Abstract
Human genetic studies have long been vastly Eurocentric, raising a key question about the generalizability of these study findings to other populations. Because humans originated in Africa, these populations retain more genetic diversity, and yet individuals of African descent have been tremendously underrepresented in genetic studies. The diversity in Africa affords ample opportunities to improve fine-mapping resolution for associated loci, discover novel genetic associations with phenotypes, build more generalizable genetic risk prediction models, and better understand the genetic architecture of complex traits and diseases subject to varying environmental pressures. Thus, it is both ethically and scientifically imperative that geneticists globally surmount challenges that have limited progress in African genetic studies to date while meaningfully including African investigators, as greater inclusivity and enhanced research capacity affords enormous opportunities to accelerate genomic discoveries that translate more effectively to all populations. We review the advantages, challenges, and examples of genetic architecture studies of complex traits and diseases in Africa. For example, with greater genetic diversity comes greater ancestral heterogeneity; this higher level of understudied diversity can yield novel genetic findings, but some methods that assume homogeneous population structure and work well in European populations may work less well in the presence of greater heterogeneity in African populations. Consequently, we advocate for methodological development that will accelerate studies important for all populations, especially those currently underrepresented in genetics.
Keywords: genetic architecture, Africa, GWAS, health disparities
Historical biases in genetic studies
Nearly a decade ago, 96% of participants in genome-wide association studies (GWAS) were of European descent [1]. While European individuals now account for 78% of GWAS participants [2], the non-European proportion has stagnated since 2014. African ancestry individuals constitute merely 2.4% of participants (although notably account for 7% of all associations) [2]. This participant bias results in interpretability gaps by ancestry with medically relevant consequences [3,4]. For example, while easily avoidable, African American patients were more likely than white Americans to be incorrectly told they have a genetic mutation that increases their risk of hypertrophic cardiomyopathy, an early-onset life-threatening heart disease, at leading genetic testing labs [5]. Additionally, drug metabolism genes such as CYP3A4 contain mutations that can alter dosage requirements, but pharmacogenetic variants are disproportionately uncatalogued among African populations [6], so genotype-based dosage guidelines are less useful. In the US, the National Human Genome Research Institute has prioritized increased diversity in genetic studies [7]. This prioritization is an important step that, if heeded, will aid interpretations in medical genomics for all ethnicities [8]. Greater inclusivity of African populations in medical genomics is important for accelerating genomic discoveries, enabling reconstruction of modern human origins, producing results that can be translated across populations more accurately, identifying genetic associations with traits for variants absent elsewhere, and building research capacity in Africa.
Genetic study biases have not happened in a vacuum, but have had widespread consequences for GWAS tools and resources in African populations. Genotyping arrays have traditionally been biased towards alleles most frequent and imputable in European populations [9,10], compounding biases in which GWAS identify variant associations most common in the study population [11,12]. In contrast, array backbones prioritizing SNPs that maximally tag variants across all populations improve imputation performance, providing more even genomic coverage [13]. Perhaps more importantly, imputation panels are vastly Eurocentric, shortchanging representation of the greater haplotypic diversity present in Africans from deeper recombination history [12,14,15]. The most widely available African sequencing resources have biased representation towards African Americans and West Africans [8,12], leaving huge swaths of African diversity uncatalogued.
Existing challenges to surmount for African genetics studies
To empower African genetic studies and build capacity for research aiding biological understanding across a diverse swath of humanity, we review challenges that need to be confronted and continually addressed.
Historical
Africa has long been subjected to a violent and oppressive colonial history that has bred suspicion and an anticipation of resource exploitation. This understandable mistrust continues to strain ongoing relations, with new actors such as China in addition to European groups scrambling for African resources [16,17]. The impact on research collaborations is evident, with some authors discussing ‘neo-colonial science’ [18]. Such strained relations are more pronounced in collaborations involving genetic studies, especially when shipping samples out of Africa and the global south [19]. Some discuss ‘genomic sovereignty’ of Africans and ownership of African genetic material [20]. Proponents of international collaborations argue that working with high income countries will eventually ensure equity, justice, and benefit to Africans, with capacity building for genomic research providing immediate benefit for African institutions [21], although concerns have been raised about the sustainability of these efforts. Ongoing tensions weigh the benefit to Africans by including more African researchers and DNA in global research against the challenges of promoting African science while integrating and importing the best science around the world into Africa.
Infrastructural
Conducting genetic studies in Africa is not an easy task. Infrastructural problems can include unreliable or no electricity in clinics and laboratories that process samples, impassable roads in some areas, and crime or political instability making some areas dangerous and/or inaccessible for researchers. Many African countries do not have sufficient laboratory equipment or facilities for genomics research, and most require imported reagents. Importing is not only time-consuming, but also costly—reagents are often many times more expensive in Africa than Western countries in real terms, not including shipment costs. Biobanks are less abundant, partially due to power interruptions affecting storage and processing of samples. Some African institutions have experience in large-scale human genetic analyses; the H3ABionet consortium has developed core bioinformatics infrastructure in Africa [22]. However, high-speed internet connections and powerful computers are not always available to access large data files. Human resource issues can also be a challenge, namely high staff turnover due to inadequate pay, competing demands for time from qualified staff, and/or too few qualified staff. Relatedly, brain drain is a major issue, as many skilled African scientists leave the continent in search of greener pastures [23,24]. To be sensitive to these challenges, some major international research initiatives such as H3Africa have required a relatively long embargo period on publication for African researchers [25]. Connecting African researchers to adequate computing power (e.g., stable wireless connections to cloud computing) may offer more direct means to facilitate research. Compared with the relative ease of acquiring samples in the global north, the focus of databanks on European/white populations is unsurprising, but it is nonetheless imperative that researchers rise to these challenges for the benefit of all.
Funding
Genetics research is expensive, and a lack of attention from African policy makers in resource-limited settings is primarily driven by competing priorities for more immediate public health concerns, including infectious diseases over inherited conditions [26,27]. Data generation is still the most expensive part of genomics, whereas data analysis is more affordable and therefore a viable option for capacity building [28]. Furthermore, journals from the developed world often exist behind expensive pay-walls that are inaccessible to some researchers and do not always encourage publication of work from the global south, often returning manuscripts without review citing a lack of “sufficient general interest.” Having fewer publications has a knock-on effect on future grant funding and attracting students.
Nearly all funding for genetics research comes from outside Africa, raising questions for African scientists about the utility of investigating disease genetics with less long-term funding security and intellectual freedom to prioritize their field of study. Incentives differ from the West, heavily favoring medicine over research training—clinical demands are heavier, PhD programs are scarce, and research often does not pay. However, some external research funding in genomics, most notably by the Human Heredity and Health in Africa (H3Africa) Initiative, are being led by African scientists. H3Africa funding by the NIH (USA) and Wellcome Trust (UK) totals more than $216 million in 2015 for 185 projects in 28 African countries [29]. Its aim is to build the capacity for African scientists to conduct genomic research on heritable diseases afflicting Africans [30,31]. This international support is essential for African geneticists to continue their research [21].
Ethical
Ethics review boards may lack familiarity with genomics research, which creates challenges for advising long-term, large-scale collaborative genetic studies that can in turn delay funded projects [28]. These challenges are partially driven by restrictive ethical guidelines and uncertainty about the benefits of such studies to African populations [28]. Unlike in the US, genetics projects are subject to ethics review both at the provincial and national levels as a legacy of colonialism, which can lead to year-long delays. Ethics approval by regulatory bodies in Africa is mostly restricted to project-specific research questions, often raising questions around ‘broad consent’ and ‘indefinite storage’ of samples that are not easy to answer. A primary concern about loss of control and ownership over the DNA samples arises when they are shipped abroad [32]. Burgeoning interest in building large-scale genomics collaborations in Africa has resulted in a recent best practices ethical framework for genomics research and biobanking in Africa [25].
Some communities have set up local councils to oversee research projects and publication of results allowed from the research [33]. While these are excellent in theory, in practice there can be long delays, misunderstanding due to unfamiliarity of lay people with jargon, and a lack of continuity in leadership. Consequently, even when extensive consultation on planned or existing research projects has taken place, this often needs to be repeated at each subsequent visit. New council leaders sometimes try to enforce sample destruction before allowing further sampling, even when consent forms specify long sample storage. A middle ground of continuous community leadership from members more familiar with research methodology and terminology that is acceptable to the council would be ideal, but is often infeasible. Furthermore, while returning scientific discoveries to communities or participants should be the norm, re-contacting study participants in communities can be challenging as people lose cell phones or move for employment opportunities.
Respect and consent
To ensure mutual respect in collaborative African genetics studies, it is important to avoid generalizing “African-ness” in such a vast continent, comprising not only more genetic diversity than the rest of the world, but also many cultures, language groups, and world views, some of which are marginalized or discriminated against. Thus, it is important to obtain perspectives from diverse continental Africans when communicating science broadly. Furthermore, meaningful engagement with African colleagues is vital to healthy collaborations and to avoid tokenism. Additionally, obtaining informed consent for genomics research can be complex in any setting, but poses more challenges where there are lower income and literacy levels or language barriers. Furthermore, some diseases such as mental illness are subject to greater stigma in some African communities, requiring cultural awareness and sensitivity to differences. Participants may misunderstand the study purpose or expect benefits that are not included, such as better disease treatment [34] or individual-level ancestry results useful for land claims. Additionally, in some African societies, decisions to participate in research studies are made collectively as well as at the individual level [35], necessitating consultation with community leaders.
Communicating science respectfully can be challenging when nomenclature is subject to sociopolitical debate, as with the descendants of the original hunter-gatherers of Southern Africa. In an attempt to be politically correct, many population geneticists use “KhoeSan” to refer to the Khoe and San groups collectively. However, the San Council of Southern Africa prefers to keep these terms separate (i.e., San and Khoe or Nama) to denote different cultures. Many “San” individuals prefer being called “Bushmen,” while others consider the word to be pejorative. Labels are only useful insofar as they are universally informative, and respect is imperative. Wide pre-publication consultation is obviously necessary [36], but complete consensus is unlikely.
GWAS design challenges in Africa
Unlike most of the GWAS and complex trait studies that have been conducted in Europe, assumptions of homogeneous population structure are more likely to be violated in Africa, as few populations have remained isolated and unchanged over the past 4000 years [37]. This higher level of diversity across African populations relative to others [38,39] creates greater challenges when attempting to balance case/control collections at the outset of many studies due to greater complexities in population structure, including variable LD patterns between study sites. Consequently, false positives are more likely to arise from confounding due to unaccounted population stratification, especially for rare variants, which are challenging to analyze [40,41]. Higher rates of genetic diversity also result in a larger number of effective tests, meaning that the standard multiple testing threshold of p < 5e-8 needs to be roughly twice as stringent in African GWAS (p < ~2.5e-8) [42]. Additional challenges arise from a dearth of large, easily accessible reference panels in Africa. While the African Genome Variation Project and related projects have worked to ameliorate this gap, data access is somewhat more challenging and slower than the publicly available 1000 Genomes Project [15].
Because some complex trait genetics methods assume homogeneity that is more often violated in African populations with higher diversity, methodological advancements that explicitly account for structure over a range of time periods will be especially useful [43]. For example, heritability estimates in the presence of admixture can be biased and inflated [44]. Alternatively, higher heritability estimates may be driven by higher relatedness among geographically proximal individuals. The presence of structure can create challenges disentangling the heritable component due to genetics versus similar environments [44]. Other methods for inferring heritability (e.g. LD score regression) are suboptimal in the presence of admixture, as LD from these populations are often not reflective of the study cohort and vary locally [45,46]. Other methods for inferring genetic architecture, including Bayesian linear mixed models (LMMs) such as the Bayesian sparse LMM (BSLMM), Bayes R, and BOLT-LMM, have been shown to be effective at controlling population stratification, cryptic relatedness, and also increase power in structured populations [47-50]. These studies demonstrate that more advanced GWAS methods may be more fruitful generally, but especially in Africa where higher rates of substructure are typical.
Successful GWAS strategies in African and African descent populations
Despite these challenges, many successful examples illuminate paths forward. Due to high prioritization of infectious disease studies, most positive examples exist for genetic susceptibility studies, including of tuberculosis [51-53], malaria [54], sickle cell disease modifiers [55], HIV [56-59], nontyphoidal Salmonella [60], and trypanosomes [61]. Significant findings were aided by simpler genetic architectures and higher genetic risk divergence between endemic cases vs high-risk controls due to natural selection. Some challenges of studying these evolutionarily important traits, however, are high levels of genetic diversity in the parasite and variable LD patterns among populations, sometimes necessitating specialized association approaches that allow for multiple independent origins of resistance loci and/or allelic heterogeneity [54]. Some anthropometric studies have faced similar challenges and advantages due to high divergence, natural selection, and genetic architectures, such as in skin pigmentation [62,63]. In smaller cohorts that are underpowered for discovering individual loci, gene-based associations can sometimes be useful in conjunction with functional follow up [64]. Studies of traits with elevated prevalence in African Americans, such as BMI, prostate cancer, and low birth weight [65-67] have analyzed genome-wide significant loci by local ancestry or more easily fine-mapped variants with narrower LD.
Additionally, multiethnic studies including African Americans have demonstrated the utility of integrative genomics approaches for fine-mapping, e.g. with pulmonary function variants [68]. Several recent GWAS reviewed here have used linear mixed models, with a random effect to account for genetic relatedness. These models are useful but can produce inflated heritability estimates, which can be corrected using a second random effect to measure spatial distance as a proxy for environmental effects [69].
Advantages and opportunities for genetic architecture studies in Africa
The opportunities for large-scale genetic studies in Africa are ample. Growing inclusion of African Americans in medical genomics studies is crucial, but still leaves behind many populations and large swaths of sub-Saharan African genetic diversity, which may greatly increase our understanding of complex trait genetic architecture [70]. There is more genetic and often phenotypic diversity in Africa that has been understudied, meaning there is considerable low-hanging fruit for novel findings and insights into the genetic architectures and etiologies of complex traits. More rapid LD decay in Africa also means there is greater fine-mapping resolution to pinpoint causal variants influencing traits than will be discovered in any other global population [71], as reviewed recently [72]. For example, several variants in TCF7L2 were associated with type 2 diabetes in European and East Asian populations in the early GWAS era, but candidate loci were narrowed considerably via comparison with more diverse West African cohorts, even with smaller cohort sizes [73].
Major opportunities also present themselves in precision medicine. For example, polygenic risk scores have been of growing interest as large-scale GWAS now offer low-cost tests that can outpace the clinical status quo [74,75]. However, these scores generalize poorly across diverse populations [11]. European GWAS results consistently predict genetic risk several-fold less accurately in non-Europeans, performing the worst in African Americans (and by extension, likely even worse in eastern, central, and southern African populations) [76-79]. A typical but somewhat misguided argument in favor of immediate translational implementation of polygenic risk scores is that standard clinical lab tests from blood panels are often differentially informative across ethnicities and more reliable in European descent populations. However, interpretability gaps for current clinical tests are less acutely and consistently worse in non-European populations than genetic risk prediction; the underlying biology remains the same, such that for all diseases, drugs do not routinely work many-fold better in European than African-descent populations. Further, new population-specific interpretation of common clinical lab tests enables better prognostic value than existing reference intervals [80]. In contrast, the most significant and highest frequency genetic variants from GWAS used to predict genetic risk are not likely to be the same across populations, even when the underlying causal variants are the same. This is due to GWAS discovery biases, as variants used to predict risk tend to explain more phenotypic variation in the study population. While improved analytical methods hold promise, the only way genetic prediction power of inherited diseases in non-Europeans can truly be made equal is with massive investments to produce similar-sized GWAS of these phenotypes in non-European populations. Additionally, discoveries based on African genetics contribute to global knowledge, but many African population groups are sufficiently different [37] that insights made from trans-ethnic studies can similarly be gained by analyzing multiple GWAS of different African populations.
As a major genetics mission is to understand the biological basis and evolutionary origins of diseases and traits and use this knowledge to perform biologically-informed drug discovery, human evolution tells us that Africa has a huge role to play. Progress so far has been slower due to a need for increased capacity and collaborative engagement with African investigators. Several outstanding examples of this potential already exist, such as the Southern African Human Genome Programme (SAHGP), one of the first genetic architecture studies of African participants fully funded and analyzed by Africans [81]. International collaborations have also blazed the trail for meaningful collaborations with deep investments in building research capacity in human genomics, such as MalariaGEN, partnerships by the African Center of Excellence for Genomics of Infectious Diseases (ACEGID), as well as the Global Initiative for Neuropsychiatric Genetics Education in Research (GINGER) program. Calls from African researchers for funding and building research capacity in genetics [70,82] should be thoughtfully heeded to ensure that those with the greatest public health needs are not the last to benefit.
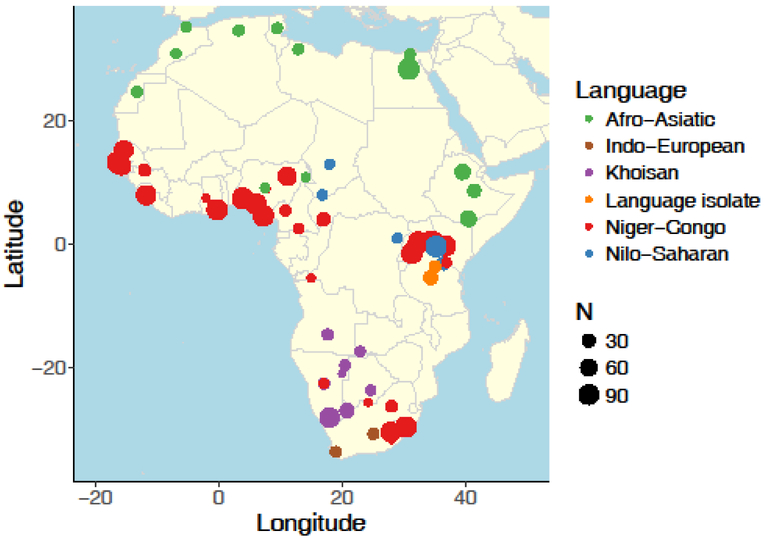
Figure 1 – Map of publicly available African samples and corresponding language families from previous studies.
Reference data comes from several previous studies [12,15,38,39,83-87]
Acknowledgments
We thank Brenna Henn and Lluis Quintana-Murci for inviting this review. We also thank Karestan Koenen, Lori Chibnik, and Anne Stevenson for their thoughtful comments. Research reported in this publication was supported by NIH K99MH117229.
Footnotes
Disclosures
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Need AC, Goldstein DB: Next generation disparities in human genomics: concerns and remedies. Trends in Genetics 2009, 25:489–494. [DOI] [PubMed] [Google Scholar]
- 2.Morales J, Welter D, Bowler EH, Cerezo M, Harris LW, McMahon AC, Hall P, Junkins HA, Milano A, Hastings E, et al. : A standardized framework for representation of ancestry data in genomics studies, with application to the NHGRI-EBI GWAS Catalog. 2018, doi: 10.1186/s13059-018-1396-2. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Provides a framework for describing ancestry in genetic studies, and describes the disproportionately high contribution of associations identified in African and Hispanic/Latin American populations.
- 3.Bustamante CD, La Vega De FM, Burchard EG: Genomics for the world. Nature 2011, 475:163–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petrovski S, Goldstein DB: Unequal representation of genetic variation across ancestry groups creates healthcare inequality in the application of precision medicine [Internet], Genome Biol. 2016, 17:157–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manrai AK, Funke BH, Rehm HL, Olesen MS, Maron BA, Szolovits P, Margulies DM, Loscalzo J, Kohane IS: Genetic Misdiagnoses and the Potential for Health Disparities. N Engl J Med 2016, 375:655–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bl Drögemöller, Wright GE, Niehaus DJ, Koen L, Malan S, Da Silva DM, Hillermann Rebello R, La Grange AM, Venter M, Warnich L: Characterization of the genetic profile of CYP2C19in two South African populations. Pharmacogenomics 2010, 11:1095–1103. [DOI] [PubMed] [Google Scholar]
- 7.Hindorff LA, Bonham VL, Brody LC, Ginoza MEC, Hutter CM, Manolio TA, Green ED: Prioritizing diversity in human genomics research. Nature Publishing Group 2017, doi: 10.1038/nrg.2017.89. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Reviews NIH NHGRI efforts to prioritize diversity in human genomics research.
- 8.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. : Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016, 536:285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Albrechtsen A, Nielsen FC, Nielsen R: Ascertainment biases in SNP chips affect measures of population divergence. Mol Biol Evol 2010, 27:2534–2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin AR, Tse G, Bustamante CD, Kenny EE: Imputation-based assessment of next generation rare exome variant arrays. Pac Symp Biocomput 2014, doi: 10.1534/g3.113.007161. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Demonstrates that genetic risk prediction from Eurocentric studies is expected to generalize poorly to non-European populations with increasing genetic divergence.
- 11.Martin AR, Gignoux CR, Walters RK, Wojcik GL, Neale BM, Gravel S, Daly MJ, Bustamante CD, Kenny EE: Human Demographic History Impacts Genetic Risk Prediction across Diverse Populations. Am. J. Hum. Genet. 2017, 100:635–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, et al. : A global reference for human genetic variation. Nature 2015, 526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wojcik GL, Fuchsberger C, Taliun D, Welch R, Martin AR, Shringarpure S, Carlson CS, Abecasis G, Kang HM, Boehnke M, et al. : Imputation aware tag SNP selection to improve power for multi-ethnic association studies. 2017, doi: 10.1101/105551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, Kang HM, Fuchsberger C, Danecek P, Sharp K, et al. : A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet 2016, 48:1279–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gurdasani D, Carstensen T, Tekola-Ayele F, Pagani L, Tachmazidou I, Hatzikotoulas K, Karthikeyan S, lies L, Pollard MO, Choudhury A, et al. : The African Genome Variation Project shapes medical genetics in Africa. Nature 2015, 517:327–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langan M: Normative Power Europe and the Moral Economy of Africa-EU Ties: A Conceptual Reorientation of “Normative Power.” New Political Economy 2012, 17:243–270. [Google Scholar]
- 17.Alden C: China in Africa. Survival 2005, 47:147–164. [Google Scholar]
- 18.Dahdouh-Guebas F, Ahimbisibwe J, Van Moll R, Koedam N: Neo-colonial science by the most industrialised upon the least developed countries in peer-reviewed publishing. Scientometrics 2003, 56:329–343. [Google Scholar]
- 19.Serwadda D, Ndebele P, Grabowski MK, Bajunirwe F, Wanyenze RK: Open data sharing and the Global South—Who benefits? Science 2018, 359:642–643. [DOI] [PubMed] [Google Scholar]
- 20.de Vries J, Pepper M: Genomic sovereignty and the African promise: mining the African genome for the benefit of Africa. J Med Ethics 2012, 38:474–478. [DOI] [PubMed] [Google Scholar]
- 21.Munung NS, Mayosi BM, de Vries J: Equity in international health research collaborations in Africa: Perceptions and expectations of African researchers. PLoS ONE 2017, 12:e0186237–17. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Collated African perspectives on genomics research equity in international collaborations.
- 22.Mulder NJ, Adebiyi E, Adebiyi M, Adeyemi S, Ahmed A, Ahmed R, Akanle B, Alibi M, Armstrong DL, Aron S, et al. : Development of Bioinformatics Infrastructure for Genomics Research. Global Heart 2017, 12:91–98. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Describes the development and impact of key bioinformatics infrastructure as part of H3Africa capacity building.
- 23.Ochola LI, Gitau E: Challenges in retaining research scientists beyond the doctoral level in Kenya. PLoS Negl Trop Dis 2009, 3:e345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adebamowo SN, Francis V, Tambo E, Diallo SH, Landouré G, Nembaware V, Dareng E, Muhamed B, Odutola M, Akeredolu T, et al. : Implementation of genomics research in Africa: challenges and recommendations. Glob Health Action 2018, 11:1419033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.A welcome framework for research in Africa. Nature 2018, 556:274. [DOI] [PubMed] [Google Scholar]; • An important ethics guideline developed through in-depth conversations with African researchers and ethics review boards to empower African researchers and communities by suggesting rules of engagement for international genomics partnerships.
- 26.Wonkam A, Mayosi BM: Genomic medicine in Africa: promise, problems and prospects. Genome Med 2014, 6:11–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uren C, Möller M, Helden PD, Henn BM, Hoal EG: Population structure and infectious disease risk in southern Africa. Molecular Genetics and Genomics 2017, 292:499–509. [DOI] [PubMed] [Google Scholar]
- 28.de Vries J, Bull SJ, Doumbo O, Ibrahim M, Mercereau-Puijalon O, Kwiatkowski D, Parker M: Ethical issues in human genomics research in developing countries. BMC Med Ethics 2011, 12:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coles E, Mensah GA: Geography of Genetics and Genomics Research Funding in Africa. Global Heart 2017, 12:173–176. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Breaks down international research funding and collaborations in Africa
- 30.H3Africa Consortium, Rotimi C, Abayomi A, Abimiku A, Adabayeri VM, Adebamowo C, Adebiyi E, Ademola AD, Adeyemo A, Adu D, et al. : Research capacity. Enabling the genomic revolution in Africa. Science 2014, 344:1346–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Vries J, Tindana P, Littler K, Ramsay M, Rotimi C, Abayomi A, Mulder N, Mayosi BM: The H3Africa policy framework: negotiating fairness in genomics. Trends in Genetics 2015, 31:117–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tindana P, Molyneux CS, Bull S, Parker M: Ethical issues in the export, storage and reuse of human biological samples in biomedical research: perspectives of key stakeholders in Ghana and Kenya. BMC Med Ethics 2014, 15:585–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nordling L: San people of Africa draft code of ethics for researchers. Science 2017, doi: 10.1126/science.aal0933. [DOI] [Google Scholar]
- 34.Masiye F, Mayosi B, de Vries J: “I passed the test!” Evidence of diagnostic misconception in the recruitment of population controls for an H3Africa genomic study in Cape Town, South Africa. 2017, doi: 10.1186/s12910-017-0175-z.• Discusses consent issues and diagnostic misconceptions in resource-limited settings in Africa.
- 35.Rotimi CN, Marshall PA: Tailoring the process of informed consent in genetic and genomic research. Genome Med 2010, 2:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chennells R, Steenkamp A: International Genomics Research Involving the San People. Springer International Publishing; 2017. [Google Scholar]
- 37.Busby GB, Band G, Si Le Q, Jallow M, Bougama E, Mangano VD, Amenga-Etego LN, Enimil A, Apinjoh T, Ndila CM, et al. : Admixture into and within sub-Saharan Africa. eLife 2016, 5: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Henn BM, Gignoux CR, Jobin M, Granka JM, Macpherson JM, Kidd JM, Rodriguez-Botigue L, Ramachandran S, Hon L, Brisbin A, et al. : Hunter-gatherer genomic diversity suggests a southern African origin for modern humans. Proc. Natl. Acad. Sci. U.S.A. 2011, 108:5154–5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Altshuler DM, Gibbs RA, Dermitzakis E, Peltonen L, Dermitzakis E, Bonnen PE, de Bakker PIW, Deloukas P, Gabriel SB, Gwilliam R, et al. : Integrating common and rare genetic variation in diverse human populations. Nature 2010, 467:52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mathieson I, McVean G: Demography and the Age of Rare Variants. PLoS Genet 2014, 10:e1004528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mathieson I, McVean G: Differential confounding of rare and common variants in spatially structured populations. Nat Genet 2012, 44:243–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pe’er I, Yelensky R, Altshuler D, Daly MJ: Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet. Epidemiol. 2008, 32:381–385. [DOI] [PubMed] [Google Scholar]
- 43.Duncan LE, Ratanatharathorn A, Aiello AE, Almli LM, Amstadter AB, Ashley-Koch AE, Baker DG, Beckham JC, Bierut LJ, Bisson J, et al. : Largest GWAS of PTSD (N=20 070) yields genetic overlap with schizophrenia and sex differences in heritability. Molecular Psychiatry 2017, doi: 10.1038/mp.2017.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Browning SR, Browning BL: Population Structure Can Inflate SNP-Based Heritability Estimates. The American Journal of Human Genetics 2011, 89:191–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bulik-Sullivan BK, Loh P-R, Finucane HK, Ripke S, Yang J, Patterson N, Daly MJ, Price AL, Neale BM: LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 2015, 47:291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park DS, Brown B, Eng C, Huntsman S, Hu D, Torgerson DG, Burchard EG, Zaitlen N: Adapt-Mix: learning local genetic correlation structure improves summary statistics-based analyses. Bioinformatics 2015, 31: i181–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lloyd-Jones LR, Robinson MR, Moser G, Zeng J, Beleza S, Barsh GS, Tang H, Visscher PM: Inference on the Genetic Basis of Eye and Skin Color in an Admixed Population via Bayesian Linear Mixed Models. Genetics 2017, 206:1113–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Applied Bayesian linear mixed models to an admixed population in Cape Verde, an island nation of West Africa, to account for population stratification, cryptic relatedness, and increase statistical power. These results have important implications for GWAS in structured populations.
- 48.Moser G, Lee SH, Hayes BJ, Goddard ME, Wray NR, Visscher PM: Simultaneous Discovery, Estimation and Prediction Analysis of Complex Traits Using a Bayesian Mixture Model. PLoS Genet 2015, 11:e1004969–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou X, Carbonetto P, Stephens M: Polygenic modeling with bayesian sparse linear mixed models. PLoS Genet 2013, 9:e1003264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Loh P-R, Tucker G, Bulik-Sullivan BK, Vilhjálmsson BJ, Finucane HK, Salem RM, Chasman DI, Ridker PM, Neale BM, Berger B, et al. : Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat Genet 2015, 47:284–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thye T, Vannberg FO, Wong SH, Owusu-Dabo E, Osei I, Gyapong J, Sirugo G, Sisay-Joof F, Enimil A, Chinbuah MA, et al. : Genome-wide association analyses identifies a susceptibility locus for tuberculosis on chromosome 18q11.2. Nat Genet 2010, 42:739–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chimusa ER, Zaitlen N, Daya M, Moller M, van Helden PD, Mulder NJ, Price AL, Hoal EG: Genome-wide association study of ancestry-specific TB risk in the South African Coloured population. Human Molecular Genetics 2014, 23:796–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Curtis J, Luo Y, Zenner HL, Cuchet-Lourenço D, Wu C, Lo K, Maes M, Alisaac A, Stebbings E, Liu JZ, et al. : Susceptibility to tuberculosis is associated with variants in the ASAP1 gene encoding a regulator of dendritic cell migration. Nat Genet 2015, 47:523–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Malaria Genomic Epidemiology Network: A novel locus of resistance to severe malaria in a region of ancient balancing selection. Nature 2015, 526:253–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mtatiro SN, Singh T, Rooks H, Mgaya J, Mariki H, Soka D, Mmbando B, Msaki E, Kolder I, Thein SL, et al. : Genome Wide Association Study of Fetal Hemoglobin in Sickle Cell Anemia in Tanzania. PLoS ONE 2014, 9:e111464–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Coussens AK, Naude CE, Goliath R, Chaplin G, Wilkinson RJ, Jablonski NG: High-dose vitamin D3 reduces deficiency caused by low UVB exposure and limits HIV-1 replication in urban Southern Africans. [Internet], Proc. Natl. Acad. Sci. U.S.A. 2015, 112:8052–8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Petrovski S, Fellay J, Shianna KV, Carpenetti N, Kumwenda J, Kamanga G, Kamwendo DD, Letvin NL, McMichael AJ, Haynes BF, et al. : Common human genetic variants and HIV-1 susceptibility: a genome-wide survey in a homogeneous African population. AIDS 2011, 25:513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carr DF, Bourgeois S, Chaponda M, Takeshita LY, Morris AP, Castro EMC, Alfirevic A, Jones AR, Rigden DJ, Haldenby S, et al. : Genome-wide association study of nevirapine hypersensitivity in a sub-Saharan African HIV-infected population. J. Antimicrob. Chemother. 2017, 27:dkw545–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Asiki G, Murphy G, Nakiyingi-Miiro J, Seeley J, Nsubuga RN, Karabarinde A, Waswa L, Biraro S, Kasamba I, Pomilla C, et al. : The general population cohort in rural south-western Uganda: a platform for communicable and non-communicable disease studies. International Journal of Epidemiology 2013, 42:129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gilchrist J J, Rautanen A, Fairfax BP, Mills TC, Naranbhai V, Trochet H, Pirinen M, Muthumbi E, Mwarumba S, Njuguna P, et al. : Risk of nontyphoidal Salmonella bacteraemia in African children is modified by STAT4. Nature Communications 2018, doi: 10.1038/s41467-017-02398-z. [DOI] [PMC free article] [PubMed] [Google Scholar]; • In a GWAS of nontyphoidal Salmonella, a common and frequently fatal cause of bacteraemia in children and HIV-infected adults in Africa, this study identified a significant susceptibility locus in STAT4 in Kenyan cases and controls with replication in Malawians. This eQTL modulates interferon-γ production in stimulated natural killer cells, highlighting the shared genetic architecture of infectious and autoimmune disease.
- 61.Genovese G, Friedman DJ, Ross MD, Lecordier L, Uzureau P, Freedman BI, Bowden DW, Langefeld CD, Oleksyk TK, Uscinski Knob AL, et al. : Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science 2010, 329:841–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martin AR, Lin M, Granka JM, Myrick JW, Liu X, Sockell A, Atkinson EG, Werely CJ, Möller M, Sandhu MS, et al. : An Unexpectedly Complex Architecture for Skin Pigmentation in Africans. Cell 2017, 171:1340–1353.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Along with [63], one of the two first studies of skin pigmentation in continental Africa, which showed that skin pigmentation is more polygenic in KhoeSan populations than described elsewhere previously.
- 63.Crawford NG, Kelly DE, Hansen MEB, Beltrame MH, Fan S, Bowman SL, Jewett E, Ranciaro A, Thompson S, Lo Y, et al. : Loci associated with skin pigmentation identified in African populations. Science 2017, 358:eaan8433–26. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Along with [62], one of the two first studies of skin pigmentation in continental Africa, which showed functional evidence of a role for MFSD12 in pigmentation.
- 64.Kilaru V, Iyer SV, Almli LM, Stevens JS, Lori A, Jovanovic T, Ely TD, Bradley B, Binder EB, Koen N, et al. : Genome-wide gene-based analysis suggests an association between Neuroligin 1 (NLGN1) and post-traumatic stress disorder. Translational Psychiatry 2016, 6:e820–e820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Monda KL, Chen GK, Taylor KC, Palmer C, Edwards TL, Lange LA, Ng MCY, Adeyemo AA, Allison MA, Bielak LF, et al. : A meta-analysis identifies new loci associated with body mass index in individuals of African ancestry. Nat Genet 2013, 45:690–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Haiman CA, Chen GK, Blot WJ, Strom SS, Berndt SI, Kittles RA, Rybicki BA, Isaacs WB, Ingles SA, Stanford JL, et al. : Genome-wide association study of prostate cancer in men of African ancestry identifies a susceptibility locus at 17q21. Nat Genet 2011, 43:570–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ochs-Balcom HM: Admixture mapping and fine-mapping of birth weight loci in the Black Women’s Health Study. Human Genetics 2018, 137:535–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wyss AB, Sofer T, Lee MK, Terzikhan N, Nguyen JN, Lahousse L, Latourelle JC, Smith AV, Bartz TM, Feitosa MF, et al. : Multiethnic meta-analysis identifies ancestry- specific and cross-ancestry loci for pulmonary function. Nature Communications 2018, doi: 10.1038/s41467-018-05369-0. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This GWAS of pulmonary function identified 60 novel genetic loci, a 50% increase in the number of known loci, with 8 identified in African ancestry and 34 from multi-ethnic meta-analyses. Notably, fine-mapping of loci was improved by incorporating LD, functional data, and/or multi-ethnic analysis.
- 69.Heckerman D, Gurdasani D, Kadie C, Pomilla C, Carstensen T, Martin H, Ekoru K, Nsubuga RN, Ssenyomo G, Kamali A, et al. : Linear mixed model for heritability estimation that explicitly addresses environmental variation. Proc Natl Acad Sci USA 2016, 113:7377–7382. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This study showed that using standard linear mixed models with kinship as a random effect to estimate heritability can produce inflated estimates, but that including a second random effect, a covariance matrix approximating geographical distance as an environmental proxy, produces less biased heritability estimates.
- 70.Dalvie S, Koen N, Duncan L, Abbo C, Akena D, Atwoli L, Chiliza B, Donald KA, Kinyanda E, Lochner C, et al. : Large Scale Genetic Research on Neuropsychiatric Disorders in African Populations is Needed. EBIOM 2015, 2:1259–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu Y, Waite LL, Jackson AU, Sheu WH-H, Buyske S, Absher D, Arnett DK, Boerwinkle E, Bonnycastle LL, Carty CL, et al. : Trans-Ethnic Fine-Mapping of Lipid Loci Identifies Population-Specific Signals and Allelic Heterogeneity That Increases the Trait Variance Explained. PLoS Genet 2013, 9:e1003379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rotimi CN, Bentley AR, Doumatey AP, Chen G, Shriner D, Adeyemo A: The genomic landscape of African populations in health and disease. Human Molecular Genetics 2017, 26:R225–R236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Helgason A, Palsson S, Thorleifsson G, Grant SFA, Emilsson V, Gunnarsdottir S, Adeyemo A, Chen Y, Chen G, Reynisdottir I, et al. : Refining the impact of TCF7L2 gene variants on type 2 diabetes and adaptive evolution. Nat Genet 2007, 39:218–225. [DOI] [PubMed] [Google Scholar]
- 74.Kullo IJ, Jouni H, Austin EE, Brown S-A, Kruisselbrink TM, Isseh IN, Haddad RA, Marroush TS, Shameer K, Olson JE, et al. : Incorporating a Genetic Risk Score Into Coronary Heart Disease Risk Estimates: Effect on Low-Density Lipoprotein Cholesterol Levels (the MI-GENES Clinical Trial). Circulation 2016, 133:1181–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Natarajan P, Young R, Stitziel NO, Padmanabhan S, Baber U, Mehran R, Sartori S, Fuster V, Reilly DF, Butterworth A, et al. : Polygenic Risk Score Identifies Subgroup With Higher Burden of Atherosclerosis and Greater Relative Benefit From Statin Therapy in the Primary Prevention SettingClinical Perspective. Circulation 2017, 135:2091–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vilhjálmsson BJ, Yang J, Finucane HK, Gusev A, Lindström S, Genovese G, Loh P-R, Bhatia G, Do R, Hayeck T, et al. : Modeling Linkage Disequilibrium Increases Accuracy of Polygenic Risk Scores. The American Journal of Human Genetics 2015, 97:576–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ware EB, Schmitz LL, Faul JD, Gard A, Mitchell C, Smith JA, Zhao W, Weir D, Kardia SL: Heterogeneity in polygenic scores for common human traits. 2017, doi: 10.1101/106062. [DOI] [Google Scholar]
- 78.Akiyama M, Okada Y, Kanai M, Takahashi A, Momozawa Y, Ikeda M, Iwata N, Ikegawa S, Hirata M, Matsuda K, et al. : Genome-wide association study identifies 112 new loci for body mass index in the Japanese population. Nat Genet 2017, 49:1458–1467. [DOI] [PubMed] [Google Scholar]
- 79.Li Z, Chen J, Yu H, He L, Xu Y, Zhang D, Yi Q, Li C, Li X, Shen J, et al. : Genome-wide association analysis identifies 30 new susceptibility loci for schizophrenia. Nature Publishing Group 2017, 49:1576–1583. [DOI] [PubMed] [Google Scholar]
- 80.Rappoport N, Paik H, Oskotsky B, Tor R, Ziv E, Zaitlen N, Butte AJ: Creating ethnicity-specific reference intervals for lab tests from EHR data. bioRxiv 2017, doi: 10.1101/213892. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This preprint demonstrates the clinical value of defining reference intervals for blood panels not by the standard small (N≈128) patient population with biased ancestry, but by very large patient samples from diverse ancestries (N>10,000). Serum creatinine and hemoglobin A1c in particular produce actionably different reference intervals across self-reported ethnicities.
- 81.Choudhury A, Ramsay M, Hazelhurst S, Aron S, Bardien S, Botha G, Chimusa ER, Christoffels A, Gamieldien J, Sefid-Dashti MJ, et al. : Whole-genome sequencing for an enhanced understanding of genetic variation among South Africans. Nature Communications 2017, doi: 10.1038/s41467-017-00663-9. [DOI] [PMC free article] [PubMed] [Google Scholar]; • A pilot study of deep whole genome sequencing of 24 southern African individuals exhibiting complex, region-specific population structure. This study was led and funded entirely by African investigators and an African government, respectively.
- 82.van der Merwe C, Mwesiga EK, McGregor NW, Ejigu A, Tilahun AW, Kalungi A, Akimana B, Dubale BW, Omari F, Mmochi J, et al. : Advancing neuropsychiatric genetics training and collaboration in Africa. Lancet Glob Health 2018, 6:e246–e247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bryc K, Velez C, Karafet T, Moreno-Estrada A, Reynolds A, Auton A, Hammer M, Bustamante CD, Ostrer H: Genome-wide patterns of population structure and admixture among Hispanic/Latino populations. Proc Natl Acad Sci USA 2010, 107:8954–8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Henn BM, Botigué LR, Gravel S, Wang W, Brisbin A, Byrnes JK, Fadhlaoui-Zid K, Zalloua PA, Moreno-Estrada A, Bertranpetit J, et al. : Genomic Ancestry of North Africans Supports Back-to-Africa Migrations. PLoS Genet 2012, 8:e1002397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pagani L, Schiffels S, Gurdasani D, Danecek P, Scally A, Chen Y, Xue Y, Haber M, Ekong R, Oljira T, et al. : Tracing the Route of Modern Humans out of Africa by Using 225 Human Genome Sequences from Ethiopians and Egyptians. The American Journal of Human Genetics 2015, 96:986–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, Cann HM, Barsh GS, Feldman M, Cavalli-Sforza LL, et al. : Worldwide Human Relationships Inferred from Genome-Wide Patterns of Variation. Science 2008, 319:1100–1104. [DOI] [PubMed] [Google Scholar]
- 87.Schlebusch CM, Skoglund P, Sjodin P, Gattepaille LM, Hernandez D, Jay F, Li S, De Jongh M, Singleton A, Blum MGB, et al. : Genomic Variation in Seven Khoe-San Groups Reveals Adaptation and Complex African History. Science 2012, 338:374–379. [DOI] [PMC free article] [PubMed] [Google Scholar]