Abstract
This particular study set out to demonstrate alterations on the microbial community of the oxic-settling-anaerobic/anoxic (OSA) process treating real domestic wastewater by changing interchange ratios (IRs). The sludge yield of systems operated at different IRs (1/13, 1/17 and 1/20) to assess sludge reduction was used to analyze microbial community composition variations. The highest IR (1/13) resulted in the highest sludge reduction (52.1%), while the OSA systems with IR of 1/17 and 1/20 reduced sludge production by 37.4% and 35.5%, respectively, in comparison to conventional systems. 16S rRNA gene amplicon sequencing analysis showed that the bacterial communities were composed of similar phylogenetic groups, Proteobacteria, Acidobacteria, and Bacteroidetes being dominant. The relative abundances differed due to the applied IRs. The highest abundance of Actinobacteria was determined at the highest IR (1/13) and increasing of the HRT to 1/20 caused a significant reduction in Actinobacteria species and the lowest abundance (6%) was determined in the OSA systems. The abundant of Thiothrix species that are boosted in the OSA trials may have a vital role in OSA systems, where its abundance was below the detection limits in the seed sludge sample. Therefore, they could be used as bioindicators in the OSA system.
Keywords: Environmental science, Engineering
1. Introduction
The current global sewage sludge production rate is about 45 million dry ton sludge per year, equivalent to about 2.0-billion population equivalent (PE) covered by full sanitation at secondary wastewater treatment facility if taking conversion factor of 70 g dry sludge produced/PE-day (Zhang et al., 2017). Although the sludge represents only 1%–2% of the treated wastewater volume, treatment and disposal of sewage sludge from wastewater treatment plants (WWTPs) account for about half, even up to 60%, of the total cost of wastewater treatment (Wei et al., 2003). Given the rising costs of and constraints associated with sludge treatment and disposal, the ideal approach to addressing problems associated with disposing or reusing excess sludge is to minimize sludge production in the first place. Additionally, there is a growing concern for the integration of sustainability issues in the new generations of wastewater treatment plants (Alyaseri and Zhou, 2017). To this end, various engineering strategies have been developed to reduce the yield coefficient and consequently sludge production in wastewater treatment plants, incorporating various microbial processes, such as lysis, cryptic growth, uncoupling metabolism, maintenance metabolism, and predation of bacteria (Liu and Tay, 2001; Low and Chase, 1999; Ratsak et al., 1996).
The oxic-settling-anaerobic (OSA) process is a modification of conventional activated sludge technology in which an anoxic/anaerobic reactor is inserted in the recycling bypass of sludge. OSA cycles equal volume of sludge between conditions that are rich (aeration tank) and deficient (external anoxic/anaerobic reactor/s) in oxygen and substrate (Semblante et al., 2014) and have many advantages, including no extra chemical or physical addition, treatment of high strength pollutants, simple configuration and flexible operation, economical efficiency also environmental friendliness (Easwaran, 2006; Novak et al., 2007). Since the OSA process is a modification of the activated sludge process, most interests till today focused on the observed yield (Novak et al., 2007; Saby et al., 2003; Sun et al., 2010), operational parameters such as HRT, ORP, temperature, IR of the OSA strategy in order to optimize and reduce the economic costs (Khursheed et al., 2015; Semblante et al., 2016a; Sun et al., 2016; Troiani et al., 2011; Yang et al., 2011), possible approaches causing sludge reduction in OSA process (Chen et al., 2003; Liu and Tay, 2001; Wei et al., 2003) or coupling sludge minimization with biological nutrient removal. However, the present level of understanding of the mechanisms involved in the OSA systems is still limited.
Molecular tools enable researchers to examine the composition of microbial populations and link them to process performance. Sludge cycling between the main reactor and external anoxic/anaerobic reactor/s has a strong influence on the microbial community composition. A previous study by Wang et al. (2008) incorporating denaturing gel gradient electrophoresis (DGGE) indicated that OSA was characterized by a wider range of bacterial species compared to conventional activated sludge (CAS) systems. Kim and co-workers showed that the external anaerobic SSR had a similar profile to a conventional anaerobic digester containing Spirochaetes, a gram-negative phylum that grows exclusively in anaerobic digesters (Kim et al., 2012). Moreover, in another study, hydrolytic-fermentative bacteria and acetogenic bacteria were found in the anaerobic compartments of a baffled reactor with repeated sequences of aerobic and anaerobic conditions (Quan et al., 2012). In the study by Zhou and co-workers, pyrosequencing results revealed that although core populations were shared with a highly functional organization, the relative abundance and stability of microbial communities in A + OSA system were higher than AO system (Zhou et al., 2015). Classes Anaerolineae and Actinobacteria responsible for hydrolysis and fermentation of organic matters were enriched in the reactors and played an important role in sludge reduction of A + OSA process. The microbial diversity and composition of a lab-scale OSA fed with real wastewater were determined by Semblante et al. (2017). A wide range of microorganisms such as hydrolyzing (e.g., phyla Bacteroidetes and Chloroflexi), fermentative (e.g., orders OP8, Firmicutes, WS3, and Spirochaetae), bacteria proliferated in the external reactors of OSA. Hydrolyzing and fermentative bacteria possibly facilitated the degradation of cellular matter. Furthermore, β- and γ-Proteobacteria were identified as the bacterial classes that primarily underwent decay in the external reactors during this research.
In this study, three OSA systems fed with real domestic wastewater that is the most common wastewater type treated in OSA systems were operated at different interchange ratios (IRs) to assess sludge reduction and analyze microbial community composition variations. Next Generation Sequencing (NGS) of bacterial 16S rRNA gene amplicons was used to analyze the change in the microbial community profile and to explore the microbial shift from conventional activated sludge systems. This study also aimed to contribute to this growing area of research by exploring relationships between microbial community structure and sludge reduction mechanism.
2. Materials and methods
2.1. The origin of activated sludge, system set-up, and operation
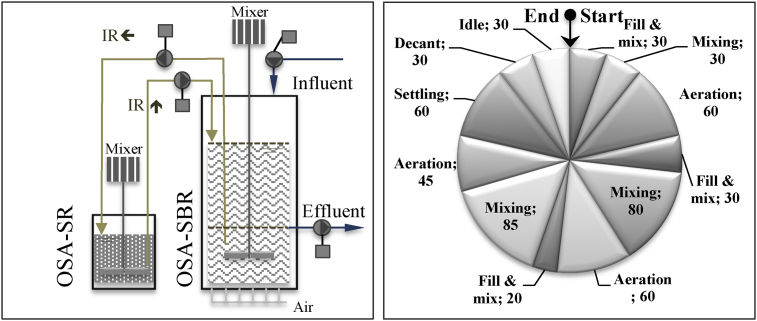
Activated sludge was used as a seed in this study, was originated from an advanced biological wastewater treatment plant in Istanbul, treating organic matter and nutrients from domestic wastewater. The systems were fed with real domestic wastewater from the outlet of a grit chamber of sewage treatment plant employing only physical treatment in Istanbul. Average total COD and TKN concentrations of domestic wastewater were analyzed as 310 ± 94, 352 ± 63 and 290 ± 113, and 47 ± 16, 53 ± 7 and 41 ± 18 mg/L during the operation of OSA1, OSA2, and OSA3, respectively, hence the required amount of glucose was added into real domestic wastewater in lab studies to achieve COD/TKN ratio of around 10 in the feed. The activated sludge collected from the plant was added into three sequencing batch reactors with a working volume of 3.4 L leaving an initial volume of 2 L (V0) after withdrawal and operates under three 8-hours cycles per day. The SBRs of OSA set-ups, namely, OSA2, and OSA3, were attached to completely closed and continuously mixed anoxic/anaerobic side-stream reactors (SR) having a total volume of 2 L. The sludge interchanged between SBR and SR was taken mixed liquor of initial volume of SBR and total volume of SR. Thus, the SR was operated under hydraulic retention time (HRT) of 13, 17 and 20 days for OSA1, OSA2, and OSA3, respectively. The corresponding IRs (Qin-SR/V0-SBR = (VSR/HRTSR)/V0-SBR) were correspondingly 1/13, 1/17 and 1/20. No sludge was wasted intentionally from the OSA-SBRs. The cycle operation of each SBR was designed to achieve nitrogen removal with a step-feeding regime and multiple anoxic/aerobic operation modes (Artan and Orhon, 2005). Description of operating conditions applied in the lab-scale SBRs and schematic diagram of the reactor set-up can be seen in Fig. 1.
Fig. 1.
Experimental set-up of the OSA systems and sequence and duration (min) of phases in each operating cycle of the SBR reactor.
2.2. Determination of observed sludge yield
A graphical method was used to determine the observed yield of sludge in each system. The Y-axis depicts cumulative daily MLVSS production, while X-axis represents cumulative daily COD consumption (Chon et al., 2011b). The daily MLVSS production was considered as daily produced mass of MLVSS in the SBR, loss of biomass from effluent stream, sampling, and daily sludge interchange between SBR and SR, while the consumed COD was the difference of COD concentrations between wastewater and effluent stream. The slope of the linear regression was used as a way of expressing Yobs. The relative sludge reduction was calculated using observed yield ratios obtained from identical systems operated in a conventional activated sludge manner with daily wastage of sludge (Karlikanovaite-Balikci and Yagci, 2019).
2.3. Analytical methods
Total and volatile suspended solids were measured gravimetrically according to the Standard Methods (APHA/AWWA/WEF, 2005). Total and soluble chemical oxygen demand (tCOD and sCOD) were quantified by means of the dichromate method using the closed reflux method (ISO 6060, 1986).
2.4. Microbial community analysis
Total genomic DNA was isolated from 500 μL of samples taken from the mixed liquor of each SBRs of OSA set-ups as well as from the seed sludge using ZymoBIOMICS DNA Miniprep kit (Zymo Research, Irvine, CA) according to manufacturer's protocol. The DNA quantification was measured by NanoPhotometer P-Class (Implen, Germany) and the extracted DNA samples were stored at -20 °C for further analysis.
Bacterial community compositions were processed and analyzed with the ZymoBIOMICS™ Service - Targeted Amplicon Sequencing (Zymo Research, Irvine, CA). 16S ribosomal RNA gene targeted sequencing was performed using the Quick-16S™ NGS Library Preparation Kit (Zymo Research, Irvine, CA). The general bacterial 16S barcoded primers used were 341f (CCTACGGGNGGCWGCAG) and 805r (GACTACHVGGGTATCTAATCC), which amplified the V3-V4 region of the 16S rRNA gene. The 16S primers used amplified the V3-V4 region of the 16S rRNA gene. The PCR products are quantified with qPCR fluorescence readings and pooled together based on equal molarity. The final pooled library was cleaned up with Select-a-Size DNA Clean & Concentrator™ (Zymo Research, Irvine, CA), then quantified with TapeStation® and Qubit®. The final library was sequenced on Illumina® MiSeq™ with a v3 reagent kit (600 cycles). The sequencing was performed with >10% PhiX mix and in paired-end mode. The sequences obtained from this study were deposited under the EMBL-EBI accession number PRJEB29107.
2.5. Bioinformatics
Amplicon sequences were inferred from raw reads using the Dada2 pipeline (Callahan et al., 2016). The number of average raw reads per sample was 183,920. Raw sequence reads were trimmed with Trimmomatic-0.33 (Bolger et al., 2014). The two paired-end reads in each pair were assembled to construct a complete amplicon sequence with SeqPrep (https://github.com/jstjohn/SeqPrep). Chimeric amplicon sequences were identified and removed with Usearch (v. 6.1) (Edgar et al., 2011) in ref mode against a curated database (http://drive5.com/uchime/rdp_gold.fa). Amplicon sequences smaller than 320 bp were removed. For each sample, up to 40,000 sequences were randomly sampled to reduce potential bias caused by uneven sampling. These amplicon sequences were compiled, clustered and analyzed with Qiime 1.9.1 (Caporaso et al., 2010). OTUs were picked by the workflow of pick_open_reference_otus.py using GreenGene database (gg_13_8) as a reference database. Singleton OTUs were removed. Taxonomy assignment, alpha-diversity, and beta-diversity analyses were performed with Qiime v.1.9.1 (Caporaso et al., 2010). The α- and β-diversities were calculated with in-house scripts. The microbial community structures were shown by Krona graphs (Ondov et al., 2011).
3. Results and discussion
3.1. Sludge reduction performance of the systems
The three systems were initially operated in general arrangement of conventional activated sludge with wastage of sludge following 44 days of microbial acclimation period. Then, SRs were attached into SBRs and continuously operated for another variable period of time (100 days for OSA1, 32 days for OSA2 and 52 days for OSA3) until the systems reached a steady-state condition. The average MLVSS concentrations of 1272 ± 2, 1649 ± 6 and 1683 ± 6 mg/L were obtained in SBRs at steady-state operation in OSA1, OSA2, and OSA3, respectively. The MLVSS concentrations of SRs were shown a similar trend at steady state and measured as 1226 ± 15, 1637 ± 38 and 1627 ± 34 mg/L for OSA1, OSA2, and OSA3, respectively.
The cumulative biomass production was plotted against cumulative substrate consumption, and observed yield values were calculated from the slope. The OSA systems presented the Yobs of 0.104, 0.117 and 0.111 gVSS/gCOD for OSA1, OSA2 and OSA3, respectively. The corresponding Yobs of conventional activated sludge were estimated as 0.217, 0.187 and 0.172, respectively. The OSA systems with HRT of 13, 17 and 20 d reduced sludge production by 52.1%, 37.4%, and 35.5%, respectively, in comparison to conventional systems. The sludge reduction efficiencies obtained in this study correspond to previous studies of the OSA (Eusebi and Battistoni, 2015; Goel and Noguera, 2006; Novak et al., 2007; Sun et al., 2016; Zhao and Wang, 2012). The total sludge age of the system was estimated as 33, 18 and 14 d as described by Chon and co-workers (Chon et al., 2011a) with a modification that assuming the only solid loss is caused by sampling and the withdrawal of treated wastewater from the system. Accordingly, the decreasing trend of sludge reduction could be explained by the decreasing total sludge age of the system. This result is in accordance with the findings by Chon and co-workers (Chon et al., 2011a). Hence, total SRT of the systems has an intrinsic relation with sludge reduction and appears as one of the most important parameters for the design and operation of these systems. Additionally, this should be also emphasized here that the effluent sCOD concentrations were in the range of 70 ± 17 mg/L during the entire experimental study and it is depicted that no significant effects on COD removal were observed by the addition of SR as shown in the previous studies given in the literature (Saby et al., 2003; Semblante et al., 2016b).
3.2. Microbial community compositions of the seed sludge
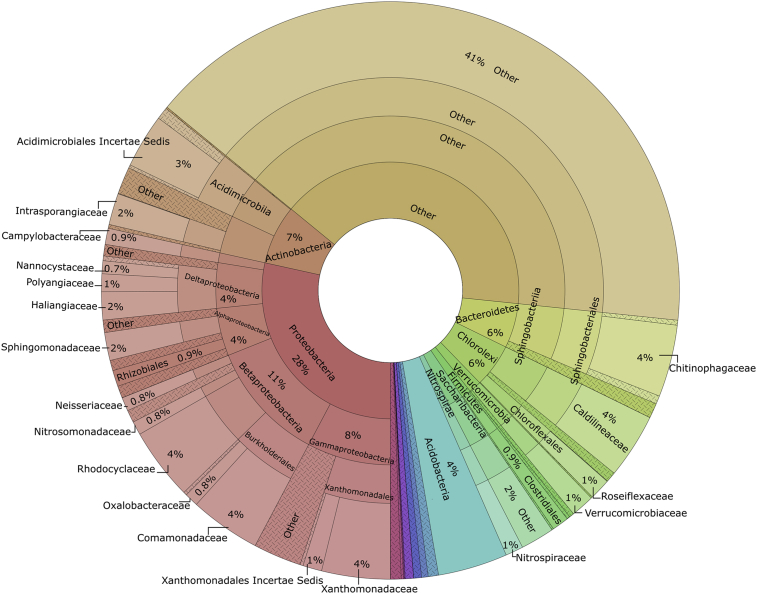
The microbial community composition of the seed sludge, which was taken from a full-scale conventional activated sludge system, was presented at the phylum, class, order and family levels in Fig. 2. From the chart, it can be seen that by far the greatest shares of the total known sequences belonged to Proteobacteria (28%) and Actinobacteria (7%) within the 14 known phyla. The remaining of the reads composed of minor phyla mainly Bacteroidetes, Chloroflexi, Acidobacteria, Verrucomicrobia, and Saccharibacteria. These results are consistent with previous observational studies, which revealed similar bacterial community patterns for a typical activated sludge process (Chen et al., 2018; Hong et al., 2016). Surprisingly, a large proportion of the reads could not be assigned to any known bacterial phylum in the present study. It may include uncultivable or unidentified novel bacterial species. Furthermore, the majority of the sequences belonged to Betaproteobacteria (11%) at the class level followed by Gammaproteobacteria (8%), Sphingobacteria (5%), Alphaproteobacteria (4%), and Deltaproteobacteria (4%).
Fig. 2.
Bacterial community composition of the seed sludge at phylum, class, order and family levels.
3.3. Microbial community dynamics of the OSA systems
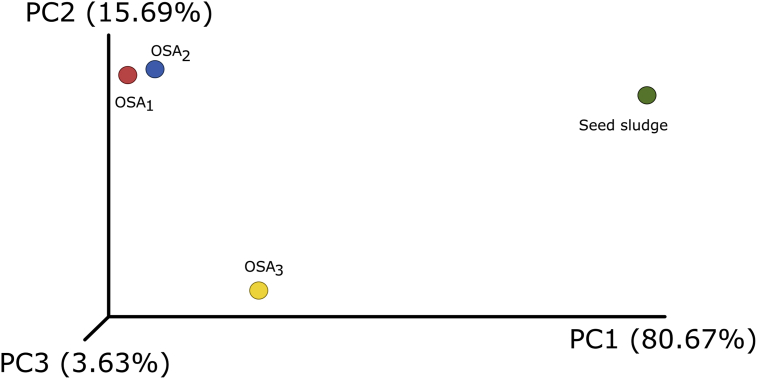
A two-dimensional plot of a 3D principal component analysis was prepared to compare the microbial communities of the different OSA trials and the seed sludge (Fig. 3). The distance between data points for the samples displays the similarity and/or dissimilarity of the microbial communities. As it is clear from the plot, OSA1 and OSA2 clustered together. However, the microbial community of the OSA3 system located distantly showing a significant difference. Moreover, a significant shift on the microbial community structure was associated with the reactor conditions compared to the seed sludge.
Fig. 3.
Two-dimensional plot of a 3D principal component analysis based on the ß-diversity of microbial communities.
Table 1 displays the estimated evenness and richness of the microbial communities by Chao1, Shannon, Simpson diversity and Pielou's eveness indices. While eveness shows abundance homogeneity in the community, richness reflects the number of different types (Lucas et al., 2017). It is apparent from the table that the seed sludge had the most diverse and even community. The reactors developed a more even community in the case of increasing HRT in which the highest eveness index calculated for OSA3 within all OSA experiments. Moreover, the OSA3 trial exhibited higher microbial evenness and diversity within all OSA systems having the highest Shannon, Chao1 and Simpson indices. Whereas, OSA1 had more diverse community compared to OSA2, the community was more even in OSA2 system.
Table 1.
Estimated richness and evenness of the microbial communities.
| Sample | IR | Chao1 | Shannon | Simpson | Pielou's evenness |
|---|---|---|---|---|---|
| OSA1 | 1/13 | 452 | 4.57 | 0.95 | 0.75 |
| OSA2 | 1/17 | 291 | 4.43 | 0.94 | 0.78 |
| OSA3 | 1/20 | 479 | 5.08 | 0.98 | 0.82 |
| Seed | - | 805 | 5.99 | 0.99 | 0.90 |
The difference in the HRT revealed a major role in the growth of various microorganisms.
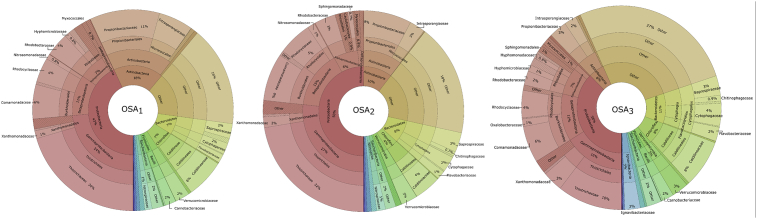
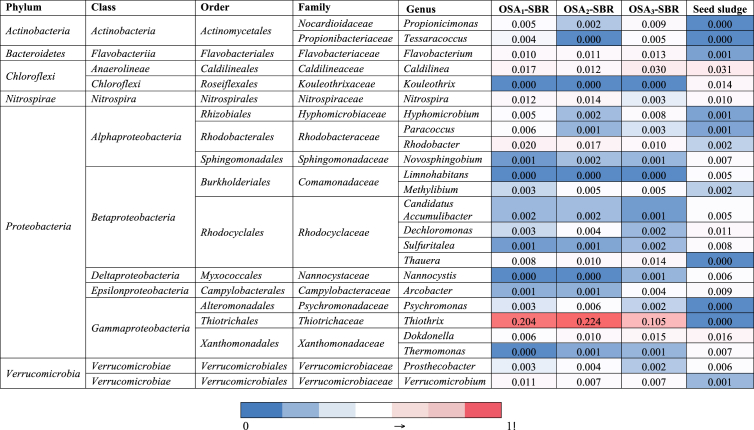
Fig. 4 displays the microbial community structures of the reactor samples from phylum to family levels. From the charts, it can be seen that by far the greatest share of the sequences annotated as Proteobacteria species as in the seed sludge sample. Furthermore, Acidobacteria, Bacteroidetes, and Chloroflexi were also predominant phyla in which their proportions in the microbial community distinguished due to the different IRs applied during the operation period. In a study carried out by Ning et al. (2014) also showed that the majority of the microbial community in an anoxic–oxic–settling–anaerobic (A + OSA) process composed of Proteobacteria members. Moreover, these results reflect those of Sun et al. (2016) who also found that Proteobacteria and Bacteroidetes species dominated the OSA process by DGGE fingerprinting method. The highest abundance of Actinobacteria was determined in the OSA1 system (18%), which has the highest IR and lowest HRT during the operation. A trend was designated between the Actinobacteria abundance and HRT in which increasing of the HRT caused a significant reduction in Actinobacteria species and the lowest abundance was determined in the OSA3 system (6%). On the other hand, an opposite profile was observed for Bacteroidetes members. An increase in the HRT resulted in the higher Bacteroidetes abundances in the systems, and the abundances were recorded as 7%, 8% and 11% for OSA1, OSA2 and OSA3 systems, respectively. These abundances were found lower than that of Ning et al. (2014). This inconsistency may be due to the different modifications on the reactor configurations and operating parameters. On the other hand, Proteobacteria members can contribute to release intracellular compounds and then Bacteroidetes may use the secondary substrate produced by those species for hydrolytic fermentation to boost their abundance (Cheng et al., 2018). Moreover, the highest abundance of Chloroflexi was found in the OSA3 system in which the reduced sludge production was the lowest (35.5%). Moreover, most of the Chloroflexi sequences belonged to the family Caldilineaceae in which the members are facultative filamentous organisms able to grow under both anaerobic and aerobic conditions (Albers and Siebers, 2014). Members of Chloroflexi can degrade various organic materials (carbohydrates) and live on the cellular materials resulted from dead biomass and metabolites (Cheng et al., 2017). Thus, it is suggested that the members may have major importance in the OSA system and the species may contribute to the sludge reduction in the system. Additionally, changes in the HRT resulted in some moderate difference in the abundance of Proteobacteria classes namely, Alphaproteobacteria, Betaproteobacteria, and Gammaproteobacteria. During the operation period, the Gammaproteobacteria species took advantage of the operation conditions and enriched in the OSA trials compared to the seed sludge. Although, there was no significant difference in the abundance of Alphaproteobacteria between the different OSA trials (average abundance 6.5%), the abundance of Gammaproteobacteria went down significantly in the OSA3 system representing 16% of the total sequences. The Betaproteobacteria members can use a wide variety of carbon sources and it is one of the major class found in wastewater treatment systems having a role in organic matter degradation (Ning et al., 2014). Alphaproteobacteria species can perform a broad range of metabolic activities including nitrogen fixation, ammonia oxidation, and methylotrophy (Williams et al., 2007). Thus, they can use a different type of organic materials commonly found in municipal wastewater treatment plants (Ning et al., 2014). Gammaproteobacteria is one of the most diverse class including more than 250 genera with a diverse range of aerobicity, trophism and of temperature adaptation (Williams et al., 2010). Sun et al. (2016) suggested that cryptic growth features of Alphaproteobacteria, Betaproteobacteria, and Gammaproteobacteria species resulted in a greater sludge reduction efficiency in OSA systems.
Fig. 4.
Bacterial community composition of the OSA systems at phylum, class, order and family levels.
Although Thiotrichaceae (phylum: Proteobacteria) species were not detected in the seed sludge sample, the majority of the total sequences were represented by Thiotrichaceae at the family level in all systems. However, a significant decrease was determined in the abundance of Thiotrichaceae members due to the higher HRT. Although, there was no significant difference between OSA1 and OSA3, changing the HRT correspondingly from 13 to 20 d, caused a remarkable effect. Most of the Thiotrichaceae species could not survive in the system and became less abundant in OSA3. Similar patterns were observed for Actinobacteria families, Propionibacteriaceae and Intrasporangiaceae as well. Whereas the abundance of Propionibacteriaceae was lower than 1% in the seed sludge, the species enriched in the OSA1 and OSA2 trials (11% and 8%, respectively). Whereas the members of Propionibacteriaceae are able to produce a large amount of propionic acids (Stackebrandt et al., 2006). Furthermore, the highest abundance of Intrasporangiaceae species was found in the OSA1 system. Some members of this family are polyphosphate accumulating organisms and some of them have denitrifying abilities (Stackebrandt et al., 2014).
Microbial community patterns at the genus level are depicted in Fig. 5. What is interesting about the data is in this heatmap that Thiothrix (phylum: Proteobacteria) was by far the most predominant genus in the three OSA trials. However, the abundance was below the detection limits in the seed sludge sample. Thiothrix species are filamentous, sulfur-oxidizing bacteria and able to grow heterotrophically, lithoautotrophically with thiosulfate or sulfide as the sole energy source and mixotrophically (Chernousova et al., 2009). The members of this genus enriched due to the OSA operating conditions. They are typically found in activated sludge systems with filamentous bulking problems (Nielsen et al., 2000). However, in the present study, sludge bulking was not observed. Whereas, the mesophilic filamentous bacteria Caldilinea (phylum: Chloroflexi) (Yoon et al., 2010), denitrifier Thauera (phylum: Proteobacteria) (Scholten et al., 1997) and Dokdonella which detected in anaerobic side-stream reactor process previously (Ferrentino et al., 2016) had relatively higher abundance in OSA3 sample, the abundance of Nitrospira (phylum: Nitrospirae) and Rhodobacter (phylum: Proteobacteria) was higher in OSA1 and OSA2 trials. Nitrospira plays a crucial role in the efficient nitrification process, oxidizing nitrite produced by the ammonia-oxidizing microorganism. Recent studies showed that some members are named as complete ammonia oxidizers (comammox) can perform both nitrification steps (Daims and Wagner, 2018).
Fig. 5.
Heatmap displaying the relative abundances of bacterial genera with a proportion of at least 1% in at least one sample.
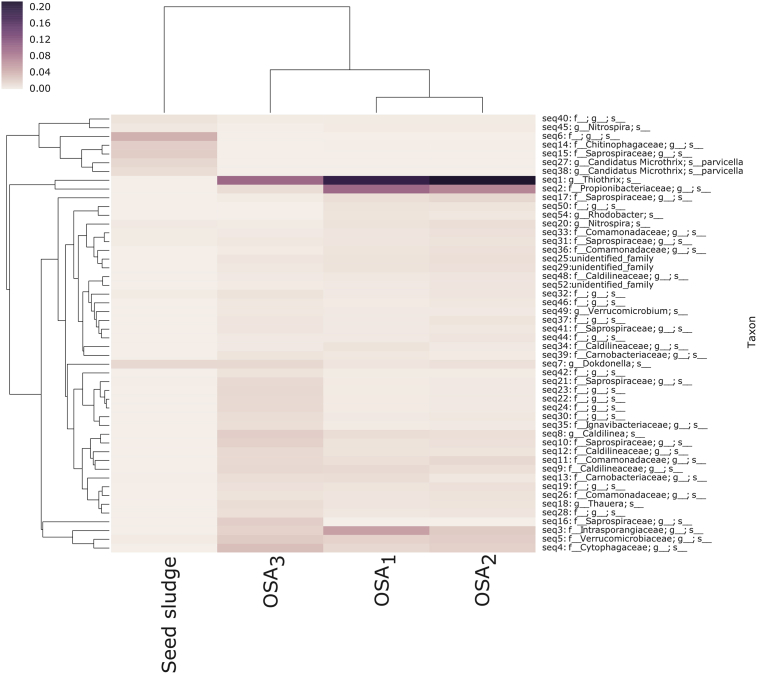
Fig. 6 shows the relative abundance of unique OTUs in all samples. The clustering of OTUs revealed two major groups. Whereas the first group included the seed sludge sample, the second group was composed of OSA trials. OTU clustering also showed that the OSA3 was separated from the other two OSA systems suggesting a distinct microbial community profile. Furthermore, OTUs named as seq1 (genus: Thiothrix) and seq2 (family: Propionibacteriaceae) were dominated the OSA trials.
Fig. 6.
Heatmap displaying the unique sequence abundance of microbial communities.
4. Conclusions
The OSA systems with HRT of 13, 17 and 20 d in the SR reduced sludge production by 52.1%, 37.4%, and 35.5%, respectively. The results of microbial community analysis were indicated that the shift in the microbial community structures of the OSA trials has an influence on process performance. Increasing of the HRT to 1/20 caused a significant reduction in Actinobacteria species. Returning to the question posed at the beginning of this study, it is now possible to state that, Thiothrix species were stimulated in OSA operating conditions and may have a key role in these systems. Moreover, minimization of the sludge production may have relevance on the abundance of Intrasporangiaceae species.
Declarations
Author contribution statement
Agne Karlikanovaite-Balikci Performed the experiments; Analyzed and interpreted the data.
Gozde Ozbayram: Analyzed and interpreted the data.
Nevin Yagci: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Orhan Ince: Contributed reagents, materials, analysis tools or data.
Funding statement
This work was supported by the Research Fund of Istanbul Technical University (Project #37722).
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
The work was supported financially by the Research Fund of Istanbul Technical University (Project #37722).
References
- Albers S.-V., Siebers B. Springer Berlin Heidelberg; Berlin, Heidelberg: 2014. The Prokaryotes, the Prokaryotes. [Google Scholar]
- Alyaseri I., Zhou J. Towards better environmental performance of wastewater sludge treatment using endpoint approach in LCA methodology. Heliyon. 2017;3 doi: 10.1016/j.heliyon.2017.e00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- APHA/AWWA/WEF . 21th ed. American Public Health Association; Washington DC: 2005. Standard Methods for the Examination of Water and Wastewater. [Google Scholar]
- Artan N., Orhon D. IWA Publishing; United Kingdom: 2005. Mechanism and Design of Sequencing Batch Reactors for Nutrient Removal. [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J.A., Holmes S.P. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Peña A.G., Goodrich J.K., Gordon J.I., Huttley G.A., Kelley S.T., Knights D., Koenig J.E., Ley R.E., Lozupone C.A., McDonald D., Muegge B.D., Pirrung M., Reeder J., Sevinsky J.R., Turnbaugh P.J., Walters W.A., Widmann J., Yatsunenko T., Zaneveld J., Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G.-H., An K.-J., Saby S., Brois E., Djafer M. Possible cause of excess sludge reduction in an oxic-settling-anaerobic activated sludge process (OSA process) Water Res. 2003;37:3855–3866. doi: 10.1016/S0043-1354(03)00331-2. [DOI] [PubMed] [Google Scholar]
- Chen Y., He H., Liu H., Li H., Zeng G., Xia X., Yang C. Effect of salinity on removal performance and activated sludge characteristics in sequencing batch reactors. Bioresour. Technol. 2018;249:890–899. doi: 10.1016/j.biortech.2017.10.092. [DOI] [PubMed] [Google Scholar]
- Cheng C., Zhou Z., Niu T., An Y., Shen X., Pan W., Chen Z., Liu J. Effects of side-stream ratio on sludge reduction and microbial structures of anaerobic side-stream reactor coupled membrane bioreactors. Bioresour. Technol. 2017;234:380–388. doi: 10.1016/j.biortech.2017.03.077. [DOI] [PubMed] [Google Scholar]
- Cheng C., Zhou Z., Qiu Z., Yang J., Wu W., Pang H. Enhancement of sludge reduction by ultrasonic pretreatment and packing carriers in the anaerobic side-stream reactor: performance, sludge characteristics and microbial community structure. Bioresour. Technol. 2018;249:298–306. doi: 10.1016/j.biortech.2017.10.043. [DOI] [PubMed] [Google Scholar]
- Chernousova E., Gridneva E., Grabovich M., Dubinina G., Akimov V., Rossetti S., Kuever J. Thiothrix caldifontis sp. nov. and Thiothrix lacustris sp. nov., gammaproteobacteria isolated from sulfide springs. Int. J. Syst. Evol. Microbiol. 2009;59:3128–3135. doi: 10.1099/ijs.0.009456-0. [DOI] [PubMed] [Google Scholar]
- Chon D.H., Rome M., Kim H.S., Park C. Investigating the mechanism of sludge reduction in activated sludge with an anaerobic side-stream reactor. Water Sci. Technol. 2011 doi: 10.2166/wst.2011.015. [DOI] [PubMed] [Google Scholar]
- Chon D.H., Rome M., Kim Y.M., Park K.Y., Park C. Investigation of the sludge reduction mechanism in the anaerobic side-stream reactor process using several control biological wastewater treatment processes. Water Res. 2011;45:6021–6029. doi: 10.1016/j.watres.2011.08.051. [DOI] [PubMed] [Google Scholar]
- Daims H., Wagner M. Nitrospira. Trends Microbiol. 2018;26:462–463. doi: 10.1016/j.tim.2018.02.001. [DOI] [PubMed] [Google Scholar]
- Easwaran S.P. Virginia Tech; 2006. Developing a Mechanistic Understanding and Optimization of the Cannibal Process: Phase II. [Google Scholar]
- Edgar R.C., Haas B.J., Clemente J.C., Quince C., Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eusebi A.L., Battistoni P. Reduction of the excess sludge production by biological alternating process: real application results and metabolic uncoupling mechanism. Environ. Technol. 2015;36:137–148. doi: 10.1080/09593330.2014.939230. [DOI] [PubMed] [Google Scholar]
- Ferrentino R., Langone M., Gandolfi I., Bertolini V., Franzetti A., Andreottola G. Shift in microbial community structure of anaerobic side-stream reactor in response to changes to anaerobic solid retention time and sludge interchange ratio. Bioresour. Technol. 2016;221:588–597. doi: 10.1016/j.biortech.2016.09.077. [DOI] [PubMed] [Google Scholar]
- Goel R.K., Noguera D.R. Evaluation of sludge yield and phosphorus removal in a Cannibal solids reduction process. J. Environ. Eng. 2006;132:1331–1337. [Google Scholar]
- Hong J. ming, Hu M.M., Sun R., Chen B.Y. Unraveling characteristics of nutrient removal and microbial community in a novel aerated landscape - activated sludge ecological system. Bioresour. Technol. 2016;212:280–288. doi: 10.1016/j.biortech.2016.04.042. [DOI] [PubMed] [Google Scholar]
- ISO 6060 . International Organization for Standardization; Geneva, Switzerland: 1986. Water Quality-Determination of the Chemical Oxygen Demand. [Google Scholar]
- Karlikanovaite-Balikci A., Yagci N. Determination and evaluation of kinetic parameters of activated sludge biomass from a sludge reduction system treating real sewage by respirometry testing. J. Environ. Manag. 2019;240:303–310. doi: 10.1016/j.jenvman.2019.03.131. [DOI] [PubMed] [Google Scholar]
- Khursheed A., Sharma M.K., Tyagi V.K., Khan A.A., Kazmi A.A. Specific oxygen uptake rate gradient – another possible cause of excess sludge reduction in oxic-settling-anaerobic (OSA) process. Chem. Eng. J. 2015;281:613–622. [Google Scholar]
- Kim Y.M., Chon D.-H., Kim H.-S., Park C. Investigation of bacterial community in activated sludge with an anaerobic side-stream reactor (ASSR) to decrease the generation of excess sludge. Water Res. 2012;46:4292–4300. doi: 10.1016/j.watres.2012.04.040. [DOI] [PubMed] [Google Scholar]
- Liu Y., Tay J.-H. Strategy for minimization of excess sludge production from the activated sludge process. Biotechnol. Adv. 2001;19:97–107. doi: 10.1016/s0734-9750(00)00066-5. [DOI] [PubMed] [Google Scholar]
- Low E.W., Chase H.A. Reducing production of excess biomass during wastewater treatment. Water Res. 1999;33:1119–1132. [Google Scholar]
- Lucas R., Groeneveld J., Harms H., Johst K., Frank K., Kleinsteuber S. A critical evaluation of ecological indices for the comparative analysis of microbial communities based on molecular datasets. FEMS Microbiol. Ecol. 2017 doi: 10.1093/femsec/fiw209. [DOI] [PubMed] [Google Scholar]
- Nielsen P.H., de Muro M.A., Nielsen J.L. Studies on the in situ physiology of Thiothrix spp. present in activated sludg. Environ. Microbiol. 2000;2:389–398. doi: 10.1046/j.1462-2920.2000.00120.x. [DOI] [PubMed] [Google Scholar]
- Ning X., Qiao W., Zhang L., Gao X. Microbial community in anoxic–oxic–settling–anaerobic sludge reduction process revealed by 454 pyrosequencing analysis. Can. J. Microbiol. 2014;60:799–809. doi: 10.1139/cjm-2014-0263. [DOI] [PubMed] [Google Scholar]
- Novak J.T., Chon D.H., Curtis B.-A., Doyle M. Biological solids reduction using the cannibal process. Water Environ. Res. 2007;79:2380–2386. doi: 10.2175/106143007x183862. [DOI] [PubMed] [Google Scholar]
- Ondov B.D., Bergman N.H., Phillippy A.M. Interactive metagenomic visualization in a Web browser. BMC Bioinf. 2011;12:385. doi: 10.1186/1471-2105-12-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan F., Anfeng Y., Libing C., Hongzhang C., Xing X.-H. Mechanistic study of on-site sludge reduction in a baffled bioreactor consisting of three series of alternating aerobic and anaerobic compartments. Biochem. Eng. J. 2012;67:45–51. [Google Scholar]
- Ratsak C.H., Maarsen K.A., Kooijman S.A.L.M. Effects of protozoa on carbon mineralization in activated sludge. Water Res. 1996;30:1–12. [Google Scholar]
- Saby S., Djafer M., Chen G.-H. Effect of low ORP in anoxic sludge zone on excess sludge production in oxic-settling-anoxic activated sludge process. Water Res. 2003;37:11–20. doi: 10.1016/s0043-1354(02)00253-1. [DOI] [PubMed] [Google Scholar]
- Scholten E., Lukow T., Auling I.G., Kroppenstedt R.M., Rainey F.A., Diekmann H. Denitrifier from a leachate treatment plant. Int. J. Syst. Bacteriol. 1997:1045–1051. doi: 10.1099/00207713-49-3-1045. [DOI] [PubMed] [Google Scholar]
- Semblante G.U., Hai F.I., Bustamante H., Guevara N., Price W.E. Biosolids reduction by the oxic-settling-anoxic process: impact of sludge interchange rate. Bioresour. Technol. 2016;210:167–173. doi: 10.1016/j.biortech.2016.01.010. [DOI] [PubMed] [Google Scholar]
- Semblante G.U., Hai F.I., Bustamante H., Price W.E., Nghiem L.D. Effects of sludge retention time on oxic-settling-anoxic process performance: biosolids reduction and dewatering properties. Bioresour. Technol. 2016;218:1187–1194. doi: 10.1016/j.biortech.2016.07.061. [DOI] [PubMed] [Google Scholar]
- Semblante G.U., Hai F.I., Ngo H.H., Guo W., You S.-J., Price W.E., Nghiem L.D. Sludge cycling between aerobic, anoxic and anaerobic regimes to reduce sludge production during wastewater treatment: performance, mechanisms, and implications. Bioresour. Technol. 2014;155:395–409. doi: 10.1016/j.biortech.2014.01.029. [DOI] [PubMed] [Google Scholar]
- Semblante G.U., Phan H.V., Hai F.I., Xu Z.-Q.Q., Price W.E., Nghiem L.D. The role of microbial diversity and composition in minimizing sludge production in the oxic-settling-anoxic process. Sci. Total Environ. 2017;607–608:558–567. doi: 10.1016/j.scitotenv.2017.06.253. [DOI] [PubMed] [Google Scholar]
- Stackebrandt E., Cummins C.S., Johnson J.L. The Prokaryotes. Springer New York; New York, NY: 2006. Family Propionibacteriaceae: the genus propionibacterium; pp. 400–418. [Google Scholar]
- Stackebrandt E., Scheuner C., Göker M., Schumann P. The Prokaryotes. Springer Berlin Heidelberg; Berlin, Heidelberg: 2014. The family Intrasporangiaceae; pp. 397–424. [Google Scholar]
- Sun L., Chen J., Wei X., Guo W., Lin M., Yu X. Study of the diversity of microbial communities in a sequencing batch reactor oxic–settling–anaerobic process and its modified process. Can. J. Microbiol. 2016;62:411–421. doi: 10.1139/cjm-2015-0194. [DOI] [PubMed] [Google Scholar]
- Sun L., Randall C.W., Novak J.T. The influence of sludge interchange times on the oxic-settling-anoxic process. Water Environ. Res. 2010;82:519–523. doi: 10.2175/106143009x12487095236711. [DOI] [PubMed] [Google Scholar]
- Troiani C., Eusebi A.L.L., Battistoni P. Excess sludge reduction by biological way: from experimental experience to a real full scale application. Bioresour. Technol. 2011;102:10352–10358. doi: 10.1016/j.biortech.2011.08.124. [DOI] [PubMed] [Google Scholar]
- Wang J., Zhao Q., Jin W., Lin J. Mechanism on minimization of excess sludge in oxic-settling-anaerobic (OSA) process. Front. Environ. Sci. Eng. China. 2008;2:36–43. [Google Scholar]
- Wei Y., Van Houten R.T., Borger A.R., Eikelboom D.H., Fan Y., Van Houten R.T., Wei Y., Eikelboom D.H. Minimization of excess sludge production for biological wastewater treatment. Water Res. 2003;37:4453–4467. doi: 10.1016/S0043-1354(03)00441-X. [DOI] [PubMed] [Google Scholar]
- Williams K.P., Gillespie J.J., Sobral B.W.S., Nordberg E.K., Snyder E.E., Shallom J.M., Dickerman A.W. Phylogeny of gammaproteobacteria. J. Bacteriol. 2010;192:2305–2314. doi: 10.1128/JB.01480-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K.P., Sobral B.W., Dickerman A.W. A robust species tree for the Alphaproteobacteria. J. Bacteriol. 2007;189:4578–4586. doi: 10.1128/JB.00269-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.-S., Guo W.-Q., Zhou X.-J., Meng Z.-H., Liu B., Ren N.-Q. Optimization of operating parameters for sludge process reduction under alternating aerobic/oxygen-limited conditions by response surface methodology. Bioresour. Technol. 2011;102:9843–9851. doi: 10.1016/j.biortech.2011.07.079. [DOI] [PubMed] [Google Scholar]
- Yoon D.N., Park S.J., Kim S.J., Jeon C.O., Chae J.C., Rhee S.K. Isolation, characterization, and abundance of filamentous members of Caldilineae in activated sludge. J. Microbiol. 2010;48:275–283. doi: 10.1007/s12275-010-9366-8. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Hu J., Lee D.-J., Chang Y., Lee Y.-J. Sludge treatment: current research trends. Bioresour. Technol. 2017;243:1159–1172. doi: 10.1016/j.biortech.2017.07.070. [DOI] [PubMed] [Google Scholar]
- Zhao Q., Wang J. Biological Sludge Minimization and Biomaterials/Bioenergy Recovery Technologies. John Wiley & Sons, Inc.; Hoboken, NJ, USA: 2012. Oxic-settling-anaerobic process for enhanced microbial decay; pp. 155–182. [Google Scholar]
- Zhou Z., Qiao W., Xing C., An Y., Shen X., Ren W., Jiang L., Wang L. Microbial community structure of anoxic–oxic-settling-anaerobic sludge reduction process revealed by 454-pyrosequencing. Chem. Eng. J. 2015;266:249–257. [Google Scholar]