Abstract
Notch (Notch 1 through 4) are transmembrane receptors that play a fundamental role in cell differentiation and function. Notch receptors are activated following interactions with their ligands in neighboring cells. There are five classic ligands termed Jagged (Jag)1 and Jag2, and Delta-like (Dll)1, Dll3 and Dll4. Recent work has established Notch as a signaling pathway that plays a critical role in the differentiation and function of cells of the osteoblast and osteoclast lineages and in skeletal development and bone remodeling. The effects of Notch are cell-context dependent, and the four Notch receptors carry out specific functions in the skeleton. Gain- and loss-of-function mutations of components of the Notch signaling pathway result in a variety of congenital disorders with significant craniofacial and skeletal manifestations. The Notch ligand Jag1 is a determinant of bone mineral density, and Notch plays a role in the early phases of fracture healing. Alterations in Notch signaling are associated with osteosarcoma and with the metastatic potential of carcinoma of the breast and of the prostate. Controlling Notch signaling could prove useful in diseases of Notch gain-of-function and in selected skeletal disorders. However, clinical data on agents that modify Notch signaling are not available. In conclusion, Notch signaling is a novel pathway that regulates skeletal homeostasis in health and disease.
Keywords: Notch, Jagged, congenital disorders, osteoblast, osteoclast
Summary:
Notch receptors are activated following interactions with their ligands (Jagged and Delta-like) in adjacent cells. Notch and ligands are expressed by osteoblasts and osteoclasts and regulate their differentiation and function. As a consequence, Notch modulates skeletal remodeling and plays an important role in bone homeostasis in health and disease.
I. NOTCH PHYSIOLOGY
Notch Receptors
Notch are transmembrane receptors that play a fundamental role in cell differentiation and function. Work conducted over the past decade has established Notch as a signaling pathway critical for bone remodeling and a determinant of skeletal homeostasis [1–7]. There are four Notch receptors (Notch1 through 4) and five classic ligands termed Jagged (Jag)1 and Jag2, and Delta-like (Dll)1, Dll3 and Dll4 [5]. Notch ligands, like Notch, are transmembrane proteins and their interactions with Notch are required to activate Notch signaling. Since Notch and its ligands are transmembrane proteins, the signaling pathway serves as a means of communication between neighboring cells.
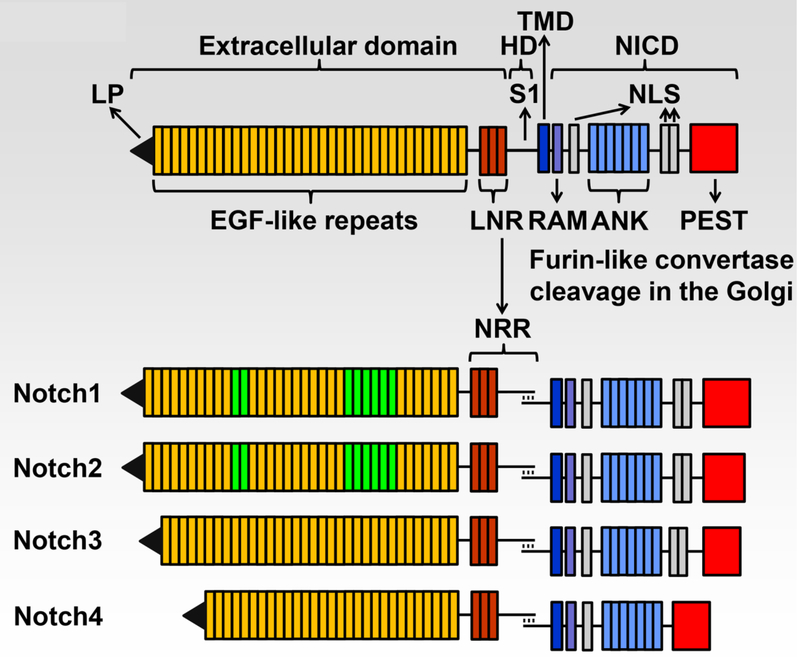
Notch receptors have a complex structure. Their extracellular domain is the site of interaction with Notch ligands, and at the junction of the extracellular and the transmembrane domain rests the negative regulatory region (NRR). This is the site of cleavage required for Notch activation, and as a consequence plays a critical regulatory role in Notch signaling [8]. The intracellular domain of Notch (NICD) consists of an RBPJκ-association module (RAM) domain, ankyrin repeats and nuclear localization sequences; these domains are required to regulate transcription. The C-terminus of Notch contains a proline (P)-, glutamic acid (E)-, serine (S)- and threonine (T)-rich (PEST) domain, which is targeted by ubiquitin ligases for the proteasomal degradation of Notch, and as such it defines the life span of Notch [5] (Fig. 1).
Fig.1.
Domains of the four Notch receptors. The upper panel shows the domain and motif organization of a generic human/murine Notch receptor before cleavage at the S1 site by furin-like convertases in the Golgi compartment. The extracellular domain contains a leader peptide (LP) and multiple epidermal growth factor (EGF)-like tandem repeats followed by Lin12-Notch repeats (LNR) and the heterodimerization domain (HD). The transmembrane domain (TMD) is located between the extracellular and intracellular domains. The Notch intracellular domain (NICD) contains an Rbpjκ-association module (RAM), a nuclear localization sequence (NLS), ankyrin (ANK) repeats and tandem NLS, which are followed by a proline (P)-, glutamic acid (E)-, serine (S)- and threonine (T)-rich (PEST) domain. The lower panel shows the domains and motifs of heterodimeric individual receptors, the negative regulatory region (NRR) is formed by the LNR and HD following cleavage at the S1 site. Notch1 and Notch2 have 36 EGF-like repeats; in green are those required for binding of Notch1 and Notch2 to cognate Delta/Serrate/Lag2 ligands. Notch1 and Notch2 have a similar NICD, and Notch3 has 34 EGF-like repeats and a shorter NICD than Notch1 and Notch2. Notch4 has 29 EGF-like repeats and an NICD that is shorter than that of other receptors and lacks the tandem NLS located between the ANK repeats and the PEST domain. Reproduced with permission from Zanotti and Canalis, Endocrine Reviews 37:223–253, 2016.
Interactions of Notch with its ligands lead to its proteolytic cleavage and release of the NICD, its translocation to the nucleus, where the NICD forms a complex with recombination signal-binding protein for Ig of κ region (RBPJκ) and mastermind-like (MAML) to regulate transcription [9, 10]. RBPJκ is also termed CBF1, Suppressor of hairless, Lag1 or CSL. RBPJκ, and not the NICD, binds to DNA and under basal conditions RBPJκ is a repressor of transcription. The translocation of the NICD to the nucleus, where it interacts with RBPJκ, leads to the displacement of transcriptional inhibitors and the recruitment of activators of transcription. As a result, the NICD, RBPJκ, MAML complex induces the transcription of target genes. Targets of this canonical signaling are members of the Hairy and enhancer of split (HES) and HES with a YRPW motif (HEY) families of transcription factors [11]. Cyclin-dependent kinases phosphorylate the PEST domain of the NICD, causing the disassembly of the NICD, RBPJκ, MAML complex followed by ubiquitination of the NICD by E3 ubiquitin ligases and the degradation of Notch [12]. A less well-characterized non-canonical pathway can operate under certain physiological conditions, and by definition it does not require RBPJκ. It is believed that in skeletal cells Notch operates through the canonical RBPJκ-dependent pathway.
It is important to recognize that there are structural and functional differences among the four Notch receptors and that their function is not redundant. Therefore, the independent study of the four Notch receptors in skeletal cells has been of utmost importance (Table 1). Functional differences among Notch receptors are related to structural distinctions in the NICD, to differential interactions of each NICD with RBPJκ, to the temporal and cellular expression of each receptor, to variations in the affinity of the extracellular domain of Notch for its ligands and to NRR sequence differences conveying specificity to the activation of Notch [13, 14]. Notch1, 2 and 3 and low levels of Notch4 are detected in skeletal cells [1, 15]. Notch 1 and 2 are expressed by cells of the osteoblast and osteoclast lineage, whereas Notch3 is mostly expressed by osteoblasts and osteocytes, and not by cells of the osteoclast lineage.
Table 1.
Functional differences among Notch receptors.
| Notch1 | Notch2 | Notch3 | Notch 4 | |
|---|---|---|---|---|
| Extracellular Domain EGF Repeats | 36 | 36 | 34 | 29 |
| Extracellular Transmembrane Junction Distinct NRR | Yes | Yes | Yes | Yes |
| Intracellular Domain Similar NICD | Yes | Yes | No | No |
| Myeloid Lineage Expression | Yes | Yes | No | Yes |
| Osteoblast Lineage Expression | Yes | Yes | Osteocyte | Yes |
| Osteoblast Replication | No effect | No effect | Increased | Not studied |
| Osteoblast Differentiation | Inhibited | Inhibited | No effect | |
| Osteoclastogenesis | Inhibited | Enhanced Direct & Indirect Effect | Enhanced | |
| Indirect Effect | ||||
| Osteoprotegerin | Induced | No effect | Suppressed | |
| RANKL Induction | Modest | Robust | Robust |
EGF, epidermal growth factor; NRR, Notch regulatory region; NICD, notch intracellular domain; RANKL, receptor activator of NF-κB ligand
Distinct cellular patterns of expression confer the Notch receptors a unique physiological role (Table 2). Moreover, the actions of Notch are cell-context dependent and cellular responses depend on the specific cell being studied and its stage of maturation at the time of Notch activation. For example, when Notch1 is activated in osteoblast precursors it inhibits osteoblast differentiation, whereas when activated in mature osteoblasts and osteocytes Notch1 inhibits bone resorption and causes an osteopetrotic phenotype [6, 7]. Depending on the cell environment, these differences in Notch activity have led to discrepant conclusions regarding the actual function of Notch in skeletal cells.
Table 2.
Expression of A. Notch receptors and their ligands, and B. Notch target genes in skeletal cells.
| Cells | Notch1 | Notch2 | Notch3 | Notch4 | Jag1 | Jag2 | Dll1 | Dll3 | Dll4 |
|---|---|---|---|---|---|---|---|---|---|
| Osteoblast | ✓ | ✓ | ✓ | ✓ | ✓ | ND | ND | ND | ND |
| Osteocytes | ✓ | ✓ | ✓ | ✓ | ✓ | ND | ND | ✓ | ✓ |
| Bone Marrow-derived Macrophages | ✓ | ✓ | ✓ | ± | ✓ | ND | ND | ND | ND |
| Osteoclast | ✓ | ✓ | ND | ✓ | ✓ | ND | ND | ND | ND |
| Cells | Hes1 | Hes3 | Hes5 | Hey1 | Hey2 | HeyL |
|---|---|---|---|---|---|---|
| Osteoblasts | ✓ | ND | ND | ✓ | ✓ | ✓ |
| Osteoclasts | ✓ | Low | Low | ND | ND | ND |
✓ = Detected by quantitative reverse transcription polymerase chain reaction
ND = Not detected
Notch, Bone Cells and Skeletal Remodeling
1. Notch and Osteoblasts
Initial studies performed in vitro demonstrated that Notch1 suppresses osteoblast cell differentiation and Wnt/β-catenin signaling [16]. Activation of Notch1 signaling in osteoblast precursors and mature osteoblasts causes profound osteopenia secondary to impaired bone formation [7].Transgenic overexpression of Notch1 in osteoblasts also causes osteopenia and the mechanism appears to be an inhibition of Wnt/β-catenin signaling [16]. In agreement with these findings, the deletion of Notch1 and Notch2 in osteoblast precursors causes an increase in osteoblast number, bone formation and cancellous bone volume [17]. It is of interest that mice harboring the activation of Notch1NICD in osteoblasts fail to form a compact cortical bone, which has the appearance of embryonic cortical bone suggesting that a compact cortex fails to develop in the presence of Notch [7]. Notch activation in the early phases of the osteoblast differentiation program prevents the maturation of cells capable to synthesize a mineralized matrix, whereas Notch induction in mature cells precludes further differentiation and causes an accumulation of dysfunctional osteoblasts [2, 4, 7]. These events are possibly mediated by suppression of RUNX2 transcription, and a decrease in Wnt signaling [2].
2. Notch and Osteocytes
Activation of canonical signaling by Notch1 in osteocytes induces osteoprotegerin and as a consequence causes an osteopetrotic phenotype. Both cancellous bone resorption and formation are reduced, so that bone remodeling is suppressed [6, 7, 18]. The phenotype is compartment-specific and cortical bone formation is increased, but cortical bone is porous, with the appearance of trabecular bone, possibly because it fails to reach maturity. The dual inactivation of Notch1 and Notch2 in osteocytes causes a modest increase in cancellous bone volume and a decrease in bone resorption [6].
Notch1 activation in osteocytes suppresses the Wnt antagonists sclerostin and dickkopf 1 (Dkk1); through this indirect mechanism Notch increases Wnt/β-catenin signaling [6, 18]. The induction of Wnt signaling seems paradoxical to the direct inhibition of Wnt signaling by Notch in osteoblasts. However, the mechanism in osteocytes is indirect; and since osteoblasts express minimal levels of Sost, an inhibition of Sost with a consequent increase in Wnt signaling cannot occur in these cells. Therefore, the direct inhibitory effect of Notch on Wnt signaling prevails and is likely to be responsible for the impaired osteoblast maturation [16].
The interactions of Notch and Wnt in osteocytes are complex. Notch receptors are induced by Wnt signaling in osteocytes creating a positive-feedback loop where Wnt enhances Notch signaling and Notch, by downregulating Sost, increases the levels of Wnt activity [19]. Mechanical forces induce Notch signaling in osteocytes either directly or indirectly through the activation of Wnt [20].
3. Notch and Osteoclasts
Notch signaling regulates osteoclast differentiation and function, but the response is highly dependent on the Notch receptor being activated. Notch1 inhibits osteoclastogenesis by direct and indirect mechanisms, including the induction of osteoprotegerin [2, 4, 6, 7]. Notch1 has direct effects on osteoclast precursors and its activation suppresses osteoclastogenesis [1, 21]. The inactivation of Rbpjκ in myeloid cell cultures leads to enhanced osteoclastogenesis, indicating that Rbpjκ is an inhibitor of osteoclastogenesis and that effects of Notch1 in osteoclast differentiation are mediated by canonical mechanisms [22].
In contrast to the inhibitory effects of Notch1 on osteoclastogenesis, Notch2 induces osteoclastogenesis by direct and indirect mechanisms. Interactions of the Notch2NICD with nuclear factor (Nf)-kB in osteoclast precursors lead to the transcription of Nfatc1, a gene critical for osteoclastogenesis [3, 23]. Notch2 also enhances osteoclastogenesis by inducing receptor activator of NF-κB ligand (RANKL) in cells of the osteoblast lineage. A similar induction of RANKL is observed with the activation of Notch3. However, Notch3 is not expressed in the myeloid lineage and does not have direct effects on osteoclast precursors. The actions of Notch4 in either the osteoblast or osteoclast lineage are less certain and low levels of Notch4 are expressed in skeletal cells.
4. Notch Target Genes and Skeletal Cells
Hes1, Hey1, Hey2 and HeyL are the Notch target genes expressed by skeletal cells (Table 2). Although HEYs are induced by Notch in skeletal cells, in vivo models of gene misexpression demonstrated modest effects of HEY1, HEY2 and HEYL on bone remodeling [24–26]. Studies conducted in our laboratory revealed that transgenics expressing Hey2 under the control of a 3.6 kilobase Col1a1 promoter develop osteopenia, but the inactivation of Hey1, Hey2 or HeyL do not result in a significant or lasting skeletal phenotype [24–26]. Hey1 and HeyL transgenics do not have an obvious skeletal phenotype. These findings indicate only a modest role of HEY proteins in bone physiology. In contrast to the modest role of Hey genes in skeletal homeostasis, initial studies from our laboratory on models of Hes1 misexpression suggest a more significant role of HES1 in bone remodeling. Hes1 inactivation in osteoblasts causes an increase in bone formation and suppressed bone resorption, and Col1a1-Hes1 transgenics are osteopenic, have decreased osteoblast function and increased bone resorption [27]. These changes are compatible with the skeletal manifestations of Notch activation in bone, making Hes1 a candidate gene responsible for the actions of Notch in the skeleton.
5. Hormonal Regulation of Notch
Parathyroid Hormone
Parathyroid hormone (PTH) induces the Notch ligand Jag1 in osteoblasts, and the effects of PTH on osteoblast differentiation are opposed by 03B3-secretase inhibitors, suggesting that Notch has the potential to mediate selected actions of PTH in osteoblasts [28]. However, γ-secretase inhibitors have nearly 100 substrates and therefore are not specific inhibitors of Notch signal activation [29]. In addition, PTH and Notch have opposite effects on bone formation and osteoblast activity. These apparently contradictory observations could be explained by additional interactions between PTH and Notch in osteoblastic cells.
PTH was examined further for its effects on Notch signaling in cells of the osteoblast lineage in vitro and in vivo. PTH was found to decrease Hey1, Hey2 and HeyL mRNA levels in Notch-activated osteoblasts and suppress the expression of Notch target genes in osteocytes [15]. In agreement with these observations in vitro, administration of PTH in vivo suppressed Hey1 and HeyL expression in bone extracts. Transactivation experiments with a Notch reporter construct and electrophoretic mobility shift assays in osteoblasts indicated that PTH acts by decreasing the capacity of Rbpjκ to bind to DNA and by interfering with the formation of an active NICD/Rbpjκ transcriptional complex.
Although Jag1 is induced by PTH, Notch signaling is downregulated by PTH in osteoblasts and osteocytes, suggesting that Jag1 induction is not sufficient to overcome the inhibitory effect of PTH on Rbpjκ-mediated signaling. Alternatively, Jag1 induced by PTH may sequester and suppress Notch receptors [30]. The inhibitory effects of PTH on Notch signaling might contribute to the mechanisms of the anabolic effects of PTH in bone.
Cortisol
Cortisol causes a time-dependent increase in Notch1 and Notch2 mRNA levels in MC3T3 osteoblast-like cells, and has no effect on the expression of Notch ligands. Cortisol acts by increasing the rate of Notch1 and Notch2 transcription [31]. Although cortisol induces Notch1 and Notch2 expression, it does not enhance Notch activity. In primary cultures of osteoblasts, cortisol decreases the transcription of Hey1, Hey2 and HeyL in Notch-activated cells. In addition, the administration of prednisolone in vivo decreases the expression of Notch target genes in bone. These results suggest that cortisol has the potential to oppose Notch activity.
II. NOTCH AND CONGENITAL DISORDERS OF THE SKELETON
Congenital Disorders Associated with a Gain-of-Notch Function
1. Hajdu Cheney Syndrome
Hajdu Cheney Syndrome (HCS) is a rare, inherited disease associated with NOTCH2 mutations (Table 3). HCS is characterized by osteoporosis with fractures, acroosteolysis of the hands and feet, craniofacial developmental defects and short stature [32–34]. HCS is associated with nonsense mutations or deletions leading to the creation of a termination codon in exon 34 of NOTCH2 upstream of the PEST domain [35–37]. Since the PEST domain is required for the degradation of NOTCH2, the mutations lead to the formation of a truncated and stable NOTCH2 protein and, as a consequence, to a NOTCH2 gain-of-function.
Table 3.
Genetic disorders associated with altered Notch signaling.
| Disorder | Associated Gene Mutation | References |
|---|---|---|
| Hajdu Cheney Syndrome | NOTCH2 | 34, 35, 36, 37 |
| Lateral Meningocele Syndrome | NOTCH3 | 52, 54 |
| Brachydactyly | CHSY1 | 56, 57 |
| Adams Oliver Syndrome | NOTCH1, DLL4, CSL, EOGT | 59, 61, 62, 63 |
| Alagille Syndrome | JAG1, NOTCH2 | 65, 66, 69, 70 |
| Spondylocostal Dysostosis | DLL3 | 74 |
| Spondylothoracic Dysostosis | MESP2, LFNG, HES7 | 76, 78, 79 |
HCS has autosomal dominant inheritance, although sporadic cases occur. There is clinical variability and evolution of the clinical manifestations, and signs of the disease can be manifested as early as in the first two years of life [32]. Acroosteolysis is frequently present and accompanied by inflammation, and lysis of the phalanges which leads to short and broad digits. Spinal abnormalities include compression fractures, kyphosis and scoliosis. Long bone deformities can occur. Platybasia and basilar invagination can result in severe neurological complications, including hydrocephalus and central respiratory arrest causing sudden death. Polycystic kidneys are present in 10% of the cases and occasionally affected subjects present with cardiac septal defects and valve abnormalities [38]. Iliac crest biopsies reveal the presence of cortical bone osteopenia, woven bone and increased osteoclast number and bone resorption [39].
Our laboratory created a Notch2 mutant mouse harboring a truncating mutation in exon 34 upstream of the PEST domain reproducing the mutation found in HCS and termed Notch2tm1.1Ecan [23]. Notch2tm1.1Ecan mutant mice exhibit cancellous and cortical bone osteopenia secondary to increased osteoclast number and bone resorption. In vitro studies demonstrated enhanced osteoclastogenesis and confirmed the stimulatory effect of Notch2 on osteoclast differentiation [3]. The mechanism involves a direct effect of Notch2 on osteoclast precursors as well as the induction of RANKL by cells of the osteoblast lineage. In an alternate murine model of HCS, an increased expression of interleukin 6 was reported in bone marrow cells, and this cytokine could contribute to the enhanced bone resorption observed [40]. Notch2tm1.1Ecan mutant mice are sensitized to the development of osteoarthritis, a mechanism that may also involve an enhanced expression of interleukin 6 by the chondrocyte of HCS mutant mice [41].
There are no controlled trials on the management of the osteoporosis in patients with HCS. Bisphosphonates alone or in combination with teriparatide have been used, but evidence of benefit is scarce [38, 42, 43]. Because Notch2 induces RANKL and enhances osteoclastogenesis, a consideration is the use of denosumab and the agent was used successfully in the treatment of a subject with osteoporosis and fractures [44]. Although PTH suppresses Notch signaling, the use of teriparatide in the treatment of HCS could pose risks [15]. There is evidence of Notch activation in osteosarcoma in humans and prolonged activation of Notch in mice can cause osteosarcoma, suggesting that Notch activation could be a risk factor for osteosarcoma [45, 46]. As such, teriparatide should be used with caution. Moreover, the mechanism responsible for the bone loss in HCS is increased bone resorption making teriparatide not an ideal choice in the management of HCS.
Notch2 itself could be a future target for the treatment of HCS, and the skeletal phenotype of Notch2tm1.1Ecan mouse mutants was reversed by anti-Notch2 antibodies that target the NRR of Notch2 and prevent the activation of Notch2 [47, 48]. There are no clinical trials on the use of this approach which carries the risk of a generalized downregulation of Notch2 with potential unwanted events.
It is of interest that somatic NOTCH2 mutations causing loss of the PEST domain exhibit enhanced Notch2 activation and have been identified in B cell lymphoma, specifically in marginal zone B lymphoma of the spleen [49]. Notch2 is required for the development of the marginal zone of the spleen, and mouse models of HCS exhibit a reallocation of marginal zone B cells at the expense of follicular cells but do not appear to develop marginal zone lymphomas [50, 51]. The reallocation of marginal zone B cells does not influence skeletal remodeling. There is no evidence of higher incidence of marginal zone B cell lymphomas in subjects with HCS.
2. Lateral Meningocele or Lehman Syndrome
Lateral Meningocele Syndrome (LMS) is a rare disorder characterized by meningoceles with relatneurological dysfunction [52]. Clinical manifestations of LMS include craniofacial developmental abnormalities, intellectual disability, hypotonia, decreased muscle mass, syringomyelia and cardiac valve abnormalities. Skeletal manifestations include short stature, scoliosis, pectus excavatum, wormian bones, increased density of the base of the skull, and increased bone remodeling and bone loss [53].
LMS is associated with point mutations or short deletions in exon 33 of NOTCH3, upstream of the PEST domain [54]. The mutations are analogous to those reported in exon 34 of NOTCH2 in HCS and result in the translation of a truncated and stable product, devoid of the PEST domain. The stabilization of the Notch3 protein likely leads to a gain-of-function and enhancement of Notch signaling. Subjects with LMS share selected clinical features with HCS [55]. Although increased bone turnover and decreased bone mineral density (BMD) may occur in LMS, acroosteolysis, a hallmark feature of HCS, has not been reported. The inheritance in LMS is not clear and autosomal dominant inheritance has been suggested [53]. However, most cases reveal de novo heterozygous mutations of NOTCH3.
3. Brachydactyly
Brachydactyly is an autosomal recessive disorder characterized by bilateral pre-axial brachydactyly or shortening of the digits of the hands and feet [56]. Affected individuals display facial dysmorphism, dental anomalies, sensory hearing loss, and growth, motor and mental retardation. A null allele of chondroitin sulfate synthase (CHSY)1 has been associated with brachydactyly and downregulation of Chsy1 results in enhanced Notch signaling, suggesting that activation of Notch signaling is responsible for aspects of the clinical syndrome [57].
Congenital Disorders Associated with a Loss-of-Notch Function
1. Adams Oliver Syndrome
Adams Oliver Syndrome (AOS), a rare congenital disorder characterized by aplasia cutis congenita and terminal transverse limb defects, is often, although not always, associated with mutations of genes encoding components of the Notch signaling pathway. These include loss-of-function mutants in NOTCH1, DLL4, CSL and EOGT, encoding for EGF-domain-specific O-linked N-acetylglucosamine transferase (O-GlcNAc). The clinical presentation of AOS is variable. The terminal limb defects include oligodactyly, syndactyly, hypoplastic nails and transverse amputations [58]. Small vessel abnormalities and vascular thrombosis during development may be responsible for the terminal transverse limb defects, and these are consistent with the established role of Notch signaling in vascular development [59, 60]. Congenital cardiac defects, including ventricular septal defects, tetralogy of Fallot, and anomalies of arteries and cardiac valves are uncommon and associated with peripheral vascular abnormalities, such as cutis marmorata telangiectatica congenita and retinal hypovascularization.
A variety of NOTCH1 mutations across the length of the receptor are found in AOS. The majority of the gene mutations occur in the extracellular domain leading to structural changes and loss-of- function [59]. Loss-of-function mutations in CSL result in decreased binding of CSL to regulatory regions of Notch target genes [61]. Missense and nonsense mutations in the coding sequence of DLL4 are predicted to cause loss-of-function [62]. Mutations in EOGT cause impaired O-GlcNAc activity and Notch1 glycosylation [63].
2. Alagille Syndrome
Alagille Syndrome is an autosomal dominant disease that presents with cardiovascular defects including tetralogy of Fallot, abnormalities of the craniofacial skeleton and vertebrae, cholestatic liver disease due to bile duct atresia and kidney anomalies causing renal failure [64]. Failure of the vertebrae to fuse ventrally during development causes a characteristic “butterfly” appearance in radiographic images [65]. Craniofacial developmental abnormalities cause craniosynostosis and characteristic facial features. Subjects have short stature, digit abnormalities and may present with osteoporosis, considered to be secondary to liver failure and malnutrition.
Alagille Syndrome is associated with loss-of-function mutations of the Notch ligand JAG1 [66–68]. Rarely, mutations of NOTCH2, isolated or in conjunction with mutations of JAG1, have been reported in Alagille Syndrome [69, 70]. Individuals with NOTCH2 null mutations frequently present with hypoplastic kidneys and renal insufficiency, since Notch2 is required for renal development [70].
Null mutations of Jag1 or inactivation of Notch2 in mice result in embryonic lethality [71, 72]. However, the combined heterozygous inactivation of Jag1 and a hypomorphic Notch2 allele in mice recapitulates features of Alagille Syndrome suggesting that the inactivation of these genes is responsible for the clinical manifestations
3. Spondylocostal and Spondylothoracic Dysostosis
Spondylocostal dysostosis and spondylothoracic dysostosis are characterized by vertebral segmentation defects and rib anomalies secondary to defective somitogenesis [73]. Dominant, recessive and sporadic loss-of-function mutations of genes encoding various components of the Notch signaling pathway are associated with these disorders. Mutations of DLL3 are found in 20–25% of affected individuals, and inactivation of Dll3 in mice recapitulates the manifestations of spondylocostal dysostosis [74, 75]. Spondylothoracic dysostosis is associated with a mutant mesoderm posterior bHLH transcription factor (MESP)2 allele [76]. MESP2 is a Notch target gene critical for somitogenesis, and Mesp2 null mice exhibit vertebral developmental defects [77]. Hes7 regulates the transcription of lunatic fringe (Lfng) which regulates the glycosylation of Notch, and loss-of-function mutations of either LFNG or HES7 also are associated with spondylocostal dysostosis [78, 79].
III. NOTCH AND ACQUIRED SKELETAL DISEASES
1. Osteoporosis
The actions of Notch signaling in skeletal physiology as well as the presence of severe bone loss in congenital disorders of Notch gain-of-function would suggest a possible role of Notch in osteoporosis. Genome-wide association studies revealed an association between JAG1, the gene encoding the Notch ligand Jag1 and BMD [80]. A SNP (rs2273061) of JAG1 is associated with high BMD at the lumbar spine and femoral neck, and low risk of osteoporotic fractures. The findings suggest that JAG1 is a candidate gene for BMD regulation and a potential factor in the pathogenesis of fractures.
2. Fractures
Fracture healing is a regenerative process that results in the formation of new bone after a fracture. The healing of a fracture occurs by either intramembranous bone formation, when the bone is mechanically stable, or by endochondral ossification, when the bone is unstable. The function of Notch in fracture healing is complex, and upregulation as well as downregulation of Notch signaling have been reported in experimental models of fracture healing [81]. Because of the known inhibitory effects of Notch on osteogenesis and chondrogenesis, downregulation of Notch signaling might be a requirement for the fracture healing process to occur. The administration of γ-secretase inhibitors, to prevent the activation of Notch signaling, accelerated fracture healing in an open mid-shaft tibial fracture mouse model [82]. Whereas these results suggest a negative role of Notch in fracture healing, it is important to note that γ-secretase inhibitors are not specific inhibitors of Notch activation. Interestingly, the downregulation of Rbpjκ, a main component of canonical Notch signaling, in chondrogenic-/osteogenic-expressing cells results in non-union fractures [83]. This suggests that a degree of Notch signaling is necessary for fracture repair.
Cell lineage tracing studies have demonstrated that osteoblast precursors move into the fracture site along with invading blood vessels, confirming the importance of vascularization for proper fracture healing [84]. Notch signaling plays a critical role in vascular development and physiology, and promotes endothelial cell proliferation and vessel growth in long bones [85]. The angiogenic effects of Notch may be necessary for the healing process of fractures to occur [86].
3. Osteosarcoma
Notch receptors and Notch target genes are upregulated in osteosarcoma [87, 88]. Gene expression analysis in tissue samples from subjects with osteosarcoma compared to unaffected human bone tissue or to normal osteoblasts has demonstrated upregulation of NOTCH1, NOTCH2 and JAG1 mRNA levels in osteosarcoma [45, 88]. Notch target genes, particularly HEY1 and HEY2, are increased in osteosarcoma, demonstrating activation of Notch canonical signaling. The growth of osteosarcoma cell lines is suppressed by γ-secretase inhibitors and by lentiviruses delivering a dominant negative MAML, to prevent Notch-dependent activation or transcription, respectively. This suggests that Notch controls the growth of osteosarcoma cells [87].
The induction of the Notch1NICD in mature osteoblasts in mice causes the spontaneous development of osteosarcoma as mice age [46]. The development of osteosarcoma requires the activation of Notch canonical signaling since it is not observed in the context of Rbpjκ inactivation. The findings in humans and mouse models demonstrate that Notch canonical signaling plays a role in the initiation as well as in the invasive potential of osteosarcoma [89]. As a result, components of the Notch signaling pathway could become future therapeutic targets in osteosarcoma.
4. Skeletal Metastases
Skeletal metastases are complications of carcinoma of the breast and prostate, and Notch has been implicated in the interactions between osteoblasts and metastatic cells [90, 91]. Breast cancer cells expressing JAG1 activate Notch signaling, which induces Interleukin 6 in osteoblasts and, as a consequence, enhances osteoclastogenesis and the formation of osteolytic bone metastases. This leads to the release of transforming growth factor (TGF)β from the bone matrix, which upregulates JAG1, causing further activation of Notch signaling and creating a positive feedback loop favoring the metastatic potential of the tumor. Downregulation of JAG1 decreases the osteolytic potential in experimental models of carcinoma of the breast.
Carcinoma of the prostate frequently metastasizes to bone. NOTCH1 and JAG1 are expressed by carcinoma of the prostate, and their expression is associated with the metastatic potential and recurrence of the tumor [92]. Downregulation of NOTCH1 by RNA interference in human prostate cancer cells decreases their invasive potential verifying a role of Notch in this malignancy.
IV. MODIFYING NOTCH SIGNALING
Notch signaling can be downregulated or tempered using a variety of approaches including the use of biochemical inhibitors of Notch activation, antibodies to Notch receptors or to their ligands, and the use of small permeable molecules that prevent the formation of an NICD/Rbpjk/Maml ternary complex [93]. γ-secretase inhibitors are used to prevent the cleavage and activation of Notch induced by presenilins [94]. Their limitation is a lack of specificity since nearly 100 substrates of the γ-secretase complex are known to exist [29]. Thapsigargin is an inhibitor of the sarco/endoplasmic reticulum Ca2+− ATPase that precludes the maturation and folding of the Notch receptor and, as a consequence, prevents Notch activation [95]. Synthetic small cell permeable molecules that prevent the assembly of an active Notch transcriptional complex can be used for the inhibition of Notch signaling, but their long-term efficacy is unknown [96]. A limitation of these agents is that they interfere with the indiscriminate activation of all four Notch receptors.
To target specific Notch receptors, antibodies to the NRR of Notch1, Notch2 and Notch3 have been developed [48, 97]. The NRR contains the initial cleavage sites of Notch required for protein maturation and signal activation (Fig. 1) [98]. The epitope for the anti-Notch NRR bridges the domain so that the antibody locks the receptor in its quiescent state preventing Notch activation [48, 99]. The targeting of the NRR prevents the activation of specific Notch isoforms, making the use of anti-Notch NRR antibodies ideal for the neutralization of individual Notch receptors.
Recently, anti-Notch2 NRR antibodies were tested for their effects on the skeletal phenotype of HCS mutant mouse models and shown to reverse the osteopenic phenotype of Notch2HCS mice [47]. However, information on the inhibition of Notch signaling in humans is quite limited, and widespread Notch neutralization is not without unwanted events; it may cause vascular tumors and gastrointestinal toxicity [100].
V. CONCLUSIONS
Notch is a determinant of cell differentiation and function in cells of the osteoblast and osteoclastlineages and plays a critical role in skeletal development and bone homeostasis. The effects of Notch are cell-context dependent, and the four Notch receptors carry out specific functions in the skeleton. Alterations in Notch signaling are associated with a variety of congenital disorders and there is evidence of altered Notch signaling in skeletal malignancies. In conclusion, Notch signaling is a novel pathway that regulates skeletal homeostasis in health and disease.
Acknowledgments
The author thanks Mary Yurczak for secretarial assistance.
Funding
This work was supported by Grants AR063049, AR068160 and AR072987 from the National Institute of Arthritis and Musculoskeletal and Skin Diseases, and Grant DK045227 from the National Institute of Diabetes and Digestive and Kidney Diseases.
Abbreviations
- ANK
Ankyrin
- BMD
bone mineral density
- CHSY
chondroitin sulfate synthase
- DII
Delta-like
- Dkk1
dickkopf 1
- EGF
epidermal growth factor
- HES
Hairy and enhancer of split
- EOGT
EGF-domain-specific O-linked N-acetylgucosamine transferase
- HCS
Hajdu Cheney Syndrome
- HEY
HES with a YRPW motif
- HD
heterodimerization domain; n
- Jag
Jagged
- MAML
mastermind-like
- LP
leader peptide
- LMS
Lateral Meningocele Syndrome
- LNR
Lin12-Notch repeats
- Lnfg
lunatic fringe
- NRR
negative regulatory region
- NICD
Notch intracellular domain
- Nf
nuclear factor
- NLS
nuclear localization sequence
- PTH
parathyroid hormone
- PEST
proline (P)-, glutamic acid (E)-, serine (S)- and threonine (T)-rich
- RANKL
receptor activator of NF-κB ligand
- RBPJκ
recombination signal-binding protein for Ig of κ region
- RAM
RBPJκ-association module
- TGF
transforming growth factor
- TMD
transmembrane domain
Footnotes
Conflict of Interest
None.
References
- 1.Bai S, Kopan R, Zou W, Hilton MJ, Ong CT, Long F, Ross FP, Teitelbaum SL (2008) NOTCH1 regulates osteoclastogenesis directly in osteoclast precursors and indirectly via osteoblast lineage cells. J Biol Chem 283:6509–6518 [DOI] [PubMed] [Google Scholar]
- 2.Engin F, Yao Z, Yang T, et al. (2008) Dimorphic effects of Notch signaling in bone homeostasis. Nat Med 14:299–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fukushima H, Nakao A, Okamoto F, Shin M, Kajiya H, Sakano S, Bigas A, Jimi E, Okabe K (2008) The association of Notch2 and NF-kappaB accelerates RANKL-induced osteoclastogenesis. Mol Cell Biol 28:6402–6412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hilton MJ, Tu X, Wu X, et al. (2008) Notch signaling maintains bone marrow mesenchymal progenitors by suppressing osteoblast differentiation. Nature medicine 14:306–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zanotti S, Canalis E (2016) Notch Signaling and the Skeleton. Endocr Rev 37:223–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canalis E, Adams DJ, Boskey A, Parker K, Kranz L, Zanotti S (2013) Notch Signaling in Osteocytes Differentially Regulates Cancellous and Cortical Bone Remodeling. J Biol Chem 288:25614–25625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Canalis E, Parker K, Feng JQ, Zanotti S (2013) Osteoblast Lineage-specific Effects of Notch Activation in the Skeleton. Endocrinology 154:623–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanchez-Irizarry C, Carpenter AC, Weng AP, Pear WS, Aster JC, Blacklow SC (2004) Notch subunit heterodimerization and prevention of ligand-independent proteolytic activation depend, respectively, on a novel domain and the LNR repeats. Mol Cell Biol 24:9265–9273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kopan R, Ilagan MX (2009) The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137:216–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siebel C, Lendahl U (2017) Notch Signaling in Development, Tissue Homeostasis, and Disease. Physiol Rev 97:1235–1294 [DOI] [PubMed] [Google Scholar]
- 11.Iso T, Kedes L, Hamamori Y (2003) HES and HERP families: multiple effectors of the Notch signaling pathway. J Cell Physiol 194:237–255 [DOI] [PubMed] [Google Scholar]
- 12.Kovall RA (2007) Structures of CSL, Notch and Mastermind proteins: piecing together an active transcription complex. Curr Opin Struct Biol 17:117–127 [DOI] [PubMed] [Google Scholar]
- 13.Wu J, Bresnick EH (2007) Bare rudiments of notch signaling: how receptor levels are regulated. Trends Biochem Sci 32:477–485 [DOI] [PubMed] [Google Scholar]
- 14.Yuan Z, Friedmann DR, Vanderwielen BD, Collins KJ, Kovall RA (2012) Characterization of CSL (CBF-1, Su(H), Lag-1) Mutants Reveals Differences in Signaling Mediated by Notch1 and Notch2. J Biol Chem 287:34904–34916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zanotti S, Canalis E (2017) Parathyroid hormone inhibits Notch signaling in osteoblasts and osteocytes. Bone 103:159–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deregowski V, Gazzerro E, Priest L, Rydziel S, Canalis E (2006) Notch 1 Overexpression Inhibits Osteoblastogenesis by Suppressing Wnt/beta-Catenin but Not Bone Morphogenetic Protein Signaling. J Biol Chem 281:6203–6210 [DOI] [PubMed] [Google Scholar]
- 17.Zanotti S, Canalis E (2014) Notch1 and Notch2 expression in osteoblast precursors regulates femoral microarchitecture. Bone 62:22–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Canalis E, Bridgewater D, Schilling L, Zanotti S (2015) Canonical Notch Activation in Osteocytes Causes Osteopetrosis. American journal of physiology Endocrinology and metabolism 310:E171–E182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tu X, Delgado-Calle J, Condon KW, Maycas M, Zhang H, Carlesso N, Taketo MM, Burr DB, Plotkin LI, Bellido T (2015) Osteocytes mediate the anabolic actions of canonical Wnt/beta-catenin signaling in bone. Proc Natl Acad Sci U S A 112:E478–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robling AG, Niziolek PJ, Baldridge LA, et al. (2008) Mechanical stimulation of bone in vivo reduces osteocyte expression of Sost/sclerostin. J Biol Chem 283:5866–5875 [DOI] [PubMed] [Google Scholar]
- 21.Yamada T, Yamazaki H, Yamane T, Yoshino M, Okuyama H, Tsuneto M, Kurino T, Hayashi S, Sakano S (2003) Regulation of osteoclast development by Notch signaling directed to osteoclastprecursors and through stromal cells. Blood 101:2227–2234 [DOI] [PubMed] [Google Scholar]
- 22.Zhao B, Grimes SN, Li S, Hu X, Ivashkiv LB (2012) TNF-induced osteoclastogenesis and inflammatory bone resorption are inhibited by transcription factor RBP-J. J Exp Med 209:319–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Canalis E, Schilling L, Yee SP, Lee SK, Zanotti S (2016) Hajdu Cheney Mouse Mutants Exhibit Osteopenia, Increased Osteoclastogenesis and Bone Resorption. J Biol Chem 291:1538–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Canalis E, Zanotti S (2017) Hairy and Enhancer of Split-Related With YRPW Motif-Like (HeyL) Is Dispensable for Bone Remodeling in Mice. J Cell Biochem 118:1819–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salie R, Kneissel M, Vukevic M, Zamurovic N, Kramer I, Evans G, Gerwin N, Mueller M, Kinzel B, Susa M (2010) Ubiquitous overexpression of Hey1 transcription factor leads to osteopenia and chondrocyte hypertrophy in bone. Bone 46:680–694 [DOI] [PubMed] [Google Scholar]
- 26.Zanotti S, Canalis E (2013) Hairy and Enhancer of Split-related with YRPW Motif (HEY)2 Regulates Bone Remodeling in Mice. J Biol Chem 288:21547–21557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zanotti S, Smerdel-Ramoya A, Canalis E (2011) Hairy and enhancer of split (HES)1 is a determinant of bone mass. J Biol Chem 286:2648–2657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Calvi LM, Adams GB, Weibrecht KW, et al. (2003) Osteoblastic cells regulate the haematopoietic stem cell niche. Nature 425:841–846 [DOI] [PubMed] [Google Scholar]
- 29.Duggan SP, McCarthy JV (2016) Beyond gamma-secretase activity: The multifunctional nature of presenilins in cell signalling pathways. Cell Signal 28:1–11 [DOI] [PubMed] [Google Scholar]
- 30.del Alamo D, Rouault H, Schweisguth F (2011) Mechanism and significance of cis-inhibition in Notch signalling. Curr Biol 21:R40–47 [DOI] [PubMed] [Google Scholar]
- 31.Zanotti S, Yu J, Adhikari S, Canalis E (2018) Glucocorticoids inhibit notch target gene expression in osteoblasts. J Cell Biochem 119:6016–6023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Descartes M, Rojnueangnit K, Cole L, Sutton A, Morgan SL, Patry L, Samuels ME (2014) Hajdu-Cheney syndrome: phenotypical progression with de-novo NOTCH2 mutation. Clin Dysmorphol 23:88–94 [DOI] [PubMed] [Google Scholar]
- 33.Hajdu N, Kauntze R (1948) Cranio-skeletal dysplasia. Br J Radiol 21:42–48 [DOI] [PubMed] [Google Scholar]
- 34.Canalis E (2018) Clinical and experimental aspects of notch receptor signaling: Hajdu-Cheney syndrome and related disorders. Metabolism 80:48–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Isidor B, Lindenbaum P, Pichon O, et al. (2011) Truncating mutations in the last exon of NOTCH2 cause a rare skeletal disorder with osteoporosis. Nat Genet 43:306–308 [DOI] [PubMed] [Google Scholar]
- 36.Simpson MA, Irving MD, Asilmaz E, et al. (2011) Mutations in NOTCH2 cause Hajdu-Cheney syndrome, a disorder of severe and progressive bone loss. Nat Genet 43:303–305 [DOI] [PubMed] [Google Scholar]
- 37.Zhao W, Petit E, Gafni RI, Collins MT, Robey PG, Seton M, Miller KK, Mannstadt M (2013) Mutations in NOTCH2 in patients with Hajdu-Cheney syndrome. Osteoporos Int 24:2275–2281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sargin G, Cildag S, Senturk T (2013) Hajdu-Cheney syndrome with ventricular septal defect. Kaohsiung J Med Sci 29:343–344 [DOI] [PubMed] [Google Scholar]
- 39.Sakka S, Gafni RI, Davies JH, et al. (2017) Bone Structural Characteristics and Response to Bisphosphonate Treatment in Children With Hajdu-Cheney Syndrome. J Clin Endocrinol Metab 102:4163–4172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vollersen N, Hermans-Borgmeyer I, Cornils K, et al. (2018) High Bone Turnover in Mice Carrying a Pathogenic Notch2 Mutation Causing Hajdu-Cheney Syndrome. J Bone Miner Res 33:70–83 [DOI] [PubMed] [Google Scholar]
- 41.Zanotti S, Yu J, Bridgewater D, Wolf JM, Canalis E (2018) Mice harboring a Hajdu Cheney Syndrome mutation are sensitized to osteoarthritis. Bone 114:198–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Galli-Tsinopoulou A, Kyrgios I, Giza S, Giannopoulou EM, Maggana I, Laliotis N (2012) Two-year cyclic infusion of pamidronate improves bone mass density and eliminates risk of fractures in a girl with osteoporosis due to Hajdu-Cheney syndrome. Minerva Endocrinol 37:283–289 [PubMed] [Google Scholar]
- 43.McKiernan FE (2008) Integrated anti-remodeling and anabolic therapy for the osteoporosis of Hajdu-Cheney syndrome: 2-year follow-up. Osteoporos Int 19:379–380 [DOI] [PubMed] [Google Scholar]
- 44.Adami G, Rossini M, Gatti D, Orsolini G, Idolazzi L, Viapiana O, Scarpa A, Canalis E (2016) Hajdu Cheney Syndrome; report of a novel NOTCH2 mutation and treatment with denosumab. Bone 92:150–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Engin F, Bertin T, Ma O, Jiang MM, Wang L, Sutton RE, Donehower LA, Lee B (2009) Notch signaling contributes to the pathogenesis of human osteosarcomas. Hum Mol Genet 18:1464–1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tao J, Jiang MM, Jiang L, et al. (2014) Notch activation as a driver of osteogenic sarcoma. Cancer Cell 26:390–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Canalis E, Sanjay A, Yu J, Zanotti S (2017) An Antibody to Notch2 Reverses the Osteopenic Phenotype of Hajdu-Cheney Mutant Male Mice. Endocrinology 158:730–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu Y, Cain-Hom C, Choy L, et al. (2010) Therapeutic antibody targeting of individual Notch receptors. Nature 464:1052–1057 [DOI] [PubMed] [Google Scholar]
- 49.Kiel MJ, Velusamy T, Betz BL, et al. (2012) Whole-genome sequencing identifies recurrent somatic NOTCH2 mutations in splenic marginal zone lymphoma. J Exp Med 209:1553–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu J, Zanotti S, Schilling L, Schoenherr C, Economides AN, Sanjay A, Canalis E (2018) Induction of the Hajdu-Cheney Syndrome Mutation in CD19 B Cells in Mice Alters B-Cell Allocation but Not Skeletal Homeostasis. The American journal of pathology 188:1430–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu J, Zanotti S, Walia B, Jellison E, Sanjay A, Canalis E (2018) The Hajdu Cheney Mutation Is a Determinant of B-Cell Allocation of the Splenic Marginal Zone. The American journal of pathology 188:149–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lehman RA, Stears JC, Wesenberg RL, Nusbaum ED (1977) Familial osteosclerosis with abnormalities of the nervous system and meninges. J Pediatr 90:49–54 [DOI] [PubMed] [Google Scholar]
- 53.Gripp KW, Scott CI Jr., Hughes HE, et al. (1997) Lateral meningocele syndrome: three new patients and review of the literature. Am J Med Genet 70:229–239 [PubMed] [Google Scholar]
- 54.Gripp KW, Robbins KM, Sobreira NL, et al. (2015) Truncating mutations in the last exon of NOTCH3 cause lateral meningocele syndrome. Am J Med Genet A 167A:271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gripp KW (2011) Lateral meningocele syndrome and Hajdu-Cheney syndrome: different disorders with overlapping phenotypes. Am J Med Genet A 155A:1773–1774; author reply 1775 [DOI] [PubMed] [Google Scholar]
- 56.Temtamy SA, Aglan MS (2008) Brachydactyly. Orphanet J Rare Dis 3:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tian J, Ling L, Shboul M, et al. (2010) Loss of CHSY1, a secreted FRINGE enzyme, causes syndromic brachydactyly in humans via increased NOTCH signaling. Am J Hum Genet 87:768–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Snape KM, Ruddy D, Zenker M, Wuyts W, Whiteford M, Johnson D, Lam W, Trembath RC (2009) The spectra of clinical phenotypes in aplasia cutis congenita and terminal transverse limb defects. Am J Med Genet A 149A:1860–1881 [DOI] [PubMed] [Google Scholar]
- 59.Stittrich AB, Lehman A, Bodian DL, et al. (2014) Mutations in NOTCH1 cause Adams-Oliver syndrome. Am J Hum Genet 95:275–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoyme HE, Jones KL, Van Allen MI, Saunders BS, Benirschke K (1982) Vascular pathogenesis of transverse limb reduction defects. J Pediatr 101:839–843 [DOI] [PubMed] [Google Scholar]
- 61.Hassed SJ, Wiley GB, Wang S, et al. (2012) RBPJ mutations identified in two families affected by Adams-Oliver syndrome. Am J Hum Genet 91:391–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meester JA, Southgate L, Stittrich AB, et al. (2015) Heterozygous Loss-of-Function Mutations in DLL4 Cause Adams-Oliver Syndrome. Am J Hum Genet 97:475–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shaheen R, Aglan M, Keppler-Noreuil K, et al. (2013) Mutations in EOGT confirm the genetic heterogeneity of autosomal-recessive Adams-Oliver syndrome. Am J Hum Genet 92:598–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Alagille D, Estrada A, Hadchouel M, Gautier M, Odievre M, Dommergues JP (1987) Syndromic paucity of interlobular bile ducts (Alagille syndrome or arteriohepatic dysplasia): review of 80 cases. J Pediatr 110:195–200 [DOI] [PubMed] [Google Scholar]
- 65.Emerick KM, Rand EB, Goldmuntz E, Krantz ID, Spinner NB, Piccoli DA (1999) Features of Alagille syndrome in 92 patients: frequency and relation to prognosis. Hepatology 29:822–829 [DOI] [PubMed] [Google Scholar]
- 66.Crosnier C, Driancourt C, Raynaud N, Dhorne-Pollet S, Pollet N, Bernard O, Hadchouel M, Meunier-Rotival M (1999) Mutations in JAGGED1 gene are predominantly sporadic in Alagille syndrome. Gastroenterology 116:1141–1148 [DOI] [PubMed] [Google Scholar]
- 67.Morrissette JD, Colliton RP, Spinner NB (2001) Defective intracellular transport and processing of JAG1 missense mutations in Alagille syndrome. Hum Mol Genet 10:405–413 [DOI] [PubMed] [Google Scholar]
- 68.Boyer-Di Ponio J, Wright-Crosnier C, Groyer-Picard MT, Driancourt C, Beau I, Hadchouel M, Meunier-Rotival M (2007) Biological function of mutant forms of JAGGED1 proteins in Alagille syndrome: inhibitory effect on Notch signaling. Hum Mol Genet 16:2683–2692 [DOI] [PubMed] [Google Scholar]
- 69.Kamath BM, Bauer RC, Loomes KM, et al. (2012) NOTCH2 mutations in Alagille syndrome. J Med Genet 49:138–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McDaniell R, Warthen DM, Sanchez-Lara PA, Pai A, Krantz ID, Piccoli DA, Spinner NB (2006) NOTCH2 mutations cause Alagille syndrome, a heterogeneous disorder of the notch signaling pathway. Am J Hum Genet 79:169–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xue Y, Gao X, Lindsell CE, Norton CR, Chang B, Hicks C, Gendron-Maguire M, Rand EB, Weinmaster G, Gridley T (1999) Embryonic lethality and vascular defects in mice lacking the Notch ligand Jagged1. Hum Mol Genet 8:723–730 [DOI] [PubMed] [Google Scholar]
- 72.McCright B, Gao X, Shen L, Lozier J, Lan Y, Maguire M, Herzlinger D, Weinmaster G, Jiang R, Gridley T (2001) Defects in development of the kidney, heart and eye vasculature in mice homozygous for a hypomorphic Notch2 mutation. Development 128:491–502 [DOI] [PubMed] [Google Scholar]
- 73.Turnpenny PD, Whittock N, Duncan J, Dunwoodie S, Kusumi K, Ellard S (2003) Novel mutations in DLL3, a somitogenesis gene encoding a ligand for the Notch signalling pathway, cause a consistent pattern of abnormal vertebral segmentation in spondylocostal dysostosis. J Med Genet 40:333–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dunwoodie SL, Clements M, Sparrow DB, Sa X, Conlon RA, Beddington RS (2002) Axial skeletal defects caused by mutation in the spondylocostal dysplasia/pudgy gene Dll3 are associated with disruption of the segmentation clock within the presomitic mesoderm. Development 129:1795–1806 [DOI] [PubMed] [Google Scholar]
- 75.Kusumi K, Sun ES, Kerrebrock AW, Bronson RT, Chi DC, Bulotsky MS, Spencer JB, Birren BW, Frankel WN, Lander ES (1998) The mouse pudgy mutation disrupts Delta homologue Dll3 and initiation of early somite boundaries. Nat Genet 19:274–278 [DOI] [PubMed] [Google Scholar]
- 76.Cornier AS, Staehling-Hampton K, Delventhal KM, et al. (2008) Mutations in the MESP2 gene cause spondylothoracic dysostosis/Jarcho-Levin syndrome. Am J Hum Genet 82:1334–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Saga Y, Hata N, Koseki H, Taketo MM (1997) Mesp2: a novel mouse gene expressed in the presegmented mesoderm and essential for segmentation initiation. Genes Dev 11:1827–1839 [DOI] [PubMed] [Google Scholar]
- 78.Sparrow DB, Chapman G, Wouters MA, Whittock NV, Ellard S, Fatkin D, Turnpenny PD, Kusumi K, Sillence D, Dunwoodie SL (2006) Mutation of the LUNATIC FRINGE gene in humans causes spondylocostal dysostosis with a severe vertebral phenotype. Am J Hum Genet 78:28–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sparrow DB, Guillen-Navarro E, Fatkin D, Dunwoodie SL (2008) Mutation of Hairy-and-Enhancer-of-Split-7 in humans causes spondylocostal dysostosis. Hum Mol Genet 17:3761–3766 [DOI] [PubMed] [Google Scholar]
- 80.Kung AW, Xiao SM, Cherny S, et al. (2010) Association of JAG1 with bone mineral density and osteoporotic fractures: a genome-wide association study and follow-up replication studies. Am J Hum Genet 86:229–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dishowitz MI, Terkhorn SP, Bostic SA, Hankenson KD (2012) Notch signaling components are upregulated during both endochondral and intramembranous bone regeneration. J Orthop Res 30:296–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang C, Shen J, Yukata K, Inzana JA, O’Keefe RJ, Awad HA, Hilton MJ (2015) Transient gamma-secretase inhibition accelerates and enhances fracture repair likely via Notch signaling modulation. Bone 73:77–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang C, Inzana JA, Mirando AJ, Ren Y, Liu Z, Shen J, O’Keefe RJ, Awad HA, Hilton MJ (2016) NOTCH signaling in skeletal progenitors is critical for fracture repair. J Clin Invest 126:1471–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Maes C, Kobayashi T, Selig MK, Torrekens S, Roth SI, Mackem S, Carmeliet G, Kronenberg HM (2010) Osteoblast precursors, but not mature osteoblasts, move into developing and fractured bones along with invading blood vessels. Dev Cell 19:329–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ramasamy SK, Kusumbe AP, Wang L, Adams RH (2014) Endothelial Notch activity promotes angiogenesis and osteogenesis in bone. Nature 507:376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dishowitz MI, Mutyaba PL, Takacs JD, Barr AM, Engiles JB, Ahn J, Hankenson KD (2013) Systemic inhibition of canonical Notch signaling results in sustained callus inflammation and alters multiple phases of fracture healing. PLoS One 8:e68726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tanaka M, Setoguchi T, Hirotsu M, Gao H, Sasaki H, Matsunoshita Y, Komiya S (2009) Inhibition of Notch pathway prevents osteosarcoma growth by cell cycle regulation. Br J Cancer 100:1957–1965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dailey DD, Anfinsen KP, Pfaff LE, Ehrhart EJ, Charles JB, Bonsdorff TB, Thamm DH, Powers BE, Jonasdottir TJ, Duval DL (2013) HES1, a target of Notch signaling, is elevated in canine osteosarcoma, but reduced in the most aggressive tumors. BMC Vet Res 9:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hughes DP (2009) How the NOTCH pathway contributes to the ability of osteosarcoma cells to metastasize. Cancer Treat Res 152:479–496 [DOI] [PubMed] [Google Scholar]
- 90.Zhang Z, Wang H, Ikeda S, Fahey F, Bielenberg D, Smits P, Hauschka PV (2010) Notch3 in human breast cancer cell lines regulates osteoblast-cancer cell interactions and osteolytic bone metastasis. Am J Pathol 177:1459–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sethi N, Dai X, Winter CG, Kang Y (2011) Tumor-derived JAGGED1 promotes osteolytic bone metastasis of breast cancer by engaging notch signaling in bone cells. Cancer Cell 19:192–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zayzafoon M, Abdulkadir SA, McDonald JM (2004) Notch signaling and ERK activation are important for the osteomimetic properties of prostate cancer bone metastatic cell lines. J Biol Chem 279:3662–3670 [DOI] [PubMed] [Google Scholar]
- 93.Ryeom SW (2011) The cautionary tale of side effects of chronic Notch1 inhibition. J Clin Invest 121:508–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.De Strooper B, Annaert W, Cupers P, et al. (1999) A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature 398:518–522 [DOI] [PubMed] [Google Scholar]
- 95.Ilagan MX, Kopan R (2013) Selective blockade of transport via SERCA inhibition: the answer for oncogenic forms of Notch? Cancer Cell 23:267–269 [DOI] [PubMed] [Google Scholar]
- 96.Moellering RE, Cornejo M, Davis TN, Del BC, Aster JC, Blacklow SC, Kung AL, Gilliland DG, Verdine GL, Bradner JE (2009) Direct inhibition of the NOTCH transcription factor complex. Nature 462:182–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li K, Li Y, Wu W, et al. (2008) Modulation of Notch signaling by antibodies specific for the extracellular negative regulatory region of NOTCH3. J Biol Chem 283:8046–8054 [DOI] [PubMed] [Google Scholar]
- 98.Zanotti S, Canalis E (2010) Notch and the Skeleton. Mol Cell Biol 30:886–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Aste-Amezaga M, Zhang N, Lineberger JE, et al. (2010) Characterization of Notch1 antibodies that inhibit signaling of both normal and mutated Notch1 receptors. PLoS One 5:e9094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yan M, Callahan CA, Beyer JC, Allamneni KP, Zhang G, Ridgway JB, Niessen K, Plowman GD (2010) Chronic DLL4 blockade induces vascular neoplasms. Nature 463:E6–7 [DOI] [PubMed] [Google Scholar]