Abstract
Staphylococcus aureus, a Gram-positive pathogen, can cause severe inflammation in humans, leading to various life-threatening diseases. The lipoprotein is a major virulence factor in S. aureus-induced infectious diseases and is responsible for excessive inflammatory mediators such as nitric oxide (NO). Short-chain fatty acids (SCFAs) including butyrate, propionate, and acetate are microbial metabolites in the gut that are known to have anti-inflammatory effects in the host. In this study, we investigated the effects of SCFAs on S. aureus lipoprotein (Sa.LPP)-induced NO production in mouse macrophages. Butyrate and propionate, but not acetate, inhibited Sa.LPP-induced production of NO in RAW 264.7 cells and bone marrow-derived macrophages. Butyrate and propionate inhibited Sa.LPP-induced expression of inducible NO synthase (iNOS). However, acetate did not show such effects under the same conditions. Furthermore, butyrate and propionate, but not acetate, inhibited Sa.LPP-induced activation of NF-κB, expression of IFN-β, and phosphorylation of STAT1, which are essential for inducing transcription of iNOS in macrophages. In addition, butyrate and propionate induced histone acetylation at lysine residues in the presence of Sa.LPP in RAW 264.7 cells. Moreover, Sa.LPP-induced NO production was decreased by histone deacetylase (HDAC) inhibitors. Collectively, these results suggest that butyrate and propionate ameliorate the inflammatory responses caused by S. aureus through the inhibition of NF-κB, IFN-β/STAT1, and HDAC, resulting in attenuated NO production in macrophages.
Keywords: Staphylococcus aureus, lipoproteins; Short-chain fatty acid; Nitric oxide; HDAC inhibitors; Macrophage
INTRODUCTION
Staphylococcus aureus is a Gram-positive pathogenic bacterium that can cause severe inflammation leading to septic shock and inflammatory bowel diseases (1,2). TLR2 ligands of Gram-positive bacteria (including S. aureus) are responsible for inducing severe inflammation in the host cell (3). In S. aureus, lipoproteins and lipoteichoic acid (LTA) are well known TLR2 ligands responsible for inducing nitric oxide (NO) and/or proinflammatory cytokine production in macrophages (4,5). Many recent studies have suggested that the bacterial lipoprotein is a potent immunostimulator in macrophages (6,7). Wild-type and LTA-deficient, but not lipoprotein-deficient, S. aureus induces NO production in macrophages (8). In addition, lipoprotein-deficient Streptococcus gordonii is less effective in inducing NO production than wild-type or LTA-deficient S. gordonii in macrophages (6). Furthermore, wild-type, but not lipoprotein-deficient, S. aureus potently induces IL-8 induction in human intestinal epithelial cells (5) and osteoclast activation (9).
NO is a small molecule that can regulate a variety of physiological functions such as innate immune responses, vascular homeostasis, and neurotransmission (10). In mammalian cells, inducible NO synthase (iNOS) can induce a micromolar level of NO by immune cell activation, which can evoke septic shock, autoimmune diseases, and chronic inflammatory diseases (11). Excessive NO production by iNOS is observed in patients with septic shock or inflammatory bowel diseases (12,13). NF-κB activation and type I IFN-mediated STAT1 phosphorylation are essential for iNOS expression in macrophages (14). In S. aureus, lipoprotein is the major cell wall component for inducing excessive NO production through TLR2-mediated iNOS induction in macrophages (8). Even though NO production induced by S. aureus lipoprotein (Sa.LPP) is known to be detrimental to the host, little is known about molecules that could potentially inhibit excessive inflammation.
Short-chain fatty acids (SCFAs) are metabolites produced by intestinal microbiota through fermentation of undigested carbohydrates and dietary fibers (15). Butyrate, propionate, and acetate are the predominant forms of SCFAs, which have anti-inflammatory properties (16,17). Butyrate has beneficial roles by having anti-inflammatory effects on diseases such as inflammatory bowel disease or sepsis (18,19). Furthermore, SCFAs regulate immune cell differentiation and function through the inhibition of histone deacetylase (HDAC) and activation of G protein-coupled receptors (20,21). SCFAs also downregulate NO production by IFN-γ through the inhibition of NF-κB and ERK signaling in macrophages (22). Although SCFAs have been suggested as anti-inflammatory molecules (23,24), it is not fully understood whether SCFAs regulate bacterial lipoprotein-mediated NO production in macrophages. In this study, we investigated whether SCFAs inhibit Sa.LPP-induced NO production in macrophages.
MATERIALS AND METHODS
Bacteria, reagents, and chemicals
S. aureus RN4220 was kindly provided by Prof. Bok Luel Lee (Pusan National University, Busan, Korea). Luria-Bertani (LB) broth was purchased from LPS Solution (Daejeon, Korea). Dulbecco's modified Eagle's medium (DMEM) and fetal bovine serum (FBS) were purchased from Welgene (Gyeongsan, Korea) and Gibco (Burlington, ON, Canada), respectively. Recombinant murine M-CSF was obtained from CreaGene (Seongnam, Korea). Sodium acetate, sodium propionate, sodium butyrate, trichostatin A (TSA), suberoylanilide hydroxamic acid (SAHA), mepenzolate bromide (MPN), pertussis toxin (PTX), Triton X-114, octyl β-D-glucopyranoside, and thiazolyl blue tetrazolium bromide were purchased from Sigma-Aldrich Inc. (St. Louis, MO, USA). Anti-iNOS rabbit polyclonal IgG antibody was obtained from Upstate Biotechnology (Lake Placid, NY, USA). Anti-acetyl-histone H3 (Lys9) polyclonal antibody was purchased from Millipore (Billerica, MA, USA). Anti-STAT1 and -phosphorylated STAT1 (P-STAT1) rabbit polyclonal antibodies were purchased from Cell Signaling Technology (Beverly, MA, USA). Anti-β-actin mouse monoclonal antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). All other reagents were purchased from Sigma-Aldrich Inc. unless indicated otherwise.
Preparation of ethanol-killed S. aureus (EKSA)
Techniques used to prepare EKSA were previously described (25). Briefly, S. aureus was cultured in LB medium at 37°C to mid-log phase. The bacterial pellet was collected, incubated, and shaken with 70% ethanol in PBS at room temperature for 2 h. After washing twice with PBS, bacterial killing was confirmed by spreading on an LB-agar plate at 37°C for 48 h. No bacterial colonies were observed.
Culture of RAW 264.7 cells
RAW 264.7 (TIB-71) was obtained from the American Type Culture Collection (Manassas, VA, USA). The cells were cultured in DMEM supplemented with 10% FBS, penicillin (100 U/ml) and streptomycin (100 μg/ml) at 37°C in a humidified incubator with 5% CO2.
Preparation of bone marrow-derived macrophages (BMDMs)
Animal experiments were conducted under the approval of the Institutional Animal Care and Use Committee of Seoul National University (SNU-170103-3). C57BL/6 mice were purchased from Orient Bio (Seongnam, Korea). Bone marrow cells were prepared from 8-week-old mice as previously described (26). Bone marrow cells were differentiated into BMDMs with DMEM supplemented with 10% FBS, penicillin (100 U/ml), streptomycin (100 μg/ml) and M-CSF (20 ng/ml) for 5 days. BMDMs (5×105 cells/ml) were stimulated with the indicated stimuli for 24 h.
Measurement of NO production
RAW 264.7 cells (3×105 cells/ml) were stimulated with the indicated stimuli for 24 h. Nitrite in cell culture supernatants was measured to determine NO as previously described (27). Briefly, an equal volume of Griess reagent (1% sulfanilamide, 0.1% naphthylethylenediamine dihydrochloride, and 2% phosphoric acid) was added to culture supernatants and incubated at room temperature for 5 min. Then, the concentration of nitrite was measured at 540 nm with a microtiter plate reader (Versamax, Molecular Devices Corporation, Sunnyvale, CA, USA) using NaNO2 as a standard.
Cell viability assay
RAW 264.7 cells (3×105 cells/ml) were stimulated with the indicated stimuli for 24 h. Then, the cells were treated with 0.5 mg/ml of 2,5-diphenyl-2H-tetrazolium bromide (MTT) reagent for 2 h. The precipitate (formazan) was dissolved with dimethyl sulfoxide and the absorbance was measured at 570 nm with a microtiter plate reader.
Western blotting
RAW 264.7 cells (3×105 cells/ml) were treated with the indicated stimuli for various time periods. After washing with PBS, the cells were lysed in a lysis buffer (50 mM Tris-HCl, 1% NP-40, 0.25% sodium deoxycholate, 1 mM EDTA, and 150 mM NaCl) containing protease/phosphatase inhibitors (1 mM PMSF, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 1 mM Na3VO4, and 1 mM NaF). The lysate was separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes (Millipore, Bedford, MA, USA). The membranes were blocked with 5% bovine serum albumin or 5% skim milk in Tris-buffered saline (TBS) at room temperature for 1 h and incubated overnight with specific antibodies against iNOS, H3K9Ac, P-STAT1, STAT1, or β-actin at 4°C. After incubation, membranes were washed three times with TBS containing 0.05% Tween 20 (TBST) and incubated with HRP-conjugated secondary antibody (Southern Biotechnology, Birmingham, AL, USA) at room temperature for 1 h. Membranes were rinsed 3 times with TBST and developed with SUPEX ECL solution (Neuronex, Pohang, Korea) as previously described (28).
RT-PCR
RAW 264.7 cells (3×105 cells/ml) were treated with the indicated stimuli for various time periods. Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instruction. Complementary DNA (cDNA) was synthesized from total RNA using M-MLV reverse transcriptase (Promega, Madison, WI, USA) and random hexamers (Roche, Basel, Switzerland). PCR was performed as previously described (29). Briefly, cDNA amplification was performed with EmeraldAmp PCR Master Mix (Takara Biomedical Inc., Osaka, Japan) and primers specific to murine iNOS (forward primer: 5′-GGA TAG GCA GAG ATT GGA GG-3′, reverse primer: 5′-AAT GAG GAT GCA AGG CTG G-3′), murine IFN-β (forward primer: 5′-AAT TCT CCA GCA CTG GGT GG-3′, reverse primer: 5′-CCA GGC GTA GCT GTT GTA CT-3′), or β-actin (forward primer: 5′-GTG GGC CCC AGG ACC A-3′, reverse primer: 5′-CTC CTT AAT GTC ACG CAC GAT TTC-3′).
Transient transfection and reporter gene assay
RAW 264.7 cells (1×106 cells/ml) were seeded on 6-well plate at 3 h before transfection. The cells were transfected with pNF-κB-Luc (Clontech, Mountain View, CA, USA) using Attractene reagent (Qiagen, Valencia, CA, USA) for 3 h according to the manufacturer's instructions. After the transfection period, the cells were harvested and plated on 96-well plates for 3 h. The cells were then stimulated with the indicated stimuli for 21 h. For reporter gene assays, the cells were lysed with Gro-lysis buffer (Promega), and the luciferase activity in supernatants was determined using Luciferase Reporter Assay System (Promega) with SPARK 10M multimode microplate reader (Tecan Group Ltd., Männedorf, Switzerland).
Preparation of lipoproteins from S. aureus
Lipoproteins were prepared from S. aureus as previously described (30). Briefly, a bacterial pellet was sonicated and vigorously mixed with 2% Triton X-114 in TBS at 4°C for 2 h. After the incubation period, debris was removed by centrifugation at 10,410 ×g for 10 min, and the supernatants containing lipoproteins were collected and mixed with an equal volume of TBS. The supernatants were incubated at 37°C for 15 min, and the lipoprotein-containing phase was obtained by centrifugation. Lipoproteins were then isolated by methanol precipitation and dissolved in 10 mM of octyl β-D-glucopyranoside in PBS.
Statistical analysis
All experiments were performed at least three times. Mean values±standard deviations were obtained from triplicate samples for each treatment group. Statistical significance was examined with ANOVA using GraphPad Prism 5 (GraphPad Software Inc., La Jolla, CA, USA). An asterisk (*) indicates treatment groups that were significantly different from the control group at p<0.05.
RESULTS
Butyrate and propionate, but not acetate, inhibit Sa.LPP-induced NO production in RAW 264.7 cells
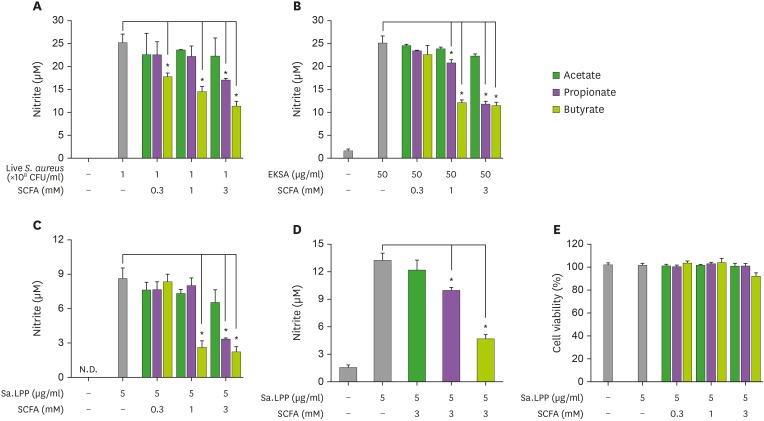
It has been reported that SCFAs have inhibitory effects against S. aureus-induced immune responses in host cells (31,32). To examine the ability of SCFAs to regulate live S. aureus- or Sa.LPP-induced NO production in macrophages, RAW 264.7 cells were stimulated with live S. aureus in the presence or absence of SCFAs for 3 h and further incubated with gentamicin to prevent bacterial growth for 21 h. Butyrate and propionate, but not acetate, remarkably inhibited live S. aureus-induced NO production (Fig. 1A). Similar to the results of live S. aureus, butyrate and propionate, but not acetate, inhibited EKSA-induced NO production (Fig. 1B). To examine if SCFAs also inhibited Sa.LPP-induced NO production, RAW 264.7 cells were stimulated with purified Sa.LPP (5 μg/ml) in the presence or absence of SCFAs. Butyrate and propionate significantly inhibited Sa.LPP-induced NO production, whereas such inhibitory effects were not observed in acetate-treated groups (Fig. 1C). Of note, butyrate potently inhibited live S. aureus-, EKSA-, or Sa.LPP-induced NO production even at 1 mM. Moreover, 3 mM of butyrate and propionate, but not acetate, significantly inhibited Sa.LPP-induced NO production in primary macrophages (Fig. 1D). To exclude the possibility that changes in cell viability had an impact on NO production, RAW 264.7 cells were stimulated with Sa.LPP in the presence or absence of SCFAs, and cell viability was determined using the MTT assay (Fig. 1E). There was no cytotoxicity even when SCFAs inhibited the Sa.LPP-induced NO production. These results suggest that butyrate and propionate inhibit Sa.LPP-induced NO production in murine macrophages.
Figure 1. Butyrate and propionate, but not acetate, inhibit S. aureus or its lipoprotein-induced NO production in murine macrophages. (A) RAW 264.7 cells (3×105 cells/ml) were treated with 1×108 CFU/ml of live S. aureus in the presence (0.3, 1, or 3 mM) or absence of acetate, propionate, or butyrate for 3 h and further incubated with 200 μg/ml of gentamicin for 21 h. (B) RAW 264.7 cells (3×105 cells/ml) were treated with 50 μg/ml of EKSA in the presence (0.3, 1, or 3 mM) or absence of acetate, propionate, or butyrate for 24 h. (C) RAW 264.7 cells (3×105 cells/ml) were stimulated with 5 μg/ml of Sa.LPP in the presence (0.3, 1, or 3 mM) or absence of acetate, propionate, or butyrate for 24 h. Nitrite in the culture supernatant was measured to determine NO concentration using the Griess reagent. (D) BMDMs (5×105 cells/ml) were stimulated with 5 μg/ml of Sa.LPP in the presence or absence of 3 mM acetate, propionate, or butyrate for 24 h, and nitrite was measured. (E) RAW 264.7 cells (3×105 cells/ml) were treated with 5 μg/ml of Sa.LPP and cultured in the presence (0.3, 1, or 3 mM) or absence of acetate, propionate, or butyrate for 24 h. Cell viability was measured by MTT assay. Data are mean values±standard deviations of 3 independent experiments.
N.D., not detected.
*Asterisks indicate significant differences (p<0.05) compared with the appropriate controls.
Butyrate and propionate, but not acetate, inhibit Sa.LPP-induced iNOS expression in RAW 264.7 cells
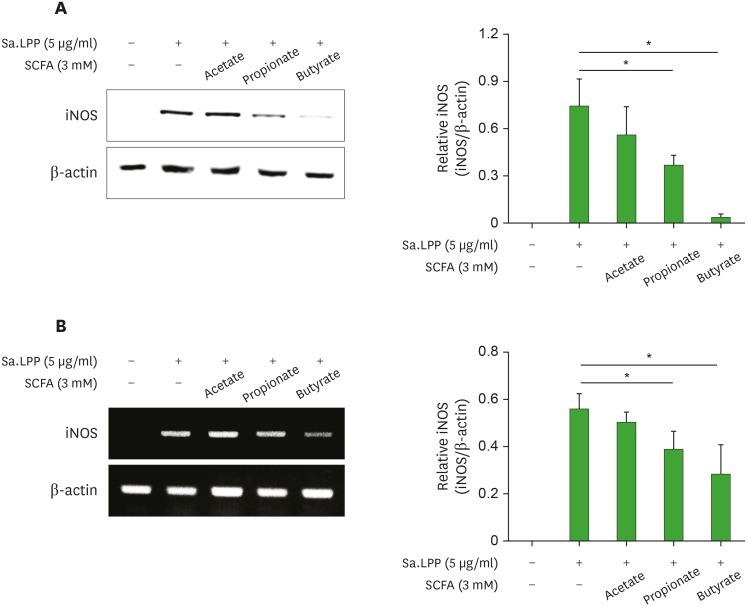
NO is produced by iNOS, which is one of the NOS isoforms (33). Therefore, we examined the inhibitory potency of SCFAs on Sa.LPP-induced iNOS expression in RAW 264.7 cells. Butyrate and propionate, but not acetate, significantly inhibited iNOS expression at the protein level (Fig. 2A). Butyrate inhibited Sa.LPP-induced iNOS protein expression more potently than propionate. Similar results were observed at the mRNA level. Butyrate and propionate, but not acetate, reduced Sa.LPP-induced iNOS mRNA expression levels in RAW 264.7 cells (Fig. 2B). These results indicate that butyrate and propionate downregulate Sa.LPP-induced iNOS expression, which is responsible for NO production in murine macrophages.
Figure 2. Butyrate and propionate, but not acetate, inhibit Sa.LPP-induced iNOS expression in RAW 264.7 cells. (A) RAW 264.7 cells (3×105 cells/ml) were treated with 5 μg/ml of Sa.LPP in the presence or absence of 3 mM acetate, propionate, or butyrate for 15 h. Cell lysates were analyzed to measure iNOS or β-actin expression by Western blotting. (B) RAW 264.7 cells (3×105 cells/ml) were treated with 5 μg/ml of Sa.LPP in the presence or absence of 3 mM acetate, propionate, or butyrate for 12 h. Total RNA was isolated, and the mRNA expression levels of iNOS or β-actin was examined by RT-PCR. Relative expression levels of iNOS protein and mRNA to those of β-actin were measured to obtain mean values±standard deviations by densitometry from three independent experiments (right panels).
*Asterisks indicate significant differences (p<0.05) between the Sa.LPP treatment group and the Sa.LPP with SCFA treatment group.
Butyrate and propionate inhibit Sa.LPP-induced NF-κB activation, STAT1 phosphorylation, and IFN-β mRNA expression in RAW 264.7 cells
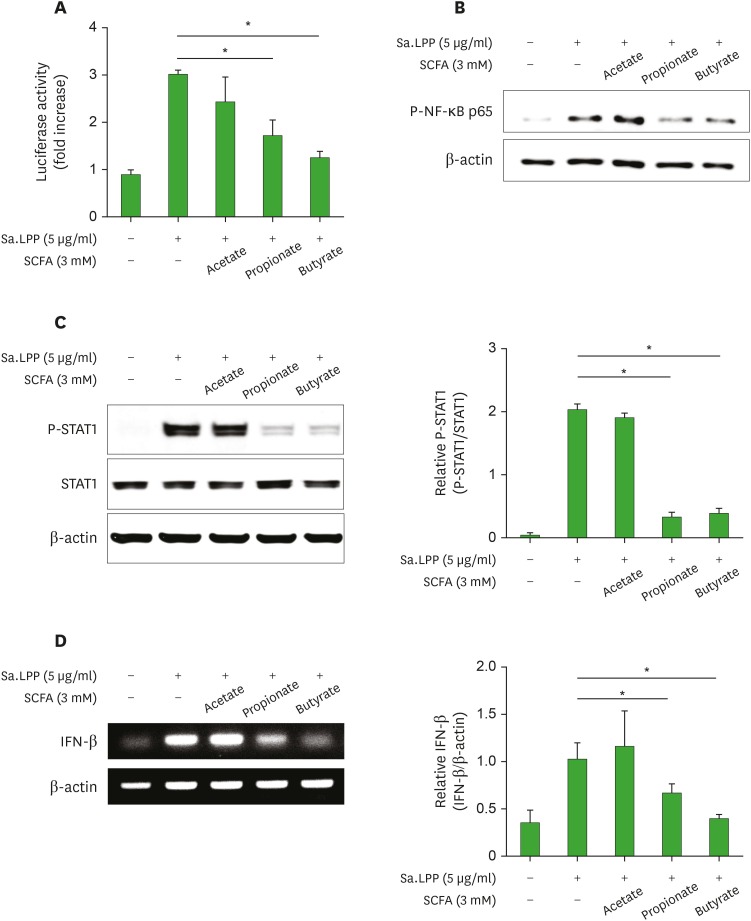
NF-κB activation through TLR2 signaling and STAT1 phosphorylation through type I IFN are essential signaling pathways responsible for the transcription of iNOS (14). To determine if SCFAs regulate the Sa.LPP-mediated NF-κB activation, RAW 264.7 cells were transfected with NF-κB reporter plasmid and treated with Sa.LPP in the presence or absence of SCFAs. NF-κB transcriptional activity was analyzed by reporter gene assay. Butyrate and propionate significantly inhibited Sa.LPP-induced NF-κB activation in RAW 264.7 cells, whereas acetate did not (Fig. 3A). In addition, butyrate and propionate, but not acetate, remarkably inhibited Sa.LPP-induced phosphorylation of NF-κB p65 (Fig. 3B). To determine whether SCFAs regulate Sa.LPP-induced STAT1 phosphorylation, RAW 264.7 cells were stimulated with Sa.LPP in the presence or absence of SCFAs for 4 h. Butyrate and propionate significantly inhibited Sa.LPP-induced STAT1 phosphorylation, whereas acetate did not (Fig. 3C). Furthermore, butyrate and propionate significantly inhibited Sa.LPP-induced IFN-β expression (Fig. 3D), which is responsible for activating STAT1 through type I IFN receptors. These results suggest that butyrate and propionate, but not acetate, significantly inhibit Sa.LPP-mediated iNOS-inducing factors including NF-κB activation, STAT1 phosphorylation, and IFN-β expression in macrophages.
Figure 3. Butyrate and propionate, but not acetate, inhibit Sa.LPP-induced NF-κB activation, STAT1 phosphorylation, and IFN-β mRNA expression in RAW 264.7 cells. (A) RAW 264.7 cells (1×106 cells/ml) were transfected with a firefly luciferase reporter plasmid regulated by NF-κB for 3 h. After the cells were harvested and plated on 96-well plates for 3 h, the cells were treated with 5 μg/ml of Sa.LPP and cultured in the presence or absence of 3 mM acetate, propionate, or butyrate for additional 21 h. Cell lysates were mixed with firefly luciferase substrate to determine luciferase activity. (B) RAW 264.7 cells (3×105 cells/ml) were treated with 5 μg/ml of Sa.LPP in the presence or absence of 3 mM acetate, propionate, or butyrate for 3 h. Cell lysates were analyzed to measure phosphorylated NF-κB p65 or β-actin by Western blotting. (C) RAW 264.7 cells (3×105 cells/ml) were treated with 5 μg/ml of Sa.LPP in the presence or absence of 3 mM acetate, propionate, or butyrate for 3 h. Cell lysates were analyzed to measure phosphorylated STAT1, nonphosphorylated STAT1 or β-actin by Western blotting. (D) RAW 264.7 cells (5×105 cells/ml) were treated with 5 μg/ml of Sa.LPP in the presence or absence of 3 mM acetate, propionate, or butyrate for 6 h. Total RNA was isolated, and the mRNA expression level of IFN-β was examined by RT-PCR. Relative expression levels of P-STAT1 to STAT1 and IFN-β to β-actin were measured to obtain mean values±standard deviations by densitometry from 3 independent experiments (right panels).
*Asterisks indicate significant differences (p<0.05) between the Sa.LPP treatment group and the Sa.LPP with SCFA treatment group.
Butyrate inhibits Sa.LPP-induced NO production by inhibiting HDAC
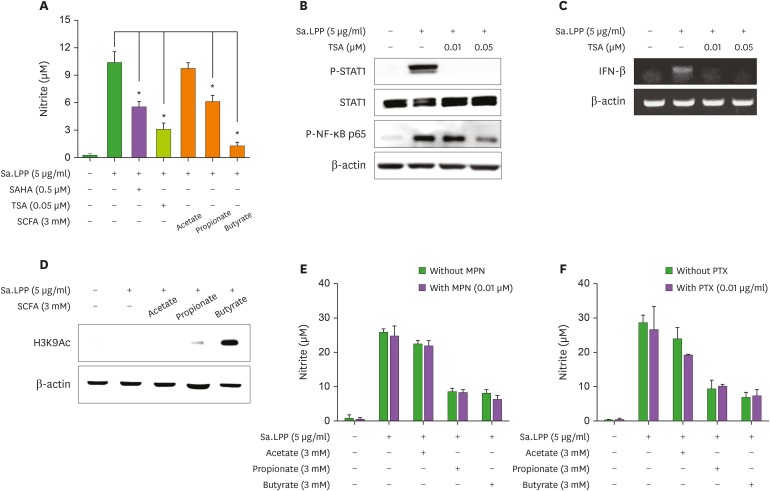
Inhibition of HDAC and activation of G protein-coupled receptor, such as GPR43 and GPR109A, are well-known immunoregulatory mechanisms of butyrate (18). To determine if HDAC inhibition is involved in the SCFA-mediated inhibitory effects on Sa.LPP-induced NO production, RAW 264.7 cells were pretreated with an HDAC inhibitor, SAHA or TSA, for 1 h and then stimulated with Sa.LPP for an additional 23 h. SAHA and TSA inhibited Sa.LPP-induced NO production in RAW 264.7 cells, to a similar extent as butyrate and propionate (Fig. 4A). TSA remarkably inhibited Sa.LPP-induced STAT1 phosphorylation and NF-κB activation (Fig. 4B). TSA also inhibited Sa.LPP-induced IFN-β mRNA expression (Fig. 4C). To confirm that butyrate and propionate induce histone acetylation at lysine residues in the presence of Sa.LPP, RAW 264.7 cells were stimulated with Sa.LPP in the presence or absence of SCFAs for 3 h. Western blotting showed that butyrate (potently) and propionate (modestly) induced histone acetylation whereas acetate hardly showed such an effect under the same conditions (Fig. 4D). Next, to determine whether signaling through GPR43 or GPR109A is involved in the inhibitory effects of SCFAs on Sa.LPP-induced NO production, RAW 264.7 cells were pretreated with inhibitors for GPR109A (MPN) or G protein-coupled receptors (PTX) for 1 h and then stimulated with Sa.LPP and SCFAs for an additional 23 h. The inhibitory effects of butyrate and propionate in Sa.LPP-induced NO production were not affected by pretreatment of those inhibitors (Fig. 4E and F). These results suggest that HDAC inhibition, but not G protein-coupled receptor signaling, is the key molecular mechanism for the inhibitory effect of butyrate and propionate on Sa.LPP-induced NO production in murine macrophages.
Figure 4. Butyrate and propionate might inhibit Sa.LPP-induced NO production by inhibiting HDAC. (A) RAW 264.7 cells (3×105 cells/ml) were pre-treated with HDAC inhibitors (0.5 μM SAHA or 0.05 μM TSA) or SCFAs (3 mM of acetate, propionate, or butyrate) for 1 h and subsequently stimulated with 5 μg/ml of Sa.LPP for additional 23 h. Nitrite in the culture supernatant was measured to determine NO concentration using the Griess reagent. (B) RAW 264.7 cells (3×105 cells/ml) were pre-treated with 0.01 or 0.05 μM of TSA for 1 h and subsequently stimulated with 5 μg/ml of Sa.LPP for additional 3 h. Cell lysates were analyzed to measure phosphorylated STAT1, nonphosphorylated STAT1, phosphorylated NF-κB p65 or β-actin by Western blotting. (C) RAW 264.7 cells (5×105 cells/ml) were pre-treated with 0.01 or 0.05 μM of TSA for 1 h and subsequently stimulated with 5 μg/ml of Sa.LPP for an additional 6 h. Total RNA was isolated, and the mRNA expression level of IFN-β was examined by RT-PCR. (D) RAW 264.7 cells (3×105 cells/ml) were treated with 5 μg/ml of Sa.LPP in the presence or absence of acetate, propionate, or butyrate for 4 h. Cell lysates were obtained and analyzed by Western blotting to measure histone acetylation at lysine residues. (E, F) RAW 264.7 cells (3×105 cells/ml) were pre-treated with (E) 0.01 μM of MPN, or (F) 0.01 μg/ml of PTX for 1 h and subsequently stimulated with 5 μg/ml of Sa.LPP for an additional 23 h. Nitrite in the culture supernatant was measured to determine NO concentration using the Griess reagent. Data are mean values±standard deviation of 3 independent experiments.
*Asterisks indicate significant differences (p<0.05) compared with the appropriate controls.
DISCUSSION
The lipoprotein of Gram-positive bacteria is a potent stimulator that can cause severe inflammation in the host cell (34). In this study, we demonstrated that butyrate and propionate inhibited Sa.LPP-induced NO production through the downregulation of iNOS expression. Butyrate and propionate also inhibited NF-κB activation, IFN-β expression, and STAT1 phosphorylation, which are essential for the induction of iNOS gene expression. Moreover, butyrate and propionate inhibited NO production, which might be via inhibition of HDAC. Butyrate was much more potent than propionate in all of the aforementioned data. Therefore, these results suggest that SCFAs butyrate and propionate are anti-inflammatory for downregulating Sa.LPP-induced NO production in macrophages.
Butyrate seems to inhibit pathogen-associated molecular pattern (PAMP)-induced inflammatory conditions in general, considering our current study and a previous study showing that butyrate inhibits peptidoglycan-induced TNF-α and IL-1β expression in THP-1 cells (35). Butyrate also inhibits Pam3CSK4 (a synthetic lipopeptide mimicking bacterial lipoproteins)-induced IL-8 production in human intestinal epithelial cells (36). Furthermore, in dendritic cells, butyrate effectively inhibits NO, IL-12p40, and IFN-γ expression by LPS, which is one of the major PAMPs of Gram-negative bacteria (37). These results indicate that butyrate could be a potent immunosuppressive molecule in various inflammatory conditions caused by PAMPs from both Gram-positive and Gram-negative bacteria.
In general, activation of NF-κB is stronger in patients who suffer from inflammatory diseases than in healthy people (38). Lipoproteins of Gram-positive bacteria including S. aureus (8), Streptococcus pneumoniae (39), and S. gordonii (6) are major TLR2 ligands. Lipoprotein-induced TLR2 activation causes MyD88-dependent NF-κB activation, which is responsible for inducing pro-inflammatory responses in human intestinal epithelial cells (5). In this study, we found that butyrate potently inhibits Sa.LPP-induced NF-κB activation. The inhibitory effect of butyrate on NF-κB activation seems to be a general phenomenon, considering that butyrate inhibits LPS-induced transcriptional activity of NF-κB in macrophages (40) and TNF-α-induced nuclear translocation of NF-κB in human colon cells (41). Therefore, butyrate has anti-inflammatory effects against inflammation caused by bacterial infections via inhibitory action on NF-κB.
HDAC activity is required for STAT1 signaling (42) and bacterial lipoprotein efficiently induces TLR2-mediated IFN-β expression in endolysosomal compartments leading to STAT1 phosphorylation in macrophages (43). Interestingly, we showed that butyrate and propionate inhibit Sa.LPP-induced IFN-β expression and STAT1 phosphorylation. HDAC inhibition seems to be important for the inhibitory effect of butyrate and propionate, considering that HDAC inhibition can induce ubiquitination and proteasomal degradation of interferon regulatory factor 1 (44), which is an essential transcription factor for inducing TLR2/MyD88-mediated IFN-β expression in macrophages (43). Furthermore, we showed that butyrate and propionate promote histone acetylation and that the inhibition of HDAC downregulates Sa.LPP-induced IFN-β expression and STAT1 phosphorylation.
Concordant with our study, Park et al. (22) demonstrated that SCFAs (butyrate, phenylbutyrate, and phenylacetate) downregulate IFN-γ-induced NO production through the inhibition of NF-κB signaling pathways in murine macrophages. However, there are some differences between their results and our results. First, they showed that not only butyrate and phenylbutyrate, but also phenylacetate, downregulate IFN-γ-induced NO production. However, we showed that acetate does not downregulate S. aureus- or Sa.LPP-induced NO production. These results suggest that the anti-inflammatory potency of SCFAs may vary depending on the subtype of SCFAs. Second, while Park et al. (22) showed that SCFAs do not affect IFN-γ-induced Jak/STAT signaling, our current study showed that butyrate and propionate regulate STAT1 signaling. Although further studies are needed to elucidate the cause of different results, this might be at least partially due to a difference in stimuli. Notably, IFN-γ (used in their study) is a sort of a host-derived cytokine that directly activates Jak/STAT signaling leading to iNOS expression. In contrast, S. aureus and its lipoproteins (used in our study) primarily stimulate pattern-recognition receptors such as TLR2, which indirectly activates Jak/STAT signaling through type I IFN responses in macrophages (43,45). Indeed, our study further demonstrated that butyrate and propionate downregulate Sa.LPP-induced IFN-β expression.
We showed that propionate also inhibits Sa.LPP-induced NO production, but its inhibitory potency is less than that of butyrate, while acetate does not inhibit production at all. Different immunomodulatory effects of SCFAs have been reported in other experimental conditions. For example, butyrate more potently inhibits LPS-induced NO, IL-6, and IL-12p40 expression than propionate or acetate in BMDMs (18). Furthermore, we have reported that only butyrate, but not propionate or acetate, potentiates oral cholera vaccine-induced CCL20 secretion in human intestinal epithelial cells (46). The different immunomodulatory potency of each SCFA might be due to differences in HDAC inhibitory activity, considering that butyrate more potently induces histone acetylation than propionate or acetate (47). The results of this study showed that butyrate more potently induces histone acetylation compared to propionate in Sa.LPP-treated RAW 264.7 cells, whereas acetate does not.
Lipoprotein is a major virulence factor in S. aureus and is responsible for inducing excessive pro-inflammatory cytokines, leading to severe sepsis in the host (48,49). Therefore, identification of an anti-inflammatory molecule is important to prevent excessive inflammation from S. aureus infections. This study suggests that butyrate is a potent anti-inflammatory molecule that can inhibit lipoprotein-induced NO production. Butyrate may act as a regulator against excessive inflammation caused by bacterial infection.
ACKNOWLEDGEMENTS
This work was supported by grants from the National Research Foundation of Korea, which is funded by the Korean government (NRF-2018R1A5A2024418).
Abbreviations
- BMDM
bone marrow-derived macrophage
- cDNA
complementary DNA
- DMEM
Dulbecco's modified Eagle's medium
- EKSA
ethanol-killed Staphylococcus aureus
- HDAC
histone deacetylase
- iNOS
inducible nitric oxide synthase
- LB
Luria-Bertani
- LTA
lipoteichoic acid
- MPN
mepenzolate bromide
- MTT
2,5-diphenyl-2H-tetrazolium bromide
- NO
nitric oxide
- PAMP
pathogen-associated molecular pattern
- P-STAT1
phosphorylated STAT1
- PTX
pertussis toxin
- Sa.LPP
Staphylococcus aureus lipoprotein
- SAHA
suberoylanilide hydroxamic acid
- SCFA
short-chain fatty acid
- TBS
Tris-buffered saline
- TBST
TBS containing 0.05% Tween 20
- TSA
trichostatin A
Footnotes
Conflict of Interest: The authors declare no potential conflicts of interest.
- Conceptualization: Park JW, Kim HY, Han SH.
- Investigation: Park JW, Kim HY, Kim MG.
- Methodology: Park JW, Kim HY, Jeong S, Han SH.
- Supervision: Yun CH, Han SH.
- Writing - original draft: Park JW, Kim HY, Jeong S, Han SH.
- Writing - review & editing: Park JW, Kim HY, Kim MG, Jeong S, Yun CH, Han SH.
References
- 1.Bettenworth D, Nowacki TM, Friedrich A, Becker K, Wessling J, Heidemann J. Crohn's disease complicated by intestinal infection with methicillin-resistant Staphylococcus aureus . World J Gastroenterol. 2013;19:4418–4421. doi: 10.3748/wjg.v19.i27.4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 3.Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 4.Han SH, Kim JH, Seo HS, Martin MH, Chung GH, Michalek SM, Nahm MH. Lipoteichoic acid-induced nitric oxide production depends on the activation of platelet-activating factor receptor and Jak2. J Immunol. 2006;176:573–579. doi: 10.4049/jimmunol.176.1.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noh SY, Kang SS, Yun CH, Han SH. Lipoteichoic acid from Lactobacillus plantarum inhibits Pam2CSK4-induced IL-8 production in human intestinal epithelial cells. Mol Immunol. 2015;64:183–189. doi: 10.1016/j.molimm.2014.11.014. [DOI] [PubMed] [Google Scholar]
- 6.Kim HY, Baik JE, Ahn KB, Seo HS, Yun CH, Han SH. Streptococcus gordonii induces nitric oxide production through its lipoproteins stimulating Toll-like receptor 2 in murine macrophages. Mol Immunol. 2017;82:75–83. doi: 10.1016/j.molimm.2016.12.016. [DOI] [PubMed] [Google Scholar]
- 7.Kim HY, Kim AR, Seo HS, Baik JE, Ahn KB, Yun CH, Han SH. Lipoproteins in Streptococcus gordonii are critical in the infection and inflammatory responses. Mol Immunol. 2018;101:574–584. doi: 10.1016/j.molimm.2018.08.023. [DOI] [PubMed] [Google Scholar]
- 8.Kim NJ, Ahn KB, Jeon JH, Yun CH, Finlay BB, Han SH. Lipoprotein in the cell wall of Staphylococcus aureus is a major inducer of nitric oxide production in murine macrophages. Mol Immunol. 2015;65:17–24. doi: 10.1016/j.molimm.2014.12.016. [DOI] [PubMed] [Google Scholar]
- 9.Kim J, Yang J, Park OJ, Kang SS, Kim WS, Kurokawa K, Yun CH, Kim HH, Lee BL, Han SH. Lipoproteins are an important bacterial component responsible for bone destruction through the induction of osteoclast differentiation and activation. J Bone Miner Res. 2013;28:2381–2391. doi: 10.1002/jbmr.1973. [DOI] [PubMed] [Google Scholar]
- 10.Tuteja N, Chandra M, Tuteja R, Misra MK. Nitric oxide as a unique bioactive signaling messenger in physiology and pathophysiology. J Biomed Biotechnol. 2004;2004:227–237. doi: 10.1155/S1110724304402034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bogdan C. Nitric oxide and the immune response. Nat Immunol. 2001;2:907–916. doi: 10.1038/ni1001-907. [DOI] [PubMed] [Google Scholar]
- 12.Avdagić N, Zaćiragić A, Babić N, Hukić M, Seremet M, Lepara O, Nakaš-Ićindić E. Nitric oxide as a potential biomarker in inflammatory bowel disease. Bosn J Basic Med Sci. 2013;13:5–9. doi: 10.17305/bjbms.2013.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Titheradge MA. Nitric oxide in septic shock. Biochim Biophys Acta. 1999;1411:437–455. doi: 10.1016/s0005-2728(99)00031-6. [DOI] [PubMed] [Google Scholar]
- 14.Farlik M, Reutterer B, Schindler C, Greten F, Vogl C, Müller M, Decker T. Nonconventional initiation complex assembly by STAT and NF-κB transcription factors regulates nitric oxide synthase expression. Immunity. 2010;33:25–34. doi: 10.1016/j.immuni.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cummings JH, Pomare EW, Branch WJ, Naylor CP, Macfarlane GT. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut. 1987;28:1221–1227. doi: 10.1136/gut.28.10.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruppin H, Bar-Meir S, Soergel KH, Wood CM, Schmitt MG., Jr Absorption of short-chain fatty acids by the colon. Gastroenterology. 1980;78:1500–1507. [PubMed] [Google Scholar]
- 17.Raqib R, Sarker P, Bergman P, Ara G, Lindh M, Sack DA, Nasirul Islam KM, Gudmundsson GH, Andersson J, Agerberth B. Improved outcome in shigellosis associated with butyrate induction of an endogenous peptide antibiotic. Proc Natl Acad Sci U S A. 2006;103:9178–9183. doi: 10.1073/pnas.0602888103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci U S A. 2014;111:2247–2252. doi: 10.1073/pnas.1322269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang F, Jin Z, Shen K, Weng T, Chen Z, Feng J, Zhang Z, Liu J, Zhang X, Chu M. Butyrate pretreatment attenuates heart depression in a mice model of endotoxin-induced sepsis via anti-inflammation and anti-oxidation. Am J Emerg Med. 2017;35:402–409. doi: 10.1016/j.ajem.2016.11.022. [DOI] [PubMed] [Google Scholar]
- 20.Thangaraju M, Cresci GA, Liu K, Ananth S, Gnanaprakasam JP, Browning DD, Mellinger JD, Smith SB, Digby GJ, Lambert NA, et al. GPR109A is a G-protein-coupled receptor for the bacterial fermentation product butyrate and functions as a tumor suppressor in colon. Cancer Res. 2009;69:2826–2832. doi: 10.1158/0008-5472.CAN-08-4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown AJ, Goldsworthy SM, Barnes AA, Eilert MM, Tcheang L, Daniels D, Muir AI, Wigglesworth MJ, Kinghorn I, Fraser NJ, et al. The orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J Biol Chem. 2003;278:11312–11319. doi: 10.1074/jbc.M211609200. [DOI] [PubMed] [Google Scholar]
- 22.Park JS, Lee EJ, Lee JC, Kim WK, Kim HS. Anti-inflammatory effects of short chain fatty acids in IFN-gamma-stimulated RAW 264.7 murine macrophage cells: involvement of NF-κB and ERK signaling pathways. Int Immunopharmacol. 2007;7:70–77. doi: 10.1016/j.intimp.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 23.Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther. 2008;27:104–119. doi: 10.1111/j.1365-2036.2007.03562.x. [DOI] [PubMed] [Google Scholar]
- 24.Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, Schilter HC, Rolph MS, Mackay F, Artis D, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hong SJ, Kim SK, Ko EB, Yun CH, Han SH. Wall teichoic acid is an essential component of Staphylococcus aureus for the induction of human dendritic cell maturation. Mol Immunol. 2017;81:135–142. doi: 10.1016/j.molimm.2016.12.008. [DOI] [PubMed] [Google Scholar]
- 26.Kim HY, Kim SK, Seo HS, Jeong S, Ahn KB, Yun CH, Han SH. Th17 activation by dendritic cells stimulated with gamma-irradiated Streptococcus pneumoniae . Mol Immunol. 2018;101:344–352. doi: 10.1016/j.molimm.2018.07.023. [DOI] [PubMed] [Google Scholar]
- 27.Ryu YH, Baik JE, Yang JS, Kang SS, Im J, Yun CH, Kim DW, Lee K, Chung DK, Ju HR, et al. Differential immunostimulatory effects of gram-positive bacteria due to their lipoteichoic acids. Int Immunopharmacol. 2009;9:127–133. doi: 10.1016/j.intimp.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 28.Song IB, Gu H, Han HJ, Lee NY, Cha JY, Son YK, Kwon J. Effects of 7-MEGA™ 500 on oxidative stress, inflammation, and skin regeneration in H2O2-treated skin cells. Toxicol Res. 2018;34:103–110. doi: 10.5487/TR.2018.34.2.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Im J, Baik JE, Kim KW, Kang SS, Jeon JH, Park OJ, Kim HY, Kum KY, Yun CH, Han SH. Enterococcus faecalis lipoteichoic acid suppresses Aggregatibacter actinomycetemcomitans lipopolysaccharide-induced IL-8 expression in human periodontal ligament cells. Int Immunol. 2015;27:381–391. doi: 10.1093/intimm/dxv016. [DOI] [PubMed] [Google Scholar]
- 30.Kim AR, Ahn KB, Kim HY, Seo HS, Kum KY, Yun CH, Han SH. Streptococcus gordonii lipoproteins induce IL-8 in human periodontal ligament cells. Mol Immunol. 2017;91:218–224. doi: 10.1016/j.molimm.2017.09.009. [DOI] [PubMed] [Google Scholar]
- 31.Alva-Murillo N, Ochoa-Zarzosa A, López-Meza JE. Short chain fatty acids (propionic and hexanoic) decrease Staphylococcus aureus internalization into bovine mammary epithelial cells and modulate antimicrobial peptide expression. Vet Microbiol. 2012;155:324–331. doi: 10.1016/j.vetmic.2011.08.025. [DOI] [PubMed] [Google Scholar]
- 32.Ochoa-Zarzosa A, Villarreal-Fernández E, Cano-Camacho H, López-Meza JE. Sodium butyrate inhibits Staphylococcus aureus internalization in bovine mammary epithelial cells and induces the expression of antimicrobial peptide genes. Microb Pathog. 2009;47:1–7. doi: 10.1016/j.micpath.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 33.Kröncke KD, Fehsel K, Kolb-Bachofen V. Inducible nitric oxide synthase in human diseases. Clin Exp Immunol. 1998;113:147–156. doi: 10.1046/j.1365-2249.1998.00648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nguyen MT, Götz F. Lipoproteins of gram-positive bacteria: key players in the immune response and virulence. Microbiol Mol Biol Rev. 2016;80:891–903. doi: 10.1128/MMBR.00028-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Foey AD. Butyrate regulation of distinct macrophage subsets: opposing effects on M1 and M2 macrophages. Int J Probiotics Prebiotics. 2011;6:147–158. [Google Scholar]
- 36.Weng M, Walker WA, Sanderson IR. Butyrate regulates the expression of pathogen-triggered IL-8 in intestinal epithelia. Pediatr Res. 2007;62:542–546. doi: 10.1203/PDR.0b013e318155a422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu L, Li L, Min J, Wang J, Wu H, Zeng Y, Chen S, Chu Z. Butyrate interferes with the differentiation and function of human monocyte-derived dendritic cells. Cell Immunol. 2012;277:66–73. doi: 10.1016/j.cellimm.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 38.Rogler G, Brand K, Vogl D, Page S, Hofmeister R, Andus T, Knuechel R, Baeuerle PA, Schölmerich J, Gross V. Nuclear factor κB is activated in macrophages and epithelial cells of inflamed intestinal mucosa. Gastroenterology. 1998;115:357–369. doi: 10.1016/s0016-5085(98)70202-1. [DOI] [PubMed] [Google Scholar]
- 39.Tomlinson G, Chimalapati S, Pollard T, Lapp T, Cohen J, Camberlein E, Stafford S, Periselneris J, Aldridge C, Vollmer W, et al. TLR-mediated inflammatory responses to Streptococcus pneumoniae are highly dependent on surface expression of bacterial lipoproteins. J Immunol. 2014;193:3736–3745. doi: 10.4049/jimmunol.1401413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Segain JP, Raingeard de la Blétière D, Bourreille A, Leray V, Gervois N, Rosales C, Ferrier L, Bonnet C, Blottière HM, Galmiche JP. Butyrate inhibits inflammatory responses through NFκB inhibition: implications for Crohn's disease. Gut. 2000;47:397–403. doi: 10.1136/gut.47.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yin L, Laevsky G, Giardina C. Butyrate suppression of colonocyte NF-κB activation and cellular proteasome activity. J Biol Chem. 2001;276:44641–44646. doi: 10.1074/jbc.M105170200. [DOI] [PubMed] [Google Scholar]
- 42.Klampfer L, Huang J, Swaby LA, Augenlicht L. Requirement of histone deacetylase activity for signaling by STAT1. J Biol Chem. 2004;279:30358–30368. doi: 10.1074/jbc.M401359200. [DOI] [PubMed] [Google Scholar]
- 43.Dietrich N, Lienenklaus S, Weiss S, Gekara NO. Murine toll-like receptor 2 activation induces type I interferon responses from endolysosomal compartments. PLoS One. 2010;5:e10250. doi: 10.1371/journal.pone.0010250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao B, Wang Y, Xu W, Li S, Li Q, Xiong S. Inhibition of histone deacetylase activity suppresses IFN-γ induction of tripartite motif 22 via CHIP-mediated proteasomal degradation of IRF-1. J Immunol. 2013;191:464–471. doi: 10.4049/jimmunol.1203533. [DOI] [PubMed] [Google Scholar]
- 45.Kang SS, Ryu YH, Baik JE, Yun CH, Lee K, Chung DK, Han SH. Lipoteichoic acid from Lactobacillus plantarum induces nitric oxide production in the presence of interferon-γ in murine macrophages. Mol Immunol. 2011;48:2170–2177. doi: 10.1016/j.molimm.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 46.Sim JR, Kang SS, Lee D, Yun CH, Han SH. Killed whole-cell oral cholera vaccine induces CCL20 secretion by human intestinal epithelial cells in the presence of the short-chain fatty acid, butyrate. Front Immunol. 2018;9:55. doi: 10.3389/fimmu.2018.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fusunyan RD, Quinn JJ, Fujimoto M, MacDermott RP, Sanderson IR. Butyrate switches the pattern of chemokine secretion by intestinal epithelial cells through histone acetylation. Mol Med. 1999;5:631–640. [PMC free article] [PubMed] [Google Scholar]
- 48.Ulloa L, Tracey KJ. The “cytokine profile”: a code for sepsis. Trends Mol Med. 2005;11:56–63. doi: 10.1016/j.molmed.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 49.Schmaler M, Jann NJ, Ferracin F, Landolt LZ, Biswas L, Götz F, Landmann R. Lipoproteins in Staphylococcus aureus mediate inflammation by TLR2 and iron-dependent growth in vivo. J Immunol. 2009;182:7110–7118. doi: 10.4049/jimmunol.0804292. [DOI] [PubMed] [Google Scholar]