ABSTRACT
In Aspergillus nidulans, nitrogen and carbon metabolism are under the control of wide-domain regulatory systems, including nitrogen metabolite repression, carbon catabolite repression and the nutrient starvation response. Transcriptomic analysis of the wild type strain grown under different combinations of carbon and nitrogen regimes was performed, to identify differentially regulated genes. Carbon metabolism predominates as the most important regulatory signal but for many genes, both carbon and nitrogen metabolisms coordinate regulation. To identify mechanisms coordinating nitrogen and carbon metabolism, we tested the role of AreB, previously identified as a regulator of genes involved in nitrogen metabolism. Deletion of areB has significant phenotypic effects on the utilization of specific carbon sources, confirming its role in the regulation of carbon metabolism. AreB was shown to regulate the expression of areA, tamA, creA, xprG and cpcA regulatory genes suggesting areB has a range of indirect, regulatory effects. Different isoforms of AreB are produced as a result of differential splicing and use of two promoters which are differentially regulated by carbon and nitrogen conditions. These isoforms are likely to be functionally distinct and thus contributing to the modulation of AreB activity.
Keywords: Nitrogen metabolite repression, carbon catabolite repression, RNA-Seq, AreB, AreA, CreA
AreB is a wide-domain regulator mediating a transcriptional response to changes in both nitrogen and carbon conditions.
INTRODUCTION
Fungi can utilize a wide variety of compounds as a source of carbon and/or nitrogen. Coordinated regulation of carbon and nitrogen metabolism is crucial for a quick adaptation of their physiology in response to the quality and concentration of available nutrients. Generally, these catabolic processes are under the control of two main global regulatory systems, carbon catabolite repression and nitrogen metabolite repression, in addition to pathway specific induction. These general regulatory systems are responsible for a preferential utilization of the most economical source of carbon or nitrogen available, thus enabling fungi to optimally utilize a wide variety of compounds (Wong, Hynes and Davis 2008; Kelly and Katz 2010), (Todd 2016).
In the model filamentous fungus, Aspergillus nidulans, carbon catabolite repression is mediated by the transcriptional repressor CreA (Dowzer and Kelly 1991; Cubero and Scazzocchio 1994), which also participates in the response to carbon starvation (Katz, Bernardo and Cheetham 2008). Ubiquitination/deubiquitination processes mediated by CreB, CreC and CreD are important in this regulation (Lockington and Kelly 2002; Boase and Kelly 2004), although CreA itself is not ubiquitinated (Alam and Kelly 2017). Response to nutrient stress, like carbon or nitrogen starvation, is also meditated by XprG, the p53-like transcription factor (Katz, Gray and Cheetham 2006; Katz et al. 2013).
Nitrogen metabolite repression modulates the expression of genes participating in uptake and catabolism of various nitrogen sources (Arst and Cove 1973). The respective genes are transcribed only when there is limiting glutamine or ammonium in the environment, with intracellular glutamine levels being a key signal (Morozov et al. 2001). In A. nidulans, nitrogen metabolite repression is mediated primarily by the GATA transcriptional activator, AreA (Kudla et al. 1990; Ravagnani et al. 1997). AreA mediates chromatin remodeling, increases histone acetylation and directly stimulates binding of specific transcriptional activators (Muro-Pastor et al. 1999; Berger et al. 2006; Berger et al. 2008). AreA activity is modulated by both posttranscriptional and posttranslational mechanisms, in response to nitrogen source and availability. The stability of the areA transcript reflects intracellular glutamine levels, resulting in instability and low levels of the transcription factor under conditions of nitrogen sufficiency (Platt et al. 1996; Morozov et al. 2001). This signaling is mediated by the RrmA protein, which regulates the rate of areA transcript deadenylation (Morozov et al. 2000; Morozov et al. 2001; Krol et al. 2013). The activity of AreA is also regulated at the protein level by a co-repressor, NmrA, and co-activator TamA. In the presence of ammonium or glutamine NmrA interacts with the zinc finger and the highly conserved C terminus of AreA, repressing its activity (Platt et al. 1996; Andrianopoulos et al. 1998; Lamb et al. 2004; Kotaka et al. 2008). TamA interacts with the same part of AreA and co-activates the expression of target genes (Small, Hynes and Davis 1999; Small et al. 2001; Downes et al. 2014). nmrA transcription is partially regulated by the bZIP transcription factor MeaB (Polley and Caddick 1996). MeaB and AreA coordinately mediate nitrogen metabolite repression, however, they can also function independently (Wong et al. 2007; Wagner et al. 2010). It is interestingly to note that for meaB the regulatory role of the antisense transcript was demonstrated (Sibthorp et al. 2013). AreA is evenly distributed in the cell except under nitrogen starvation, when it accumulates in the nucleus. It exits the nucleus when nitrogen is added to the growth medium, or under carbon starvation, and this is mediated by multiple nuclear localization signals (Todd et al. 2005; Hunter et al. 2014).
AreB is the second GATA factor participating in nitrogen metabolite repression in A. nidulans, containing an N-terminal GATA type DNA binding domain and C-terminal leucine zipper-dimerization domain (Tollervey and Arst 1982; Conlon et al. 2001). The areB gene encodes three protein isoforms, all of which include both the GATA and dimerization domain. These three AreB variants differ at their N-termini, as a result of differential splicing and the utilization of two different promoter regions. The two areB promoters are differentially regulated in response to nitrogen regime, and this is probably mediated, at least in part, by GATA factors as GATA elements are present in both areB promoters (Conlon et al. 2001). Similarly in, F. fujikuroi areB encodes three protein isoforms. These differ in their subcellular localization, depending on nitrogen conditions and the longest AreB isoform was shown to interact with AreA in the nucleus under nitrogen starvation (Michielse et al. 2014).
Many AreA target genes have been identified and their regulation characterized (Caddick 1994). Much less is known about the role of AreB in nitrogen metabolism. fmdS, coding for formamidase (Wong et al. 2009), and the arginine catabolism genes, agaA and otaA (Dzikowska et al. 2003; Macios et al. 2012), were the first AreB target genes identified in A. nidulans. In these cases AreB functions as a repressor, however, the resulting regulation is different: in the presence of ammonium AreB represses agaA and otaA expression while the expression of the fmdS gene is repressed by AreB under nitrogen limiting and nitrogen starvation conditions.
In the presence of ammonium, the expression of arginine catabolism genes is also negatively regulated by AreA, which is at variance with most other characterized examples where AreA functions as the activator under nitrogen derepressing conditions. With respect to arginine catabolism, the activity of both AreA and AreB was shown to depend on carbon source: AreA being necessary for the ammonium repression of agaA and otaA under carbon repressing conditions while AreB is primarily responsible under non-repressing, carbon-limiting conditions. Carbon signaling via AreA and AreB does not depend on the main carbon catabolite repressor CreA (Macios et al. 2012).
In F. fujikuroi both GATA factors, AreA and AreB, were shown to function as positive and negative transcriptional regulators, participating not only in regulating nitrogen metabolism but also secondary metabolism (Mihlan et al. 2003; Michielse et al. 2014; Pfannmuller et al. 2017). Nitrogen metabolism is also subjected to transcriptional regulation by CpcA, which mediates cross-pathway control in response to amino acid limitation and stress (Hoffmann et al. 2001; Busch et al. 2003).
In most cases, carbon and nitrogen regulation are studied separately. In order to define how both carbon and nitrogen conditions influence the expression profile of A. nidulans genes, we undertook transcriptomic analysis of the wild type strain grown under different carbon and nitrogen regimes. We tested the role of AreB as a potential global transcription factor linking nitrogen and carbon metabolism and investigated the differential regulation of its isoforms.
MATERIALS AND METHODS
Aspergillus nidulans strains and growth condition
biA1 (wild type), areBΔ (areBΔ::Af-pyrG, (pyrG89), argB2, pabaB22, nkuAΔ::argB, riboB2) (Macios et al. 2012) and creA1, paba1 (Shroff, Lockington and Kelly 1996) strains were employed. Strains were grown for 10–12 hours in 37°C in minimal medium, under the following nitrogen and carbon regimes:
carbon and nitrogen repression (CR/NR), comprised 1% glucose, 10 mM ammonium tartrate (GNH4)
carbon and nitrogen derepression (CD/ND), comprised 0.1% fructose, 10 mM urea (FU)
carbon repression and nitrogen derepression (CR/ND), comprised 1% glucose, 10 mM urea (GU)
carbon derepression and nitrogen repression (CD/NR), comprised 0.1% fructose,10 mM ammonium tartrate (FNH4)
Glucose and fructose were filter sterilised and added to the medium after autoclaving. Carbon derepression observed on 0.1% fructose results from the low concentration of the sugar. Growth tests were performed using 5 mM allyl alcohol which was added to the cooled minimal medium.
To compare the growth of areBΔ and the wild type on different carbon sources, conidia of both strains were suspended in 0.25% Phytagel, 0.03% Tween 20 (final concentration 104/ml) and grown on FF MicroPlateTM (BIOLOG) microtiter plates, comprising 126 selected carbon sources. This is a standard biology plate used as a fungi identification test panel. Strains were grown at 30°C in VICTOR3™ Multilabel Counter (Perkin Elmer). Turbidity was measured at 650 nm after 40 hours. Three independent biological experiments were performed.
Preparation of RNA-Seq libraries and Illumina HiSeq 2000 sequencing
Total RNA for RNA-Seq was isolated from the wild type strain using mirVanaTM miRNA Isolation Kit (Ambion) according to the protocol for total RNA isolation. Ribosomal RNA was depleted with RiboZero Magnetic kit from Epicentre and, additionally, Ribo MinusTM Concentrarion Module (Invitrogen). The success of depletion and sample quality was assessed using an Agilent 2100 Bioanalyzer.
RNA-Seq was performed by the Centre for Genomic Research, The University of Liverpool. RNA–Seq libraries were prepared using the Epicentre ScriptSeq v2 RNA-Seq Library Preparation Kit. 17.5 ng of rRNA-depleted RNA, according to quantification by Bioanalyzer, was used as input and following 10 cycles of amplification, libraries were purified using AMPure XP beads. Each library was quantified using a Qubit fluorimeter and the size distribution assessed using the Bioanalyzer. Libraries were pooled in equimolar amounts, based on the Qubit and Bioanalyzer data. The quantity and quality of each pool was assessed by Bioanalyzer and subsequently by qPCR using the Illumina Library Quantification Kit from Kapa on a Roche Light Cycler LC480II. The pool of libraries was sequenced on one lane of the HiSeq 2000 at 2 × 100 bp paired-end sequencing with v3 chemistry.
Bioinformatics and statistical analysis
Paired-end reads were aligned to the A. nidulans reference genome sequence A_nidulans_FGSC_A4 version ‘s09-m04-r07’, downloaded from the Aspergillus Genome Database (AspGD) website (http://www.aspergillusgenome.org/). Alignment was done using Tophat v1.3.2 (Trapnell, Pachter and Salzberg 2009) with default parameters except for those regarding mapping across spliced introns, where the minimum (option ‘-i’) and maximum (option ‘-I’) allowed intron sizes were set to 10 and 4000 nucleotides, respectively.
Mapped reads were filtered to retain only those where both reads of the pair (R1 and R2) aligned in the correct relative orientation. These were used to estimate expression levels for annotated loci by counting fragments (two reads, R1 and R2, represent one fragment of a transcript) mapped to each locus. Fragment counting was done using htseq-count (http://www-huber.embl.de/users/anders/HTSeq/doc/count.html#count). Alignment data is summarized in Table S1 (Supporting Information).
Fragment counts were used to assess differential gene expression, after normalization for library size, using the R package edgeR (Robinson and Oshlack 2010; Robinson, McCarthy and Smyth 2010). As there was only one sequence library for each carbon/nitrogen regime (although it consisted of three-pooled replicates), normalisation was done using a nominal value of 0.3 for the ‘biological coefficient of variation’ (BCV). Gene expression was regarded as significantly changed if the P-value was < 0.05 and the |FC| > 2.The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (Edgar, Domrachev and Lash 2002) and are accessible through GEO Series accession number GSE115021 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE115021). Gene Ontology (GO) enrichment analysis was performed using the ‘GO-term-finder’ tool on the AspGD website and protein function annotation was performed using the AspGD (Cerqueira et al. 2013) and FungiDB (http://fungidb.org/fungidb/) (Stajich et al. 2012; Basenko et al. 2018) databases. Venn diagrams were prepared using BioVenn (Hulsen, de Vlieg and Alkema 2008).
RT-qPCR analysis
For RT-qPCR analysis, total RNA was isolated from areBΔ and the wild type strains (Schmitt, Brown and Trumpower 1990),using the FastPrep®-24 instrument (MP Biomedicals) for mycelium homogenization. RNA was treated with Turbo DNase (Thermo Fisher Scientific). RNA quality and concentration were measured using an RNA Nano chip on Bioanalyzer. cDNA was synthesized using 2 μg of total RNA using SuperScript® III Reverse Transcriptase (Invitrogen, Life Technologies) and a mixture of oligo-dT and random hexamer primers. Real-time RT-PCR was performed using the LightCycler® 480 II System (Roche Diagnostics) with specific primers for creA, tamA, areA, meaB, cpcA, xprG; or areBα, areBβ, areBγ transcripts (Table S2, Supporting Information) and LightCycler®480 SYBR Green I Master mix (Roche Laboratories). Three biological replicates were analyzed for both strains with three technical replicates for each. Efficiency (E) and specificity of each pair of primers were tested in RT-qPCR reactions using 6-point standard curves of 5-fold diluted cDNA of the control strain. E-value for all primer pairs used was in the range of 1.89–2.00. Cp values were calculated using LightCycler®480 Software 1.5 (Roche Diagnostics), based on the Second Derivative Maximum Method. Cp values were normalized using 18S rRNA as an endogenous control. Expression levels were compared between the wild type and areBΔ strains.
RESULTS AND DISCUSSION
Carbon- and nitrogen-regulated genes in the wild type strain
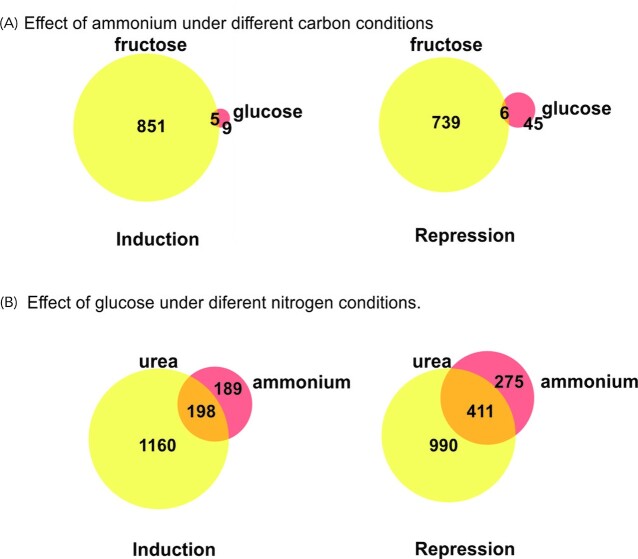
To identify genes regulated in response to different carbon and nitrogen regimes, we conducted global transcriptomic analysis of the wild type strain grown under four conditions, combining nitrogen metabolite repression (ammonium as N source) or derepression (urea as N source) with carbon catabolite repression (glucose as C source) or derepression (fructose as C source). This analysis showed that the expression of 3491, out of 10 987 annotated A. nidulans genes are differentially regulated by carbon (fructose v glucose) and/or nitrogen (urea v ammonium) source, under at least one of the four growth conditions tested (Table S3A-D, Supporting Information). We identified genes regulated by ammonium under different carbon conditions and/or regulated by glucose under different nitrogen conditions (Fig. 1A). 53% (1842) of the differentially regulated genes identified in the wild type responded specifically to changes in the carbon source while 39% (1357) responded to both carbon and nitrogen source. Genes regulated only by nitrogen source represent merely 8% (292) of all differentially regulated genes.
Figure 1.
Nitrogen and carbon regulated genes in A. nidulans wild type strain. A. Effect of ammonium or glucose on the wild type strain transcriptome under carbon or nitrogen repressing/derepressing conditions, respectively. Expression of genes under two different carbon/nitrogen conditions was compared. Number of up or down regulated genes is shown. CR—carbon repression; CD—carbon derepression; NR—nitrogen repression; ND—nitrogen derepression. For detailed lists of differentially expressed genes see Table S3A–D (Supporting Information). B. Expression profile codes for groups of genes regulated by nitrogen and/or carbon source in the wild type strain. Expressions decreased (↓), increased (↑) or not changed (−) in the presence of ammonium or glucose, respectively. GU vs. GNH4–effect of ammonium under carbon repressing conditions (glucose as a carbon source); FU vs. FNH4–effect of ammonium under carbon derepressing conditions (fructose as a carbon source); FU vs. GU—effect of glucose under nitrogen derepressing conditions (urea as a nitrogen source); FNH4 vs. GNH4–effect of glucose under nitrogen repressing conditions (ammonium as a nitrogen source). Number of genes in each group is shown. For detailed lists of genes in each group see Table S4 (Supporting Information).
We classified all differentially regulated genes into 34 groups defined by their expression profile, i.e. up or down regulation under specific conditions (Fig. 1B). 16 of these groups have at least 10 genes. Table S4 (Supporting Information) comprises a full list of genes in each group.
For several previously characterized genes, these expression profiles correlate well with published data. For example, ureA (Abreu et al. 2010), mepA (Monahan et al. 2002), agtA (Apostolaki et al. 2009), otaA (Dzikowska et al. 1999) were all repressed by ammonium; alcR, alcA (Fillinger et al. 1995), acuG (Hynes et al. 2007), xlnD (Tamayo et al. 2008) and creA (Strauss et al. 1999) were all subject to carbon catabolite repression and amdS was repressed by both ammonium and glucose (Hynes 1994).
To assess the interdependence of the nitrogen and carbon regulatory mechanisms, we compared the groups of genes repressed or derepressed by ammonium and under the two carbon regimes (Fig. 2A), and similarly those repressed or derepressed by glucose under the two nitrogen regimes (Fig. 2B). Results show that for the vast majority of genes, both positive and negative effects of ammonium depend on the carbon regime, with the majority only being observed under carbon derepressing conditions (fructose). Similarly, although to a lesser degree, the effects of glucose depend on nitrogen regime and were observable mainly under nitrogen derepressing conditions (urea).
Figure 2.
Effect of ammonium and glucose in the wild type strain grown under different carbon or nitrogen conditions, respectively. (A), Number of genes induced or repressed by ammonium under carbon derepressing (yellow), repressing (pink) or independently on carbon conditions (orange). (B), Number of genes induced or repressed by glucose under nitrogen derepressing (yellow), repressing (pink) or independently on nitrogen conditions (orange)
Functional enrichment analysis was performed for all genes differentially expressed under the four carbon/nitrogen conditions tested. For genes affected by ammonium, the analysis indicated an over representation of genes involved in nitrogen and/or carbon metabolism, amino acids and nucleotides metabolism, ribosome biogenesis and metal ion homeostasis (Fig. S1 and Table S5A–B, Supporting Information). Genes participating in carbon and/or nitrogen metabolism, organic acids and carbohydrate metabolism, ribosome assembly, RNA metabolism and translation are over-represented among glucose regulated genes (Fig. S2 and Table S5C–D, Supporting Information).
AreB is involved in both nitrogen and carbon regulation
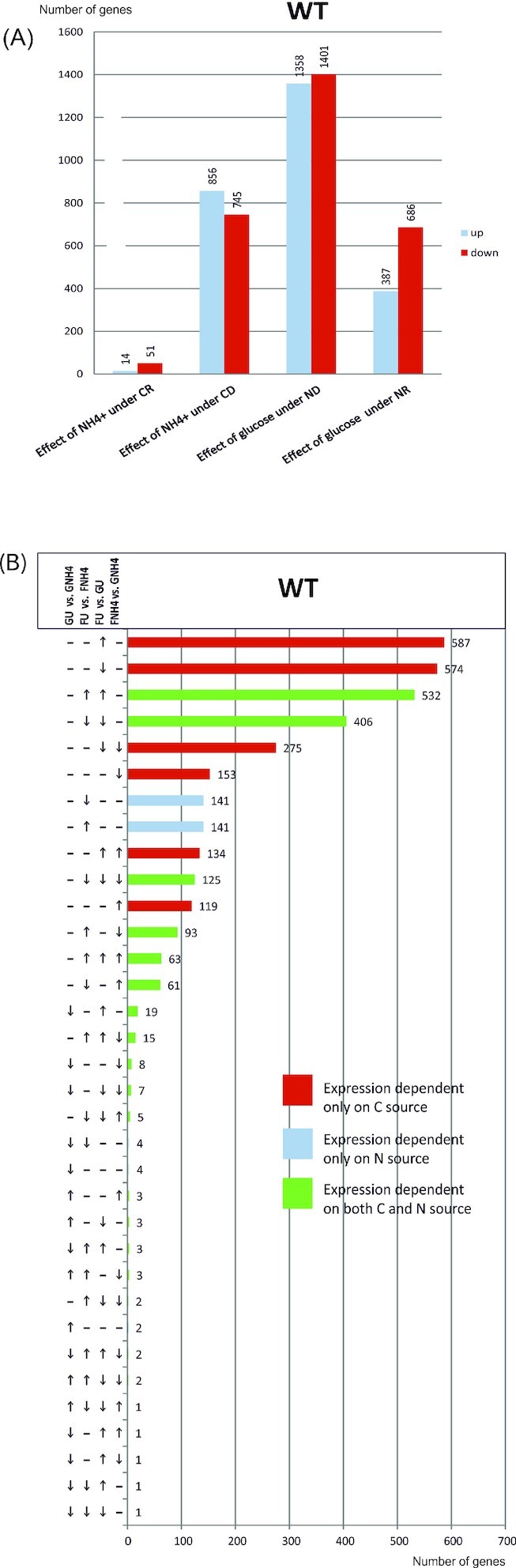
Transcriptomic analysis of the wild type strain showed that nitrogen regulation of several A. nidulans genes depends on carbon conditions. As the previously published results suggest that AreB might be involved in the regulation of not only nitrogen but also carbon metabolism (Macios et al. 2012), we utilized growth profiling using the Biolog FF MicroPlate to determine if deletion of areB changes the ability of the fungus to utilize different carbon sources. As shown in Fig. 3 and Fig. S3 (Supporting Information), the growth of the areBΔ strain was significantly different from wild type on several carbon sources tested. The mutant strain grew more poorly than wild type on 63 carbon sources tested, with only three exceptions of better growth. The reduced growth of the mutant on several amino acids suggests that AreB might regulate their catabolism, as shown previously for arginine (Macios et al. 2012). Among compounds for which deletion of areB most dramatically impaired growth are four tricarboxylic acids from the TCA cycle: α-ketoglutaric, succinic, fumaric and malic acid. Reduced efficiency of the TCA cycle might explain the previously described phenotype of slower growth of areBΔ (Wong et al. 2009; Macios et al. 2012). These data confirm that AreB plays an important role in carbon metabolism. It is worth noting that in the plant pathogen, F. fujikuroi, analysis of areB deletion mutant also showed that AreB is involved in regulation of not only nitrogen, but also carbon and secondary metabolism (Pfannmuller et al. 2017).
Figure 3.
Deletion of areBΔ influences the growth on several carbon sources. Growth of areBΔ and the wild type strain on glucose, fructose and some other selected carbon sources. Amino acids (in green); tricarboxylic acids from TCA (Krebs) cycle (in blue); compounds utilised by areBΔ as a carbon source better than by the wild type (in red). For detailed lists of carbon sources utilised less effectively by areBΔ see Fig. S3 (Supporting Information).
Deletion of areB affects the expression of several downstream regulators
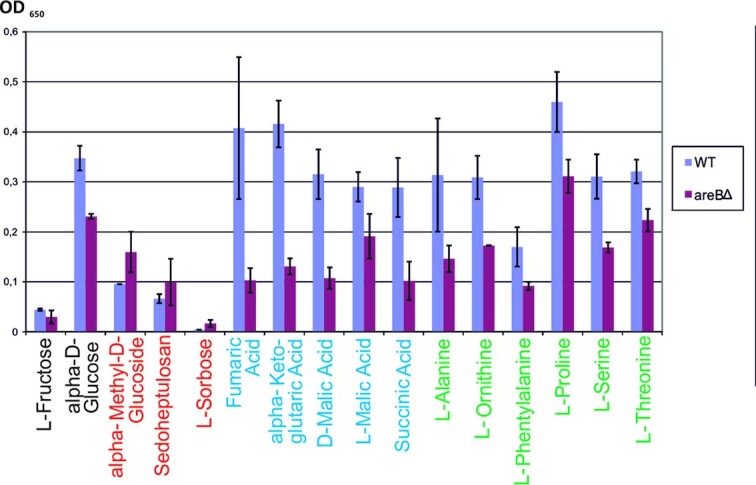
To assess the potential role of AreB in the coordination of carbon and nitrogen metabolism we performed the transcriptional analysis of genes coding for key carbon/nitrogen regulators: areA, tamA, meaB, creA, xprG and cpcA (Fig. 4B). qPCR analysis was performed using total RNA from mycelia of the areBΔ and wild type strains grown under the four carbon/nitrogen regimes used for the transcriptomic analysis. With only one exception, meaB, deletion of areB results in significant changes in transcript levels (Fig. 4A). These effects depend on carbon/nitrogen conditions and are specific for each gene analyzed. Decreased level of expression in areBΔ strain was observed for areA (about 0.5 fold under all conditions tested except carbon repressing and nitrogen derepressing conditions), tamA (till 3-fold under carbon and nitrogen derepressing conditions), creA (about 2-fold under carbon derepressing conditions) and xprG (about 2-fold under carbon repressing conditions), suggesting a positive regulatory function for AreB. For cpcA its function is negative as deletion of areB results in increased level of expression (till 3-fold under carbon and nitrogen repressing conditions). Potential GATA factor binding sites were found in promoter regions of all these five genes (Fig. 4B), suggesting that AreB might directly regulate their transcription.
Figure 4.
Deletion of areB influences the expression of the wide-domain regulatory genes areA, tamA, creA, xprG and cpcA. (A), Quantitative transcriptional analysis of areA, tamA, creA, xprG and cpcA in areB deletion strain. Relative expression in areBΔ mutant in comparison with the control wild type strain was calculated by RT-qPCR analysis. *—P-value < 0.1; **—P-value < 0.05; FC—fold change areBΔ/wt. (B), Number of GATA sites in potential promoter regions of areA, tamA, creA, xprG and cpcA. GATA pair was defined as two sites at a distance of less than 30 bp. Potential promoter region was defined as 1 kb upstream of ATG for tamA, creA and xprG or 1.5 kb for areA and cpcA that comprise much longer 5' UTR. C. Deletion of areB decreases the expression of the CreA, the carbon catabolite repressor. Sensitivity of the wild type, areBΔ and creA1 strains was tested on minimal medium with 10 mM sodium nitrate and 1% glucose (MM) or with other carbon/nitrogen sources, as described in Materials and Methods.
A strong decrease in tamA transcript level in the areBΔ strain grown under nitrogen repressing conditions was also shown in F. fujikuroi (Pfannmuller et al. 2017), suggesting that AreB might also regulate AreA activity indirectly by modulating the level of its co-activator, TamA. However, it is worth noting, that in a few cases TamA was shown to activate gene expression as a DNA-binding activator (Downes et al. 2014).The activity of xprG might be regulated by AreB both directly and indirectly, as xprG was proposed to be regulated by CreA (Katz, Bernardo and Cheetham 2008).
Deletion of areB results also in increased sensitivity to allyl alcohol, similarly as in the case of the creA1 mutant (Fig. 4C), what is consistent with decreased level of creA transcription in areBΔ (Fig. 4A). This creA mutation relieves the catabolite repression of several genes, including alcA, which codes for catabolic alcohol dehydrogenase converting allyl alcohol to a toxic acrolein (Bailey and Arst 1975; Shroff, Lockington and Kelly 1996). This growth effect doesn't depend on nitrogen source but is not observable under carbon derepressing conditions (on fructose) where derepression of alcA results in similar sensitivity also of the wild type.
All these data suggest an important, global regulatory function for AreB which appears to act both positively and negatively. A significant proportion of the regulatory effects are likely to be indirect, as AreB plays a significant role in regulating key wide domain transcriptional factors.
Analysis of areB transcripts
areB encodes three different protein isoforms which differ at their N-terminus. This results from alternative splicing, the presence of two different promoter regions and three start codons (Fig. 5A) (Conlon et al. 2001). Such variation in a transcript sequence of a specific gene is rather rare in A. nidulans (Sibthorp et al. 2013). The longest transcript, areBγ, is synthesized after three introns are removed and it can potentially produce two different proteins (AreBγ and/or AreBβ) depending on which start codon is used. Translation from the first AUG would produce AreBβ (320 aa), whereas translation from a highly conserved, non-canonical GUG start codon would result in AreBγ (436 aa), with a unique 116 amino-acid extension at the N terminus. The areBβ transcript is obtained when only two introns are removed. Retention of the first intron disrupts the ORF of the AreBγ isoform. Consequently, areBβ mRNA can produce only the AreBβ protein. areBα is the shortest areB transcript resulting from the selection of an alternative downstream promoter, located within the second intron of the longer transcripts. This transcript includes an alternative AUG start codon which would produce AreBα (312 aa), the shortest version of the protein with a unique 14 amino-acids at the N-terminus.
Figure 5.
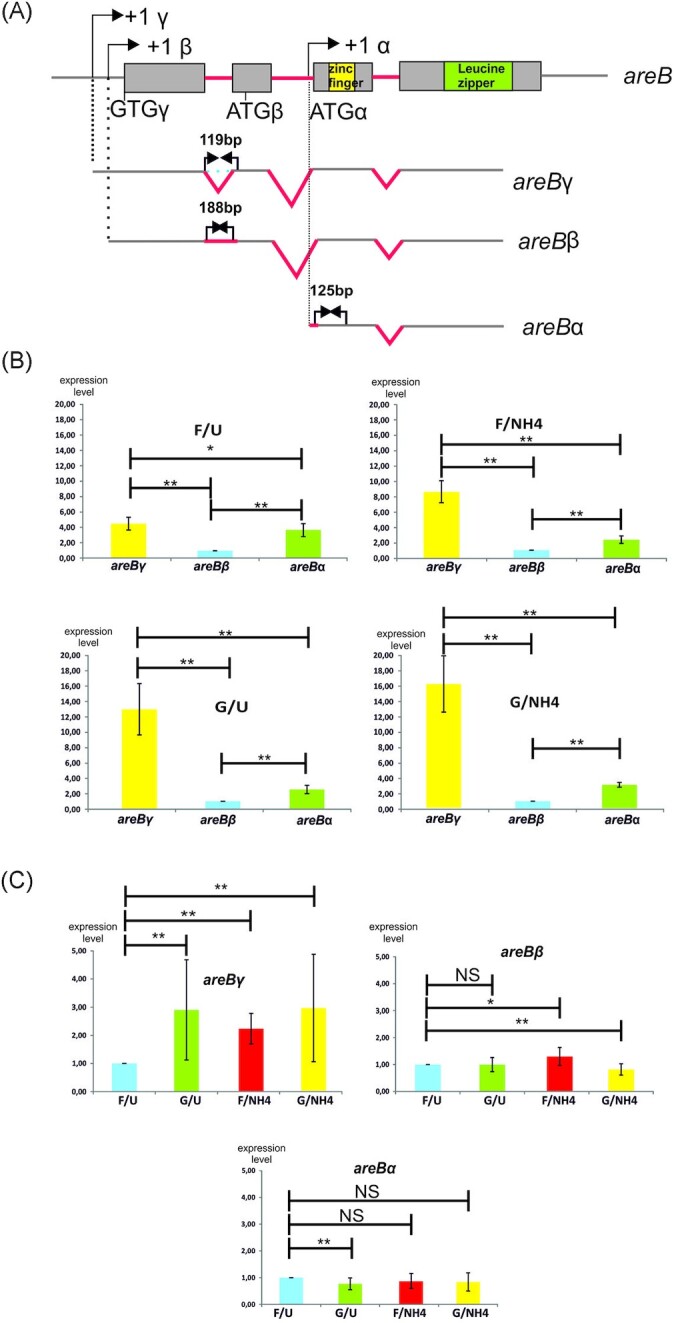
Transcriptional analysis of the areB gene under different carbon and nitrogen regimes. (A), Structure of areB gene and its three transcripts (based on Conlon et al. 2001). Differential RT-qPCR analysis of areB transcripts was performed using primers specific for each mRNA. Introns are marked in red. Amplified fragments, specific for α, β or γ areB mRNA are marked with blue arrows. Left primer for areBγ mRNA amplification is complementary to exon—exon junction in the spliced transcript. (B), Comparison of expression levels of three areB transcripts in the wild type strain grown under specific carbon/nitrogen repressing and/or derepressing conditions. The level of areBβ mRNA was arbitrarily set to 1. *- P-value < 0.1; **- P-value < 0.05. (C), Comparison of expression levels of each specific areB transcript, in the wild type strain grown under different carbon/nitrogen repressing and/or derepressing conditions. The level of each mRNA under nitrogen and carbon de-repressing conditions (FU) was arbitrarily set to 1. *- P-value < 0.1; **- P-value < 0.05. NS—difference not significant.
RT-qPCR analysis was performed to analyze proportions of the three areB transcripts under the four carbon/nitrogen regimes used for the transcriptomic analysis. We compared levels of these three transcripts under specific conditions (Fig. 5B) and the levels of each specific transcript under different conditions (Fig. 5C). Under all conditions tested the areBγ mRNA is synthesized at the highest level (Fig. 5B). This transcript has a capacity to code for two AreB isoforms, differing at the N terminus that creates an additional possibility of regulation at the level of translational initiation via selection of the standard AUG or the non-canonical GUG start codon.
Only under carbon and nitrogen derepressing conditions (F/U), was the level of areBγ and areBα mRNA comparable (Fig. 5B). Moreover, under these conditions, the expression level of areBγ transcript is lowest, while areBα transcript levels were similar under all the conditions tested (Fig. 5C). This shows that the differential expression of the three AreB isoforms is regulated by carbon and nitrogen regimes. It is possible that the function of these proteins will differ due to the variation at the N-terminus, as was suggested for AreB isoforms in F. fujikuroi (Michielse et al. 2014). Consequently, this variation may have a direct role in the observed differences in AreB functionality under different growth regimes.
CONCLUSIONS
Transcriptome analysis of the wild type strain showed that carbon metabolism predominates as the most important regulatory signal but for a large proportion of genes both carbon and nitrogen metabolism coordinates regulation. The nitrogen regime affects the expression of far more genes under carbon derepressing conditions. Genes affected by nitrogen metabolite repression independently of the carbon regime, are relatively rare. Similarly, the effect of carbon was more prevalent under nitrogen derepressing conditions.
AreB is the wide-domain regulator in A. nidulans, involved in mediating a regulatory response to changes in both nitrogen and carbon conditions. Although originally considered as a gene involved in regulation of nitrogen metabolism, we have shown that the deletion of areB has significant phenotypic effects on carbon utilisation, consistent with a broader role.
AreB positively regulates key regulatory genes mediating nitrogen metabolite repression (areA, tamA), carbon catabolite repression (creA), response to nutrient starvation (xprG) and negatively—cpcA, the main regulator of amino acid biosynthesis. Consequently, the regulatory effect of AreB on the transcription is likely to be extended by a range of indirect effects mediated by these wide-domain regulators which, in turn, regulate the expression of several pathway specific transcription factor.
We confirmed that the three transcripts encoded by areB are differentially regulated in response to carbon and nitrogen regime. We propose that three AreB isoforms differ in their specific functions or activity, contributing to modulation of AreB activity. These results underline the complex interplay between carbon and nitrogen regulation and the potential for the differential use of transcription factors under different regimes.
Supplementary Material
ACKNOWLEDGEMENTS
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE115021 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE115021). AspGD, SGD and FungiDB provided resources for genome analysis.
FUNDING
This work was supported by the Polish National Science Centre [grant numbers 2011/01/N/NZ2/04861 and 2014/15/B/NZ3/02396] and the project financing agreements CePT [POIG.02.02.00–14-024/08–00].
Conflicts of interest . None declared.
REFERENCES
- Abreu C, Sanguinetti M, Amillis Set al.. UreA, the major urea/H+ symporter in Aspergillus nidulans. Fungal Genet Biol. 2010;7:1023–33. [DOI] [PubMed] [Google Scholar]
- Alam MA, Kelly JM. Proteins interacting with CreA and CreB in the carbon catabolite repression network in Aspergillus nidulans. Curr Genet. 2017;3:669–83. [DOI] [PubMed] [Google Scholar]
- Andrianopoulos A, Kourambas S, Sharp JAet al.. Characterization of the Aspergillus nidulans nmrA gene involved in nitrogen metabolite repression. J Bacteriol. 1998;80:1973–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolaki A, Erpapazoglou Z, Harispe Let al.. AgtA, the dicarboxylic amino acid transporter of Aspergillus nidulans, is concertedly down-regulated by exquisite sensitivity to nitrogen metabolite repression and ammonium-elicited endocytosis. Eukaryot Cell. 2009;8:339–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arst HN Jr., Cove DJ. Nitrogen metabolite repression in Aspergillus nidulans. Mol Gen Genet. 1973;126:111–41. [DOI] [PubMed] [Google Scholar]
- Bailey C, Arst HN Jr.. Carbon catabolite repression in Aspergillus nidulans. Eur J Biochem. 1975;51:573–7. [DOI] [PubMed] [Google Scholar]
- Basenko EY, Pulman JA, Shanmugasundram Aet al.(FungiDB: an integrated bioinformatic resource for fungi and Oomycetes. J Fungi Basel. 2018;4:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger H, Basheer A, Bock Set al.. Dissecting individual steps of nitrogen transcription factor cooperation in the Aspergillus nidulans nitrate cluster. Mol Microbiol. 2008;69:1385–98. [DOI] [PubMed] [Google Scholar]
- Berger H, Pachlinger R, Morozov Iet al.. The GATA factor AreA regulates localization and in vivo binding site occupancy of the nitrate activator NirA. Mol Microbiol. 2006;59:433–46. [DOI] [PubMed] [Google Scholar]
- Boase NA, Kelly JM. A role for creD, a carbon catabolite repression gene from Aspergillus nidulans, in ubiquitination. Mol Microbiol. 2004;53:929–40. [DOI] [PubMed] [Google Scholar]
- Busch S, Bode HB, Brakhage AAet al.. Impact of the cross-pathway control on the regulation of lysine and penicillin biosynthesis in Aspergillus nidulans. Curr Genet. 2003;42:209–19. [DOI] [PubMed] [Google Scholar]
- Caddick MX. Nitrogen metabolite repression. In:Martinelli BSand Kinghorn JR(eds). Aspergillus: 50 Years on. Amsterdam-London-New York-Tokyo: Elsevier, 1994;Vol. 29, 323–53. [PubMed] [Google Scholar]
- Cerqueira GC, Arnaud MB, Inglis DOet al.. The Aspergillus Genome Database: multispecies curation and incorporation of RNA-Seq data to improve structural gene annotations. Nucleic Acids Res. 2013;42:D705–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon H, Zadra I, Haas Het al.. The Aspergillus nidulans GATA transcription factor gene areB encodes at least three proteins and features three classes of mutation. Mol Microbiol. 2001;40:361–75. [DOI] [PubMed] [Google Scholar]
- Cubero B, Scazzocchio C. Two different, adjacent and divergent zinc finger binding sites are necessary for CREA-mediated carbon catabolite repression in the proline gene cluster of Aspergillus nidulans. Embo J. 1994;13:407–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes DJ, Davis MA, Wong KHet al.. Dual DNA binding and coactivator functions of Aspergillus nidulans TamA, a Zn(II)2Cys6 transcription factor. Mol Microbiol. 2014;92:1198–211. [DOI] [PubMed] [Google Scholar]
- Dowzer CE, Kelly JM. Analysis of the creA gene, a regulator of carbon catabolite repression in Aspergillus nidulans. Mol Cell Biol. 1991;11:5701–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzikowska A, Kacprzak M, Tomecki Ret al.. Specific induction and carbon/nitrogen repression of arginine catabolism gene of Aspergillus nidulans-functional in vivo analysis of the otaA promoter. Fungal Genet Biol. 2003;38:175–86. [DOI] [PubMed] [Google Scholar]
- Dzikowska A, Swianiewicz M, Talarczyk Aet al.. Cloning, characterisation and regulation of the ornithine transaminase (otaA) gene of Aspergillus nidulans. Curr Genet. 1999;35:118–26. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillinger S, Panozzo C, Mathieu Met al.. The basal level of transcription of the alc genes in the ethanol regulon in Aspergillus nidulans is controlled both by the specific transactivator AlcR and the general carbon catabolite repressor CreA. FEBS Lett. 1995;368:547–50. [DOI] [PubMed] [Google Scholar]
- Hoffmann B, Valerius O, Andermann Met al.. Transcriptional autoregulation and inhibition of mRNA translation of amino acid regulator gene cpcA of filamentous fungus Aspergillus nidulans. Mol Biol Cell. 2001;12:2846–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulsen T, de Vlieg J, Alkema W. BioVenn - a web application for the comparison and visualization of biological lists using area-proportional Venn diagrams. BMC genomics. 2008;9:488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter CC, Siebert KS, Downes DJet al.. Multiple nuclear localization signals mediate nuclear localization of the GATA transcription factor AreA. Eukaryot Cell. 2014;13:527–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes MJ, Szewczyk E, Murray SLet al.. Transcriptional control of gluconeogenesis in Aspergillus nidulans. Genetics. 2007;176:139–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes MJ. Regulatory circuits of the amdS gene of Aspergillus nidulans. Antonie Van Leeuwenhoek. 1994;65:179–82. [DOI] [PubMed] [Google Scholar]
- Katz ME, Bernardo SM, Cheetham BF. The interaction of induction, repression and starvation in the regulation of extracellular proteases in Aspergillus nidulans: evidence for a role for CreA in the response to carbon starvation. Curr Genet. 2008;54:47–55. [DOI] [PubMed] [Google Scholar]
- Katz ME, Braunberger K, Yi Get al.. A p53-like transcription factor similar to Ndt80 controls the response to nutrient stress in the filamentous fungus. Aspergillus nidulans F1000Res. 2013;2:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz ME, Gray KA, Cheetham BF. The Aspergillus nidulans xprG (phoG) gene encodes a putative transcriptional activator involved in the response to nutrient limitation. Fungal Genet Biol. 2006;43:190–9. [DOI] [PubMed] [Google Scholar]
- Kelly JM, Katz ME. Glucose. In:Borkovich KAand Ebbole DJ(eds). Cellular and Molecular Biology of Filamentous Fungi. Washington: ASM Press, 2010, 291–311. [Google Scholar]
- Kotaka M, Johnson C, Lamb HKet al.. Structural analysis of the recognition of the negative regulator NmrA and DNA by the zinc finger from the GATA-type transcription factor AreA. J Mol Biol. 2008;381:373–82. [DOI] [PubMed] [Google Scholar]
- Krol K, Morozov IY, Jones MG. et al. RrmA regulates the stability of specific transcripts in response to both nitrogen source and oxidative stress. Mol Microbiol. 2013;89:975–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla B, Caddick MX, Langdon Tet al.. The regulatory gene areA mediating nitrogen metabolite repression in Aspergillus nidulans. Mutations affecting specificity of gene activation alter a loop residue of a putative zinc finger. Embo J. 1990;9:1355–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb HK, Ren J, Park Aet al.. Modulation of the ligand binding properties of the transcription repressor NmrA by GATA-containing DNA and site-directed mutagenesis. Protein Sci. 2004;13:3127–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockington RA, Kelly JM. The WD40-repeat protein CreC interacts with and stabilizes the deubiquitinating enzyme CreB in vivo in Aspergillus nidulans. Mol Microbiol. 2002;43:1173–82. [DOI] [PubMed] [Google Scholar]
- Macios M, Caddick MX, Weglenski Pet al.. The GATA factors AREA and AREB together with the co-repressor NMRA, negatively regulate arginine catabolism in Aspergillus nidulans in response to nitrogen and carbon source. Fungal Genet Biol. 2012;49:189–98. [DOI] [PubMed] [Google Scholar]
- Michielse CB, Pfannmuller A, Macios Met al.. The interplay between the GATA transcription factors AreA, the global nitrogen regulator and AreB in Fusarium fujikuroi. Mol Microbiol. 2014;91:472–93. [DOI] [PubMed] [Google Scholar]
- Mihlan M, Homann V, Liu TWet al.. AREA directly mediates nitrogen regulation of gibberellin biosynthesis in Gibberella fujikuroi, but its activity is not affected by NMR. Mol Microbiol. 2003;47:975–91. [DOI] [PubMed] [Google Scholar]
- Monahan BJ, Fraser JA, Hynes MJet al.. Isolation and characterization of two ammonium permease genes, meaA and mepA, from Aspergillus nidulans. Eukaryot Cell. 2002;1:85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozov IY, Galbis-Martinez M, Jones MGet al.. Characterization of nitrogen metabolite signalling in Aspergillus via the regulated degradation of areA mRNA. Mol Microbiol. 2001;42:269–77. [DOI] [PubMed] [Google Scholar]
- Morozov IY, Martinez MG, Jones MGet al.. A defined sequence within the 3' UTR of the areA transcript is sufficient to mediate nitrogen metabolite signalling via accelerated deadenylation. Mol Microbiol. 2000;37:1248–57. [DOI] [PubMed] [Google Scholar]
- Muro-Pastor MI, Gonzalez R, Strauss Jet al.. The GATA factor AreA is essential for chromatin remodelling in a eukaryotic bidirectional promoter. Embo J. 1999;18:1584–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannmuller A, Leufken J, Studt Let al.. Comparative transcriptome and proteome analysis reveals a global impact of the nitrogen regulators AreA and AreB on secondary metabolism in Fusarium fujikuroi. PLoS One. 2017;12:e0176194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt A, Langdon T, Arst HN Jr.et al.. Nitrogen metabolite signalling involves the C-terminus and the GATA domain of the Aspergillus transcription factor AREA and the 3' untranslated region of its mRNA. Embo J. 1996;15:2791–801. [PMC free article] [PubMed] [Google Scholar]
- Polley SD, Caddick MX. Molecular characterisation of meaB, a novel gene affecting nitrogen metabolite repression in Aspergillus nidulans. FEBS Lett. 1996;388:200–5. [DOI] [PubMed] [Google Scholar]
- Ravagnani A, Gorfinkiel L, Langdon Tet al.. Subtle hydrophobic interactions between the seventh residue of the zinc finger loop and the first base of an HGATAR sequence determine promoter-specific recognition by the Aspergillus nidulans GATA factor AreA. Embo J. 1997;16:3974–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt ME, Brown TA, Trumpower BL. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroff RA, Lockington RA, Kelly JM. Analysis of mutations in the creA gene involved in carbon catabolite repression in Aspergillus nidulans. Can J Microbiol. 1996;42:950–9. [DOI] [PubMed] [Google Scholar]
- Sibthorp C, Wu H, Cowley Get al.. Transcriptome analysis of the filamentous fungus Aspergillus nidulans directed to the global identification of promoters. BMC genomics. 2013;14:847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small AJ, Hynes MJ, Davis MA. The TamA protein fused to a DNA-binding domain can recruit AreA, the major nitrogen regulatory protein, to activate gene expression in Aspergillus nidulans. Genetics. 1999;153:95–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small AJ, Todd RB, Zanker MCet al.. Functional analysis of TamA, a coactivator of nitrogen-regulated gene expression in Aspergillus nidulans. Mol Genet Genomics. 2001;265:636–46. [DOI] [PubMed] [Google Scholar]
- Stajich JE, Harris T, Brunk BPet al.. FungiDB: an integrated functional genomics database for fungi. Nucleic Acids Res. 2012;40:D675–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss J, Horvath HK, Abdallah BMet al.. The function of CreA, the carbon catabolite repressor of Aspergillus nidulans, is regulated at the transcriptional and post-transcriptional level. Mol Microbiol. 1999;32:169–78. [DOI] [PubMed] [Google Scholar]
- Tamayo EN, Villanueva A, Hasper AAet al.. CreA mediates repression of the regulatory gene xlnR which controls the production of xylanolytic enzymes in Aspergillus nidulans. Fungal Genet Biol. 2008;45:984–93. [DOI] [PubMed] [Google Scholar]
- Todd RB, Fraser JA, Wong KHet al.. Nuclear accumulation of the GATA factor AreA in response to complete nitrogen starvation by regulation of nuclear export. Eukaryot Cell. 2005;4:1646–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd RB. Regulation of fungal nitrogen metabolism. In: [Google Scholar]; Hoffmeister D(ed). The Mycota III: Biochemistry and molecular biology, Third Edition. Switzerland: Springer International Publishing, 2016, 281–303. [Google Scholar]
- Tollervey DW, Arst HN Jr.. Domain-wide, locus-specific suppression of nitrogen metabolite repressed mutations in Aspergillus nidulans. Curr Genet. 1982;6:79–85. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D, Schmeinck A, Mos Met al.. The bZIP transcription factor MeaB mediates nitrogen metabolite repression at specific loci. Eukaryot Cell. 2010;9:1588–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KH, Hynes MJ, Davis MA. Recent advances in nitrogen regulation: a comparison between Saccharomyces cerevisiae and filamentous fungi. Eukaryot Cell. 2008;7:917–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KH, Hynes MJ, Todd RBet al.. Deletion and overexpression of the Aspergillus nidulans GATA factor AreB reveals unexpected pleiotropy. Microbiology. 2009;155:3868–80. [DOI] [PubMed] [Google Scholar]
- Wong KH, Hynes MJ, Todd RBet al.. Transcriptional control of nmrA by the bZIP transcription factor MeaB reveals a new level of nitrogen regulation in Aspergillus nidulans. Mol Microbiol. 2007;66:534–51. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.