Abstract
This is a protocol for a Cochrane Review (Intervention). The objectives are as follows:
To assess the effects of bisphosphonates and RANK‐ligand (RANKL)‐inhibitors for supportive treatment in prostate cancer with bone metastases and to generate a clinically meaningful treatment ranking according to their safety and efficacy.
Background
Description of the condition
Prostate cancer is the second most commonly diagnosed form of cancer and the sixth leading cause of cancer‐related death among men worldwide (Jin 2011). Over the past few decades, improved early stage disease detection and advances in medical treatments have decreased the overall mortality rate of prostate cancer, but its metastatic progression has been found to be the major cause of prostate cancer‐associated morbidity and mortality (Thobe 2011). Researchers have shown that men with prostate cancer metastases have a 29.8% five‐year survival rate as compared to 100% survival rate in men with localised or regional prostate cancer (Howlader 2013). Similar to other cancer diseases, prostate cancer can metastasise to organs like the liver, lungs and brain, but it has a very high affinity for bone metastases which was found to have 80% prevalence in men who have died from prostate cancer (Jin 2011). Bone metastasis affects quality of life; it is painful and causes pathological fractures, spinal cord compression and high calcium levels in the blood (Coleman 1997). Androgen deprivation therapy (ADT), being the mainstay of treatment for men with prostate cancer, has been reported to contribute to skeletal morbidity by causing an annual 3% to 5% decrease in bone mineral density, putting men at a higher risk for ADT‐induced osteoporosis and bone fractures (Sountoulides 2013). As a result, treatments that specifically target bone metastasis have been established and are being used as supplementary therapies to reduce or prevent the occurrence of skeletal‐related events.
Description of the intervention
Palliative treatments with bone‐modifying agents, such as bisphosphonates and receptor activator of nuclear factor‐kappa B ligand (RANKL)‐inhibitors are widely used to prevent bone resorption (Macherey 2017). When prostate cancer cells metastasise to bone, cancer cells produce parathyroid hormone‐related protein that stimulates the osteoblasts to produce RANKL, which in turn binds and activates the RANK receptor on osteoclast precursors, leading to their growth and maturation (Ramaswamy 2003). Osteoclasts are multinucleated cells of haematopoietic origin, capable of bone resorption, and play a major role in bone‐related conditions, such as rheumatoid arthritis, Paget's disease and osteoporosis (Soysa 2012).
Bisphosphonates prevent osteoclastic bone resorption by inducing osteoclast apoptosis (Oades 2002). Recent studies have furthermore shown evidence supporting direct antitumour activity of bisphosphonates by inhibiting tumour self‐seeding, tumour‐associated angiogenesis and recruitment of tumour‐associated macrophages (TAMs) to tumours (Clezardin 2013). On the contrary, RANKL‐inhibitors work by binding to RANKL, effectively preventing it from binding to receptor activator of nuclear factor‐kappa B (RANK) in osteoclasts and osteoclast precursors, thus blocking the transduction pathway that stimulates osteoclast formation, activation and survival (Gomez‐Veiga 2013). RANKL has also been shown to mediate increased invasion and migration of RANK‐expressing cancer cells, therefore, pharmacological inhibition of RANKL not only prevents osteolysis but also reduces bone and lung metastasis (Dougall 2014).
Adverse events of the intervention
Skeletal‐related adverse events such as osteonecrosis of the jaw, an adverse event directly mediated by bone remodeling inhibition, was reported in 0.1% of participants receiving bisphosphonates treatment and in 1.7% of participants receiving denosumab (RANKL‐inhibitor) treatment (Hellstein 2011; Qi 2014).
A number of non‐skeletal adverse events associated with the interventions have been reported to affect the gastrointestinal tract (Bartl 2007; Bartl 2008; Reyes 2016). Nausea, emesis, diarrhoea or gastric pain have been reported in 2% to 10% of men receiving bisphosphonates (Bartl 2008). Additionally, reported gastrointestinal complications include oesophagitis, gastrointestinal bleeding or ulcera (Bartl 2008; Reyes 2016). Other non‐skeletal adverse events caused by bisphosphonates and RANKL‐inhibitors include hypocalcaemia and reduction of renal function (Bartl 2008; Gartrell 2014). In particular, intravenous administration of bisphosphonates has been reported to be associated with an increased risk of renal impairment and requires haemostasis of the patient's fluid balance (Bartl 2008). Furthermore, RANKL is a co‐stimulatory cytokine for T‐cell activation and its inhibition with denosumab has been found to be associated with increased infection rates in men receiving the intervention (Anastasilakis 2009).
How the intervention might work
Over the past two decades, several randomised controlled trials (RCTs) have demonstrated the effectiveness of bisphosphonates in reducing bone pain and skeletal morbidity caused by breast cancer and multiple myeloma (Coleman 2008). Usage of the most potent bisphosphonate, zoledronic acid, has reduced the risk of skeletal complications by 30% to 50% (Neville‐Webbe 2010). This reduction was reported across a range of solid tumours affecting the bone and as a result, bisphosphonates are increasingly being used in parallel with specific anticancer treatments to prevent skeletal complications.
Bisphosphonates are analogues of pyrophosphate that are subgrouped to either amino‐bisphosphonates or non‐amino‐bisphosphonates, and target osteoclastic cells (Reyes 2016). Examples of amino‐bisphosphonates are zoledronate, risedronate, pamidronate or alendronate. They affect the osteoclast metabolism by targeting the farnesyl diphosphate synthase, which is responsible for post‐translational modification of guanosine‐5'‐triphosphate‐binding proteins (Reyes 2016). The group of non‐amino‐bisphosphonates includes etidronate, clodronate or tiludronate. These substances function by forming an analogue of adenosine triphosphate. The resulting metabolite has toxic properties and induces apoptosis of osteoclasts (Reyes 2016). Both groups of bisphosphonates, amino‐ and non‐amino bisphosphonates, inhibit the effect of prostacyclines and cytokines in bone tissue and reduce the number of osteoclasts by down‐regulation of the reticuloendothelial system (Bartl 2007). They also bind hydroxyapatite in bone matrix (Gartrell 2015).
Denosumab, a fully humanised monoclonal antibody, functions by targeting and neutralising RANKL, which has been found to be a major contributor to the progression of bone metastases (Hanley 2012). In a phase III clinical trial conducted for men with prostate cancer receiving ADT in parallel with 60 mg denosumab (Prolia) administered subcutaneously every six months, it was reported that participants had a 5.6% increase in bone mass density in the lumbar spine and a decreased incidence of 1.5% vertebral fractures when compared to the placebo group, which had a 3.9% incidence rate (Smith 2009). Similarly, a phase III clinical trial of participants with metastatic castration‐resistant prostate cancer, receiving 120 mg denosumab (Xgeva) administered subcutaneously every four weeks, showed that denosumab treatment could significantly lower the risk of developing symptomatic skeletal events, in addition to reducing bone turnover markers (Fizazi 2011). These findings have led to the approval of denosumab by both the US Food and Drug Adminstration (FDA) and European Medicines Agency (EMA) to be used as an osteoprotective agent for treatment of ADT‐induced osteoporosis (Hegemann 2017).
Why it is important to do this review
Although bone‐targeted therapy is common in men with prostate cancer at risk of skeletal complications, recommendations in current guidelines are rare and inconsistent. The guidelines by the European Association of Urology (EAU) and by the German Guideline Program in Oncology (GGPO) recommend the use of zoledronic acid (bisphosphonate) or the RANKL‐inhibitor, denosumab in men with advanced, relapsed or castration‐resistant prostate cancer, without evidence to demonstrate greater efficacy of one drug over another (Mottet 2017). The guidelines by the European Society of Medical Oncology (ESMO) suggest denosumab or zoledronic acid for men with bone metastases from castration‐resistant prostate carcinoma at high‐risk for clinically relevant skeletal‐related events (Parker 2015). Neither the National Comprehensive Cancer Network (NCCN) nor the European Organisation for Research and Treatment of Cancer (EORTC) give strong recommendations to use denosumab or bisphosphonates for handling skeletal‐related events in men with prostate cancer (Fitzpatrick 2014; Mohler 2016). Despite extensive research efforts in this field, sufficient evidence from randomised head‐to‐head comparisons of the efficacy of various types of bisphosphonates or compared to RANKL‐inhibitors is lacking. Therefore, this review aims to provide the highest level of evidence for treatment decisions and a hierarchy of treatment options via a network meta‐analysis that summarises the direct and indirect evidence.
Objectives
To assess the effects of bisphosphonates and RANK‐ligand (RANKL)‐inhibitors for supportive treatment in prostate cancer with bone metastases and to generate a clinically meaningful treatment ranking according to their safety and efficacy.
Methods
Criteria for considering studies for this review
Types of studies
We will include studies if they are randomised controlled trials (RCTs). We require full journal publication, with the exception of online clinical trial results and summaries of otherwise unpublished clinical trials and abstracts with sufficient data for analysis. In the case of cross‐over trials, we will only analyse the first period of the trial. We will not impose any limitation with respect to the length of follow‐up. We will include studies regardless of their publication status or language of publication. We will exclude studies that were non‐randomised, case reports and clinical observations.
Types of participants
We will include studies involving adult participants according to the definition in the studies (usually ≥ 18 years of age), with a confirmed diagnosis of prostate cancer and bone metastases, irrespective of stage of disease or type of therapy. We will include studies in the analysis involving both hormone‐sensitive and castrate‐refractory participants receiving either bisphosphonates or RANKL‐inhibitors.
Should we identify studies in which only a subset of participants are relevant to this review, we will include such studies if data are available separately for the relevant subset.
Types of interventions
We will include trials comparing bisphosphonates or RANKL‐inhibitors to control regimens for the treatment of bone metastases from prostate cancer. We will consider any type of bisphosphonate or RANKL‐inhibitor, apart from radioactive bisphosphonates. We will not impose any restriction on the dose, route, frequency or duration of bisphosphonate treatment, nor duration of follow‐up. We plan to investigate the following comparisons of experimental interventions versus comparator interventions. Concomitant interventions will have to be the same in the experimental and comparator groups to establish fair comparisons.
Experimental interventions
Bisphosphonates
RANK‐ligand (RANKL)‐inhibitors
Comparator interventions
Bisphosphonates
RANKL‐inhibitors
Placebo/no further treatment
Comparisons
Bisphosphonates versus placebo/no further treatment
RANKL‐inhibitors versus placebo/no further treatment
Bisphosphonates versus RANKL‐inhibitors
Bisphosphonate A versus Bisphosphonate B
RANKL‐inhibitor A versus RANKL‐inhibitor B
We will compare combinations of these interventions at any dose and by any route to each other in a full network meta‐analysis. We will include all RCTs comparing at least two study arms for the intervention of interest, either bisphosphonates with placebo, RANKL‐inhibitors with placebo, or bisphosphonates with RANKL‐inhibitors for a full network of direct and indirect comparisons (Figure 1). Participants who fulfil the inclusion criteria are, in principle, equally likely to be randomised to any of the eligible interventions. We plan to perform two separate analyses: one by merging doses according to the product characteristics, the other one with the exact doses as described in the individual studies without merging.
Figure 1.
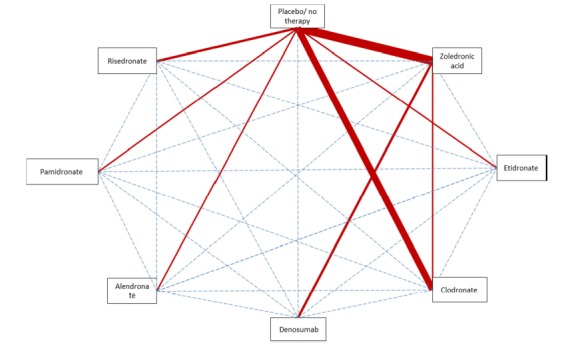
Direct and indirect comparisons of interventions (strength of line represents number of trials evaluating the comparison; dotted lines are indirect comparisons).
Types of outcome measures
We will include all trials fitting the inclusion criteria mentioned above, irrespective of reported outcomes. We will estimate the relative ranking of the competing interventions according to each of the following outcomes.
Primary outcomes
-
Proportion of participants with pain response
We will consider all trials reporting on the proportion of participants with pain response; we will not impose restrictions on pain assessment tools or the definition of pain response in the trials
-
Adverse events
-
Renal adverse events
We will consider all trials reporting renal adverse events; as drugs might be described with nephrotoxicity with variable expression, we consider creatinine elevation and renal failure as renal adverse events
Osteonecrosis of the jaw
-
Secondary outcomes
-
Skeletal‐related events
Any skeletal‐related event
Pathological fractures (in total and subgrouped by vertebral or non‐vertebral fractures)
Spinal cord compression
Bone radiotherapy
Bone surgery
-
Overall survival or mortality
If we are unable to retrieve the necessary information to analyse time‐to‐event outcomes, we will attempt to assess the number of events per total for dichotomised outcomes
-
Quality of life
If measured by validated instruments
Further adverse events
Method and timing of outcome measurement
Proportion of participants with pain response: assessed using validated generic and disease‐specific questionnaires; measured at baseline, six months, one year, two years, or at the longest reported follow‐up
Adverse events (renal adverse events, osteonecrosis of the jaw and further adverse events): grade 3 and 4 according to the common terminology criteria for adverse events (CTCAE) or as defined in the trial, measured at any time after participants were randomised to intervention/comparator groups
Skeletal‐related events: combined outcome evaluating pathological fractures (in total and subgrouped by vertebral or non‐vertebral fractures), spinal cord compression, bone radiotherapy and bone surgery at any time after participants were randomised to intervention/comparator groups
Mortality: defined as the time from randomisation to the date of death. If we are unable to retrieve the necessary information to analyse time‐to‐event outcomes, we will assess the number of events per treatment group for these outcomes at six months, one year, two years, or at the longest reported follow‐up
Quality of life: assessed using validated generic and disease‐specific questionnaires; measured at baseline, six months, one year, two years, or at the longest reported follow‐up
We will compare and analyse separately each of these measures. To determine the validity of data synthesis across separate studies, the review author extracted definitions used by each study to describe all outcomes of interest.
Main outcomes for 'Summary of findings' table
We will present a 'Summary of findings' table reporting the following outcomes, listed according to priority.
Proportion of participants with pain response.
Renal adverse events.
Adverse event: osteonecrosis of the jaw.
Total number of skeletal‐related events.
Overall survival/mortality.
Quality of life.
Search methods for identification of studies
We will perform a comprehensive search with no restrictions on the language of publication or publication status. We plan to rerun searches within three months prior to anticipated publication of the review and will include all studies fitting our inclusion criteria in the analyses.
Electronic searches
We will search the following sources from inception of each database.
-
Cochrane Library (until present) (via Wiley.com; see Appendix 1)
Cochrane Database of Systematic Reviews (CDSR)
Cochrane Central Register of Controlled Trials (CENTRAL)
Database of Abstracts of Reviews of Effects (DARE).
MEDLINE (via Ovid, 1946 to present) (see Appendix 2).
-
Embase (via Ovid, 1988 to present) databases of ongoing trials
World Health Organization (WHO) International Clinical Trials Registry Platform (ICTRP) search portal (who.int/trialsearch)
EU clinical trials register (www.clinicaltrialsregister.eu)
ClinicalTrials.gov (www.clinicaltrials.gov/)
UMIN clinical trial registration (www.umin.ac.jp).
-
Handsearching of references
We will check references of all identified trials, relevant review articles and current treatment guidelines for further literature.
-
Personal contacts
We will contact experts in the field in order to retrieve other trials.
We will use medical subject headings (MeSH) or equivalent and text word terms. We will not impose any language restrictions. We will tailor searches to individual databases.
Searching other resources
We will try to identify other potentially eligible trials or ancillary publications by searching the reference lists of retrieved included trials, reviews, meta‐analyses and health technology assessment reports. We will also contact study authors of included trials to identify any further studies that we may have missed. We will contact drug/device manufacturers for ongoing or unpublished trials.
We will search abstract proceedings of relevant meetings of the last five years if they are not included in CENTRAL.
American Society of Clinical Oncology (ASCO).
Prostate Cancer World Congress.
European Society of Medical Oncology (ESMO).
Multinational Association of Supportive Care in Cancer (MASCC).
Data collection and analysis
Selection of studies
Two review authors (YMT, TJ) will independently screen the results of the search strategies for eligibility for this review by reading the abstracts using Covidence software (Covidence 2017). We will code the abstracts as either 'retrieve' or 'do not retrieve'. In the case of disagreement or if it is unclear whether we should retrieve the abstract or not, we will obtain the full‐text publication for further discussion. Independent review authors will eliminate studies that clearly do not satisfy inclusion criteria, and obtain full copies of the remaining studies. Two review authors (YMT, TJ) will read these studies independently to select relevant studies, and in the event of disagreement, a third review author (NS) will adjudicate. We will not anonymise the studies in any way before assessment. We will include a PRISMA flow chart in the full review (Moher 2009), which will show the status of identified studies, as recommended in Part 2, Section 11.2.1 of the Cochrane Handbook for Systematic Reviews of Interventions (Schünemann 2011a). We will include studies in the review irrespective of whether measured outcome data are reported in a 'usable' way. We will use reference management software (Endnote 2016) to identify and remove potential duplicate records. We will document reasons for exclusion of studies that may have reasonably been expected to be included in the review in a 'Characteristics of excluded studies' table.
Data extraction and management
Two review authors (YMT, TJ) will extract data independently using a standardised data extraction form developed in Covidence (Covidence 2017). We will pilot this data extraction form for two included trials and adapt if necessary. If the review authors are unable to reach a consensus, we will consult a third review author (NS) for final decision. If required, we will contact the authors of specific studies for supplementary information (Higgins 2011a).
After agreement we will enter data into Review Manager 5 (Review Manager 2014). We will extract the following information.
General information: author, title, source, publication date, country, language, duplicate publications.
Quality assessment: sequence generation, allocation concealment, blinding (participants, personnel, outcome assessors), incomplete outcome data, selective outcome reporting, other sources of bias.
Study characteristics: trial design, aims, setting and dates, source of participants, inclusion/exclusion criteria, comparability of groups, subgroup analysis, statistical methods, power calculations, treatment cross‐overs, compliance with assigned treatment, length of follow‐up, time point of randomisation.
Participant characteristics: participant details, baseline demographics, age, ethnicity, number of participants recruited/allocated/evaluated, participants lost to follow‐up, cancer type and stage, additional diagnoses, type and intensity of pain, skeletal‐related events risk.
Interventions: type and dosage of drugs used, route, frequency, duration of prophylaxis, duration of follow‐up.
Outcomes: proportion of participants with pain response, renal adverse events, adverse event (osteonecrosis of the jaw), total number of skeletal‐related events, overall survival/mortality, quality of life, proportion of participants with disease progression). Where possible, we will extract data at the arm level, not summary effects.
Notes: sponsorship/funding for trial and notable conflicts of interest of review authors.
We will collect multiple reports of the same study, so that each study, rather than each report, is the unit of interest in the review. We will collect characteristics of the included studies in sufficient detail to populate a 'Characteristics of included studies' table in the full review.
We will extract outcome data relevant to this Cochrane Review, as needed for calculation of summary statistics and measures of variance. For dichotomous outcomes, we will attempt to obtain numbers of events and totals for population of a two‐by‐two table, as well as summary statistics with corresponding measures of variance. For continuous outcomes, we will attempt to obtain means and standard deviations or data necessary to calculate this information. We will provide information, including trial identifier, about potentially relevant ongoing studies in the 'Characteristics of ongoing studies' table.
Data on potential effect modifiers
We will extract from each included study, the following information that may act as effect modifiers.
Year of publication.
Type of anticancer drug used for treatment.
Intervention
Population characterisitcs
Dealing with duplicate and companion publications
In the event of duplicate publications, companion documents or multiple reports of a primary study, we will maximise yield of information by mapping all publications to unique studies and collating all available data. We will use the most complete data set aggregated across all known publications. If in doubt, we will give priority to the publication reporting the longest follow‐up associated with our primary or secondary outcomes.
Assessment of risk of bias in included studies
We will complete a 'Risk of bias' table for each included study using the 'Risk of bias' tool in Review Manager 5 (Review Manager 2014). Two review authors (YMT, TJ) will independently assess risk of bias for each study and if they are unable to reach a consensus, we will consult a third review author (NS) for a final decision. We will assess the following criteria as outlined in Chapter Eight of the Cochrane Handbook for Systematic Reviews of Interventions (Higgins 2011b).
Sequence generation.
Allocation concealment.
Blinding (participants, personnel, outcome assessors).
Incomplete outcome data.
Selective outcome reporting.
Other sources of bias.
We will make a judgement for each criterion, using one of the following categories.
'Low risk': if the criteria are adequately fulfilled in the study (i.e. the study is at low risk of bias for the given criteria).
'High risk': if the criteria are not fulfilled in the study (i.e. the study is at high risk of bias for the given criteria).
'Unclear': if the study report does not provide sufficient information to allow a clear judgement, or if risk of bias is unknown for one of the criterion listed above.
For performance bias (blinding of participants and personnel) and detection bias (blinding of outcome assessment), we will evaluate the risk of bias separately for each outcome, and we will group outcomes according to whether measured subjectively or objectively when reporting our findings in the 'Risk of bias' tables.
We define the following endpoint as not being influenced by blinding (objective outcome).
Overall survival/mortality.
We define the following endpoints as subjective outcomes.
Proportion of participants with pain response.
Quality of life.
Total number of skeletal‐related events.
Adverse events: renal adverse events, osteonecrosis of the jaw, further adverse events.
We will also assess attrition bias (incomplete outcome data) on an outcome‐specific basis, and will present the judgement for each outcome separately when reporting our findings in the 'Risk of bias' tables.
We will further summarise the risk of bias across domains for each outcome in each included study, as well as across studies and domains for each outcome, in accordance with the approach for summary assessments of the risk of bias presented in the Cochrane Handbook for Systematic Reviews of Interventions (Higgins 2011b). In sensitivity analyses, we will compare trials with at least two criteria assessed as being at high risk of bias with those with no, or only one criterion, being high risk of potential bias.
Measures of treatment effect
Relative treatment effect
We will use intention‐to‐treat data. For binary outcomes, we will use risk ratios (RRs) with 95% confidence intervals (CIs) as the measure of treatment effect. We will calculate continuous outcomes as mean differences (MDs) with 95% CI. In case different instruments are used to assess effects in continuous outcomes, we will use standardised mean differences (SMD) with 95% CI. If participant‐related outcomes are reported both as binary and continuous outcomes, we will analyse binary outcomes in one analysis and continuous outcomes in another analysis. For time‐to‐event outcomes, we will use hazard ratios (HRs) and their 95% CIs. We will extract data from publications according to Parmar 1998 and Tierney 2007. In addition to pooled estimates with CIs, we will report prediction intervals.
Relative treatment ranking
We will obtain a treatment hierarchy using P‐scores (Rücker 2015). P‐scores allow ranking treatments on a continuous 0 to 1 scale in a frequentist network meta‐analysis.
Unit of analysis issues
The unit of analysis will be the individual participant. Should we identify cross‐over trials, cluster‐randomised trials, or trials with more than two intervention groups for inclusion in the review, we will handle these in accordance with guidance provided in the Cochrane Handbook for Systematic Reviews of Interventions (Higgins 2011a).
Studies with multiple treatment groups
As recommended in Chapter 16.5.4 of the Cochrane Handbook for Systematic Reviews of Interventions (Higgins 2011c), for studies with multiple treatment groups we will combine arms, as long as they can be regarded as subtypes of the same intervention.
When arms can not be pooled this way, we will compare each arm with the common comparator separately. For pairwise meta‐analysis, we will split the 'shared' group into two or more groups with smaller sample sizes, and include two or more (reasonably independent) comparisons. For this purpose, for dichotomous outcomes, we will divide up both the number of events and the total number of participants, and for continuous outcomes, we will divide up the total number of participants with unchanged means and standard deviations. For network meta‐analysis, instead of subdividing the common comparator, we will use an approach that accounts for the within‐study correlation between the effect sizes by re‐weighting all comparisons of each multiple‐arm study (Rücker 2012; Rücker 2014).
Dealing with missing data
As suggested in Chapter 16 of theCochrane Handbook for Systematic Reviews of Interventions (Higgins 2011c), we will take the following steps to deal with missing data.
Whenever possible, we will contact the original investigators to request relevant missing data. If the number of participants evaluated for a given outcome is not reported, we will use the number of participants randomised per treatment arm as the denominator. If only percentages, but no absolute number of events are reported for binary outcomes, we will calculate numerators using percentages. If estimates for mean and standard deviations are missing, we will calculate these statistics from reported data whenever possible, using approaches described in Chapter 7.7 of the Cochrane Handbook for Systematic Reviews of Interventions (Higgins 2011d). If standard deviations are missing and we are not able to calculate them from reported data, we will calculate values according to a validated imputation method (Furukawa 2006). If data are not reported numerically, but graphically, we will estimate missing data from figures. We will perform sensitivity analyses to assess how sensitive results are to imputing data in some way. We will address in the Discussion section the potential impact of missing data on findings of the review.
Assessment of heterogeneity
Pairwise meta‐analyses
For each direct comparison, we will use visual inspection of the forest plots as well as Cochran's Q based on a Chi2 statistic and the I2 statistic in order to detect the presence of heterogeneity. We will interpret I2 values according to Chapter 9.5.2 of the Cochrane Handbook for Systematic Reviews of Interventions (Deeks 2011) as follows.
0% to 40%, may not be important.
30% to 60%, represents moderate heterogeneity.
50% to 90%, represents substantial heterogeneity.
75% to 100%, represents considerable heterogeneity.
We will use the P value of the Chi2 test only for describing the extent of heterogeneity and not for determining statistical significance. In addition, we will report Tau2, the between‐study variance in random‐effects meta‐analysis. When we find heterogeneity, we will attempt to determine possible reasons for it by examining individual study and subgroup characteristics. In the event of excessive heterogeneity, unexplained by subgroup analyses, we will not report outcome results as the pooled effect estimate in a meta‐analysis, but provide a narrative description of the results of each study.
Network meta‐analysis
A very important presupposition for using network meta‐analysis is to make sure that the network is consistent, meaning that direct and indirect evidence on the same comparisons agree. Inconsistency can be caused by incomparable inclusion and exclusion criteria of the trials in the network.
We will evaluate the assumption of transitivity epidemiologically by comparing the distribution of the potential effect modifiers across the different pairwise comparisons. For each set of studies, grouped by treatment comparison, we will create a table of important clinical and methodological characteristics. We will visually inspect the similarity of these factors, including the inclusion and exclusion criteria of every trial in the network.
To evaluate the presence of inconsistency locally, we will compare direct and indirect treatment estimates of each treatment comparisons. This can serve as a check for consistency of a network meta‐analysis (Dias 2010). For this purpose, we will use the netsplit command in the R package netmeta, which enables the splitting of the network evidence into direct and indirect contributions (Netmeta 2017; R 2017). For each treatment comparison, we will present direct and indirect treatment estimates plus the network estimate using forest plots. In addition, for each comparison we will give the z‐value and P value of test for disagreement (direct versus indirect). It should be noted that in a network of evidence there may be many loops and with multiple testing there is an increased likelihood that we might find an inconsistent loop by chance. Therefore, we will be cautious deriving conclusions from this approach.
To evaluate the presence of inconsistency in the entire network, we will give the generalised heterogeneity statistic Qtotal and the generalised I2 statistic, as described in Schwarzer 2015. We will use the decomp.design command in the R package netmeta for decomposition of the heterogeneity statistic into a Q statistic for assessing the heterogeneity between studies with the same design and a Q statistic for assessing design inconsistency to identify the amount of heterogeneity/inconsistency within, as well as between, designs (Netmeta 2017; R 2017). Furthermore, we will create a net heat plot (Krahn 2013), a graphical tool for locating inconsistency in network meta‐analysis, using the command netheat in the R package netmeta (Netmeta 2017). We will use Qtotal and its components as well as netheat plots based on fixed‐effect and random‐effects models to identify differences between these approaches. For random‐effects models, we will report Tau2.
If we find substantive heterogeneity,or inconsistency, or both, we will explore possible sources by performing prespecified sensitivity and subgroup analyses (see below). In addition, we will review the evidence base, reconsider inclusion criteria, and discuss the potential role of unmeasured effect modifiers to identify further sources.
Assessment of reporting biases
In pairwise comparisons with at least 10 trials, we will examine the presence of small study effects graphically by generating funnel plots. We will use linear regression tests to test for funnel plot asymmetry (Egger 1997). We will consider a P value less than 0.1 to be significant for this test (Sterne 2011). We will examine the presence of small study effects for the primary outcome only. Moreover, we will search study registries to identify completed but not published trials.
Data synthesis
Methods for direct treatment comparisons
We will perform analyses according to recommendations provided in Chapter 9 of the Cochrane Handbook for Systematic Reviews of Interventions (Deeks 2011), and we will use Review Manager 2014 and R 2017 for analyses.
Pairwise comparisons are part of the network meta‐analysis. However, in order to outline available direct evidence, we will provide forest plots for pairwise comparisons with at least 10 trials, and if trials are clinically homogenous. We will perform these standard pairwise meta‐analyses using a random‐effects model. We will calculate corresponding 95% CIs as well as prediction intervals for all analyses, and will graphically present the results using forest plots. When trials are clinically too heterogenous to be combined, we will perform only subgroup analyses without calculating an overall estimate.
Methods for indirect and mixed comparisons
Should we consider the data to be sufficiently similar to be combined, we will perform a network meta‐analysis using the frequentist weighted least‐squared approach described by Rücker 2012. We will use a random‐effects model, taking into account the correlated treatment effects in multiple‐arm studies. We will assume a common estimate for the heterogeneity variance across the different comparisons. To evaluate the extent to which treatments are connected, we will give a network plot for our primary and secondary outcomes. For each comparison, we will give the estimated treatment effect along with its 95% CI and prediction interval. We will graphically present the results using forest plots, with placebo/no treatment as reference. We will use the R package netmeta for statistical analyses (Netmeta 2017; R 2017).
GRADE
Quality of the evidence
Two review authors (YMT, TJ) will independently rate the quality of each outcome. We will use the GRADE system to rank the quality of the evidence using the GRADEprofiler Guideline Development Tool software (GRADEpro GDT 2015), and the guidelines provided in Chapter 12.2 of the Cochrane Handbook for Systematic Reviews of Interventions (Schünemann 2011b), and specifically for network meta‐analyses (Puhan 2014).
The GRADE approach uses five considerations (study limitations, consistency of effect, imprecision, indirectness and publication bias) to assess the quality of the body of evidence for each outcome. The GRADE system uses the following criteria for assigning quality of evidence.
High‐quality: we are very confident that the true effect lies close to that of the estimate of the effect;
Moderate‐quality: we are moderately confident in the effect estimate; the true effect is likely to be close to the estimate of effect, but there is a possibility that it is substantially different;
Low‐quality: our confidence in the effect estimate is limited; the true effect may be substantially different from the estimate of the effect;
Very low‐quality: we have very little confidence in the effect estimate; the true effect is likely to be substantially different from the estimate of effect.
The GRADE system uses the following criteria for assigning a quality level to a body of evidence (Schünemann 2011b).
High: randomised trials; or double‐upgraded observational studies.
Moderate: downgraded randomised trials; or upgraded observational studies.
Low: double‐downgraded randomised trials; or observational studies.
Very low: triple‐downgraded randomised trials; or downgraded observational studies; or case series/case reports.
We will decrease the quality level if:
serious (‐1) or very serious (‐ 2) limitation to study quality;
important inconsistency (‐ 1);
some (‐1) or major (‐ 2) uncertainty about directness;
imprecise or sparse data (‐ 1);
high probability of reporting bias (‐ 1).
'Summary of findings' table
We will include one 'Summary of findings' table to present the main findings in a transparent and simple tabular format. In particular, we will include key information concerning the quality of evidence, the magnitude of effect of the interventions examined, and the sum of available data on the outcomes mentioned above. In case data are too heterogenous to meta‐analyse or meta‐analysis is not possible, we will present results in a narrative way.
Subgroup analysis and investigation of heterogeneity
We will perform subgroup analyses related to participant characteristics which might have an effect on the outcomes.
Participant age (due to age‐related decreases in bone marrow density).
Tumor status of the cohorts (TNM classification of malignant tumours (TNM) and grading, as well as castration resistance).
We consider performing subgroup analyses according to the type of bisphosphonate and the route of administration.
As previously described (see How the intervention might work), amino‐bisphosphonates and non‐amino‐bisphosphonates work through similar but also different mechanisms of action. Subgroup analysis is intended to reveal whether these differences in mechanism of action might affect participants' outcome.
Amino‐bisphosphonates: alendronate, ibandronate, pamidronate, risedronate, zoledronate.
Non‐amino‐bisphosphonate: clodronate, etidronate.
Bisphosphonates are potentially nephrotoxic substances. There are hints in the literature that intravenously administered bisphosphonates increased the risk of nephrotoxicity in comparison with oral application (Bartl 2007). Moreover, Lee 2014 found participants on intravenously administered bisphosphonates to be at higher risk for osteonecrosis of the jaw.
Intravenous administration.
Oral administration.
Sensitivity analysis
To test the robustness of the results, we will conduct fixed‐effect pairwise and network meta‐analyses. We will report the estimates of the fixed‐effect only if they show a difference to the random‐effects model. We will explore the influence of quality components with regard to low and high risk of bias (see Assessment of risk of bias in included studies: we will evaluate trials being at risk of potential bias in at least two criteria versus those with one or no criterion being at high risk of potential bias).
Acknowledgements
We would like to thank members of Cochrane Urology for their comments and improving the review.
This project is funded by the Federal Ministry for Education and Research, grant no. 01KG1702.
The views and opinions expressed therein are those of the authors and do not necessarily reflect those of the Systematic Reviews Programme.
Appendices
Appendix 1. Cochrane Central Register of Controlled Trials search strategy
ID Search
#1 MeSH descriptor: [Prostatic Neoplasms] explode all trees
#2 (prostat* near/3 (cancer* or carcinoma* or malignan* or tumor* or tumour* or neoplas* or intraepithelial* or adenocarcinoma*))
#3 MeSH descriptor: [Prostatitis] explode all trees
#4 (prostatitis or prostatitides or prostatosis)
#5 #1 or #2 or #3 or #4
#6 MeSH descriptor: [Diphosphonates] explode all trees
#7 (diphosphonate* or diphosph*nate*)
#8 (bisphosph*nate* or biphosph*nate*)
#9 #6 or #7 or #8
#10 MeSH descriptor: [Alendronate] explode all trees
#11 (alendronat* or aledronic*)
#12 (fosamax* or binosto* or adronat* or alendros* or onclast*)
#13 #10 or #11 or #12
#14 MeSH descriptor: [Clodronic Acid] explode all trees
#15 (clodronic* or clodronat*)
#16 (bonefos* or clasteon* or difosfonal* or ossiten* or mebonat* or loron* or ostac*)
#17 Cl2MDP
#18 #14 or #15 or #16 or #17
#19 MeSH descriptor: [Etidronic Acid] explode all trees
#20 (etidronic* or etidronat*)
#21 (didronel* or xidifon* or dicalcium or xidiphon*)
#22 (HEDP or EHDP)
#23 #19 or #20 or #21 or #22
#24 MeSH descriptor: [Technetium Tc 99m Medronate] explode all trees
#25 (medronat* or medronic*)
#26 (Technetium near/2 Tc 99m near/2 Medronat*)
#27 #24 or #25 or #26
#28 (pamidronat* or pamidronic* or amidronat*)
#29 (aredia* or ADP sodium* or aminomux*)
#30 (GCP23339A or GCP‐23339A or YM529 or YM‐529)
#31 #28 or #29 or #30
#32 (zoledronic* or zoledronat*)
#33 (zometa* or zomera* or aclasta* or reclast* or aredia* or zoldron*)
#34 (m05BA08 or CGP‐42446* or CGP42446* or zol‐446 or zol446)
#35 #32 or #33 or #34
#36 (ibandronic* or ibandrovic* or ibandronat*)
#37 (bon*iva* or bondronat* or bondranat* or adronil*)
#38 (RPR102289A or RPR‐102289A)
#39 (BM210955 or BM‐210955)
#40 #36 or #37 or #38 or #39
#41 (risedronic* or risedronat*)
#42 (actonel* or atelvia* or benet*)
#43 (NE58095 or NE‐58095)
#44 #41 or #42 or #43
#45 (neridronat* or neridronic*)
#46 (AHHexBP or 6AHHDP or 6‐AHHDP)
#47 #45 or #46
#48 MeSH descriptor: [RANK Ligand] explode all trees
#49 (rank near/3 ligand*)
#50 RANK ligand inhibitor*
#51 (protein* near/2 (RANKL or TRANCE))
#52 Tumor Necrosis Factor‐Related Activation‐Induced Cytokin*
#53 #48 or #49 or #50 or #51 or #52
#54 MeSH descriptor: [Denosumab] explode all trees
#55 denosumab*
#56 (xgeva* or prolia*)
#57 (AMG162 or AMG‐162)
#58 #55 or #56 or #57
#59 tiludronat* or tiludronic* or skelid*
#60 Incadronat* or YM175 or YM‐175
#61 olpadronat* or olpadronic*
#62 #9 or #13 or #18 or #23 or #27 or #31 or #35 or #40 or #44 or #47 or #53 or #58 or #59 or #60 or #61
#63 #5 and #62 in Trials
Appendix 2. MEDLINE search strategy
| # | Searches |
| 1 | exp PROSTATIC NEOPLASMS/ |
| 2 | (prostat$ adj3 (cancer$ or carcinoma$ or malignan$ or tumo?r$ or neoplas$ or intraepithelial or adenocarcinoma$)).tw. |
| 3 | (prostat$ adj6 (cancer$ or carcinoma$ or tumo?r$ or neoplas$ or adenocarcinoma$)).ab,ti. |
| 4 | PROSTATITIS/ or prostatitis.tw. or prostatitides.tw. or prostatosis.tw. |
| 5 | or/1‐4 |
| 6 | exp DIPHOSPHONATES/ |
| 7 | (diphosphonate$ or diphosph#nate$).tw,kf,ot,nm. |
| 8 | (bisphosph#nate$ or biphosph#nate$).tw,kf,ot,nm. |
| 9 | or/6‐8 |
| 10 | ALENDRONATE/ |
| 11 | (alendronat$ or aledronic$).tw,kf,ot,nm. |
| 12 | (fosamax$ or binosto$ or adronat$ or alendros$ or onclast$).tw,kf,ot,nm. |
| 13 | or/10‐12 |
| 14 | CLODRONIC ACID/ |
| 15 | (clodronic$ or clodronat$).tw,kf,ot,nm. |
| 16 | (bonefos$ or clasteon$ or difosfonal$ or ossiten$ or mebonat$ or loron$ or ostac$).tw,kf,ot,nm. |
| 17 | Cl2MDP.tw,kf,ot,nm. |
| 18 | or/14‐17 |
| 19 | ETIDRONIC ACID/ |
| 20 | (etidronic$ or etidronat$).tw,kf,ot,nm. |
| 21 | (didronel$ or xidifon$ or dicalcium$ or didrocal$ or xidiphon$).tw,kf,ot. |
| 22 | (HEDP or EHDP).tw,kf,ot. |
| 23 | or/19‐22 |
| 24 | TECHNETIUM TC 99M MEDRONATE/ |
| 25 | (medronat$ or medronic$).tw,kf,ot,nm. |
| 26 | (Technetium adj2 Tc 99m adj2 Medronat$).tw,kf,ot,nm. |
| 27 | or/24‐26 |
| 28 | (pamidronat$ or pamidronic$ or amidronat$).tw,kf,ot,nm. |
| 29 | (aredia$ or ADP sodium$ or incadron$ or aminomux$).tw,kf,ot,nm. |
| 30 | (GCP23339A or GCP‐23339A or YM529 or YM‐529).tw,kf,ot,nm. |
| 31 | or/28‐30 |
| 32 | (zoledronic$ or zoledronat$).tw,kf,ot,nm. |
| 33 | (zometa$ or zomera$ or aclasta$ or zoldron$ or reclast$ or aredia$).tw,kf,ot,nm. |
| 34 | (m05BA08 or CGP‐42446$ or CGP42446$ or zol‐446 or zol446).tw,kf,ot,nm. |
| 35 | or/32‐34 |
| 36 | (ibandronic$ or ibandrovic$ or ibandronat$).tw,kf,ot,nm. |
| 37 | (bon?iva$ or bondronat$ or bondranat$ or adronil$).tw,kf,ot,nm. |
| 38 | (RPR102289A or RPR‐102289A).tw,kf,ot,nm. |
| 39 | (BM210955 or BM‐210955).tw,kf,ot,nm. |
| 40 | or/36‐39 |
| 41 | (risedronic$ or risedronat$).tw,kf,ot,nm. |
| 42 | (actonel$ or atelvia$ or benet$).tw,kf,ot,nm. |
| 43 | (NE58095 or NE‐58095).tw,kf,ot,nm. |
| 44 | or/41‐43 |
| 45 | (neridronat$ or neridronic$).tw,kf,ot,nm. |
| 46 | (AHHexBP or 6AHHDP or 6‐AHHDP).tw,kf,ot,nm. |
| 47 | or/45‐46 |
| 48 | RANK Ligand/ |
| 49 | (rank$ adj3 ligand$).tw,kf,ot,nm. |
| 50 | RANK ligand inhibitor$.tw,kf,ot,nm. |
| 51 | (protein$ adj2 (RANKL or TRANCE)).tw,kf,ot,nm. |
| 52 | Tumor Necrosis Factor‐Related Activation‐Induced Cytokin$.tw,kf,ot,nm. |
| 53 | or/48‐51 |
| 54 | DENOSUMAB/ |
| 55 | denosumab$.tw,kf,ot,nm. |
| 56 | (xgeva$ or prolia$).tw,kf,ot,nm. |
| 57 | (AMG162 or AMG‐162).tw,kf,ot,nm. |
| 58 | or/54‐57 |
| 59 | (tiludronat$ or tiludronic$ or skelid$).tw,kf,ot,nm. |
| 60 | (Incadronat$ or YM175 or YM‐175).tw,kf,ot,nm. |
| 61 | (olpadronat$ or olpadronic$).tw,kf,ot,nm. |
| 62 | 9 or 13 or 18 or 23 or 27 or 31 or 35 or 40 or 44 or 47 or 53 or 58 or 59 or 60 or 61 |
| 63 | 5 and 62 |
| 64 | randomized controlled trial.pt. |
| 65 | controlled clinical trial.pt. |
| 66 | randomi?ed.ab. |
| 67 | placebo.ab. |
| 68 | drug therapy.fs. |
| 69 | randomly.ab. |
| 70 | trial.ab. |
| 71 | groups.ab. |
| 72 | or/64‐71 |
| 73 | exp animals/ not humans/ |
| 74 | 72 not 73 |
| 75 | 63 and 74 |
Contributions of authors
Yonas Mehari Tesfamariam: wrote the protocol.
Sascha Macherey: content input.
Kathrin Kuhr: methods input.
Ingrid Becker: methods input.
Ina Mosef: search strategy development.
Tina Jakob: wrote the protocol.
Axel Heidenreich: clinical expertise.
Nicole Skoetz: wrote the protocol.
Sources of support
Internal sources
No sources of support supplied
External sources
-
Federal Ministry for Education and Research (BMBF), Germany.
Grant no: 01KG1702
Declarations of interest
Yonas Mehari Tesfamariam: none known.
Sascha Macherey: none known.
Kathrin Kuhr: none known.
Ingrid Becker: none known.
Ina Monsef: none known.
Tina Jakob: none known.
Axel Heidenreich: none known.
Nicole Skoetz: none known.
Notes
We have based parts of the Methods section of this protocol on a standard template developed by the Cochrane Metabolic and Endocrine Disorders Group, which has been modified and adapted for use by the Cochrane Urology Group. In addition, this protocol is partly based on suggested wording from the Pain, Palliative and Supportive Care Review Group (PaPaS CRG), the Cochrane Heamatological Malignancies Review Group and the protocol templates for a Cochrane intervention review that compares multiple interventions (Chaimani 2014; Chaimani 2017).
New
References
Additional references
- Anastasilakis AD, Toulis KA, Goulis DG, Polyzos SA, Delaroudis S, Giomisi A, et al. Efficacy and safety of denosumab in postmenopausal women with osteopenia or osteoporosis: a systematic review and a meta‐analysis. Hormone and Metabolic Research 2009;41:721‐9. [DOI] [PubMed] [Google Scholar]
- Bartl R, Frisch B, Tresckow E, Bartl C. Bisphosphonates in Medical Practice ‐ Actions, Side effects, Indications, Strategies. 1st Edition. Dordrecht: Springer, 2007. [Google Scholar]
- Bartl R, Bartl C, Gradinger R. Use of bisphosphonates in orthopedic surgery [Einsatz der Bisphosphonate in derOrthopädie und Unfallchirurgie]. Der Orthopäde 2008;37(6):595‐614. [DOI] [PubMed] [Google Scholar]
- Chaimani A, Salanti G, Becker L, Caldwell D, Higgins J, Li T. Protocol template for a Cochrane intervention review that compares multiple interventions. methods.cochrane.org/sites/methods.cochrane.org.cmi/files/public/uploads/Protocol%20for%20Cochrane%20Reviews%20with%20Multiple%20Interventions.pdf2014.
- Chaimani A, Caldwell DM, Li T, Higgins JP, Salanti G. Additional considerations are required when preparing a protocol for a systematic review with multiple interventions. Journal of Clinical Epidemiology 2017;83:65‐74. [PUBMED: 28088593] [DOI] [PubMed] [Google Scholar]
- Clezardin P. Mechanisms of action of bisphosphonates in oncology: a scientific concept evolving from antiresorptive to anticancer activities. BoneKEy Reports 2013;2:267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman RE. Skeletal complications of malignancy. Cancer 1997;80(8 Suppl):1588‐94. [DOI] [PubMed] [Google Scholar]
- Coleman RE. Risks and benefits of bisphosphonates. British Journal of Cancer 2008;98(11):1736‐40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veritas Health Innovation. Covidence. Version accessed 21 July 2017. Melbourne, Australia: Veritas Health Innovation.
- Deeks JJ, Higgins JP, Altman DG, editor(s). Chapter 9: Analysing data and undertaking meta‐analyses. In: Higgins JP, Green S, editor(s). Cochrane Handbook for Systematic Reviews of Interventions Version 5.1.0 (Updated March 2011). The Cochrane Collaboration, 2011. Available from handbook.cochrane.org.
- Dias S, Welton NJ, Caldwell DM, Ades AE. Checking consistency in mixed treatment comparison meta‐analysis. Statistics in Medicine 2010;29(7‐8):932‐44. [PUBMED: 20213715] [DOI] [PubMed] [Google Scholar]
- Dougall WC, Holen I, González Suárez E. Targeting RANKL in metastasis. BoneKEy Reports 2014;3:519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta‐analysis detected by a simple, graphical test. BMJ 1997;315(7109):629‐34. [PUBMED: 9310563] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick JM, Bellmunt J, Fizazi K, Heidenreich A, Sternberg CN, Tombal B, et al. Optimal management of metastatic castration‐resistant prostate cancer: highlights from a European Expert Consensus Panel. European Journal of Cancer 2014;50:1617‐27. [DOI] [PubMed] [Google Scholar]
- Fizazi K, Carducci M, Smith M, Damiao R, Brown J, Karsh L, et al. Denosumab versus zoledronic acid for treatment of bone metastases in men with castration‐resistant prostate cancer: a randomised, double‐blind study. Lancet 2011;377(9768):813‐22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa TA, Barbui C, Cipriani A, Brambilla P, Watanabe N. Imputing missing standard deviations in meta‐analyses can provide accurate results. Journal of Clinical Epidemiology 2006;59(1):7‐10. [PUBMED: 16360555] [DOI] [PubMed] [Google Scholar]
- Gartrell BA, Coleman RE, Fizazi K, Miller K, Saad F, Sternberg CN, et al. Toxicities following treatment with bisphosphonates and receptor activator of nuclear factor‐kB ligand inhibitors in patients with advanced prostate cancer. European Urology 2014;65(2):278‐86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartrell BA, Coleman R, Efstathiou E, Fizazi K, Logothetis CJ, Smith MR, et al. Metastatic prostate cancer and the bone: significance and therapeutic options. European Urology 2015;68(5):850‐8. [DOI] [PubMed] [Google Scholar]
- Gomez‐Veiga F, Ponce‐Reixa J, Martinez‐Breijo S, Planas J, Morote J. Advances in prevention and treatment of bone metastases in prostate cancer. Role of RANK/RANKL inhibition. Actas Urologicals Espanolas 2013;37(5):292‐304. [DOI] [PubMed] [Google Scholar]
- McMaster University (developed by Evidence Prime). GRADEpro GDT. Version accessed 21 July 2017. Hamilton (ON): McMaster University (developed by Evidence Prime), 2015.
- Hanley DA, Adachi JD, Bell A, Brown V. Denosumab: mechanism of action and clinical outcomes. International Journal of Clinical Practice 2012;66(12):1139‐46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegemann M, Bedke J, Stenzl A, Todenhöfer T. Denosumab treatment in the management of patients with advanced prostate cancer: clinical evidence and experience. Therapeutic Advances in Urology 2017;9(3‐4):81‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellstein JW, Adler RA, Edwards B, Jacobsen PL, Kalmar JR, Koka S, et al. Managing the care of patients receiving antiresorptive therapy for prevention and treatment of osteoporosis: executive summary of recommendations from the American Dental Association Council on Scientific Affairs. The Journal of American Dental Association 2011;142(11):1243‐51. [DOI] [PubMed] [Google Scholar]
- Higgins JP, Green S, editor(s). Cochrane Handbook for Systematic Reviews of Interventions Version 5.1.0 (updated March 2011). The Cochrane Collaboration, 2011. Available from handbook.cochrane.org.
- Higgins JP, Altman DG, Sterne JA, editor(s). Chapter 8: Assessing risk of bias in included studies. In: Higgins JP, Green S, editor(s). Cochrane Handbook for Systematic Reviews of Interventions Version 5.1.0 (updated March 2011). The Cochrane Collaboration, 2011. Available from handbook.cochrane.org.
- Higgins JP, Deeks JJ, Altman DG, editor(s). Chapter 16: Special topics in statistics. In: Higgins JP, Green S, editor(s). Cochrane Handbook for Systematic Reviews of Interventions Version 5.1.0 (updated March 2011). The Cochrane Collaboration, 2011. Available from handbook.cochrane.org.
- Higgins JP, Deeks JJ, editor(s). Chapter 7: Selecting studies and collecting data. In: Higgins JP, Green S, editor(s). Cochrane Handbook for Systematic Reviews of Interventions Version 5.1.0 (updated March 2011). The Cochrane Collaboration, 2011. Available from handbook.cochrane.org. The Cochrane Collaboration.
- Howlader N, Noone AM, Krapcho M, Garshell J, Neyman N, Altekruse SF, et al. National Cancer Institute. SEER Cancer Statistics Review, 1975‐2010. www.seer.cancer.gov/csr/1975_2010 (accessed 23 June 2015).
- Jin JK, Dayyani F, Gallick GE. Steps in prostate cancer progression that lead to bone metastasis. International Journal of Cancer 2011;128(11):2545‐61. [PUBMED: 21365645] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahn U, Binder H, König J. A graphical tool for locating inconsistency in network meta‐analyses. BMC Medical Research Methodology 2013;13:35. [PUBMED: 23496991] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Chan RC, Chang SS, Tan YL, Chang KH, Lee MC, et al. Use of bisphosphonates and the risk of osteonecrosis among cancer patients: a systemic review and meta‐analysis of the observational studies. Supportive Care in Cancer 2014;22(2):553‐60. [DOI] [PubMed] [Google Scholar]
- Macherey S, Monsef I, Jahn F, Jordan K, Yuen KK, Heidenreich A, et al. Bisphosphonates for advanced prostate cancer. Cochrane Database of Systematic Reviews 2017, Issue 12. [DOI: 10.1002/14651858.CD006250.pub2] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group. Preferred reporting items for systematic reviews and meta‐analyses: the PRISMA statement. Journal of Clinical Epidemiology 2009;62(10):1006‐12. [DOI] [PubMed] [Google Scholar]
- Mohler JL, Armstrong AJ, Bahnson RR, D'Amico AV, Davis BJ, Eastham JA, et al. Prostate Cancer, Version 1.2016. Journal of the National Comprehensive Cancer Network 2016;14:19‐30. [DOI] [PubMed] [Google Scholar]
- Mottet N, Bellmunt J, Bolla M, Briers E, Cumberbatch MG, Santis M, et al. EAU‐ESTRO‐SIOG Guidelines on Prostate Cancer. Part 1: screening, diagnosis, and local treatment with curative intent. European Urology 2017;71(4):618‐29. [DOI] [PubMed] [Google Scholar]
- Rücker G, Schwarzer G, Krahn U, König J. Netmeta [Network Meta‐Analysis using Frequentist Methods]. Version R package version 0.9‐7. CRAN.R‐project.org/package=netmeta, 2017.
- Neville‐Webbe HL, Coleman RE. Bisphosphonates and RANK ligand inhibitors for the treatment and prevention of metastatic bone disease. European Journal of Cancer 2010;46(7):1211‐22. [DOI] [PubMed] [Google Scholar]
- Oades GM, Coxon J, Colston KW. The potential role of bisphosphonates in prostate cancer. Prostate Cancer and Prostatic Diseases 2002;5(4):264‐72. [DOI] [PubMed] [Google Scholar]
- Parker C, Gillessen S, Heidenreich A, Horwich A, ESMO Guidelines Committee. Cancer of the prostate: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow‐up. Annals of Oncology 2015;26 Suppl 5:v69‐77. [DOI] [PubMed] [Google Scholar]
- Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta‐analyses of the published literature for survival endpoints. Statistics in Medicine 1998;17(24):2815‐34. [DOI] [PubMed] [Google Scholar]
- Puhan MA, Schunemann HJ, Murad MH, Li T, Brignardello‐Petersen R, Singh JA, et al. A GRADE Working Group approach for rating the quality of treatment effect estimates from network meta‐analysis. BMJ (Clinical research ed.) 2014;349:g5630. [PUBMED: 25252733] [DOI] [PubMed] [Google Scholar]
- Qi WX, Tang LN, He AN, Yao Y, Shen Z. Risk of osteonecrosis of the jaw in cancer patients receiving denosumab: a meta‐analysis of seven randomised controlled trials. International Journal of Clinical Oncology 2014;19(2):403‐10. [DOI] [PubMed] [Google Scholar]
- R Foundation for Statistical Computing. R: A language and environment for statistical computing. Version 3.4.2. Vienna, Austria: R Foundation for Statistical Computing, 2017.
- Ramaswamy B, Shapiro CL. Bisphosphonates in the prevention and treatment of bone metastases. Oncology 2003;17:1261‐70; discussion 1270‐2, 1277‐8, 1280. [PubMed] [Google Scholar]
- Nordic Cochrane Centre, The Cochrane Collaboration. Review Manager 5 (RevMan 5). Version 5.3. Copenhagen: Nordic Cochrane Centre, The Cochrane Collaboration, 2014.
- Reyes C, Hitz M, Prieto‐Alhambra D, Abrahamsen B. Risks and benefits of bisphosphonate therapies. Journal of Cellular Biochemistry 2016;117(1):20‐8. [DOI] [PubMed] [Google Scholar]
- Rücker G. Network meta‐analysis, electrical networks and graph theory. Research Synthesis Methods 2012;3(4):312‐24. [PUBMED: 26053424] [DOI] [PubMed] [Google Scholar]
- Rücker G, Schwarzer G. Reduce dimension or reduce weights? Comparing two approaches to multi‐arm studies in network meta‐analysis. Statistics in Medicine 2014;33(25):4353‐69. [PUBMED: 24942211] [DOI] [PubMed] [Google Scholar]
- Rücker G, Schwarzer G. Ranking treatments in frequentist network meta‐analysis works without resampling methods. BMC Medical Research Methodology 2015;15:58. [PUBMED: 26227148] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer G, Carpenter JR, Rücker G. Chapter 8: Network meta‐analysis. Meta‐Analysis with R. Springer International Publishing Switzerland, 2015. [Google Scholar]
- Schünemann HJ, Oxman AD, Higgins JPT, Vist GE, Glasziou P, Guyatt GH. Chapter 11: Presenting results and 'Summary of findings' tables. In: Higgins JP, Green S, editor(s), Cochrane Handbook for Systematic Reviews of Interventions Version 5.1.0 (updated March 2011). The Cochrane Collaboration, 2011. Available from handbook.cochrane.org.
- Schünemann HJ, Oxman AD, Higgins JP, Vist GE, Glasziou P, Guyatt GH. Chapter 12: Interpreting results and drawing conclusions. In: Higgins JP, Green S, editor(s), Cochrane Handbook for Systematic Reviews of Interventions Version 5.1.0 (updated March 2011). The Cochrane Collaboration, 2011. Available from handbook.cochrane.org.
- Smith MR, Egerdie B, Hernandez Toriz N, Feldman R, Tammela TLJ, Saad F, et al. Denosumab in men receiving androgen‐deprivation therapy for prostate cancer. New England Journal of Medicine 2009;361(8):745‐55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sountoulides P, Rountos T. Adverse effects of androgen deprivation therapy for prostate cancer: prevention and management. ISRN Urology 2013;2013:240108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soysa NS, Alles N, Aoki K, Ohya K. Osteoclast formation and differentiation: an overview. Journal of Medical and Dental Sciences 2012;59(3):65‐74. [PUBMED: 23897045] [PubMed] [Google Scholar]
- Sterne JAC, Egger M, Moher D, editor(s). Chapter 10: Addressing reporting biases. In: Higgins JP, Green S, editor(s). Cochrane Handbook for Systematc Reviews of Interventions Version 5.1.0 (updated March 2011). The Cochrane Collaboration, 2011. Available from handbook.cochrane.org. The Cochrane Collaboration.
- Thobe MN, Clark RJ, Bainer RO, Prasad SM, Rinker‐Schaeffer CW. From prostate to bone: key players in prostate cancer bone metastasis. Cancers 2011;3(1):478‐93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time‐to‐event data into meta‐analysis. Trials 2007;8:16. [PUBMED: 17555582] [DOI] [PMC free article] [PubMed] [Google Scholar]


