Abstract
The purpose of this study was to determine the impact of acute ethanol administration on the major signal transduction pathways in skeletal muscle responsible for regulating the protein synthetic and degradative response to refeeding. Adult male C57Bl/6 mice were fasted overnight; mice were then either refed normal rodent chow for 30 min or a separate group of mice remained food deprived (i.e., fasted). Thereafter, mice were administered either 3 g/kg ethanol or saline. Gastrocnemius/plantaris was collected 1 hour later and analyzed. Acute ethanol decreased basal and prevented the refeeding-induced increase in muscle protein synthesis. While ethanol prevented a nutrient-stimulated increase in S6K1 phosphorylation, it did not alter the increase in 4E-BP1 phosphorylation. Downstream of S6K1, ethanol also attenuated the refeeding-induced increase in S6 and eIF4B phosphorylation, as well as the decrease in eEF2 phosphorylation. Although ethanol decreased ERK and p90 RSK phosphorylation, activation of this signaling pathway was not altered by refeeding in either control or ethanol-treated mice. Related to protein degradation, in vitro-determined proteasome activity and the content of total ubiquinated proteins were not altered by ethanol and/or refeeding. Control mice appeared to exhibit a refeeding-induced decreased in autophagy as suggested by the increased FoxO3 and ULK1 phosphorylation and total p62 protein as well as decreased LC3B-II; however, ethanol blunted these refeeding-induced changes. These data suggest that ethanol can acutely prevent the normally observed mTOR-dependent increase in protein synthesis and reduction in autophagy in response to nutrient stimulation, but does not appear to alter proteasome activity.
Keywords: muscle, alcohol, protein synthesis, mTOR, proteasome, autophagy
INTRODUCTION
Ethanol, both acute intoxication and chronic consumption, has profound effects on a variety of organs, including skeletal muscle, adversely affecting morbidity [1]. Alcoholic skeletal muscle myopathy caused by heavy, prolonged ethanol consumption is manifested by a reduction in lean body mass and decrease in muscle cross-sectional area [2, 3], although a single episode of ethanol intoxication can also acutely impair various aspects of muscle protein metabolism [4, 5]. As a result, changes that are first manifested after acute ethanol exposure may, if sustained or repeated over time, contribute to the development of alcoholic myopathy. In this regard, acute ethanol decreases skeletal muscle protein synthesis independent of a change in the number of ribosomes and dependent, at least in part, on a reduction in translation initiation [6]. In the basal condition, ethanol acutely inhibits translation via suppression of mTOR (mechanistic target of rapamycin) kinase activity as evidenced by the reduction in S6 kinase (S6K)-1 and eukaryotic initiation factor 4E binding protein (4E-BP)-1 phosphorylation [6]. Moreover, in separate studies, acute ethanol also has been reported to prevent or antagonize mTOR-dependent increases in muscle protein synthesis stimulated by pharmacological doses of anabolic mediators, such as leucine, insulin and insulin-like growth factor (IGF)-I [7–9]. In large part, these ethanol-induced changes are also seen in animals that consume ethanol more chronically over a period of weeks or months [3].
While there are some conflicting data, the consensus from the literature is that ethanol does not acutely alter protein degradative pathways in skeletal muscle under basal conditions [as reviewed in [10]]; however, the impact of ethanol on anabolic-induced changes in protein breakdown have not been previously investigated. Likewise, there is little information on the interaction of ethanol with nutrients in regulating other proteolytic pathways, such as autophagy and the inflammasome in muscle.
Muscle protein homeostasis is dynamic and fluctuations occur throughout the day in response to feeding and fasting [1, 11]. Previous attempts to mimic isolated components of refeeding have used a bolus gavage of the branched-chain amino acid leucine or the anabolic hormone IGF-I. As noted above, acute ethanol can blunt the leucine or IGF-I induced increase in mTOR-dependent signal transduction in muscle [7, 8]. However, these previous studies are constrained by the non-physiological and pharmacological nature of the stimuli used that may not faithfully duplicate the protein metabolic response seen after consumption of a complete nutrient load or meal. Hence, the purpose of this study was to determine the impact of acute ethanol administration on the major signal transduction pathways in skeletal muscle that are responsible for regulating the protein synthetic and degradative response to refeeding.
MATERIALS and METHODS
Acute ethanol and refeeding protocols
Male 12-week old C57BL/6 mice (Charles River Breeding Laboratories; Cambridge, MA) were acclimated for at least 1 wk prior to the start of the experiment. Throughout, mice were individually housed in shoe-box cages with corn cob bedding under environmental controlled conditions (22 ± 1 °C; 12 h;12 h light/dark cycle). Mice were provided standard rodent chow (Envigo Global no. 8604 diet; percent calories from protein 32%, from fat 14%, and from carbohydrates 54%; Envigo Teklad, Boston, MA) and water ad libitum. All mice were fasted beginning at 2000 hours but continued to have free access to water. The next morning, mice were randomly assigned to either the: a) refed group and provided free access to food placed within the confines of the cage for 30 minutes or b) fasted group that remained without food. At the conclusion of refeeding, food intake for the refed group was gravimetrically determined. Thereafter, mice in the refed and fasted groups were randomly divided to receive an intraperitoneal (IP) injection of ethanol (3 g/kg) or an equal volume of 0.9% saline (i.e., “control” group), and euthanized 60 min thereafter. As previously reported, there was no significant difference in the body weight at time of euthanasia or the amount of food consumed during the refeeding period between control and ethanol-treated mice, or the blood ethanol concentration between mice in the refed + ethanol group and those in the fasted + ethanol group [12]. All experimental procedures were performed in accordance with the National Institutes of Health guidelines for the use of experimental animals and were approved by the Institutional Animal Care and Use Committee of Penn State College of Medicine.
In vivo protein synthesis
The rate of global in vivo protein synthesis was determined by injecting mice with puromycin (IP; 0.04 μmol/g body weight) 30 minutes prior to sacrifice [4]. Mice were anesthetized using isoflurane (3-5% in O2), and the gastrocnemius/plantaris (here after referred to as muscle) complex was excised, frozen between liquid nitrogen precooled aluminum clamps, and weighed. Muscles were homogenized in ice-cold homogenization buffer consisting of (in mM) 48.3 HEPES (pH 7.4), 4 EGTA, 10 EDTA, 15 sodium pyrophosphate, 100 β-glycerophosphate, 25 sodium fluoride, 5 sodium vanadate, 0.1% Triton X-100, and 1 μl/ml protease inhibitor. The protein content in each tissue was determined using the BioRad protein assay kit (Bio-Rad; Hercules, CA). Western blotting (see below) was performed on equal amounts of total protein per sample. An anti-puromycin antibody (Kerafast, Boston, MA) was used for immunological detection of puromycin-labeled peptides, and the relative rate of protein synthesis determined by quantitating (see details below) all puromycin-labeled peptides with molecular weights between 20-100 kD.
Western blotting
Homogenates were clarified by centrifugation and mixed with 2× Laemmli SDS sample buffer. Equal amounts of protein per sample were subjected to electrophoresis on 4-20% SDS-PAGE, and Western blotting was performed using antibodies (unless otherwise noted from Cell Signaling Technology, Danvers, MA) for: total and phosphorylated S6K1 (Thr389), S6 (Ser240/244), FoxO1/FoxO3 (Thr24/Thr32), and 4E-BP1 (Ser65; Bethyl Laboratories, Montgomery, TX), total and phosphorylated p44/42 MAPK ERK (1/2), total and phosphorylated eIF4B (Ser422), total and phosphorylated eIF2α (Ser51), total and phosphorylated AKT (both Thr308 and Ser473). To assess autophagy, Western analysis was performed using antibodies against LC3B, p62, Atgl2, Beclin-1, and total and phosphorylated ULK (Ser757). Proteins were transferred onto polyvinylidene fluoride (PVDF; Immobilon P) membranes and incubated with a primary antibody overnight at 4 °C. Blots were developed using enhanced chemiluminescence (Amersham ECL; GE Healthcare Bio-Sciences, RPN2106, Pittsburgh, PA) and then exposed to X-ray film in a cassette equipped with a DuPont Lightening Plus intensifying screen. The film was scanned and analyzed using NIH Image 1.6 software.
Protein degradation
Skeletal muscle was homogenized in lysis buffer containing (in mM) 25 HEPES, 5 MgCl2, 5 EDTA, 5 DTT, pH 7.5 at 4 °C followed by centrifugation at 14,000 rpm for 2 min at 4 °C . Proteasome 20S enzymatic activity was determined fluorometrically by measuring the proteasome-dependent hydrolysis of the fluorogenic peptidyl substrate Suc-Leu-Leu-Val-Tyr-AMC (AMC: 7-amino-4-methylcoumarin) and release of free AMC (Enzo Life Sciences, Farmingdale, New York) [13]. Data were recorded and chymotrypsin-like activity was calculated as AFU/min/mg protein over the linear range. Similarly, Cathepsin L activity (nmol/min/mg protein) was determined in muscle homogenates using a commercially available kit (Abcam, Cambridge, MA, USA) following instructions from the manufacturer.
RNA extraction and real-time quantitative PCR
To assess the mRNA content for two muscle-specific E3 ligases in muscle, total RNA was extracted using Tri-reagent (Molecular Research Center, Inc., Cincinnati, OH) and an RNeasy mini kit (Qiagen, Valencia, CA), according to the manufacturer’s instructions, exactly as previously described [4]. RNA was eluted from the column with RNase-free water and an aliquot was used for quantitation (NanoDrop 2000, Thermo Fischer Scientific, Waltham, MA). Total RNA was reversed transcribed to cDNA using superscript III reverse transcriptase (Invitrogen, Carlsbad, CA) following instructions from the manufacturer. RT-qPCR was performed on the reversed transcribed reaction mix in a StepOnePlus system using TaqMan gene expression assays (Applied Biosystems, Foster City, CA) for the following: atrogin-1 (NM_026346.2) and muscle RING-finger 1 (MuRFl; NM_001039048.2). The comparative quantitation method 2-ΔΔCt was used in presenting gene expression, normalized to L32 mRNA, and all data were referenced to the average value of the fasted saline-injected control (no ethanol) group which was set at 1.0 arbitrary units (AU).
Statistics
Values were presented as means ± standard error of the mean (SEM). In general, the sample size varied between 10-12 mice per group, and the specific number of mice per group is indicated in each figure legend. Data were analyzed using two-way analysis of variance with post hoc Student-Newman-Keuls test to determine significant differences among the four experimental groups. Differences were considered significant when p<0.05.
RESULTS
Protein synthesis and signal transduction
Acute ethanol decreased global muscle protein synthesis by 60% in the fasted condition (Figure 1). While refeeding increased protein synthesis in muscle of control mice, no increase in skeletal muscle protein synthesis was detected in ethanol-treated mice.
Fig 1.
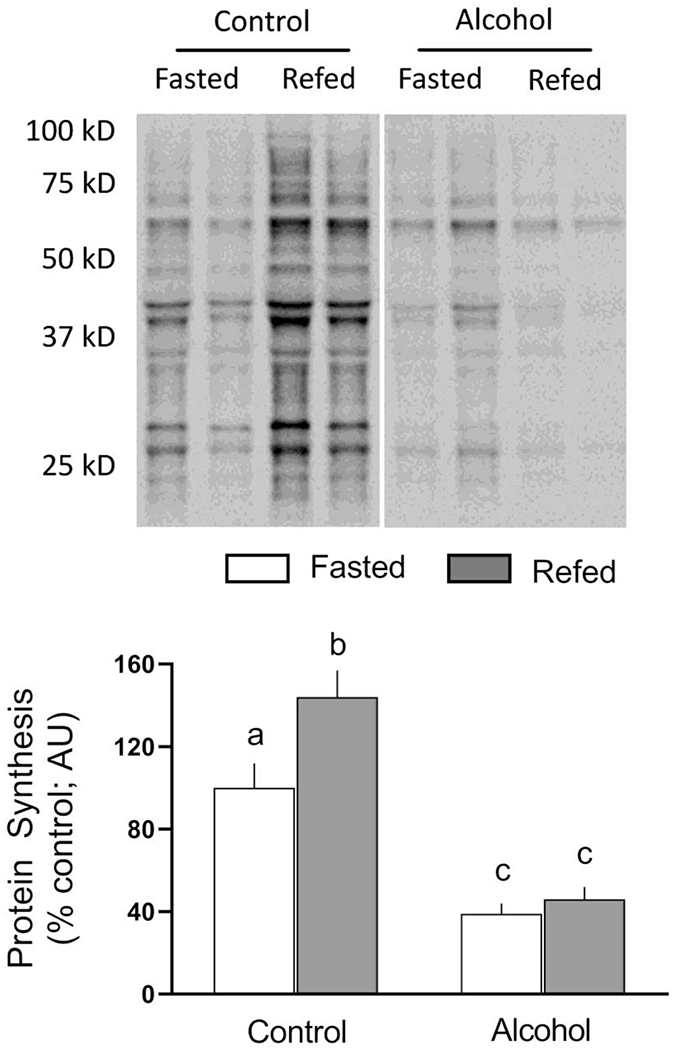
Refeeding-induced changes in global protein synthesis in gastrocnemius/plantaris from saline-treated control and ethanol-treated mice. A representative puromycin Western blot is shown in the top panel. Samples from all four experimental groups were run on the same gel and the vertical white line indicates that intervening lanes of the gel have been removed for presentation purposes. Bar graph represents densitometric analysis for puromycin-labeled proteins with molecular weights between approximately 100 and 20 kD. All immunoblots where quantified and the average value from fasted saline-treated control mice was set at 100 AU. Values are means ± SEM; n=10, 10, 12 and 12, respectively. Different letters above bars (i.e., a vs b; a vs c; or b vs c) indicate that mean values are statistically different from each other (P<0.05), whereas groups with means having the same letter (i.e., c vs c) are not statistically different
Under basal conditions and in response to nutrients, mTOR complex 1 (mTORC1) is a central regulator of muscle protein synthesis. Hence, we examined AKT phosphorylation which is a key upstream regulator of mTORC1 as well as the phosphorylation of 4E-BP1 and S6K1 as they are downstream substrates for the kinase activity of mTORC1 [14]. Figure 2 illustrates the extent of T308 and S473 AKT phosphorylation did not differ under fasted conditions in ethanol-treated and time-matched saline-treated control mice. Refeeding increased phosphorylation at both sites in control muscle. Ethanol selectively prevented the refeeding-induced increase in AKT phosphorylation at S473; however, the nutrient-stimulated increase in T308 phosphorylation did not differ between control and ethanol-treated mice. Given the discordant results for AKT phosphorylation, we also determined the phosphorylation of 4E-BP1 and S6K1 as these proteins are direct substrates for mTORCl and are typically used to assess its activity in vivo. Refeeding increased 4E-BP1 S65 phosphorylation, compared to time-matched fasted control values, to the same extent in control and ethanol-treated mice (Figure 2). In contrast, ethanol prevented the refeeding-induced increase in T389 phosphorylated S6K1 (Figure 2). Refeeding did not alter the total amount of AKT, 4E-BP1 or S6K1.
Fig 2.
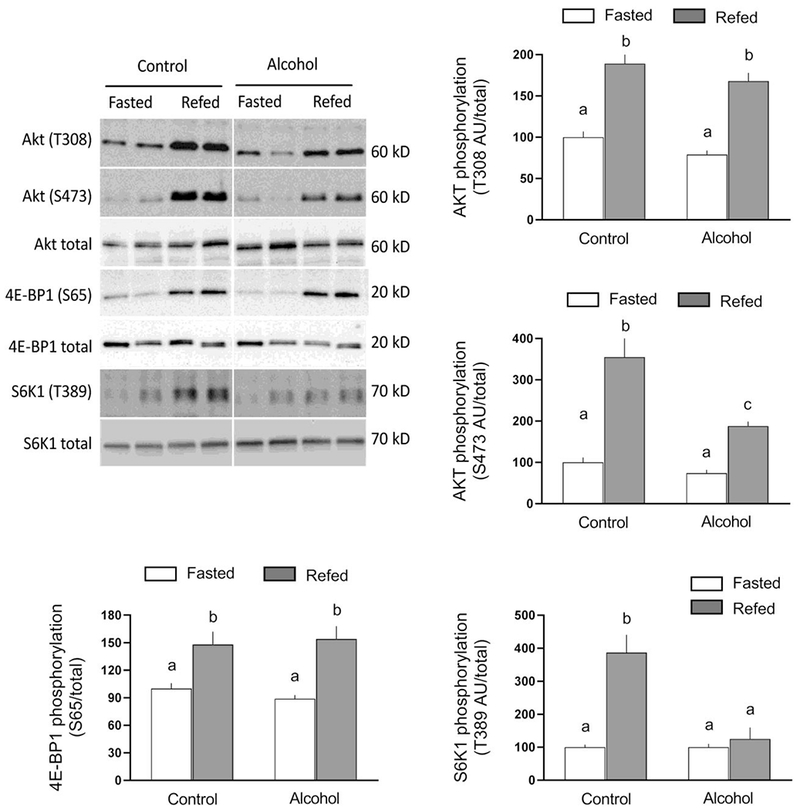
Refeeding-induced changes in 4E binding protein-1 (4E-BP1), ribosomal S6 kinase (S6K)-1, and AKT phosphorylation in gastrocnemius/plantaris from saline-treated control and ethanol-treated mice. A representative Western blot for each protein of interest is shown in the top left panel. The approximate molecular weight in kD for the protein of interest is shown to the right of each blot. Samples from all four experimental groups were run on the same gel and the vertical white line indicates that intervening lanes of the gel have been removed for presentation purposes. Bar graphs represent densitometric analysis of all immunoblots, where the value from fasted saline-treated control mice was set at 100 AU. Values are means ± SEM; n=10, 10, 12 and 12, respectively. Different letters above bars (i.e., a vs b; a vs c; or b vs c) indicate that mean values are statistically different from each other (P<0.05), whereas group means having the same letter (i.e., a vs a; or b vs b) are not statistically different
To assess whether the ethanol-induced inhibition of S6K1 phosphorylation is physiological relevant, we determined downstream protein substrates for this kinase. In control mice, refeeding increased the phosphorylation of ribosome protein S6 (S240/244) and eIF4B (S422), and decreased the phosphorylation of eEF2 (T56) (Figure 3). In contrast, these refeeding-induced changes in phosphorylation were either absent or blunted in ethanol-treated mice, and were independent of changes in the total amount of the respective protein. Neither ethanol nor refeeding produced a consistent change in the total amount of eIF4G or its phosphorylation (Figure 3). There were no significant ethanol- and/or refeeding-induced changes in translation initiation factors that regulate the formation of the 43S preinitiation complex [15], including the amount of total or phosphorylated eIF2α (S51) or total eIF2Bε (data not shown),
Fig 3.
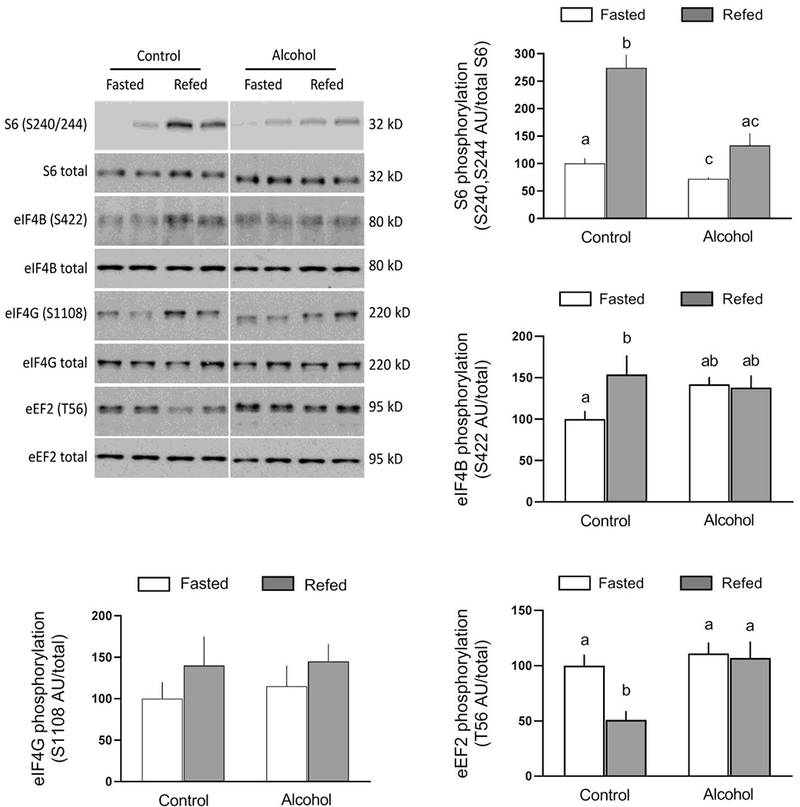
Refeeding-induced changes in the phosphorylation of S6, eukaryotic initiation factor (eIF) 4B, eIF4G and eukaryotic elongation factor (eEF) 2 in gastrocnemius/plantaris from saline-treated control and ethanol-treated mice. A representative Western blot for each protein of interest is shown in the top left panel. The approximate molecular weight in kD for the protein of interest is shown to the right of each blot. Samples from all four experimental groups were run on the same gel and the vertical white line indicates that intervening lanes of the gel have been removed for presentation purposes. Bar graphs represent densitometric analysis of all immunoblots, where the value from fasted saline-treated control mice was set at 100 AU. Values are means ± SEM; n=10, 10, 12 and 12, respectively. Different letters above bars (i.e., a vs b; a vs c; or b vs c) indicate that mean values are statistically different from each other (P<0.05), whereas group means having the same letter (i.e., a vs a; or b vs b) are not statistically different; where there are no letters above bars, there were no statistically differences detected
The MEK/ERK/RSK signal transduction pathway modulates a variety of cellular functions, including protein synthesis [16]. There was no refeeding-induced changes in the phosphorylation of ERK1/2 (T202/Y204) or two of its downstream protein substrates [p90 RSK (S380) and MNK1 (T197/202)] in saline-treated control mice (Figure 4). However, acute ethanol decreased phosphorylation of ERK1/2 and p90 RSK, but not MNK1 in muscle, and there were no subsequent changes in the phosphorylation state of these three proteins in ethanol-treated mice following refeeding. MNK1 can phosphorylate eIF4E (S209) producing hypertrophy [17]; however, we detected no significant ethanol- or refeeding-induced changes in the phosphorylation of this initiation factor (data not shown).
Fig 4.
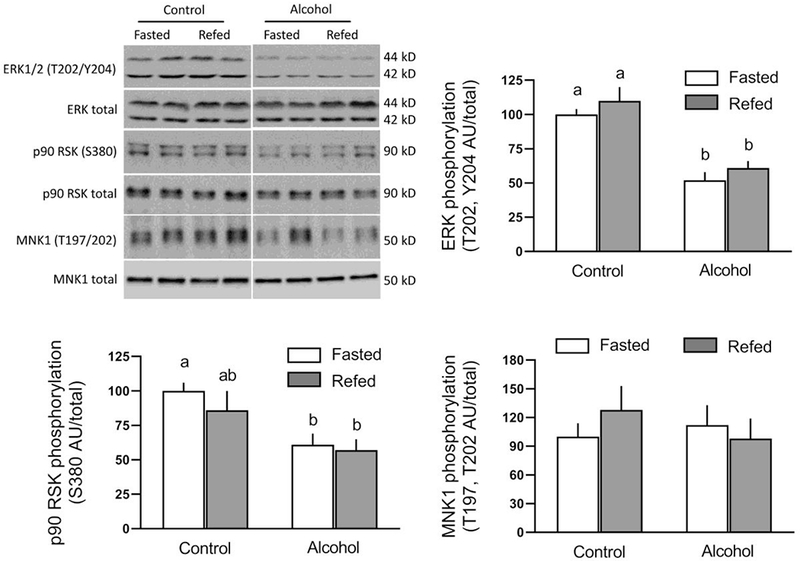
Refeeding-induced changes in the phosphorylation of ERK-/2, p90 RSK and MNK1 in gastrocnemius/plantaris from saline-treated control and ethanol-treated mice. A representative Western blot for each protein of interest is shown in the top left panel. The approximate molecular weight in kD for the protein of interest is shown to the right of each blot. Samples from all four experimental groups were run on the same gel and the vertical white line indicates that intervening lanes of the gel have been removed for presentation purposes. Bar graphs represent densitometric analysis of all immunoblots, where the value from fasted saline-treated control mice was set at 100 AU. Values are means ± SEM; n=10, 10, 12 and 12, respectively. Different letters above bars (i.e., a vs b) indicate that mean values are statistically different from each other (P<0.05), whereas group means having the same letter (i.e., a vs a; or b vs b) are not statistically different; where there are no letters above bars, there were no statistically differences detected
Autophagy
Autophagy is a degradative process necessary for the turnover of cytoplasmic proteins and organelles [18]. Autophagy can be suppressed by activation of mTORC1 and the subsequent increase in ULK1 S757 phosphorylation, which disrupts the interaction between AMPK and ULK1 [19]. Moreover, FoxO3/1 have been reported to regulate autophagy in skeletal muscle [20]. In the present study, refeeding increased FoxO3/O1 phosphorylation in saline-treated control muscle (Figure 5). While alcohol did not alter the extent of FoxO3/1 phosphorylation in the fasted condition, compared to the control fasted state, alcohol did blunt the refeeding-induced increase in FoxO3/1 phosphorylation. Refeeding also increased ULK1 S757 phosphorylation almost 3-fold in muscle from saline-treated control mice (Figure 5). Alcohol not only decreased ULK1 phosphorylation under fasted conditions, compared to control, but also attenuated the magnitude of the increased by refeeding (Figure 5). LC3B-II, a surrogate marker of autophagy, was decreased in saline-treated control mice after refeeding, and this was associated with an increase in the protein level for p62, a protein that is consumed when autophagy is stimulated. In contrast, the relatively amount of LC3B-II and p62 were not altered by alcohol in either the fasted or refed state. Additionally, Atg12 and Beclin-1 were not altered by refeeding in either control or alcohol-treated mice (Figure 5).
Fig 5.
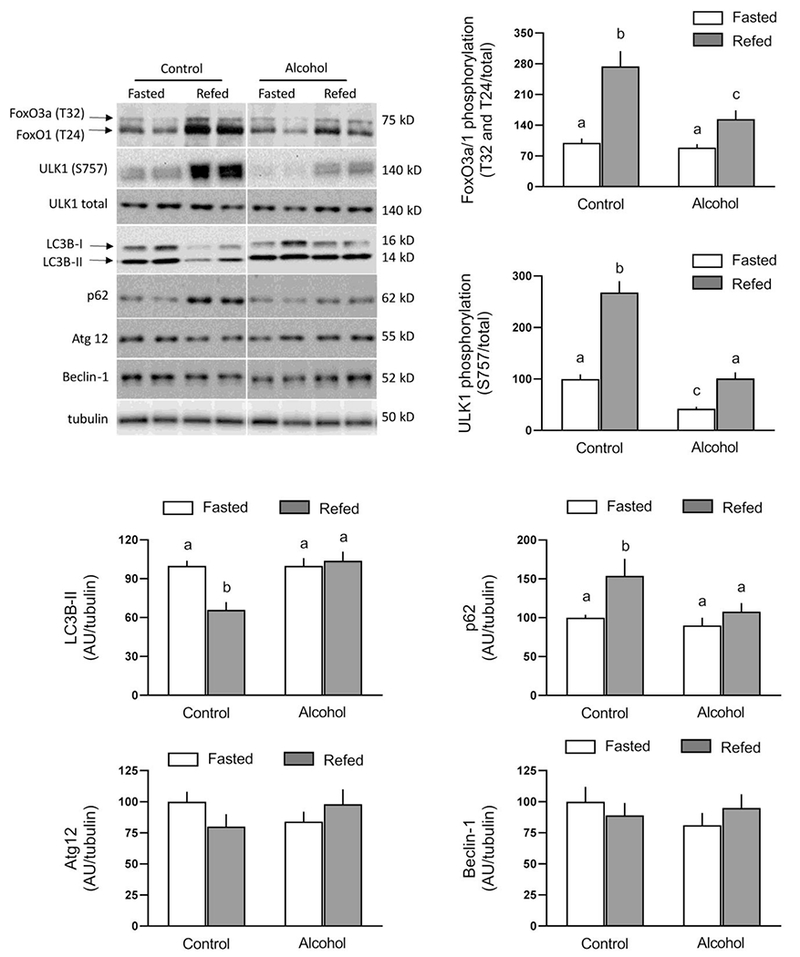
Refeeding-induced changes in FoxO3/FoxO1 and ULK1 phosphorylation as well as total LC3B-II, p62, Atg12, and Beclin-1 in gastrocnemius/plantaris from saline-treated control and ethanol-treated mice. A representative Western blot for each protein of interest is shown in the top left panel. The approximate molecular weight in kD for the protein of interest is shown to the right of each blot. Samples from all four experimental groups were run on the same gel and the vertical white line indicates that intervening lanes of the gel have been removed for presentation purposes. Bar graphs represent densitometric analysis of all immunoblots, where the value from fasted saline-treated control mice was set at 100 AU. Values are means ± SEM; n=10, 10, 12 and 12, respectively. Means with different letters (a vs b) are statistically different from each other (P<0.05), whereas means with the same letter are not statistically different
Proteasome and cathepsin L activity
Intracellular protein homeostasis is also regulated by the ubiquitin (Ub)-proteasome pathway (UPP) that is activated in many pathological conditions exhibiting muscle wasting [21]. We detected no ethanol- and/or refeeding-induced changes in the amount of total ubiquitinated proteins or amount of the various molecular weight forms of calpastatin (data not shown). Further, we detected no ethanol- and/or refeeding-induced change in the mRNA content for atrogin-1 or MuRF1 (Table 1), two muscle-specific Ub E3 ligases that are typically elevated in catabolic disease [22]. Lastly, in vitro-determination of proteasome activity and cathepsin L activity was also not altered by ethanol and/or refeeding (Table 1).
Table 1.
Effect of ethanol and/or refeeding on mRNA content for ubiquitin E3 ligases, and proteasome and cathepsin L activity in skeletal muscle.
| Saline-treated Control | Ethanol-treated | |||
|---|---|---|---|---|
| Fasted | Refed | Fasted | Refed | |
| Proteasome activity, nmol/min/μg protein | 2.8 ± 0.4 | 2.6 ± 0.3 | 2.7 ± 0.4 | 2.5 ± 0.4 |
| Cathepsin L activity, nmol/min/ mg protein | 5.7 ± 0.8 | 5.5 ± 0.6 | 5.8 ± 0.7 | 5.7 ± 0.4 |
| Atrogin-1 mRNA, AU/L32 | 100 ± 14 | 95 ± 11 | 114 ± 12 | 109 ± 8 |
| MuRF1 mRNA, AU/L32 | 100 ± 13 | 91 ± 10 | 111 ± 14 | 105 ± 9 |
Values are means ± SEM; n=10-12 per group. For each endpoint determined, there was no statistical difference determined among the four experimental groups.
Apoptosis and inflammasome pathway
Surrogate markers of apoptosis as well as the canonical and non-canonical inflammasome pathway were also assessed. There were no statistically significant ethanol or refeeding effects on the protein expression of cleaved caspase 3 and cleaved PARP (e.g., apoptosis), NLRP3 or cleaved caspase 1 (canonical inflammasome), or caspase 11 and Gasdermin D (non-canonical inflammasome) (data not shown).
DISCUSSION
Herein we present a comprehensive description of the effect of acute ethanol on signal transduction pathways regulating protein synthesis and degradation in skeletal muscle as they are modulated by a physiologically relevant refeeding stimulus. Our data demonstrate refeeding increases protein synthesis and decreases autophagy in association with activation of the AKT-mTORC1 pathway in saline-treated control mice, without apparent alteration of protein breakdown via the Ub-proteasome. In contrast, acute ethanol suppresses or blunts, at least in part, AKT-mTORC1 activity stimulated by refeeding indicating that alcohol prevents the nutrient-induced stimulation of protein synthesis as well as impairs the anticipated decrease in autophagy in skeletal muscle.
In control fasted mice, refeeding for 30 min was sufficient to stimulate the AKT-mTORCl pathway, as evidenced by the coordinate increase in the phosphorylation of AKT (T308 and S473), S6K1 (T389), 4E-BP1 (S65) and ULK1 (S757). The functional importance of the refeeding-induced increase in S6K1 can be inferred by the increased phosphorylation of downstream substrates S6 and eIF4B, which would be anticipated to increase mRNA translation initiation [23]. Further, S6K1 phosphorylates eEF2 kinase which subsequently inhibits eEF2 phosphorylation, and hence the refeeding response was also consistent with an enhanced elongation phase of mRNA translation [24]. Overall, the activation of the AKT-mTORCl pathway was consistent with the increase in global protein synthesis we observed in skeletal muscle, and these data are consistent with previous reports of the anabolic response of refeeding in rodents [25–27]
Acute ethanol, in contrast, produced a profound nutrient-resistant state as it completely prevented the refeeding-induced increase in muscle protein synthesis as well as blunted the increased phosphorylation of S473 AKT and S6K1 phosphorylation. It is noteworthy that ethanol did not alter the phosphorylation state of 4E-BP1 another downstream target of mTORCl, suggesting that the suppression of translation initiation and protein synthesis was not due to a redistribution of eIF4E between the inactive eIF4E•4EBP1 complex and the active eIF4E•eIF4G complex [28]. The underlying mechanism by which ethanol preferentially inhibits the refeeding-induced increase in mTORCl kinase activity directed toward S6K1 (and its downstream substrates) compared to 4E-BP1 is not known, but has been previously reported in studies examining the interaction of ethanol and either IGF-I or leucine [7, 8]. Additionally, there was no refeeding-induced decrease in eEF2 phosphorylation in ethanol-treated mice suggesting that the rate of translational elongation was not increased, as seen in control mice. This impairment in elongation by alcohol has also been observed previously in response to the anabolic stimulus of muscle contraction when completed prior to acute alcohol intoxication [29]. We have previously determined the plasma concentration for insulin under the same experimental conditions and found no difference in the prevailing concentration of this anabolic hormone between control and ethanol-treated mice after refeeding [12]. Similarly, both groups exhibited a comparable increase in the plasma leucine concentration after refeeding. Hence, differences in the absorption and/or whole-body clearance of these anabolic mediators appears unlikely contributors to the observed nutrient resistance. This conclusion is consistent with data from in vitro studies using the isolated epitrochlearis muscle preparation and cultured myocytes indicating ethanol directly blunts nutrient or hormone stimulated muscle protein synthesis and S6K1 activity [30, 31]. Similarly, the ability of ethanol to impaired muscle protein synthesis in response to other anabolic stimuli (e.g., muscle contraction) also appears more related to its capacity to suppress S6K1 as opposed to 4E-BP1 phosphorylation [32]. Furthermore, this differential mTORCl phosphorylation of sites on S6K1 and 4E-BP1 has been proposed to modulate the many effects of this kinase [33] As refeeding has been previously shown to increase in protein synthesis in cardiac muscle [12], the observed nutrient resistance in skeletal muscle suggests a tissue- or muscle type-specific effect.
The ability of ethanol to prevent the above mentioned refeeding-induced changes in AKT-mTORCl signaling occurred independent of ethanol’s effect on signaling through this pathway under basal fasted conditions. Moreover, while ethanol did not alter the phosphorylation state of ART, 4E-BP1, S6K1 or eEF2 in the fasted condition, it did inhibit muscle protein synthesis. These data are consistent with previously published results [32]. These data obtained in mice suggest an mTORCl-independent mechanism in the fasted condition. However, in contrast, data from rat have consistently demonstrated a reduction in 4E-BP1 phosphorylation as well as an increase in inactive eIF4E•4EBP1 complex and decrease in the active eIF4E•eIF4G complex [3, 7], suggesting and mTORCl dependent mechanism. The reason for this apparent species-specific effect of alcohol in skeletal muscle is not known.
As noted above, we also observed that refeeding increased ULKl S757-phosphorylation in control mice. Such an increase, coupled with the decrease in LC3B-II and increase in p62, is consistent with decreased autophagy [19]. A comparable refeeding-induced decrease in LC3B-II and increase in p62 in muscle has been previously reported [34] and is, at least in part, rapamycin (mTORCl)-dependent [26]. Moreover, the elevation in insulin as opposed to amino acids appears primarily responsible for the inhibition of LC3B-II in muscle [26]. While we detected no evidence of a change in surrogate markers of autophagy with ethanol in the fasted state, the typical refeeding-induced inhibition of autophagy was not detected in ethanol-treated mice. This failure to suppress autophagy was associated with a diminished phosphorylation of ULKl and FoxO3/1 in ethanol-fed mice, which is consistent with the coordinate decrease in the activation of the AKT-mTOR signaling pathway.
In contrast to the rapid refeeding-induced changes in autophagy, we failed to detect consistent changes in the amount of total ubiquitinated proteins, the mRNA content for the muscle-specific E3 ligases MuRFl and atrogin-1, and the in vitro-determined proteasome activity and cathepsin L activity either in response to ethanol alone or in combination with refeeding. This lack of response was not anticipated based on the refeeding-induced increase in FoxO3/1 phosphorylation in the current study and the data from previous studies demonstrating that FoxO3 regulates the ubiquitin-proteasome pathway [20]. However, our current data are consistent with the inability of ethanol to alter the IGF-I-induced suppression of myofibrillar degradation based on 3-methylhistidine and tyrosine release by the isolated perfused hindlimb [35]. Collectively, these data suggest that refeeding did not alter proteasome-dependent protein degradation at the time point examined. While we have limited data pertaining to lysosomal proteolysis, cathepsin L activity was not altered by ethanol intoxication or acutely by refeeding. Similarly, cathepsins B and D have also been reported to be unchanged by acute ethanol intake [36]. However, again, we cannot exclude the possibility that refeeding and/or alcohol could alter proteasomal and/or lysosomal proteolysis at later time points. Such a possibility would be consistent with previous studies indicating that the nutrient-induced inhibition of proteasome and lysosomal/calcium-dependent proteolysis are only manifested many hours after refeeding [37].
Finally, we also examined the effect of ethanol and/or refeeding on surrogate markers of apoptosis as well as the canonical and non-canonical inflammasome pathway as possible signaling pathways that regulate protein degradation. In this regard, there was no statistically significant ethanol and/or refeeding effect on the relative amount of key proteins known to regulate apoptosis (i.e., cleaved caspase 3 and cleaved PARP), or the canonical inflammasome pathway (i.e., NLRP3 or cleaved caspase 1), or the non-canonical inflammasome pathway (i.e., caspase 11 and Gasdermin D). As described above in relation to proteasome activity, we cannot exclude the possibility that ethanol and/or refeeding influenced one or more of these pathways at a later time point.
In summary, acute ethanol intoxication suppressed the normal refeeding-induced increase in mTORC1 activity, which was manifested by a reduction in S6K1 activity and ULK1 phosphorylation. This suppressive effect of ethanol likely accounts for the failure of ethanol-treated mice to increase protein synthesis and inhibit autophagy in skeletal muscle in response to nutrient intake (i.e., a meal). Future studies are needed to determine whether chronic ethanol consumption produces comparable alterations, and whether the resulting imbalance in proteostasis is casually related to the development of alcoholic myopathy.
Acknowledgements
The authors would like to acknowledge the expert technical assistance of Anne Pruznak and Maithili Navarantnarajah. Research reported in this publication was supported by the National Institutes of Health under award numbers R37 AA011290 (CHL) and F32 AA023422 (JLS).
Compliance with Ethical Standards
Conflict of interest The authors declare that they have no competing interest
REFERENCES
- 1.Kimball SR and Lang CH (2018) Mechanisms Underlying Muscle Protein Imbalance Induced by Alcohol. Annu Rev Nutr 38:197–217. doi: 10.1146/annurev-nutr-071816-064642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thapaliya S, Runkana A, McMullen MR, Nagy LE, McDonald C, Naga Prasad SV and Dasarathy S (2014) Alcohol-induced autophagy contributes to loss in skeletal muscle mass. Autophagy 10:677–90. doi: 10.4161/auto.27918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lang CH, Wu D, Frost RA, Jefferson LS, Kimball SR and Vary TC (1999) Inhibition of muscle protein synthesis by alcohol is associated with modulation of eIF2B and eIF4E. Am J Physiol 277:E268–76. [DOI] [PubMed] [Google Scholar]
- 4.Steiner JL, Kimball SR and Lang CH (2016) Acute Alcohol-Induced Decrease in Muscle Protein Synthesis in Female Mice Is REDD-1 and mTOR-Independent. Alcohol Alcohol 51:242–50. doi: 10.1093/alcalc/agv105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Korzick DH, Sharda DR, Pruznak AM and Lang CH (2013) Aging accentuates alcohol-induced decrease in protein synthesis in gastrocnemius. Am J Physiol Regul Integr Comp Physiol 304:R887–98. doi: 10.1152/ajpregu.00083.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lang CH, Frost RA, Kumar V, Wu D and Vary TC (2000) Impaired protein synthesis induced by acute alcohol intoxication is associated with changes in eIF4E in muscle and eIF2B in liver. Alcohol Clin Exp Res 24:322–31. [PubMed] [Google Scholar]
- 7.Kumar V, Frost RA and Lang CH (2002) Alcohol impairs insulin and IGF-I stimulation of S6K1 but not 4E-BP1 in skeletal muscle. Am J Physiol Endocrinol Metab 283:E917–28. doi: 10.1152/ajpendo.00181.2002 [DOI] [PubMed] [Google Scholar]
- 8.Lang CH, Frost RA, Deshpande N, Kumar V, Vary TC, Jefferson LS and Kimball SR (2003) Alcohol impairs leucine-mediated phosphorylation of 4E-BP1, S6K1, eIF4G, and mTOR in skeletal muscle. Am J Physiol Endocrinol Metab 285:E1205–15. doi: 10.1152/ajpendo.00177.2003 [DOI] [PubMed] [Google Scholar]
- 9.Sneddon AA, Koll M, Wallace MC, Jones J, Miell JP, Garlick PJ and Preedy VR (2003) Acute alcohol administration inhibits the refeeding response after starvation in rat skeletal muscle. Am J Physiol Endocrinol Metab 284:E874–82. doi: 10.1152/ajpendo.00209.2002 [DOI] [PubMed] [Google Scholar]
- 10.Steiner JL and Lang CH (2015) Dysregulation of skeletal muscle protein metabolism by alcohol. Am J Physiol Endocrinol Metab 308:E699–712. doi: 10.1152/ajpendo.00006.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garlick PJ, Millward DJ and Waterlow JC (1973) Protein turnover in cardiac and skeletal muscle. J Physiol 231:101P–102P. [PubMed] [Google Scholar]
- 12.Mekheal M SJaLC (2018) Acute alcohol prevents the refeeding-induced decrease in autophagy but does not alter the increased protein synthetic response in heart. Alcohol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lang CH and Korzick DH (2014) Chronic alcohol consumption disrupts myocardial protein balance and function in aged, but not adult, female F344 rats. Am J Physiol Regul Integr Comp Physiol 306:R23–33. doi: 10.1152/ajpregu.00414.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manning BD and Toker A (2017) AKT/PKB Signaling: Navigating the Network. Cell 169:381–405. doi: 10.1016/j.cell.2017.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Proud CG (2005) eIF2 and the control of cell physiology. Semin Cell Dev Biol 16:3–12. doi: 10.1016/j.semcdb.2004.11.004 [DOI] [PubMed] [Google Scholar]
- 16.Zhang Z, Liu R, Townsend PA and Proud CG (2013) p90(RSK)s mediate the activation of ribosomal RNA synthesis by the hypertrophic agonist phenylephrine in adult cardiomyocytes. J Mol Cell Cardiol 59:139–47. doi: 10.1016/j.yjmcc.2013.03.006 [DOI] [PubMed] [Google Scholar]
- 17.Das F, Ghosh-Choudhury N, Bera A, Kasinath BS and Choudhury GG (2013) TGFbeta-induced PI 3 kinase-dependent Mnk-1 activation is necessary for Ser-209 phosphorylation of eIF4E and mesangial cell hypertrophy. J Cell Physiol 228:1617–26. doi: 10.1002/jcp.24327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen-Kaplan V, Livneh I, Avni N, Cohen-Rosenzweig C and Ciechanover A (2016) The ubiquitin-proteasome system and autophagy: Coordinated and independent activities. Int J Biochem Cell Biol 79:403–418. doi: 10.1016/j.biocel.2016.07.019 [DOI] [PubMed] [Google Scholar]
- 19.Gallagher LE, Williamson LE and Chan EY (2016) Advances in Autophagy Regulatory Mechanisms. Cells 5. doi: 10.3390/cells5020024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, Burden SJ, Di Lisi R, Sandri C, Zhao J, Goldberg AL, Schiaffino S and Sandri M (2007) FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab 6:458–71. doi: 10.1016/j.cmet.2007.11.001 [DOI] [PubMed] [Google Scholar]
- 21.Bilodeau PA, Coyne ES and Wing SS (2016) The ubiquitin proteasome system in atrophying skeletal muscle: roles and regulation. Am J Physiol Cell Physiol 311:C392–403. doi: 10.1152/ajpcell.00125.2016 [DOI] [PubMed] [Google Scholar]
- 22.Bodine SC, Latres E, Baumhueter S, Lai VK, Nunez L, Clarke BA, Poueymirou WT, Panaro FJ, Na E, Dharmarajan K, Pan ZQ, Valenzuela DM, DeChiara TM, Stitt TN, Yancopoulos GD and Glass DJ (2001) Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 294:1704–8. doi: 10.1126/science.1065874 [DOI] [PubMed] [Google Scholar]
- 23.Magnuson B, Ekim B and Fingar DC (2012) Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks. Biochem J 441:1–21. doi: 10.1042/BJ20110892 [DOI] [PubMed] [Google Scholar]
- 24.Proud CG (2015) Regulation and roles of elongation factor 2 kinase. Biochem Soc Trans 43:328–32. doi: 10.1042/BST20140323 [DOI] [PubMed] [Google Scholar]
- 25.Dardevet D, Sornet C, Bayle G, Prugnaud J, Pouyet C and Grizard J (2002) Postprandial stimulation of muscle protein synthesis in old rats can be restored by a leucine-supplemented meal. J Nutr 132:95–100. doi: 10.1093/jn/132.1.95 [DOI] [PubMed] [Google Scholar]
- 26.Naito T, Kuma A and Mizushima N (2013) Differential contribution of insulin and amino acids to the mTORC1-autophagy pathway in the liver and muscle. J Biol Chem 288:21074–81. doi: 10.1074/jbc.M113.456228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gordon BS, Williamson DL, Lang CH, Jefferson LS and Kimball SR (2015) Nutrient-induced stimulation of protein synthesis in mouse skeletal muscle is limited by the mTORC1 repressor REDD1. J Nutr 145:708–13. doi: 10.3945/jn.114.207621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin TA, Kong X, Haystead TA, Pause A, Belsham G, Sonenberg N and Lawrence JC Jr. (1994) PHAS-I as a link between mitogen-activated protein kinase and translation initiation. Science 266:653–6. [DOI] [PubMed] [Google Scholar]
- 29.Steiner JL and Lang CH (2015) Alcohol intoxication following muscle contraction in mice decreases muscle protein synthesis but not mTOR signal transduction. Alcohol Clin Exp Res 39:1–10. doi: 10.1111/acer.12600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lang CH, Pruznak AM, Deshpande N, Palopoli MM, Frost RA and Vary TC (2004) Alcohol intoxication impairs phosphorylation of S6K1 and S6 in skeletal muscle independently of ethanol metabolism. Alcohol Clin Exp Res 28:1758–67. [DOI] [PubMed] [Google Scholar]
- 31.Hong-Brown LQ, Brown CR, Kazi AA, Navaratnarajah M and Lang CH (2012) Rag GTPases and AMPK/TSC2/Rheb mediate the differential regulation of mTORC1 signaling in response to alcohol and leucine. Am J Physiol Cell Physiol 302:C1557–65. doi: 10.1152/ajpcell.00407.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steiner JL and Lang CH (2014) Alcohol impairs skeletal muscle protein synthesis and mTOR signaling in a time-dependent manner following electrically stimulated muscle contraction. J Appl Physiol (1985) 117:1170–9. doi: 10.1152/japplphysiol.00180.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kang SA, Pacold ME, Cervantes CL, Lim D, Lou HJ, Ottina K, Gray NS, Turk BE, Yaffe MB and Sabatini DM (2013) mTORC1 phosphorylation sites encode their sensitivity to starvation and rapamycin. Science 341:1236566. doi: 10.1126/science.1236566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ning L, Xu Z, Furuya N, Nonaka R, Yamada Y and Arikawa-Hirasawa E (2015) Perlecan inhibits autophagy to maintain muscle homeostasis in mouse soleus muscle. Matrix Biol 48:26–35. doi: 10.1016/j.matbio.2015.08.002 [DOI] [PubMed] [Google Scholar]
- 35.Vary TC, Frost RA and Lang CH (2008) Acute alcohol intoxication increases atrogin-1 and MuRF1 mRNA without increasing proteolysis in skeletal muscle. Am J Physiol Regul Integr Comp Physiol 294:R1777–89. doi: 10.1152/ajpregu.00056.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koll M, Ahmed S, Mantle D, Donohue TM, Palmer TN, Simanowski UA, Seltz HK, Peters TJ and Preedy VR (2002) Effect of acute and chronic alcohol treatment and their superimposition on lysosomal, cytoplasmic, and proteosomal protease activities in rat skeletal muscle in vivo. Metabolism 51:97–104. [DOI] [PubMed] [Google Scholar]
- 37.Kee AJ, Combaret L, Tilignac T, Souweine B, Aurousseau E, Dalle M, Taillandier D and Attaix D (2003) Ubiquitin-proteasome-dependent muscle proteolysis responds slowly to insulin release and refeeding in starved rats. J Physiol 546:765–76. [DOI] [PMC free article] [PubMed] [Google Scholar]


