Abstract
The Arp2/3 complex, a seven-subunit protein complex containing two actin-related proteins, Arp2 and Arp3, initiates formation of actin filament networks in response to intracellular signals. The molecular mechanism of filament nucleation, however, is not well understood. Arp2 and Arp3 are predicted to bind ATP via a highly conserved nucleotide-binding domain found in all members of the actin superfamily and to form a heterodimer than mimics a conventional actin dimer. We show here that adenosine nucleotides bind with micromolar affinity to both Arp2 and Arp3 and that hydrolyzable ATP is required for actin nucleation activity. Binding of N-WASP WA increases the affinity of both Arp2 and Arp3 for ATP but does not alter the stoichiometry of nucleotides bound in the presence of saturating concentrations of ATP. The Arp2/3 complex bound to ADP or the nonhydrolyzable ATP analogue AMP-PNP cannot nucleate actin filaments, but addition of the phosphate analogue BeF3 partially restores activity to the ADP–Arp2/3 complex. Bound nucleotide also regulates the affinity of the Arp2/3 complex for its upstream activators N-WASP and ActA. We propose that the active nucleating form of the Arp2/3 complex is the ADP-Pi intermediate in the ATPase cycle and that the ATPase activity of the Arp2/3 complex controls both nucleation of new filaments and release of the Arp2/3 complex from membrane-associated activators.
The actin cytoskeleton determines the shape, motility, and internal organization of eukaryotic cells. Many actin-based structures, especially those involved in membrane protrusion, are assembled by coordinated polymerization and crosslinking of new actin filaments from actin monomers into either orthogonal or parallel filament networks (1). In these structures, work is accomplished by the free energy of polymerization. Subsequent ATP hydrolysis by filamentous actin then allows the networks to be disassembled. The rapid and regulated assembly and disassembly of actin filament networks lies at the heart of many cellular processes that involve membrane protrusion such as cell locomotion, endocytosis, and phagocytosis (2).
The Arp2/3 complex plays a central role in the regulated assembly of actin-based structures. The Arp2/3 complex nucleates formation of new actin filaments in response to upstream signaling events and simultaneously crosslinks them into orthogonal networks. Activation of Rho-family G proteins, including Rac and Cdc42, leads to dramatic reorganization of the actin cytoskeleton (3, 4). Rac and Cdc42 promote activation of members of the WASP family of proteins (5–7) that include WASP, N-WASP, and several isoforms of Scar (8). WASP family proteins, in turn, recruit and activate the Arp2/3 complex (7, 9, 10). The Arp2/3 complex nucleates formation of new actin filaments from the sides of older filaments, creating a dendritic network of crosslinked actin filaments in vitro (11, 12) and in vivo (13).
We proposed that activation of Arp2/3 complex involves the two actin-related proteins, Arp2 and Arp3, forming a heterodimer that mimics a conventional actin dimer and nucleates a new actin filament (14, 15). Actin-related proteins and other members of the actin superfamily, such as molecular chaperones like Hsc70, apyrases like ecto-apyrase HB6, and sugar kinases like hexokinase and glycerokinase, share a core structural fold, the actin fold, which forms an adenosine nucleotide-binding site (16). In these proteins, the actin fold couples ATP hydrolysis and phosphate release to conformational changes that control, for example, actin filament stability (17–19) and the affinity of Hsc70 for unfolded proteins (20). Actin monomer–monomer interactions within a filament are stronger in the ATP and the ADP-Pi states (21, 22) than in the ADP state. In contrast, Hsp70 binds its substrates weakly in the ATP state but tightly in the ADP state (23). Bound nucleotide regulates the affinities of several members of the actin superfamily for their substrates, therefore we speculated that bound nucleotide might also regulate the interaction of Arp2 and Arp3 and control the formation of an Arp2–Arp3 nucleus. To address these questions, we characterized nucleotide binding and hydrolysis by the Arp2/3 complex and investigated the role of the bound nucleotide in the biochemical activity of the Arp2/3 complex.
Materials and Methods
Purification of Proteins.
We purified the Arp2/3 complex from Acanthamoeba castellanii by a combination of conventional and affinity chromatography. We resuspended 75 g of Acanthamoeba in two volumes of lysis buffer (11% sucrose/10 mM Tris, pH 8.0/5 mM DTT/1 mM ATP/1 mM EGTA/2 mM PMSF/0.1 mM benzamidine/1 mg leupeptin/4 mg soybean trypsin inhibitor/1 mg pepstatin A/1 mg aprotinin), and lysed cells in a dounce homogenizer. We spun the lysate at 13,000 rpm in a Sorvall GSA rotor for 20 min to remove cell debris, and then in a Ti50.2 rotor at 38,000 rpm for 90 min. We collected the layer of supernatant between the pellet and lipid layer and bound it in batch for 20 min to 100 ml of DEAE resin equilibrated in column buffer (100 μM ATP/1 mM Ca2+/2 mM Tris, pH 8.0/1 mM DTT). DEAE flow-through was loaded on a 15-ml C200 column (Cellufine 200(m); Millipore). We collected the flow-through, added 25 mM KCl, and passed it over a 2-ml CH-Sepharose (Activated CH-Sepharose 4B; Amersham Pharmacia Biosciences, Uppsala, Sweden) N-WASP WA (398) affinity column. We washed the resin with 10 volumes each of 25 mM and 100 mM KCl in column buffer. The Arp2/3 complex was eluted with 400 mM MgCl2 and passed over a 400 μl phenyl-Sepharose column (no. 6 Fast-Flow low sub; Amersham Pharmacia Biosciences) then either gel filtered on a 30-ml Superdex S-200 column equilibrated with KMEI buffer or exchanged into KMEI by using a PD-10 desalting column (Amersham Pharmacia Biosciences). The complex was concentrated by dialysis against sucrose and dialyzed overnight into KMEI buffer (50 mM KCl/1 mM MgCl2/1 mM EGTA/0.5 mM TCEP/10 mM imidazole, pH 7.0) + 100 μM ATP for use in assays. Typical yields were ≈6 mg of pure Arp2/3 complex from 75 g of packed cells. The protein was never freeze-thawed or stored for more than 1 week. Typically it was used within 24 h of purification.
Actin was purified from Acanthamoeba by the method of MacLean-Fletcher and Pollard (24). Actin filaments used as seeds for polymerization or in ATP hydrolysis experiments were polymerized for 30 min at room temperature by adding of 0.1 volume of 10× KMEI to monomeric actin, followed by the addition of 1.2 molar excess phalloidin. Phalloidin–actin was allowed to continue polymerizing overnight on ice, and nucleotide was removed the following day by 2 treatments with 5% Dowex resin (AG 1-X8 resin, Bio-Rad) at 4°C. Residual denatured actin monomers were removed by centrifugation at 20,000 × g for 10 min.
Recombinant rat N-WASP-WA (398) with an N-terminal 6His tag was purified by nickel-affinity chromatography, using standard methods.
Assay Conditions.
All assays were performed in KMEI buffer at 25°C. Free nucleotide was removed from the Arp2/3 complex before assays by 2–4 treatments for 3 min each with 5% volumes of Dowex resin.
UV Crosslinking Studies.
The Arp2/3 complex was used at 1–2 μM, F-actin was used at 10 μM, and N-WASP WA was used at 25 μM. For normal ATP crosslinking, a stock solution of 300 μM ATP in KMEI was doped with 150 μCu [α-32P]ATP. For azido-ATP crosslinking, we used a stock of [α-32P]azido-ATP at 500 μM. Free nucleotide was removed from the Arp2/3 complex by 4 treatments with 5% Dowex resin. We incubated the Arp2/3 complex in KMEI for 20 min with labeled ATP and then photo-crosslinked bound nucleotide to the protein by exposing the mixture to UV light. For ATP, the UV exposure was 30 s, and for azido-ATP 1.5–19 s (312 nm UV handlamp, Fisher Scientific) (25). Subunits were separated by SDS/PAGE. 32P labeling was quantified by using a PhosphorImager (Storm 840, Molecular Dynamics) and data fit to a binding quadratic with dynafit (26) to obtain dissociation constants.
HPLC.
The Arp2/3 complex in KMEI plus 100 μM ATP was exchanged into KMEI plus 0 μM, 1.4 μM, or 10 μM ATP by using a PD-10 buffer-exchange column (Amersham Pharmacia Biosciences). Exchange took less than 2 min, and flow-through was collected in 150 μl fractions. Arp2/3 complex concentration was determined by A290 (7.14 μM/OD). The peak fraction (≈5 μM) was heat-denatured, and buffer plus released nucleotide was separated from denatured protein by filtration through a 10-kDa MWCO filter (UltraFree-MC, Millipore). Nucleotide was quantified by HPLC in 75 mM KH2PO4, pH 4.0/1.5 mM tetrabutylammonium phosphate/12% methanol on a C18 column (Waters DeltaPak 15 μM 100A 300 × 3.9 mm) at 1 ml/min (27). The concentration of nucleotide in the column buffer was subtracted from that in the sample and this was divided by the Arp2/3 complex concentration to calculate the number of bound nucleotides per complex. For the N-WASP WA condition, the PD-10 column was preequilibrated in 25 μM N-WASP WA, and 25 μM N-WASP WA was added to the Arp2/3 complex before loading onto the column.
Etheno-ATP Affinity Measurement.
Free nucleotide was removed from the Arp2/3 complex by 4 treatments with 5% Dowex resin. Etheno-ATP fluorescence was measured (λex = 340 nm, λem = 420 nm) for 100 μl of KMEI + 0.8 μM Arp2/3 complex + 30 μM etheno-ATP + 200 mM acrylamide [as a dynamic quencher (28)]. This solution was serially diluted with 50% volumes of 0.8 μM Arp2/3 complex + 200 mM acrylamide in KMEI to titrate down etheno-ATP concentrations. Etheno-ATP + Arp2/3 complex fluorescence was linear with etheno-ATP concentration, for concentrations greater than 10 μM, and we fit this linear portion of the data to f = a + b[eATP] to find b, the contribution of unbound etheno-ATP to the measured fluorescence. We subtracted background protein fluorescence and free etheno-ATP fluorescence from the measured fluorescence to obtain the enhancement due to etheno-ATP binding and data fit to the binding quadratic to obtain dissociation constants.
Measurement of ATP Hydrolysis.
We determined steady-state ATP hydrolysis rates at 25°C in KMEI buffer. The Arp2/3 complex was used at 4.4 μM, N-WASP WA was used at 25 μM, and F-actin was used at 15 μM. Free nucleotide was removed from the Arp2/3 complex by 2 treatments with 5% Dowex resin, and the Arp2/3 complex was brought up to room temperature. [α-32P]ATP (50 μCi) and 500 μM ATP concentrations were added to start the reaction. Aliquots (5 μl) were quenched with equal volumes of 5 M formic acid, spotted on polyethyleneimine-cellulose TLC plates, and separated by chromatography with 0.6 M KH2PO4 (pH 3.4). [α-32P]ATP and product [α-32P]ADP were quantified by using a PhosphorImager and normalized by the starting ATP concentration.
Kinetic Assays.
We measured the time course of nucleotide release from the Arp2/3 complex by the increase in fluorescence intensity of etheno-ATP (λex = 360 nm, λem > 389 nm) on binding to Arp2/3 complex, using a hand-operated rapid mixer (SFA-20; HiTech Scientific, Salisbury, U.K.) and fluorimeter (K2; ISS, Champagne, IL). Samples were excited at 360 nm, and emission was measured at >389 nm by using a long pass filter (KV389; Schott, Duryea, PA). We used 1–2 μM Arp2/3 complex in KMEI buffer, treated 2 times with 5% Dowex just before use to remove free nucleotide. For ATP and ADP, off-rate measurements, 100 μM nucleotide was added to the buffer, excess removed once more by using Dowex, and 5 μM nucleotide added immediately before use. The dissociation rate for nucleotide was measured by competing off the bound nucleotide with 50 μM etheno-ATP. Fluorescence data were analyzed by nonlinear least squares fitting to a single exponential function.
Polarization Anisotropy.
N-WASP WA (398) was labeled on cysteine 427 in 2 mM Hepes/500 mM KCl plus 4-fold molar excess of rhodamine maleimide (Molecular Probes) overnight at 4°C. The sample was centrifuged at 350,000 × g for 20 min, and the supernatant gel was filtered on a Sephacryl S-300 column to remove free dye and denatured protein. ActA (30) was labeled as in Zalevsky et al. (29). By this procedure, proteins were ≈25% labeled as determined by A280 and A554. Polarization anisotropy was measured for 250 nM labeled protein in the presence of various concentrations of the Arp2/3 complex in KMEI plus 1 mM nucleotide (ATP, ADP, or AMP-PNP), and the Arp2/3 complex concentration was titrated by serial dilution. Relative affinities of labeled and unlabeled protein were measured by monitoring anisotropy of labeled protein in competition with unlabeled protein. Labeled N-WASP WA was determined to bind 4-fold tighter than unlabeled N-WASP WA in the presence of ATP and ADP. Anisotropy data were fit to a model accounting for fraction of unlabeled protein and their different affinities by using dynafit (26).
Actin Polymerization Assays.
Acanthamoeba actin doped with 10% pyrene-labeled actin was used to monitor actin polymerization by fluorescence (λex = 365 nm, λem = 407 nm) (30, 31). ATP- and ADP-actin were prepared on ice by incubating in respective nucleotides for >10 min before briefly treating twice with 5% Dowex resin to remove unbound nucleotide. Polymerization reactions were performed at room temperature. We started seeded polymerization assays by mixing prepolymerized filamentous actin seeds in 1.1× KMEI plus 110 μM nucleotide with 10 μl ATP- or ADP-actin. The Arp2/3 complex-nucleated polymerization was started by mixing 90 μl of 11 nM Arp2/3 complex plus 1.2 μM N-WASP WA plus 110 μM nucleotide in 1.1× KMEI (± filamentous actin seeds) with 10 μl doped ATP- or ADP-actin. Kinetic modeling was done by using the Macintosh version of berkeley madonna (R. Macey, T. Zahnley, and G. Oster, Univ. of California, Berkeley, CA).
Results and Discussion
The Arp2 and Arp3 Subunits of the Arp2/3 Complex Bind Nucleotide with Micromolar Affinity.
To identify the subunits of the Arp2/3 complex that bind nucleotide, we crosslinked the radiolabeled photoreactive ATP derivative [α-32P]azido-ATP to the Arp2/3 complex by using UV light. Samples were allowed to equilibrate for several minutes before crosslinking. For short crosslinking times, Arp2 is preferentially labeled, and for longer crosslinking times, labeled azido-ATP specifically crosslinks to both the Arp2 and Arp3 subunits (Fig. 1A). This finding supports the prediction of Kelleher et al. (14) that Arp2 and Arp3 contain nucleotide-binding sites similar to the nucleotide-binding pocket of conventional actin.
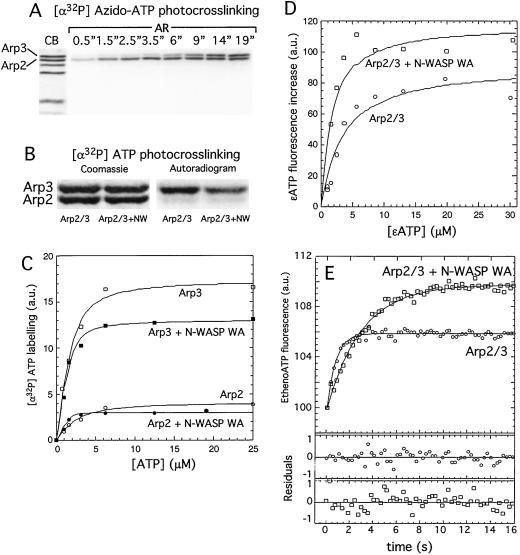
Figure 1.
Both the Arp2 and Arp3 subunits of the Arp2/3 complex bind nucleotide with micromolar affinity. (A) UV crosslinking of 5 μM [α-32P]azido-ATP to 1 μM Arp2/3 complex shows both Arp2 and Arp3 bind nucleotide and that [α-32P]azido-ATP crosslinking efficiency is greater on Arp2. (B) Crosslinking of ATP doped with [α-32P]ATP to 2 μM Arp2/3 complex by using UV light. In the presence of 100 μM ATP, radiolabeling occurs mainly on Arp3 with less efficient labeling on Arp2 both in the absence and presence of 25 μM N-WASP WA. (C) Quantitation of B for a range of ATP concentrations fit to the binding quadratic to obtain dissociation constants: Arp3 Kd = 0.6 μM; Arp2 Kd = 1.3 μM; Arp3 + N-WASP WA Kd = 0.25 μM; Arp2 + N-WASP WA Kd = 0.5 μM. Binding of N-WASP WA decreases the crosslinking efficiency. (D) Affinity of the Arp2/3 complex for etheno-ATP. Etheno-ATP fluorescence increase upon binding was measured for a range of etheno-ATP concentrations in the presence of a constant concentration of Arp2/3 complex by subtracting the contribution of free etheno-ATP fluorescence from the total measured intensity (see Materials and Methods). Arp2/3 complex alone binds etheno-ATP with an affinity of 3.1 μM, and the N-WASP–Arp2/3 complex binds etheno-ATP with an affinity of 1.3 μM. Binding of N-WASP WA enhances the etheno-ATP fluorescence signal. (E) ATP dissociation kinetics. In 5 μM ATP, nucleotide was driven from 1 μM Arp2/3 complex by mixing with 50 μM etheno-ATP, and binding kinetics were measured by fluorescence enhancement of etheno-ATP on binding. koff MgATP = 2.6 s−1. Dissociation slows in the presence of N-WASP WA koff MgATP = 0.7 s−1 and enhances the etheno-ATP fluorescence signal.
We measured the affinity of the Arp2/3 complex for nucleotide by three methods, UV crosslinking, etheno-ATP fluorescence, and nucleotide dissociation kinetics, and obtained essentially identical results. Radiolabeled nucleotides lacking the photoreactive azido group can be crosslinked to both Arp2 and Arp3 but with greatly reduced crosslinking efficiency. We measured the affinities of each of the actin-related proteins to ATP by analysis of the photocrosslinking with [α-32P]ATP. This method works because photocrosslinking with ATP crosslinks only a small fraction of the total protein (see Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org). We crosslinked [α-32P]ATP to the Arp2/3 complex with UV light and quantified labeling of Arp2 and Arp3 with increasing concentrations of [α-32P]ATP to construct a binding isotherm (Fig. 1 B and C). At saturation, Arp2 labels only 20% as strongly as Arp3, presumably reflecting a lower relative crosslinking efficiency because of different amino acid residues contacting the nucleotide (32). From this binding isotherm we determined that Arp3 binds ATP with a Kd of 0.6 μM and Arp2 with a Kd of 1.3 μM (Fig. 1C; Table 1). In the presence of the Arp2/3 complex activator N-WASP WA, the nucleotide affinity increases ≈3-fold on both subunits to 0.25 μM on Arp3 and to 0.5 μM on Arp2. In addition, the efficiency of Arp3 labeling decreases in the presence of N-WASP WA, suggesting that it induces a change in the structure of the nucleotide-binding pocket.
Table 1.
Kinetic and equilibrium constants of Arp2/3 complex for adenosine nucleotides at 25°C
| Nucleotide | Arp2/3
|
Arp2/3
+ N-WASP WA
|
Method | ||
|---|---|---|---|---|---|
| koff (s−1) | Kd (μM) | koff (s−1) | Kd (μM) | ||
| ATP | 2.6 | 0.5* | 0.7 | 0.15* | Stopped flow |
| ATP (Arp2) | 0.6 | 0.25 | 32P UV crosslinking | ||
| ATP (Arp3) | 1.3 | 0.5 | 32P UV crosslinking | ||
| ADP | 0.7 | 0.14* | 0.3 | 0.06* | Stopped flow |
| EthenoATP | 3.1 | 1.3 | Fluorescence | ||
Equilibrium constants calculated assuming kon = 5 μM−1s−1 (34).
We also measured the affinity of the entire Arp2/3 complex for the fluorescent nucleotide analog, etheno-ATP, by monitoring the increase in etheno-ATP fluorescence on binding to the Arp2/3 complex (Fig. 1D). Consistent with the UV crosslinking results, the affinity of Arp2/3 complex for etheno-ATP increases in the presence of NWASP WA from 3.1 μm to 1.3 μm. There is also an increase in the magnitude of the etheno-ATP fluorescence change in the presence of N-WASP WA, further evidence that N-WASP alters the structure of the nucleotide-binding pocket of Arp2 and/or Arp3. To test this hypothesis, we added 1.0 μM etheno-ATP to 6 μM nucleotide-free Arp2/3 complex. Under these conditions, 90% of the etheno-ATP will be bound to Arp2/3 complex. Subsequent addition of N-WASP increased etheno-ATP fluorescence 2-fold further (unpublished observations), indicating that the fluorescence enhancement is due entirely to a change in the local environment of the nucleotide. The binding of N-WASP WA has been shown to cause a conformational change on the Arp2/3 complex (33). We would expect that a conformational change might alter the residues that contact the adenosine ring of ATP or the etheno group of etheno-ATP. Such changes would explain the observed changes in the efficiency of crosslinking of [α-32P]ATP and the degree of fluorescence enhancement for etheno-ATP.
We measured the dissociation rates for nonfluorescent nucleotides by competing off bound unlabeled nucleotide with etheno-ATP (Fig. 1E), and assuming association rate constants equal to actin (34), calculated the dissociation rate constants summarized in Table 1. The Arp2/3 complex binds MgATP with an affinity of 0.5 μM, 400 times less tightly than actin binds MgATP [1.2 nM (34)], and binds MgADP 3-fold tighter than it binds MgATP. In the presence of N-WASP, the affinity for ATP increases 3–4-fold, and we note an enhancement of the fluorescence change of ≈2-fold.
The efficiency of UV crosslinking of ATP and the fluorescence of etheno-ATP both change on the binding of N-WASP to the Arp2/3 complex. To be certain that the number of nucleotides bound is the same in the presence and absence of N-WASP WA, we used HPLC to quantitate the total amount of nucleotide bound by the complex (Table 2). In buffer containing enough free ATP to saturate both the binding sites (10 μM), the Arp2/3 complex binds two nucleotides per complex, and this number does not change in the presence or absence of N-WASP WA.
Table 2.
Number of nucleotides bound per Arp2/3 complex
| Exchange buffer | Stoichiometry of bound nucleotide
|
||||
|---|---|---|---|---|---|
| Measured
|
Normalized to
actin
|
||||
| Actin (n=4) | Arp2/3 | Arp2/3 +N-WASP | Arp2/3 | Arp2/3 +N-WASP | |
| No added ATP | 0.24 | 0.21 | 0.45 | 0.4 | |
| 1.4 μM ATP | 0.76 | 0.83 | 1.4 | 1.6 | |
| 10.0 μM ATP | 0.53 | 1.2 | 1.1 | 2.3 | 2.1 |
We investigated the total number of nucleotides bound to the Arp2/3 complex by using a buffer-exchange column to rapidly exchange the Arp2/3 complex into a low concentration of ATP and measure the total nucleotide present. We heat-denatured the Arp2/3 complex to release bound nucleotide into solution, removed protein by filtration through a 10-kD MWCO filter, and analyzed nucleotide content of the filtrate by HPLC, subtracting the contribution caused by buffer alone. To correct for systematic error, results were normalized to actin.
Bound Nucleotide Affects the Affinity of the Arp2/3 Complex for Its Upstream Activators.
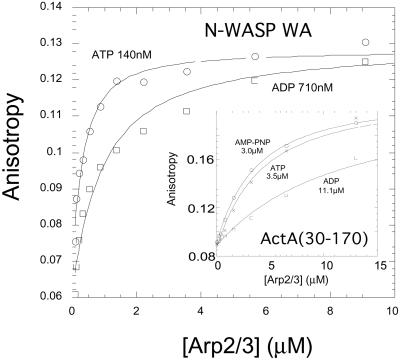
The Arp2/3 complex binds its activators N-WASP WA and ActA 3–4-fold more tightly in saturating concentrations of ATP than in ADP (Fig. 2). Rapid release of the Arp2/3 complex from its activators is a general requirement for efficient motility. In the case of Listeria motility, the Arp2/3 complex activator, ActA, is permanently attached to the bacterium while the Arp2/3 complex remains part of the actin network, which treadmills away from the bacterium surface during motility. High-resolution positional measurements (35) suggest that the speed of Listeria movement is limited by release of attachments between the bacterium and actin filaments in the tail. We know of two such attachments (i) via VASP and (ii) via the Arp2/3 complex. VASP binds to the sides of filaments with 10 nM affinity (36) and to ActA with ≈10 μM affinity (37). The Arp2/3 complex binds to the pointed ends of new filaments with 10–50 nM affinity (11) and to full-length ActA with an affinity of 0.6 μM (29). If these are the only connections between the bacterium and the actin comet tail, ActA binding to the ATP–Arp2/3 complex would be the rate-limiting interaction. If the ATP on the Arp2/3 complex is hydrolyzed to ADP on nucleation, ActA would bind the Arp2/3 complex less tightly (Fig. 2 Inset) and its effect on motility would be decreased. By this mechanism, an Arp2/3 complex activator could bind and recruit the Arp2/3 complex with high affinity and then release it before the binding retards motility. We propose that the same may be true for the endogenous WASP-family proteins. WASP and N-WASP associate with the membrane-anchored G protein Cdc42, and the affinity of N-WASP for the Arp2/3 complex also depends on the bound nucleotide (Fig. 2). Although these proteins are not permanently membrane-anchored like the transmembrane ActA protein, we hypothesize that these upstream activators remain at the membrane after activating the Arp2/3 complex, released by a decrease in affinity after nucleotide hydrolysis.
Figure 2.
The affinity of N-WASP WA and ActA for Arp2/3 complex depends on the bound nucleotide. We used polarization anisotropy to measure the affinity of nucleation promoting factors to the Arp2/3 complex in the presence of 1 mM ATP, ADP, and AMP-PNP. Rhodamine ActA (250 nM) (30) binds MgATP-Arp2/3 complex with a kd of 3.5 μM, MgADP-Arp2/3 complex with a kd of 11.1 μM, and MgAMP-PNP-Arp2/3 complex with a kd of 3.0 μM. (Inset) Rhodamine N-WASP WA (250 nM) binds MgATP–Arp2/3 complex with a kd of 140 nM and MgADP–Arp2/3 complex with a kd of 710 nM.
The Arp2/3 Complex Requires Hydrolyzable ATP to Nucleate New Actin Filaments.
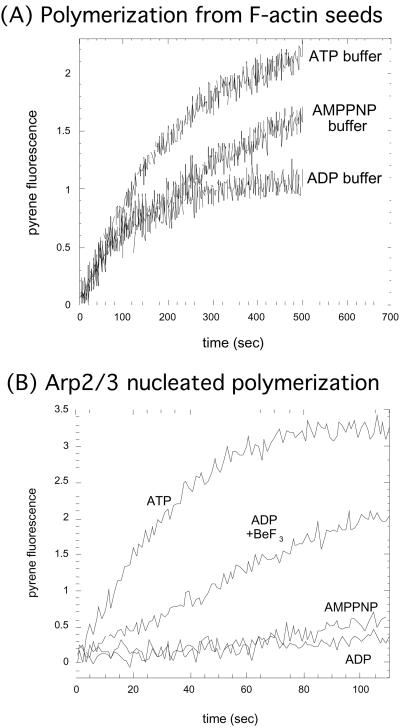
We carried out pyrene-actin polymerization assays to test the ability of N-WASP-stimulated Arp2/3 complex to nucleate new filaments when bound to different nucleotides or nucleotide analogues. Because actin itself binds nucleotide, we needed to be certain that we were observing an effect of nucleotide on Arp2/3 complex nucleation and not an effect that nucleotide in solution might have on actin polymerization. We therefore measured the elongation of 2 μM MgATP-actin with no free ATP from phalloidin-stabilized actin seeds in the presence of 100 μM ATP, ADP, and AMP-PNP (Fig. 3A). Kinetics of polymerization are independent of added nucleotide within the first 100 s of starting the reaction but diverge soon after this, because of the exchange of nucleotide on the monomeric actin with nucleotide in solution (38). Therefore, in subsequent experiments with the Arp2/3 complex, we limited our observations to the first 100 s of the reaction. We used N-WASP WA at 3 times its saturating concentration for the ATP-Arp2/3 complex to be certain that we were not simply seeing an effect of nucleotide on N-WASP affinity. As expected in this 100-s time window, the ATP–Arp2/3 complex activated by N-WASP WA rapidly nucleates new actin filaments (Fig. 3B). Our data show that when bound to ADP, the Arp2/3 complex is unable to nucleate new actin filaments (Fig. 3B). We next wanted to test the ability of the Arp2/3 complex to nucleate new filaments in the presence of the nonhydrolyzable ATP analog, AMP-PNP. AMP-PNP-bound Arp2/3 complex has an affinity for ActA (30) that is similar to that of the ATP-bound Arp2/3 complex (Fig. 2). We found that AMP-PNP-bound Arp2/3 complex is unable to nucleate new filaments (Fig. 3B). This finding is consistent with our previous results showing that G protein-stimulated Arp2/3 complex-dependent actin polymerization in Acanthamoeba extracts requires hydrolyzable ATP (39). We then tested whether ADP-BeF3, which simulates an ADP-Pi conformation for actin (21, 40), supports Arp2/3 complex activation. Adding BeF3 to the ADP–Arp2/3 complex partially restores nucleation activity (Fig. 3B). These results suggest either (i) that the ADP-Pi-bound Arp2/3 complex is the active form and AMP-PNP Arp2/3 complex is inactive, because AMP-PNP cannot be hydrolyzed, or (ii) that the ATP-bound Arp2/3 complex is the active form and that ADP-BeF3 looks more like ATP to the complex than AMP-PNP. We favor the first possibility because mathematical modeling of the kinetics of Arp2/3 complex activation seem to require a hydrolysis step (below).
Figure 3.
Actin nucleation by Arp2/3 complex requires hydrolyzable ATP. (A) As a control, we tested elongation of 2 μM MgATP-actin from F-actin seeds in the presence of 100 μM ATP, ADP, and AMP-PNP. Kinetics of polymerization are independent of solution nucleotide within the first 100 s of starting the reaction but diverge soon after this, because of the exchange of nucleotide with solution. We therefore limited our observations to the first 100 s when using ATP-actin. (B) The Arp2/3 complex induces polymerization of MgATP-actin in the presence of ATP or ADP-BeF3 but not AMP-PNP or ADP. We used the fact that nucleotide in solution exchanges onto Arp2/3 complex within seconds (Fig. 1E) but takes much longer to exchange onto actin monomers (38) to load one type of nucleotide onto the Arp2/3 complex, but keep ATP bound to actin. We mixed 2 μM MgATP-actin with a solution of 10 nM Arp2/3 complex and 1.2 μM N-WASP WA (3-fold above saturating concentration) with 100 μM ATP, ADP, ADP + 2 mM BeF3, or AMP-PNP.
The Hydrolysis of ATP by the Arp2/3 Complex May Account for the First-Order Activation Step in Arp2/3 Complex-Dependent Actin Assembly.
We propose that the Arp2/3 complex must hydrolyze ATP to nucleate a new filament. Marchand et al. (41) and Zalevsky et al. (33) noticed that affinity of wild-type and mutant WASP family proteins for the Arp2/3 complex does not correlate with ability to activate the Arp2/3 complex and suggested that some further activation step(s) must occur between activator binding and filament nucleation (41). ATP hydrolysis may be the rate-limiting activation step in nucleation. ATP hydrolysis is a first-order reaction and we can incorporate this hydrolysis step into a kinetic scheme for filament formation:
 |
where F is F-actin, A is Arp2/3 complex, N is N-WASP, G is G-actin, and the F.AADP-Pi.N.G form is the active nucleus. We fixed the association constants for the components in the reaction (here grouped together as ka) to determined values (11). We fit this scheme to polymerization data collected during N-WASP-stimulated pyrene-actin polymerization experiments and calculated the hydrolysis rate, khyd, for the Arp2/3 complex during the polymerization reaction to be 0.02 s−1. To achieve this ATP hydrolysis rate, the Arp2/3 complex must be stimulated by some combination of filamentous actin, monomeric actin, and N-WASP.
N-WASP and F-Actin Do Not Stimulate ATP Hydrolysis by Arp2/3 Complex.
To investigate the basal ATP hydrolysis rate of the Arp2/3 complex, we incubated the Arp2/3 complex with 500 μM ATP doped with [α-32P]ATP and quantified the amount of [α-32P]ADP produced over time by TLC. We were unable to detect ATP hydrolysis within the limits of our measurement, suggesting that the basal Arp2/3 complex ATP hydrolysis rate is less than 0.1 × 10−3 s−1 (data not shown). We could not use the TLC method to directly measure the ATPase rate of Arp2/3 complex in the presence of F-actin, N-WASP, and actin monomers, because the actin monomers themselves would hydrolyze ATP as they polymerize. We measured Arp2/3 complex ATPase activity in the presence of N-WASP and F-actin in the absence of the actin monomers but found no significant ATPase activity above control levels (data not shown). Because adding F-actin and N-WASP to Arp2/3 complex does not change the hydrolysis rate, and hydrolysis seems to occur during activation, we suggest that actin monomers may be required to stimulate Arp2/3 complex ATPase activity. All Arp2/3 complex activators that promote rapid nucleation contain actin monomer-binding WH2 domains in close proximity to an Arp2/3 complex-binding domain. We suggest the role of the WH2 domain may be to place a monomer in contact with a filament-bound Arp2/3 complex and stimulate ATP hydrolysis. Hydrolysis of ATP to an ADP-Pi state would then cause a conformational change that allows Arp2, and an actin monomer Arp3, to associate and form a nucleus.
Model for the Link Between ATP Hydrolysis and Actin Nucleus Formation.
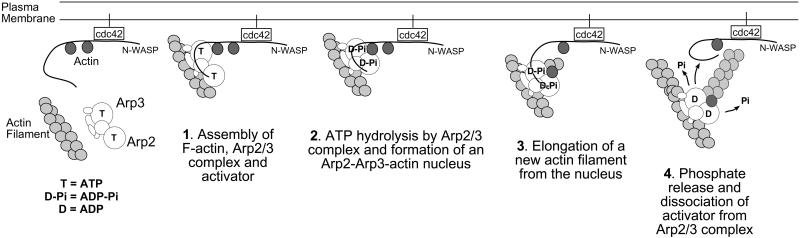
Based on our data, we propose a model (Fig. 4) relating Arp2/3 complex ATPase activity to its actin filament nucleation activity. Step 1: Arp2/3 complex binds to the activator with high affinity, because Arp2/3 complex is ATP-bound. Binding N-WASP brings the actin monomer attached to the WH2 domain of N-WASP in contact with the Arp2/3 complex and this stimulates ATP hydrolysis. Step 2: Hydrolyzing ATP to ADP-Pi causes a conformational change on the complex, forming a stable nucleus among Arp3, Arp2, and the conventional actin monomer. Step 3: A new actin filament polymerizes from this nucleus. Step 4: Phosphate release from Arp2/3 complex decreases the affinity for N-WASP and allows the Arp2/3 complex to release its membrane-associated activator.
Figure 4.
Model for the role of Arp2/3 complex nucleotide binding and hydrolysis in the formation of new actin filaments. Step 1: Assembly of F-actin, Arpd/3 complex, and activator. The Arp2/3 complex binds to the activator, in this case N-WASP, with high affinity because the Arp2/3 complex is ATP-bound. Binding N-WASP brings the actin monomer attached to the WH2 domain of N-WASP in contact with the Arp2/3 complex and this stimulates ATP hydrolysis. Step 2: Hydrolyzing ATP to ADP-Pi causes a conformational change on the complex forming a stable nucleus among Arp3, Arp2, and the conventional actin monomer. Step 3: A new actin filament polymerizes from this nucleus. Step 4: Phosphate release from the Arp2/3 complex decreases its affinity for N-WASP and allows the Arp2/3 complex to release its membrane-associated activator.
Supplementary Material
Acknowledgments
We are grateful to members of the Mullins lab for helpful discussions. Special thanks to L. Lempert for the continuous supply of media and reagents, K. Guy, T. Geistlinger, and J. Taunton for use of the HPLC, and to J. Iwasa for help harvesting Amoeba and also to O. Akin and H. Kranitz. R.D.M. dedicates this work to the memories of his friend David Scott Henderson (1964–2001) and to Edith Van Hook (1905–2001). An early version of this work was first presented in a poster at the ASCB annual meeting (2000). This work was supported by National Institutes of Health Grant GM61010-01, Pew Charitable Trust Grant P0325SC, and by Human Frontiers in Science Program Grant RG0111/2000-M (to R.D.M.). E.A.H. is supported by a National Institutes of Health postdoctoral fellowship. This work was also supported by Research Resources Program Grant 53000284 from the Howard Hughes Medical Institute to the Univ. of California at San Francisco School of Medicine.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Pollard T, Blanchoin L, Mullins R. Annu Rev Biophys Biomol Struct. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- 2.Taunton J. Curr Opin Cell Biol. 2001;13:85–91. doi: 10.1016/s0955-0674(00)00178-2. [DOI] [PubMed] [Google Scholar]
- 3.Ridley A J, Paterson H F, Johnston C L, Diekmann D, Hall A. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- 4.Ridley A J, Hall A. Cell. 1992;70:389–399. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- 5.Miki H, Suetsugu S, Takenawa T. EMBO J. 1998;17:6932–6941. doi: 10.1093/emboj/17.23.6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miki H, Sasaki T, Takai Y, Takenawa T. Nature (London) 1998;391:93–96. doi: 10.1038/34208. [DOI] [PubMed] [Google Scholar]
- 7.Rohatgi R, Ma L, Miki H, Lopez M, Kirchhausen T, Takenawa T, Kirschner M W. Cell. 1999;97:221–231. doi: 10.1016/s0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- 8.Mullins R. Curr Opin Cell Biol. 2000;12:91–96. doi: 10.1016/s0955-0674(99)00061-7. [DOI] [PubMed] [Google Scholar]
- 9.Machesky L M, Insall R H. Curr Biol. 1998;8:1347–1356. doi: 10.1016/s0960-9822(98)00015-3. [DOI] [PubMed] [Google Scholar]
- 10.Yarar D, To W, Abo A, Welch M D. Curr Biol. 1999;9:555–558. doi: 10.1016/s0960-9822(99)80243-7. [DOI] [PubMed] [Google Scholar]
- 11.Mullins R D, Heuser J A, Pollard T D. Proc Natl Acad Sci USA. 1998;95:6181–6186. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blanchoin L, Amann K J, Higgs H N, Marchand J B, Kaiser D A, Pollard T D. Nature (London) 2000;404:1007–1011. doi: 10.1038/35010008. [DOI] [PubMed] [Google Scholar]
- 13.Svitkina T M, Borisy G G. J Cell Biol. 1999;145:1009–1026. doi: 10.1083/jcb.145.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelleher J F, Atkinson S J, Pollard TD. J Cell Biol. 1995;131:385–397. doi: 10.1083/jcb.131.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mullins R D, Pollard T D. Curr Opin Struct Biol. 1999;9:244–249. doi: 10.1016/s0959-440x(99)80034-7. [DOI] [PubMed] [Google Scholar]
- 16.Bork P, Sander C, Valencia A. Proc Natl Acad Sci USA. 1992;89:7290–7294. doi: 10.1073/pnas.89.16.7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Korn E D, Carlier M F, Pantaloni D. Science. 1987;238:638–644. doi: 10.1126/science.3672117. [DOI] [PubMed] [Google Scholar]
- 18.Maciver S K, Zot H G, Pollard T D. J Cell Biol. 1991;115:1611–1620. doi: 10.1083/jcb.115.6.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Belmont L D, Orlova A, Drubin D G, Egelman E H. Proc Natl Acad Sci USA. 1999;96:29–34. doi: 10.1073/pnas.96.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liberek K, Skowyra D, Zylicz M, Johnson C, Georgopoulos C. J Biol Chem. 1991;266:14491–14496. [PubMed] [Google Scholar]
- 21.Orlova A, Egelman E H. J Mol Biol. 1992;227:1043–1053. doi: 10.1016/0022-2836(92)90520-t. [DOI] [PubMed] [Google Scholar]
- 22.Otterbein L R, Graceffa P, Dominguez R. Science. 2001;293:708–711. doi: 10.1126/science.1059700. [DOI] [PubMed] [Google Scholar]
- 23.Takeda S, McKay D B. Biochemistry. 1996;35:4636–4644. doi: 10.1021/bi952903o. [DOI] [PubMed] [Google Scholar]
- 24.MacLean-Fletcher S, Pollard T D. Cell. 1980;20:329–341. doi: 10.1016/0092-8674(80)90619-4. [DOI] [PubMed] [Google Scholar]
- 25.Biswas S B, Kornberg A. J Biol Chem. 1984;259:7990–7993. [PubMed] [Google Scholar]
- 26.Kuzmic P. Anal Biochem. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- 27.Childs K F, Ning X H, Bolling S F. J Chromatogr B Biomed Appl. 1996;678:181–186. doi: 10.1016/0378-4347(95)00474-2. [DOI] [PubMed] [Google Scholar]
- 28.Rosenfeld S S, Taylor E W. J Biol Chem. 1984;259:11920–11929. [PubMed] [Google Scholar]
- 29.Zalevsky J, Grigorova I, Mullins R. J Biol Chem. 2001;276:3468–3475. doi: 10.1074/jbc.M006407200. [DOI] [PubMed] [Google Scholar]
- 30.Mullins R, Machesky L. Methods Enzymol. 2000;325:214–237. doi: 10.1016/s0076-6879(00)25445-1. [DOI] [PubMed] [Google Scholar]
- 31.Cooper J A, Pollard T D. Methods Enzymol. 1982;85:182–210. doi: 10.1016/0076-6879(82)85021-0. [DOI] [PubMed] [Google Scholar]
- 32.Bai R, Choe K, Ewell J B, Nguyen N Y, Hamel E. J Biol Chem. 1998;273:9894–9897. doi: 10.1074/jbc.273.16.9894. [DOI] [PubMed] [Google Scholar]
- 33.Zalevsky, J., Lempert, L., Kranitz, H. & Mullins, R. D. (2001) Curr. Biol., in press. [DOI] [PubMed]
- 34.De La Cruz E, Pollard T D. Biochemistry. 1995;34:5452–5461. doi: 10.1021/bi00016a016. [DOI] [PubMed] [Google Scholar]
- 35.Kuo S C, McGrath J L. Nature (London) 2000;407:1026–1029. doi: 10.1038/35039544. [DOI] [PubMed] [Google Scholar]
- 36.Laurent V, Loisel T P, Harbeck B, Wehman A, Grobe L, Jockusch B M, Wehland J, Gertler F B, Carlier M F. J Cell Biol. 1999;144:1245–1258. doi: 10.1083/jcb.144.6.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ball L J, Kuhne R, Hoffmann B, Hafner A, Schmieder P, Volkmer-Engert R, Hof M, Wahl M, Schneider-Mergener J, Walter U, et al. EMBO J. 2000;19:4903–4914. doi: 10.1093/emboj/19.18.4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Selden L A, Kinosian H J, Estes J E, Gershman L C. Biochemistry. 1999;38:2769–2778. doi: 10.1021/bi981543c. [DOI] [PubMed] [Google Scholar]
- 39.Mullins R D, Pollard T D. Curr Biol. 1999;9:405–415. doi: 10.1016/s0960-9822(99)80187-0. [DOI] [PubMed] [Google Scholar]
- 40.Combeau C, Carlier M-F. J Biol Chem. 1988;263:17429–17436. [PubMed] [Google Scholar]
- 41.Marchand J B, Kaiser D A, Pollard T D, Higgs H N. Nat Cell Biol. 2001;3:76–82. doi: 10.1038/35050590. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.