAVID functional clusters associated with these differentially expressed genes were also related to oxidative, mitochondrial, fatty acid, lipid, and bile acid metabolism in agreement with the GSEA results (Supplementary Table S1). Thus, FChHIGH tumors appear to have more transcriptional features of hepatocyte differentiation than FChLOW tumors.
In an exploratory analysis that included 2701 gene sets covering a broad variety of diseases and conditions, 68 gene sets demonstrated enrichment by FChHIGH tumors at FDR q < 0.25 (Supplementary Table S2). Consistent with the findings obtained by GSEA using the Hallmarks Collection, the top scoring gene sets included those related to oxidative phosphorylation and mitochondrial metabolism. In addition, 33 HCC or liver related gene sets were identified, including the ones found significant by GSEA using only the 70 HCC-related gene sets.
DISCUSSION
Molecular imaging holds the potential to non-invasively identify the molecular vulnerabilities of a tumor, to possibly enable greater precision in treatment. In this study, we tested the hypothesis that an imaging phenotype defined by FCh PET/CT identifies a subset of HCC tumors already characterized by previously-published molecular signatures. To test this hypothesis, we applied GSEA to identify sets of genes that were significantly enriched in tumors displaying high FCh uptake from a collection of a priori defined gene sets. This approach obviated the need to develop new molecular signatures to explain imaging phenotypes, and instead leveraged the work of many who have previously developed and validated gene signatures for HCC. By conducting analyses at the gene set level rather than the gene level, statistical power was also conserved, and biological interpretation streamlined.
This study is the first to report significant associations between tumor FCh avidity and enrichment by sets of genes comprising multiple clinically relevant molecular signatures for HCC. The gene set associated with the highest degree of enrichment in our study was HOSHIDA_LIVER_CANCER_SUBCLASS_S3. As part of a tumor classification signature defining 3 HCC sub-types (S1, S2, and S3)(5), this gene set defines a sub-type (S3) that, in 8 different patient cohorts, was found to be the most common sub-type and the one associated with the most favorable clinical features(5, 8). Consistent with these findings, patients with S3 tumors formed the largest sub-group and experienced the longest survival in the present study. Ironically, S3 tumors have proven the most difficult to recapitulate in pre-clinical models. One analysis of 25 different HCC cell lines found all to be sub-type S1 or S2, suggesting either that S3 cells cannot be immortalized or that they change into S1 or S2 in-vitro(34, 35). Therefore, non-invasive techniques such as FCh PET/CT may have value for studying this predominant HCC sub-type in-vivo.
When the Hoshida signature was applied, it was found that all S3 tumors in this study displayed the FChHIGH phenotype. Interestingly, all four of the S2 tumors in this study displayed a FChLOW phenotype. While inferences should not be made based on a small number of tumors, it is worth noting that S2 is associated with the most aggressive biological traits and poorest survival of the tumor sub-types(35). Since S2 tumors may be selectively vulnerable to specific molecularly-targeted agents(8, 35), the ability to non-invasively identify this sub-type could have clinical importance. However, S1 tumors were found capable of demonstrating either low or high FCh uptake, limiting the specificity of the FChLOW phenotype as an indicator of tumor sub-type. As a potential explanation for why S1 tumors may be associated with either imaging phenotype, it is possible that the Hoshida molecular classification system does not incorporate every gene influencing tumor choline metabolism (or alternatively, FCh uptake). S1 may also be the most molecularly heterogeneous among the sub-types(5), with heat maps from our analysis also showing the greatest heterogeneity in gene expression corresponding to S1 tumors (Supplementary Figures S2, S3, and S4). Conceivably, it may be possible to further sub-divide the S1 sub-type using additional defining genes, including those relevant to FCh or choline metabolism. The performance of FCh PET/CT for tumor classification based on the Hoshida system will hopefully be refined by further studies.
GSEA was also performed using canonical gene set collections curated by the Broad Institute to provide a wider survey of molecular features and traits. The significant gene sets identified from the Hallmarks collection conveyed particular metabolic traits to FChHIGH tumors, including those associated with oxidative, fatty acid, bile acid, and xenobiotic metabolism. GSEA using the CGP collection also identified significant gene sets associated with oxidative and mitochondrial metabolism, while showing that HCC-related gene sets prevailed over gene sets of other diseases with regards to their associations with this imaging phenotype. Functional annotation clustering based on differential gene expression between FChHIGH and FChLOW tumors implicated these metabolic features as well, which notably are also constitutively expressed by hepatocytes. This appears to indicate that cellular differentiation is a prevailing characteristic of FChHIGH tumors. Interestingly, the S3 tumor sub-type defined by Hoshida et al. is also distinctly associated with hepatocyte differentiation(5). Consistent with these observations, tumors displaying FChHIGH phenotype were found on histopathology to be significantly comprised of moderate and well differentiated tumors.
Another notable result was the finding of poorer overall survival in patients with FChLOW tumors compared to patients with FChHIGH tumors (Figure 3). Consistent with this finding, a previous pilot study of patients who underwent FCh PET/CT prior to hepatectomy reported earlier tumor recurrence among patients with tumors showing low FCh uptake(36). Similarly, a study that used 11C-acetate, another lipid metabolism tracer, found that tumors demonstrating high uptake of this tracer required a lower radiation dose (152–174 Gy vs. 262 Gy) to achieve good therapeutic response following yttrium-90 glass microsphere radioembolization(37). The possibility of enabling clinical risk stratification and further optimizing treatment for HCC using FCh PET/CT will hopefully be studied prospectively in larger cohorts.
Several previously published gene signatures corresponding to gene sets enriched by FChHIGH tumors also discriminated survival in our cohort, including interestingly the molecular classification signature developed by Hoshida et al(5). Patients with S3 tumors identified by this signature experienced significantly lower mortality in our study (Table 3). Although S3 tumors are known to harbor fewer aggressive features compared to S1 or S2 tumors(34), to the best of our knowledge, the S3 sub-type has not before been associated with better survival in a prospective study. However, Hoshida et al. did report that S3 tumors were co-enriched by genes from a previously reported clinical prognostic signature for HCC(5). In our study, over-expressed genes from that co-enriched signature (LEE_LIVER_CANCER_SURVIVAL_UP)(31), as well as genes from another HCC survival signature (KIM_LIVER_CANCER_POOR_SURVIVAL_DN) developed by Villanueva et al.(32), demonstrated significant enrichment by FChHIGH tumors (Table 2). Furthermore, univariate survival analyses revealed significantly lower mortality among the good survival groups defined by these signatures (Table 3). These associations with survival, in addition to supporting speculation that FCh PET/CT has prognostic value for HCC, constitute an additional validation of these previously published prognostic signatures in an independent albeit relatively small cohort of HCC patients.
Phosphatidylcholines are bilayer forming phospholipids comprising the most abundant constituent of cell and sub-cellular membranes(38). Both cell and mitochondrial proliferation depend on its supply (20). Phosphatidylcholine synthesis from choline via the intermediate phosphorylcholine (called the Kennedy pathway) involves esterification with two fatty acids, linking this pathway to lipid metabolism. Approximately 50% of de-novo synthesized phosphiatdylcholine is consumed by mitochondria where membrane function plays a critical role in oxidative phosphorylation and electron transport(20, 38). In keeping with these roles, gene set enrichment by tumors exhibiting the FChHIGH phenotype was associated with oxidative phosphorylation, lipid metabolism, and mitochondrial function, implying that oxidative phosphorylation is active in the tumors exhibiting high FCh uptake. Gene sets associated with the FChHIGH phenotype also overlapped significantly with gene sets from MSigDB associated with fatty acid metabolism, oxidative phosphorylation, and mitochondrial function (Supplementary Tables S3 to S8).
With regards to energy metabolism, cancers can also turn to aerobic glycolysis (ie. the Warburg phenomenon), which can potentially be monitored by FDG PET/CT(39). In contrast to the favorable association between tumor FCh uptake and survival, tumor FDG uptake has been associated with a worse prognosis in HCC(39). Although FDG uptake was not investigated in the present study, previous diagnostic trials comparing FDG PET/CT and FCh PET/CT for HCC revealed discordance between these tracers (ie. high tumor FDG uptake was associated with low FCh uptake, and vice versa). Pathobiologically, decreased phosphatidylcholine biosynthesis might interfere with mitochondrial function, turning tumors toward aerobic glycolysis and high FDG uptake. Alternatively, aerobic glycolysis may not support the Kennedy pathway, leading Warburg-phenotype tumors to demonstrate low FCh uptake. This radiologic discordance has led some centers to adopt clinical protocols involving two PET/CT scans for HCC, one performed with a lipid based tracer such as FCh and a second performed with FDG(17, 18, 22, 23).
At present, no second cohort is available to confirm our reported associations between imaging, gene signatures, and clinical outcomes. Despite its increasing incidence, HCC is still relatively uncommon in the US compared to other countries, and this study was conducted at only one institution. This constitutes an important limitation of our study. Another potential limitation stems from the inclusion of only early-stage surgically treated HCC patients, as it is possible that genomic and other molecular alterations will be more varied in patients with advanced tumors. However, previous studies that involved only patients with early-stage HCC have been successful at linking different clinical outcomes to specific molecular profiles(5, 31–33, 40). Although several predictors of survival were identified, this study was also limited in its power to detect and compare significant predictors due to its sample size. It is important to note however that this study was adequately powered for GSEA(27). While additional studies are needed, especially to confirm prognostic associations, the present study did accomplish its goal of providing a biologically cohesive framework for interpreting the two imaging phenotypes of HCC detected on FCh PET/CT.
To our knowledge, few studies have reconciled molecular imaging with genomic findings to the extent presented, even though the bioinformatics tools and resources necessary are widely accessible, well-established, and reproducible. We believe that the methods of this study form an under-utilized approach to gaining insight on tumor pathobiology. Such an approach may also aid in identifying and developing new clinical applications for emerging or established molecular imaging techniques. Although further studies are needed, the multiple significant associations with previously validated gene signatures found in this study support FCh PET/CT as a non-invasive tool for molecularly categorizing HCC. Associations between FChHIGH tumor phenotype and favorable molecular and clinical features have the potential to impact clinical decision-making. For example, using FCh PET/CT to determine tumor-associated prognosis may help in identifying the most favorable candidates for surgery and liver transplantation, or the need for adjuvant treatments or heightened surveillance. Non-invasive tumor phenotyping based on this form of molecular imaging may also be of value in selecting patients for targeted therapies and clinical trials.
Transcriptome-based signatures for molecularly classifying HCC and other cancers abound. This study is the first in providing a cohesive biological framework that explains specific molecular imaging findings of HCC through the analysis of tumor enrichment for genes from previously reported gene signatures. FCh PET/CT is currently being used in Europe as a clinical diagnostic and staging tool for HCC. Following the wake of failures of large clinical trials for HCC(41, 42), there is growing interest in tumor molecular sub-classification to better match patients to potential treatments(8, 34, 43). Our findings, if further validated, suggest that FCh PET/CT may be useful as an in-vivo imaging-based biomarker for molecular tumor classification and patient risk stratification in support of precision medicine and biomarker-based clinical trial enrichment.
Supplementary Material
STATEMENT OF SIGNIFICANCE:
A pathobiological framework for hepatocellular carcinoma brings together multiple prognostically relevant gene signatures via convergence with 18F-fluorocholine PET/CT imaging phenotype.
ACKNOWLEDGEMENT:
This work was supported by United States National Institutes of Health grant R01CA161209-06.
Footnotes
CONFLICT OF INTEREST: LLW reports receiving a speakers bureau honoraria from Eisai. No potential conflicts of interest were disclosed by the other authors.
REFERENCES
- 1.Marrero JA, Kulik LM, Sirlin CB, Zhu AX, Finn RS, Abecassis MM, et al. Diagnosis, Staging, and Management of Hepatocellular Carcinoma: 2018 Practice Guidance by the American Association for the Study of Liver Diseases. Hepatology 2018;68:723–50. [DOI] [PubMed] [Google Scholar]
- 2.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer research 2014;74:2913–21. [DOI] [PubMed] [Google Scholar]
- 3.Yamashita T, Forgues M, Wang W, Kim JW, Ye Q, Jia H, et al. EpCAM and alpha-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma. Cancer research 2008;68:1451–61. [DOI] [PubMed] [Google Scholar]
- 4.Boyault S, Rickman DS, de Reynies A, Balabaud C, Rebouissou S, Jeannot E, et al. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology 2007;45:42–52. [DOI] [PubMed] [Google Scholar]
- 5.Hoshida Y, Nijman SM, Kobayashi M, Chan JA, Brunet JP, Chiang DY, et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer research 2009;69:7385–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiang DY, Villanueva A, Hoshida Y, Peix J, Newell P, Minguez B, et al. Focal gains of VEGFA and molecular classification of hepatocellular carcinoma. Cancer research 2008;68:6779–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nault JC, Galle PR, Marquardt JU. The role of molecular enrichment on future therapies in hepatocellular carcinoma. Journal of hepatology 2018;69:237–47. [DOI] [PubMed] [Google Scholar]
- 8.Tan PS, Nakagawa S, Goossens N, Venkatesh A, Huang T, Ward SC, et al. Clinicopathological indices to predict hepatocellular carcinoma molecular classification. Liver Int 2016;36:108–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan SL, Wong AM, Lee K, Wong N, Chan AK. Personalized therapy for hepatocellular carcinoma: Where are we now? Cancer Treat Rev 2016;45:77–86. [DOI] [PubMed] [Google Scholar]
- 10.Pompili M, Rapaccini GL, Covino M, Pignataro G, Caturelli E, Siena DA, et al. Prognostic factors for survival in patients with compensated cirrhosis and small hepatocellular carcinoma after percutaneous ethanol injection therapy. Cancer 2001;92:126–35. [DOI] [PubMed] [Google Scholar]
- 11.Wong LL, Hernandez BY, Shvetsov YB, Kawano Y, Tang ZY, Ji JF. Liver resection for early hepatocellular cancer: Comparison of centers in 3 different countries. World J Hepatol 2016;8:1327–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma C, Chi M, Su H, Cheng X, Chen L, Kan Y, et al. Evaluation of the clinical features of HCC following hepatectomy for different stages of HCC. Hepato-gastroenterology 2012;59:2104–11. [DOI] [PubMed] [Google Scholar]
- 13.Benson AB 3rd, D’Angelica MI, Abbott DE, Abrams TA, Alberts SR, Saenz DA, et al. NCCN Guidelines Insights: Hepatobiliary Cancers, Version 1.2017. J Natl Compr Canc Netw 2017;15:563–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heimbach JK, Kulik LM, Finn RS, Sirlin CB, Abecassis MM, Roberts LR, et al. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology 2018;67:358–80. [DOI] [PubMed] [Google Scholar]
- 15.Lopez KT, Kuwada SK, Wong LL. Consequences of needle tract seeding of hepatocellular cancer after liver transplant. Clinical transplantation 2013;27:E400–6. [DOI] [PubMed] [Google Scholar]
- 16.Kwee SA, Sato MM, Kuang Y, Franke A, Custer L, Miyazaki K, et al. [(18)F]Fluorocholine PET/CT Imaging of Liver Cancer: Radiopathologic Correlation with Tissue Phospholipid Profiling. Molecular imaging and biology : MIB : the official publication of the Academy of Molecular Imaging 2017;19:446–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Talbot JN, Fartoux L, Balogova S, Nataf V, Kerrou K, Gutman F, et al. Detection of hepatocellular carcinoma with PET/CT: a prospective comparison of 18F-fluorocholine and 18F-FDG in patients with cirrhosis or chronic liver disease. Journal of nuclear medicine : official publication, Society of Nuclear Medicine 2010;51:1699–706. [DOI] [PubMed] [Google Scholar]
- 18.Chalaye J, Costentin CE, Luciani A, Amaddeo G, Ganne-Carrie N, Baranes L, et al. Positron emission tomography/computed tomography with 18F-fluorocholine improve tumor staging and treatment allocation in patients with hepatocellular carcinoma. Journal of hepatology 2018;69:336–44. [DOI] [PubMed] [Google Scholar]
- 19.Glunde K, Bhujwalla ZM, Ronen SM. Choline metabolism in malignant transformation. Nature reviews Cancer 2011;11:835–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plagemann PG. Choline metabolism and membrane formation in rat hepatoma cells grown in suspension culture. II. Phosphatidylcholine synthesis during growth cycle and fluctuation of mitochondrial density. J Cell Biol 1969;42:766–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuang Y, Salem N, Tian H, Kolthammer JA, Corn DJ, Wu C, et al. Imaging lipid synthesis in hepatocellular carcinoma with [methyl-11c]choline: correlation with in vivo metabolic studies. Journal of nuclear medicine : official publication, Society of Nuclear Medicine 2011;52:98–106. [DOI] [PubMed] [Google Scholar]
- 22.Talbot JN, Michaud L, Grange JD, Rosmorduc O, Balogova S. Use of choline PET for studying hepatocellular carcinoma. Clin Transl Imaging 2014;2:103–13. [Google Scholar]
- 23.Ho CL, Yu SC, Yeung DW. 11C-acetate PET imaging in hepatocellular carcinoma and other liver masses. Journal of nuclear medicine : official publication, Society of Nuclear Medicine 2003;44:213–21. [PubMed] [Google Scholar]
- 24.Salem N, Kuang Y, Wang F, Maclennan GT, Lee Z. PET imaging of hepatocellular carcinoma with 2-deoxy-2[18F]fluoro-D-glucose, 6-deoxy-6[18F] fluoro-D-glucose, [1–11C]-acetate and [N-methyl-11C]-choline. The quarterly journal of nuclear medicine and molecular imaging : official publication of the Italian Association of Nuclear Medicine 2009;53:144–56. [PubMed] [Google Scholar]
- 25.Tang A, Bashir MR, Corwin MT, Cruite I, Dietrich CF, Do RKG, et al. Evidence Supporting LI-RADS Major Features for CT- and MR Imaging-based Diagnosis of Hepatocellular Carcinoma: A Systematic Review. Radiology 2018;286:29–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adler S, Seidel J, Choyke P, Knopp MV, Binzel K, Zhang J, et al. Minimum lesion detectability as a measure of PET system performance. EJNMMI Phys 2017;4:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu L, Shen SS, Hoshida Y, Subramanian A, Ross K, Brunet JP, et al. Gene expression changes in an animal melanoma model correlate with aggressiveness of human melanoma metastases. Mol Cancer Res 2008;6:760–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44–57. [DOI] [PubMed] [Google Scholar]
- 30.Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, et al. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol 2007;8:R183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee JS, Chu IS, Heo J, Calvisi DF, Sun Z, Roskams T, et al. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology 2004;40:667–76. [DOI] [PubMed] [Google Scholar]
- 32.Villanueva A, Hoshida Y, Battiston C, Tovar V, Sia D, Alsinet C, et al. Combining clinical, pathology, and gene expression data to predict recurrence of hepatocellular carcinoma. Gastroenterology 2011;140:1501–12 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang SM, Ooi LL, Hui KM. Identification and validation of a novel gene signature associated with the recurrence of human hepatocellular carcinoma. Clin Cancer Res 2007;13:6275–83. [DOI] [PubMed] [Google Scholar]
- 34.Hirschfield H, Bian CB, Higashi T, Nakagawa S, Zeleke TZ, Nair VD, et al. In vitro modeling of hepatocellular carcinoma molecular subtypes for anti-cancer drug assessment. Exp Mol Med 2018;50:e419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidt B, Wei L, DePeralta DK, Hoshida Y, Tan PS, Sun X, et al. Molecular subclasses of hepatocellular carcinoma predict sensitivity to fibroblast growth factor receptor inhibition. Int J Cancer 2016;138:1494–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fartoux L, Balogova S, Nataf V, Kerrou K, Huchet V, Rosmorduc O, et al. A pilot comparison of 18F-fluorodeoxyglucose and 18F-fluorocholine PET/CT to predict early recurrence of unifocal hepatocellular carcinoma after surgical resection. Nucl Med Commun 2012;33:757–65. [DOI] [PubMed] [Google Scholar]
- 37.Ho CL, Chen S, Cheung SK, Leung YL, Cheng KC, Wong KN, et al. Radioembolization with (90)Y glass microspheres for hepatocellular carcinoma: significance of pretreatment (11)C-acetate and (18)F-FDG PET/CT and posttreatment (90)Y PET/CT in individualized dose prescription. European journal of nuclear medicine and molecular imaging 2018. [DOI] [PubMed]
- 38.Morton R, Cunningham C, Jester R, Waite M, Miller N, Morris HP. Alteration of mitochondrial function and lipid composition in Morris 7777 hepatoma. Cancer research 1976;36:3246–54. [PubMed] [Google Scholar]
- 39.Takeuchi S, Rohren EM, Abdel-Wahab R, Xiao L, Morris JS, Macapinlac HA, et al. Refining prognosis in patients with hepatocellular carcinoma through incorporation of metabolic imaging biomarkers. European journal of nuclear medicine and molecular imaging 2017;44:969–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nault JC, De Reynies A, Villanueva A, Calderaro J, Rebouissou S, Couchy G, et al. A hepatocellular carcinoma 5-gene score associated with survival of patients after liver resection. Gastroenterology 2013;145:176–87. [DOI] [PubMed] [Google Scholar]
- 41.Scudellari M Drug Development: Try and Try Again. Nature 2014;516:S4–6. [DOI] [PubMed] [Google Scholar]
- 42.Nault JC. The end of almost 10 years of negative RCTs in advanced hepatocellular carcinoma. Lancet 2017;389:4–6. [DOI] [PubMed] [Google Scholar]
- 43.Goossens N, Sun X, Hoshida Y. Molecular classification of hepatocellular carcinoma: potential therapeutic implications. Hepat Oncol 2015;2:371–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.