Abstract
Multipass membrane proteins perform critical signal transduction and transport across membranes. How transmembrane helix (TMH) sequences encode the topology and conformational flexibility regulating these functions remains poorly understood. Here we describe a comprehensive analysis of the sequence-structure relationships at multiple interacting TMHs from all membrane proteins with structures in the Protein Data Bank (PDB). We found that membrane proteins can be deconstructed in interacting TMH trimer units, which mostly fold into six distinct structural classes of topologies and conformations. Each class is enriched in recurrent sequence motifs from functionally unrelated proteins, revealing unforeseen consensus and evolutionary conserved networks of stabilizing interhelical contacts. Interacting TMHs’ topology and local protein conformational flexibility were remarkably well predicted in a blinded fashion from the identified binding-hotspot motifs. Our results reveal universal sequence-structure principles governing the complex anatomy and plasticity of multipass membrane proteins that may guide de novo structure prediction, design, and studies of folding and dynamics.
Membrane proteins represent ~30% of all genome-encoded proteins and perform critical cellular functions, from signal transduction to the transport of diverse molecules across lipid membranes. This large spectrum of highly specialized functions is reflected by the diversity of topology adopted by membrane proteins despite the physical constraints imposed by the membrane environment1. Additionally, most membrane proteins perform their functions through conformational changes ranging from small local TMH movements to large inter-domain movements involving the formation of distinct sets of intraprotein and interprotein contacts2,3. By perturbing these interactions, missense substitutions can critically impact membrane protein functions and cause serious diseases4,5. A large fraction of disease-associated mutations also impair proper folding of large multipass membrane proteins, which often involve complex cooperative interhelical interactions and assembly during translocation and membrane insertion6. Therefore, understanding how transmembrane protein amino acid sequences encode protein topology, conformational stability and plasticity has critical biomedical relevance.
The availability of many experimental structures for water-soluble proteins has enabled studies of sequence-structure relationships that uncovered important determinants underlying the structure, stability, folding and design of those proteins. For example, fundamental principles governing secondary-structure formation and stability7,8, loop-turn conformations9, protein folds and plasticity10,11 have been established through the discovery of recurrent amino acid sequence-structure relationships. More recently, such rules have been expanded to the de novo design of water-soluble protein folds with precise topologies, a step toward rational engineering of novel synthetic enzymes or protein-based nanomaterials12,13.
Until recently, similar investigations on membrane proteins have been hampered by the scarcity of membrane protein structures and the strong bias toward hydrophobic amino acids in TMHs. Structural characterization of membrane proteins remains very challenging owing to the high intrinsic flexibility and instability of these proteins in nonnative traditional crystallization conditions14.
Additionally, requirements for high overall hydrophobicity to promote effective translocation of TMHs in lipid membranes leads to strong enrichment of hydrophobic amino acids6,15. TMHs lack the periodicity in structure distribution of polar/non polar amino acids that have facilitated the discovery of critical sequence-structure relationships in water-soluble proteins16. As a consequence, such studies in membrane proteins have been limited to the analysis of the minimal structural TMH dimer unit16–21. Compared to water-soluble proteins, TMH interacting pairs are enriched in small residues that promote packing through side chain−mediated and backbone-mediated hydrogen bonds, especially at right-handed dimer interfaces16. However, a large fraction of TMHs in multipass membrane proteins are tightly packed and form interacting surfaces with multiple TMHs that cannot easily be deconstructed in TMH dimers (Supplementary Results, Supplementary Fig. 1). Hence, interaction networks stabilizing multipass membrane proteins likely involve cooperatively multiple TMHs and are likely more complex than those identified by analyzing TMH dimers only. Sequence-structure relationships governing the folding, topology and stability of large membrane proteins with complex topologies therefore remain largely unknown.
Owing to recent advances in genome sequencing, analysis of sequence covariation has become more reliable for protein families with large numbers of homologous sequences, which allows for the identification of strongly coevolving residue networks. These residues often form contacts critical for protein structure, stability and function, and can in principle guide our understanding of sequence-structure relationships22,23. Nevertheless, sufficient numbers of homologous sequences are available only for a fraction of the membrane proteome24. Random or saturation mutagenesis coupled with directed-evolution techniques can also be used to explore the sequence space compatible with specific protein structures and functions. In principle, these methods can be used to identify particular constraints and emerging rules in sequence- structure relationships. Although these approaches have been quite successful for water-soluble proteins, their application to membrane proteins has been largely restricted to multipass membrane protein model systems that can fold properly in bacterial outer membranes or to simple self-associating TMH domains25–27. Despite their promise, these studies have yet to uncover sequence-structure principles governing multipass membrane protein topology and stability.
Here we developed a multifaceted approach to investigate the sequence-structure relationships encoding the anatomy and plasticity of multiple contacting TMHs from all membrane proteins with structures in the PDB. We took advantage of the recent increased number of high-resolution X-ray crystallography structures to uncover sequence-structure rules of large TMH assemblies. Specifically, we show that multipass membrane proteins can be deconstructed into interacting TMH trimer packing units that mostly fold into six structural classes with well-defined topologies and conformations. Each structural class is strongly enriched in sequence motifs forming consensus networks of three-dimensional (3D) interhelical contacts across functionally unrelated protein families. We constructed a predictor of trimer TMH topology from sequence solely trained on the class-specific sequence motifs, which validate the universality of the uncovered sequence-structure relationships. We also found that the motifs are enriched in strongly covarying residues and stabilizing physical interactions, consistent with their location in the least dynamic regions of membrane proteins. These unforeseen consensus hotspot-binding motifs define a new universal sequence−3D structure alphabet encoding the topology, packing and local conformational stability across the entire multipass transmembrane helical proteome. Our findings set the stage for a better under-standing of the determinants of membrane protein folding, and should guide the de novo structure prediction and design of membrane proteins with complex topologies and conformational dynamics from sequence.
RESULTS
Strategy overview
To identify sequence-structure rules governing the topology, conformations and stability of interacting TMHs in multipass membrane proteins, we first analyzed how TMHs interact in these proteins, and found that they are often tightly packed and share the same interacting surfaces with two other helices (Supplementary Fig. 1). Hence we reasoned that cooperative interactions at these interfaces might not be correctly recapitulated by analyzing simple TMH dimers. Therefore, we analyzed interhelical interactions at the interfaces of higher-order TMH trimer units extracted from multipass membrane proteins. If universal sequence-structure relationships govern the topology and packing of TMH trimers, these should be readily identified by gathering trimer data from evolutionary unrelated proteins into classes of similar structures. Within each class, sequence motifs enriched compared to a random distribution of amino acids and forming consensus network of interhelical interactions should define important sequence−3D structure determinants of TMH trimer topology and conformation (Supplementary Fig. 2a). If these emerging rules define a universal sequence-structure alphabet, they should guide the prediction of membrane protein topology and conformational stability (Supplementary Fig. 2b). We describe first a method to identify sequence-structure relationships at interacting TMH trimers, and then validation of those rules in topology and conformational flexibility predictions.
TMH trimers pack with a small number of topologies
To investigate how helices interact in TMH trimer units embedded in large multipass membrane proteins, we constructed a library of 1,027 helical trimer units extracted from 203 nonredundant transmembrane domain structures. For each trimer we identified regions displaying the highest density of interhelical contacts between the three helices and clustered them based on structural similarity. Despite the large number of combinations of possible helical orientations, we found that a majority of TMH trimer units (56%) could be clustered into six well-defined structure classes with distinct overall topology and geometry (Fig. 1 and Supplementary Fig. 3). This small number of classes was representative of the membrane protein structures in our library and comprises structures from 61 different protein families (i.e., 90% of all families in the library) and 41 superfamilies (i.e., 87% of all superfamilies in the library). In helical dimers, helices usually interact nearly parallel or antiparallel to each other to maximize interhelical interactions. For each of these orientations, helix-helix packing is typically observed for conformations characterized by small crossing angles (i.e., angles between helical axes) ranging from −45° (right-handed conformation) to +45° (left-handed conformation)21.
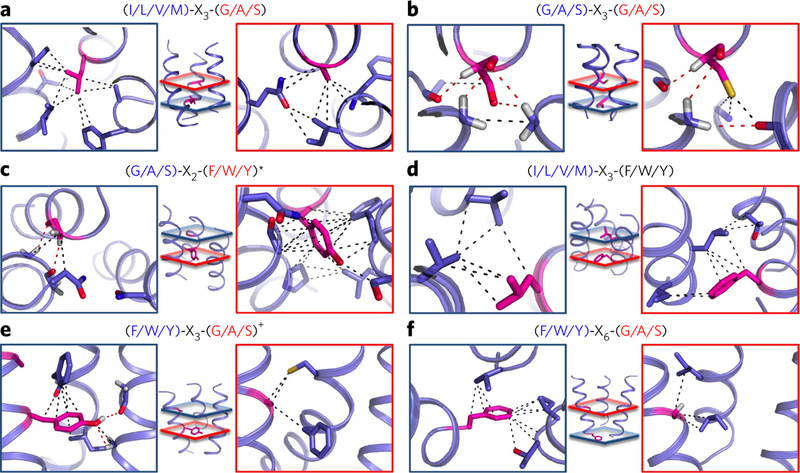
Figure 1. TMH trimers cluster in six major structure classes with enriched sequence motifs.
(a−f) Descriptions of six largest classes of TMH trimer structure with specific sequence motifs enriched at the interhelical interface. Sequence motifs are constituted by pairs of amino acids belonging to three chemical classes (G/A/S, small: glycine, alanine or serine; I/L/V/M, large: isoleucine, leucine, valine or methionine; F/W/Y or aromatic: phenylalanine, tryptophan or tyrosine). Residues in the motif are aligned along an ideal helix following their sequence separation (register). The helix number describes on which helix or helices of a reference trimer are found the motifs. For example, if a motif is found on helices 1 and 2 in a given trimer, the corresponding helix numbers are given as: ½. The topology and helix number of the reference trimer for each class are described in Supplementary Table 1. Each helix is colored using a blue to red spectrum from N to Cterminus. The reported motifs were found in at least 20% of trimer interacting regions in a given class and are ordered from left to right according to their frequency of occurrence in that class.
We separated each TMH trimer structural class (first through sixth, most to least populated) by defining the geometry of the three helical pairs forming the trimer. Consistent with antiparallel packing being more favored than parallel packing in helical protein folds21,28, two pairs of helices were antiparallel and only one pair was parallel in the trimers (Fig. 1 and Supplementary Table 1). The first and second classes comprised all left-handed and all right-handed helix pairs, respectively (Fig. 1a,b and Supplementary Fig. 3). The third class (‘left and right-handed’) and fourth class (‘left and right-handed II’) were more similar to each other with two helices packing to the same helix in left-handed and right-handed conformation, respectively (Fig. 1c,d). The fifth class (‘parallel and left-handed’) and sixth class (‘parallel and right-handed’) comprised TMH trimers with one topologically parallel helical pair characterized by small interhelical angle (13.9° and −18.5°, respectively; Fig. 1e,f and Supplementary Table 1) and two left-handed (fifth class) or right-handed (sixth class) helical pairs. The topologically parallel helical pairs displayed larger interhelical distances and were not as closely packed as the other pairs in these trimers (11.3 Å compared to 9.3 Å on average), suggesting that multiple interacting TMH assemblies might stably accommodate relatively loosely packed helical pairs.
Enrichment in trimer structure classes
If protein sequences determine specific structure motifs, they should be enriched within the amino acid sequence profile of functionally unrelated protein families sharing the same local structures. In each class of trimers, we analyzed the sequence profiles at multiple positions of the trimer interhelical interface to identify combinations of enriched residues along that interface. Specifically, we built one sequence library for each helix in the trimers and identified frequently occurring sequence motifs using a variant of the statistical method TMSTAT20 adapted for TMH trimers. Owing to the limited number of trimer structures in each cluster, we grouped the amino acids with similar chemical properties into a simplified alphabet (for example, we grouped G, A and S in the ‘small’ amino acid category) and limited our search to pairs of residues. We found 25 significantly overrepresented pairs of amino acids, compared to their expected random distribution (P values in Supplementary Table 2). Seventeen of these motifs were still found to be significantly enriched in a highly homology-reduced data set composed of only one protein structure per protein superfamily (Supplementary Table 2). We observed 13 ‘major’ motifs in at least three protein super- families, in three protein families and in more than 20% of the trimer interacting units in each class (Fig. 1 and Supplementary Fig. 4). 11 of these 13 enriched sequence motifs had not been reported in previous TMH interaction studies and were unique to TMH trimer interfaces, except for the (G/A/S)-X3-(G/A/S) and (G/A/S)-X6-(G/A/S) motifs. The (G/A/S)-X3-(G/A/S) repeat is common to both ‘all right-handed’ trimers and right-handed dimers17,20,29 and the (G/A/S)-X6-(G/A/S) motif found at ‘all left-handed’ trimers is reminiscent of heptad repeats identified in TMH dimers and water-soluble coiled coils with left-handed conformations21. Sequence patterns combining small and large residues were the most enriched motifs in four of the six largest trimer classes (for example, (I/L/V/M)-X3-(G/A/S) and (G/A/S)-X2-(F/W/Y) motifs in the ‘all left-handed’ and the ‘parallel and left-handed’ classes, respectively). The absence of enrichment of these motifs at TMH dimers likely reflects the need for multiple helices to accommodate large amino acids at TMH interacting interfaces. Eleven among 13 sequence patterns were unique to a specific trimer topology and conformation, suggesting a potential role as strong determinants of trimer structures. To understand the role of these sequence motifs in specific trimer assemblies, we studied the network of contacts that 2 they establish across TMHs.
A consensus networks of contacts
We reasoned that if recurrent sequence motifs determine trimer topology and conformations, they may create similar interhelical interaction patterns across protein families. To address this, we analyzed the 13 highly enriched sequence motifs that we identified. We clustered the networks of interhelical contacts emerging from each residue in the motifs and identified common patterns of residue-residue interactions for all motifs (Fig. 2 and Supplementary Table 3). We highlight four examples of consensus contacts (found in more than 50% of the trimers bearing the same motif) involving two-residue sequence motifs found in structurally similar trimers from three unrelated protein superfamilies in Figure 2. Both residues often point to the center of the helical trimer and form similar interaction networks connecting the three helices together across protein families. Although the (G/A/S)-X3-(G/A/S) motif is common to TMH dimers and trimers, it forms a unique trimer-specific interaction network dominated by backbone−backbone or backbone−small side chain contacts connecting the three helices simultaneously in the ‘all right-handed’ class (Fig. 2a). In the ‘all left-handed’ family, the G, A or S residue in the (I/L/V/M)-X3-(G/A/S) motif forms to our knowledge a previously unidentified set of interactions involving a side chain of larger residues; these interactions differ from those in other trimers or dimer assemblies, highlighting the cooperativity between the pair of residues in the motifs (Fig. 2b). In the ‘parallel and left’ trimer family, the G, A or S residue in the (G/A/S)-X2-(F/W/Y) motif forms close contacts with only one helix, and the large residue at the second position docks at the center of the trimer interface connecting the three helices using an extensive network of consensus contacts (Fig. 2c). In the ‘parallel and right’ trimer family, each residue of the (I/L/V/M)-X3-(F/W/Y) motif contacts the side chains of relatively large residues from two cognate helices simultaneously. As a result, the side chains do not always point toward the center of the trimer interface and display greater variability in orientation than other sequence-structure motifs (Fig. 2d). Our results indicate that the motifs create trimer-specific networks of consensus contacts recurrent across protein families that are directly involved in determining packing of the three helices.
Figure 2. Recurrent sequence motifs create consensus interhelical trimer interactions across protein families.
(a−d) Consensus interaction networks created by each of the two sequence motif residues enriched at specific TMH trimer structure class labeled in the figure. A dashed red line indicates a consensus interhelical interaction between a motif residue (magenta) and a residue on an adjacent helix (cyan). Interaction networks are highlighted for a given sequence motif in three functionally unrelated membrane proteins.
Consensus contacts form unique atomic interaction motifs
Consistent with most sequence motifs interacting with residues from multiple helices, the combinations of atomic interactions involved in the consensus contact maps are often specific to trimers. A notable example concerns the (G/A/S)-X3-(G/A/S) motif over-represented in ‘all right-handed’ trimers and dimers. The combination of two small residues in trimers promotes very tight packing of the three helices, allowing backbone alpha hydrogen and carbonyl oxygens in their direct vicinity to form weak hydrogen bonds with two helices simultaneously (Fig. 3a). These bifurcated weak hydrogen bonds are unique to ‘all right-handed’ trimers and represent to our knowledge a previously unidentified atomic interaction motif that can connect and bridge multiple helices. In the most enriched (I/L/V/M)-X3-(G/A/S) motif in the ‘all left-handed’ trimers, the long hydrophobic side chain on the first position is fully buried at the center of the trimer and forms an extensive network of van der Waals interactions with often more than four residues from the other two helices (Supplementary Table 3). The small G, A or S side chain in the motif slightly deviates from the trimer interface center and forms exclusively van der Waals interactions with the side chains of one residue from each cognate helix (Fig. 3b). In the (G/A/S)-X2-(F/W/Y) motif selectively observed in ‘parallel and left-handed’ helical trimers, the small (G/A/S) amino acid in the first position forms a key docking site for another helix, allowing the two helices to tightly pack in a left-handed conformation by forming numerous close backbone-backbone (including weak hydrogen bonds between alpha hydrogens and carbonyl oxygens) and backbone−small side chain contacts. The second motif residue packs its large side chain at the interhelical interface of the two neighboring helices through extensive side chain−backbone contacts. This large side chain anchors the third helix to the left-handed helix dimer through parallel interhelical interfaces dominated by contacts between aromatic residues (Fig. 3c). In contrast, in the (I/L/V/M)-X3-(F/W/Y) motif unique to the ‘parallel and right-handed’ family, the first residue anchors the three helices together through extensive van der Waals interactions, and the second aromatic residue packs on the edge of the parallel interhelical dimer interface of the trimer (Fig. 3d). In both (F/W/Y)-X3-(G/A/S) and (F/W/Y)-X6-(G/A/S) motifs from the ‘left- and right-handed’ I and II classes, respectively, the large aromatic residue is fully buried at the center of the trimer interface and connects the three helices through extensive van der Waals interactions, whereas the small G, A or S residue contacts only one helix (Fig. 3e,f). Overall, the large diversity of unique trimer-specific networks of atomic interactions involving all classes of amino acids strongly support the TMH trimer as a critical elementary structural and folding unit in multipass membrane proteins.
Figure 3. Consensus patterns of contacts display unique combinations of atomic interactions.
(a−f) Enriched sequence motifs create unique trimer-specific interatomic van der Waals (black dotted line) and hydrogen bonding (red dotted line) interactions in local regions of the trimers defined by red and blue planes at the center of each panel. Interatomic interactions are defined for any pair of atoms distant by <5 Å belonging to one of the two motif residues (magenta) and a residue on an adjacent helix. The red and blue boxes highlight the atomic contacts from the corresponding colored planes created by each residue of the following motifs and trimer structure classes: (G/A/S)-X3-(G/A/S) motif specific to all right-handed trimers (a); (I/L/V/M)-X3-(G/A/S) motif specific to all left-handed trimers (b); (G/A/S)-X2-(F/W/Y/(M)) motif specific to parallel and left-handed trimers (c); (I/L/V/M)-X3-(F/W/Y) motif specific to parallel and right-handed trimers (d); (F/W/Y)-X3-(G/A/S) motif specific to left- and right-handed trimers (e); (F/W/Y)-X6-(G/A/S) motif specific to left- and right-handed IItrimers (f). *, methionine also found in the second position with similar contacts; +, histidine also found in the first position with similar contacts.
Sequence motifs largely determine TMH trimer topology
If the sequence motifs enriched in specific classes of trimer structures are important determinants of their topology, we should be able to predict the geometry of a trimer using information from sequence only. We trained a support vector classification method30,31 to predict TMH trimer topology from the enriched sequence motifs identified in the six largest classes of trimer structures. To eliminate any possible bias toward particular protein families, we performed training and testing using structures and enriched motifs extracted from a highly homology-reduced data set consisting of only one protein per superfamily (Supplementary Table 2). We tested the predictor in a fivefold cross-validation approach that classified trimer structures from sequence, with greater accuracy compared to a random assignment (i.e., without predictor). The predictor achieved 41.2% accuracy in correctly assigning trimer sequences to the six largest classes of structures, compared to only 16.7% accuracy in a random assignment. Up to 83.5% classification accuracy was achieved in assigning trimer sequences between two structural classes, with an average of 73% accuracy over all 15 possible class pairs, compared to only 50% accuracy in a random assignment (Supplementary Table 4). The results obtained using this simple predictor indicate that the sequence motifs are important determinants of TMH trimer topology and may guide the de novo structure prediction of multipass membrane proteins from sequence.
Sequence motifs are enriched in coevolving residues
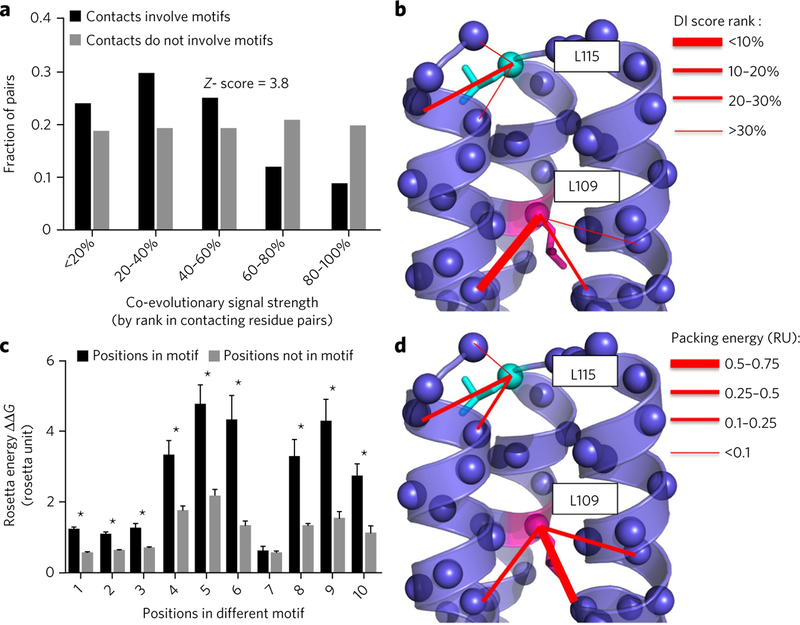
If the sequence motifs are strong structure determinants, they may promote critical stabilizing interactions and be under high evolutionary pressure of selection. To address this question, we first systematically compared the strength of evolutionary coupling between pairs of residues in or outside the contact network created by the motifs. We analyzed all trimers from 15 different major protein families, with enough homologous sequences to reliably calculate evolutionary coupling score using the method EV-fold23. The results indicate that contacting residue pairs involving motif residues are substantially more enriched in highly coevolving residues pairs than nonmotif residue pairs (Fig. 4a,b; Z score = 3.8).
Figure 4. Sequence motifs create evolutionary conserved networks of interhelical stabilizing contacts.
(a) Distribution of residue pairs as a function of their coevolutionary strengths measured by the method EVfold23. The coevolutionary strength of a pair is reported relative to the distribution of coevolutionary scores for all residue pairs as a rank (the lower the rank, the higher the relative coevolutionary strength). Selected residue pairs are in contact across the trimer interhelical interface and involve motif residues (black) or do not involve motif residues (gray). The Z score describes the statistical significance of the difference in rank distribution between the two classes of residue pairs. (b) Example of coevolutionary scores (direct interaction (DI) score) for interresidue contacts involving either a residue in a motif (magenta) or the same residue type not in a motif (cyan) at a trimer interface. (c) Comparison between the energy contribution to the trimer stability of a residue in a motif (black) and that of the same residue type not in a motif (gray). The energy contribution was calculated by alanine scanning, and the comparison was performed for ten different motif residues and reported as mean values + s.d. *P < 0.01, comparison between motif and nonmotif residues’ energetic contributions (Welch’s two-sided t-test). (d) Example of interaction energy between contacts involving a motif residue (magenta) and contacts not involving motif residues (cyan) at a trimer interface.
Sequence motifs are enriched in stabilizing residues
We then calculated the energetic contribution to the trimer stability of each residue in the motif (‘motif residue’) using in silico alanine scanning32,33. We compared these energies to those obtained for the same residue type at the trimer interface but not part of the motif (‘nonmotif residue’). A majority of motif residues had significantly larger contribution to the trimer stability than non-motif residues (Fig. 4c,d). Our findings indicate that the interactions promoted by the motifs contribute substantially to the stability of the trimers and are more likely to be maintained in evolution, implying their critical roles in maintaining the structural integrity of the TM helical trimer.
Motifs are found in the least flexible protein regions
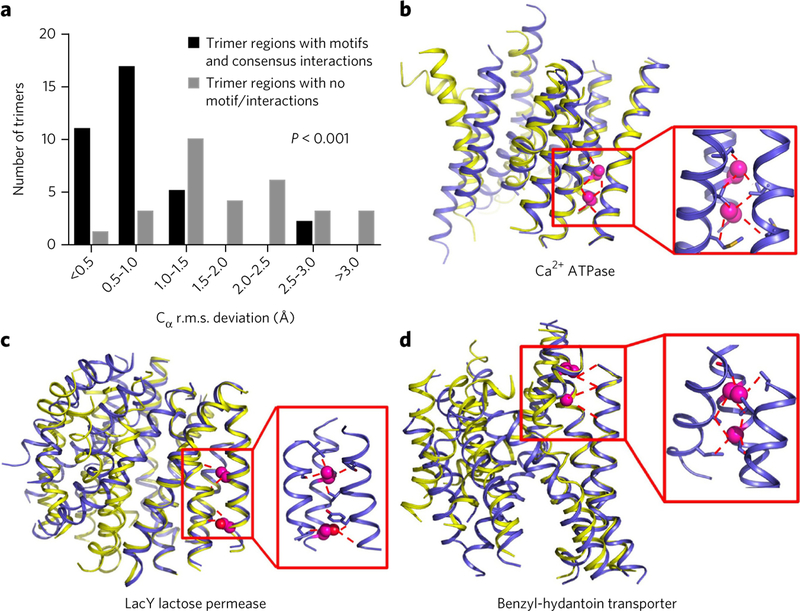
If the sequence motifs and associated interaction networks are important stability determinants of TMH trimers, they should be enriched in regions displaying low conformational flexibility in multipass membrane proteins. We identified 17 large membrane proteins from the protein structure database crystallized in multiple conformations differing by Cα r.m.s. deviation > 0.5 Å and comprising a total of 65 TMH trimer units (Supplementary Table 5). We compared the Cα r.m.s. deviation between two protein conformations of trimers bearing motifs/interaction with that of trimers not bearing any motifs. We found that the trimers with sequence motifs and corresponding interaction patterns had substantially smaller Cα r.m.s. deviation (P < 0.001, Welch’s t-test) between distinct protein conformations and were therefore significantly more rigid than the trimers without such sequence−3D contacts (Fig. 5). We obtained similar results when we selected only one protein per superfamily (Supplementary Table 6 and Supplementary Fig. 5). Our findings further demonstrate the critical role of the enriched sequence and interaction motifs in controlling conformational stability and/or flexibility in multipass membrane proteins.
Figure 5. Sequence−3D contact motifs are strong predictors of local conformational stability.
(a) Distribution of trimer unit structural changes (measured by Cα r.m.s. deviation in Å) in multipass membrane proteins crystallized in distinct conformations. The statistical significance in the distribution of Cα r.m.s. deviation between trimers bearing or not sequence−3Dcontact motifs was calculated using Welch’s two-sided t-test. (b−d) Examples of multipass membrane protein X-ray crystallography structures crystallized in two distinct conformations (superimposed backbone representations in blue and yellow). The trimer units containing a sequence-contact motif (red window) do not change conformations.
DISCUSSION
We found that large multipass membrane proteins can be deconstructed in elementary interacting TMH trimer units that mostly belong to only six classes of topology and conformations (Fig. 1). Each structure class is characterized by a few enriched sequence motifs specifying the packing of TMHs through consensus networks of interhelical contacts recurrent among many functionally unrelated proteins (i.e., from distinct protein superfamilies; Figs. 1–3). As such, these sequence−3D contact motifs largely contribute to the overall TMH trimer stability and remain also under evolutionary pressure of selection in protein families (Fig. 4). Information encoded in this sequence-structure alphabet was sufficient to predict in a blinded fashion interacting TMH trimer topology from sequence (Supplementary Table 4) and local conformational flexibility in large multipass membrane proteins (Fig. 5). Our results demonstrate the existence of strong and selective sequence-structure relationships that govern the anatomy and plasticity of multiple interacting TMHs units in multipass membrane proteins, which is to our knowledge unprecedented.
So far, sequence-structure relationship studies in membrane proteins have been limited to the interactions between pairs of helices16,21. From those studies, the recurrent sequence contact (G/A/S)-X3-(G/A/S) motif was identified at right-handed dimer interfaces and thoroughly characterized17,20. Our analysis of large multipass membrane proteins indicated that most TMHs are tightly packed and share a binding interface with often two helices at a time, making TMH trimers the relevant elementary TMH packing unit in such proteins (Supplementary Fig. 1). We identified 13 frequent sequence−3D contact motifs specifying the structure of these trimer units. Additionally, the majority of the enriched residue motifs at trimer interhelical binding surfaces formed cooperative interaction networks bridging the three helices, but they were not identified in TMH dimers (Figs. 2 and 3, and Supplementary Fig. 3). Even the (G/A/S)-X3-(G/A/S) motif initially characterized at right-handed TMH dimer interfaces creates a unique trimer-specific atomic interaction network in the ‘all right-handed’ trimer family. Despite the strongly biased amino acid alphabet toward hydrophobic and small amino acids in TMH proteins, most of the enriched residue pair sequence motifs were unique to each trimer structure class. This observation may actually not be that surprising considering the large number of combinations available to three hydrophobic amino acid chemical classes (small, large and aromatic) at all possible positions available to residue pairs along the same helical surface. Our results suggest that many more motifs may be identified in the future if the size of the membrane protein structure database continues to steadily increase. We did not identify any polar or charged residues in highly enriched trimer-specific sequence motifs, suggesting that they may either be only accommodated at larger TMH assemblies or more frequently encode functional and/or metastable protein family−specific properties. Overall, our results suggest that hydrophobic and weak polar interactions define a sufficient structure and chemical space to encode the structural specificity and stability of the elementary trimer units composing multipass membrane proteins.
Our findings have important implications for the de novo structure prediction of large multipass membrane proteins, which is an important alternate approach to their difficult experimental structure determination. Although accurate residue contacts can now be extracted from amino acid sequence covariations and effectively guide protein structure prediction, this approach relies on large number of homologous sequences that are not consistently available across the membrane proteome24. Our newly identified universal sequence−3D consensus contact motifs would be straightforward to implement as structural constraints in folding simulations34. By specifying both topology and local contact networks bridging simultaneously three helices, these features provide a larger spectrum of structural constraints than residue pairwise contacts predicted from sequence.
With the protein design field moving toward engineering novel artificial nanomaterials and molecular devices12,35, extending the capabilities of de novo protein design approaches is becoming necessary. Because our sequence and 3D contact motifs are strong determinants of TMH trimer conformation but also stability, they would provide ideal hotspot anchoring motifs for TMH trimer units from which novel complex membrane protein folds could be constructed de novo.
Our results have also important implications for better understanding the processes of multipass membrane protein folding. Increasing evidence suggest that critical interhelical associations can take place during translocation to facilitate proper membrane insertion of helices with low overall hydrophobicity6. On the basis of our findings, we propose that the topology of the contact networks characterizing each sequence motif may encode the assembly of trimer TMHs during folding. Motifs constituted by cooperative interactions bridging three helices simultaneously could promote the spontaneous assembly of trimer units. Conversely, motifs combining dimer- and trimer-specific interactions (found in four of the six largest families of trimer units) may promote the sequential assembly of dimers following by the docking of the third helix. As such, our library of motifs provides unprecedented structural entry points and testable hypothesis to guide the ab initio understanding of membrane protein folding pathways.
Lastly, the strong anticorrelation between trimer conformational flexibility and motif-encoded interhelical contact networks indicates that these features are key signatures of local conformational stability. As such, they may guide the biasing of atomistic molecular dynamic simulations that are often difficult to perform on large membrane protein systems36 and the selection of sites to couple experimental probes for monitoring membrane protein dynamics. Ultimately, knowledge of flexible protein regions may also guide the rational design of drugs targeting specific functional states that are often characterized by unique transient conformations in membrane proteins.
In summary, we uncovered a universal sequence-structure alphabet encoding important topology determinants and conformational regulatory properties of multipass membrane protein functions that may guide future de novo membrane protein structure prediction, design and folding studies.
METHODS
Methods and any associated references are available in the online version of the paper.
ONLINE METHODS
Library of interacting TMH trimer structures.
Multipass membrane protein structures with resolution lower than 3.5 Å were extracted from the Protein Data Bank (PDB) and filtered by 60% sequence identity threshold to remove close homologs, which generated a data set of 203 protein structures. At that stage, more than one protein per superfamily was considered to ensure that the diversity in sequence motifs and interhelical contacts for a given trimer structure class could be identified. In Supplementary Figure 4 we highlight two examples of proteins from the same superfamily bearing two distinct enriched sequence motifs at the same trimer interface structure. For topology prediction from sequence, the data set was further homology-reduced to one protein per superfamily (see below and Supplementary Table 2).
Each protein was dissected into TMHs using either the MPtopo or PDBTM libraries37,38, which specify the topology and location of the TMHs. Interacting TMH trimers were identified as any set of three helices where each pair of helices interacts through more than five contacts, which were defined by residue pairs separated by <9 Å between their Cα atoms. Such coarse-grained contact definition is justified at this stage because it was solely used to identify interacting transmembrane helices in protein structures. Similar metrics have been used in previous studies to extract interacting TMHs from protein structures21,29. More sophisticated contact map definition and all-atom contacts were used later in the study to compare trimer structures and identify all-atom contact maps, respectively (see below). A total of 896 trimer structures were defined from the selected 203 membrane protein structures.
Library of interacting TMH trimer local regions to identify sequence/structure motifs.
Local interhelical interacting regions containing the highest density of contacts were then identified in each TMH trimer structure by scanning a 10-amino-acid window along the trimer interface. Previous studies indicated that such window length is enough to cover relevant interaction regions in TMH packing21. We developed the following method to identify the interacting regions. First, for each helix in the trimer, we constructed two one-dimensional distance vectors, each of which stands for the distance relationship to one of the other two helices in the trimer. The length of the distance vectors is equal to the number of residues in the helix of interest (target). Each element in the vector equates the sum of weighted distances between a given position on the target helix and all the residues in the other helix. For a trimer composed by helix T (target), helix A and helix B, the score for one position in the target helix T to helix A is given by equation (1):
| (1) |
where dn stands for the Cα distance between target position to nth position in helix A.
The weight function is defined by equation (2):
| (2) |
Then, a trimer specific score is calculated for each position in helix T using equation (3):
| (3) |
The 10-amino-acid window in the target helix with the highest sum of position specific scores was selected. This procedure was then repeated for the two other helices leading to two additional windows that share the most contacts with the target helix selected window. By penalizing positions weakly interacting with one of the two adjacent helices, equation (3) ensures that the selected window comprises residues that form large number of contacts with the two other helices simultaneously. Also, the weight function for distance is a fall-off function, which gives weights close to 1 for distances <8 Å and weights close to 0 for distances >14 Å. This weight factor reflects the fact that residues in close contacts are highly important to determine the structure of interacting regions, whereas residues that are far away are relatively unimportant. By using this method, we selected the most relevant interhelical interacting regions of each trimer structure. In principle, multiple interacting regions can be defined for a given trimer, but because we restricted window overlap to a maximum of three positions, typically up to three interacting regions could be identified per trimer structure. A total of 1,027 trimer interacting regions were identified before clustering.
TMH trimer structure clustering method.
We used Cα r.m.s. deviation clustering algorithms to group the helical trimer regions in structurally similar families. First, helical trimers were aligned along their interacting regions by overlaying their respective contact map defined by the distance profiles between each pair of helix as described above. Second, the alignment was used to calculate the r.m.s. deviation of each pair of helical trimers and generate an all-against-all r.m.s. deviation matrix. Third, we used the average linkage clustering algorithm to separate the library in different clusters of structurally similar trimer regions21. The results were also confirmed by the k-means clustering method to ensure that no bias was introduced by the choice of clustering algorithm.
Specifically, we simplified the problem of aligning trimers by finding the best alignment for each of the three helical dimers that compose the trimer and adopted the alignment strategy of helical dimers guided by contact map described previously21. Briefly, all possible dimer alignments were tried to generate trimer region alignments. The trimer alignments giving the lowest Cα r.m.s. deviation were kept as final alignment from which the all-against-all r.m.s. deviation matrix was constructed. Average linkage clustering method was used to cluster the helical trimers based on their Cα r.m.s. deviation, and the results were confirmed by the k-means clustering method. Trimers were clustered such that each member of a given cluster would be within 2.0 Å r.m.s. deviation of a central reference trimer (the centroid). 2.0 Å was selected because smaller r.m.s. deviation cutoffs would lead to trimers bearing the same topology, conformation and sequence/structure features being split into multiple distinct smaller clusters. We characterized each cluster by calculating the common topological features (interhelical angles and inter-helical distance, etc.) of the corresponding trimer structures.
Identification of enriched sequence motifs at TMH trimer interfaces.
We adapted the approach TMSTAT used previously to detect frequently occurring residue pair motifs (for example, (G/A/S)-X3-(G/A/S)) in transmembrane domain sequence library20. Instead of assuming that all sequences derive from a homogeneous population and calculating expectation based on overall amino acid composition in the entire TMH database, TMSTAT considers the composition of each individual sequence and calculates expectation from all theoretical permutations of that sequence.
The implementation of TMSTAT for TMH trimers starts with creating the sequence library for each trimer structure cluster. The sequence of each helix in the TMH trimer interaction window extended by one helical turn on each side (i.e., 18 positions total) was used to construct a library of sequences. The sequences of the three helices in the trimers were considered separately as they may have different roles in trimer-structure assembly. Owing to the limited number of sequences, we grouped the amino acids with similar structural and chemical features into a simplified alphabet composed of 5 classes (‘small’: G, A, S, C and T; ‘hydrophobic’: I, L, V and M; ‘aromatic’: F, W, Y and H; ‘polar’: N and Q; ‘charged’: D, E, R and K). Next, we calculated the expected distribution of a pair of residue classes in each sequence library. Consider a pair of residues class A and B with a position difference of k as our target motif. The chance of observing the residue pair NABk times in a single sequence is given by equation (4).
| (4) |
Equation (4) depends on four parameters: the length of the sequence l (18 in our case), the register k and how many A (NA) and B residues (NB) are in the sequence. The overall chance (i.e., probability distribution) of observing such residue pair in a sequence library of n sequences, PDB, can be calculated iteratively following equation (5).
| (5) |
NA,n and NB,n are the number of A and B residue type in sequence n.
Once all the sequence in each library has been analyzed, average expected values and s.d. were computed from probability distribution curve according to equations (6) and (7):
| (6) |
| (7) |
The two-tailed integral of PDB was used to calculate the significance (P) for the observed occurrences of a given residue pair. Enriched residue pairs with P < 0.05 and located at the trimer interface were selected as trimer-specific sequence motifs.
Identification of consensus atomic contact maps.
Inspired by previous work on identifying interaction patterns in proteins39,40, we developed a method to identify geometrically similar consensus interhelical contact maps created by common sequence motifs across different protein families and superfamilies. The first step was alignment of the trimer regions on the helix that carry the motif of interest. The second step was identification of the interhelical atomic contacts involving each motif residue merged into interaction vector features. The third step was clustering of the interaction vector features in all the trimers to find consensus contact patterns.
In step 1, trimers containing the same motif were all aligned to the local region of the helix that carries the sequence motif of interest. In step 2, vectors were constructed to recapitulate interhelical atomic contacts involving the motif of interest. At this stage, contacts were defined between two heavy atoms separated by a distance <5.0 Å to select physical atomic interaction networks. When a motif residue interacted with another residue on one of the adjacent helix, a Cartesian vector was constructed for each pair of interacting heavy atoms. To identify consensus interaction patterns for motif residue classes composed of distinct side-chain structures and lengths, the origin of the vector was defined at the center of mass of the residue side chain (i.e. centroid) instead of the exact heavy atom involved in the contact. However, the end point of the vector was defined as the midpoint of the line connecting the two contacting heavy atoms. This hybrid centroid-atom vector allows accommodating multiple chemical structures of the motif residue while keeping the exact atomic information of the contact at its destination on the adjacent helices. As the density of contacts is largely dependent on the number of heavy atoms on the motif residue, and many of these hybrid contact vectors colocalize in three-dimensional space, ensemble of vectors were simplified and clustered into geometrically similar vector features to allow for reliable comparison of contact maps in different trimers as described previously40. The hybrid centroid-atom vectors were clustered following a two-step average linkage hierarchical clustering method. Vectors were first pre-clustered based on the location of the vector end points to generate groups of vectors with similar spatial location; then, the vectors within each group were sub-clustered based on the direction of the exact heavy atom contacts. Optimal clustering thresholds were determined using the elbow method41, and were found to lie within 2−3 Å for the location distance range of the vector end points and 17.4°− 20.5° for the angle range of the vector directions. The results were also confirmed by the complete linkage clustering method to ensure that no bias was introduced by the choice of clustering algorithm. Lastly, the feature vectors across different trimers sharing the same motif were clustered in the same manner. The consensus contact map was defined by the ensemble of interaction vector features shared by more than half of the trimers with the same motif.
Support vector machine−based training of a TMH trimer topology predictor from sequence.
Two-class or multiclass support vector classification methods were implemented to train a predictor of TMH trimer topology/conformation from sequence motifs only for classifying trimer structure families. We used the support vector machine−based training algorithm called ‘libsvm’31 together with the Radial Basis kernel function (RBF).
To stringently remove any evolutionary bias in the prediction, distant homologs were removed from structure database, leaving one protein per superfamily. A data set was constructed in such way that no proteins in this set can find hits with E value < 1 to any other proteins in the same set. The homology reduction was achieved by performing a jackhmmer search (HMMER 3.1b2 (February 2015); http://hmmer.org/) against all protein sequences in the PDB library, and we further refined our data set using the SCOP and CATH superfamily databases to ensure that only one protein structure per superfamily was selected. From the data set of 203 membrane protein structures generated by homology reduction at 60% sequence identity, we selected 47 proteins belonging to 47 distinct superfamilies. A total of 267 trimer interacting regions were extracted from this stringent homology-reduced data set. Trimer regions were described as a 17-dimensional vector (17 refers to the total number of sequence motifs included in the training and found enriched in the homology-reduced protein structure data set; Supplementary Table 2) with each element in the vector having values from 0 to 3, which correspond to the number of helices that contain the same sequence motif. This value turned out to be an important discriminator between trimer classes with right-handed helical dimer interfaces. Indeed, the ‘all right-handed’ class contained often more than one (G/A/S)- X3-(G/A/S) motif per trimer, whereas the ‘parallel and right-handed’ class contained only one such motif. The training and validation of the predictor was performed using the fivefold cross-validation technique. Two metrics of accuracy were calculated: (i) the accuracy of assigning trimer sequences into one of six classes of trimer structures (the accuracy over six classes) calculated using the structure data set of the six major clusters; (ii) the accuracy of assigning a trimer into one of two selected clusters. In this reduced-discrimination test, the fivefold cross-validation was carried out using the structure data set of the two selected clusters.
Residue-level energetic contribution to the trimer stability.
A representative set of 393 trimers from the six largest classes of trimer structures was selected to calculate residue contributions to the trimer stability. To remove partial atomic overlaps and idealize non-optimal bond length and angles in PDB structures42, trimers extracted from X-ray structures were relaxed with tight constraints on all heavy atoms and side chain conformations using the Rosetta protocol (https://www.rosettacommons.org/docs/latest/rosetta_basics/preparation/preparing-structures). All relaxed structures remained within 0.2 Å Cα r.m.s. deviation to the original PDB structure and kept most, if not all, atomic interaction details. The structural quality of the relaxed models was confirmed by Molprobity42. The contribution of one position to the TMH association energy of the trimer was calculated in two steps. First, the association energy of the native trimer (ΔG) was calculated by subtracting the sum of each individual helix free energy from the trimer free energy. Second, to calculate the contribution of one residue to the trimer association, that residue was changed to alanine using the design mode of the software RosettaMembrane32,33. The free energy of trimer association (ΔGˊ) was calculated as described above for the alanine variant. The specific energy contribution of the side chain of the target residue was determined by calculating ΔΔG (ΔG − ΔGˊ). The statistical significance of the differences between motif and nonmotif residues’ energetic contributions was calculated using Welch’s two-sided t-test. The trimer class and motif positions used to calculate the energetic contribution were: all left-handed (I/L/V/M)-X3-(G/A/S) first position; all left-handed trimer (I/L/V/M)- X6-(I/L/V/M) both positions; all left-handed (I/L/V/M)-(X)6-(F/W/Y) first position; all left-handed (I/L/V/M)-(X)6-(F/W/Y) second position; parallel and left-handed trimer (G/A/S)-X3-(F/W/Y) second position; parallel and right-handed (F/W/Y)-(X)2-(F/W/Y) both positions; parallel and right-handed trimer (I/L/V/M)-(X)3-(F/W/Y) first position; parallel and right-handed trimer (I/L/V/M)-(X)3-(F/W/Y) second position; left- and right-handed type I trimer (F/W/Y)-(X)3-(G/A/S) first position; and left- and right-handed type II trimer (F/W/Y)-(X)6-(G/A/S) first position.
Calculation of coevolutionary strength for residue pairs.
Co-evolutionary signals for residue pairs were calculated from sequence covariation by the method EV-fold23. 15 multipass membrane protein families were selected that had enough homolog sequences to generate large multiple sequence alignments. The selected families were sensory rhodopsin, nitric oxide reductase, mitochondrial cytochrome c oxidase, ApcT amino acid transporter, cytochrome bc1, cation diffusion facilitator (CDF) family zinc transporter, metal-chelate-type ABC transporter, major facilitator superfamily (MFS) Glp transporters, ammonia channel, rhesus proteins, class A GPCR, bacterial multi-drug efflux transporter P type ATPase, xanthorhodopsin, lactose permease. Residue pairs making contacts at the trimer interhelical interface were selected and ranked by their coevolutionary direct interaction (DI) score (the higher the DI score, the stronger the pressure of selection for coevolution). The rank difference between pairs involving motif residues and those not involving motif residues were calculated using rank-based t-test.
Trimer motif plasticity in different conformations of multipass membrane proteins.
17 proteins composed of multiple interacting TMH trimer units and crystallized in distinct conformations differing by more than 0.5 Å Cα r.m.s. deviation in the transmembrane region were identified and selected from the PDB. The Cα r.m.s. deviation along the interacting interhelical region of each trimer was used to measure the local trimer-specific structural variations in different protein conformational states. The statistical significance in the distribution of Cα r.m.s. deviation between trimers bearing or not sequence−3D contact motifs was calculated using Welch’s two-sided t-test.
Supplementary Material
Acknowledgments
We thank the members of the Barth laboratory for insightful discussions during this study and critical comments on the manuscript. This work was supported by a grant from the US National Institute of Health (1R01GM097207–01A1) and by a supercomputer allocation from XSEDE (Extreme Science and Engineering Discovery Environment; MCB120101) to P.B.
Footnotes
Competing financial interests
The authors declare no competing financial interests.
Additional information
Any supplementary information, chemical compound information and source data are available in the online version of the paper. Reprints and permissions information is available online at http://www.nature.com/reprints/index.html. Correspondence and requests for materials should be addressed to P.B.
Code availability.
The scripts developed to analyze the structure database in this study will be made available and released through a Baylor College of Medicine server.
References
- 1.von Heijne G Membrane-protein topology. Nat. Rev. Mol. Cell Biol 7, 909–918 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Matthews EE, Zoonens M & Engelman DM Dynamic helix interactions in transmembrane signaling. Cell 127, 447–450 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Krishnamurthy H, Piscitelli CL & Gouaux E Unlocking the molecular secrets of sodium-coupled transporters. Nature 459, 347–355 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rakoczy EP, Kiel C, McKeone R, Stricher F & Serrano L Analysis of disease-linked rhodopsin mutations based on structure, function, and protein stability calculations. J. Mol. Biol 405, 584–606 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Partridge AW, Therien AG & Deber CM Missense mutations in transmembrane domains of proteins: phenotypic propensity of polar residues for human disease. Proteins 54, 648–656 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Cymer F, von Heijne G & White SH Mechanisms of integral membrane protein insertion and folding. J. Mol. Biol 427, 999–1022 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petukhov M, Muñoz V, Yumoto N, Yoshikawa S & Serrano L Position dependence of non-polar amino acid intrinsic helical propensities. J. Mol. Biol 278, 279–289 (1998). [DOI] [PubMed] [Google Scholar]
- 8.Minor DL Jr. & Kim PS Measurement of the beta-sheet-forming propensities of amino acids. Nature 367, 660–663 (1994). [DOI] [PubMed] [Google Scholar]
- 9.Bystroff C & Baker D Prediction of local structure in proteins using a library of sequence-structure motifs. J. Mol. Biol 281, 565–577 (1998). [DOI] [PubMed] [Google Scholar]
- 10.Wolf E, Kim PS & Berger B MultiCoil: a program for predicting two- and three-stranded coiled coils. Protein Sci 6, 1179–1189 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng F, Zhang J & Grigoryan G Tertiary structural propensities reveal fundamental sequence/structure relationships. Structure 23, 961–971 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Koga N et al. Principles for designing ideal protein structures. Nature 491, 222–227 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.King NP et al. Accurate design of co-assembling multi-component protein nanomaterials. Nature 510, 103–108 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bill RM et al. Overcoming barriers to membrane protein structure determination. Nat. Biotechnol 29, 335–340 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Liu Y, Engelman DM & Gerstein M Genomic analysis of membrane protein families: abundance and conserved motifs. Genome Biol 3, h0054 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang SQ et al. The membrane- and soluble-protein helix-helix interactome: similar geometry via different interactions. Structure 23, 527–541 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mueller BK, Subramaniam S & Senes A A frequent, GxxxG-mediated, transmembrane association motif is optimized for the formation of interhelical Cα-H hydrogen bonds. Proc. Natl. Acad. Sci. USA 111, E888–E895 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langosch D & Arkin IT Interaction and conformational dynamics of membrane-spanning protein helices. Protein Sci 18, 1343–1358 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneider D Rendezvous in a membrane: close packing, hydrogen bonding, and the formation of transmembrane helix oligomers. FEBS Lett 577, 5–8 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Senes A, Gerstein M & Engelman DM Statistical analysis of amino acid patterns in transmembrane helices: the GxxxG motif occurs frequently and in association with beta-branched residues at neighboring positions. J. Mol. Biol 296, 921–936 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Walters RF & DeGrado WF Helix-packing motifs in membrane proteins. Proc. Natl. Acad. Sci. USA 103, 13658–13663 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nugent T & Jones DT Accurate de novo structure prediction of large transmembrane protein domains using fragment-assembly and correlated mutation analysis. Proc. Natl. Acad. Sci. USA 109, E1540–E1547 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hopf TA et al. Three-dimensional structures of membrane proteins from genomic sequencing. Cell 149, 1607–1621 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marks DS, Hopf TA & Sander C Protein structure prediction from sequence variation. Nat. Biotechnol 30, 1072–1080 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sarkar CA et al. Directed evolution of a G protein-coupled receptor for expression, stability, and binding selectivity. Proc. Natl. Acad. Sci. USA 105, 14808–14813 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cammett TJ et al. Construction and genetic selection of small transmembrane proteins that activate the human erythropoietin receptor. Proc. Natl. Acad. Sci. USA 107, 3447–3452 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gurezka R & Langosch D In vitro selection of membrane-spanning leucine zipper protein-protein interaction motifs using POSSYCCAT. J. Biol. Chem 276, 45580–45587 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Bowie JU Helix packing in membrane proteins. J. Mol. Biol 272, 780–789 (1997). [DOI] [PubMed] [Google Scholar]
- 29.Wang Y & Barth P Evolutionary-guided de novo structure prediction of self-associated transmembrane helical proteins with near-atomic accuracy. Nat. Commun 6, 7196 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cortes C & Vapnik V Support-vector networks. Mach. Learn 20, 273–297 (1995). [Google Scholar]
- 31.Chang C-C & Lin C-J LIBSVM: a library for support vector machines. ACM Trans. Intell. Syst. Technol 2, 1–27 (2011). [Google Scholar]
- 32.Barth P, Schonbrun J & Baker D Toward high-resolution prediction and design of transmembrane helical protein structures. Proc. Natl. Acad. Sci. USA 104, 15682–15687 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen KY, Zhou F, Fryszczyn BG & Barth P Naturally evolved G protein-coupled receptors adopt metastable conformations. Proc. Natl. Acad. Sci. USA 109, 13284–13289 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barth P, Wallner B & Baker D Prediction of membrane protein structures with complex topologies using limited constraints. Proc. Natl. Acad. Sci. USA 106, 1409–1414 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joh NH et al. De novo design of a transmembrane Zn2-transporting four-helix bundle. Science 346, 1520–1524 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dror RO et al. Activation mechanism of the β2-adrenergic receptor. Proc. Natl. Acad. Sci. USA 108, 18684–18689 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jayasinghe S, Hristova K & White SH MPtopo: a database of membrane protein topology. Protein Sci 10, 455–458 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kozma D, Simon I & Tusnády GE PDBTM: Protein Data Bank of transmembrane proteins after 8 years. Nucleic Acids Res 41, D524–D529 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andreani J, Faure G & Guerois R Versatility and invariance in the evolution of homologous heteromeric interfaces. PLoS Comput. Biol 8, e1002677 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu H, Sommer I, Lengauer T & Domingues FS Alignment of non- covalent interactions at protein-protein interfaces. PLoS One 3, e1926 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tibshirani R, Walther G & Hastie T Estimating the number of data clusters via the Gap statistic. J. Roy. Stat. Soc. B 63, 411–423 (2001). [Google Scholar]
- 42.Chen VB et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.