Abstract
Isomerization of a closed to open complex of a promoter upon RNA polymerase binding involves base unpairing at the −10 region. After potassium permanganate sensitivity of unpaired thymine residues, we studied base unpairing at the −10 region during isomerization upon RNA polymerase binding at the P1 and P3 promoters of the gal operon. Substitution of adenine by 2-amino purine (2-AP) at the invariable A⋅T base pair at the −11 position of P1 and P3 prevented unpairing not only at that position but also at the other downstream positions, suggesting a “master” role of the adenine base at −11 of the template strand in overall base unpairing. 2-AP at −11 did not inhibit the formation of RNA polymerase⋅promoter complex and subsequent isomerization of the polymerase. Substitution of adenine by 2-AP at several other positions did not affect thymine unpairing. Changing the position of the amino group from C6 in adenine to C2 in 2-AP is mutational only at the master switch position, −11.
Under ordinary physiological conditions, transcription in Escherichia coli is executed by the RNA polymerase holoenzyme comprising α2ββ′σ70 subunits. Transcription initiation follows specific binding of RNA polymerase to the promotor. The initial binary complex (closed complex) undergoes structural changes (isomerization) in both proteins and DNA to make an open complex that is competent for subsequent template-dependent polymerization of ribonucleotides (1, 2). Whether isomerization of RNA polymerase and DNA occurs simultaneously or one after the other is unknown. The σ70 subunit plays a key role in recognizing and making specific contacts with base pairs at the −35 and −10 regions of the promoter (3). In the open complex, DNA at the −10 region presumably becomes single-stranded for RNA polymerase to start reading the exposed template strand and polymerizing ribonucleotides (4–10). The bases in the nontemplate strand make specific interactions with amino acid residues of σ70 (11). The consequence of these interactions is believed to stabilize the unstable nature of the unpaired DNA region (11–13). The consensus DNA sequence of the −10 region is 5′-TATAAT-3′ in the nontemplate strand, with the first T being at the −12 position (1). To elucidate the molecular procedures of base unpairing during open complex formation, altered forms of DNA (depurinated, nicked, forked, or with single-stranded gap) in the −10 region were used as templates (10, 14–19). These studies suggested that base unpairing involves an initial distortion or localized melting event around −11 (16, 20). In this paper, we provide results by using intact duplex DNA to demonstrate that the adenine residue itself at the position −11 of the nontemplate strand plays a master role in the initiation of base unpairing.
Materials and Methods
Materials.
E. coli RNA polymerase holoenzyme was purchased from Epicentre Technologies (Madison, WI). cAMP receptor protein (CRP), purified to 98% homogeneity by FPLC (Amersham Pharmacia) from an E. coli strain carrying the crp gene in a multicopy plasmid pHA5, was a gift from S. Garges (National Cancer Institute, Bethesda). Synthetic oligodeoxynucleotides containing 2-amino purine (2-AP) substitutions (106-bases long) were obtained from Genosys (The Woodlands, TX) or Trilink BioTechnologies (San Diego). The final product of synthetic oligonucleotides was purified by PAGE.
Preparations of DNA Templates Containing 2-AP.
The 2-AP-containing DNA templates were prepared by PCR in which one of the primers was 2-AP containing synthetic oligo. The second primer corresponded to the segment downstream to a transcription terminator (Fig. 1). The DNA template for PCRs was plasmid pSA852. We prepared 358-bp-long PCR DNA fragments that generate a 125-nt-long RNA transcript from the gal P1 promoter in in vitro transcription reactions. After the PCR, the 358-bp DNA fragments were purified by electroelution (Bio-Rad) followed by ethanol precipitation.
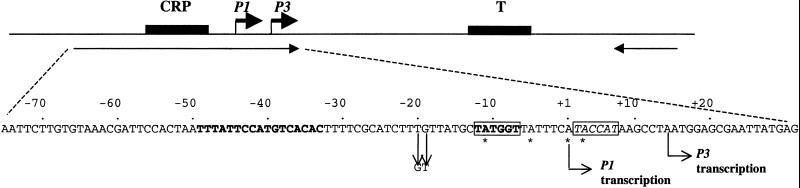
Figure 1.
The gal promoter region of E. coli showing the P1 and P3 promoters in the plasmid pSA852 (D. Lewis, unpublished data). pSA852 contains a 288-bp segment of the gal control region (−197 to +91) followed by a transcription terminator (21). The CRP binding site is in bold. The transcription start site of P1 is denoted as +1. The −10 region, the P1 promoter, is in bold and boxed, and that of P3 is boxed. The adenine residues with the asterisks were replaced individually by 2-AP in the 106-base-long rightward primer. DNA templates (385 bp) for in vitro transcription containing 2-AP were generated by PCR using the rightward and leftward primers (shown by arrows) from pSA852. The 106-base-long primer also contained −20T to G and −19T to G changes to inactivate the P2 promoter. T, transcription termination signal.
In Vitro Transcription.
In vitro transcription reactions were performed at 25°C in a 50-μl volume following the procedure of Choy and Adhya (21). The initial reaction mixture (45 μl) contained 10 nM DNA template (2 nM for supercoiled pSA852), 20 nM RNA polymerase, 20 mM Tris acetate, pH 7.5, 10 mM magnesium acetate, 200 mM potassium glutamate, and 40 units of rRNasin (Promega). CRP, when present, was added to the initial reaction mixture at 50 nM. The cAMP contraction was 100 μM. The mixture was incubated at 25°C for 20 min, and 5 μl of NTP (2 mM of ATP, GTP, and CTP, 0.2 mM UTP, and 5 μCi [α-32P]UTP) was added. The mixture was incubated further for 10 min at 25°C, and the reaction was terminated by the addition of 50 μl of STOP solution (BRL). Samples were boiled for 2 min, and 3 μl of each sample was loaded onto 8% polyacrylamide-urea sequencing gels to analyze the RNA. Quantitation of the transcripts was done by using a PhosphorImager (Molecular Dynamics).
DNaseI Footprinting Assay.
The complex formation reaction was performed in a 50-μl transcription buffer at 25°C for 20 min. The final concentrations of RNA polymerase and DNA were 50 nM and 4 nM, respectively. This reaction was treated with DNase I (1.5 units, BRL) for 4 min at 25°C. DNase I was inactivated by a phenol/chloroform (1:1) extraction followed by a G-50 spin column (Amersham Pharmacia). The pattern was generated by PCR using a 32P-labeled single primer followed by a DNA-sequencing gel electrophoresis.
Electrophoretic Mobility-Shift Assay.
The complex formation was identical to that of the DNase I footprinting assay. One microliter of properly diluted RNA polymerase was mixed with 10 μl of 32P-labeled 358-bp DNA template. The final concentrations of RNA polymerase for each reaction are shown in Fig. 4. The DNA concentration was 200 pM. The reaction was incubated at 25°C for 20 min followed by the addition of heparin to a final concentration of 50 μg/ml. Five microliters of the reaction was analyzed by 4% PAGE.
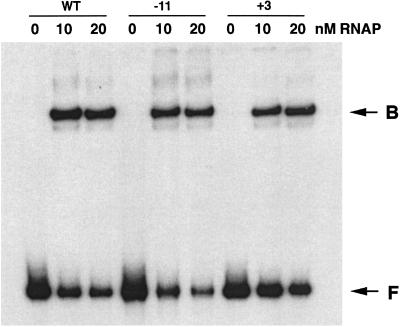
Figure 4.
Electrophoretic mobility-shift assays for RNA polymerase (RNAP) binding to 2-AP-containing DNA templates in the presence of heparin. WT, DNA containing no 2-AP; −11 and +3, the positions of 2-AP in DNA, when present; B, the RNA polymerase⋅DNA binary complex; F, unbound DNA.
Potassium Permanganate Assay.
The complex formation was identical to that of the DNase I footprinting assay. The reaction was treated with 5 μl of 80 mM KMnO4 and quenched by the addition of 5 μl of 14.4 M β-mercaptoethanol followed by a phenol/chloroform (1:1) extraction. The DNA was cleaned further by G-50 spin column and precipitated by ethanol. The modified resides were identified by PCR using 32P-labeled primers followed by electrophoresis in DNA-sequencing gel.
Results and Discussion
2-AP is an adenine analog in which the amino group is present at the C2 of the purine ring instead of at C6. 2-AP can substitute for adenine and pair with thymine without changing much of the physicochemical properties of double-stranded polynucleotide (22–26). 2-AP acts as an adenine and pairs with thymine when present in the template strand during polynucleotide synthesis by DNA polymerase (see the experiments mentioned in the Fig. 5 legend). Yet as shown below, 2-AP has an effect on transcription initiation only when present in a specific position in the −10 region of the P1 and P3 promoters of the gal operon of E. coli. 2-AP-containing DNA (Fig. 1) was generated by PCR for use as templates of transcription by RNA polymerase in vitro. The gal operon has been shown to contain four promotors: P1, P2, P3, and P4¶ (27–29). Of these, P2 was inactivated by substitution mutations (Fig. 1), and P4, which is located further upstream, was left out in these templates made by PCR. Thus, these templates showed synthesis of 135- and 121-nt-long RNA molecules corresponding to the P1 and P3 promoters as described below. We replaced the four adenine residues at positions −11, −5, +1, and +3 in the −10 region of the nontemplate strand of P1 by 2-AP individually. Wild-type DNA and DNA containing 2-AP at position −5 or +3 produced as much P1 RNA as from wild type (Fig. 2). The template with 2-AP at position +1 made P1 RNA at 10% of the wild-type level. However, the RNA molecules from P1 were conspicuously absent from the template with 2-AP at position −11. The results of defective P1 RNA synthesis with 2-AP at the −11 position were interesting considering the minor structural difference between adenine and 2-AP and similarities in the physical and biochemical properties of adenine and 2-AP-containing DNA as mentioned. When the adenine at position −11 of the nontemplate strand of the template was replaced by guanine, cytosine, or thymine, the resulting P1 promoter also became totally defective in the corresponding RNA synthesis (data not shown). An adenine → thymine transversion at the −11 position was shown previously to inactivate the P1 promoter both in vivo and in vitro (30, 31). Together, these results showed that the adenine at the −11 position of P1 is critical for transcription initiation and are consistent with adenine being the most invariant at this position in the consensus sequence (32).
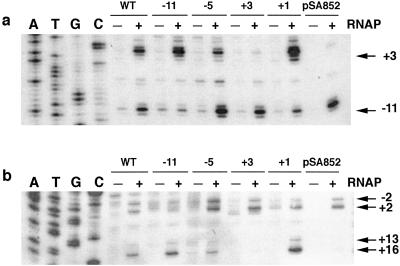
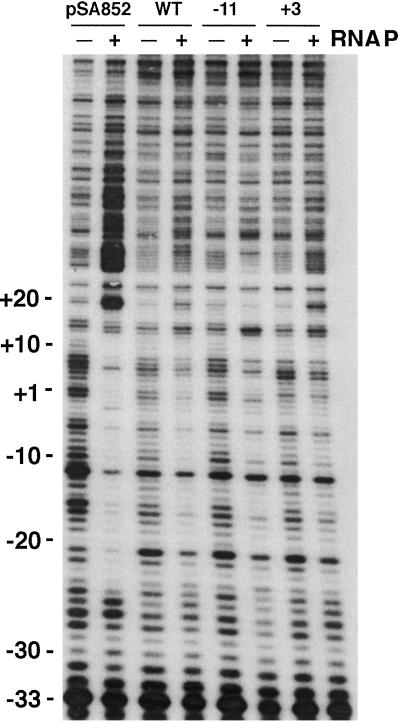
Figure 5.
Probing of thymine unpairing in DNA by KMnO4. (a) Nontemplate strand. (b) Template strand. WT, DNA containing no 2-AP. Numbers on the top indicate the positions of 2-AP in DNA. RNAP, RNA polymerase. pSA852 is supercoiled plasmid DNA. The positions of each KMnO4-reacted unpaired thymine are indicated by the numbers on the right with arrows. DNA sequencing ladders for A, T, G, and C were generated by using the DNA sequencing kit (BRL). The DNA template used in sequencing was the 358-bp PCR DNA fragment containing 2-AP at −11 position; the results show that the complementary base for 2-AP after amplification is T.
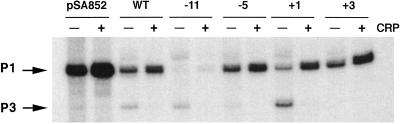
Figure 2.
In vitro transcription of the P1 promoter containing 2-AP at various positions are indicated in the figure in the absence (−) or presence (+) of CRP. pSA852, supercoiled DNA template containing no 2-AP; WT, DNA containing no 2-AP; arrows, P1 and P3 transcripts.
The 121-nt-long RNA from the P3 promoter was not observed in wild-type supercoiled DNA template but was found only with PCR-generated linear templates (Fig. 2). In the latter examples, P3 was made generally in lower amounts relative to P1 RNA both from wild type and 2-AP-containing DNA templates, as was observed previously (33), except when the 2-AP was present at position +3. With 2-AP at +3, no P3 RNA was observed. The +3 position of P1 corresponds to the −11 position of P3 (33). Thus similar to P1, transcription initiation from P3 was abolished when 2-AP replaced adenine at the corresponding −11 position.
CRP in conjunction with cAMP enhances the P1 promoter and represses P3 (21, 31, 33). Consistently, the presence of cAMP and CRP stimulated transcription 3–4-fold from P1 and inhibited the same from P3 with DNA templates without 2-AP (Fig. 2). The cAMP- and CRP-medicated stimulation of P1 RNA synthesis was observed also with templates having 2-AP present at position −5, +1, or +3 by the same amount as was observed with DNA without 2-AP. Although the intrinsic amount of P1 RNA synthesis was significantly lower in the template with 2-AP at the +1 position, CRP resurrected this P1 defect to the wild-type level. Unlike 2-AP at +1, cAMP and CRP barely helped the defect of P1 transcription in DNA containing 2-AP at position −11. cAMP and CRP inhibited transcription from P3 not only with the wild-type DNA, as shown previously (33), but also in DNA containing 2-AP at position −11, −5, or +1.
The binding of RNA polymerase to wild type and 2-AP-containing DNA templates was followed by DNase I protection experiments and electrophoretic mobility-shift assays. The binding of RNA polymerase to the P3 promoter was not detectable under our experimental conditions, perhaps because of a very low affinity of P3. The results of DNase I protection showed that the presence of RNA polymerase affected the digestion pattern of the DNA segment from the −28 to +14 position of P1 not only in DNA without 2-AP (both supercoiled and PCR-generated), as expected, but also in DNA containing 2-AP at position −11 (Fig. 3). The effect was less pronounced for DNA with 2-AP at position +3. The results suggested RNA polymerase binds to the P1 promoter with 2-AP at position −11. The binding was confirmed by gel electrophoresis experiments in the presence of heparin (Fig. 4). The formation of a heparin-resistant complex demonstrates structural changes, i.e., isomerization of RNA polymerase. DNA without 2-AP and DNA with 2-AP at position −11 or +3 complexed with RNA polymerase, thus retarding the mobility of the bound DNA in the same amount and to the same extent relative to free DNA during electrophoresis. Thus, 2-AP substitution at position −11, although it did not allow transcription, did not prevent binding and isomerization of RNA polymerase.
Figure 3.
DNase I footprinting of the DNA templates in the absence (−) or presence (+) of RNA polymerase (RNAP). pSA852 is supercoiled plasmid DNA. WT, DNA containing no 2-AP; −11 and +3, the positions of 2-AP in the DNA, when present.
We studied base unpairing at the −10 region of DNA containing 2-AP at different positions. Base unpairing was monitored by the treatment of the DNA⋅protein complexes with KMnO4 (34). KMnO4 generates hydroxy radicals, which detect unpaired thymines in DNA duplexes. The location of the unpaired thymines in both template and nontemplate strands was detected by primer extension (35). The unpairing of thymine residues in open complexes at the P1 promoter has been identified previously by KMnO4 reaction (36, 37). These authors showed that the major opening of thymines occurred at the −11, −5, and +3 positions in the nontemplate strand. Fig. 5 shows the results of thymine unpairing with or without 2-AP-containing DNA. Thymine unpairing was observed under our assay conditions at positions −11 and +3 in the nontemplate strand and at positions −2, +2, +13, and +16 in the template strands of DNA without 2-AP. Of these, thymines at position −11 in the nontemplate strand and at −2 and +2 in the template strand were unpaired during open complex formation at the P1 promoter. The thymine unpairing at +3 in the nontemplate strand and at +13 and +16 in the template strand were caused by open complex formation at P3; unpairing at these positions was not observed in supercoiled DNA as expected from the transcription results described above. Note that the +3, +13, and +16 positions correspond to the −11, −2, and +2 positions of P3. We investigated the effect of 2-AP substitutions of adenine opposite to thymines at positions −11, −5, +1, and +3 on thymine unpairing by KMnO4 treatment (Fig. 5). It is clear that 2-AP at positions −5, +1, and +3 did not affect the thymine unpairing pattern at the P1 promoter; thymine residues at positions −11, −2, and +2 were sensitive to KMnO4 as with DNA without 2-AP. Interestingly, the presence of 2-AP at position −11 of P1 has a pleiotropic effect; it not only abolished unpairing of thymine opposite to 2-AP at −11 but also failed to show unpairing at positions −2 and +2 of P1. The presence of 2-AP at +3 also showed the same pleiotropic effect in P3; it not only abolished unpairing of thymine opposite to 2-AP at +3 but also failed to show unpairing at positions +13 and +16. These results clearly showed that when 2-AP resides at the corresponding −11 position of P1 and P3, none of the thymines unpaired.
Despite the fact that adenine and 2-AP are isomers that are indistinguishable in several physical and biochemical properties as discussed, 2-AP behaved differently from adenine when it replaced the latter at the invariant −11 position in the P1 and P3 promoters of gal. Although DNA containing 2-AP at −11 did bind RNA polymerase, it prevented unpairing and subsequent transcription, perhaps because the amino group at C6 of the purine makes a critical contact with the σ subunit, which is essential for unpairing. It has been suggested that the adenine at −11 interacts with the σ70 subunit by becoming stacked between two aromatic amino acid residues, Trp-433 and Tyr-430 (12, 38, 39). It is noteworthy that 2-AP at several other positions of the two gal promoters did not show any mutational effect. Unlike at other positions, 2-AP at position −11 at P1 not only prevented unpairing at that position but also prevented base unpairing at other positions located downstream, thus showing a polar effect. These results suggest that unpairing of the invariant A⋅T base pair −11 is a forerunner event and acts as the “master” of unpairing of bases. It has been suggested that DNA “melting” by RNA polymerase starts with a nucleation in the −12 to −4 region and extends up to +2 position downstream (18). Our finding of −11 A⋅T being the master position further pinpoints the actual start of the process of the nucleation. There may be a hidden rule in the pattern of base unpairing in a promoter guided by overall DNA sequence of the region; nevertheless, base unpairing shows polarity starting at a master position and moving downstream. Although no information is available about how RNA polymerase opens base pairs at a promoter, our demonstration of formation of heparin-resistant complex without base unpairing indicates that isomerization of RNA polymerase precedes that of DNA.
Acknowledgments
We thank Dale Lewis for the gift of several plasmids, Angela Fox for technical assistance, and Susan Garges and Jay Gralla for critical suggestions.
Abbreviations
- CRP
cAMP receptor protein
- 2-AP
2-amino purine
Footnotes
The newly discovered P4 promoter also was named as P3 in the original publication (27). We have changed that to P4 in this report, with the permission of the authors, to avoid confusion.
References
- 1.McClure W R. Annu Rev Biochem. 1985;54:171–204. doi: 10.1146/annurev.bi.54.070185.001131. [DOI] [PubMed] [Google Scholar]
- 2.Record M T J, Reznikoff W S, Craig M L, McQuade K L, Schlax P J. In: Escherichia Coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, editor. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 792–820. [Google Scholar]
- 3.Gross C A, Lonetto M, Losick R. In: Transcriptional Regulation. Yamamoto K, McKnight S, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. pp. 129–176. [Google Scholar]
- 4.Buc H, McClure W R. Biochemistry. 1985;24:2712–2722. doi: 10.1021/bi00332a018. [DOI] [PubMed] [Google Scholar]
- 5.Craig M L, Suh W-C, Record M T J. Biochemistry. 1995;34:15624–15632. doi: 10.1021/bi00048a004. [DOI] [PubMed] [Google Scholar]
- 6.deHaseth P L, Helmann J D. Mol Microbiol. 1995;16:817–824. doi: 10.1111/j.1365-2958.1995.tb02309.x. [DOI] [PubMed] [Google Scholar]
- 7.Roe J H, Burgess R R, Record M T. J Mol Biol. 1984;176:495–521. doi: 10.1016/0022-2836(84)90174-8. [DOI] [PubMed] [Google Scholar]
- 8.Roe J H, Burgess R R, Record M T. J Mol Biol. 1985;184:441–453. doi: 10.1016/0022-2836(85)90293-1. [DOI] [PubMed] [Google Scholar]
- 9.Severinov K, Darst S A. Proc Natl Acad Sci USA. 1997;94:13481–13486. doi: 10.1073/pnas.94.25.13481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zaychikov E, Denissova L, Meier T, Gotte M, Heumann H. J Biol Chem. 1997;272:2259–2267. doi: 10.1074/jbc.272.4.2259. [DOI] [PubMed] [Google Scholar]
- 11.Roberts C W, Roberts J W. Cell. 1996;86:495–501. doi: 10.1016/s0092-8674(00)80122-1. [DOI] [PubMed] [Google Scholar]
- 12.Helmann J D, Chamberlin M J. Annu Rev Biochem. 1988;57:839–872. doi: 10.1146/annurev.bi.57.070188.004203. [DOI] [PubMed] [Google Scholar]
- 13.Juang Y-L, Helmann J D. J Mol Biol. 1994;235:1470–1488. doi: 10.1006/jmbi.1994.1102. [DOI] [PubMed] [Google Scholar]
- 14.Burns H, Minchin S. Nucleic Acids Res. 1994;22:3840–3845. doi: 10.1093/nar/22.19.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Y-F, Helmann J D. J Mol Biol. 1997;267:47–59. doi: 10.1006/jmbi.1996.0853. [DOI] [PubMed] [Google Scholar]
- 16.Helmann J D, deHaseth P L. Biochemistry. 1999;38:5959–5967. doi: 10.1021/bi990206g. [DOI] [PubMed] [Google Scholar]
- 17.Juang Y-L, Helmann J D. Biochemistry. 1995;34:8465–8473. doi: 10.1021/bi00026a030. [DOI] [PubMed] [Google Scholar]
- 18.Li X-Y, McClure W R. J Biol Chem. 1998;273:23558–23566. doi: 10.1074/jbc.273.36.23558. [DOI] [PubMed] [Google Scholar]
- 19.Meier T, Schickor P, Wedel A, Cellai L, Heumann H. Nucleic Acids Res. 1995;23:988–994. doi: 10.1093/nar/23.6.988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomsic M, Tsujikawa L, Panaghie G, Wang Y, Azok J, deHaseth P L. J Biol Chem. 2001;274:31891–31896. doi: 10.1074/jbc.M105027200. [DOI] [PubMed] [Google Scholar]
- 21.Choy H E, Adhya S. Proc Natl Acad Sci USA. 1993;90:472–476. doi: 10.1073/pnas.90.2.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McLaughlin L W, Benseler F, Graeser E, Piel N, Scholtissek S. Biochemistry. 1987;26:7238–7245. doi: 10.1021/bi00397a007. [DOI] [PubMed] [Google Scholar]
- 23.Nordlund T M, Andersson S, Nilsson L, Rigler R, Graslund A, McLaughlin L W. Biochemistry. 1989;28:9095–9103. doi: 10.1021/bi00449a021. [DOI] [PubMed] [Google Scholar]
- 24.Sowers L C, Fazakerley G V, Eritja R, Kaplan B E, Goodman M F. Proc Natl Acad Sci USA. 1986;83:5434–5438. doi: 10.1073/pnas.83.15.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu P, Nordlund T M, Gildia B, McLaughlin L W. Biochemistry. 1990;29:6508–6514. doi: 10.1021/bi00479a024. [DOI] [PubMed] [Google Scholar]
- 26.Xu D, Evans K O, Nordlund T M. Biochemistry. 1994;16:9592–9595. doi: 10.1021/bi00198a027. [DOI] [PubMed] [Google Scholar]
- 27.Sur R, Debnath D, Mukhopadhya J, Parrack P. Eur J Biochem. 2001;268:2344–2350. doi: 10.1046/j.1432-1327.2001.02114.x. [DOI] [PubMed] [Google Scholar]
- 28.Adhya S, Miller W. Nature (London) 1979;279:492–494. doi: 10.1038/279492a0. [DOI] [PubMed] [Google Scholar]
- 29.Ponnambalam S, Chan B, Busby S. Mol Microbiol. 1988;2:165–172. doi: 10.1111/j.1365-2958.1988.tb00018.x. [DOI] [PubMed] [Google Scholar]
- 30.Busby S. J Mol Biol. 1982;154:211–227. doi: 10.1016/0022-2836(82)90061-4. [DOI] [PubMed] [Google Scholar]
- 31.Musso R E, Di Lauro R, Adhya S, de Crombrugghe B. Cell. 1977;12:847–854. doi: 10.1016/0092-8674(77)90283-5. [DOI] [PubMed] [Google Scholar]
- 32.Hawley D K, McClure W R. Nucleic Acids Res. 1983;11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ponnambalam S, Spassky A, Busby S. FEBS Lett. 1987;219:189–196. doi: 10.1016/0014-5793(87)81214-0. [DOI] [PubMed] [Google Scholar]
- 34.Sasse-Dwight S, Gralla J D. J Biol Chem. 1989;264:8074–8081. [PubMed] [Google Scholar]
- 35.Sasse-Dwight S, Gralla J D. Methods Enzymol. 1991;208:146–168. doi: 10.1016/0076-6879(91)08012-7. [DOI] [PubMed] [Google Scholar]
- 36.Burns H D, Ishihama A, Minchin S D. Nucleic Acids Res. 1999;27:2051–2056. doi: 10.1093/nar/27.9.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chan B, Minchin S, Busby S. FEBS Lett. 1990;267:46–50. doi: 10.1016/0014-5793(90)80284-p. [DOI] [PubMed] [Google Scholar]
- 38.Fenton M S, Lee S J, Gralla J D. EMBO J. 2000;19:1130–1137. doi: 10.1093/emboj/19.5.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Panaghie G, Aiyar S E, Bobb K L, Hayward R S, de Haseth P L. J Mol Biol. 2000;299:1217–1230. doi: 10.1006/jmbi.2000.3808. [DOI] [PubMed] [Google Scholar]