Abstract
Analysis of entire transparent rodent bodies after clearing could provide holistic biological information in health and disease, but reliable imaging and quantification of fluorescent protein signals deep inside the tissues remained a challenge. Here, we developed vDISCO, a pressure driven, nanobody based whole-body immunolabeling technology to enhance the signal of fluorescent proteins by up to two orders of magnitude. This allowed us to image and quantify subcellular details through bones, skin and highly autofluorescent tissues of intact transparent mice. For the first time, we visualized whole-body neuronal projections in adult mice. We assessed CNS trauma effects in the whole-body and found degeneration of peripheral nerve terminals in the torso. Furthermore, vDISCO revealed short vascular connections between skull marrow and brain meninges, which were filled with immune cells upon stroke. Thus, our new approach enables unbiased comprehensive studies of the interactions between the nervous system and the rest of the body.
Most diseases, even when arising in a specific site, eventually affect the entire organism. Histological techniques developed in the last century have been the standard procedure for charting pathology, yet, a more complete understanding of biological mechanisms requires an unbiased exploration of the whole organism, not just pre-defined tissues.
However, mammalian tissues are naturally opaque, hindering high-resolution imaging in any tissue deeper than a few hundred micrometers– a major reason why sectioning is needed for histological examination of target organs1. Recent innovations in tissue clearing technology now allow 3D histological examination of intact organs2–12. Tissue clearing is a chemical process aiming to match refractive indices throughout intact tissues, thus rendering them transparent and allowing deep tissue fluorescent microscope imaging. Most initial applications of tissue clearing relied on the assessment of endogenous fluorescent proteins expressed in the mouse central nervous system (CNS). More recently, deep tissue immunolabeling methods have enhanced the quality of imaging for whole rodent organs as well as human embryos thanks to the conjugation of bright fluorescent dyes with secondary antibodies4,13,14. A few studies have even rendered entire adult mouse bodies transparent after removal of the skin3,15–18, and allowed head-to-toe light-sheet microscopy imaging of intact adult mice17,18. All whole-body clearing and imaging methods up to date rely on transgenic expression of fluorescent proteins such as EGFP, EYFP and mCherry17,18. While these fluorescent proteins emit light in the visible spectrum, skeletal muscles and other bodily tissues possess obstructive autofluorescence in this range19. In addition, fluorescent proteins are often less bright compared to many synthetic fluorophores and their signal intensity is further attenuated during the clearing and imaging procedure. Together, these bottlenecks hinder reliable detection and quantification of subcellular details in centimeters-thick transparent mice and therefore rely on imaging of dissected organs for quantifications, compromising the benefits of whole-body transparency.
Here, we developed a whole-body immunolabeling method named vDISCO, to boost the signal of fluorescent proteins using nanobodies, which consist of the “variable domain of heavy chain antibodies” (VHH)20. This technology can enhance fluorescent signals more than 100 times, and thereby allows head-to-toe light sheet microscopy scanning of transparent mice (panoptic imaging) and quantification of subcellular details. We used this technology to construct the first neuronal projection map of an adult mouse and to reveal far-reaching changes in neuronal projections and inflammatory processes following acute CNS injuries. Panoptic imaging also revealed short vascular connections between the skull marrow and meninges (both at the brain surface and sagittal sinus), which may serve as immune gateways following stroke.
Results
vDISCO principles and signal enhancement
Quantitative assessments of intact transparent mouse bodies remained a major challenge impeding the study of neurological diseases at the whole-body scale. We reasoned that fluorescent proteins such as EGFP and RFP are relatively dim compared to bright fluorochromes (e.g., Alexa and Atto dyes) after tissue clearing, making them difficult to be detected in centimeters-thick mouse bodies especially through intact thick bones (e.g., skull and vertebra) and skin. Therefore, we set out to enhance the signal of fluorescent proteins via whole-body immunolabeling with brighter and more stable fluorescent dyes to provide a much higher contrast (signal-to-background ratio = SBR) in cleared mice. Furthermore, using fluorescent dyes in the far-red spectrum could help to overcome tissue autofluorescence, thus allowing reliable detection of subcellular details in all tissues4,19 (Supplementary Fig. 1). We reasoned that nanobodies are particularly suited to achieve a thorough immunolabeling throughout entire adult mouse bodies because of their small molecular weight (12–15 kDa) compared to that of conventional antibodies (~150 kDa)3,20. Indeed, nanobodies were more efficient to label large tissues compared to conventional antibodies (Supplementary Figs. 2,3).
First, we tested the signal quality of nanobody labeling in dissected mouse brains from Thy1-GFPM mice, in which a subset of neurons express EGFP21. Following the nanoboosting, the tissues were cleared using organic solvents6,7. We found that nanoboosting enhanced the signal intensity by one to two orders of magnitude compared to direct imaging of fluorescent proteins (Fig. 1a-i). In the nanoboosted samples, fine details of neurons were evident even in the deep brain regions of Thy1-GFPM mice (Fig. 1b,c vs. Fig. 1e,f). We also obtained a similar signal intensity increase in the cerebellum of Thy1-GFPM brains (Fig. 1g vs. Fig. 1h), which is notoriously difficult to clear due to the high lipid content.
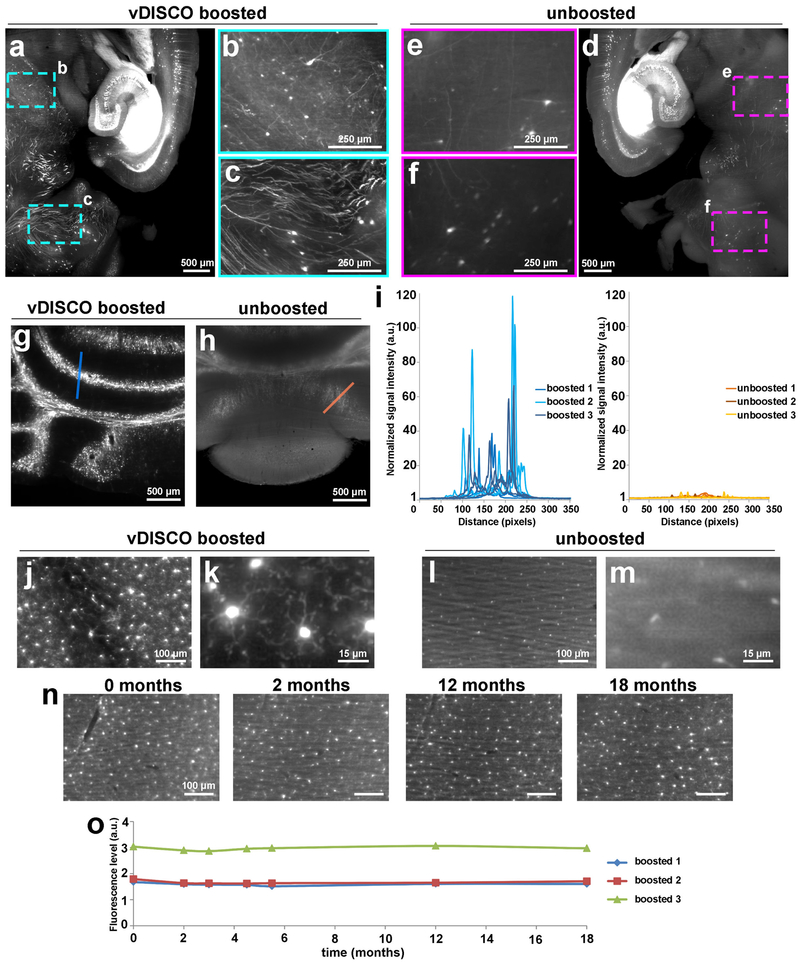
Figure 1. Enhancement and permanent preservation of the fluorescence signal with vDISCO.
Signal quality comparison between vDISCO boosted (a) and unboosted (d) half brain samples coming from the same 11 months old Thy1-GFPM mouse. To achieve the best comparison between the two procedures, we divided the mouse brains in two halves for light-sheet microscopy imaging (one hemisphere boosted and imaged in far-red channel, the other hemisphere unboosted and imaged in the green channel for endogenous EGFP). The boosted hemisphere showed highly distinguishable cellular details (b,c) such as axonal projections not visible in the unboosted hemisphere (e,f) especially in the regions with dim GFP labeling in Thy1-GFPM mice, such as mid brain (b,e) and cerebellum (c,f) (representative images, repeated at least on 3 different samples). (g,h) Comparison of signal quality in cerebellum from the boosted (g) vs. unboosted (h) samples (repeated at least with 3 mice per group). (i) Plots of signal intensity profiles from boosted samples (left) vs. unboosted samples (right) along the blue and orange lines in panels g and h, respectively; n=3 brains for each group (2-6 months old Thy1-GFPM mice). (j-m) 4x and 25x magnification light-sheet microscopy images of the microglia from CX3CR1GFP/+ in boosted (j,k) vs. unboosted samples (l,m) showing the fine details of microglia ramifications obtained with vDISCO boosting (similar results repeated at least with 3 mice per group). (n) Representative light-sheet microscopy images of one of 3 CX3CR1GFP/+ mouse brains at 0, 2, 12 and 18 months after boosting, showing the preservation of the fluorescence signal over 18 months. (o) Fluorescence level quantifications in CX3CR1GFP/+ brains after boosting at different time points post-clearing (n=3 mice, 2-6 months old).
A major aim of tissue clearing approaches is to perform automated quantifications in large imaging datasets in an unbiased and timely way. Towards this goal, we used the NeuroGPS-Tree algorithm, a robust automated neuron-tracing tool that was recently developed for tracing neurons in scans of cortical regions obtained by high-resolution confocal microscopy22. We found that virtually all of the neuronal cell bodies and neurites were detected and linked to each other as complete neurons upon vDISCO nanoboosting in light-sheet microscopy scans (Supplementary Figs. 4, 5). In contrast, in unboosted samples or in standard IgG antibody boosted samples, many fine extensions of neurons were not identified or not connected to somas and neuronal trees (Supplementary Fig. 4). Nanoboosting allowed imaging of not only neuronal details, but also smaller individual cells such as microglia cells and immune cells (Fig. 1j-m, Supplementary Fig. 6). Compared to unboosted samples, we could resolve fine details of microglia cells in intact transparent brains of CX3CR1GFP/+ mice using light-sheet microscopy (Fig. 1j-m, Supplementary Video 1). Nanoboosting also enabled automated quantification of CX3CR1 GFP+ cells using the ClearMap algorithms23: we found approximately 2.3 million microglia cells in the adult mouse brain (Supplementary Fig. 7a). We also automatically quantified microglia in all brain regions annotated by the Allen brain atlas. Thereby we found for example ~150,000 microglia in the hippocampus and ~50,000 in the thalamus of adult mice (Supplementary Fig. 7b-k). Furthermore, we found no significant decrease in signal intensity over time of the same samples, suggesting that vDISCO nanoboosting stabilizes the fluorescent signal (Fig. 1n,o) allowing long-term preservation and re-imaging of the samples. Thus, owing to the enormous enhancement and stabilization of fluorescent signals using nanobodies, we could use available computational tools to automatically trace neurons and count cell numbers in images acquired by light-sheet microscopy.
vDISCO allows panoptic imaging of intact adult mouse bodies
To apply whole-body clearing technology at the systems level, it is critical to obtain information not only from dissected organs but also from intact transparent bodies. Towards this goal, we aimed at establishing an approach to achieve nanoboosting in the entire mouse body. We reasoned that vDISCO amplification of the fluorescent signals would allow reliable quantification of cells through intact thick bones and highly autofluorescent muscles. To ensure homogenous delivery of nanobodies into all body regions, we used high-pressure cardiac perfusion with two- to three-fold increased pressure compared to standard perfusion protocols (160–230 mmHg vs. 70–110 mmHg24, respectively). We also illustrated the effect of high-pressure delivery compared to standard pressure using Methylene blue. This novel high-pressure delivery approach facilitated a consistent distribution of the dye into deep tissues (Supplementary Fig. 8). Assessing the tissue integrity upon high-pressure delivery, we did not observe any structural changes (Supplementary Fig. 9). Furthermore, we used a permeabilization solution containing Triton X-100, methyl-β-cyclodextrin (to extract the cholesterol from membranes), and trans-1-acetyl-4-hydroxy-L-proline (to loosen the collagen network)12. To further reduce the background signal caused by the residual blood and to decalcify the bones, we treated whole mouse bodies with aminoalcohols5,15 and EDTA25, before the whole-body immunolabeling step. Combination of these diverse improvements allowed us to achieve the first whole-body immunolabeling in adult mice with a high degree of transparency (Fig. 2a, Supplementary Fig. 10) and to obtain quantitative information on single cells in deep brain regions through the intact skull compared to other methods3,15–18,23 (Supplementary Fig. 11,12). Here, we used anti-XFP nanobodies conjugated with bright Atto dyes (called nanoboosters) to boost the fluorescent protein signal in transparent mice. Moreover, we used anti-vimentin nanobodies to label activated glia cells in the brain upon trauma demonstrating that our approach also works with nanobodies targeting endogenously expressed proteins (Supplementary Fig. 13).
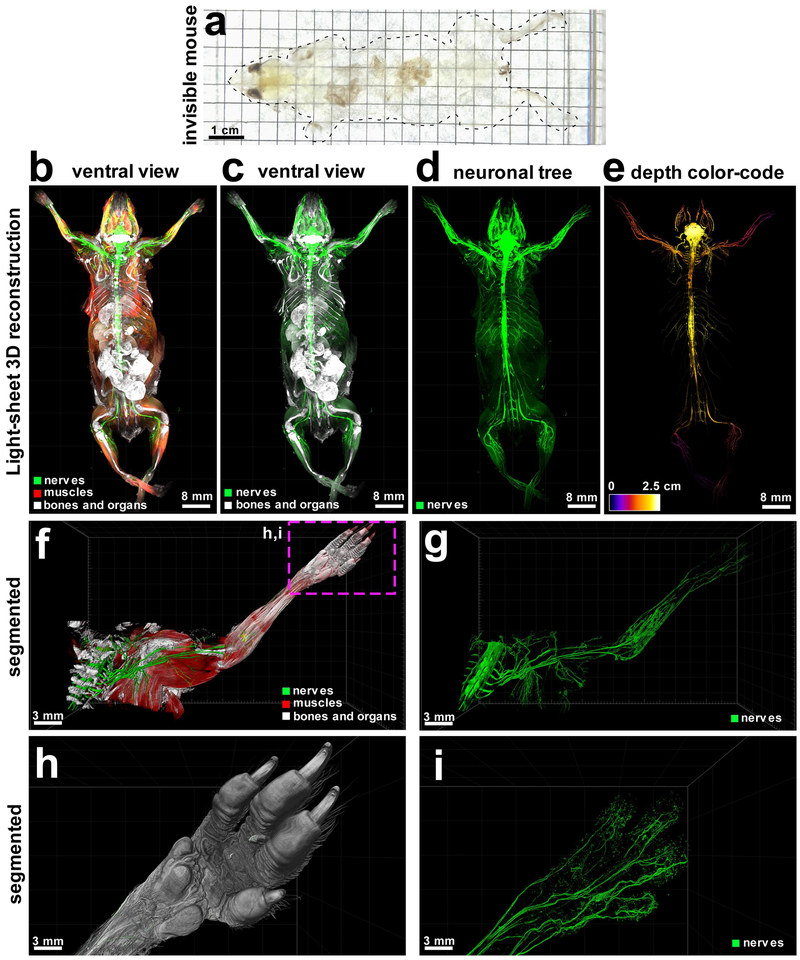
Figure 2. vDISCO panoptic imaging uncovers neuronal projections in intact mice.
(a) An example of a transparent 6 weeks old mouse generated by vDISCO (similar results were observed from at least 20 independent mice). (b-e) 3D reconstruction of complete neuronal projections of a 6-weeks old Thy1-GFPM mouse obtained by light-sheet microscopy imaging (similar labelling and imaging results were achieved at least in 5 different mice; whole body reconstruction was performed on 2 mice). vDISCO boosted GFP+ neuronal structures are shown in green, bones and internal organs are prominent with PI labeling in white, and the muscles visualized by autofluorescence background imaging are in red (b-d). The depth color-coding shows the neuronal projections at different z-levels in the 2.5 cm thick whole mouse body (e). (f,g) High resolution 3D reconstruction views of the left torso and forelimb from the same animal in b-e. Details of innervation throughout muscles and bones are evident. (h,i) Surface reconstruction of the paw (h) and its nerves (i) from the marked region in f. See also Supplementary Video 2-4.
In addition to the specific boosted signal, we also visualized and segmented other major tissues in the transparent body: muscles by their autofluorescence within the blue-green spectrum, bones and internal organs by propidium iodide (PI) labeling (Fig. 2b,c, Supplementary Fig. 14a-d). Usage of organic solvents, which induce isotropic tissue shrinkage17 allowed us to perform head-to-toe panoptic imaging of entire mice by light-sheet microscopy.
Being able to image subcellular details of neurons through intact bones and highly autofluorescent muscles in the whole mouse body using vDISCO panoptic imaging, we constructed the first whole-body neuronal projection map of a Thy1-GFPM transgenic mouse (Fig. 2d,e, Supplementary Fig. 14, Supplementary Video 2). We noticed that in the peripheral nervous system (PNS) mainly axons innervating muscles were labeled in this mouse line, which enabled us to investigate the neuromuscular junctions in greater details. We also observed fluorescent labeling of the internal organs such as kidneys of Thy1-YFPH mice, which was not reported before21 (Supplementary Fig. 15). Owing to the great increase in SBR by vDISCO, we could readily visualize axonal projections from the spinal cord through intact vertebra, to their terminals into the muscles even in very remote locations such as toes (Fig. 2f-i, Supplementary Videos 3,4). Panoptic imaging of individual neuronal projections through intact bones can now provide more details on neuroanatomy. For example, how axons coming from consecutive roots enter the spinal cord i.e., in an overlapping or non-overlapping manner has been unclear26. Using our technology, we observed that in mice, the axonal bundles coming from different roots enter the spinal cord at non-overlapping territories (Supplementary Fig. 16). Thus, the vDISCO approach provides a holistic view of the intact mouse, which should lead to novel discoveries on how interconnected organ systems function in health and what happens during their perturbation in disease.
Clearing of the whole-body without removal of the skin
The skin is the largest organ, forming ~15–20% of the body. Among many other components, it consists of nerve endings, glands, immune cells, blood and lymphatic vessels signifying its diverse biological roles in addition to being a physical barrier against the external environment27. However, clearing and imaging whole mice with the skin has not been reported yet. Towards this goal, we adopted the vDISCO protocol to clear and image mice with skin. We observed that vDISCO could efficiently clear whole nude mice without removal of the skin (Fig. 3a,b). In addition, we cleared C57BL/6J (BL6) background mice after shaving the fur. In CX3CR1GFP/+ mice, we could readily image immune cells through the intact skin owing to the increase of fluorescent signal by vDISCO (Fig. 3c-h, yellow arrowheads, Supplementary Video 5). In Thy1-GFPM mice, we observed the axonal innervations in the skin in addition to Thy1 GFP+ immune cells28 located under the skin (Fig. 3i-k, yellow and white arrowheads, respectively). These results demonstrate that vDISCO allows detailed imaging of intact mice with the skin, which may be of great value for studying skin related pathologies.
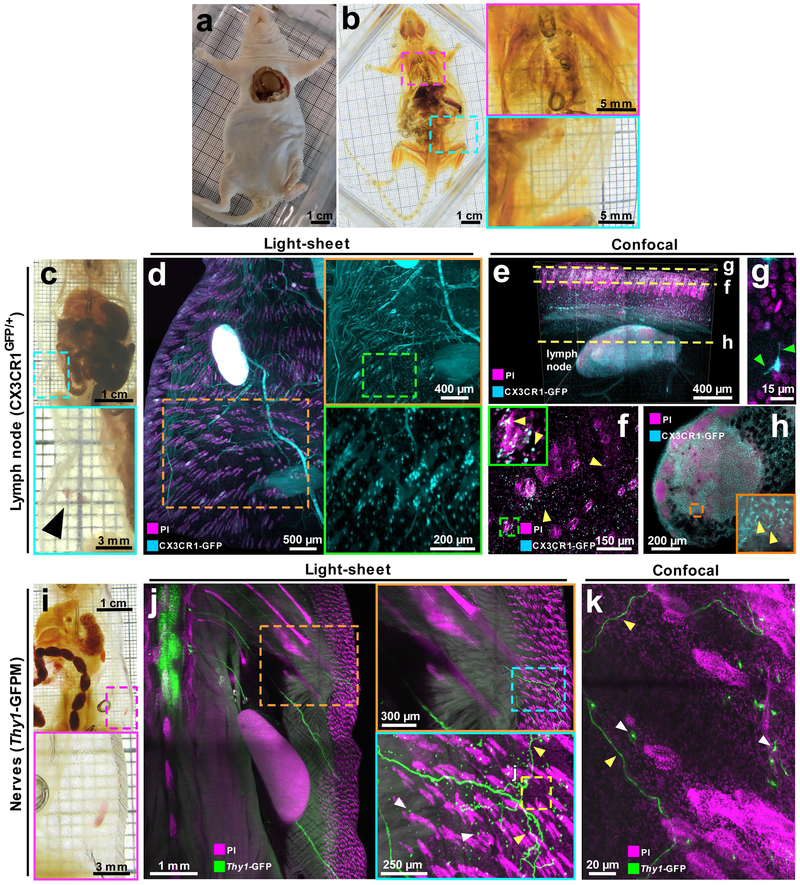
Figure 3. vDISCO panoptic imaging on mice with the intact skin.
Adult NMRI nu/nu nude mouse (3-4 months old) (a) cleared with vDISCO method (b): the colored rectangles show zoom-in of the cleared lungs (magenta) and skin (cyan) in b (similar results were observed from 3 independent NMRI nu/nu mice). (c-h) vDISCO on 6 weeks old CX3CR1GFP/+ mouse with intact skin: (c) cleared body with the cyan rectangle showing visible inguinal lymph nodes (black arrow-head) through the transparent skin; (d) light-sheet images of CX3CR1GFP/+ mouse boosted with anti-GFP 647-nanobooster (cyan) and labeled with PI (magenta) showing CX3CR1 GFP+ cells (cyan) densely located in the lymph node and sparsely located in and under the skin such as around hair follicles (magenta). (e-h) confocal 1mm z-scan of the skin and lymph node in d with visible CX3CR1 GFP+ cells (cyan) and nuclei (magenta): (e) 3D rendering of the stack showing the skin and the lymph node beneath; (f) 2D image at the level of the skin with single CX3CR1 GFP+ cells in the tissue and in the hair follicles (yellow arrow-heads): the fine details of the ramifications of the immune cells at the surface of the skin are visible (green arrow-heads) (g); (h) 2D image of the lymph node with visible immune cells (yellow arrow-heads) in the organ. See also Supplementary Video 5. (i-k) vDISCO on 2 months old Thy1-GFPM mouse with intact skin: (i) cleared body with the magenta rectangle showing a visible inguinal lymph node through the skin; (j) light-sheet images of Thy1-GFPM mouse boosted with anti-GFP 647-nanobooster (green) and labeled with PI (magenta) showing Thy1-GFP expressing immune cells (white arrow-heads) and neuronal projections coming from the spinal cord and projecting to the rest of the body including the skin (green), the colored rectangles show zoom-in details of a neuronal projection (green fibers and yellow arrow-heads) projecting into the skin; (k) high magnification confocal image of the region in j (yellow dashed rectangle) showing details of the immune cells (white arrow-heads) and neuronal projections (green, yellow arrow-heads) into the skin of the animal. All the results from c to k were similarly observed in 2 independent mice per each mouse line.
Degeneration of peripheral nerve terminals after brain injury
Traumatic brain injury (TBI) is a major cause of death and disability and currently there is no disease modifying treatment available to combat especially chronic sequelae. In addition to its acute consequences, TBI often leads to chronic focal and global neurological impairments, such as dementia, epilepsy, progressive motor decline, peripheral neuropathy and dystonia29,30. These impairments could be due to degeneration of the corresponding brain regions and/or the dysfunction of nerves outside of the brain. However, so far, the impact of TBI on long-range neuronal projections outside of the brain has not been assessed. For example, while TBI-induced sensory-motor dysfunction has been observed both in animal models and human pathologies31–34, the underlying mechanisms remain unknown, hampering the development of effective therapeutic approaches. Here, imaging the whole CNS, we found an extensive neurodegeneration of the descending motor axons in the brainstem and spinal cord upon TBI over the right somatosensory and motor cortices (Supplementary Fig. 17). However, how TBI alters neuronal projections in the PNS remains elusive. We used panoptic imaging to examine neuronal changes throughout the body more than a month after the brain injury (chronic stage) in comparison to control animals. As mainly axons innervating muscles are labelled in Thy1-GFPM mice, we primarily focused on these nerve terminals. Our quantifications conducted in intact cleared mouse bodies demonstrated that the complexity of nerve terminals at the neuromuscular junctions was reduced after TBI, especially in the upper torso compared to unlesioned control mice (Fig. 4a-d). We observed that nerve endings were reduced in complexity at the contralateral body regions, with fewer axonal ramifications left, implying partial degeneration of these axon terminals (Fig. 4e,f). Interestingly, we also observed changes at the ipsilateral axons albeit to a smaller degree with respect to the contralateral body region (Fig. 4e). Overall, these data demonstrate that light-sheet microscopy imaging of entire transparent mouse bodies by vDISCO holds the great promise of discovering hitherto unknown biological and pathophysiological processes.
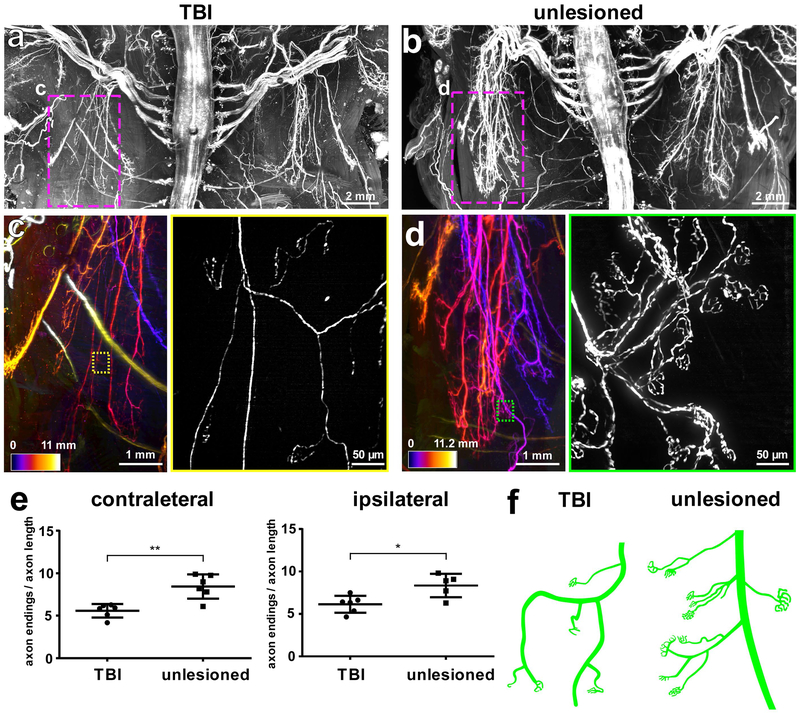
Figure 4. TBI-induced peripheral nerve degeneration revealed by vDISCO panoptic imaging.
Background equalized light-sheet microscopy maximum intensity projections of the torso from TBI-induced (a) vs. unlesioned control (b) Thy1-GFPM mice. (c,d) High magnification views of marked regions in a and b, showing the left thoracic peripheral nerve projections in the TBI (c) vs. control animal (d), the color-code indicates the z-depth of anatomical regions as given in the scale bars. The colored rectangles show the high magnification images of the marked regions in c and d, respectively, demostrating fewer intact peripheral nerve endings in TBI mice compared to controls (similar results were observed from 6 independent mice per group). (e) Quantification of axonal projection complexity expressed as number of peripheral nerve endings over length of axonal ramifications in TBI vs. control mice (3-5 months old) at the level of the contralateral side (left) and ipsilateral side (right) (mean ± s.d.; for the contralateral side: n=6 animals per group; for the ipsilateral side: n=6 and 5 mice for the TBI group and for the unlesioned group, respectively; statistical significance (**p = 0.003, *p = 0.03) was assessed by two tailed t-test). (f) Representative illustration showing the peripheral nerve ending morphology in TBI vs. control mice.
Visualizing new routes for immune cell trafficking in the brain and spinal cord
The lymphatic system, which connects various lymphatic organs in the body, is crucial for immune responses. Until recently, the brain was considered to be devoid of any lymphatic vessels. In recent years, brain lymphatic vessels were re-discovered and considered to play critical roles in brain pathologies35–38. As these lymphatic vessels are located between the brain and skull39, their connections are largely destroyed when the brain is harvested for standard histology.
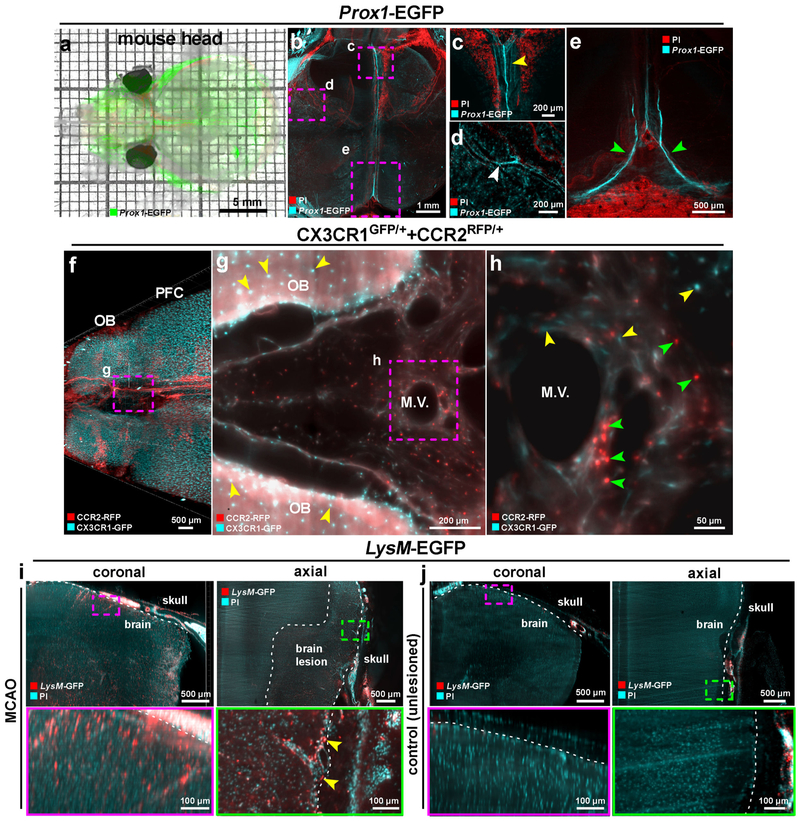
We utilized panoptic imaging to overcome this hurdle and imaged details of meningeal vessels in intact transparent mice. We used Prox1-EGFP reporter mice, in which lymphatic endothelial cells express EGFP40. We readily observed previously described brain lymphatic structures, particularly along the sagittal sinus, pterygopalatine artery and transverse sinus (Fig. 5a-e, Supplementary Fig. 18) in addition to many other body regions (Supplementary Fig. 19, Supplementary Video 6). Next, we imaged multiple subtypes of immune cells within and outside the meningeal vessels using the CX3CR1GFP/+ x CCR2RFP/+ mice by multiplexing with two different nanoboosters (anti-GFP conjugated to Atto-647N and anti-RFP conjugated to Atto-594N). The CX3CR1 GFP+ microglia cells in the brain parenchyma (Fig. 5f,g, yellow arrowheads) and CCR2 RFP+ peripheral immune cells in the meningeal vessels were clearly identifiable (Fig. 5g,h, green arrowheads, Supplementary Video 7). As expected, we also observed CX3CR1 GFP+ immune cells in meningeal vessels, which likely represent meningeal monocytes and macrophages (Fig. 5h, yellow arrowheads). It remains unclear, how the immune cells within meningeal vessels contribute to the pathology of acute brain injuries, as they may be in an advantageous position to invade the brain. To start addressing this query, we used the middle cerebral artery occlusion (MCAO) model of stroke, which was performed in LysM-EGFP mice, a transgenic mouse line expressing EGFP in myeloid cells (mainly in neutrophils and monocytes), but not in microglial cells41. The mice with MCAO showed an invasion of LysM GFP+ cells into the brain parenchyma especially in the peri-infarct region compared to sham-operated controls (Fig. 5i,j). LysM GFP+ cell also increased in the meningeal structures after MCAO (Fig. 5i yellow arrowheads), suggesting that the meninges might play a role as entry and/or exit routes of the brain in addition to the disrupted blood brain-barrier and the choroid plexus42. Thus, vDISCO panoptic imaging of transparent mice is a powerful tool to study the anatomy and cellular repertoire of meningeal vessels in health and disease.
Figure 5. Visualizing meningeal vessels through intact skull by vDISCO panoptic imaging.
(a) A representative 4 weeks old transparent Prox1-EGFP mouse head from 5 independent animals showing the labeled vessels underneath the skull. (b-e) The Prox1-EGFP mouse head showing the brain lymphatic vessels along the sagittal sinus, pterygopalatine artery and transverse sinus (yellow, white and green arrowheads in c, d and e, respectively). Bone structures become prominent with PI labeling (red). Single experiment. (f) 3D visualization of prefrontal cortex (PFC) and olfactory bulb (OB) in a 2 months old CX3CR1GFP/+ (cyan) and CCR2RFP/+ (red) double transgenic mouse (similar results were observed from 3 independent double transgenic mice). (g,h) High-magnification image of marked region in f showing CCR2 RFP+ cells (green arrowheads) and CX3CR1 GFP+ cells (yellow arrowheads) in meningeal vessels. See also Supplementary Video 7. (i,j) 6 months old LysM-EGFP transgenic mice with MCAO vs. unlesioned control showing the infiltration of immune cells in MCAO (similar results were observed from 4 independent mice per group). LysM GFP+ cells are shown in red and nucleus labeling by PI in cyan. Immune cells in the meningeal vessels of the injured mice were observed (yellow arrowheads). All images were obtained by light-sheet microscopy.
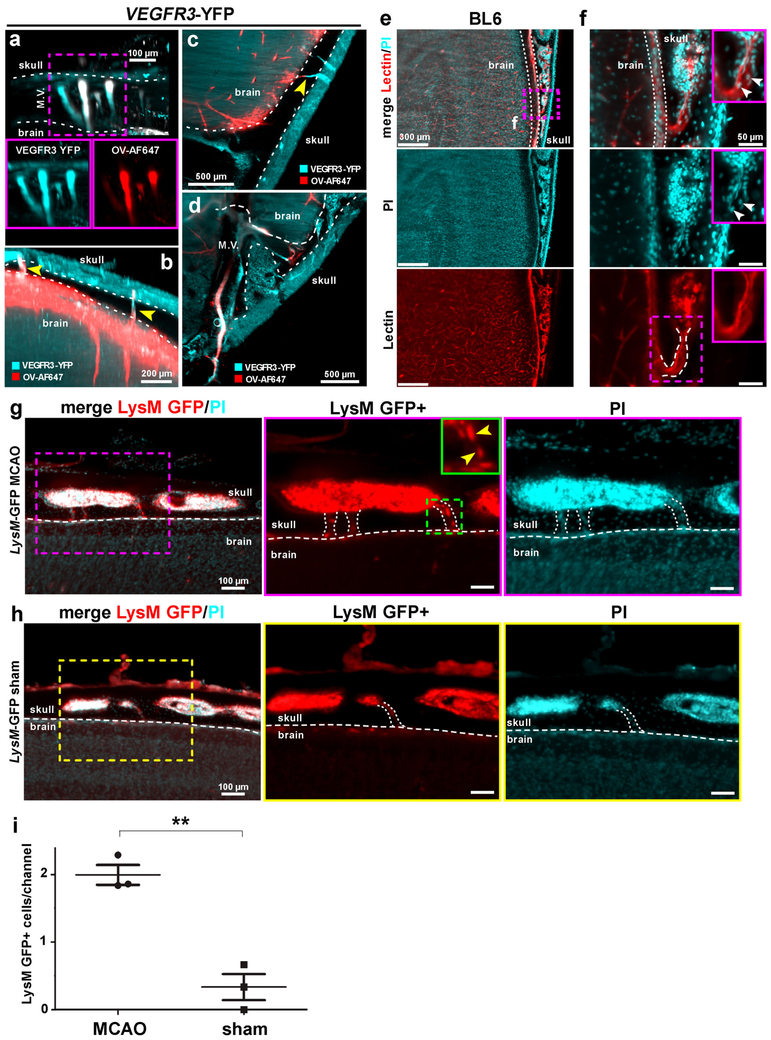
Next, we aimed at investigating vascular connections between the skull and the meninges in greater detail. Towards this goal, we injected a cerebrospinal fluid (CSF) tracer (fluorophore-conjugated ovalbumin) into the cisterna magna (CM) in VEGFR3-YFP mice, another commonly used reporter line for meningeal vessels. We observed that the CSF tracer was quickly transported into the VEGFR3+ meningeal vessels (Fig. 6a-d). Interestingly, we also observed that the tracer labelled vessels extended into the skull (Fig. 6b,c yellow arrowheads). We further investigated the details of these new connections using lectin– another vessel tracer binding to endothelial cells. We found that these vascular connections are mostly located between the skull marrow and the outer surface of the meninges (both at the brain and sagittal sinus interfaces, Fig. 6e,f, Supplementary Fig. 20, Supplementary Videos 8,9). We measured that they have an average size of 46.9 ± 5.8 µm in length and 19.2 ± 2.5 µm in width at the brain interface and of 83.7 ± 0.3 µm in length and 28.5 ± 1.8 µm in width at the sagittal sinus interface (mean ± SEM, n = 3 mice, Supplementary Fig. 20), therefore, we named them as short skull – meninges connections (SMCs). They also occasionally contain cells (Fig. 6e,f, white arrowheads, Supplementary Fig. 20). To address whether the SMCs might play a role in neuropathological conditions, we studied the distribution of LysM GFP+ immune cells in these connections after MCAO. The mice with MCAO showed significantly more LysM GFP+ cells in the SMCs compared to sham-operated controls (Fig. 6g-i, Supplementary Video 10) suggesting that they might represent an entry route for myeloid cells from the skull marrow to the meningeal compartments after brain injury. Thus, vDISCO imaging revealed connections between skull and meninges, which might be important for immune cell trafficking especially during brain pathologies.
Figure 6. Uncovering skull – meninges connections (SMCs) through intact skull by vDISCO panoptic imaging.
(a-d) A 6 months old VEGFR3-YFP mouse head in sagittal (a,b) and axial views (c,d) showing that injected CSF tracer (Ovalbumin-Alexa647, red) fills not only meningeal vessels but also some short skull-meninges connections (SMCs) between skull marrow and brain surface (b,c, yellow arrowheads) (representative images, single experiment). (e,f) Details of SMCs after whole mouse body PI and lectin labeling of a 3 weeks old C57BL/6 mouse processed by vDISCO: the SMCs from skull marrow to brain surface with a funnel-shape opening are marked with dashed lines in f. Cells in the channel are indicated by white arrow heads (similar results were observed from 5 independent mice). (g,h) 6 months old LysM-EGFP transgenic mice after MCAO (g) vs. sham control (h) showing increased numbers of LysM GFP+ cells (yellow arrow-heads) in the SMCs (marked with dashed lines) in MCAO (similar results were observed from 3 independent mice per group). LysM GFP+ cells are shown in red and nucleus labeling by PI in cyan. See also Supplementary Video 10. (i) Quantification of LysM GFP+ cells in the SMCs in MCAO vs. sham mice (mean ± SEM; n=3 animals per group; statistical significance (**p = 0.003) was assessed by two tailed t-test).
Finally, we explored the immune cell trafficking after spinal cord injury (SCI) using vDISCO panoptic imaging. To this end, we induced a SCI in CD68-EGFP transgenic mice expressing EGFP in monocytes and macrophages43. Upon SCI, we observed an influx of CD68 GFP+ cells at the lesion site as well as a rather widespread increase in CD68 GFP+ cells around the spinal cord (Supplementary Fig. 21, Supplementary Video 11). Indeed, only 3.5% of these phagocytes were seen in the injured spinal cord, while ~96.5% of the CD68 GFP+ cells were located in the surrounding tissues of the spinal cord including adjacent muscles, spinal cord roots and meningeal vessels similar to sham mice (Supplementary Fig. 22). Thus, vDISCO is a powerful tool to image and quantify the trafficking of immune cells to and within the injured central nervous system.
Discussion
Panoptic imaging of transparent adult mouse bodies from head to toe holds the promise of providing an unbiased and highly resolved view of entire organ systems in health and disease. Here, we developed a new whole-body nanobody labeling method in conjunction with whole-body tissue clearing, enabling a reliable visualization and quantification of subcellular details throughout centimeters-thick tissues of intact mouse bodies. This new panoptic imaging technology is straightforward in application, and suitable for systemic analysis of a vast range of biomedical inquires, as demonstrated here in visualization of the neuronal projections in whole mouse bodies, the description of vascular pathways between the skull and meninges, and the visualization of remote neuronal and immune consequences of acute CNS injury models. It is notable that, while we developed vDISCO for panoptic imaging of the whole mouse body, it is readily applicable for individual organs, using a simplified protocol to drastically increase and stabilize the signal contrast, as we show for mouse brains imaged more than a year after boosting and clearing (Supplementary Video 12, Supplementary Table 1).
The panoptic imaging of nervous and immune systems in intact mouse bodies was achieved owing to the massive enhancement of the signal intensity by one to two orders of magnitude via whole-body immunolabeling employing nanobodies conjugated to bright dyes in the far-red spectrum. Therefore, the strong autofluorescence of tissues in shorter wavelength spectra, in which traditional fluorescent proteins such as GFP are excited and imaged, is avoided. Here, we primarily used Atto dyes at 647 nm, and additional Atto 594 nm for multiplexing 2 different cell types. In the future, usage of nanobodies conjugated to dyes that emit in the near infrared range44 can further facilitate multiplexed detection of more than 2 targets. While the number of commercially available nanobodies is still limited, the nanoboosters used in this study can stain a broad selection of 21 different fluorescent proteins including EGFP, YFP, Venus, mCherry, and mRFP. Our method can also be used with nanobodies against endogenous proteins as we demonstrated using anti-vimentin nanobodies labeling activated glia cells in the brain upon trauma45,46. Currently, nanobodies are developed mainly for in vivo applications due to their small size (e.g., for crossing the blood-brain barrier)47, therefore, they are mostly selected for in vivo epitopes but not for fixed tissues. We believe that our study will encourage the production of diverse nanobodies for deep tissue immunolabeling.
Here, we used transgenic mice endogenously expressing fluorescent proteins, however, the labeling can also be achieved by rabies virus retrograde tracing, systemic AAV injections48 or transplantation of genetically engineered cells (such as stem cells or adoptive transfer of immune cells). Another key advantage of vDISCO panoptic imaging is that upon staining with Atto dyes through nanoboosting the signal becomes permanent, permitting long-term imaging sessions and also future re-analysis of the same sample if needed. This also enables high-resolution imaging of desired body parts by confocal or 2-photon microscopy. Indeed, the samples from this study are available to the scientific community upon request for further studies. We believe making vDISCO treated samples accessible to other labs for further studies can save time and resources, reduce the number of mice used in experimental research and create more powerful data sets by allowing direct correlation of various inquiries by multiple studies from the same animals.
A current limitation is the absence of light-sheet microscope systems that could image the entire body of a mouse in one session without any displacement and turning of the sample. Construction of such new light-sheet microscopes should further decrease the time needed for data acquisition and simplify data processing for 3D reconstruction. In fact, as shown here, optics and light sources of current commercial light-sheet microscopes are sufficient to image intact mouse bodies at subcellular resolution, albeit the whole body has to be manually moved in the imaging chamber to achieve the scanning of different body regions. In addition, development of machine learning-based (artificial intelligence) algorithms to automatically identify and quantify cellular changes on the whole organism scale will be important to further scale up this technology in the future.
Using panoptic imaging, we constructed a neuronal projection map for the Thy1-GFPM transgenic line, showing subcellular details of long-range neuronal connections from the CNS to the distal extremities. Here, we visualized neuronal details in Thy1-GFPM mice, in which only a subset of CNS neurons and PNS neurons innervating skeletal muscles are labelled. In the future, panoptic imaging of other transgenic lines labeling a greater and more diverse subset of neurons will allow the exploration of other biological systems such as the autonomic innervation of internal organs.
In recent years, researchers have been providing more evidence on post TBI changes that seem to form the basis of chronic complications, such as epilepsy, neuropsychiatric disorders, dementia and progressive motor decline among others29. However, the effects of a localized brain lesion on the rest of the body have been poorly understood mainly due to the technical challenges in studying long-range neuronal projections. Because vDISCO fluorescent signal boosting allows both imaging and quantifications of light-sheet microscopy images of transparent mouse bodies, we identified TBI-induced changes at the peripheral axonal projections innervating the skeletal musculature of the torso. These data are in line with recent work by others and us demonstrating the functional deficits in fine motor movements in mice and the presence of sensory-motor dysfunctions in TBI patients29,30,33. Our data provide a possible cellular mechanism for these functional impairments, suggesting that they could arise due to degeneration of long-range axons throughout the spinal cord and PNS. This discovery opens alternate avenues for the identification of therapeutic targets. For instance, recent literature showed that microtubule stabilization is a powerful way to block degeneration of axons in spinal cord injury49. In this regard, it would be important to assess if microtubule stabilization would also counteract axon degeneration outside of the brain and circumvent the development of chronic sensory-motor dysfunctions. Here, we also demonstrated that meningeal vessels extending between the cranium and CNS tissue and their cellular content can readily be imaged by vDISCO panoptic imaging in naïve animals and acute CNS injury models. Importantly, we observed short vascular connections between the skull and meninges both at the brain interface (recently seen also by others50) and at the sagittal sinus, which were filled with immune cells following ischemic stroke injury. Most of the SMCs seem to be open both at the skull marrow and meningeal ends without being connected to the vascular system of the brain/head (Supplementary Video 9). In the future, it will be important to identify to which meningeal compartment/s (subdura, subarachnoid or subpial) the SMCs are connected to. In addition, due to the present lack of appropriate reporter mice, it was not possible to distinguish the type of SMCs (blood or lymphatic vessels) with increased LysM+ cells after stroke. In the future, vDISCO imaging of immune cells in a lymphatic vessel reporter mouse line with different fluorescent proteins (e.g., LysM-EGFP + Prox1-RFP double reporter) could reveal more details of this new route for immune cell infiltration. As neuroinflammation is a major determinant of neuronal function and survival following CNS injuries of various aetiologies, our technology should accelerate investigation of these novel immune cell infiltration routes in various CNS pathologies.
In conclusion, vDISCO enhancement and stabilization of fluorescent proteins combined with signal acquisition in far-red spectra should facilitate discovery of previously inaccessible biological information. For example, imaging and quantification of neuronal projections, vascular organizations, and immune cell populations in the entire mouse body should contribute to a more comprehensive understanding of the initiation, progression and extent of neurological conditions.
MATERIALS AND METHODS
Animals:
We used the following mixed gender animals in the study: CX3CR1GFP/+ (B6.129P-Cx3cr1tm1Litt/J, Jackson Laboratory strain code: 005582)51, Thy1-GFPM and Thy1-YFPH21, Prox1-EGFP (Tg(Prox1-EGFP)KY221Gsat/Mmucd, MMRRC strain code: 031006-UCD), VEGFR3-YFP, CX3CR1GFP/+ x CCR2RFP/+ (B6.129(Cg)-Ccr2tm2.1Ifc/J, Jackson Laboratory strain code: 017586), LysM-EGFP (Lyz2tm1.1Graf, MGI: 2654931), CD68-EGFP (C57BL/6-Tg(CD68-EGFP)1Drg/j, Jackson Laboratory strain code: 026827), C57BL/6J, NMRI nu/nu mice. The animals were housed under a 12/12 hours light/dark cycle. The animal experiments were conducted according to institutional guidelines: Klinikum der Universität München / Ludwig Maximilian University of Munich and after approval of the Ethical Review Board of the Government of Upper Bavaria (Regierung von Oberbayern, Munich, Germany) and the Animal Experiments Council under the Danish Ministry of Environment and Food (2015–15-0201–00535) and in accordance with the European directive 2010/63/EU for animal research. Ages of the animals are indicated in the figure legends. All data are reported according to the ARRIVE criteria. Sample sizes were chosen based on prior experience with similar models and are specified in figure legends. Within each strain, animals were randomly selected. Animals that resulted negative for the expression of fluorescent proteins by genotyping were excluded from the study. No statistical methods were used to pre-determine sample sizes but our sample sizes are similar to those reported in previous publications17.
Traumatic brain injury
Traumatic brain injury was performed using a Controlled Cortical Impact (CCI) device (Leica Benchmark Stereotaxic Impactor, 39463923). 30 minutes before surgery, we administered Carprofen (4 mg/kg) and Buprenorphin (0.05 mg/kg) to the animals via subcutaneous injection. Then, anaesthesia was induced in animals with 4% isoflurane in N2O/O2 (70%/30%) mixture and afterwards maintained with 1.5% isoflurane in the same mixture for the whole surgery. As soon as the animals did not show any pedal reflex, they were placed in the associated stereotaxic apparatus and their body temperature was kept at 37°C using a heating pad for the whole surgery procedure. Next, the scalp of the animals was shaved, aseptically prepared by wiping with Octenisept (Schülke, 22580-A) as disinfectant and the skin of the scalp was incised longitudinally between the occiput and forehead. We identified the target area of the injury, which was the right somatosensory cortex extending to the motor cortex, using the stereotaxic frame (bregma coordinates: 2-mm posterior, 5-mm right lateral). The injury was then triggered via CCI machine using the following parameters: impact speed: 6.9 m/s; impact duration: 400 ms and impact depth: 2 mm. With these parameters, the injury resulted severe with cracks of the skull, bleeding and exposed brain tissue. After the impact, the skin was sutured with metallic wound closure clips (VWR, 203–1000) and the animals were kept at 31°C in a recovery chamber (Mediheat, 34–0516) for at least 30 minutes until they recovered from the anesthesia. In the following days, animals were subcutaneously injected with Carprofen (4 mg/kg) once every day for 4 days and sacrificed at > 1 month post injury by transcardial perfusion according to the ‘perfusion and tissue preparation’ section below.
MCAO model
Experimental stroke was induced using the intraluminal filament model of middle cerebral artery occlusion (fMCAO) occlusion for transient, focal brain ischemia. Mice were anesthetized with isoflurane delivered in a mixture of 30% O2 and 70% N2O. A heat-blunted nylon suture (6/0) was inserted into the external carotid artery and advanced until it obstructed the MCA together with the ligation of the common carotid artery for 30 min. Regional cerebral blood flow (CBF, bregma coordinates: 2-mm posterior, 5-mm lateral) was continuously recorded by transcranial laser Doppler flowmetry from the induction of ischemia until 10 min after reperfusion. Following fMCAO, mice were placed in temperature-controlled recovery cages for 2 h to prevent post-surgery hypothermia. For the survival period (3 days), the mice were kept in their home cage with facilitated access to water and food. Sham-operated mice received the same surgical procedure without insertion of the filament. Body temperature was maintained throughout surgery using a feedback-controlled heating pad and kept constant (37.0 ± 0.5 °C). Exclusion criteria were as follows: insufficient MCA occlusion (a reduction in blood flow to 15% of the baseline value) and blood flow recovery >80% within 10 min of reperfusion. Mice were sacrificed at 3 days post injury by transcardial perfusion according to the ‘perfusion and tissue preparation’ section below.
CM injection for meningeal vessel labeling
For the cisterna magna injections, mice (VEGFR3-YFP; 6 months old) were anesthetized with a mixture of ketamine and xylazine (100 mg/kg; 10 mg/kg, respectively) via intraperitoneal (i.p.) injection. After toe pinch reflexes ceased, mice were fixed in a stereotaxic frame by the zygomatic arch, with the head slightly tilted to form an angle of 120° in relation to the body. The head and neck regions were shaved to expose the neck muscles, which were bluntly dissected to expose the cisterna magna (CM). Cannulas composed of a dental needle (SOPIRA® Carpule 30G 0.3 × 12mm; Kulzer; AA001) and polyethylene tubing (0.024” OD x 0.011” ID; Scandidact; PE10-CL-500) were used to perform the CM injections. A cannula filled with cerebrospinal fluid (CSF) tracer (2% ovalbumin 45 kDa – Alexa Fluor 647 conjugated; Thermo Fisher Scientific, O34784, diluted in artificial CSF: 126 mM NaCl, 2.5 mM KCl, 1.25 mM NaH2PO4, 2 mM Mg2SO4, 2 mM CaCl2, 10 mM glucose, 26 mM NaHCO3; pH 7.4 when gassed with 95% O2 and 5% CO2) was inserted into the CM. With the aid of an injection pump (LEGATO 130 Syringe pump, KD Scientific, 788130), 10 µl of CSF tracer was injected into the CM at a rate of 1 µl/min. At the end of the injection, CSF tracer was allowed to circulate in the subarachnoid and paravascular spaces for 1 hour. Mice were then transcardially perfused according to the ‘perfusion and tissue preparation’ section below.
Spinal cord injury model
Mice were deeply anaesthetized by i.p. injection of a combination of midazolam (5 mg/kg body weight), medetomidine (0.5 mg/kg) and fentanyl (0.05 mg/kg). The mid-thoracic spinal cord of anaesthetized CD68-EGFP mice was surgically exposed by a dorsal laminectomy as previously described52. A hemisection of the spinal cord was performed using fine-tip surgical scissors (F.S.T 15000–08 spring scissor 2.5 mm cutting edge). For sham surgery, the dorsal laminae of mid-thoracic vertebrae were surgically exposed in anaesthetized CD68-EGFP mice. After spinal cord injury or sham surgery, muscle tissue and skin were sutured with a surgical thread (Ethilon suture 6–0, 667H) and animals were allowed to recover on a heating pad. After 48 hours, mice were transcardially perfused according to the ‘perfusion and tissue preparation’ section below.
Perfusion and tissue preparation
Mice were deeply anesthetized using a combination of midazolam, medetomidine and fentanyl (MMF) (1ml/100g of body mass for mice; i.p.). As soon as the animals did not show any pedal reflex, they were intracardially perfused with heparinized 0.1 M PBS (10 U/ml of Heparin, Ratiopharm; ~110 mmHg pressure using a Leica Perfusion One system) for 5–10 minutes at room temperature until the blood was washed out, followed by 4% paraformaldehyde (PFA) in 0.1 M PBS (pH 7.4) (Morphisto, 11762.01000) for 10–20 minutes Next, optionally skin and eyes, premaxilla and maxilla bones were carefully removed and the palate of the animal was opened (without damaging the tissue beneath), and the feces were gently washed out from the intestine with 0.1 M PBS through small cuts using a syringe. For the animals cleared with skin: the skin was not removed from the bodies and in case of BL6 background animals, the fur was shaved off using a razor blade (Personna, 604305–001001). Then, the bodies were postfixed in 4% PFA for 1 day at 4 °C and later washed with 0.1 M PBS for 10 minutes 3 times at room temperature. The whole-body nanoboosting procedure was started immediately or whole mouse bodies were stored in PBS at 4 °C for up to 4 weeks or in PBS containing 0.05% sodium azide (Sigma, 71290) for up to 6 months.
For the collection of dissected tissues such as brains, mice were perfused with PBS and PFA as described above. Subsequently, the organs of interest were dissected out and post-fixed in 4% PFA overnight at 4°C, washed with 0.1 M PBS for 10 minutes 3 times at room temperature and kept in PBS plus 0.05% sodium azide up to 3 weeks.
Clearing of unboosted samples
For the quantification of fluorescence signal after clearing, without boosting, we followed the uDISCO passive clearing protocol as described in Pan et al., 201617. In brief, dissected brains were placed in 5 ml tubes (Eppendorf, 0030 119.401) and covered with 4.5 ml of clearing solution. All incubation steps were performed in a fume hood with gentle shaking or rotation, with the samples covered with aluminum foil to keep them in dark. To clear the samples, we incubated them in a gradient of tert-butanol (Sigma, 360538): 30 Vol%, 50 Vol%, 70 Vol%, 80 Vol%, 90 Vol%, 96% Vol% (in distilled water), 100 Vol% twice at 35°C for 12 hours each step, followed by immersion in dichloromethane (DCM, Sigma, 270997) for 45–60 minutes at room temperature and finally incubated with the refractive index matching solution BABB-D15 containing 15 parts BABB (benzyl alcohol + benzyl benzoate 1:2, Sigma, 24122 and W213802), 1 part diphenyl ether (DPE) (Alfa Aesar, A15791) and 0.4% Vol vitamin E (DL-alpha-tocopherol, Alfa Aesar, A17039), for at least 6 hours at room temperature until achieving transparency.
Nanobodies, antibodies and fluorescent dyes used in this study
Atto594 conjugated anti-RFP nanobooster (Chromotek, rba594–100), Atto647N conjugated anti-GFP nanobooster (Chromotek, gba647n-100), Atto488N conjugated anti-GFP nanobooster (Chromotek, gba488–100), Atto488 conjugated anti-Vimentin nanobooster (Vimentin-Label_Atto488, Chromotek, vba488–100), Alexa647 conjugated anti-GFP antibody (Invitrogen, A31852), Propidium iodide (PI, Sigma, P4864). For the nanobody and antibody validation information, see the Life Sciences Reporting Summary.
Validation of nanoboosters for vDISCO
Post-fixed brains from fluorescent protein expressing mice (e.g. Thy1-GFPM) were cut into 400 µm slices using a vibratome (Leica, VT1200S). The slices were imaged before staining with Zeiss AxioZoom EMS3/SyCoP3 fluorescence stereomicroscope to make sure that the samples have fluorescent protein expression. All the following steps were done with gentle shaking. The slices were first incubated for 3 hours in a 24 multiwell (Falcon, 353504) at 37°C with 1 ml of permeabilization solution containing 1.5% goat serum (Gibco, 16210072), 0.5% Triton X-100 (AppliChem, A4975,1000), 0.5 mM of Methyl-beta-cyclodextrin (Sigma, 332615), 0.2% trans-1-Acetyl-4-hydroxy-L-proline (Sigma, 441562) and 0.05% Sodium Azide (Sigma, 71290) in 0.1 M PBS. Subsequently, they were incubated overnight at 37°C with the same permeabilization solution adding the nanobooster of interest with dilution 1:500 considering final volume = 500 µl. The multiwell was covered with aluminium foil to keep the slices in dark and was sealed well with parafilm (Bemis, PM-992) to prevent the solution to dry out. One slice was kept as negative control where there was no adding of nanobooster. After the incubation, slices were washed with the washing solution (1.5% goat serum, 0.5% Triton X-100, 0.05% of sodium azide in 0.1 M PBS) at room temperature for 15 minutes, 4 times and then washed with 0.1 M PBS at room temperature for 10 minutes, 3 times. Then the slices were imaged again after staining with the Axiozoom microscope to check whether the staining worked. In the end, slices were moved to 5 ml Eppendorf tubes wrapped with aluminium foil to keep them in dark and cleared with 3DISCO protocol: first they were incubated at room temperature in the following gradient of tetrahydrofuran (THF) (Sigma, 186562) in distilled water (45 minutes for each step): 50 Vol% THF, 70 Vol% THF, 80 Vol% THF, 100 Vol% THF and 1hour 100 Vol% THF; after dehydration, 15 minutes in DCM, and finally in BABB until transparency. Finally, the slices were imaged again with the AxioZoom microscope to make sure that the staining worked even after clearing. For more details of the AxioZoom imaging setup see “Fluorescence stereomicroscopy imaging” paragraph.
vDISCO whole-body immunostaining, PI labeling and clearing
In order to remove remaining blood and heme after PFA perfusion, and to decalcify the bones, the animals were subjected to perfusion with decolorization solution and decalcification solution before immunostaining. The decolorization solution was made with 25–30 Vol% dilution of CUBIC reagent 15 in 0.1 M PBS. CUBIC reagent 1 was prepared with 25 wt% urea (Carl Roth, 3941.3), 25 wt% N,N,N´,N´-tetrakis (2-hydroxypropyl)ethylenediamine (Sigma, 122262) and 15 wt% Triton X-100 in 0.1 M PBS. The decalcification solution consisted of 10 wt/Vol% EDTA (Carl Roth, 1702922685) in 0.1 M PBS adjusting the pH to 8–9 with sodium hydroxide NaOH (Sigma, 71687).
The solutions for the immunolabeling pipeline were pumped inside the body of the animal by transcardial-circulatory perfusion exploiting the same entry point hole into the heart created during the PBS and PFA perfusion step (see above, perfusion and tissue preparation paragraph) and following the procedure already described in Pan et al., 2016. In brief, the mouse body was placed in a 300 ml glass chamber (Omnilab, 5163279) filled with 250–300 ml of appropriate solution, which covered the body completely. Next, the transcardial-circulatory system was established involving a peristaltic pump (ISMATEC, REGLO Digital MS-4/8 ISM 834; reference tubing, SC0266) keeping the pressure at 160–230 mmHg (45–60 rpm). One channel from the pump, made by a single reference tube, was set for circulation of the solution through the heart into the vasculature: one ending of the tube was connected to the tip of a syringe (cut from a 1 ml syringe-Braun, 9166017V) which held the perfusion needle (Leica, 39471024) and the other ending was immersed in the solution chamber where the animal was placed. The perfusion needle pumped the appropriate solution into the mouse body, and the other ending collected the solution exiting from the mouse body in order to recirculate the solution, pumping it back into the animal. To fix the needle tip in place and to ensure extensive perfusion, we put a drop of super glue (Pattex, PSK1C) at the level of the hole where the needle was inserted inside the heart. Using the setting explained above, after post-fixation and PBS washing, the mice were first perfused with 0.1 M PBS overnight at room temperature, then the animals were perfused with 250 ml of decolorization solution for 2 days at room temperature, exchanging with fresh decolorization solution every 6–12 hours until the solution turned from yellowish to clear and the spleen became lighter colour (indicating that the blood heme was extracted). Then, they were perfused with 0.1 M PBS, washing for 3 hours 3 times, followed by perfusion with 250 ml of decalcification solution for 2 days at room temperature and again perfused/washed with 0.1 M PBS for 3 hours 3 times. After this, the animals were perfused with 250 ml of permeabilization solution containing 1.5% goat serum, 0.5% Triton X-100, 0.5 mM of Methyl-beta-cyclodextrin, 0.2% trans-1-Acetyl-4-hydroxy-L-proline and 0.05% Sodium Azide in 0.1 M PBS for half a day at room temperature. Subsequently, the perfusion proceeded further, connecting a 0.20 µm syringe filter (Sartorius, 16532) to the ending of the tube not holding the needle, in order to efficiently prevent accumulation of dye aggregates into the sample. At the same time, from this step we used an infrared lamp (Beuer, IL21) directed to the chamber to heat up the solution to 26–28°C. With this setting, the animals were perfused for 6 days with 250 ml of the same permeabilization solution containing 35 µl of nanobooster, which is 20–35 µg in 250 ml (0.08– 0.14 µg/ml), 1:7000 in dilution, (stock concentration 0.5 – 1 mg/ml) (the amount of nanobody was adjusted depending on the expected presence of fluorescent protein in the mouse body) and 290 µl of propidium iodide (stock concentration 1 mg/ml). Next, we removed the animals from the chamber and with fine scissors we removed a tiny piece of skull from the back of the skull (above the cerebellum) at the level of the occipital bone, and we placed the bodies in a 50 ml tube (Falcon, 352070), filled with the same permeabilization solution, containing an extra 5 µl of nanobooster and incubated the tubes at 37°C with gentle shaking for an additional 2–3 days of labeling. After that, the mice were placed back in the perfusion system and labeling solution was washed out by perfusing with washing solution (1.5% goat serum, 0.5% Triton X-100, 0.05% of sodium azide in 0.1 M PBS) for 3 hours three times at room temperature and 0.1 M PBS for 3 hours three times at room temperature.
After the staining, the animals were cleared using a 3DISCO based passive whole-body clearing protocol optimized for big samples. The mice were incubated at room temperature in dehydration and clearing solutions inside a 300 ml glass chamber, kept with gentle rotation on top of a shaking rocker (IKA, 2D digital) inside a fume hood. For dehydration, mice bodies were incubated in 200 ml of the following gradient of THF in distilled water (12 hours for each step): 50 Vol% THF, 70 Vol% THF, 80 Vol% THF, 100 Vol% THF and again 100 Vol% THF, followed by 3 hours in dichloromethane and finally in BABB. During all incubation steps, the glass chamber was sealed with parafilm and covered with aluminum foil. See Supplementary Table 1 and www.discotechnologies.org/vDISCO.
vDISCO whole-mount immunolabeling of individual organs
vDISCO was performed on dissected organs as well: dissected brains were stained using the immunolabeling protocol for dissected organs. First, the post-fixed brains were pretreated, incubating them for 2 days at 37°C with gentle shaking in 4.5 ml of same solution used at the permeabilization step (see paragraph above) (1.5% goat serum, 0.5% Triton X-100, 0.5 mM of Methyl-beta-cyclodextrin, 0.2% trans-1-Acetyl-4-hydroxy-L-proline, 0.05% sodium azide in 0.1 M PBS). Subsequently, the brains were incubated in 4.5 ml of this same permeabilization solution plus the nanobooster of interest with the concentration adjusted to expression of the target (for example Atto647N conjugated anti-GFP nanobooster dilution 1:600, which is ~5–8µg of nanobooster in 4.5 ml (1.1–1.8 µg/ml) (stock concentration = 0.5 – 1 mg/ml) for Thy1-GFPM brains) for 12–14 days at 37°C with gentle shaking, then brains were washed for 2 hours 3 times and once overnight with the washing solution (1.5% goat serum, 0.5% Triton X-100, 0.05% of sodium azide in 0.1 M PBS) at room temperature and in the end washed for 2 hours 4 times with 0.1 M PBS at room temperature. The immunostained brains were cleared with 3DISCO clearing: first they were put in the Eppendorf 5 ml tubes and then incubated at room temperature with gentle shaking in 4.5 ml of the following gradient of THF in distilled water (2 hours for each step): 50 Vol% THF, 70 Vol% THF, 80 Vol% THF, 100 Vol% THF and overnight 100 Vol% THF; after dehydration, the samples were incubated for 1 hour in dichloromethane, and finally in BABB until transparency. During all the clearing steps, the tubes were wrapped with aluminum foil to keep them in dark. See Supplementary Table 1.
For the comparison of deep tissue staining efficiency in terms of penetration of the staining of nanobody vs. conventional antibody (Supplementary Figure 2), we followed the same protocol described above in this section for both nanobody and antibody experimental groups. The only exception was that for the antibody group, in the immunostaining step, we replaced the nanobooster with Alexa647 conjugated anti-GFP antibody (Invitrogen, A31852) with the same concentration (see above).
Pumping pressure measurement
Kkmoon Digital Manometer Pressure Gauge Manometer (HT-1891) was used to measure the pressure. To achieve one additional measuring channel to connect with Manometer, a 2 heads connector (B.Braun Discofix® C Dreiwegehahn, 16494C) was inserted into the pumping channel and the second head was further connected to the Manometer with extended PVC tubing. The pumping channel was set in the standard way as described including a transcardiac perfusion needle. Firstly, the switch on the 2 heads connector was set without connecting the measuring tubing and pumping was started until all the air bubbles were excluded from the pumping channel. Subsequently, the switch was set to connect the measuring tubing while pumping was continued and the pressure was measured when the Manometer readouts stabilized. Three measurements were made at different time points including morning, afternoon and evening to exclude the variation of room temperature and the data were quantified by averaging.
High pressure low pressure comparison with methylene blue
The diffusion of methylene blue (Sigma, M9140), perfused into the body of the animals, was used to compare the efficiency of high pressure vs. low pressure perfusion (Supplementary Fig. 8). For this purpose, we used 3 BL6 animals (4 months old) per each experimental group: post-fixed whole bodies were perfused for 15 minutes with 0.05% of methylene blue in 0.1 M PBS, at either 50 rpm (~180 mmHg) for the high-pressure group or 25 rpm (~70 mmHg) for the low-pressure group. The same pump setting explained in “vDISCO whole-body immunostaining, PI labeling and clearing” section was used, including the usage of super glue to fix the needle in place inside the hole in the heart.
iDISCO+
Antibody validation: methanol compatibility test
Before performing the comparison of the efficiency between vDISCO nanobody staining vs. iDISCO+ antibody staining on whole brains, we verified the compatibility of the chosen antibody with the iDISCO+ protocol (Supplementary Figure 3a-d). This was done performing the antibody validation step, which tested the compatibility of the antibody with the methanol pretreatment required by iDISCO+. To do so, we followed the original iDISCO+ publication23 and the latest protocol updates from https://idisco.info. Briefly, post-fixed brains from Thy1-GFPM and CX3CR1GFP/+ mice (2–3 months old) were cut into 600 µm slices with a vibratome. First, the slices were incubated overnight in 100% methanol (Roth, 4627.6) at room temperature; then, after rehydrating and washing with 0.1 M PBS, the iDISCO+ immunostaining protocol was done as normally using 3 hours as permeabilization time, 6 hours as blocking time and overnight as immunolabeling time, adding 1:850 of Alexa647 conjugated anti-GFP antibody (already used in the previous section) in 500 µl of incubation solution. Non methanol treated slices were used as positive controls. In the end, the slices were imaged using the Zeiss AxioZoom stereomicroscope (see “Fluorescence stereomicroscopy imaging” section below).
iDISCO+ on dissected brains
After having verified that the previously mentioned conventional anti-GFP antibody is compatible with iDISCO+, we performed comparison of staining efficiency between vDISCO with nanobody vs. iDISCO+ with conventional antibody (Supplementary Fig. 3e-i). For this purpose, 3 CX3CR1GFP/+ post-fixed brains (6–7 months old) were used per each experimental group. The brains allocated into the vDISCO group were processed following the vDISCO protocol for dissected brains (see “vDISCO whole-mount immunolabeling of individual organs” section); while the brains allocated into the iDISCO+ group were processed using 1:730 of the above mentioned Alexa647 conjugated anti-GFP antibody following the iDISCO+ protocol and its updates from https://idisco.info23.
Different whole-body labeling protocols comparison
To show that vDISCO with nanobody is the only method that can efficiently boost the GFP signal in mouse whole bodies, we performed the following comparison using 3 GFPM animals (2–3 months old) per each experimental group: (1) high pressure active perfusion vDISCO with nanobody, (2) passive incubation vDISCO with nanobody, (3) high pressure active perfusion vDISCO with conventional antibody, (4) high pressure active perfusion iDISCO+ with conventional antibody, (5) high pressure active perfusion iDISCO+ with nanobody. The group (1) was processed using the already described vDISCO whole-body immunostaining and clearing method (see the related section above); the group (2) was processed following all the steps of the vDISCO whole-body immunostaining and clearing protocol described above, but replacing all the original steps done in perfusion with passive incubation; the group (3) was processed using the same vDISCO whole-body immunostaining and clearing protocol but replacing the nanobooster with Alexa647 conjugated anti-GFP antibody (already mentioned before) at the same concentration of the nanobooster during the staining step; the group (4) and (5) were processed performing the iDISCO+ methanol pretreatment on the animals and followed by sequential perfusion of all the iDISCO+ solutions23 (except that the clearing was done with passive incubation): the timing of each iDISCO+ step was adjusted considering the size of a whole body compared to a dissected brain (for example during the methanol dehydration, each step was increased to 5 hours vs. 1 hour for dissected brains). The antibody used for (4) is the Alexa647 conjugated anti-GFP antibody already mentioned before.
Previously published whole-body clearing methods for comparison
For PARS clearing, 10% EDTA adopted from Bone-CLARITY25 was transcardially perfused through the whole mouse bodies after PFA fixation for 5–7 days to decalcify the bones. Then, the standard PARS procedure was followed53. CUBIC18, uDISCO17 and PEGASOS16 were conducted based on the original publications. It is worth noting that among all the clearing methods tested, PARS, PEGASOS and vDISCO were characterized by a decalcification step using EDTA solutions.
Light-sheet microscopy imaging
Single plane illuminated (light-sheet) image stacks were acquired using an Ultramicroscope II (LaVision BioTec), featuring an axial resolution of 4 μm with following filter sets: ex 470/40 nm, em 535/50 nm; ex 545/25 nm, em 605/70 nm; ex 560/30 nm, em 609/54 nm; ex 580/25 nm, em 625/30 nm; ex 640/40 nm, em 690/50 nm. For low magnification-whole-body imaging of the Thy1-GFPM mouse, we used a 1x Olympus air objective (Olympus MV PLAPO 1x/0.25 NA [WD = 65mm]) coupled to an Olympus MVX10 zoom body, which provided zoom-out and -in ranging from 0.63x up to 6.3x. Using 1x objective and 0.63x of zoom, we imaged a field of view of 2 × 2.5 cm, covering the entire width of the mouse body. Tile scans with 60% overlap along the longitudinal y-axis of the mouse body were obtained from ventral and dorsal surfaces up to 13 mm in depth, covering the entire volume of the body using a z-step of 8 µm. Exposure time was 120 ms, laser power was adjusted depending on the intensity of the fluorescent signal (in order to never reach the saturation) and the light-sheet width was kept at maximum. After tile imaging of the sample within the entire field of view, already scanned regions were cut using a thin motorized dental blade (0.2 mm) (Dremel 8200) for further imaging. After low magnification imaging of the whole body, a forelimb of the Thy1-GFPM animal was imaged with a 2x objective (Olympus MVPLAPO2XC/0.5 NA [WD = 6 mm]) coupled with the same Olympus MVX10 zoom body at zoom magnification 1.6x. Moreover, the same 2x objective with different zooms was used to perform high magnification imaging of specific body regions (e.g back of the animal at the level of lumbar vertebra, inguinal lymph node area,or the head). Individual organs (including brain, lungs, intestine and thymus) were imaged individually using high magnification objectives: 2x objective (Olympus MVPLAPO2XC/0.5 NA [WD = 6 mm]) coupled with the same Olympus MVX10 zoom body, 4x objective (Olympus XLFLUOR 4x corrected/0.28 NA [WD = 10 mm]), 25x objective (Olympus XLPLN 25x/0.95 NA [WD 4mm]) and 20x objective (Zeiss 20x Clr Plan-Neofluar/0.1 NA [WD 5.6 = mm]) coupled to an Olympus revolving zoom body unit (U-TVCAC) kept at 1x. High magnification tile scans were acquired using 8–30% overlap and the light-sheet width was reduced to obtain maximum illumination in the field. For the specific imaging settings used in each Figure panel, see Supplementary Table 2.
Fluorescence stereomicroscopy imaging
For images in Supplementary Figure 3, brain slices were kept immersed in PBS. For nanobody validation, cleared slices were put on a glass lid and kept immersed in BABB. The imaging was performed with Zeiss AxioZoom EMS3/SyCoP3 fluorescence stereomicroscope using Zen 2 software (Version 2.0.0.0, Carl Zeiss AG) using a 1x long working distance air objective lens (Plan Z 1×, 0.25 NA, WD = 56 mm; Supplementary Table 2). For the comparison of YFP-H mouse tissues after high pressure pumping versus low pressure pumping in Supplementary Figure 9, the mouse brains were cut into 1 mm slices using a vibratome, imaged with magnification 7x for overview and with magnification 112x in GFP channel for their detailed cell structures. See Supplementary Table 2.
Laser-scanning confocal microscopy imaging
After imaging with light-sheet microscopy, areas of interest from the cleared specimens such as whole heads and body parts were imaged with an inverted laser-scanning confocal microscope (Zeiss, LSM 880) using Zen 2 software (Version 10.0.4.910, Carl Zeiss AG). Before imaging, samples were mounted by placing them onto the glass surface of a 35 mm glass-bottom petri dishes (MatTek, P35G-0–14-C) and adding a few drops of BABB to make sure that the imaging region was immersed in BABB. Closing or sealing of the petri dish was not necessary because BABB has a high evaporation point and doesn´t dry out for several days. The only precaution taken was to check the resistance of the dish against BABB, by testing that the glue sealing the glass to the dish and the plastic parts were not dissolved after several hours of exposure to BABB.
The imaging was done using a 40x oil-immersion objective lens (Zeiss, ECPlan-NeoFluar 40x/1.30 Oil DIC M27, 1.3 NA, WD = 0.21 mm) and a 25x water-immersion long working distance objective lens (Leica, 0.95 NA, WD = 2.5 mm) mounted on a custom mounting thread. See Supplementary Table 2.
Reconstructions of whole-mouse body scans
We acquired light-sheet microscope stacks using ImSpector (Version 5.295, LaVision BioTec GmbH) as 16-bit grayscale TIFF images for each channel separately. In each pair of neighbouring stacks, alignment was done by manually selecting 3 to 4 anatomic landmarks from the overlapping regions, then the stitching was done sequentially with the Scope Fusion module of the Vision4D (Version 2.12.6 ×64, Arivis AG) software. Landmarks were mainly chosen from the skeletal bones or fewer from the neuronal structures based on visual inspection of the anatomical features. After completing the 3D reconstructions, the data visualization was done with Amira (Version 6.3.0, FEI Visualization Sciences Group), Imaris (Version 9.1, Bitplane AG) and Vision4D in both volumetric and maximum intensity projection color mapping. Depth coding was done using Temporal-Color Code plugin in Fiji (ImageJ2, Version 1.51, https://fiji.sc/).
Image processing
Processing, data analysis, 3D rendering and video generation for the rest of the data were done on an HP workstation Z840, with 8 core Xeon processor, 196 GB RAM, and Nvidia Quadro k5000 graphics card and HP workstation Z840 dual Xeon 256 GB DDR4 RAM, nVidia Quadro M5000 8GB graphic card. We used Imaris, Amira, and Fiji (ImageJ2) for 3D and 2D image visualization. Tile scans were stitched by Fiji’s stitching plugin49. Stitched images were saved in tiff format and optionally compressed in LZW format to enable fast processing. We removed tiles with acquisition errors using Fiji’s TrakEM2 plugin and Imglib2library. In case of tiling errors in z dimension, we used TeraStitcher (Version, 1.10, https://abria.github.io/TeraStitcher/) with its default settings to globally optimize the tiled volumes and reconstruct the entire dataset. To increase the quality of the images we used the following functions in Fiji: to enhance the contrast of microglia cells (Fig. 1n and Supplementary Fig. 2a-f) we used “Enhance Local Contrast (CLAHE)”, to equalize the images (Fig. 4a,b and Supplementary Fig. 2a-f) we used “Pseudo Flat-Field Correction”, to enhance the contrast over the background of the axonal terminals in Fig 4c,d (yellow and green boxes) we used the custom-made macro for Fiji that we utilized to generate the pre-processed data for NeuroGPS-Tree (see next paragraph: Neuron tracing)
Neuron tracing
For automated neuron tracing in our light-sheet datasets obtained with Zeiss 20x Clr Plan-Neofluar/0.1 NA [WD = 5.6 mm] objective, we used the NeuroGPS-Tree algorithm22. NeuroGPS-Tree was developed for tracing relatively small volumes of confocal microscopy data, therefore, we initially reduced the file size to under 1 GB (approximate maximum data size for NeuroGPS-Tree computation) by using Fiji scale function. Due to high signal intensity discrepancy between soma and neurites, we next pre-processed the data with a custom-made macro in Fiji (available upon request), which consisted of background removal, pseudo background correction, noise filtering and sharpening. Next, the pre-processed data was loaded and analysed first in NeuroGPS for soma detection and later in NeuroGPS-Tree for neurite detection (both steps are part of the same algorithm package). The best parameters of soma and neurite detection were chosen following the original publication. To quantify the features (such as number of neurites per cell and the number of somas with at least one neurite) of these detected neuronal cells, we used Amira software: we chose the 10 neurons with the biggest file size per each group and we analyzed them using the Spatial Graph Statistics function of the software.
Quantifications
All the details of imaging settings used to acquire the data for the quantifications described in this section are available in Supplementary Table 2
Analysis of fluorescence signal profiles from light-sheet images:
The fluorescence signal profiles from each channel (excitation 470 nm, 560 nm and 647 nm) were plotted in the same z-stack and normalized as percentage over the maximum peak using Fiji. (Supplementary Fig. 1).
To compare vDISCO boosted vs. unboosted protocols and consequently the reduction of the background and the improvement of the signal over background ratio in far-red channel, we analyzed neurons and axonal bundles expressing GFP imaged with excitation at 470 nm, and neurons and axonal bundles labelled with anti-GFP nanobody conjugated with Atto647N imaged with excitation at 640 nm at the same anatomic region. First, the scans were taken with the light-sheet microscope using the 4x objective (Olympus XLFLUOR 4x corrected/0.28 NA [WD = 10 mm]). Then, the signal profiles were analysed in Fiji and measured from a defined straight line covering the neuronal structure and surrounding tissue background, and the normalized plots of the signal profile (Fig. 1i) were calculated by normalizing the plots of neuronal structures obtained as described above over the average signal intensity of the respective surrounding background. Each experimental group consisted of 3 animals and per each animal at the same anatomic region we plotted 3 profiles.
The same analysis of signal profile was used to compare the labeling efficiency of vDISCO with nanobody vs. iDISCO+ with antibody (Supplementary Figure 3g-i): in this case the signal of interest was the one coming from microglia. Each experimental group consisted of 3 animals and per each animal at the same anatomic region we plotted 3 profiles from 3 different microglia. The same analysis of signal profile was used to compare different clearing methods in Supplementary Figure 12.
Fluorescence level:
Fluorescence level quantification was expressed as a signal-to-background ratio and was calculated using Fiji17 at the following time points: 0, 2, 3, 4 and half, 5 and half, 12 and 18 months after nanoboosting (Fig. 1n,o). Each 4x light-sheet microscopy brain scan was taken with the same imaging parameters, and an image in Tiff format of the same anatomic region for all the samples was quantified. The mean value of the background for each image was obtained by averaging the background values of 12–40 regions from equally sized areas of the image in regions of the sample without signal. To calculate the mean value of the signal per each image, we used the threshold function of the software: the threshold was adjusted to consider the fluorescence signal visible in the image. After adjusting the threshold, only the sharp signal from specific cellular structures was analyzed per image. To this end, we used Fiji’s “analyse particles” function to measure the signal intensity only of particles sized between 5–10 and 100–150 pixels (visible fluorescent cells), and calculated the average value from all the particles. Next, this value was divided by the mean value of the background of the respective image, obtaining the fluorescence level over the background. The corresponding images, visually showing the preservation of signal over time in relation to the respective fluorescence levels, were processed using Enhance Local Contrast (CLAHE) function in Fiji to increase the contrast of fluorescent cells in the tissue.
Quantification of deep tissue staining efficiency of nanobody vs. antibody
To quantify the depth of staining and therefore to compare whole organ staining efficiency of nanobody vs. antibody labeling (Supplementary Fig.2), we used CX3CR1GFP/+ animals (3–7 months old), 3 whole brains per each experimental group. The tile scans of the brains were taken with light-sheet microscopy in axial view using the 4x objective (Olympus XLFLUOR 4x corrected/0.28 NA [WD = 10 mm]). After stitching we chose the z-plane showing the same anatomic structures from all brains for comparison. These axial view 2D images of whole brains were analyzed in Fiji: the distance of staining depth was quantified using the “line” function in the z-plane at the greatest width of the brain, measuring from the surface until the last visible boosted microglia. Then, per each image, this staining depth value was normalized over the total thickness of the brain at the considered z-level: measuring a line drawn from the same point of the surface until the axial middle line of the brains.
Quantification of whole-body labeling efficiency of different whole-body labeling protocols
To quantify the rate of staining of the different whole-body labeling protocols in Supplementary Figure 11, the tile scans of 3 whole heads per method were taken with light-sheet microscopy in axial view using the 2x objective described in “light-sheet microscopy imaging section” (Olympus MVPLAPO2XC/0.5 NA objective coupled with the Olympus MVX10 zoom body) with a total magnification of 1.26x. For the analysis, we used Fiji to choose 3 regions in z in the 2D visualization, in order to have comparative data in 3 different depths (see Supplementary Fig. 2a,b). For each region, we chose the z-plane displaying the same anatomic structures in each sample. Next, the 2D images were opened in Fiji and, after adjusting the threshold to the fluorescence signal visible in the image, the stained area in the brain per each image was quantified using “freehand selections” function. Next, this stained area value was normalized over the total area of the brain visible in that 2D image. This total area was quantified using the same “freehand selections” function in Fiji, following the brain outline. Statistical comparison was performed considering each set of corresponding regions from the different samples separately.
ClearMap
To quantify microglia distribution we used ClearMap23. Since the script was originally developed for quantification of the cFos+ cells, to comply with the offered method, we did the following pre-processing steps on our microglia data using Fiji before ClearMap:
Background equalization to homogenize intensity distribution and appearance of the microglia cells over different regions of the brain, using pseudo flat-field correction function from Bio-Voxxel toolbox.
Convoluted background removal, to remove all particles bigger than relevant cells. This was done with the median option in Bio-Voxxel toolbox.
2D median filter to remove remaining noise after background removal. The filter radius was chosen to ensure the removal of all particles smaller than microglia cells.
Unshapen mask to amplify the high-frequency components of a signal and increase overall accuracy of the cell detection algorithm of ClearMap.
After pre-processing, ClearMap was applied by following the original publication and considering the threshold levels that we obtained from the pre-processing steps. As soon as the quantification was completed, the data was exported as excel file for further analysis. For example, the cellular density per each brain region was obtained considering the absolute number of cells detected by ClearMap and the volume of that specific brain region, which was calculated using a custom script (available upon request) based on ClearMap (Elastix registration).
Quantification of peripheral neuronal degeneration in acute brain injury
Peripheral neuronal degeneration in TBI animals versus unlesioned control animals was done in the Thy1-GFPM line and was assessed considering the complexity of axonal ramifications that projected from the left (left= contralateral side) cervical and thoracic vertebra to the left muscles of the back at the level of the torso of the mouse, including the left spinotrapezius and latissimus dorsi. The complexity was expressed as number of axonal endpoints (nerve terminals that appear as button-like shape (See Fig. 4) over the total length of axonal ramifications that were protruding from a main branch. To calculate this index, first a 3×3 tile z-stacks of this anatomic region was taken from the animals by light-sheet microscopy using the 2x objective described in “light-sheet microscopy imaging section” (Olympus MVPLAPO2XC/0.5 NA objective coupled with the Olympus MVX10 zoom body) with a total magnification of 5x, in order to have enough resolution to manually trace the axonal ramifications and axonal end-feet. Then, the analysis was done over the max intensity projections of the tile scans with Fiji software. To measure the length of the ramifications, we used the “free hand line” function and the “ROI manager tool” of Fiji, in order to record all the traced axonal ramifications of interest, which were coming from a main branch; later we calculated the sum of the length of all of the recorded ramifications with “measure” function of Fiji. To count the nerve terminals, we used the “point tool” function and the same “ROI manager tool” in order to record all the visible axonal endpoints protruding from the traced ramifications. The analysis was performed over 2–4 branches from the same anatomic region per each animal, in 6 animals (3–6 months old) per each experimental group. The same quantification procedure was followed to analyse the complexity at the equivalent ipsilateral (right side from dorsal view) side of the animals, in 6 and 5 animals for the TBI and unlesioned group, respectively.
Quantification of CD68 GFP+ cells with spinal cord injury
The spine areas of interest from CD68-EGFP line mice with SCI, sham surgery and from naïve animals were imaged with light-sheet microscopy using the 2x objective described in “light-sheet microscopy imaging section” (Olympus MVPLAPO2XC/0.5 NA objective coupled with the Olympus MVX10 zoom body) with a total magnification of 3.2x and with 2×3 tile z-stacks. After stitching, the entire z-stacks of scans were loaded in Fiji software with the ‘use virtual stack’ mode. In order to segment the spinal cord tissue from surrounding tissue including bone and muscles and count the cell numbers separately, the border of the spinal cord was marked by using the “free hand line” function in Fiji as a manual selection. These manual selections were done in every 6–9 image slices and were recorded by the “ROI manager tool” function. The complete selections for the entire z-stacks were then achieved by using “Interpolate ROIs” function and were saved as “.zip” files. Then, the entire z-stacks of scans were loaded in Fiji software again, without selecting the ‘use virtual stack’ mode. By loading the complete selections generated from former steps respectively, each spinal cord was segmented by using a custom-made macro with ‘clear outside’ function and saved as a new “Image Sequence”. In order to keep the integrity of the raw data, it is worth noting that one should not save the changes when closing the window after running the macro. Then, the intact original z-stacks and complete selections were loaded again in the same way and surrounding tissue including bone and muscles were segmented by using another custom-made macro with ‘clear’ function. All the custom-made macros are available upon request. Next, the segmented image stacks of spinal cord or surrounding tissue were loaded with Imaris respectively and the ‘Surface’ function was used to segment the CD68 GFP+ cells. By generating the ‘Surface’ selection, the ‘Statistic’ function was available to count the number of cells in the region of interest.
Quantification of short skull marrow – meninges vascular connections (SMCs) with MCAO
The transparent skulls from LysM-EGFP line mice with MCAO or sham were imaged with intact heads from the sagittal view with light-sheet microscopy using the 2x objective described in “light-sheet microscopy imaging section” (Olympus MVPLAPO2XC/0.5 NA objective coupled with the Olympus MVX10 zoom body) with a total magnification of 3.2x and with 2×4 tile z-stacks. After stitching, the entire z-stacks of scans were loaded in Imaris and the length, width and cell numbers in the SMCs were manually calculated by using the ‘Measure-line’ function. Then, the length and width of the channels were corrected by a shrinkage rate of DISCO clearing17.
Statistical analysis
Data collection and analysis were not performed blind to the conditions of the experiments. Data distribution was assumed to be normal but this was not formally tested. Data are presented as mean ± s.d except for Fig.6 and Supplementary Fig.22 where they are presented as mean ± S.E.M. Sample sizes are indicated in the figure legends. Statistical analysis was performed using Prism GraphPad software (Version 6, GraphPad) using 95% confidence interval. P values were calculated using two tailed unpaired t-test to compare data between two groups. P values were calculated using one-way ANOVA followed by Dunnett´s post-hoc test to compare data in Supplementary Figure 4 and 11. For CD68-EGFP quantification in Supplementary Figure 22, p values were calculated using one-way ANOVA followed by Tukey multiple comparison test to compare data between SCI, sham and naïve groups. A p value <0.05 was considered statistically significant. In ANOVA analysis the reported p values are adjusted in GraphPad prism to account for multiple comparisons. The protocols described in this study were replicated successfully more than 5 times in independent experiments and they were also reproduced at least by 3 different operators. See also the Life Sciences Reporting Summary.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Code availability
The custom codes used to analyze the data in this study are available from the corresponding author upon request.
Supplementary Material
Supplementary Video 1 vDISCO reveals individual microglia in CX3CR1GFP/+ mouse brain 3D brain reconstruction of a vDISCO boosted CX3CR1GFP/+ mouse brain, in which microglia express GFP, imaged with 4x objective by light-sheet microscopy. After vDISCO, all labelled microglia became evident, allowing quantification of their numbers in each brain region and assessment of the details of the microglia ramifications. Similar results were observed in 2 independent animals.
Supplementary Video 2 vDISCO reveals whole body neuronal projections in Thy1-GFPM mouse 3D reconstruction of neuronal projections in a Thy1-GFPM mouse scanned by light-sheet microscopy. The muscles (red) are visualized in autofluorescent channel (blue-green spectra). The bones and internal organs (white) are prominent with PI labeling. The GFP expressing neurons are boosted with nanobody conjugated with Atto 647N and imaged in far-red channel. The overall view of the entire labeled nervous system in the Thy1-GFPM mouse and fine details of neuronal connections are evident throughout the whole body. Comparable labeling and imaging results were achieved in 5 independent animals, whole body reconstruction was done in 2 mice.
Supplementary Video 3 Neuronal projections from spinal cord to right forelimb in Thy1-GFPM mouse 3D visualization obtained by light-sheet microscopy of neuronal projections from spinal cord to right forelimb of a Thy1-GFPM mouse. The muscles are shown in red, bones in white and the neurons in green. The fine details of axonal extensions and their endings at neuromuscular junctions are visible. Similar results were observed in 2 independent animals.
Supplementary Video 4 vDISCO imaging of neuronal projections in the spinal cord and muscles The first part of the video is the 2D orthoslicing of the spinal cord of an intact Thy1-GFPM mouse in dorso-ventral orientation. The muscles are shown in red, bones in white and the neurons in green. The details of neuronal cell bodies in ganglia embedded in the spinal cord vertebra and their axonal extensions into the CNS and PNS are visible. In the second part, neuronal connections (green) from spinal cord to muscles are shown in 3D and 2D. Similar results were observed in 2 independent animals.
Supplementary Video 5 vDISCO imaging of CX3CR1GFP/+ mouse with intact skin The first part of the video is the 3D reconstruction of the inguinal area from a CX3CR1GFP/+ mouse cleared with intact skin, showing inguinal lymph nodes and surrounding tissues (skin and muscles). CX3CR1 GFP+ immune cells are shown in cyan and cell nuclei labeled by PI are shown in magenta. The second part is the 2D orthoslicing visualization of a confocal scan of the same area, showing the subcellular details of CX3CR1 GFP+ cells in the lymph node and around the hair follicles. Similar results were observed in 2 independent animals.
Supplementary Video 6 vDISCO reveals lymphatic vessels of different organs in Prox1-EGFP mouse After applying vDISCO whole-body labeling of a Prox1-EGFP line mouse, the lungs and intestine were further imaged using high-magnification light-sheet microscopy. Prox1-EGFP lymphatic vessels (green) are evident throughout the tissues. Single experiment.
Supplementary Video 7 Microglia and peripheral immune cells in CX3CR1GFP/+ x CCR2RFP/+ mouse Multiplexed visualization of CX3CR1GFP/+ x CCR2RFP/+ transgenic mouse head after panoptic imaging with two different nanoboosters (anti-GFP conjugated to Atto647N and anti-RFP conjugated to Atto594N). The CX3CR1 GFP+ microglia cells in the brain parenchyma vs. CCR2 RFP+ peripheral immune cells in the meningeal vessels were clearly visible in 3D reconstruction and 2D orthoslicing. Similar results were observed from 3 independent double transgenic mice.
Supplementary Video 8 Revealing short skull meninges connections (SMCs) in intact CX3CR1GFP/+ mouse heads 2D orthoslicing of transparent head from the sagittal view of a CX3CR1GFP/+ line mouse imaged by light-sheet microscope. The vasculature was labelled by Lectin (red) and CX3CR1 GFP+ immune cells and microglia cells (green) were boosted by vDISCO. Short skull-meninges connections (SMCs) containing CX3CR1 GFP+ immune cells are evident. Similar results were observed in 3 independent animals.
Supplementary Video 9 Cellular details of SMCs in intact mouse heads 3D visualization of skull and brain interface in a CX3CR1GFP/+ line mouse imaged by confocal microscope. Vasculature was labelled with lectin (magenta) and CX3CR1 GFP+ cells (green) were boosted by vDISCO. Short vascular connections between skull marrow and meninges at the sagittal sinus and brain interfaces are clearly visualized. We also observed occasional connections between neighbouring skull marrow regions. Note that lectin dye is taken up by phagocytic cells in the skull marrow similar to dextran54. Similar results were observed in 3 independent animals.
Supplementary Video 10 LysM GFP+ immune cells observed in SMCs upon ischemic stroke lesion 2D orthoslicing of LysM-EGFP mouse head with MCAO. LysM GFP+ neutrophils and monocytes are shown in red and cell nuclei labelled by PI are shown in green. LysM GFP+ cells increased in the lesion site indicating the infiltration of immune cells after MCAO. In addition, many LysM GFP+ cells are observed in the vascular connections to the meninges suggesting that SMCs can be a potential cell trafficking route and play a role in the neuroinflammatory process. Similar results were observed from 4 independent mice per group.
Supplementary Video 11 Widespread inflammation upon SCI assessed by panoptic vDISCO imaging Panoptic imaging of CD68-EGFP transgenic mouse upon SCI. 2D orthoslicing in horizontal and sagittal views clearly shows the activated immune cells (green) in the muscles, spinal cord roots and meningeal compartments. Vertebra, which become prominent with PI labelling, are shown in white. Similar results were observed from 3 independent mice per group.
Supplementary Video 12 Extensive details of neuronal connections in the mouse brain revealed by vDISCO A Thy1-GFPM mouse brain was imaged by light-sheet microscopy more than a year after vDISCO. The details of the neuronal structures down to the subcellular level were imaged by Zeiss 20x objective on the light-sheet microscope. Similar results were observed in 2 independent animals.
ACKNOWLEDGMENTS:
This work was supported by the Vascular Dementia Research Foundation, Synergy Excellence Cluster Munich (SyNergy), ERA-Net Neuron (01EW1501A, A.E.), Fritz Thyssen Stiftung (A.E., Ref. 10.17.1.019MN), DFG (A.E., Ref. ER 810/2–1), (NIH A.E. and M.N.), Helmholtz ICEMED Alliance (A.E.), the Novo Nordisk Foundation (M.N.), the Howard Hughes (B.T.K.), and the Lundbeck Foundation (to A.L.R.X. and M.N.). We thank A. Weingart for illustrations, F. Hellal for technical advice and critical reading of the manuscript, and F. P. Quacquarelli, and G. Locatelli for help during initial optimization. A.E, C.P., R.C., A.L. and M.I.T. are members of the Graduate School of Systemic Neurosciences (GSN) at the Ludwig-Maximilian University of Munich.
Footnotes
COMPETING FINANCIAL INTERESTS
A.E. filed a patent on some of the technologies presented in this work.
Reference:
- 1.Tuchin VV & Tuchin V Tissue optics: light scattering methods and instruments for medical diagnosis. Vol. 642 (SPIE press; Bellingham, 2007). [Google Scholar]
- 2.Chung K et al. Structural and molecular interrogation of intact biological systems. Nature 497, 332–337, doi: 10.1038/nature12107 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang B et al. Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell 158, 945–958, doi: 10.1016/j.cell.2014.07.017 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Renier N et al. iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 159, 896–910, doi: 10.1016/j.cell.2014.10.010 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Susaki EA et al. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell 157, 726–739, doi: 10.1016/j.cell.2014.03.042 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Erturk A et al. Three-dimensional imaging of solvent-cleared organs using 3DISCO. Nat Protoc 7, 1983–1995, doi: 10.1038/nprot.2012.119 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Erturk A et al. Three-dimensional imaging of the unsectioned adult spinal cord to assess axon regeneration and glial responses after injury. Nat Med 18, 166–171, doi: 10.1038/nm.2600 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Belle M et al. A simple method for 3D analysis of immunolabeled axonal tracts in a transparent nervous system. Cell Rep 9, 1191–1201, doi: 10.1016/j.celrep.2014.10.037 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Costantini I et al. A versatile clearing agent for multi-modal brain imaging. Scientific reports 5, 9808, doi: 10.1038/srep09808 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ke MT, Fujimoto S & Imai T SeeDB: a simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat Neurosci 16, 1154–1161, doi: 10.1038/nn.3447 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Dodt HU et al. Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nat Methods 4, 331–336, doi:nmeth1036 [pii] 10.1038/nmeth1036 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Hama H et al. ScaleS: an optical clearing palette for biological imaging. Nat Neurosci 18, 1518–1529, doi: 10.1038/nn.4107 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Belle M et al. Tridimensional Visualization and Analysis of Early Human Development. Cell 169, 161–173 e112, doi: 10.1016/j.cell.2017.03.008 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Murray E et al. Simple, Scalable Proteomic Imaging for High-Dimensional Profiling of Intact Systems. Cell 163, 1500–1514, doi: 10.1016/j.cell.2015.11.025 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tainaka K et al. Whole-body imaging with single-cell resolution by tissue decolorization. Cell 159, 911–924, doi: 10.1016/j.cell.2014.10.034 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Jing D et al. Tissue clearing of both hard and soft tissue organs with the PEGASOS method. Cell Res, doi: 10.1038/s41422-018-0049-z (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan C et al. Shrinkage-mediated imaging of entire organs and organisms using uDISCO. Nat Methods, doi: 10.1038/nmeth.3964 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Kubota SI et al. Whole-Body Profiling of Cancer Metastasis with Single-Cell Resolution. Cell Rep 20, 236–250, doi: 10.1016/j.celrep.2017.06.010 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Tuchin VV Editor’s Introduction: Optical Methods for Biomedical Diagnosis. (2016). [Google Scholar]
- 20.Muyldermans S Nanobodies: natural single-domain antibodies. Annu Rev Biochem 82, 775–797, doi: 10.1146/annurev-biochem-063011-092449 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Feng G et al. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron 28, 41–51, doi:S0896–6273(00)00084–2 [pii] (2000). [DOI] [PubMed] [Google Scholar]
- 22.Quan T et al. NeuroGPS-Tree: automatic reconstruction of large-scale neuronal populations with dense neurites. Nat Methods 13, 51–54, doi: 10.1038/nmeth.3662 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Renier N et al. Mapping of Brain Activity by Automated Volume Analysis of Immediate Early Genes. Cell 165, 1789–1802, doi: 10.1016/j.cell.2016.05.007 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gage GJ, Kipke DR & Shain W Whole animal perfusion fixation for rodents. J Vis Exp, doi: 10.3791/3564 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greenbaum A et al. Bone CLARITY: Clearing, imaging, and computational analysis of osteoprogenitors within intact bone marrow. Sci Transl Med 9, doi: 10.1126/scitranslmed.aah6518 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Leijnse JN & D’Herde K Revisiting the segmental organization of the human spinal cord. J Anat 229, 384–393, doi: 10.1111/joa.12493 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gimenez-Arnau A Standards for the Protection of Skin Barrier Function. Curr Probl Dermatol 49, 123–134, doi: 10.1159/000441588 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Haeryfar SM & Hoskin DW Thy-1: more than a mouse pan-T cell marker. J Immunol 173, 3581–3588 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Smith DH, Johnson VE & Stewart W Chronic neuropathologies of single and repetitive TBI: substrates of dementia? Nature reviews. Neurology 9, 211–221, doi: 10.1038/nrneurol.2013.29 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frei K Posttraumatic dystonia. J Neurol Sci 379, 183–191, doi: 10.1016/j.jns.2017.05.040 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Williams G, Schache A & Morris ME Running abnormalities after traumatic brain injury. Brain Inj 27, 434–443, doi: 10.3109/02699052.2012.750754 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Chen Y, Constantini S, Trembovler V, Weinstock M & Shohami E An experimental model of closed head injury in mice: pathophysiology, histopathology, and cognitive deficits. J Neurotrauma 13, 557–568, doi: 10.1089/neu.1996.13.557 (1996). [DOI] [PubMed] [Google Scholar]
- 33.Erturk A et al. Interfering with the Chronic Immune Response Rescues Chronic Degeneration After Traumatic Brain Injury. J Neurosci 36, 9962–9975, doi: 10.1523/JNEUROSCI.1898-15.2016 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Evans TM et al. The effect of mild traumatic brain injury on peripheral nervous system pathology in wild-type mice and the G93A mutant mouse model of motor neuron disease. Neuroscience 298, 410–423, doi: 10.1016/j.neuroscience.2015.04.041 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Louveau A et al. Structural and functional features of central nervous system lymphatic vessels. Nature 523, 337–341, doi: 10.1038/nature14432 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nedergaard M Neuroscience. Garbage truck of the brain. Science 340, 1529–1530, doi: 10.1126/science.1240514 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Da Mesquita S et al. Functional aspects of meningeal lymphatics in ageing and Alzheimer’s disease. Nature, doi: 10.1038/s41586-018-0368-8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andres KH, von During M, Muszynski K & Schmidt RF Nerve fibres and their terminals of the dura mater encephali of the rat. Anat Embryol (Berl) 175, 289–301 (1987). [DOI] [PubMed] [Google Scholar]
- 39.Louveau A et al. Understanding the functions and relationships of the glymphatic system and meningeal lymphatics. J Clin Invest 127, 3210–3219, doi: 10.1172/JCI90603 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choi I et al. Visualization of lymphatic vessels by Prox1-promoter directed GFP reporter in a bacterial artificial chromosome-based transgenic mouse. Blood 117, 362–365, doi: 10.1182/blood-2010-07-298562 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Faust N, Varas F, Kelly LM, Heck S & Graf T Insertion of enhanced green fluorescent protein into the lysozyme gene creates mice with green fluorescent granulocytes and macrophages. Blood 96, 719–726 (2000). [PubMed] [Google Scholar]
- 42.Llovera G et al. The choroid plexus is a key cerebral invasion route for T cells after stroke. Acta Neuropathol 134, 851–868, doi: 10.1007/s00401-017-1758-y (2017). [DOI] [PubMed] [Google Scholar]
- 43.Iqbal AJ et al. Human CD68 promoter GFP transgenic mice allow analysis of monocyte to macrophage differentiation in vivo. Blood 124, e33–44, doi: 10.1182/blood-2014-04-568691 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hong G, Antaris AL & Dai H Near-infrared fluorophores for biomedical imaging. Nature Biomedical Engineering 1, 0010 (2017). [Google Scholar]
- 45.Noristani HN et al. RNA-Seq Analysis of Microglia Reveals Time-Dependent Activation of Specific Genetic Programs following Spinal Cord Injury. Front Mol Neurosci 10, 90, doi: 10.3389/fnmol.2017.00090 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Villapol S, Byrnes KR & Symes AJ Temporal dynamics of cerebral blood flow, cortical damage, apoptosis, astrocyte-vasculature interaction and astrogliosis in the pericontusional region after traumatic brain injury. Front Neurol 5, 82, doi: 10.3389/fneur.2014.00082 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leslie M Small but mighty. Science 360, 594–597, doi: 10.1126/science.360.6389.594 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Deverman BE et al. Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat Biotechnol 34, 204–209, doi: 10.1038/nbt.3440 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hellal F et al. Microtubule stabilization reduces scarring and causes axon regeneration after spinal cord injury. Science 331, 928–931, doi: 10.1126/science.1201148 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Herisson F et al. Direct vascular channels connect skull bone marrow and the brain surface enabling myeloid cell migration. Nat Neurosci 21, 1209–1217, doi: 10.1038/s41593-018-0213-2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Niess JH et al. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science 307, 254–258, doi: 10.1126/science.1102901 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Nikic I et al. A reversible form of axon damage in experimental autoimmune encephalomyelitis and multiple sclerosis. Nat Med 17, 495–499, doi: 10.1038/nm.2324 (2011). [DOI] [PubMed] [Google Scholar]
- 53.Treweek JB et al. Whole-body tissue stabilization and selective extractions via tissue-hydrogel hybrids for high-resolution intact circuit mapping and phenotyping. Nat Protoc 10, 1860–1896, doi: 10.1038/nprot.2015.122 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bixel MG et al. Flow Dynamics and HSPC Homing in Bone Marrow Microvessels. Cell Rep 18, 1804–1816, doi: 10.1016/j.celrep.2017.01.042 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Video 1 vDISCO reveals individual microglia in CX3CR1GFP/+ mouse brain 3D brain reconstruction of a vDISCO boosted CX3CR1GFP/+ mouse brain, in which microglia express GFP, imaged with 4x objective by light-sheet microscopy. After vDISCO, all labelled microglia became evident, allowing quantification of their numbers in each brain region and assessment of the details of the microglia ramifications. Similar results were observed in 2 independent animals.
Supplementary Video 2 vDISCO reveals whole body neuronal projections in Thy1-GFPM mouse 3D reconstruction of neuronal projections in a Thy1-GFPM mouse scanned by light-sheet microscopy. The muscles (red) are visualized in autofluorescent channel (blue-green spectra). The bones and internal organs (white) are prominent with PI labeling. The GFP expressing neurons are boosted with nanobody conjugated with Atto 647N and imaged in far-red channel. The overall view of the entire labeled nervous system in the Thy1-GFPM mouse and fine details of neuronal connections are evident throughout the whole body. Comparable labeling and imaging results were achieved in 5 independent animals, whole body reconstruction was done in 2 mice.
Supplementary Video 3 Neuronal projections from spinal cord to right forelimb in Thy1-GFPM mouse 3D visualization obtained by light-sheet microscopy of neuronal projections from spinal cord to right forelimb of a Thy1-GFPM mouse. The muscles are shown in red, bones in white and the neurons in green. The fine details of axonal extensions and their endings at neuromuscular junctions are visible. Similar results were observed in 2 independent animals.
Supplementary Video 4 vDISCO imaging of neuronal projections in the spinal cord and muscles The first part of the video is the 2D orthoslicing of the spinal cord of an intact Thy1-GFPM mouse in dorso-ventral orientation. The muscles are shown in red, bones in white and the neurons in green. The details of neuronal cell bodies in ganglia embedded in the spinal cord vertebra and their axonal extensions into the CNS and PNS are visible. In the second part, neuronal connections (green) from spinal cord to muscles are shown in 3D and 2D. Similar results were observed in 2 independent animals.
Supplementary Video 5 vDISCO imaging of CX3CR1GFP/+ mouse with intact skin The first part of the video is the 3D reconstruction of the inguinal area from a CX3CR1GFP/+ mouse cleared with intact skin, showing inguinal lymph nodes and surrounding tissues (skin and muscles). CX3CR1 GFP+ immune cells are shown in cyan and cell nuclei labeled by PI are shown in magenta. The second part is the 2D orthoslicing visualization of a confocal scan of the same area, showing the subcellular details of CX3CR1 GFP+ cells in the lymph node and around the hair follicles. Similar results were observed in 2 independent animals.
Supplementary Video 6 vDISCO reveals lymphatic vessels of different organs in Prox1-EGFP mouse After applying vDISCO whole-body labeling of a Prox1-EGFP line mouse, the lungs and intestine were further imaged using high-magnification light-sheet microscopy. Prox1-EGFP lymphatic vessels (green) are evident throughout the tissues. Single experiment.
Supplementary Video 7 Microglia and peripheral immune cells in CX3CR1GFP/+ x CCR2RFP/+ mouse Multiplexed visualization of CX3CR1GFP/+ x CCR2RFP/+ transgenic mouse head after panoptic imaging with two different nanoboosters (anti-GFP conjugated to Atto647N and anti-RFP conjugated to Atto594N). The CX3CR1 GFP+ microglia cells in the brain parenchyma vs. CCR2 RFP+ peripheral immune cells in the meningeal vessels were clearly visible in 3D reconstruction and 2D orthoslicing. Similar results were observed from 3 independent double transgenic mice.
Supplementary Video 8 Revealing short skull meninges connections (SMCs) in intact CX3CR1GFP/+ mouse heads 2D orthoslicing of transparent head from the sagittal view of a CX3CR1GFP/+ line mouse imaged by light-sheet microscope. The vasculature was labelled by Lectin (red) and CX3CR1 GFP+ immune cells and microglia cells (green) were boosted by vDISCO. Short skull-meninges connections (SMCs) containing CX3CR1 GFP+ immune cells are evident. Similar results were observed in 3 independent animals.
Supplementary Video 9 Cellular details of SMCs in intact mouse heads 3D visualization of skull and brain interface in a CX3CR1GFP/+ line mouse imaged by confocal microscope. Vasculature was labelled with lectin (magenta) and CX3CR1 GFP+ cells (green) were boosted by vDISCO. Short vascular connections between skull marrow and meninges at the sagittal sinus and brain interfaces are clearly visualized. We also observed occasional connections between neighbouring skull marrow regions. Note that lectin dye is taken up by phagocytic cells in the skull marrow similar to dextran54. Similar results were observed in 3 independent animals.
Supplementary Video 10 LysM GFP+ immune cells observed in SMCs upon ischemic stroke lesion 2D orthoslicing of LysM-EGFP mouse head with MCAO. LysM GFP+ neutrophils and monocytes are shown in red and cell nuclei labelled by PI are shown in green. LysM GFP+ cells increased in the lesion site indicating the infiltration of immune cells after MCAO. In addition, many LysM GFP+ cells are observed in the vascular connections to the meninges suggesting that SMCs can be a potential cell trafficking route and play a role in the neuroinflammatory process. Similar results were observed from 4 independent mice per group.
Supplementary Video 11 Widespread inflammation upon SCI assessed by panoptic vDISCO imaging Panoptic imaging of CD68-EGFP transgenic mouse upon SCI. 2D orthoslicing in horizontal and sagittal views clearly shows the activated immune cells (green) in the muscles, spinal cord roots and meningeal compartments. Vertebra, which become prominent with PI labelling, are shown in white. Similar results were observed from 3 independent mice per group.
Supplementary Video 12 Extensive details of neuronal connections in the mouse brain revealed by vDISCO A Thy1-GFPM mouse brain was imaged by light-sheet microscopy more than a year after vDISCO. The details of the neuronal structures down to the subcellular level were imaged by Zeiss 20x objective on the light-sheet microscope. Similar results were observed in 2 independent animals.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.