Abstract
The two most common polymorphisms of the human DRD4 gene encode a dopamine D4 receptor (D4R) with four or seven repeats of a proline-rich sequence of 16 amino acids (D4.4R or D4.7R). Although the seven-repeat polymorphism has been repeatedly associated with attention deficit hyperactivity disorder and substance use disorders, the differential functional properties between D4.4R and D4.7R remained enigmatic until recent electrophysiological and optogenetic-microdialysis experiments indicated a gain of function of D4.7R. Since no clear differences in the biochemical properties of individual D4.4R and D4.7R have been reported, it was previously suggested that those differences emerge upon heteromerization with dopamine D2 receptor (D2R), which co-localizes with D4R in the brain. However, contrary to a gain of function, experiments in mammalian transfected cells suggested that heteromerization with D2R results in lower MAPK signaling by D4.7R as compared to D4.4R. In the present study, we readdressed the question of functional differences of D4.4R and D4.7R forming homomers or heteromers with the short isoform of D2R (D2SR), using a functional bioluminescence resonance energy transfer (BRET) assay that allows the measurement of ligand-induced changes in the interaction between G protein coupled receptors (GPCRs) forming homomers or heteromers with their cognate G protein. Significant functional and pharmacological differences between D4.4R and D4.7R were only evident upon heteromerization with the short isoform of D2R (D2SR). The most dramatic finding was a significant increase and decrease in the constitutive activity of D2S upon heteromerization with D4.7R and D4.4R, respectively, providing the first clear mechanism for a functional difference between both products of polymorphic variants and for a gain of function of the D4.7R.
Keywords: Dopamine D4 receptor, dopamine D2 receptor, gene polymorphisms, G protein-coupled receptor heteromers, constitutive activity, bioluminescence resonance energy transfer
Background
The human DRD4 gene displays a high number of polymorphisms in its coding sequence. The most extensive polymorphism is found in exon 3, a region that encodes the third intracellular loop of the receptor [1–3]. This polymorphism consists of a variable number of tandem repeats (VNTR) in which a 48-base pair sequence exists as 2- to 11-fold repeats. The two most common polymorphisms contain four and seven TRs (with allelic frequencies of about 60% and 20%, respectively) [2] and encode a dopamine D4 receptor (D4R) with four and seven repeats of a proline-rich sequence of 16 amino acids (D4.4R and D4.7R) [1–3]. DRD4 polymorphisms have been suggested to associate with numerous behavioral individual differences and neuropsychiatric disorders. The most consistent associations are the link between the gene encoding D4.7R and attention deficit hyperactivity disorder (ADHD) [1, 4–6] or substance use disorders (SUDs) [7].
The functional significance of the products of DRD4 polymorphisms remained enigmatic until recent results from electrophysiological experiments in mouse cortical slices with viral infection of human D4.4R and D4.7R [8] and immuno-histochemical and optogenetic-microdialysis experiments in a D4.7 knock-in mouse, expressing a humanized D4R with the long intracellular domain of the human D4.7R [9]. D4.7 or the humanized D4R demonstrated a pronounced gain of function when respectively compared with D4.4 or the wild-type (WT) mouse D4R [8, 9], which expresses a D4R with a shorter third intracellular loop comparable to the human D4.2 receptor [10]. In the brain, D4R is particularly expressed in the prefrontal cortex, particularly in the pyramidal glutamatergic neuron, where D4R exerts a significant inhibitory control at the somatodendritic as well as at its striatal projections [8–10]. Electrophysiological recording in cortical slices from DRD4 knockout mice, showed an increased ability of a D4R agonist to suppress glutamatergic excitatory network bursts upon viral infection with human D4.7 as compared with D4.4 receptor cDNA [8]. D4.7 knock-in mouse showed a blunting of methamphetamine-induced cortical activation and optogenetically- and methamphetamine-induced corticostriatal glutamate release [9].
The question now remains about the biochemical mechanism responsible for the gain of function of D4.7R. In fact, initial and still canonically cited studies seemed to indicate that the D4.7R signals with less efficiency than D4.4R [11]. Nevertheless, in a recent study using functional bioluminescence resonance energy transfer (BRET) experiments with biosensors fused to the different Gαi/o subtypes and to the Gγ subunit, we could not find significant differences in the ability of D4.4R or D4.7R to promote dopamine-induced G protein activation [12]. It has also been suggested that the potential functional and pharmacological differences between D4.4R and D4.7R signaling become evident upon heteromerization with the dopamine D2 receptor (D2R). In fact, D4R and the short isoform of the D2R (D2SR) are co-localized in striatal glutamatergic terminals and found to contribute to the inhibitory control of glutamate release exerted by dopamine and dopamine receptor agonists [10, 13]. Two separate studies, based on BRET and co-immunoprecipitation techniques in transfected mammalian cells suggested that D4.7R establishes weaker intermolecular and functional interactions with D2R (both isoforms, D2LR and D2SR) than D4.4R [10, 14]. At the functional level, a significantly stronger MAPK activation could only be observed upon simultaneous activation of D2R and D4.4R [10, 14], which seems opposite to what it would be expected from a gain of function of D4.7R over D4.4R. In the present study, we readdressed the question of functional differences of D4.4R and D4.7R forming homomers or heteromers with D2SR, using a functional BRET assay that allows the measurement of ligand-induced changes in the interaction between G protein coupled receptors (GPCRs) forming homomers or heteromers with their cognate G protein [15 16].
Materials and Methods
DNA Constructs and Transfection
Sequences encoding amino acid residues 1–229 and 230–311 of Renilla luciferase (RLuc) protein were subcloned in the pcDNA3.1 vector to obtain complementary hemitruncated RLuc proteins (nRLuc and cRLuc, respectively). D4.4R and D4.7R human cDNAs cloned into pcDNA3.1 were amplified without their stop codons using sense and antisense primers (CCGCTAGCATGAAGACGATCATCGCCCTGA and GACTCGAGGCAGCAAGCACGTAGAGCCTT, respectively) harboring unique NheI and XhoI restriction sites. The amplified fragment corresponding to D4.4R or D4.7R was subcloned to be in-frame with restriction sites of pcDNA3.1-nRLuc and pcDNA3.1-cRLuc to yield plasmids that express D4.4R or D4.7R fused to the corresponding hemitruncated RLuc on the N-or C-terminal end of the receptor (D4.4R-nRLuc, D4.4R-cRLuc, D4.7R-nRLuc and D4.7R-cRLuc). To obtain D4.4R and D4.7R fused to full-length RLuc on the C-terminal end of the receptor (D4.4R-RLuc and D4.7R-RLuc), D4.4R or D4.7R cDNAs were amplified without their stop codons using sense and antisense primers (see sequences above) harboring unique NheI and XhoI restriction sites and the amplified fragment was subcloned to be in-frame with restriction sites of a RLuc-expressing vector (pRLuc-N1; PerkinElmer, Wellesley, MA). The following human G protein constructs were also used: Gαil fused to mVenus, a derivative of yellow fluorescence protein (YFP) inserted at position 91 (Gαi-YFP), untagged Gβ1, and untagged Gγ2. All the constructs were confirmed by sequencing analysis. Plasmid cDNAs were transfected into human embryonic kidney (HEK-293T) cells (ATCC, Manassas, VA; RRID: SCR_001672) using polyethylenimine (PEI, Sigma-Aldrich, St. Louis, MO) with a 1 to 2 ratio in 10-cm dishes. Cells were maintained in culture with Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum. The amount and proportion of transfected receptor-nRLuc, receptor-cRLuc, Gα1-YFP, Gβ1 and Gγ2 were optimized by testing various proportions of plasmids encoding the different sensors. Experiments were performed ~48 hours post-transfection. D2SR-RLuc, D2SR-nRLuc, D2SR-cRLuc and Gαi1-YFP were kindly provided by J. A. Javitch (Columbia University, New York).
Complemented Donor Acceptor Functional Bioluminescence Resonance Energy Transfer (CODA-RET) assay
In the CODA-RET assay, two complementary halves of the bioluminescent biosensor, such as RLuc (nRLuc and cRLuc) are separately fused to two different receptor molecules putatively able to oligomerize and a fluorescent biosensor (such as YFP) is fused to a G protein subunit, in this case Gαi1 [15, 16]. Cells were harvested, washed and resuspended in phosphate-buffered saline (PBS). About 200,000 cells/well were distributed in 96-well plates, and 5 μM coelenterazine H (substrate for BRET1) was added to each well. One minute after addition of coelenterazine, different concentrations of dopamine, pramipexole or ropinirole (or L745–870, when analyzing its possible inverse agonism) were added to each well. In the experiments testing for the effect of antagonists of dopamine, L745–870 or raclopride were added 12 min before the addition of dopamine. Fluorescence of the acceptor was quantified (excitation at 500 nm and emission at 540 nm for 1 s recording) in Mithras LB940 (Berthold technologies, Bad Wildbad Germany) to confirm the constant expression level across experiments. In parallel, BRET signal from the same batch of cells was determined as the ratio of the light emitted by YFP (mVenus variant; 510–540 nm) over RLuc (485 nm). Results were calculated for the BRET change (BRET ratio for the corresponding drug minus BRET ratio in the absence of the drug) 10 minutes after addition of the agonists (or L745–870, when indicated). Data manipulations and statistical analyses (described in the Table legends) were performed with Prism 4 (GraphPad Software).
Statistical Information
One-way ANOVA followed by Dunnett’s multiple comparison tests (when all groups were compared versus one group) or by Tukey’s multiple comparison tests (when all possible comparisons between different groups were analyzed) were used for statistical comparisons between different groups of results (GraphPad Prism7 software, San Diego, CA). Number of experiments and replications as well as the statistical results are shown in the corresponding Fig. and Table legends.
Results
Ligand-Induced Changes in the Interaction between Gi Protein and D4.4R, D4.7R or D2SR Homomers
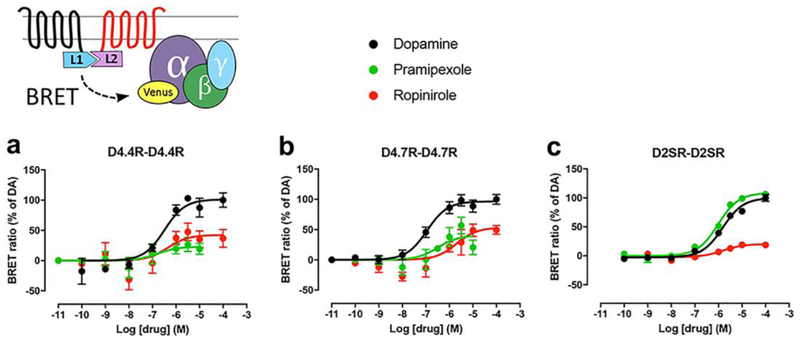
Ligand-induced changes in CODA-RET measurements imply, first, a successful complementation of Rluc and, therefore, oligomerization of the corresponding GPCR units. Second, although CODA-RET does not provide an estimate of the degree of oligomerization (affinity, stoichiometry), it represents the reading of a specific signaling through the GPCR homo- or heterodimer [15, 16]. Concentration-response curves of ligand-induced changes in BRET were then determined with CODA-RET experiments. The ligands used were dopamine and the clinically used D2-like receptor agonists pramipexole and ropinirole. For each group of experiments with the same transfected cDNA constructs, EC50 and Emax values were calculated from concentration-response curves of BRET values expressed as the percentage of the maximal effect of dopamine, considering dopamine as full agonist. Functional D4.4R-D4.4R and D4.7R-D4.7R homodimers could be demonstrated with CODA-RET, and the pattern of response to the different ligands was qualitatively similar (Fig. 1 and Table 1). Pramipexole and ropinirole were partial agonists as compared to dopamine and the efficacy of pramipexole was mostly undetectable for the D4.4R-D4.4R homomer (Fig. 1 and Table 1). On the other hand, pramipexole behaved as a full agonist on the D2R-D2R homodimer, while ropinirole was partial agonist with very low efficacy. With dopamine, the qualitative results were the same as those obtained in a previous study where we analyzed ligand-induced BRET between different Gαi/o subunits fused to RLuc and Gγ fused to YFP [12]. In both assays, dopamine showed a very similar potency for D4.4R and D4.7R and a significantly lower potency for D2SR, which would agree with the general hypothesis that homodimers represent a main functional population of GPCR [17].
Fig. 1. Ligand-induced changes in the interaction between Gi protein and D4.4R-D4.4R, D4.7R-D4.7R or D2SR-D2SR homomers.
On top, scheme of the constructs used for the CODA-RET experiments, where two complementary halves of RLuc (nRLuc and cRLuc, L1 and L2, respectively) are fused to the corresponding receptors and YFP (mVenus variant) is fused to the Gαi1 subunit. (a-c) Concentration-response experiments of changes in BRET ratio induced by dopamine, pramipexole or ropinirole, determined by changes in the interaction of D4.4R-D4.4R homomers (a), D4.7R-D4.7R homomers (b) or D2SR-D2SR homomers (c) with Gαi1 protein. HEK-293T cells were transiently transfected with D4.4R-nRLuc and D4.4R-cRLuc (a), D4.7R-nRLuc and D4.7R-cRLuc (b) or D2SR-nRLuc and D2SR-cRLuc (c), the G protein subunits Gαi1-YFP and unfused β1 and γ2. Cells were treated with coelenterazine H followed by increasing concentrations of the ligand. After 10 minutes, BRET between the corresponding complemented RLuc receptor and Gαi1-YFP was measured as described in the Methods. BRET values in the absence of ligands were subtracted from the BRET values for each ligand concentration. BRET values expressed as the percentage of the maximal effect of dopamine (% of DA), considering dopamine as full agonist. Data were fit by nonlinear regression to a sigmoidal concentration-response curve against the agonist concentration and are shown as a percentage of the maximal dopamine effect. Data represent the mean ± S.E.M. of 6–9 experiments performed in triplicate (see Table 1 for EC50 and Emax values and statistical comparisons).
Table 1.
Ligand-induced changes in the interaction between Gi protein and D4.4R-D4.4R, D4.7R-D4.7R or D2SR-D2SR homomers
| EC50 (nM) | Emax (%) | ||
|---|---|---|---|
| D4.4R-D4.4R | Dopamine | 240 ± 60 ** | 100 ± 10 |
| Pramipexole | NA | NA | |
| Ropinirole | 400 ± 200 ** | 53 ± 4 # | |
| D4.7R-D4.7R | Dopamine | 130 ± 30 ** | 100 ± 5 |
| Pramipexole | 500±350 | 65 ± 5 ## | |
| Ropinirole | 1300 ± 300 | 73 ± 8 | |
| D2SR-D2SR | Dopamine | 1600 ± 200 | 100 ± 4 |
| Pramipexole | 1200 ± 300 | 109 ± 2 | |
| Ropinirole | 2200 ± 500 | 28 ± 3 ### |
EC50 and Emax (% of dopamine) values (mean ± SEM) from concentration-response experiments of changes in BRET ratio induced by dopamine, pramipexole or ropinirole mediated by D4.4R-D4.4R, D4.7R-D4.7R or D2SR-D2SR homomers in HEK-293T cells transiently expressing D4.4R-nRLuc and D4.4R-cRLuc, D4.7R-nRLuc and D4.7R-cRLuc or D2SR-nRLuc and D2SR-cRLuc, the G protein subunits Gαi1-YFP and unfused β1 and γ2. Statistical differences between the EC50 value of each ligand for D4.4R-D4.4R or D4.7R-D4.7R homomers as compared to the EC50 value of the same ligand for D2SR-D2SR were calculated by one-way ANOVA followed by Dunnett post-hoc test; **: p<0.01, respectively. Statistical differences between the Emax value of each ligand for each receptor homomer as compared to the Emax values of dopamine for the same receptor homomer were calculated by one-way ANOVA followed by Dunnett post-hoc test; #, ## and ###: p<0.05, p<0.01 and p<0.001 respectively.
Ligand-Induced Changes in the Interaction between Gi Protein and D4.4R-D2S or D4.7R-D2SR Heteromers
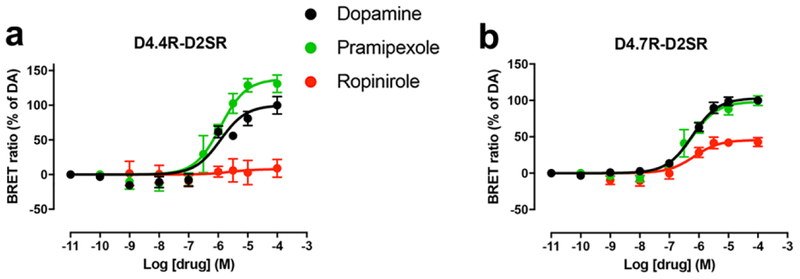
Next, we addressed the question about D4R-D2S heteromerization. CODA-RET experiments demonstrated that both D4.4R-nRLuc and D4.7R-nRLuc can form functional heteromers with D2SR-cRLuc and, importantly, with significantly different pharmacological properties (Figs. 2a and 2b and Table 2). First, compared with D2SR-D2SR homomers, dopamine showed a small but significant increase in potency for D4.7R-D2SR, but not for D4.4R-D2SR heteromers (Figs. 2a and 2b and Table 2). Second, pramipexole showed full efficacy for D4.7R-D2SR heteromers and significant higher efficacy than dopamine for the D4.4R-D2SR heteromers. On the other hand, ropinirole was a partial agonist at D4.7R-D2SR heteromers and it lost its efficacy at D4.4R-D2SR heteromers (Figs. 2a and 2b). To our knowledge, these results provide the first clear demonstration of pharmacodynamic differences of D4R ligands that are dependent on the most common DRD4 polymorphic variants and which are disclosed by heteromerization with D2SR.
Fig. 2. Ligand-induced changes in the interaction between Gi protein and D4.4R-D2SR or D4.7R-D2SR heteromers.
(a-b) Concentration-response experiments of changes in BRET ratio induced by dopamine, pramipexole or ropinirole determined by changes in the interaction of D4.4R-D2SR or D4.7R-D2SR heteromers with Gαi1 protein. HEK-293T cells were transiently transfected with D4.4R-nRLuc and D2SR-cRLuc (a) or D4.7R-nRLuc and D2SR-cRLuc (b), the G protein subunits Gαi1-YFP and unfused β1 and γ2. Cells were treated with coelenterazine H followed by increasing concentrations of the ligand. After 10 minutes, BRET between the corresponding complemented RLuc receptor and Gαi1-YFP was measured as described in the Methods. BRET values in the absence of ligands were subtracted from the BRET values for each ligand concentration. BRET values expressed as the percentage of the maximal effect of dopamine (% of DA), considering dopamine as full agonist. Data were fit by nonlinear regression to a sigmoidal concentration-response curve against the agonist concentration and are shown as a percentage of the maximal dopamine effect. Data represent the mean ± S.E.M. of 5–10 experiments performed in triplicate (see Table 2 for EC50 and Emax values and statistical comparisons).
Table 2:
Ligand-induced changes in the interaction between Gi protein and D4.4R-D2SR or D4.7R-D2SR heteromers
| EC50 (nM) | Emax (%) | ||
|---|---|---|---|
| D4.4R-D2SR | Dopamine | 1000 ± 200 | 100 ± 6 |
| Pramipexole | 1100 ± 200 | 125 ± 5 # | |
| Ropinirole | NA | NA | |
| D4.7R-D2SR | Dopamine | 700 ±100 * | 100 ± 4 |
| Pramipexole | 625 ±196 | 94 ± 5 | |
| Ropinirole | 600 ± 150 ** | 49 ± 5 ## |
EC50 and Emax (% of dopamine) values (mean ± SEM) from concentration-response experiments of changes in BRET ratio induced by dopamine, pramipexole or ropinirole mediated by D4.4R-D2SR or D4.7R-D2SR heteromers in HEK-293T cells transiently expressing D4.4R-nRLuc and D2SR-cRLuc or D4.7R-nRLuc and D2SR-cRLuc, the G protein subunits Gαi1-YFP and unfused β1 and γ2. Statistical differences between the EC50 value of each ligand for D4.4R-D2SR or D4.7R-D2SR heteromers as compared to the EC50 value of the same ligand for the D2SR-D2SR homomer (Table 2) were calculated by one-way ANOVA followed by Dunnett post-hoc test; * and **: p<0.05 and p<0.01, respectively. Statistical differences between the Emax value of each ligand for each receptor heteromer as compared to the Emax values of dopamine for the same receptor heteromer were calculated by one-way ANOVA followed by Dunnett post-hoc test; # and ##: p<0.05 and p<0.01, respectively.
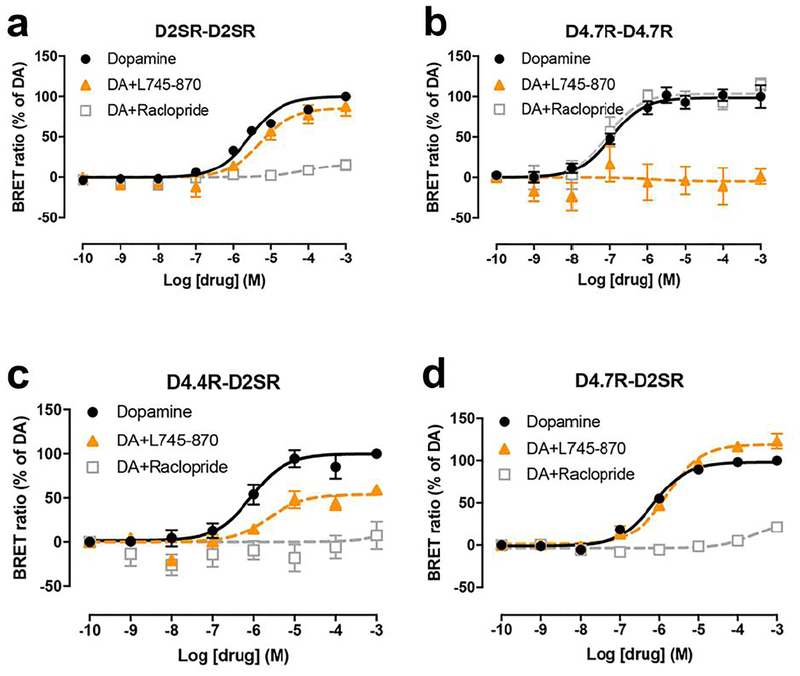
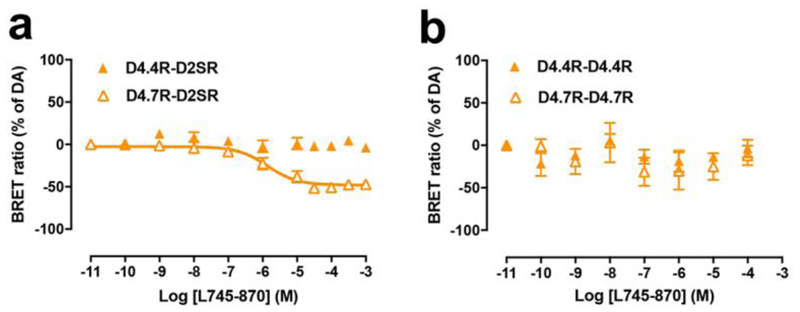
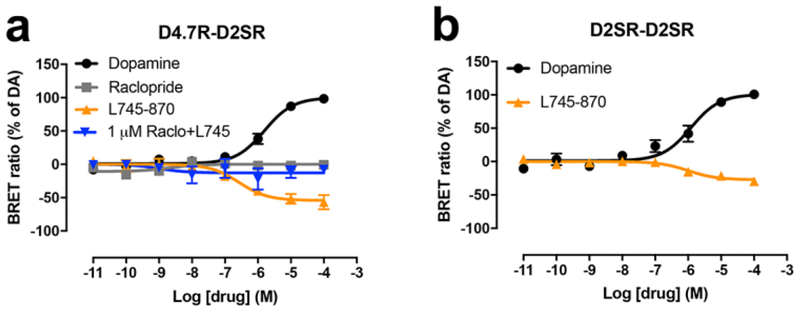
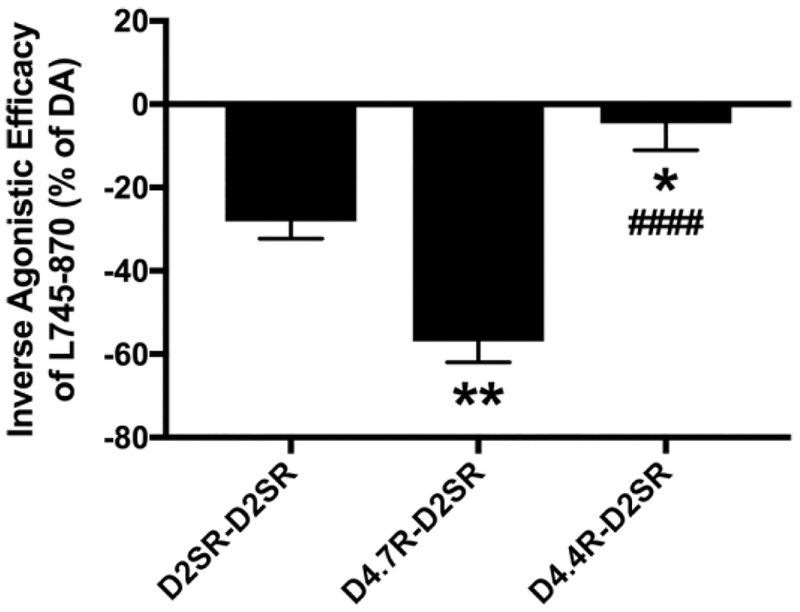
In addition, a very differential response of D4.4R-D2SR and D4.7R-D2SR heteromers to raclopride (D2-like receptor antagonist with very low affinity for D4R) and the selective D4R antagonist L745–870 was observed. Raclopride, at a concentration (100 nM) that completely antagonized dopamine-mediated G protein activation by D2SR-D2SR homomers and was completely inefficient at D4.7R-D4.7R homomers (Figs. 3a and 3b), completely antagonized the effect of dopamine at D4.4R-D2SR and D4.7R-D2SR heteromers (Figs. 3c and 3d). On the other hand, L745–870, at a concentration (1 μM) that completely antagonized dopamine-mediated G protein activation by D4.7R-D4.7R homomers and was mostly inefficient at D2SR-D2SR homomers (Figs. 3a and 3b), produced a significant but partial counteraction of the effect of dopamine at D4.4R-D2SR, but a small but consistent increase at D4.7R-D2SR (Figs. 3c and 3d). This increase could nevertheless be artefactual, since basal levels before normalization were slightly lower for D4.7R-D2SR heteromers in the presence of L745–870. We therefore investigated the possibility that L745–870 could selectively act as an inverse agonist on the D4.7R-D2SR heteromer, indicating the existence of a specific constitutive activity of the D4.7R-D2SR heteromer. In fact, L745–870 induced a significant concentration-dependent decrease in BRET values in CODA-RET experiments in cells transfected with D4.7R-nRLuc and D2SR-cRLuc, but not in cells transfected with D4.4R-nRLuc and D2SR-cRLuc, D4.4R-nRLuc and D4.4R-cRLuc or D4.7R-nRLuc and D4.7R-cRLuc (Figs. 4a and 4b). However, concentrations of L745–870 higher than 1 μM were necessary to clearly disclose the constitutive activity of the D4.7R-D2SR heteromer (Fig. 4a). Since at these concentrations L745–970 also binds to D2R [18], the results suggested that D2SR provides the constitutive activity in the D4.7R-D2S heteromer. In fact, the effect of L745–870 was counteracted by raclopride, which by itself did not show any inverse agonistic activity (Fig. 5a), as previously described in mammalian cell lines selectively expressing D2SR [19]. Finally, and also at micromolar concentrations, L745–870 behaved as an inverse agonist in the D2SR-D2SR homomer, although with less efficacy than in the D4.7R-D2SR heteromer (Fig. 5b). As shown in Fig. 6, the maximal inverse agonistic efficacy of L745–870 was significantly increased in the D4.7R-D2SR heteromers and significantly decreased in the D4.4R-D2SR heteromers as compared to the D2SR-D2SR homomers.
Fig. 3. Effect of L745–870 and raclopride on dopamine-induced changes in the interaction between Gi protein and D2SR-D2SR or D4.7R-D4.7R homomers and between Gi protein and D4.4R-D2SR or D4.7R-D2SR heteromers.
(a-d) Concentration-response experiments of dopamine-induced changes in BRET ratio that measure changes in the interaction of D2SR-D2SR homomers, D4.7R-D4.7R homomers, D4.4R-D2SR heteromers or D4.7R-D2SR heteromers with Gαi1 protein. HEK-293T cells were transiently transfected with D2SR-nRLuc and D2SR-cRLuc (a), D4.7R-nRLuc and D4.7R-cRLuc (b), D4.4R-nRLuc and D2SR-cRLuc (c) or D4.7R-nRLuc and D2SR-cRLuc (d), the G protein subunits Gαi1-YFP and unfused β1 and γ2. Cells were treated with coelenterazine H followed by increasing concentrations of dopamine alone or with the presence of the selective D4R antagonist L745–870 or raclopride, a D2-like receptor antagonist with very low affinity for D4R. After 10 minutes, BRET between the corresponding complemented RLuc receptor and Gαi1-YFP was measured as described in the Methods. BRET values in the absence of ligands were subtracted from the BRET values for each ligand concentration. BRET values expressed as the percentage of the maximal effect of dopamine (% of DA), considering dopamine as full agonist. Data were fit by nonlinear regression to a sigmoidal concentration-response curve against the agonist concentration and are shown as a percentage of the maximal dopamine effect. Data represent the mean ± S.E.M. of 5–13 experiments performed in triplicate.
Fig. 4. Inverse agonism of L745–870 on D4.7R-D2SR heteromers.
(a-b) Concentration-response experiments of the selective D4R antagonist L745–870-induced changes in BRET ratio that measure changes in the interactions of D4.4R-D2SR or D4.7R-D2SR heteromers (a) and D2SR-D2SR or D4.7R-D4.7R homomers (b) with the Gαi1 protein. HEK-293T cells were transiently transfected with D4.4R-RLuc or D4.7R-RLuc (a), D4.4R-nRLuc and D4.4R-cRLuc or D4.7R-nRLuc and D4.7R-cRLuc (b), the G protein subunits Gαi1-YFP and unfused β1 and γ2. Cells were treated with coelenterazine H followed by increasing concentrations of L745–870. After 10 minutes, BRET between the corresponding RLuc- or complemented RLuc receptor and Gαi1-YFP was measured as described in the Methods. BRET values in the absence of ligands were subtracted from the BRET values for each ligand concentration. BRET values expressed as the percentage of the maximal effect of dopamine (% of DA), considering dopamine as full agonist. Data were fit by nonlinear regression to a sigmoidal concentration-response curve against the agonist concentration and are shown as a percentage of the maximal dopamine effect. Data represent the mean ± S.E.M. of 4–8 experiments performed in triplicate.
Fig. 5. Constitutive activity of D2SR-D2SR homomers and D4.7R-D2SR heteromers.
(a-b) Concentration-response experiments of changes in BRET ratio induced by dopamine, the selective D4R antagonist L745–870 or raclopride which measure changes in the interactions of D4.7R-D2SR heteromers (a) or D2SR-D2SR homomers (b) with the Gαi1 protein. HEK-293T cells were transiently transfected with D4.7R-nRLuc and D2SR-cRLuc (a) or D2SR-nRLuc and D2SR-cRLuc (b), the G protein subunits Gαi1-YFP and unfused β1 and γ2. Cells were treated with coelenterazine H followed by increasing concentrations of L745–870. After 10 minutes, BRET between the corresponding RLuc- or complemented RLuc receptor and Gαi1-YFP was measured as described in the Methods. BRET values in the absence of ligands were subtracted from the BRET values for each ligand concentration. BRET values expressed as the percentage of the maximal effect of dopamine (% of DA), considering dopamine as full agonist. Data were fit by nonlinear regression to a sigmoidal concentration-response curve against the agonist concentration and are shown as a percentage of the maximal dopamine effect. Data represent the mean ± S.E.M. of 4–8 experiments performed in triplicate.
Fig. 6. Differences between the constitutive activity of D2SR-D2SR homomers, D4.7R-D2SR heteromers and D4.4R-D2SR heteromers.
Comparison of the maximal inverse agonistic efficacy of L745–870 obtained in cells expressing D2SR-D2SR homomers (results from Fig. 5b), D4.7R-D2SR heteromers (results from Fig. 5a) and D4.4R-D2SR heteromers (results from Fig. 4a). Oneway ANOVA followed by Tukey’s multiple comparison test: * and **: p<0.05 and p<0.01 as compared to D2SR-D2SR; ####: p<0.0001 as compared to D4.7R-D2SR.
In summary, the experiments with antagonists provide an additional demonstration that both D4.4R and D4.7R form functional heteromers with D2SR with significantly different functional properties. D2SR determines dopamine-mediated Gi-dependent signaling in both heteromers. Within the D4.4R-D2SR heteromer, D4.4R participates in the dopamine-mediated Gi-dependent activation of D4.4R-D2SR and decreases the constitutive activity of D2SR. Within the D4.7R-D2SR heteromer, D4.7R confers an increase in the potency of dopamine and a significant increase in the constitutive activity of the D2SR.
Discussion
In the present study, using functional BRET assays in mammalian transfected cells, we could demonstrate significant functional and pharmacological differences between D4.4R and D4.7R only evident upon heteromerization with D2SR, providing a plausible mechanism for the gain of function of D4.7R versus D4.4R recently demonstrated in vitro, in mouse cortical slices with viral infection of human D4.4R and D4.7R [8], and in vivo, in optogenetic-microdialysis experimentsin the D4.7R knock-in mouse [9]. The most dramatic finding was the significant increase in the constitutive activity that D4.7R confers to the D2SR upon heteromerization. To our knowledge, this is the first reported example of changes in the constitutive activity of a GPCR upon heteromerization, which adds to the list of new properties associated with GPCR oligomerization [17]. More significant is the fact that this new property was specifically associated to the product of a DRD4 polymorphism, conferring a gain of function to the D4.7R versus the D4.4R variant. Previous studies already suggested that functional and pharmacological differences between D4.4R and D4.7R could depend on heteromerization with D2R [10, 14]. However, those studies found a stronger agonist-mediated MAPK signaling in cells co-transfected with D4.4R and D2R versus D4.7R and D2R, suggesting the opposite, a loss of function of D4.7R upon heteromerization with D2R and a reduced ability of D4.7R to heteromerize with D2R [10, 14]. Nevertheless, the results of those studies did not demonstrate that the observed differences on MAPK signaling depended on receptor heteromerization. On the other hand, in the present study, with the use of the CODA-RET technique, we could demonstrate the existence of functional D4.7R-D2SR heteromers in transfected cells and also dissect the pharmacological profile of D4.4R and D4.7 homodimers and heteromers. The fact that a gain of function of D4.7R versus D4.4R could only be demonstrated upon heteromerization with D2SR, strongly suggests that D4R-D2R heteromers represent a significant receptor population that modulates the function of cortico-striatal glutamatergic neurons.
Another significant finding, with possible translational implications, was the differential profile of the clinically used dopamine agonists pramipexole and ropinirole, with their more pronounced relative effect in the D4.4R-D2SR and D4.7R-D2SR, respectively. We have in fact recently suggested that the therapeutic effect of both compounds in Restless Legs Syndrome is most probably related to their ability to activate D4R and D2SR localized in cortico-striatal glutamatergic terminals [13]. That being the case, we should expect different clinical efficacies of pramipexole and ropinirole depending on the predominant expression of the products of DRD4 polymorphisms.
Acknowledgements
We thank Dr. J. A. Javitch (Columbia University, New York) for kindly providing some of the DNA constructs (D2SR-nRLuc, D2SR-cRLuc and Gαi1-YFP).
Funding Work supported by the intramural funds of the National Institute on Drug Abuse and a grant from the Spanish “Ministerio de Economía y Competitividad” and the European Regional Development Funds of the European Union (SAF2014–54840-R).
Footnotes
Conflict of Interest The authors declare no conflict of interest.
References
- 1.LaHoste GJ, Swanson JM, Wigal SB, Glabe C, Wigal T, King N, Kennedy JL (1996) Dopamine D4 receptor gene polymorphism is associated with attention deficit hyperactivity disorder. Mol Psychiatry 1:121–124. [PubMed] [Google Scholar]
- 2.Chang FM, Kidd JR, Livak KJ, Pakstis AJ, Kidd KK (1996) The world-wide distribution of allele frequencies at the human dopamine D4 receptor locus. Hum Genet 98:91–101. [DOI] [PubMed] [Google Scholar]
- 3.Wang E, Ding YC, Flodman P, Kidd JR, Kidd KK, Grady DL, Ryder OA, Spence MA (2004) The genetic architecture of selection at the human dopamine receptor D4 (DRD4) gene locus. Am J Hum Genet 74:931–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Faraone SV, Perlis RH, Doyle AE, Smoller JW, Goralnick JJ, Holmgren MA, Sklar P (2005) Molecular genetics of attention-deficit/hyperactivity disorder. Biol Psychiatry 57:13131323. [DOI] [PubMed] [Google Scholar]
- 5.Li D, Sham PC, Owen MJ, He L (2006) Meta-analysis shows significant association between dopamine system genes and attention deficit hyperactivity disorder (ADHD). Hum Mol Genet 15:2276–2284. [DOI] [PubMed] [Google Scholar]
- 6.Gizer IR, Ficks C, Waldman ID (2009) Candidate gene studies of ADHD: a meta-analytic review. Hum Genet 126:51–90. [DOI] [PubMed] [Google Scholar]
- 7.Belcher AM, Volkow ND, Moeller FG, Ferré S (2014) Personality traits and vulnerability or resilience to substance use disorders. Trends Cogn Sci 18:211–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhong P, Liu W, Yan Z (2016) Aberrant regulation of synchronous network activity by the attention-deficit/hyperactivity disorder-associated human dopamine D4 receptor variant D4.7 in the prefrontal cortex. J Physiol 594:135–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonaventura J, Quiroz C, Cai NS, Rubinstein M, Tanda G, Ferré S (2017) Key role of the dopamine D(4) receptor in the modulation of corticostriatal glutamatergic neurotransmission. Sci Adv 3:e1601631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.González S, Rangel-Barajas C, Peper M, Lorenzo R, Moreno E, Ciruela F, Borycz J, Ortiz J, Lluís C, Franco R, McCormick PJ, Volkow ND, Rubinstein M, Floran B, Ferré S (2012) Dopamine D4 receptor, but not the ADHD-associated D4.7 variant, forms functional heteromers with the dopamine D2S receptor in the brain. Mol Psychiatry 17:650–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Asghari V, Sanyal S, Buchwaldt S, Paterson A, Jovanovic V, Van Tol HH (1995) Modulation of intracellular cyclic AMP levels by different human dopamine D4 receptor variants. J Neurochem 65:1157–1165. [DOI] [PubMed] [Google Scholar]
- 12.Sánchez-Soto M, Bonifazi A, Cai N-S, Ellenberger MP, Newman AH, Ferré S, and Yano H (2016) Evidence for Noncanonical Neurotransmitter Activation: Norepinephrine as a dopamine D2-like receptor agonist. Mol Pharmacol 89:457–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yepes G, Guitart X, Rea W, Richard PA, Earley CJ, Quiroz C, Ferré S (2017) Targeting hypersensitive corticostriatal terminals in Restless Legs Syndrome. Ann Neurol 82:951960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borroto-Escuela DO, Van Craenenbroeck K, Romero-Fernandez W, Guidolin D, Woods AS, Rivera A, Haegeman G, Agnati LF, Tarakanov AO, Fuxe K (2011) Dopamine D2 and D4 receptor heteromerization and its allosteric receptor-receptor interactions. Biochem Biophys Res Commun 404:928–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Urizar E, Yano H, Kolster R, Galés C, Lambert N, Javitch JA (2011) CODA-RET reveals functional selectivity as a result of GPCR heteromerization. Nat Chem Biol 7:624–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guitart X, Navarro G, Moreno E, Yano H, Cai NS, Sánchez-Soto M, Kumar-Barodia S, Naidu YT, Mallol J, Cortés A, Lluís C, Canela EI, Casadó V, McCormick PJ, Ferré S (2014) Functional selectivity of allosteric interactions within G protein-coupled receptor oligomers: the dopamine D1–D3 receptor heterotetramer. Mol Pharmacol 86:417–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferré S, Casadó V, Devi LA, Filizola M, Jockers R, Lohse MJ, Milligan G, Pin J-P, Guitart X (2014) G protein-coupled receptor oligomerization revisited: functional and pharmacological perspectives. Pharmacol Rev 66:413–434., [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patel S, Freedman S, Chapman KL, Emms F, Fletcher AE Knowles M, Marwood R, Mcallister G, Myers J, Curtis N, Kulagowski JJ, Leeson PD, Ridgill M, Graham M, Matheson S, Rathbone D, Watt AP, Bristow LJ, Rupniak NM, Baskin E, Lynch JJ, Ragan CI (1997) Biological profile of L-745,870, a selective antagonist with high affinity for the dopamine D4 receptor. J Pharmacol Exp Ther 283:636–647. [PubMed] [Google Scholar]
- 19.Nilsson CL, Ekman A, Hellstrand M, Eriksson E (1996) Inverse agonism at dopamine D2 receptors. Haloperidol-induced prolactin release from GH4C1 cells transfected with the human D2 receptor is antagonized by R(−)-n-propylnorapomorphine, raclopride, and phenoxybenzamine. Neuropsychopharmacology 15:53–61. [DOI] [PubMed] [Google Scholar]