Abstract
Neurodevelopmental disorders offer insight into synaptic mechanisms. To unbiasedly uncover these mechanisms, we studied the 22q11.2 syndrome, a recurrent copy number variant, which is the highest schizophrenia genetic risk factor. We quantified the proteomes of 22q11.2 mutant human fibroblasts from both sexes and mouse brains carrying a 22q11.2-like defect, Df(16)A+/−. Molecular ontologies defined mitochondrial compartments and pathways as some of top ranked categories. In particular, we identified perturbations in the SLC25A1-SLC25A4 mitochondrial transporter interactome as associated with the 22q11.2 genetic defect. Expression of SLC25A1-SLC25A4 interactome components was affected in neuronal cells from schizophrenia patients. Furthermore, hemideficiency of the Drosophila SLC25A1 or SLC25A4 orthologues, dSLC25A1-sea and dSLC25A4-sesB, affected synapse morphology, neurotransmission, plasticity, and sleep patterns. Our findings indicate that synapses are sensitive to partial loss of function of mitochondrial solute transporters. We propose that mitoproteomes regulate synapse development and function in normal and pathological conditions in a cell-specific manner.
SIGNIFICANCE STATEMENT We address the central question of how to comprehensively define molecular mechanisms of the most prevalent and penetrant microdeletion associated with neurodevelopmental disorders, the 22q11.2 microdeletion syndrome. This complex mutation reduces gene dosage of ∼63 genes in humans. We describe a disruption of the mitoproteome in 22q11.2 patients and brains of a 22q11.2 mouse model. In particular, we identify a network of inner mitochondrial membrane transporters as a hub required for synapse function. Our findings suggest that mitochondrial composition and function modulate the risk of neurodevelopmental disorders, such as schizophrenia.
Keywords: 22q11.2 microdeletion, mitochondria, schizophrenia, SLC25A1, SLC25A4, synapse
Introduction
Single gene defects associated with neurodevelopmental disorders provide a fertile ground to uncover fundamental synaptic mechanisms. For example, mutations in FMR1, MECP2, DISC1, or NRXN1 associate with diverse mental and/or behavioral disorders, including autism spectrum disorder and schizophrenia. Understanding molecular mechanisms linking these single gene defects with pathways that impinge on synapse function has been significantly advanced (Ishizuka et al., 2006; Santoro et al., 2012; Bena et al., 2013; Wen et al., 2014; Sztainberg and Zoghbi, 2016). This fact is founded on well established experimental paradigms that identify and test causality between a single gene defect, its downstream molecular mechanisms, and phenotypes. In contrast, there are a great number of neurodevelopmental disorders that associate with chromosomal microdeletions, in particular, hemizygous deletions containing multiple contiguous genes. Microdeletions have received great attention as they are the most penetrant and frequent genetic defects linked to neurodevelopmental disorders (Girirajan et al., 2011; Malhotra and Sebat, 2012; Sullivan et al., 2012; Kirov, 2015; Rutkowski et al., 2017). Compared with monogenic disorders, the study of microdeletions is impeded by the lack of experimental paradigms that comprehensively capture contributions of all genes within the hemideletion to downstream molecular mechanisms and phenotypes (Iyer et al., 2018). Thus, the identity of molecular mechanisms downstream a whole microdeletion and their phenotypic impact in synapses remains elusive. Here we address this issue focusing on the 22q11.2 microdeletion syndrome.
The 22q11.2 microdeletion syndrome (OMIM #192430, #188400; McDonald-McGinn et al., 2015) is the strongest and most prevalent genetic risk factor for schizophrenia increasing the overall risk of psychiatric pathology 20- to 25-fold compared with the general population (Bassett et al., 2000; Hodgkinson et al., 2001; Bassett and Chow, 2008). Twenty five percent of 22q11.2 patients develop schizophrenia. In addition, the 22q11.2 microdeletion is the most common genetic defect found in sporadic cases of schizophrenia (Bassett et al., 2003; Bassett and Chow, 2008; International Schizophrenia Consortium, 2008; Karayiorgou et al., 2010; Jonas et al., 2014; Schneider et al., 2014; Hoeffding et al., 2017; Marshall et al., 2017). The strong association of mental and/or behavioral disorders with the 22q11.2 genetic defect makes this syndrome a robust model to test new experimental paradigms to identify molecular pathways and synaptic mechanisms downstream complex neurodevelopmental genetic defects.
We studied the most prevalent 22q11.2 microdeletion in humans, which encompasses three megabases. This microdeletion creates an haploinsufficiency of 46 protein coding genes and 17 regulatory small RNAs, thus opening the door for multiple pathways and organelles that could be affected downstream (Guna et al., 2015). We reasoned that top-ranked molecular ontologies associated with the 22q11.2 genetic defect should enrich pathways and organelles implicated in mechanisms affecting synapse function and thus contribute to psychiatric phenotypes in humans. Using genealogical and integrated mass spectrometry-based proteomics, we report the unbiased and statistically prioritized identification of pathways and organelles affected by the 22q11.2 microdeletion syndrome. Our comparative systems biology studies interrogated the proteome of fibroblasts from human pedigrees, genealogical proteomics, and the brain of a mouse model that genotypically and phenotypically mimics the 22q11.2 syndrome, the Df(16)A+/− deficiency (Karayiorgou et al., 2010). We conclude that the mitochondrion is a top-ranked organelle affected in the 22q11.2 microdeletion syndrome. We propose that mitoproteomes modulate synapse development and function in normal and pathological states.
Materials and Methods
Cell lines and culture conditions
Pedigrees of Ch22q11.2 fibroblasts were obtained from RUCDR Infinite Biologics repository (RUID:MH0162519; RUID:MH0162508, MH0162509; RUID:MH0162499; RUID:MH0162510; RUID:MH0162511; RUID:MH0162626; RUID:MH0162636; RUID:MH0162627; RUID:MH0162628; RUID:MH0162673, MH0162674; RUID:MH0162675, MH0162676; RUID:MH0162677; RUID:MH0162678). The fibroblasts were grown according to supplier recommendations in DMEM (Corning, 10-013-CV) media supplemented with 15% fetal bovine serum (FBS; Atlanta Biologicals, S12450) and 100 μg/ml penicillin and streptomycin (Hyclone, SV30010) at 37°C in 5% CO2. SH-SY5Y cells (ATCC, CRL-2266; RRID:CVCL_0019) were cultured in DMEM media supplemented with 10% FBS and 100 μg/ml penicillin and streptomycin at 37°C in 10% CO2. The SH-SY5Y cells were stably transfected either with a control empty vector (GeneCopoeia, EX-NEG-Lv102) or ORF expression clone for N terminally tagged FLAG-SLC25A1 (GeneCopoeia, EX-A1932-Lv1020GS) and grown in a selection media containing DMEM media supplemented with 10%FBS and Puromycin 2 μg/ml (Invitrogen, A1113803). HEK293-Flp-In-pCDNA5/FRT-CNAP-Ant1/Ant2 (SLC25A4/SLC25A5) cells were previously described (Lu et al., 2017). The cells were grown in DMEM media with 10%FBS and 100 μg/ml hygromycin (Invitrogen, 10687010). HAP1 cell lines– Control (C631), SLC25A1 knock-out cell lines (HZGHC001753c003 and HZGHC001753c010), and SLC25A4 knock-out cell line (HZGHC000778c011) were obtained from Horizon (RRID:CVCL_5G07; RRID:CVCL_TM04; RRID:CVCL_TM05; RRID:CVCL_TM45). HAP1 cells were cultured in IMDM media (Lonza, 12-722F) supplemented with 10% FBS and 100 μg/ml penicillin and streptomycin at 37°C in 10% CO2.
Drosophila husbandry and stocks
Drosophila stocks were reared at 25°C in a humidified incubator (Shel Lab, SR120PF) with a 12 h light/dark cycle and fed standard molasses food (900 ml milli-Q water, 48 g active dry yeast, 120 g cornmeal, 9 g agar, 120 g molasses, 2.4 g tegosept, 9ml propionic acid). The following stocks were used w[1118] (BDSC, catalog #3605; RRID:BDSC_3605), C155-GAL4 (P{w[+mW.hs]=GawB}elav[C155]; RRID:BDSC_458), Ddc-GAL4 (w[1118];P{w[+mC]=Ddc-GAL4.L}Lmpt[4.36]; RRID:BDSC_7009) were obtained from the Bloomington Drosophila Stock Center. Gal4 lines used: c739 (α/βKC), NP1131 (γKC), R27G01 (MBON-γ5β'2a), R71D08(V2), G0431 (DAL). R27G01 (RRID:BDSC_49233), G0239 (12639), G0431 (12837), and UAS-2xeGFP (RRID:BDSC_6874) were ordered from the Bloomington Stock Center. NP1131-Gal4 was ordered from the DGRC Stock center. R71D08 was a kind gift from Dr. H. Tanimoto, Max-Planck-Institut für Neurobiologie, 3-86-Gal4 was a kind gift from Dr. U. Heberlein, HHMI Janelia, and c739-Gal4 was a kind gift from Dr. A. Sehgal, University of Pennsylvania. All Gal4 lines were crossed with UAS-2xeGFP to allow for cell harvesting.
yw;;UAS-GFP-AP4mito55 (BSC 25748) and;;seaΔ24/TM3, Sb,Dfd::YFP (gift from Giovanni Cenci, Università dell'Aquila and Jason Tennessen, Indiana University). c155-GAL4;; flies were crossed to yw;;UAS-GFP-AP4mito55. Progeny were then crossed to either:;;seaΔ24/TM3,Sb,Dfd::YFP or sesBorg;; and larval progeny selected for GFP expression before dissection.
Human subjects
Seventy-seven patients with a molecularly confirmed diagnosis of 22q11DS and 50 unaffected, demographically matched healthy controls who were part of an ongoing longitudinal study at the University of California, Los Angeles (UCLA) were included in the current analyses. 22q11DS participants were recruited from posts to 22q11DS/Velocardiofacial online foundations and flyers through contacts with local craniofacial or genetics clinics. Controls were recruited from flyers posted at local schools and community centers. The study was approved by the UCLA Institutional Review Board and performed in accordance with the Declaration of Helsinki. All subjects or their legal guardians provided written informed consent and/or assent. This cohort has been previously published (Jalbrzikowski et al., 2015)
All data from the studies performed in postmortem human brain tissue have been previously published (Arion et al., 2015; Enwright Iii et al., 2018), and all methods and materials descriptions and data are publicly available (Arion et al., 2015; Enwright Iii et al., 2018).
Antibodies
Antibodies used for immunoblots were as follows: SLC25A1 (Proteintech, 15235-1-AP; RRID:AB_2254794), SLC25A4 (1F3F11, a gift from the Claypool laboratory, Johns Hopkins University), β-Actin (Sigma-Aldrich, A5441; RRID:AB_476744), HSP90 (BD Biosciences, 610418; RRID:AB_397798), TFRC (Invitrogen, 13-6800; RRID:AB_86623), FLAG (Bethyl, A190-102A; RRID:AB_67407). Blotting secondary antibodies were against mouse or rabbit conjugated to HRP (ThermoFisher Scientific, A10668; RRID:AB_2534058; RRID:AB_2536530).
Cell lysis and immunoprecipitation
Cells intended for immunoprecipitation (Control HAP1 cells, HAP1 with SLC25A1/SLC25A4 knockdowns, SH-SY5Y empty vector or SH-SY5Y transfected with FLAG-SLC25A1 or HEK293-Flp-In-pCDNA5/FRT-CNAP-Ant1/Ant2 cells) were placed on ice, rinsed twice with ice-cold PBS (Corning, 21-040-CV) containing 0.1 mm CaCl2 and 1.0 mm MgCl2. The cells were then rinsed twice with PBS and lysed in buffer A (in mm: 150 NaCl, 10 HEPES, 1 EGTA, and 0.1 MgCl2, pH 7.4) with 0.5% Triton X-100 and Complete anti-protease (Roche, 11245200). Cells were scraped from the dish, placed in Eppendorf tubes, and followed by incubation for 30 min on ice. Cell homogenates were then centrifuged at 16,100 × g for 10 min and the clarified supernatant was recovered. Protein concentration determined using the Bradford Assay (Bio-Rad, 5000006). For immunoprecipitation, 500 μg of protein extract was incubated with 30 ml Dynal magnetic beads (Invitrogen, 110.31) coated with antibody, and incubated for 2 h at 4°C. In some cases, immunoprecipitations were done in the presence of the antigenic 3× FLAG peptide (340 μm; Sigma-Aldrich, F4799) as a control. The beads were then washed 4–6 times with buffer A with 0.1% Triton X-100. Proteins were eluted from the beads with Laemmli buffer. Samples were resolved by SDS-PAGE and contents analyzed by immunoblot described below.
Quantitative mass spectrometry
Proteomic services were provided by MS Bioworks (http://www.msbioworks.com/) or the Emory Integrated Proteomics Core (https://www.cores.emory.edu/eipc/).
Stable isotope labeling of amino acids
Ch22q11.2 fibroblasts were labeled using published protocols. Cells were cultured in DMEM with either “light” unlabeled arginine and lysine amino acids (R0K0; Dundee Cell Products, LM014) “medium” 13C- and 15N-labeled arginine, and 13C- and 15N-labeled lysine amino acids (R6K4; Dundee Cell Products, LM016) or “heavy” 13C- and 15N-labeled arginine, and 13C- and 15N-labeled lysine amino acids (R10K8; Dundee Cell Products, LM015) supplemented with 15% FBS (Dundee Cell Products, DS1003) and 100 μg/ml penicillin and streptomycin. Each cell line was grown for seven passages allowing maximum incorporation (at least 97.5%) of the amino acids in the total cellular pool. Cellular lysate samples were prepared, as described in the previous section. Quantitative mass spectrometry was performed as described previously using the services of MS Bioworks and the Emory Integrated Proteomics Core.
The stable isotope-labeled amino acids in cell culture (SILAC)-labeled samples were pooled 1:1:1 and 20 μg of this mix was resolved on a 4–12% Bis-Tris Novex mini-gel (Invitrogen) using the MOPS buffer system. The gel was stained with Coomassie and the lanes excised into 40 equal sections using a grid. Gel pieces were robotically processed (ProGest, Digilab) by first washing with 25 mm ammonium bicarbonate (ABC) followed by acetonitrile, followed by reduction with 10 mm dithiothreitol at 60°C, alkylation with 50 mm iodoacetamide at room temperature. Pieces were digested with trypsin (Promega) at 37°C for 4 h and quenched with formic acid. The supernatant was analyzed directly without further processing. Gel digests were analyzed by nano LC/MS/MS with a Waters NanoAcquity HPLC system interfaced to a ThermoFisher Q Exactive. Peptides were loaded on a trapping column and eluted over a 75 μm analytical column at 350 nl/min; both columns were packed with Jupiter Proteo resin (Phenomenex). The mass spectrometer was operated in data-dependent mode, with MS and MS/MS performed in the Orbitrap at 70,000 FWHM resolution and 17,500 FWHM resolution, respectively. The 15 most abundant ions were selected for MS/MS. Data were processed through the MaxQuant software 1.4.1.2 which served the following functions: (1) Recalibration of MS data. (2) Filtering of database search results at the 1% protein and peptide false discovery rate (FDR). (3) Calculation of SILAC heavy–light ratios. Data were searched using a local copy of Andromeda with the following parameters: enzyme, trypsin; database: Swissprot (concatenated forward and reverse plus common contaminants); fixed modification, Carbamidomethyl (C); variable modifications: oxidation (M), acetyl (protein N-term), 13C6/15N2 (K), 13C6/15N4 (R), 4H2 (K), 13C6 (R); fragment mass tolerance: 20 ppm. Pertinent MaxQuant settings were as follows: peptide FDR 0.01, protein FDR 0.01, min peptide length 7, min unique peptides 0, min ratio count 2, re-quantify TRUE, second peptide TRUE.
Label-free quantitation and cellular preparation
Cells were grown in 10 cm dishes to 85–90% confluency. On the day of the experiment the cells were placed on ice and washed three times with PBS supplemented with 10 mm EDTA (Sigma-Aldrich, 150-38-9) for 3 min each. After the third wash, the cells were incubated with PBS and 10 mm EDTA for 30 min on ice. Cells were then lifted with mechanical agitation using a 10 ml pipette and collected in a 15 ml falcon tube. Cells were then spun at 800 × g for 5 min at 4°C. The supernatant was then aspirated out and the remaining pellet was washed with ice-cold PBS. The resuspended cells were then centrifuged at 16,100 ×g for 5 min. The supernatant was discarded and the resulting pellet was immediately frozen on dry ice for at least 5 min and stored at −80°C for future use.
Cell pellets were lysed in 200 μl of urea lysis buffer (8 m urea, 100 mm NaH2PO4, pH 8.5), supplemented with 2 μl (100× stock) HALT protease and phosphatase inhibitor cocktail (Pierce). Lysates were then subjected to three rounds of probe sonication. Each round consisted of 5 s of activation at 30% amplitude and 15 s of rest on ice. Protein concentration was determined by bicinchoninic acid analysis and 100 μg of each lysate was aliquoted and volumes were equilibrated with additional lysis buffer. Aliquots were diluted with 50 mm ABC and was treated with 1 mm DTT and 5 mm iodoacetamide (IAA) in sequential steps. Both steps were performed at room temperature with end to end rotation for 30 min. The alkylation step with IAA was performed in the dark. Lysyl endopeptidase (Wako) was added at a 1:50 (w/w) enzyme–protein ratio and the samples were digested for overnight. The following morning, a 50 μg aliquot was taken out, acidified to a final concentration of 1% formic acid and stored. Trypsin (Promega) was added to the residual 50 μg aliquot at a 1:100 (w/w) and digestion was allowed to proceed overnight again. Resulting peptides from both digestions rounds were desalted with a Sep-Pak C18 column (Waters).
Dried peptide fractions were resuspended in 100 μl of peptide loading buffer (0.1% formic acid, 0.03% trifluoroacetic acid, 1% acetonitrile). Peptide mixtures were separated on a self-packed C18 (1.9 μm Dr. Maisch, Germany) fused silica column (25 × 75 μm internal diameter; New Objective) by mass spectrometer platforms: (1) Dionex Ultimate 3000 RSLCNano coupled to a Fusion orbitrap tribrid mass spectrometer (ThermoFisher Scientific) and (2) Waters NanoAcquity coupled to a Q-Exactive Plus hybrid mass spectrometer (ThermoFisher Scientific). For the Fusion system, 2 μl was loaded and elution was performed over a 140 min gradient at a rate of 300 nl/min with buffer B ranging from 3% to 99% (buffer A: 0.1% formic acid in water; buffer B: 0.1% formic in acetonitrile). The mass spectrometer cycle was programmed to collect at the top speed for 5 s cycles consisting of 1 MS scan (400–1600 m/z range; 200,000 AGC; 50 ms maximum ion time) were collected at a resolution of 120,000 at m/z 200 in profile mode followed by ion trap collected HCD MS/MS spectra (0.7 m/z isolation width; 30% collision energy; 10,000 AGC target; 35 ms maximum ion time). Dynamic exclusion was set to exclude previous sequenced precursor ions for 20 s within a 10 ppm window. Precursor ions with +1 and +8 or higher charge states were excluded from sequencing. For the Q-Exactive Plus system, 4 μl was loaded and elution was performed over a 140 min gradient at a rate of 250 nl/min with buffer B ranging from 3 to 80% ACN. The mass spectrometer was set to acquire 1 MS scan (70,000 resolution at 200 m/z in profile mode; 300–1800 m/z range; 1,000,000 AGC; 100 ms maximum ion time) followed by at most 10 MS/MS scans (17,500 resolution at 200 m/z; 2.0 m/z isolation width with an offset of 0.5 m/z; 50,000 AGC; 50 ms maximum ion time). Dynamic exclusion was for 30 s with a 10 ppm window.
All spectra from both platforms were loaded into Maxquant v1.5.2.8 and searched against a database downloaded from the NCBI's REFSEQ v54 with common contaminants appended. Search parameter included fully tryptic (or lysyl endopeptidase) cleavage, variable modifications for protein N-terminal acetylation and methionine oxidation, static modifications for cysteine carbamindomethyl, 20 ppm precursor mass tolerance, 0.5 Da for ion trap and 0.05 Da for Orbitrap product ion tolerances, FDR at 1% for all levels including protein, peptide, and peptide-spectrum match.
Tandem mass tagging
Cell pellets were lysed, reduced, alkylated and digested similarly as with the label-free quantitation (LFQ) protocol with the only differences being that 50 mm triethylammonium bicarbonate (TEAB) was used for dilution and only lysyl endopeptidase was used for digestion. An aliquot equivalent to 10 μg of total protein was taken out of each sample and combined to obtain a global internal standard (GIS) use later for tandem mass tagging (TMT) labeling.
TMT labeling was performed according to the manufacturer's protocol. Briefly (Ping et al., 2018), the reagents were allowed to equilibrate to room temperature. Dried peptide samples (90 μg each) were resuspended in 100 μl of 100 mm TEAB buffer (supplied with the kit). Anhydrous acetonitrile (41 μl) was added to each labeling reagent tube and the peptide solutions were transferred into their respective channel tubes. The reaction was incubated for 1 h and quenched for 15 min afterward with 8 μl of 5% hydroxylamine. All samples were combined and dried down. Peptides were resuspended in 100 μl of 90% acetonitrile and 0.01% acetic acid. The entire sample was loaded onto an offline electrostatic repulsion–hydrophilic interaction chromatography fractionation HPLC system and 40 fractions were collected over a time of 40 min. The fractions were combined into 10 and dried down. Dried peptide fractions were resuspended in 100 μl of peptide loading buffer (0.1% formic acid, 0.03% trifluoroacetic acid, 1% acetonitrile). Peptide mixtures (2 μl) were separated on a self-packed C18 (1.9 μm Dr. Maisch, Germany) fused silica column (25 × 75 μm internal diameter; New Objective) by a Dionex Ultimate 3000 RSLCNano and monitored on a Fusion mass spectrometer (ThermoFisher Scientific). Elution was performed over a 140 min gradient at a rate of 300 nl/min with buffer B ranging from 3% to 80% (buffer A: 0.1% formic acid in water; buffer B: 0.1% formic in acetonitrile). The mass spectrometer cycle was programmed to collect at the top speed for 3 s cycles in synchronous precursor selection mode (SPS-MS3). The MS scans (380–1500 m/z range, 200,000 AGC, 50 ms maximum ion time) were collected at a resolution of 120,000 at m/z 200 in profile mode. CID MS/MS spectra (1.5 m/z isolation width, 35% collision energy; 10,000 AGC target; 50 ms maximum ion time) were detected in the ion trap. HCD MS/MS/MS spectra (2 m/z isolation width; 65% collision energy; 100,000 AGC target; 120 ms maximum ion time) of the top 10 MS/MS product ions were collected in the Orbitrap at a resolution of 60,000. Dynamic exclusion was set to exclude previous sequenced precursor ions for 30 s within a 10 ppm window. Precursor ions with +1 and +8 or higher charge states were excluded from sequencing.
MS/MS spectra were searched against human database from REFSEQ v54 and Uniprot (downloaded on 03/06/2015) with Proteome Discoverer 1.4 and 2.0 (ThermoFisher Scientific), respectively. Methionine oxidation (+15.9949 Da), asparagine, and glutamine deamidation (+0.9840 Da) and protein N-terminal acetylation (+42.0106 Da) were variable modifications (up to 3 allowed per peptide); static modifications included cysteine carbamidomethyl (+57.0215 Da), peptide N-terminus TMT (+229.16293 Da), and lysine TMT (+229.16293 Da). Only fully cleaved lysyl endopeptidase peptides were considered with up to two miscleavages in the database search. A precursor mass tolerance of ±20 ppm and a fragment mass tolerance of 0.6 Da were applied. Spectra matches were filtered by Percolator to a peptide-spectrum matches FDR of <1%. Only razor and unique peptides were used for abundance calculations. Ratio of sample over the GIS of normalized channel abundances were used for comparison across all samples.
Electrophoresis and immunoblotting
For Western blot, lysate was reduced and denatured in Laemmli buffer containing SDS and 2-mercaptoethanol and heated for 5 min at 75°C. Equal amounts of cellular lysates were loaded onto 4–20% Criterion gels (Bio-Rad, 5671094) for electrophoresis and transferred to PVDF (Millipore, IPFL00010) using the semidry transfer method. The PVDF membranes were blocked with Tris-buffered saline containing 5% nonfat milk and 0.05% Triton X-100 (TBST), rinsed and incubated overnight in presence of appropriately diluted primary antibody in antibody base solution (PBS with 3% bovine serum albumin, 0.2% sodium azide). Membranes were then washed multiple times in TBST and incubated in HRP-conjugated secondary antibody diluted 1:5000 in the blocking solution above. Following multiple washes, the membranes were then exposed to GE Healthcare Hyperfilm ECL (28906839) with Western Lightning Plus ECL reagent (PerkinElmer, NEL105001EA).
Cell line RNA extraction and quantitative RT-PCR
RNA extraction for cells and tissues was done using Trizol Reagent (Invitrogen, 15596026) following the published protocol. Total amount, concentration and purity of RNA were determined using the Bio-Rad SmartSpec Plus Spectrophotometer. First strand synthesis was completed using the Superscript III First Strand Synthesis System Kit (Invitrogen, 18080-051) using 5 μg total RNA per reaction and random hexamer primers following the manufacturer's protocol. RT-PCR was done with 1 μl cDNA from first strand synthesis in LightCycler 480 SYBR Green I Master (Roche, 04707516001) according to the manufacturer's protocol on a LightCycler 480 Instrument with 96-well format. RT-PCR protocol included an initial denaturation at 95°C for 5 min, followed by 45 cycles of amplification with a 5 s hold at 95°C ramped at 4.4°C/s to 55°C. Temperature was then held for 10 s at 55°C and ramped up to 72°C at 2.2°C/s. Temperature was held at 72°C for 20 s were a single acquisition point was collected and then ramped at 4.4°C/s to begin the cycle anew. A melting curve was collected following amplification. The temperature was then held at 65° for 1 min and ramped to 97°C at a rate of 0.11°C/s. Five acquisition points were collected per °C. Primers were designed using the IDT Real-Time qPCR Assay Entry site using site recommended parameters. Primers were obtained from Sigma-Aldrich Custom DNA Oligo service. Melting curves were used to confirm primer specificity to single transcripts. The primer list is provided in Table 1. For quantification, standard curves for each primer were applied to all samples using LightCycler 480 software. Ratios of experimental to control samples, normalized to reference genes, are reported.
Table 1.
Primers used in these studies
| Transcript | Species | Forward | Reverse |
|---|---|---|---|
| SLC25A1 | Hs | GTGTGGAAGACGGACTAAGC | ACTGGAATCGTGAGACAAAGG |
| SLC25A3 | Hs | AGGATGGTGTTCGTGGTTTG | TGTGCGCCAGAGATAAGTATTC |
| SLC25A4 | Hs | AGGGTTTCAACGTCTCTGTC | GTCACACTCTGGGCAATCAT |
| SLC25A5 | Hs | CTGATGGGATTAAGGGCCTG | ACGATGTGAGTGTTCTTGGG |
| SLC25A10 | Hs | ACTTGGTCAACGTCAGGATG | TTGCACCCGAGAACAGTC |
| SLC25A11 | Hs | CCTAAGTCCGTCAAGTTCCTG | AGCTGGTTTTGTACTCTCGAG |
| SLC25A12 | Hs | ACGCTATGGACAAGTCACAC | AGTTCTGCCAGGTTGTAAGG |
| SLC25A20 | Hs | ATCAGCCCGCTCAAGAAC | GTCAAAGGTCCCAGAGTACATG |
| SLC25A24 | Hs | TCTCGAACAAGCACTGCC | TGTACCATTTCCCCTCCAAAG |
| SLC25A25 | Hs | GGCTGGTGTTTAAGAGTTTGG | TGGTCATCGTGCCGTTTT |
| VAMP3 | Hs | TTGAGGTAGACTCTGACCGTCTC | GCTGGAGTCCACAGCTGATAAT |
| VIM | Hs | CGTGAATACCAAGACCTGCTC | GGAAAAGTTTGGAAGAGGCAG |
| αTub84B | Dm | TGTCGCGTGTGAAACACTTC | AGCAGGCGTTTCCAATCTG |
| Sea | Dm | CCTAAGTCAGCGGCGAGA | CCACGATGGCCTCACATAC |
| SesB | Dm | TTGTCTACCCCTTGGACTTTG | CTGACCACCCTTGCCAGT |
| MCU | Dm | TCCTGCACCATCGAAAGC | CCAATGTGCGATTGTTGATT |
| SERCA | Dm | CGAAATGTTGAACGCAATGA | AAGGAGAGTGCCATTGATCC |
Drosophila neuromuscular microscopy
Neuromuscular junction staining was performed using late third instar larvae. Larval body wall dissections using a dorsal incision were performed with 10 mm cell culture dishes partially filled with charcoal infused SYLGARD, microdissection pins, forceps, and microdissection scissors. Drosophila were dissected using in standard Ca2+ free HL3 Ringer's solution (in mm:70 NaCl, 5 KCl, 21.5 MgCl2, 10 NaHCO3, 5 trehalose, 115 sucrose, 5 BES, pH 7.2–7.3), fixed using 4% paraformaldehyde for 45 min to 1 h at room temp, rinsed 10 min with PBS-T (PBS+.15% Triton), incubated in FITC-HRP conjugate (MP Biomedicals, 0855977) overnight at 4C. Rinses followed the next day in PBS-T at 3× 1 min rinse then 3× 10 min rinse, and finished with a 3× 1 min rinse in PBS. Larval body wall preparations were then placed on slides with a drop of VECTASHIELD and coverslip. Nail polish was used to seal the edges of the coverslip in place and samples were stored at 4°C until imaged. Confocal images were obtained using a Zeiss LSM 510 microscope and Zen 2009 software. NMJs from 6/7 muscles of the third or fourth segments were identified and z-stack images collected with a continuous wave 458,488 nm argon laser at 200 mW. Z-stacks were converted to jpegs using FIJI software and blinded for bouton quantification.
For mitochondrial stainings, third instar larvae were dissected in normal HL3 (Ca2+ free) and fixed in 4% paraformaldehyde at 25°C for 45 min to 1 h. Samples were briefly rinsed with PBS-T (PBS + 0.15% Triton-X) for 10 min. Primary antibody was then applied to the samples overnight at 4°C. Primary antibody consisted of rabbit anti-GFP at 1:1000 (Sysy 132002; RRID:AB_887725), Cy3-HRP at 1:1000 (Santa Cruz Biotechnology, 166894 HRP; RRID:AB_10614143), and AlexaFluor 633-phalloidin at 1:500 (Invitrogen, A22284) in PBS-T. Samples were then rinsed in PBS-T. Secondary antibody was applied for 2 h at 25°C. Secondary antibody consisted of AlexaFluor 488-goat anti-rabbit at 1:1000 (Invitrogen, 10453272) plus additional Cy3-HRP at 1:1000 and AlexaFluor 633-phalloidin at 1:500 in PBS-T. NMJs were imaged on an Olympus FV1000 Confocal Microscope at 20×. Oib files were converted to jpegs using Fiji software (RRID:SCR_002285). Eight-bit jpegs were cropped to selected ROIs. Threshold was then adjusted to accurately highlight the fluorescent signal of both of the mitochondrial (GFP) and neuronal (Cy3) images for each NMJ. The Create Selection command was then used to outline the fluorescence in each image and measured using the ROI Manager tool. GFP/Cy3 signal ratios were then calculated for each genotype.
Drosophila electrophysiology
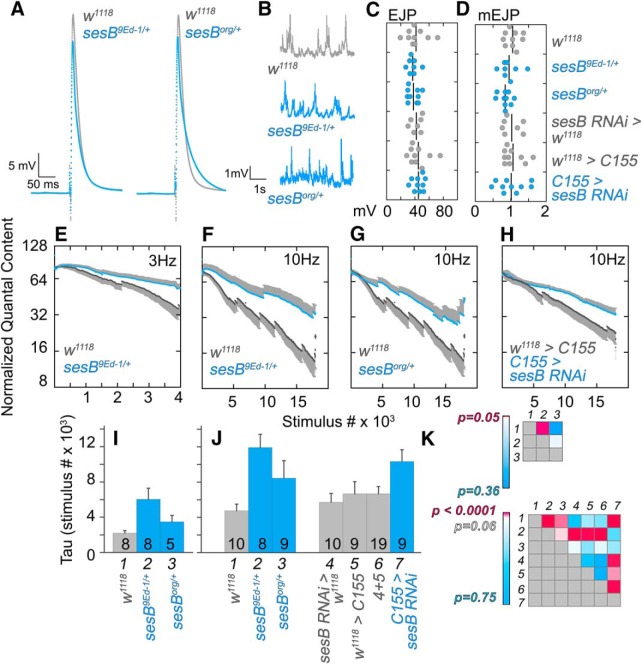
NMJ dissections of third instar, female larvae were performed in ice-cold, calcium-free HL-3 Ringer's solution (in mm: 70 NaCl, 5 KCl, 21.5 MgCl2, 10 NHCO3, 5 trehalose, 115 sucrose, 5 BES in ddH2O, pH 7.2–7.3). After the dissection, the filleted preparation was rinsed twice in low-Ca2+ Ringer's solution (in mm:70 NaCl, 5 KCl, 1 CaCl2, 20 MgCl2, 10 NHCO3, 5 trehalose, 115 sucrose, 5 BES in ddH2O, pH 7.2–7.3) and the low-Ca2+ Ringer's solution was used throughout the remainder of the experiment. Motor axons were severed close to the ventral ganglion and were taken up into a borosilicate glass capillary suction electrode with a firepolished tip (Microforge MF-830, Narishige). Recording electrodes were prepared using borosilicate glass capillary tubes (1 mm outer diameter, 0.58 internal diameter; A-M Systems), which were pulled to a fine tip (PN-3, Narishige) with 25–50 MΩ resistance and backfilled with 3M KCl. All recordings were obtained from muscle 6 in the second or third abdominal sections (A2 or A3, respectively). Stimulations were delivered using a Model 2100 Isoplated Pulse Stimulator (A-M Systems) and recordings were acquired with an AxoClamp 900A amplifier (Molecular Devices). pClamp 10 software (Molecular Devices; RRID:SCR_011323) was used to collect data and analyzed excitatory junction potential (EJP) amplitude and membrane potential and Mini Analysis Program (Synaptosoft; RRID:SCR_002184) was used to analyze miniature EJP (mEJP) frequency and amplitude.
Paired pulse facilitation (PPF) and mEJP data were collected from muscle 6 of segments 3 or 4 in late third instar female larvae of the indicated genotypes (see Fig. 8). Recordings had a resting membrane potential between −55 and −90 mV and a muscle input resistance of >10 MΩ. Recordings were performed in normal HL3 with pH of 7.2 with either low Ca2+ (in mm): 70 NaCl, 5 KCl, 2.1 MgCl2, 11.5 sucrose, 0.5 CaCl, 10 NaHCO3, 5 trehalose, and 5 BES; or high Ca2+ (in mm): 70 NaCl, 5KCl, 1.95 MgCl2, 11.5 sucrose, 2 CaCl, 10 NaHCO3, 5 trehalose, and 5 BES. For all PPF recordings, signals were acquired with AxoClamp 900A, digitized with Digidata 1440A and recorded with Clampfit 10.1. mEJPs were recorded by stimulating at 1 Hz for 50 s. mEJP analysis was performed with Mini Analysis (Synaptosoft; RRID:SCR_002184) and Microsoft Excel (RRID:SCR_016137). PPF recordings were recorded by stimulating paired EJPs at 10 Hz with a 2 ms pulse duration and a 25 ms interval between stimuli. PPF analysis was done with Axograph 1.7.2 (RRID:SCR_014284), Clampfit 10.7 (RRID:SCR_011323), and Microsoft Excel.
Figure 8.
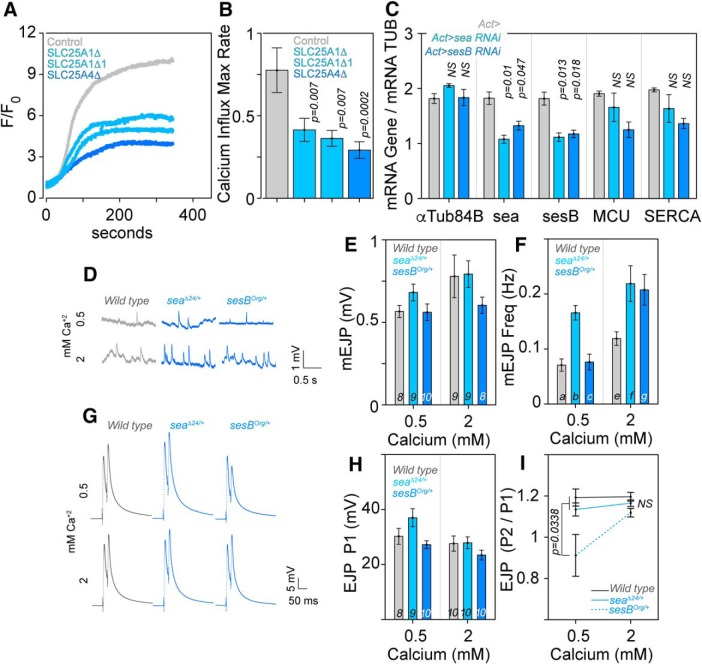
SLC25A1 and SLC25A4 are necessary for calcium homeostasis. A, Representative traces of mitochondrial Ca2+ uptake in permeabilized Hap1 cells challenged with 5 μm free Ca2+. Mitochondrial Rhod2 fluorescence (F/F0) was measured as function of time (seconds). B, Quantification of the maximal rates of mitochondrial calcium influx ΔF/F0/s in control and mutant Hap1 cell lines. Comparisons between genotypes were performed with one-way ANOVA followed by Fisher's multiple comparisons. C, qRT-PCR quantification of transcripts (x-axis) in Drosophila heads of control animals carrying the Actin-Gal4 driver alone or in combination with the UAS-RNAi for SLC25A1-sea or SLC25A4-sesB transgenes (n = 4, one-way ANOVA followed by Fisher's least significant difference comparison). D, mEJPs traces, (E) amplitudes, and (F) frequency at low and high extracellular calcium. The number of animals listed at the base of columns in E applies to F, and genotypes are listed on top of traces. E, P values for columns listed with italics at the base were determined by between-subjects ANOVA followed by Bonferroni–Dunn test: comparison a and b = 0.0014, a to c = 0.8421, e to f = 0.0008, e to g = 0.0026, f to g = 0.6967. G, PPF traces at low and high calcium concentrations. H, Amplitude of the first EJP (P1). The number of animals listed at the base of columns in H applies to I, and genotypes are listed on top of traces. I, Ratios of the two pulses. P values were determined by between-subjects ANOVA followed by Bonferroni–Dunn test.
Drosophila behavior (sleep)
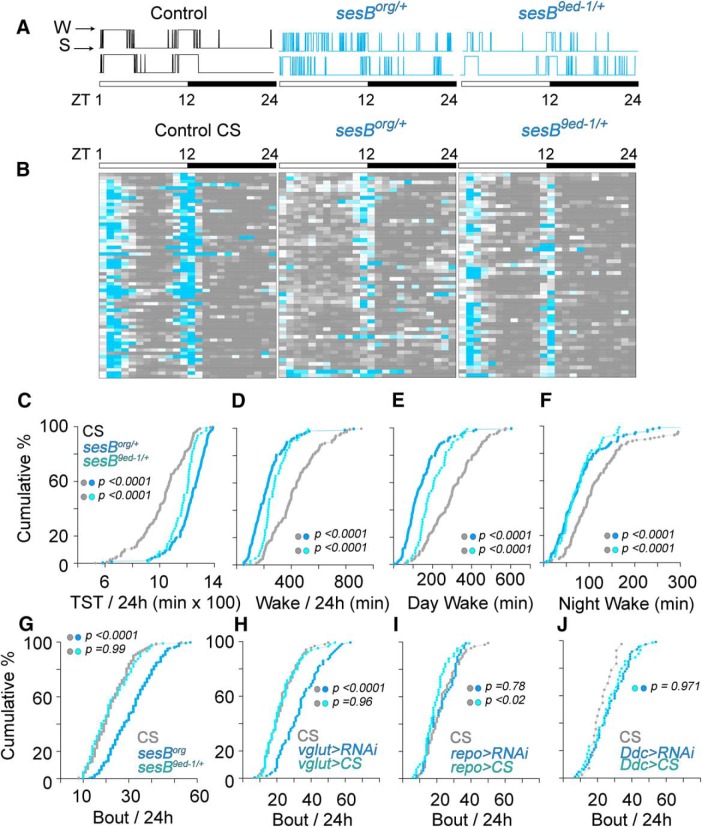
Female flies were collected under CO2 anesthesia within 72 h of eclosure. Twenty-four hours later, flies were briefly cooled on ice to allow mouth pipetting of individual flies into polycarbonate tubes (5 mm external diameter × 65 mm; TriKinetics). One end of the tube contained a 5% (w/v) sucrose and 2% (w/v) agarose medium, whereas the other end was sealed with Parafilm perforated with an 18 gauge needle to allow air circulation. Tubes were placed in the Drosophila Acitivity Monitoring System (DAM2, TriKinetics), which was housed in a light-controlled cabinet with a 12 h light/dark cycle at room temperature.
Data were collected in 15 s intervals using the DAMSystem308 acquisition software (TriKinetics) and analysis was based upon 1 min bins across 6 d of data collection, starting at Lights ON the day after the animals were placed in the tubes. Periods of inactivity lasting >5 min were scored as sleep (Hendricks et al., 2000; Shaw et al., 2000) and sleep duration, bout number, and bout length were calculated using a custom created analysis in Excel. All genotypes were compared with Canton S. Each UAS- and Gal4- line was crossed to Canton S to verify that neither the presence of the transgenes nor the genetic background of these individual lines altered the sleep/wake phenotype. Sleep–wake phenotypes of the sesB mutants were assessed based upon homozygous populations of the hypomorphic sesBorg mutation and heterozygous populations of the lethal sesB9Ed-1mutation (sesB9Ed-1/FM7a;;).
Drosophila RNAseq library generation
Cell harvesting.
GFP-labeled cells were handpicked in vivo through suction into a pipette. Cells designated for sequencing were harvested into 0.5 μl nuclease-free water in the pipette tip and then the tip was broken into a 96-well PCR tube containing RNase inhibitors and buffer as described by Clontech's ultra-low HV SMARTer Ultra Low RNAseq kit (catalog #634823) resulting in the lysing of cells without mechanical means. Amplification was performed following the Clontech Ultra-Low volume SMARter RNAseq Protocol. For the DAL neuron, the MBONα3 neurons, the MBON-γ5β'2a, and MBON-β2β'2a neurons, four cells were pooled into each tube, thus these samples contained cells from more than one fly. For the V2, α/βKCs and γKCs all cells were taken from one animal per sample. V2 samples contained 14 cells and the α/βKC and γKC samples contained ∼100 cells. Fifteen rounds of PCR amplification were performed using the Clontech SMARTer Ultra-Low RNAseq Kit. For this work only cells collected from animals that had undergone unpaired odor and shock presentation were used.
Following amplification samples were selected if there was a peak ∼7 kb and 0.4–2 ng/μl of product between the range 400 bp to 10 kb. Samples were then sheared using a Covaris LE220 sonicator to 200 bp. The libraries were made using the IntegenX automated library prep system. The PrepX Illumina DNA library prep kit/ PrepX CHIPseq kit (WaferGen Biosystems) was used with an amplification of 17–22 cycles. They were multiplexed using Bioo Scientific barcodes, and then cleaned using the IntegenX PCR cleanup kit. Libraries were run on the Illumina HiSeq2500, 12 samples per lane, and each sample run across two lanes, resulting in a sequencing depth of 30 million reads. Sequencing was all done single end.
Analysis of sequencing reads.
FastQC (Andrews, 2010) was performed to remove samples of poor quality. Samples all contained a bias for polyA and T sequences. This was uniform across all samples and was removed from sequences before mapping. GC content was not flagged on samples used in the study. All mapping was performed using Princeton University's Galaxy server running TopHat 2 with Bowtie2 (Langmead and Salzberg, 2012). The Ensembl build of the reference sequence (BDGF 5.25) and the GTF files were used and can acquired from iGenome (Illumina). The aligned SAM/BAM file were processed using HTseq-count (Intersection mode -strict; Anders et al., 2015). HTseq Counts output files and raw illumine read files are publicly available (GEO with accession GSE4989). The HTseq Counts compiled file is GSE74989_HTseqCountscompiledData.txt.gz
Calculating normalized gene counts.
The GSE74989_HTseqCountscompiledData.txt.gz dataset was used for analysis. In R, all genes with counts <2 counts per million (8 counts) across all samples independent of cell type were considered noise and removed from analysis. Gene counts were normalized using DESeq2 (Love et al., 2014) followed by a regularized log transformation. Genes with <2 counts per million within cell type were recoded as zero. Principal component analysis was performed on this processed dataset in R. R function prcomp was used to generate the principle components and gene loading values.
Drosophila transcriptome encoding mitochondrial proteins
All data were acquired from the GEO dataset GSE74989, which is publicly available. From this dataset only control animals were used to generate the figure and cell-type results. Thus 5 DAL samples, 5 V2 samples, 5 a/b KC samples, 5 gKC samples, 5 MBON b2b′2a, and 4 MBON g5b′2a.
Human postmortem RNA analysis
All data from the studies performed in postmortem human brain tissue have been previously published. All tissue sample collection and RNA sequencing (RNAseq) details are publicly available (https://www.synapse.org/#!Synapse:syn2759792/wiki/). All tissue sample collection and microarray analysis details were described in detail (Arion et al., 2015; Enwright Iii et al., 2018), and the data are publicly available upon request.
22q11DS patient and control RNAseq
RNA was extracted from whole blood using the PAXgene extraction kit (Qiagen), then stored at −80°C for subsequent analysis. RNA quantity was assessed with NanoDrop (NanoDrop Technologies) and quality with the Agilent Bioanalyzer (Agilent Technologies). Gene expression profiling was performed using Illumina HT-12 v4 microarrays. Two-hundred nanograms of total RNA were amplified, biotinylated, and hybridized to Illumina Human V4-HT-12 Beadchips, including ∼47,000 probes, following the manufacturer's recommendations. Slides were scanned using Illumina BeadStation, and the signal was extracted by using Illumina BeadStudio software.
Raw data were analyzed using Bioconductor packages in the R statistical environment. Only samples with an RNA integrity number (RIN) of 7 or greater were included in the analyses. Gene expression variance was normalized using variance stabilized transformation. Quality assessment was performed by examining the inter-array biweight midcorrelation; samples >3 SD from the mean were excluded. Batch effects were removed using ComBat. Differential gene expression analysis used the limma package in R to implement general linear model fit, with batch correction, age, sex, and RIN as covariates.
ATP and ADP determinations
HAP1 SLC25A1 and SLC25A4 knock-out and the parental control line were grown as described above. On the day of harvest, cells were washed at 37°C twice with prewarmed PBS. Cells were then scraped up from culture plates in 0.1 m perchloric acid. For Drosophila, 10 late third instar larvae or 24- to 48-h-old adults from each genotype were selected and placed into microcentrifuge tubes, flash frozen for 5 min on dry ice, and stored at −80°C. The day before purine analysis, the samples were placed in liquid nitrogen and mechanically crushed using a straight pick awl (Husky, 60004H) to break down the cuticle. 400 μl of ice-cold 0.1 m perchloric acid was added to each sample and briefly vortexed to mix contents. Cell culture and Drosophila samples were kept on ice and sonicated using a (Sonic Dismembrator, Fisher Scientific). Ten microliters of 2.5% 3.5 m K2CO3 were then added to restore pH to 7 or higher and samples stored on ice for 10 min. After 10 min, the homogenates were centrifuged at 10,000 rpm for 10 min at 4°C. Supernatant was collected in a 0.45 μm PVDF microcentrifuge filter tube and centrifuged at 10,000 rpm for 10 min at 4°C. Protein pellets and supernatants were flash frozen for 5 min on dry ice and stored at −80°C. Before HPLC analysis, supernatants were slowly thawed on ice. The supernatant was filtered again in 0.22 μm PVDF microcentrifuge filter tubes at 5000 rpm for 5 min at 4°C, transferred to Waters Vials with Caps (Waters, 22476) and stored on ice.
Purines were measured by HPLC with photodiode array ultraviolet detection. A Waters HPLC system consisting of model 717-plus autosampler, model 1525 binary pump and model 2996 photodiode array detector was used. Analytes were separated using reverse-phase ion-pair chromatography on an Atlantis T3 column (3 μm particle size 4.6 × 150 mm; Waters). Elution was conducted at 1 ml/min with a stepped gradient of buffer A (10 mm ammonium acetate and 2 mm tetrabutylammonium phosphate, pH 5.0) and buffer B (10 mm ammonium phosphate, 2 mm TBAP, 25% acetonitrile, pH 7.0 before adding acetonitrile). The gradient consistent of the following sequence: 100% buffer A for 10 min; a linear gradient to 75% buffer B over 15 min, 10 min at 75% buffer B, a linear gradient to 100% buffer B over 5 min, 100% buffer B for 15 min, and a linear gradient to 0% buffer A over 5 min. The column was then re-equilibrated with 100% buffer A for 15 min before next run. Purines were identified by comparing their retention times and spectral profiles to known standards, quantified at a detection wavelength of 254 nm.
Mitochondrial Ca2+ measurements
Mitochondrial Ca2+ measurements using the Ca2+ sensitive dye Rhod-2/AM (Invitrogen) were conducted as described previously (Maxwell et al., 2018). Briefly, Hap1 cells were plated on glass coverslips and loaded with 10 μm Rhod-2/AM with 0.25% Pluronic F-127 (Invitrogen) in Tyrode's solution for 30 min at room temperature followed by a 30 min de-esterification. Cells were permeabilized with 0.005% saponin to remove the non-mitochondrial Rhod-2 dye. Time-lapse laser scanning confocal microscopy (Olympus, FV1000) was used to image Rhod-2 fluorescence (543 nm excitation/575–675 nm emission). Baseline mitochondrial Ca2+ measurements were taken in Ca2+-free internal solution (in mm: 100 potassium acetate, 15 KCl, 0.35 EGTA, 0.75 MgCl2, 10 HEPES, pH 7.2), and then cells were perfused with internal solution containing 5 μm free Ca2+, calculated using MaxChelator (Bers et al., 2010). Rhod-2 fluorescence values (F) were normalized to initial fluorescence values (F0) and plotted as a function of time.
Quantification and statistical analysis
Experimental conditions were compared using Synergy Kaleida-Graph v4.1.3 (RRID:SCR_014980) or Aabel NG2 v5 ×64 by Gigawiz as specified in each figure. No outlier exclusion was performed.
Results
Genealogical and comparative proteomics prioritize mitochondrial targets in 22q11.2 microdeletions
We quantified proteome differences cosegregating with the 3 Mb microdeletion in 22q11.2 affected human fibroblasts and in brains from mice carrying a syntenic microdeletion in chromosome 16, Df(16)A+/−. We used human fibroblasts from pedigrees where one of the individuals was affected by 22q11.2 microdeletion syndrome and childhood psychosis, and compared affected subjects to their disease-free relatives. This strategy, termed genealogical proteomics, minimizes genetic variability between individuals and offers molecular insight into disease mechanisms despite limited subject number (Zlatic et al., 2018). We compared genealogical proteomic outcomes with Df(16)A+/− hippocampal and prefrontal cortex proteomes to identify universal mechanisms downstream of the 22q11.2 microdeletion.
We studied proteomes from the following families: one family where all members are disease-free (Fig. 1A), three families harboring one member affected by the 3 Mb 22q11.2 microdeletion (Fig. 1A,E,G), and two isolated 3 Mb 22q11.2 microdeletion patients (Fig. 1A,G). Proteomes were quantified with three mass spectrometry approaches: isobaric TMT (Fig. 1B, F), triple SILAC (Fig. 1F,H), and LFQ (Fig. 1F and Fig. 1-2). The discriminatory power of genealogical proteomics was tested by comparing the cellular proteomes from nine individuals within a single multiplexed TMT experiment. These nine individuals are organized in a disease-free family (Fig. 1A, Subjects 11–16), a pedigree with one 22q11.2 affected subject (Fig. 1A, Subjects 2–3), and an isolated 22q11.2 patient (Fig. 1A, Subject 1). Hierarchical clustering of 4264 proteins quantified in all nine subjects (Fig. 1B) segregated within a cluster all, but one, members of the unaffected family from unrelated subjects (Fig. 1C, Subjects 11, 12, 14–16). This dataset contained the quantification of 10 of the 46 proteins encoded within the 3 Mb 22q11.2 locus (Fig. 1D). Of these proteins SLC25A1, SEPT5, TXNRD2, COMT, RANBP1, and SNAP29 were predictably and significantly reduced by ∼50% (Fig. 1D). Thus, genealogical proteomics discriminates genealogical relationships among a limited number of subjects and identifies expected protein expression levels in genes encoded within the 22q11.2 locus. Proteomic analysis of an independent 22q11.2 pedigree using three quantitative mass spectrometry approaches in independent experiments identified partially overlapping proteins whose expression was sensitive to the 22q11.2 microdeletion. However, these three datasets produced convergent and similarly ranked ontology terms [gene ontology cellular component (GO CC); see Canvas depiction in Fig. 1-1A and Fig. 1-3]. These results indicate that similar ontological inferences can be obtained from proteomic datasets produced by different quantitation methods, highlighting the rigor and reproducibility of our integrated proteomics approach.
Figure 1.
Genealogical proteomics of 22q11.2 pedigrees fibroblasts using quantitative mass spectrometry. Human pedigrees of a control family (A) and families where one of the subjects is affected by 22q11.2 microdeletion syndrome and early childhood psychosis (A, E, G, blue numbers). Experimental design is designated at the top left corner of dot plots (B, F, H). For example, B shows a TMT experiment where proteomes from probands 1 and 2 were compared against unaffected individuals, Subjects 3, 11–16. C, Hierarchical clustering analysis of the proteome in Subjects 1–3 and 11–16. Euclidian distance clustering of columns and rows (4264 TMT protein quantitations) shows segregation of related family members. D, Dot plot of proteins encoded within the 22q11.2 chromosomal segment quantitated in TMT Experiment B. Asterisks denote significant differences p = 0.04146 to p < 0.0001, t test. B, F, H, Depictions of all mass spectrometry quantifications where the color code denotes individuals being compared (blue symbols are proteins whose expression is changed, gray symbols are unaffected proteins). Significant protein expression changes for: TMT and SILAC were considered to be >2 or <0.5, whereas in LFQ a −log(p) value threshold of 1.3 was used. I, The Venn diagram summarizes proteins with significant expression changes in B, F, and H. Asterisk denotes proteins whose expression changed in all patients. Bold font depicts proteins encoded within the 22q11.2 segment. Blue color fonts are proteins contained in the human Mitocarta 2.0 dataset. Individual MS/MS data can be found in Figure 1-2. Ontological comparisons among pedigrees and proteomic platforms can be found in Figure 1-1, and Figure 1-3.
22q11.2 Microdeletion Genealogical Proteome Comparisons Among Pedigrees and Mass Spectrometry Quantitation Strategies. A-D) Cellular Component Gene ontologies (GO:CC) obtained using the ENRICHR engine. Data are depicted as canvases where every tile is occupied by an individual GO category whose p value significance is depicted by color intensity. A) First canvas to the left depicts a comparison for genealogical proteomes obtained in one pedigree by TMT (green) and to Label Free Quantification (LFQ, red). Other canvases in A show experiments comparing TMT with two independent SILAC experiments and combinations of TMT, LFQ and SILAC. Overlap of GO:CC terms is presented as yellow. B) Comparison of GO:CC terms obtained by genealogical proteomics from three pedigrees. C) Represents GO:CC term tiles overlapping among pedigrees in B). D) Shows gene ontology terms obtained by pooling into one dataset the proteomes from all pedigrees in B). E. Presents relevant GO terms in D. Individual pedigree and collective bioinformatics data can be found in Fig. 1-3. Download Figure 1-1, TIF file (1.2MB, tif)
Quantitative Mass Spectrometry Data for 22q11.2 Genealogical Proteomic Studies. Download Figure 1-2, XLSB file (10.7MB, xlsb)
Comparative Bioinformatic Analysis of 22q11.2 Genealogical Proteomes. Download Figure 1-3, XLSX file (68.7KB, xlsx)
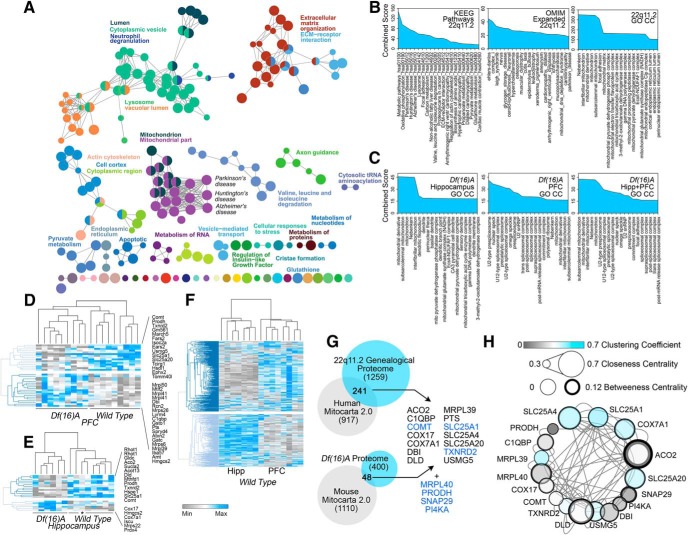
Genealogical proteomics of the three 22q11.2 pedigrees (Fig. 1A,E-G) collectively identified 1500 proteins whose expression was altered in 22q11.2 microdeletion cells (Fig. 1I, and Fig. 1-2). Of these proteins, only 18 polypeptides were common to all of the 22q11.2 affected individuals (Fig. 1I), including five polypeptides contained in the 22q11.2 locus and 13 polypeptides previously not implicated in 22q11.2 syndrome (Fig. 1I). Independent gene ontology analysis of each one of these three pedigree datasets converged on partially overlapping gene ontology categories (Fig. 1-1B and C and Fig. 1-3). We inferred ontological categories with the 1500 proteins whose expression co-segregated with the 22q11.2 microdeletion, hereafter referred as the 22q11.2 proteome. We used three bioinformatic algorithms that produced similarly ranked ontological categories. We queried the GO CC, REACTOME, and KEGG pathways simultaneously with the ClueGo algorithm to discern statistically ranked organelles, pathways, and associated pathologies downstream of the 22q11.2 microdeletion (Bindea et al., 2009). The top ontology categories/pathways were all related to mitochondrial compartments (Fig. 2A and Fig. 2-3; group q value 1.05E-38), as well as diseases where mitochondria are implicated in pathogenesis such as Parkinson's and Huntington's diseases (Fig. 2A and Fig. 2-3; group q value 3.93E-37; Lin and Beal, 2006). Additionally, the 22q11.2 proteome was enriched in extracellular matrix, lysosome, and actin cytoskeleton components and pathways (Fig. 2A and Fig. 2-3; group q values 3.23E-23, 6.61E-18, and 7.93E-16, respectively). We confirmed these bioinformatic results with the ENRICHR engine to interrogate the KEGG, OMIM, and GO CC databases (Chen et al., 2013). Mitochondrial compartments and pathways, Parkinson's, Huntington's, and other diseases where mitochondria are affected were enriched in the 22q11.2 proteomic dataset (Fig. 2B and Fig. 2-3; q values 3.3E-36, 1.2E-21, and 5.1 E-12, respectively).
Figure 2.
Comparative bioinformatic analysis of the 22q11.2 and the Df(16)A+/− mouse brain proteomes. A, The 22q11.2 Proteome was analyzed with the engine ClueGo integrating the Cellular Component gene ontology GO CC, REACTOME, and KEGG databases. Functionally grouped network was built with terms as nodes and edges based on their term–term similarity statistics. The node size represents the term enrichment significance (p < 0.015 Bonferroni corrected). B, ENRICHR analysis of the 22q11.2 proteome querying GO CC, KEEG, and OMIM databases. C, ENRICHR analysis of the Df(16)A+/− mouse hippocampus and prefrontal cortex (PFC) proteomes as in B. Mouse brain proteomes were quantified using TMT mass spectrometry (Figure 2-4; n = 6 mutant and 5 control mice). D–F, Differences in the mitoproteomes of wild-type and Df(16)A+/− mitoproteomes. Hierarchical clustering analysis of the Df(16)A+/− mouse hippocampus and prefrontal cortex mitochondrial proteome hits (D, E) compared with the wild-type mitoproteomes quantified in hippocampus (Hipp) and PFC. Kendall's tau distance clustering of columns and rows. E, The asterisk marks a wild-type animal. Figure 2-1 presents a similar analysis of mitochondrial transcriptomes in different Drosophila neurons. G, Venn diagrams present overlapping protein hits between the 22q11.2 and Df(16)A+/− proteome with the human and mouse Mitocarta 2.0 datasets. Listed proteins correspond to mitochondrial proteins whose expression is sensitive to the microdeletion in human and mouse (top two columns). Lower column and blue font proteins are encoded in the 22q11.2 chromosomal segment. Comparisons with previous Df(16)A+/− proteome are depicted in Figure 2-2. H, SLC25A1 and SLC25A4 are high-connectivity nodes in a discrete 22q11.2 and Df(16)A+/− mitoproteome interactome. In silico interactome of protein hits listed in G. Interactome was analyzed with graph theory to determine high-connectivity nodes predictive of essential genes. Additional bioinformatic data and MS/MS data can be found in Figure 2-3, and Figure 2-4.
The Drosophila Transcriptome Encoding Mitochondrial Proteins is Cell Type Specific. A-B) mRNA from single neuron types isolated from Drosophila mushroom bodies were analyzed by RNAseq. The transcriptome encoding mitochondrial proteins, as defined by Chen et al (Chen et al., 2015), was analyzed by principal component analysis (A) and hierarchical clustering using 1-Pearson correlation clustering (B) of columns (cells) and rows (transcripts). Cell types were identified as in Crocker et al (Crocker et al., 2016). Note the robust segregation of Kenyon cells from other cell types by the expression of the transcriptome encoding mitochondrial proteins. Download Figure 2-1, TIF file (673.8KB, tif)
Comparative Bioinformatics of the 22q11.2 Proteome and Two Independent Df(16)A-/+ Brain Proteomes. A) Venn diagrams depict from top to bottom: a comparison of common hits between our Df(16)A-/+ brain proteome and the Df(16)A-/+ brain proteome reported by Wesseling et al. PMID: 27001617. The Wesseling Df(16)A-/+ brain proteome and our 22q11.2 proteome. The Wesseling Df(16)A-/+ brain proteome and the mouse Mitocarta 2.0 dataset. B) Cellular Component gene ontology analysis of GO CC generated with the ENRICHR engine using the Wesseling Df(16)A-/+ brain proteome dataset and a similarly sized random mouse gene dataset. Random gene list was generated with the engine RandomGeneSetGenerator. C) Cellular Component gene ontology analysis (GO CC) was performed with the ENRICHR engine using the Wesseling Df(16)A-/+ brain proteome dataset either by itself, or in combination with our 22q11.2 proteome, or with 1500 (1x) or 3000 (2x) randomly generated genes. See discussion. Download Figure 2-2, TIF file (883.6KB, tif)
Bioinformatic Analysis of 22q11.2 Genealogical Proteomes and Df(16)A-/+ brain proteomes. Download Figure 2-3, XLSX file (146KB, xlsx)
Quantitative Mass Spectrometry Data for Df(16)A-/+ brains. Download Figure 2-4, XLSX file (3.7MB, xlsx)
We examined ontology terms inferred from a brain proteome sensitive to the syntenic Df(16)A+/− deficiency in mice, hereafter referred as the Df(16)A+/− brain proteome (Fig. 2C and Fig. 2-3). We reasoned that overlapping ontological categories between the 22q11.2 proteome and the Df(16)A+/− brain proteome would point to robust and universal mechanisms downstream of the 22q11.2 microdeletion. We profiled by TMT the hippocampus and prefrontal cortex proteomes of control and Df(16)A+/− mouse brains. We quantified 6419 proteins and identified 110 hippocampal and 365 prefrontal cortex proteins whose expression was sensitive to the Df(16)A+/− microdeletion. ENRICHR bioinformatic analysis indicated that mitochondrial terms were top ranked in the Df(16)A+/− hippocampus proteome (Fig. 2C and Fig. 2-3; q value 0.0018 and combined score of 45.77). In contrast, the spliceosome ranked first in the Df(16)A+/− prefrontal cortex proteome (Fig. 2C and Fig. 2-3; q value 5.01E-06 and combined score of 41.16) with mitochondrial ontological categories scoring in the sixteenth place (Fig. 2C and Fig. 2-3; p value 0.012 and combined score of 16.52). These ranking differences among ontological hits were due to different mitochondrial polypeptides being affected by the Df(16)A+/− microdeletion in the hippocampus and prefrontal cortex mitoproteomes (Fig. 2D,E and Fig. 2-3). We attribute these differences to distinctive mitoproteome stoichiometries that distinguish the hippocampus and prefrontal cortex in control mouse brain (Fig. 2F and Fig. 2-4). These regional mouse brain mitoproteome differences were also observed in flies were distinct neurons of the Drosophila mushroom body, the fly hippocampus equivalent (Campbell and Turner, 2010), can be segregated away just based on stoichiometric differences in the transcriptome encoding the fly mitoproteome (Fig. 2-1; Chen et al., 2015; Crocker et al., 2016). We conclude that the proteomes sensitive to either the 22q11.2 or the Df(16)A+/− hemideficiencies enrich components of the mitoproteome in a brain region-specific manner.
Identification and prioritization of key mitochondrial proteins within the 22q11.2 proteome
We used the Mitocarta 2.0 mitoproteome dataset as a reference to identify mitochondrial proteins among the 22q11.2 and Df(16)A+/− proteomes (Pagliarini et al., 2008; Calvo et al., 2016). We identified 241 mitochondrial proteins sensitive to the 22q11.2 microdeletion and 48 mitochondrial proteins sensitive to the Df(16)A+/− deficiency (Fig. 2G). Expression of 14 mitochondrial proteins was affected either in all human pedigrees (Fig. 1I) or simultaneously in human and mouse cells with the microdeletion (Fig. 2G). We merged these 14 mitochondrial proteins with four additional proteins encoded within the 22q11.2 chromosomal segment which are also part of the Mitocarta 2.0 datasets (Fig. 2G, blue font represents 22q11.2 encoded proteins). A network of protein-protein interactions constrained to these 18 polypeptides was subjected to graph theory analysis to unbiasedly determine node relevance within this network (Fig. 2H). We used clustering, closeness centrality, and betweenness centrality coefficients to measure node relevance (del Rio et al., 2009). The gene products with the highest relevance scores within this interactome were SLC25A1 and SLC25A4 (Fig. 2H). SLC25A1 and SLC25A4 are encoded in the 22q11.2 and 4q35.1 cytogenetic bands. These two transporters participate in central inner mitochondrial solute transport mechanisms and are widely expressed in multiple tissues (Palmieri and Monné, 2016; Taylor, 2017). Thus, we selected these two inner mitochondrial transporters as candidate genes whose disruption would maximize network perturbation.
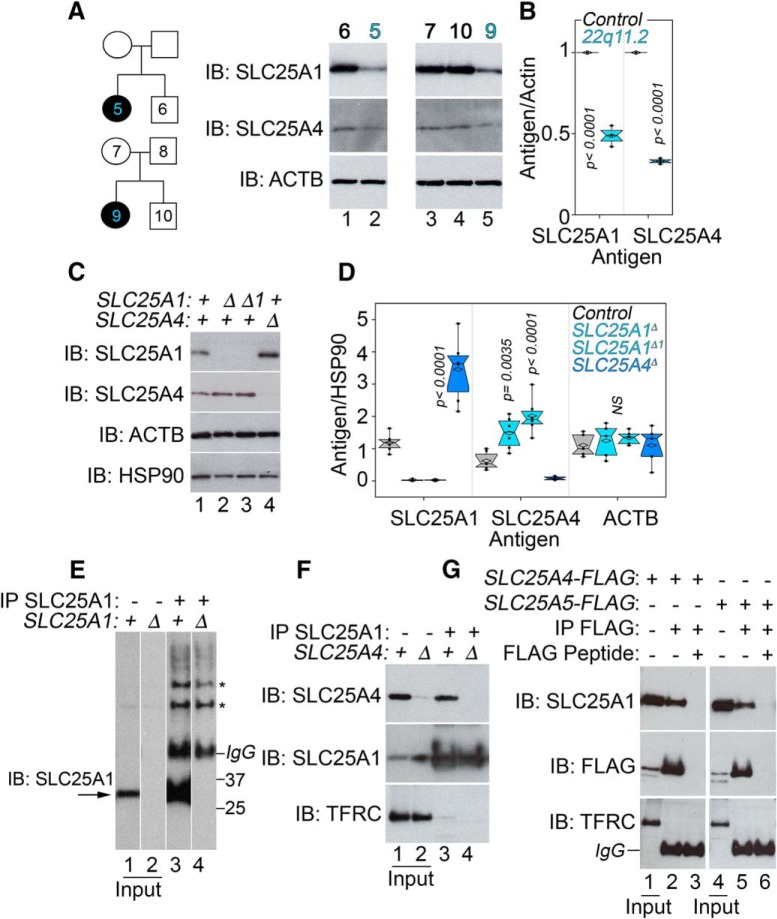
We confirmed that SLC25A1 and SLC25A4 expression was altered in 22q11.2 fibroblasts as compared with non-affected family members. Both transporters were decreased at least by 50% in 22q11.2 affected fibroblasts compared with unaffected family members (Fig. 3A, compare lanes 1 and 2, 3–4 and 5, quantified in Fig. 3B). We hypothesized that coexpression changes observed in microdeletion patient cells may be the result of biochemical/metabolic interactions between SLC25A1 and SLC25A4. We used two approaches to address this question. First, we tested whether SLC25A1 and SLC25A4 influenced each other's expression, a common occurrence in proteins that physically interact or belong to a pathway (Wu et al., 2013). We used cells where SLC25A1 or SLC25A4 expression was abrogated by CRISPR-Cas9 genome editing (Fig. 3C,D). Cells lacking SLC25A1 significantly increased the expression of SLC25A4 ∼1.5–2-fold, whereas SLC25A4-null cells upregulated SLC25A1 3.6 times demonstrating a genetic interaction between these two transporters (Fig. 3C,D). Second, we performed immunomagnetic isolation of SLC25A1 from detergent soluble extracts from wild-type and either SLC25A1 or SLC25A4 mutant cells. An SLC25A1 antibody robustly immunoprecipitated a SLC25A1-immunoreactive band absent in SLC25A1-null cells (Fig. 3E, compare lanes 3–4). This SLC25A1 antibody also coimmunoprecipitated SLC25A4 from wild-type cell extracts but not from SLC25A4-null cells (Fig. 3F, compare lanes 3–4). We determined coprecipitation selectivity by blotting for transferrin receptor, a transmembrane protein absent from Mitocarta 2.0 (Fig. 3F, TFRC; Pagliarini et al., 2008; Calvo et al., 2016). Reverse immunomagnetic isolations with FLAG-tagged SLC25A4 and its paralog SLC25A5 recovered endogenous SLC25A1 (Fig. 3G, lanes 2 and 5). SLC25A1 co-isolation with tagged SLC25A4 and 5 was prevented by FLAG peptide competition (Fig. 3G, lanes 3 and 6). Collectively, these findings demonstrate that SLC25A1 and SLC25A4 genetically and biochemically interact.
Figure 3.
SLC25A1 and SLC25A4 expression is affected by the 22q11.2 microdeletion and these transporters biochemically and genetically interact. A, Human pedigrees of families affected by 22q11.2 microdeletion syndrome. Immunoblots of total cellular lysates from fibroblasts obtained from individuals in pedigrees. B, Quantitation of results shown in A. P values, one-way ANOVA followed by Dunnett's multiple comparisons; n = 3. C, SLC25A1 and SLC25A4 expression changes in cells carrying null mutations (Δ) in SLC25A1 or SLC25A4 clonal cell lines. Detergent-soluble cell extracts were blotted with indicated antibodies. Actin (ACTB) and HSP90 were used as controls. D, Depicts quantitation of expression levels compared with wild-type cells. P values, one-way ANOVA followed by Dunnett's multiple comparisons; n = 5. E, SLC25A1 antibody precipitates an SLC25A1-immunoreactive band (lane 3) absent from SLC25A1-null cells (lane 4). Asterisks denote nonspecific bands recognized by the antibody. F, SLC25A1 antibody precipitates an SLC25A4-immunoreactive band (lane 3) absent from SLC25A4-null cells (lane 4). G, FLAG-tagged SLC25A4 or SLC25A5 precipitate SLC25A1 (lames 2 and 5). Lanes 1 and 3 correspond to inputs. Lanes 4 and 6 correspond to immunoprecipitation where an excess FLAG peptide was used for out-competition. F, G, Transferrin receptor (TFRC) was used as a control for nonspecific membrane protein precipitation.
Expression of SLC25A family of mitochondrial transporters is altered in 22q11.2 fibroblasts and schizophrenia patient neurons
We created a comprehensive ab initio SLC25A1-SLC24A4 interactome using as building blocks a SLC25A4-focused interactome plus all SLC25A1 and SLC25A4 interactions curated from seven proteome-wide physical interaction datasets (Fig. 4A and Fig. 4-1; Havugimana et al., 2012; Hein et al., 2015; Huttlin et al., 2015, 2017; Wan et al., 2015; Floyd et al., 2016; Lu et al., 2017). The ab initio SLC25A1-SLC25A4 interactome contained 106 nodes encompassing mitochondrial respiratory chain components and 12 SLC25A transporter family members (Fig. 4A and Fig. 4-1). The SLC25A1 and SLC25A4 nodes maintained their relative relevance within the ab initio network, as ascertained by SLC25A1 and SLC25A4 centrality coefficients (Fig. 4A and Fig. 4-1). Forty five of the 106 ab initio SLC25A1-SLC25A4 interactome nodes were represented in the human 22q11.2 proteome indicating a convergence of the 22q11.2 proteome mitochondrial hits and the ab initio network (Fig. 4A and Fig. 4-1, gray nodes).
Figure 4.
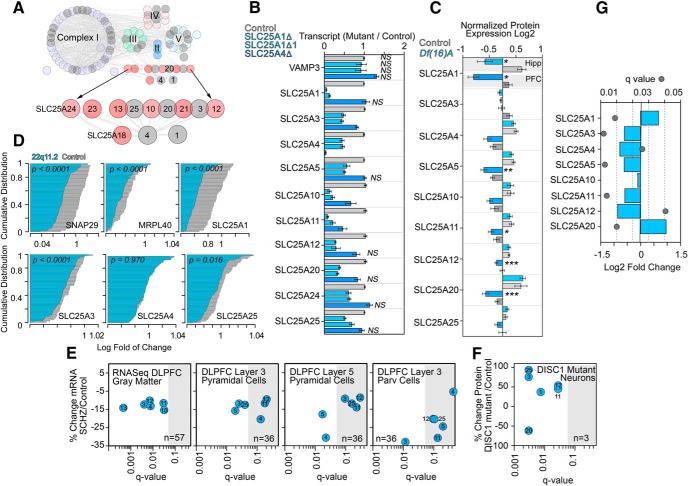
Expression of components of the SLC25A1-SLC25A4 interactome is affected in neurodevelopmental disorders. A, Comprehensive in silico interactome of the SLC25A1 and SLC25A4 mitochondrial transporters. Complexes I to V of the respiratory chain as well as SLC25A transporter family members are color coded. All nodes colored gray represents hits in the 22q11.2 proteome. Additional details can be found in Figure 4-1. B, Expression of SLC25A transporter family member transcripts is altered in SLC25A1 or SLC25A4-null cells. Transcript quantification by qRT-PCR is expressed as a ratio to vimentin mRNA. VAMP3 was used as control. n = 4; one-way ANOVA followed by Fisher's least significant difference comparison. All nonsignificant comparisons are marked (NS). C, Expression of SLC25A transporter family member polypeptides is altered in Df(16)A+/− mouse hippocampus (Hipp) or prefrontal cortex (PFC). SLC25A transporters were quantitated by TMT mass spectrometry. n = 6 mutant and 5 control mice; one-way ANOVA followed by Fisher's least significant difference comparison. *p ≤ 0.0001, **p = 0.0098, ***p ≤ 0.028. D, Expression of SLC25A family member mRNAs is reduced in whole blood from unaffected and 22q11.2 patients. Probability plots of mRNA quantified by microarray on 50 unaffected (gray) and 77 22q11.2 patients (blue). SNAP29, MRPL40, and SLC25A1 reside in the 22q11.2 microdeletion locus and were used as controls to determine the range of expression change attributable to the microdeletion. SLC25A3 and SLC25A25 expression is modified within this range. P values were calculated using Kolmogorov–Smirnov test. E, mRNA expression of SLC25A transporters in gray matter or single cells isolated from unaffected and schizophrenia cases. Gray matter mRNA quantitations were performed by RNAseq, whereas single-cell mRNA quantitations were performed by microarray in dorsolateral prefrontal cortex (DLPFC) samples. F, Proteomic quantitation of SLC25A transporters in iPSC-derived cortical neurons from DISC-1 mutant patient and isogenic controls. E, F, SLC25A transporter family members SLC25An where n correspond to the number on blue circle. Gray box denotes nonsignificant changes in expression after multiple corrections. G, mRNA expression of SLC25A transporter family members is altered in schizophrenia brains. Meta-analysis data obtained from Gandal et al. (2018).
Comprehensive in silico Interactome of the SLC25A1 and SLC25A4 Transporters. A) Comprehensive in silico interactome of the SLC25A1 and SLC25A4 mitochondrial transporters. Complexes I to V of the respiratory chain as well as SLC25A transporter family members are color coded. All nodes colored gray represent hits in the 22q11.2 proteome. B) The comprehensive interactome was analyzed with graph theory to determine high connectivity nodes predictive of essential genes using the closeness and betweeness centrality coefficients. Note the high connectivity of SLC25A4 in the comprehensive interactome. Download Figure 4-1, TIF file (4.4MB, tif)
We selected the SLC25A family member transporters to test the reliability of the ab initio network (Fig. 4A). We asked whether members of the SLC25A transporter family genetically interacted as inferred from the ab initio SLC25A1-SLC25A4 interactome. We first investigated whether mRNA levels of SLC25A transporter family members were altered in SLC25A1- or SLC25A4-null cells. We measured transcripts of 10 of the 12 ab initio network SLC25A transporters in both SLC25A1 and SLC25A4 knock-out cells by qRT-PCR. SLC25A1-null cells significantly altered the expression of 9 of the 10 measured SLC25A transporters, whereas SLC25A4-null cells affected three transporters (Fig. 4B). These changes in transcript content were selective as evidenced by unaltered levels of the housekeeping genes VAMP3 and VIM (Fig. 4B). We further analyzed whether these SLC25A network transporters were affected in other 22q11.2 and syntenic microdeletion tissues. Quantitative mass spectrometry of SLC25A family transporters showed an anticipated decrease of ∼50% in SLC25A1 in prefrontal cortex and hippocampus of Df(16)A+/− mice (Fig. 4C). Additionally, expression of five of nine SLC25A family transporters was decreased in prefrontal cortex (Fig. 4C). We extended these observations to lymphoblasts from 77 22q11.2 microdeletion patients and compared mRNA levels to 50 unaffected subjects. Expression of genes within the 22q11.2 locus significantly reduced their expression (Fig. 4D; SNAP29, MRPL40, and SLC25A1). Messenger RNA levels of three of 10 SLC25A family transporters were significantly reduced in 22q11.2 cells (Fig. 4D). We conclude that components of the ab initio SLC25A1-SLC25A4 interactome are affected in tissues from human and mouse carrying 22q11.2 and syntenic microdeletions, respectively. These findings validate the ab initio SLC25A1-SLC25A4 interactome for studies of neurodevelopmental and synaptic mechanisms.
We hypothesized that if components of the ab initio SLC25A1-SLC25A4 interactome were to participate in the pathogenesis or phenotypic expression of 22q11.2 neuropsychiatric nosology, then neurons from patients affected by schizophrenia or neurons from patients with mutations in the schizophrenia risk gene DISC1 would alter the expression of SLC25A transporters. Expression of SLC25A transcripts was measured by RNAseq in dorsolateral prefrontal cortex gray matter of 57 age- and sex-matched pairs of unaffected comparison and schizophrenia subjects (Fig. 4E). Of the six SLC25A family transporters with detectable levels of expression, all had mean mRNA levels that were lower in subjects with schizophrenia (Fig. 4E; q < 0.05 SLC25A3, SLC25A4, SLC25A11, and SLC25A12). Next, pools of layer 3 and layer 5 pyramidal cells and layer 3 parvalbumin cells were individually collected via laser capture microdissection in a subset of subjects (N = 36 pairs), and SLC25A transcripts were measured via microarray (Arion et al., 2015; Enwright Iii et al., 2018). Expression of each of the six SLC25A transcripts that were detectable by microarray was lower in all three cell types, though not all met statistical significance (Fig. 4E). The most affected transporter in schizophrenia layer 5 pyramidal neurons was SLC25A4 (Fig. 4E; 30.6% reduction, q value = 0.0222) while in parvalbumin neurons it was SLC25A3 (Fig. 4E; 32.9% reduction, q value = 0.0116). Changes in the expression of SLC25A family transporters was also observed in iPSC-derived human prefrontal neurons carrying a frameshift mutation of DISC1 compared with isogenic controls generated by editing of the DISC1 gene defect (Wen et al., 2014). Proteomics identified significant changes in the expression of six of 12 ab initio network SLC25A transporters with the most pronounced effects on SLC25A3, SLC25A20, and SLC25A25 (Fig. 4F; q values < 0.003), yet the magnitude and direction of these changes were different to those in sporadic cases of schizophrenia (Fig. 4E). These findings are also reproduced in a comprehensive meta-analysis of mRNA expression changes in 159 cortical schizophrenia patient samples compared with 293 unaffected subjects (Fig. 4G; Gandal et al., 2018). These results indicate that components of the ab initio SLC25A1-SLC25A4 interactome are altered in tissue samples from patients affected by neurodevelopmental disorders sharing phenotypes with the 22q11.2 syndrome. Collectively, these findings demonstrate that SLC25A1 and SLC25A4 are principal nodes within a mitochondrial interactome. Our results suggest that partial downregulation of these mitochondrial transporters and their interactome may participate in mechanisms necessary for synapse function and behavior.
SLC25A1 and SLC25A4 are required for normal synapse development and to maintain the synaptic mitochondrial pool
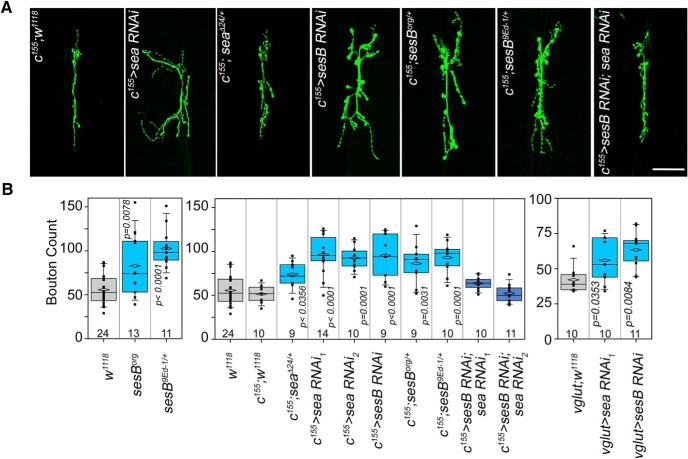
To test the consequences of a partial decrease in the expression of SLC25A mitochondrial transporters on neuronal function and behavior, we selected Drosophila because of genetic tools that allow precise control of gene expression in a developmental-, cell-, and tissue-restricted fashion. We focused on studying synaptic and behavioral phenotypes caused by SLC25A1 and SLC25A4 haploinsufficiencies in Drosophila. SLC25A1 and SLC25A4 orthologues are encoded by the gene scheggia (sea, CG6782, dSLC25A1) and stress-sensitive B genes (sesB, CG16944, dSLC25A4), respectively. We examined morphological and electrophysiological phenotypes in the third instar Drosophila neuromuscular junction synapse, which is a reliable model to assess synaptic developmental phenotypes associated with neurodevelopmental gene defects (Frank et al., 2013). We controlled the expression of dSLC25A1-sea with UAS-RNAi reagents and the null allele seaΔ24 (Morciano et al., 2009). dSLC25A4-sesB expression was modified with UAS-RNAi as well as two genomic alleles of sesB, sesBorg and sesB9Ed-1. sesBorg is a thermosensitive allele that decreases the ADP-ATP transport activity by 60%, offering a functional haploinsufficiency model (Rikhy et al., 2003). In contrast, sesB9Ed-1 is a strong lethal null allele that is viable as single copy deficiency (Y. Q. Zhang et al., 1999). We confirmed that UAS-sea or UAS-sesB RNAi transgenes driven by actin-GAL4 indeed reduced mRNA expression of their target genes by 50% in Drosophila heads (Fig. 8C). Therefore, these fly reagents mimic the partial reduction in the expression of SLC25A family members observed in 22q11.2 human and mouse mutant tissues, as well as samples from patients diagnosed with schizophrenia (Figs. 3, 4).
We used genomic alleles and RNAi transgenes driven by neuronal-specific elavc155-Gal4 and VGlut-Gal4 transgenic drivers and analyzed the morphology of the larval neuromuscular junction (Fig. 5). Reducing the expression of sea or sesB by half increased the number of boutons and/or branches per synapse, regardless of whether genomic alleles or neuronal-specific RNAi decreased transporter expression (Fig. 5A,B). These synaptic morphological phenotypes could be simply attributed to reduced mitochondrial function caused by these haploinsufficiencies. We scrutinized this hypothesis by genetic and biochemical approaches. First, we compared outcomes of single or double neuronal-specific dSLC25A1 and dSLC25A4 RNAi on synapse morphology. If reduced mitochondrial function accounts for phenotypes observed in single transporter genetic defects, we reasoned that combined RNAi of these two transporters should enhance synaptic morphological phenotypes compared to single RNAi synapses. Contrary to this prediction, double dSLC25A1-sea plus dSLC25A4-sesB RNAi rescued synapse morphology (Fig. 5A,B). Second, we measured ATP–ADP ratios in Drosophila dSLC25A1-sea and dSLC25A4-sesB RNAi haploinsufficient larvae and Drosophila heads (Fig. 5-1). ATP/ADP ratios were not significantly and predictably modified in both tissues. The same is the case in mutant human SLC25A1 cells, even though the mutation completely abrogated SLC25A1 expression (Fig. 3C,D and Fig. 5-1). However, the ATP–ADP ratio was decreased by one-half in SLC25A4-null human cells (Fig. 5-1). This last result is expected and confirms the sensitivity of our assay (Klingenberg, 2008). These genetic and biochemical findings strongly argue against loss of mitochondrial function caused by dSLC25A1-sea or dSLC25A4-sesB RNAi haploinsufficiencies.
Figure 5.
Reduced expression of Drosophila dSLC25A1-dSLC25A4 alters synapse morphology. A, B, Muscle VI–VII third instar neuromuscular junctions were stained with antibodies against the neuronal marker HRP. Expression of SLC25A1 (sea, scheggia) was downregulated with two RNAi transgenes or the null allele seaΔ24/+. dSLC25A4 expression was reduced with a RNAi transgene or two genomic alleles (sesBorg and sesB9Ed-1/+). Neuronal-specific expression of RNAi regents was driven by the elavc155-GAL4 (c155) or Vglut-GAL4 drivers. The w1118, w1118; elavc155-GAL4 or w1118; Vglut-GAL4 animals were used as controls. Scale bar, 50 μm. B, Shows quantitation of bouton counts per synapse. Counts were performed blind to the animal genotype. All comparisons in B were performed with one-way ANOVA followed by Bonferroni's multiple comparison. Number of animals is at the bottom of each box. Analysis of ATP–ADP ratios in tissues from mutant animals can be found in Figure 5-1.
Reduced Expression of Drosophila dSLC25A1-dSLC25A4 does not Affect Cellular ATP/ADP ratios. ATP/ADP ratios were measured in third instar larvae, adult heads, and human Hap1 cells of the indicated genotypes. n=3 for Drosophila Tissues and n=6 for Hap1 cells, One Way ANOVA followed by Fisher’s Least Significant Difference Comparison. Download Figure 5-1, TIF file (227.2KB, tif)
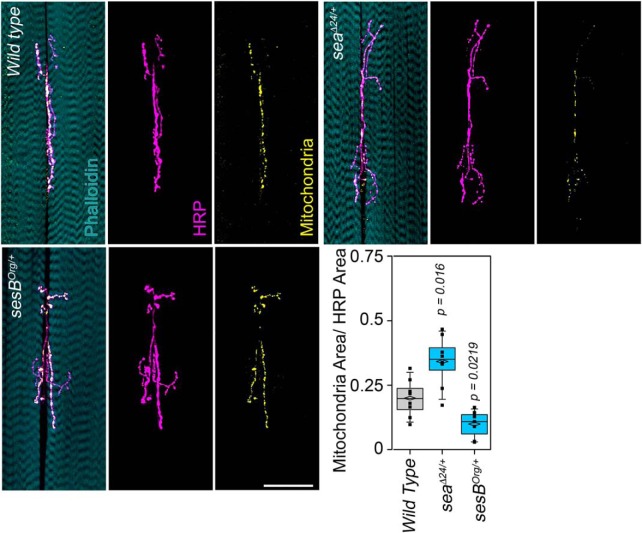
Mutations in the GTPase miro-deplete synapses of mitochondria and increase the number of boutons at the Drosophila neuromuscular synapse (Guo et al., 2005). Moreover, mutations in miro and Drp1 that severely deplete synaptic mitochondria also selectively compromise neurotransmission at high frequencies while sparing the amplitude of evoked and spontaneous neurotransmission events (Guo et al., 2005; Verstreken et al., 2005). This prompted us to ask whether the increased synaptic branching in dSLC25A1-sea or dSLC25A4-sesB haploinsufficiencies could be associated to synaptic mitochondrial depletion and impaired high-frequency neurotransmission. We quantified mitochondria in larval neuromuscular junctions by neuronally expressing a UAS-mitochondria-GFP transgene in animals harboring either UAS-sea or UAS-sesB RNAi transgenes. Downregulation of dSLC25A1-sea increases mitochondrial content at the synapse 1.8 times, whereas downregulation of dSLC25A4-sesB decreases mitochondrial content by half (Fig. 6). These results demonstrate that dSLC25A1-sea and dSLC25A4-sesB are required to maintain the synaptic pool of mitochondria and exclude mitochondrial depletion as a cause of the synapse branching phenotype.
Figure 6.
Drosophila SLC25A1-sea and SLC25A4-sesB are required for maintaining the synapse mitochondrial pool. Triple-stained muscle VI–VII third instar neuromuscular junctions from elavc155-GAL4;w1118 wild-type larvae (n = 8) or elavc155-GAL4 crossed to either seaΔ24/+ (n = 8) or sesBOrg/+ (n = 7) were imaged by confocal microscopy. Cyan marks muscle with phalloidin, magenta marks neuronal plasma membrane with HRP antibodies, and yellow mitochondria with the UAS-mitochondria-GFP transgene. Boxplot depicts the ratio between the mitochondrial signal and the HRP signal. Comparisons between genotypes were performed with one-way ANOVA followed by Fisher's multiple comparison. Scale bar, 50 μm.
We next asked whether the partial reduction in mitochondrial content in dSLC25A4-sesB haploinsufficient synapses was consequential for neurotransmission. Evoked neurotransmission, measured as evoked EJP amplitude (Figs. 7A,C, 8A,B), and spontaneous mEJP amplitude (Figs. 7B,D, 8D,E), remained unaffected in all sesB genotypes tested. We examined and compared neurotransmission elicited at 3 Hz and at high-frequency (10 Hz) on wild-type and sesB haploinsufficient synapses (Fig. 7E–K). We incubated neuromuscular junctions in the presence of bafilomycin A1, a vacuolar ATPase inhibitor, to prevent neurotransmitter vesicle reloading after a round of vesicle fusion (Delgado et al., 2000; Kim et al., 2009; Mullin et al., 2015). This strategy leads to synapse fatigue in wild-type larvae because of synaptic vesicle depletion (Fig. 7E–H, gray curves). Synaptic transmission at low-frequency stimulation was normal in sesB deficient synapses (Fig. 7E,I,K). However, neurotransmission at high-frequency was consistently increased in all sesB alleles as expressed by synaptic resilience to fatigue (10 Hz; Fig. 7F,G,J,K). These effects were because of changes in the expression of sesB in neurons and not the muscle, as demonstrated by downregulation of sesB with neuronal-specific RNAi (Fig. 7H,J,K). These results demonstrate that dSLC25A1-sea and dSLC25A4-sesB haploinsufficiencies differentially control the synaptic pool of mitochondria, regardless of the synaptic bouton count, and in the case of dSLC25A4-sesB, without impairing high-frequency neurotransmission.
Figure 7.
Hemideficiency of the Drosophila SLC25A4-sesB alters synapse function. Muscle VI–VII third instar neuromuscular junctions from w1118 control (gray traces) and sesB mutants (blue traces) were analyzed for evoked (A, EJP) and spontaneous neurotransmission (B, mEJP). C, D, Amplitudes as dot plots with each dot corresponding to one animal. Lines depict the mean of the sample. E, Neuromuscular junctions stimulated at low-frequency (3 Hz; E) and high-frequency (10 Hz; F–H) in the presence of 1 μm bafilomycin A1 to assess recycling and reserve pools of synaptic vesicles. E–H, Control animals shown as black symbols (E-G, w1118; H, w1118>C155), blue symbols show sesB mutants (F, G), and neuronal-specific sesB RNAi (H, C155>sesB RNAi). Average ± SEM. I, J, Quantitation of graphs E–H as time (measured as stimulus number) to 50% depletion (Tau) compared with response at time/stimulus 0. I, Corresponds to synapses stimulated at 3 Hz, recycling pool of vesicles, whereas J shows results for synapses stimulated at 10 Hz, reserve pool of vesicles. Number of animal is shown in the bottom bar. Average ± SEM. K, Tau statistical differences among genotypes at 3 Hz (top) and 10 Hz (bottom) represented as heat maps. Italic numbers depict genotypes in I and J. All comparisons in I and J were performed with one-way ANOVA followed by Fisher's multiple comparison.
SLC25A1 and SLC25A4 differentially modulate calcium-dependent synaptic activity
Calcium and mitochondrial calcium stores modulate spontaneous and evoked synaptic transmission as well as synaptic plasticity (Vos et al., 2010). To determine whether dSLC25A1-sea and dSLC25A4-sesB haploinsufficiencies affect mitochondrial calcium storage, we measured the message levels of calcium pumps and transporters residing in mitochondria and endoplasmic reticulum, spontaneous and evoked neurotransmission, and PPF at low and high extracellular calcium. We chose PPF because its sensitivity to the functional integrity of mitochondria (Weeber et al., 2002; Levy et al., 2003; Regehr, 2012).
We first sought to established whether complete knock-out of any of these SLC25A transporters could compromise mitochondrial calcium buffering mechanisms. We used human cells rendered SLC25A1- or SLC25A4-null by CRISPR-Cas9 genome editing. Mitochondria were labeled with the Ca2+-sensitive dye Rhod-2/AM, followed by perfusion with a calcium-containing buffer. Wild-type mitochondria rapidly increased their fluorescence (Fig. 8A). In contrast, the calcium influx rate was similarly reduced in either SLC25A1- or SLC25A4-null mitochondria to ∼50% (Fig. 8A,B). Expression of the mitochondrial calcium uniporter (MCU) and endoplasmic reticulum calcium ATPases (SERCA1–3) mRNAs was unaltered in both null cells (data not shown). These results raise the possibility that dSLC25A1-sea and dSLC25A4-sesB haploinsufficiencies may modulate calcium-dependent neurotransmission and plasticity.
The expression of the MCU and the endoplasmic reticulum calcium ATPase (SERCA) mRNAs was normal in dSLC25A1-sea and dSLC25A4-sesB haploinsufficient Drosophila heads (Fig. 8C). Thus, we inquired whether dSLC25A1-sea and dSLC25A4-sesB haploinsufficiencies could affect spontaneous neurotransmission in a calcium-sensitive manner (Fig. 8D–F). The amplitude of spontaneous fusion events (mEJPs) was neither altered by dSLC25A1-sea nor dSLC25A4-sesB haploinsufficiency, nor by the extracellular calcium concentration (Fig. 8D,E). However, the frequency of the mEJPs was increased in seaΔ24/+ synapses both at low and high extracellular calcium while sesBorg/+ synapses increased their frequency only at high calcium concentrations (Fig. 8D,F). These results suggest an impaired calcium buffering capacity at haploinsufficient synapses. These different mEJP frequency responses between dSLC25A1-sea and dSLC25A4-sesB haploinsufficiencies could reflect the same mechanism affected to a different extent or divergent calcium buffering mechanisms affected by dSLC25A1-sea and dSLC25A4-sesB. To resolve between these hypotheses, we analyzed PPF (Fig. 8G–I). We reasoned that paired-pulsed facilitation should be similarly perturbed in seaΔ24/+ and sesBorg/+ synapses under a common mechanism model. sesBorg/+ synapses have reduced paired-pulsed facilitation at low extracellular calcium concentrations but normal at high extracellular calcium (Fig. 8G,I). In contrast, PPF was normal at both extracellular calcium concentrations in seaΔ24/+ synapses (Fig. 8G,I). These results support a model whereby different calcium buffering mechanisms are perturbed in dSLC25A1-sea and dSLC25A4-sesB heterozygotic synapses.
SLC25A1 and SLC25A4 haploinsufficiencies differentially modulate behavior
To assess the behavioral consequences of partial loss of function in components of the SLC25A1-SLC25A4 interactome, we analyzed sleep patterns in wild-type animals and mutants carrying sesBorg, sesB9Ed-1, and sesB RNAi (Fig. 9). We contrasted sesB-dependent phenotypes with sleep phenotypes induced by downregulation of dSLC25A1-sea (Fig. 9-1). We chose sleep as a non-invasive and high-throughput analysis of an adult behavior that is entrained by environmental cues (Hendricks et al., 2000; Freeman et al., 2012). Moreover, sleep alterations are frequent in neurodevelopmental disorders (Chouinard et al., 2004; Monti and Monti, 2004; Krakowiak et al., 2008; Petrovsky et al., 2014; Kamath et al., 2015). We measured locomotor activity using the Drosophila Activity Monitoring system to quantify episodes of activity and sleep in a 12 h light/dark cycle. Wild-type Canton S animals demonstrated the highest density of sleep activity during the dark period (Fig. 9A,B and Fig. 9-1; zeitgeber times ZT12–ZT24) with an increased number of sleep–wake transitions at the beginning and end of the light cycle (Fig. 9A,B and Fig. 9-1; zeitgeber times ZT1 and ZT12). This pattern was disrupted in sesBorg and sesB9Ed-1 animals, which exhibit increased sleep–wake transitions throughout the 24 h period (Fig. 9A,B, zeitgeber times ZT1–ZT24). Most sleep events occurred at night (Fig. 9A,B and Fig. 9-1; zeitgeber times ZT12– ZT24). This pattern was disrupted in sesBorg and sesB9Ed-1 animals, which experienced increased awake–sleep transitions throughout the whole day (Fig. 9A,B; zeitgeber times ZT1– ZT24). sesB-deficient animals slept more (Fig. 9C,D,G), a phenotype that was evident during the day [Fig. 9A,B (zeitgeber times ZT1– ZT12), E] and night [Fig. 9A,B, (zeitgeber times ZT12–ZT24) F]. The sleep increase phenotype observed in sesB alleles was selectively phenocopied only by glutamatergic neuron-specific sesB RNAi (Fig. 9H, Vglut driver). Neither glial-specific (Fig. 9I, repo driver) nor dopaminergic neuron-specific sesB RNAi elicited any sleep phenotypes (Fig. 9J, Ddc driver). These sesB-dependent phenotypes were in sharp contrast with the sea-dependent traits in two key aspects. First, sea RNAi decreased, rather than increased, total sleep duration, but only during the light period (Fig. 9-1A,B; zeitgeber times ZT1–ZT12; and Fig. 9-1 compare C,D with E,F). Second, this light-selective phenotype was only induced by downregulation of dSLC25A1-sea in dopaminergic neurons but not in glutamatergic neurons (Fig. 9-1, compare C,D with I,J). These results demonstrate that partial loss of function in dSLC25A1-sea or dSLC25A4-sesB produce neuronal cell-type-specific alterations of sleep.
Figure 9.
The Drosophila SLC25A4 orthologue, sesB, is required in glutamatergic neurons for sleep. A, Individual hypnograms of two Canton S control and two sesB mutant flies illustrate sleep–wake activity patterns across the 12 h light (zeitgeber times ZT1–ZT12) and 12 h dark (zeitgeber times ZT12–ZT24) periods. B, Heat map of sleep–wake activity (gray and teal, respectively) in Canton S control (n = 229), sesorg (n = 234), and sesB9ed-1/+ (n = 53) depict the activity for each animal averaged across 1 h bins. Each column is 1 zeitgeber hour and each row an animal. C–G, Probability plots of sleep parameters per 24 h (C, D, G) or 12 h light/dark periods (E, F) from animals depicted in B. TST, Total sleeping time. H, The number of sleep bouts per 24 h is increased by sesB RNAi targeted to glutamatergic neurons (CS = 78, VGlut>CS = 72, VGlut>RNAi = 82 animals) but neither in (I) glial cells (CS = 78, repo>CS = 53, repo>RNAi = 59 animals), nor (J) catecholaminergic neurons (CS = 21, Ddc>CS = 37, Ddc>RNAi = 56 animals). C–J, P values were estimated with the Kolmogorov–Smirnov test. Similar analysis in sea RNAi animals is presented in Figure 9-1.
Drosophila SLC25A1 Orthologue Sea is Required in Catecholaminergic Neurons for Sleep. A) Individual hypnograms of Canton S control, sea RNAi controls, and catecholaminergic-specific sea RNAi (Ddc>RNAi) flies (n=2 each) illustrates sleep-wake activity patterns across the 12:12 hour light (zeitgeber times ZT1 to 12) and dark (zeitgeber times ZT12 to 24) periods. B) Heat maps of sleep-wake activity (gray and teal, respectively) in Ddc driver control (Ddc>CS, n=56), sea RNAi control (n=40), and catecholaminergic-specific sea RNAi animals (Ddc>RNAi, n=40) depict activity for each animal averaged across one hour bins. Each column is one zeitgeber hour and each row is one animal. C-H) Probability plots of sleep parameters per 24 hours (C, D and G) or 12 hours light/dark periods (E, F and H) from animals depicted in B. TST is total sleeping time. G-H) The number of sleep bouts is decreased in catecholaminergic-specific sea RNAi animals. I-J) No effect of glutamatergic-specific sea RNAi (VGlut>CS=38, sea RNAi=40, VGlut>RNAi= 44 animals) p values were estimated with the Kolmogorov–Smirnov test. Download Figure 9-1, TIF file (2.6MB, tif)
Collectively, our findings demonstrate that synaptic morphology, plasticity, and complex behaviors are sensitive to partial loss of function of mitochondrial solute transporters. We propose that cell-type-specific mitoproteomes tune synapse development and function in normal and disease states.
Discussion
We identified mitochondrial pathways as statistically prioritized ontological terms in the 22q11.2 and the Df(16)A+/− proteomes. Our results recapitulate a previous proteomic study of Df(16)A+/− brains, which is also enriched in mitochondrial targets (Fig. 2-2; Wesseling et al., 2017). Here, we expanded this prior study by demonstrating first, that mitoproteomes are affected in neuronal and non-neuronal cells carrying this microdeletion. Second, we identified a novel interaction between the inner mitochondrial transporters SLC25A1 and SLC25A4; this physical, functional, and genetic interaction forming a high connectivity hub downstream to the 22q11.2 microdeletion (Fig. 4A and Fig. 4-1). The proper gene dosage of these mitochondrial transporters is required for normal synapse morphology, function, plasticity, and behavior as demonstrated by Drosophila haploinsufficiencies in dSLC25A1-sea and dSLC25A4-sesB (Figs. 5–9). We further curated a SLC25A1-SLC25A4 interactome using comprehensive in silico tools and found that the expression of other SLC25A transporters belonging to this interactome is altered in neurons from schizophrenia cases where the genetic risk factor is other than the 22q11.2 microdeletion (Fig. 4E–G). Our systems analysis of the 22q11.2 and the Df(16)A+/− proteomes intersects with studies where changes in mitochondrial ontologies strongly associate with psychiatric disease (Gandal et al., 2018) and with reports of alterations in mitochondrial transcripts, protein composition, function, and morphology in brains from patients with psychotic disorders (Middleton et al., 2002; Rosenfeld et al., 2011; Enwright Iii et al., 2018; Norkett et al., 2017). Although our studies were performed in a single Drosophila peripheral synapse and await confirmation in mammalian central synapses; our results support the hypothesis that the mitochondrion, in particular components of the SLC25A1-SLC25A4 interactome (Fig. 4A and Fig. 4-1), modulate the risk of neurodevelopmental disorders.
The robustness of our gene ontology conclusions is founded on the strong agreement between the 22q11.2 and the Df(16)A+/− proteomes. However, we deem necessary to discuss limitations inherent to the genealogical proteomic approach used here to generate the 22q11.2 proteome. First, the use of fibroblasts limits proteomic surveys to uncover systemic rather than neuronal-specific molecular phenotypes. Second, we observed that gene ontologies derived from different families are not overlapping despite the consistency of ontologies obtained from the same family yet with different quantitative proteomics methods (Fig. 1-1). Although we minimized the noise introduced by genetic variability among subjects by comparing proteomes within a family; noise introduced by variables like age, sex, cell passage, and epigenetic modifications because of possible drug use still contribute to our datasets. To circumvent these limitations, we reasoned that if 22q11.2 proteomes from different families contained a majority of 22q11.2-specific hits plus different sources of random noise, then the addition of these family-specific proteomes into one dataset should enrich 22q11.2-specific gene ontologies while degrading ontologies due to random noise. We empirically tested this idea by adding the 22q11.2 and the Df(16)A+/− proteomes, which resulted in improved statistical scores for the ontological categories associated to each one of these two datasets while maintaining their overall priority rank (Fig. 2-2C). In contrast, addition of a random gene dataset of increasing size progressively degraded statistical scores and/or ranking of microdeletion-specific ontological categories (Fig. 2-2C). These in silico data analyses support our approach of adding 22q11.2 family-specific proteomes to enrich gene ontologies affected by a genetic defect while diluting those generated by random noise. However, the best evidence supporting genealogical proteomics are the two independent Df(16)A+/− brain proteomes, which validate our studies (Fig. 2 and 2–2). All three datasets, one in human and two in mouse, converge on similar ontological categories and rankings despite differences in tissues, species, and proteomic platforms used. The use of isogenic model systems is a way to circumvent random noise introduced by limited number of families in genealogical proteomics. Alternatively, either increasing the number of families analyzed or using different biological samples from the same family (fibroblasts, lymphoblasts, and IPSCs) should minimize the effects of noise on a dataset because of unforeseen variables or independent variables out of experimental reach.
The 22q11.2 locus encodes seven proteins contained in the Mitocarta 2.0 mitoproteome (COMT, MRPL40, PI4K, PRODH, SLC25A1, SNAP29, and TXNRD2; Pagliarini et al., 2008; Calvo et al., 2016). Some of these loci have been documented or proposed to be required for normal synaptic function (Paterlini et al., 2005; Maynard et al., 2008; Crabtree et al., 2016; Devaraju et al., 2017). The mitoproteome of 22q11.2 mutant cells and Df(16)A+/− brain likely reflect the collective effect of some or all these seven hemideficient genes (Devaraju and Zakharenko, 2017; Devaraju et al., 2017). We argue that these seven genes may not be the only 22q11.2 loci contributing to the alterations in the mitoproteome. For example, the DGCR8 gene, controlling microRNA-production, and seven miRNAs present in the 22q11.2 chromosomal segment could modulate the mitochondrial proteome acting both in nuclear and mitochondrial encoded RNAs (Stark et al., 2008; Chan et al., 2009; Bandiera et al., 2011; Miñones-Moyano et al., 2011; X. Zhang et al., 2014). The seven 22q11.2 genes, which are part of Mitocarta 2.0, are differentially expressed in different brain regions and cell types. Thus, their expression could influence the extent and quality of changes in mitochondrial proteomes from different cell types and brain regions in normal and disease states. We found that the stoichiometry of the mitoproteome or mitotranscriptome is different between two brain regions in normal mouse brain and between different cell types within Drosophila mushroom bodies (Fig. 2 and Fig. 2-1). We believe that these regional and cellular differences in mitochondrial composition stoichiometry are consequential because the impact of dSLC25A1-sea or dSLC25A4-sesB haploinsufficiencies on Drosophila neurotransmission, synaptic plasticity, and sleep patterns is differential. The clearest example is the disruption of sleep by dSLC25A1-sea or dSLC25A4-sesB haploinsufficiencies, where a phenotype depends on whether glutamatergic or catecholaminergic cells are targeted. It remains to be confirmed whether these effects are phenocopied by cell-type-specific downregulation or knock-out of other components of the SLC25A1-SLC25A4 interactome in vertebrate and invertebrate brains. However, it is reasonable to propose that differences in mitochondrial composition stoichiometry in different neuronal cell types could explain why, of the SLC25A transcripts analyzed, SLC25A3 was the most affected in layer 3 parvalbumin cells and SLC25A4 the most affected in layers 3 and 5 pyramidal cells in subjects with schizophrenia (Fig. 4E).
22q11.2 microdeletion syndrome increases the risk of developing schizophrenia or Parkinson's disease by 20-fold (Bassett et al., 2000; Hodgkinson et al., 2001; Bassett and Chow, 2008; Zaleski et al., 2009; Butcher et al., 2013, 2017; Mok et al., 2016). This observation prompted us to ask about the identity of candidate pathways capable of contributing to both psychiatric and neurodegenerative phenotypes. Complex I and other respiratory complex subunits are prominently represented in the 22q11.2 and Df(16)A+/− proteomes and the SLC25A1-SLC25A4 interactome (Fig. 4 and Fig. 4-1). Respiratory chain complexes could contribute to the expression of psychiatric and/or neurodegenerative pathologies. Our contention is founded on the capacity of complex I chemical inhibitors to either cause Parkinson's-like phenotypes (MPP+ and rotenone) or ameliorate psychosis symptoms (haloperidol, chlorpromazine, risperidone; Burkhardt et al., 1993; Prince et al., 1997; Modica-Napolitano et al., 2003; Rosenfeld et al., 2011; Elmorsy and Smith, 2015). Moreover, primarily mitochondrial diseases that affect the activity of the respiratory chain complexes, such as Leigh syndrome, can cause neurodegeneration and psychiatric symptoms (DiMauro and Schon, 2008; Anglin et al., 2012a,b; Sheng and Cai, 2012). Although still speculative, we put forward a testable model where changes in the expression of the mitoproteome modulate the expression of neurodevelopmental and/or neurodegenerative phenotypes.
Footnotes
This work was supported by grants from the National Institutes of Health R56 MH111459 to V.F. and Emory Catalyst Grant, Fondecyt-Chile Grant 1171014 to G.M.R., R01HL108882 to S.M.C., R01MH097879 to J.A.G., and by the FIRST postdoctoral Fellowship NIH 5K12GM000680 to C.H., in part by the Emory HPLC Bioanalytical Core (EHBC) and the Emory Imaging Core, and EHBC is supported by the Department of Pharmacology, Emory University School of Medicine and the Georgia Clinical and Translational Science Alliance of the National Institutes of Health under Award Number UL1TR002378.We thank members of the Faundez Laboratory for their comments and insight. Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study.
The authors declare no competing financial interests.
References
- Anders S, Pyl PT, Huber W (2015) HTSeq: a python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews S. (2010) FASTQC: a quality control tool for high throughput sequence data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- Anglin RE, Tarnopolsky MA, Mazurek MF, Rosebush PI (2012a) The psychiatric presentation of mitochondrial disorders in adults. J Neuropsychiatry Clin Neurosci 24:394–409. 10.1176/appi.neuropsych.11110345 [DOI] [PubMed] [Google Scholar]
- Anglin RE, Garside SL, Tarnopolsky MA, Mazurek MF, Rosebush PI (2012b) The psychiatric manifestations of mitochondrial disorders: a case and review of the literature. J Clin Psychiatry 73:506–512. 10.4088/JCP.11r07237 [DOI] [PubMed] [Google Scholar]
- Arion D, Corradi JP, Tang S, Datta D, Boothe F, He A, Cacace AM, Zaczek R, Albright CF, Tseng G, Lewis DA (2015) Distinctive transcriptome alterations of prefrontal pyramidal neurons in schizophrenia and schizoaffective disorder. Mol Psychiatry 20:1397–1405. 10.1038/mp.2014.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandiera S, Rüberg S, Girard M, Cagnard N, Hanein S, Chrétien D, Munnich A, Lyonnet S, Henrion-Caude A (2011) Nuclear outsourcing of RNA interference components to human mitochondria. PloS One 6:e20746. 10.1371/journal.pone.0020746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett AS, Chow EW (2008) Schizophrenia and 22q11.2 deletion syndrome. Curr Psychiatry Rep 10:148–157. 10.1007/s11920-008-0026-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett AS, Chow EW, Weksberg R (2000) Chromosomal abnormalities and schizophrenia. Am J Med Genet 97:45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett AS, Chow EW, AbdelMalik P, Gheorghiu M, Husted J, Weksberg R (2003) The schizophrenia phenotype in 22q11 deletion syndrome. Am J Psychiatry 160:1580–1586. 10.1176/appi.ajp.160.9.1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bena F, Bruno DL, Eriksson M, van Ravenswaaij-Arts C, Stark Z, Dijkhuizen T, Gerkes E, Gimelli S, Ganesamoorthy D, Thuresson AC, Labalme A, Till M, Bilan F, Pasquier L, Kitzis A, Dubourgm C, Rossi M, Bottani A, Gagnebin M, Sanlaville D, et al. (2013) Molecular and clinical characterization of 25 individuals with exonic deletions of NRXN1 and comprehensive review of the literature. Am J Med Genet B Neuropsychiatr Genet 162B:388–403. 10.1002/ajmg.b.32148 [DOI] [PubMed] [Google Scholar]
- Bers DM, Patton CW, Nuccitelli R (2010) A practical guide to the preparation of Ca2+ buffers. Methods Cell Biol 99:1–26. 10.1016/B978-0-12-374841-6.00001-3 [DOI] [PubMed] [Google Scholar]
- Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z, Galon J (2009) ClueGO: a cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25:1091–1093. 10.1093/bioinformatics/btp101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhardt C, Kelly JP, Lim YH, Filley CM, Parker WD Jr (1993) Neuroleptic medications inhibit complex I of the electron transport chain. Ann Neurol 33:512–517. 10.1002/ana.410330516 [DOI] [PubMed] [Google Scholar]
- Butcher NJ, Kiehl TR, Hazrati LN, Chow EW, Rogaeva E, Lang AE, Bassett AS (2013) Association between early-onset Parkinson disease and 22q11.2 deletion syndrome: identification of a novel genetic form of Parkinson disease and its clinical implications. JAMA Neurol 70:1359–1366. 10.1001/jamaneurol.2013.3646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher NJ, Marras C, Pondal M, Rusjan P, Boot E, Christopher L, Repetto GM, Fritsch R, Chow EWC, Masellis M, Strafella AP, Lang AE, Bassett AS (2017) Neuroimaging and clinical features in adults with a 22q11.2 deletion at risk of Parkinson's disease. Brain 140:1371–1383. 10.1093/brain/awx053 [DOI] [PubMed] [Google Scholar]
- Calvo SE, Clauser KR, Mootha VK (2016) MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res 44:D1251–1257. 10.1093/nar/gkv1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell RA, Turner GC (2010) The mushroom body. Curr Biol 20:R11–R12. 10.1016/j.cub.2009.10.031 [DOI] [PubMed] [Google Scholar]
- Chan SY, Zhang YY, Hemann C, Mahoney CE, Zweier JL, Loscalzo J (2009) MicroRNA-210 controls mitochondrial metabolism during hypoxia by repressing the iron-sulfur cluster assembly proteins ISCU1/2. Cell Metab 10:273–284. 10.1016/j.cmet.2009.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CL, Hu Y, Udeshi ND, Lau TY, Wirtz-Peitz F, He L, Ting AY, Carr SA, Perrimon N (2015) Proteomic mapping in live Drosophila tissues using an engineered ascorbate peroxidase. Proc Natl Acad Sci U S A 112:12093–12098. 10.1073/pnas.1515623112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma'ayan A (2013) Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14:128. 10.1186/1471-2105-14-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouinard S, Poulin J, Stip E, Godbout R (2004) Sleep in untreated patients with schizophrenia: a meta-analysis. Schizoph Bull 30:957–967. 10.1093/oxfordjournals.schbul.a007145 [DOI] [PubMed] [Google Scholar]
- Crabtree GW, Park AJ, Gordon JA, Gogos JA (2016) Cytosolic accumulation of l-proline disrupts GABA-ergic transmission through GAD blockade. Cell Rep 17:570–582. 10.1016/j.celrep.2016.09.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker A, Guan XJ, Murphy CT, Murthy M (2016) Cell-type-specific transcriptome analysis in the Drosophila mushroom body reveals memory-related changes in gene expression. Cell Rep 15:1580–1596. 10.1016/j.celrep.2016.04.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Rio G, Koschützki D, Coello G (2009) How to identify essential genes from molecular networks? BMC Syst Biol 3:102. 10.1186/1752-0509-3-102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado R, Maureira C, Oliva C, Kidokoro Y, Labarca P (2000) Size of vesicle pools, rates of mobilization, and recycling at neuromuscular synapses of a Drosophila mutant, shibire. Neuron 28:941–953. 10.1016/S0896-6273(00)00165-3 [DOI] [PubMed] [Google Scholar]
- Devaraju P, Zakharenko SS (2017) Mitochondria in complex psychiatric disorders: lessons from mouse models of 22q11.2 deletion syndrome: hemizygous deletion of several mitochondrial genes in the 22q11.2 genomic region can lead to symptoms associated with neuropsychiatric disease. Bioessays 39:1600177. 10.1002/bies.201600177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaraju P, Yu J, Eddins D, Mellado-Lagarde MM, Earls LR, Westmoreland JJ, Quarato G, Green DR, Zakharenko SS (2017) Haploinsufficiency of the 22q11.2 microdeletion gene Mrpl40 disrupts short-term synaptic plasticity and working memory through dysregulation of mitochondrial calcium. Mol Psychiatry 22:1313–1326. 10.1038/mp.2016.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMauro S, Schon EA (2008) Mitochondrial disorders in the nervous system. Annu Rev Neurosci 31:91–123. 10.1146/annurev.neuro.30.051606.094302 [DOI] [PubMed] [Google Scholar]
- Elmorsy E, Smith PA (2015) Bioenergetic disruption of human micro-vascular endothelial cells by antipsychotics. Biochem Biophys Res Commun 460:857–862. 10.1016/j.bbrc.2015.03.122 [DOI] [PubMed] [Google Scholar]
- Enwright Iii JF, Huo Z, Arion D, Corradi JP, Tseng G, Lewis DA (2018) Transcriptome alterations of prefrontal cortical parvalbumin neurons in schizophrenia. Mol Psychiatry 23:1606–1613. 10.1038/mp.2017.216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd BJ, Wilkerson EM, Veling MT, Minogue CE, Xia C, Beebe ET, Wrobel RL, Cho H, Kremer LS, Alston CL, Gromek KA, Dolan BK, Ulbrich A, Stefely JA, Bohl SL, Werner KM, Jochem A, Westphall MS, Rensvold JW, Taylor RW, Prokisch H, et al. (2016) Mitochondrial protein interaction mapping identifies regulators of respiratory chain function. Mol Cell 63:621–632. 10.1016/j.molcel.2016.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank CA, Wang X, Collins CA, Rodal AA, Yuan Q, Verstreken P, Dickman DK (2013) New approaches for studying synaptic development, function, and plasticity using Drosophila as a model system. J Neurosci 33:17560–17568. 10.1523/JNEUROSCI.3261-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman A, Pranski E, Miller RD, Radmard S, Bernhard D, Jinnah HA, Betarbet R, Rye DB, Sanyal S (2012) Sleep fragmentation and motor restlessness in a Drosophila model of restless legs syndrome. Curr Biol 22:1142–1148. 10.1016/j.cub.2012.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandal MJ, Haney JR, Parikshak NN, Leppa V, Ramaswami G, Hartl C, Schork AJ, Appadurai V, Buil A, Werge TM, Liu C, White KP, CommonMind C, Psych EC, i PBWG, Horvath S, Geschwind DH (2018) Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science 359:693–697. 10.1126/science.aad6469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girirajan S, Campbell CD, Eichler EE (2011) Human copy number variation and complex genetic disease. Annu Rev Genet 45:203–226. 10.1146/annurev-genet-102209-163544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guna A, Butcher NJ, Bassett AS (2015) Comparative mapping of the 22q11.2 deletion region and the potential of simple model organisms. J Neurodev Disord 7:18. 10.1186/s11689-015-9113-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Macleod GT, Wellington A, Hu F, Panchumarthi S, Schoenfield M, Marin L, Charlton MP, Atwood HL, Zinsmaier KE (2005) The GTPase dMiro is required for axonal transport of mitochondria to Drosophila synapses. Neuron 47:379–393. 10.1016/j.neuron.2005.06.027 [DOI] [PubMed] [Google Scholar]
- Havugimana PC, Hart GT, Nepusz T, Yang H, Turinsky AL, Li Z, Wang PI, Boutz DR, Fong V, Phanse S, Babu M, Craig SA, Hu P, Wan C, Vlasblom J, Dar VU, Bezginov A, Clark GW, Wu GC, Wodak SJ, et al. (2012) A census of human soluble protein complexes. Cell 150:1068–1081. 10.1016/j.cell.2012.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein MY, Hubner NC, Poser I, Cox J, Nagaraj N, Toyoda Y, Gak IA, Weisswange I, Mansfeld J, Buchholz F, Hyman AA, Mann M (2015) A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell 163:712–723. 10.1016/j.cell.2015.09.053 [DOI] [PubMed] [Google Scholar]
- Hendricks JC, Finn SM, Panckeri KA, Chavkin J, Williams JA, Sehgal A, Pack AI (2000) Rest in Drosophila is a sleep-like state. Neuron 25:129–138. 10.1016/S0896-6273(00)80877-6 [DOI] [PubMed] [Google Scholar]
- Hodgkinson KA, Murphy J, O'Neill S, Brzustowicz L, Bassett AS (2001) Genetic counselling for schizophrenia in the era of molecular genetics. Can J Psychiatry 46:123–130. 10.1177/070674370104600202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffding LK, Trabjerg BB, Olsen L, Mazin W, Sparsø T, Vangkilde A, Mortensen PB, Pedersen CB, Werge T (2017) Risk of psychiatric disorders among individuals with the 22q11.2 deletion or duplication: a Danish nationwide, register-based study. JAMA Psychiatry 74:282–290. 10.1001/jamapsychiatry.2016.3939 [DOI] [PubMed] [Google Scholar]
- Huttlin EL, Ting L, Bruckner RJ, Gebreab F, Gygi MP, Szpyt J, Tam S, Zarraga G, Colby G, Baltier K, Dong R, Guarani V, Vaites LP, Ordureau A, Rad R, Erickson BK, Wühr M, Chick J, Zhai B, Kolippakkam D, et al. (2015) The BioPlex network: a systematic exploration of the human interactome. Cell 162:425–440. 10.1016/j.cell.2015.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin EL, Bruckner RJ, Paulo JA, Cannon JR, Ting L, Baltier K, Colby G, Gebreab F, Gygi MP, Parzen H, Szpyt J, Tam S, Zarraga G, Pontano-Vaites L, Swarup S, White AE, Schweppe DK, Rad R, Erickson BK, Obar RA, et al. (2017) Architecture of the human interactome defines protein communities and disease networks. Nature 545:505–509. 10.1038/nature22366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Schizophrenia Consortium (2008) Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature 455:237–241. 10.1038/nature07239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka K, Paek M, Kamiya A, Sawa A (2006) A review of disrupted-in-schizophrenia-1 (DISC1): neurodevelopment, cognition, and mental conditions. Biol Psychiatry 59:1189–1197. 10.1016/j.biopsych.2006.03.065 [DOI] [PubMed] [Google Scholar]
- Iyer J, Singh MD, Jensen M, Patel P, Pizzo L, Huber E, Koerselman H, Weiner AT, Lepanto P, Vadodaria K, Kubina A, Wang Q, Talbert A, Yennawar S, Badano J, Manak JR, Rolls MM, Krishnan A, Girirajan S (2018) Pervasive genetic interactions modulate neurodevelopmental defects of the autism-associated 16p11.2 deletion in Drosophila melanogaster. Nat Commun 9:2548. 10.1038/s41467-018-04882-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalbrzikowski M, Lazaro MT, Gao F, Huang A, Chow C, Geschwind DH, Coppola G, Bearden CE (2015) Transcriptome profiling of peripheral blood in 22q11.2 deletion syndrome reveals functional pathways related to psychosis and autism spectrum disorder. PloS One 10:e0132542. 10.1371/journal.pone.0132542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas RK, Montojo CA, Bearden CE (2014) The 22q11.2 deletion syndrome as a window into complex neuropsychiatric disorders over the lifespan. Biol Psychiatry 75:351–360. 10.1016/j.biopsych.2013.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath J, Virdi S, Winokur A (2015) Sleep disturbances in schizophrenia. Psychiatr Clin North Am 38:777–792. 10.1016/j.psc.2015.07.007 [DOI] [PubMed] [Google Scholar]
- Karayiorgou M, Simon TJ, Gogos JA (2010) 22q11.2 microdeletions: linking DNA structural variation to brain dysfunction and schizophrenia. Nat Rev Neurosci 11:402–416. 10.1038/nrn2841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SM, Kumar V, Lin YQ, Karunanithi S, Ramaswami M (2009) Fos and jun potentiate individual release sites and mobilize the reserve synaptic vesicle pool at the Drosophila larval motor synapse. Proc Natl Acad Sci U S A 106:4000–4005. 10.1073/pnas.0806064106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov G. (2015) CNVs in neuropsychiatric disorders. Hum Mol Genet 24:R45–R49. 10.1093/hmg/ddv253 [DOI] [PubMed] [Google Scholar]
- Klingenberg M. (2008) The ADP and ATP transport in mitochondria and its carrier. Biochim Biophys Acta 1778:1978–2021. 10.1016/j.bbamem.2008.04.011 [DOI] [PubMed] [Google Scholar]
- Krakowiak P, Goodlin-Jones B, Hertz-Picciotto I, Croen LA, Hansen RL (2008) Sleep problems in children with autism spectrum disorders, developmental delays, and typical development: a population-based study. J Sleep Res 17:197–206. 10.1111/j.1365-2869.2008.00650.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with bowtie 2. Nat Methods 9:357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy M, Faas GC, Saggau P, Craigen WJ, Sweatt JD (2003) Mitochondrial regulation of synaptic plasticity in the hippocampus. J Biol Chem 278:17727–17734. 10.1074/jbc.M212878200 [DOI] [PubMed] [Google Scholar]
- Lin MT, Beal MF (2006) Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 443:787–795. 10.1038/nature05292 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YW, Acoba MG, Selvaraju K, Huang TC, Nirujogi RS, Sathe G, Pandey A, Claypool SM (2017) Human adenine nucleotide translocases physically and functionally interact with respirasomes. Mol Biol Cell 28:1489–1506. 10.1091/mbc.e17-03-0195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra D, Sebat J (2012) CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell 148:1223–1241. 10.1016/j.cell.2012.02.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CR, Howrigan DP, Merico D, Thiruvahindrapuram B, Wu W, Greer DS, Antaki D, Shetty A, Holmans PA, Pinto D, Gujral M, Brandler WM, Malhotra D, Wang Z, Fajarado KVF, Maile MS, Ripke S, Agartz I, Albus M, Alexander M, et al. (2017) Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat Genet 49:27–35. 10.1038/ng.3725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell JT, Tsai CH, Mohiuddin TA, Kwong JQ (2018) Analyses of mitochondrial calcium influx in isolated mitochondria and cultured cells. J Vis Exp 27:e57225. 10.3791/57225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard TM, Meechan DW, Dudevoir ML, Gopalakrishna D, Peters AZ, Heindel CC, Sugimoto TJ, Wu Y, Lieberman JA, Lamantia AS (2008) Mitochondrial localization and function of a subset of 22q11 deletion syndrome candidate genes. Mol Cell Neurosci 39:439–451. 10.1016/j.mcn.2008.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald-McGinn DM, Sullivan KE, Marino B, Philip N, Swillen A, Vorstman JA, Zackai EH, Emanuel BS, Vermeesch JR, Morrow BE, Scambler PJ, Bassett AS (2015) 22q11.2 deletion syndrome. Nat Rev Dis Primers 1:15071. 10.1038/nrdp.2015.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton FA, Mirnics K, Pierri JN, Lewis DA, Levitt P (2002) Gene expression profiling reveals alterations of specific metabolic pathways in schizophrenia. J Neurosci 22:2718–2729. 10.1523/JNEUROSCI.22-07-02718.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miñones-Moyano E, Porta S, Escaramís G, Rabionet R, Iraola S, Kagerbauer B, Espinosa-Parrilla Y, Ferrer I, Estivill X, Martí E (2011) MicroRNA profiling of Parkinson's disease brains identifies early downregulation of miR-34b/c which modulate mitochondrial function. Hum Mol Genet 20:3067–3078. 10.1093/hmg/ddr210 [DOI] [PubMed] [Google Scholar]
- Modica-Napolitano JS, Lagace CJ, Brennan WA, Aprille JR (2003) Differential effects of typical and atypical neuroleptics on mitochondrial function in vitro. Arch Pharm Res 26:951–959. 10.1007/BF02980205 [DOI] [PubMed] [Google Scholar]
- Mok KY, Sheerin U, Simón-Sánchez J, Salaka A, Chester L, Escott-Price V, Mantripragada K, Doherty KM, Noyce AJ, Mencacci NE, Lubbe SJ, Lubbe SJ, Williams-Gray CH, Barker RA, van Dijk KD, Berendse HW, Heutink P, Corvol JC, Cormier F, Lesage S, et al. (2016) Deletions at 22q11.2 in idiopathic Parkinson's disease: a combined analysis of genome-wide association data. Lancet Neurol 15:585–596. 10.1016/S1474-4422(16)00071-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monti JM, Monti D (2004) Sleep in schizophrenia patients and the effects of antipsychotic drugs. Sleep Med Rev 8:133–148. 10.1016/S1087-0792(02)00158-2 [DOI] [PubMed] [Google Scholar]
- Morciano P, Carrisi C, Capobianco L, Mannini L, Burgio G, Cestra G, De Benedetto GE, Corona DF, Musio A, Cenci G (2009) A conserved role for the mitochondrial citrate transporter Sea/SLC25A1 in the maintenance of chromosome integrity. Hum Mol Genet 18:4180–4188. 10.1093/hmg/ddp370 [DOI] [PubMed] [Google Scholar]
- Mullin AP, Sadanandappa MK, Ma W, Dickman DK, VijayRaghavan K, Ramaswami M, Sanyal S, Faundez V (2015) Gene dosage in the dysbindin schizophrenia susceptibility network differentially affect synaptic function and plasticity. J Neurosci 35:325–338. 10.1523/JNEUROSCI.3542-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norkett R, Modi S, Kittler JT (2017) Mitochondrial roles of the psychiatric disease risk factor DISC1. Schizophr Res 187:47–54. 10.1016/j.schres.2016.12.025 [DOI] [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, Hill DE, Vidal M, Evans JG, Thorburn DR, Carr SA, Mootha VK (2008) A mitochondrial protein compendium elucidates complex I disease biology. Cell 134:112–123. 10.1016/j.cell.2008.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri F, Monné M (2016) Discoveries, metabolic roles and diseases of mitochondrial carriers: a review. Biochim Biophys Acta 1863:2362–2378. 10.1016/j.bbamcr.2016.03.007 [DOI] [PubMed] [Google Scholar]
- Paterlini M, Zakharenko SS, Lai WS, Qin J, Zhang H, Mukai J, Westphal KG, Olivier B, Sulzer D, Pavlidis P, Siegelbaum SA, Karayiorgou M, Gogos JA (2005) Transcriptional and behavioral interaction between 22q11.2 orthologs modulates schizophrenia-related phenotypes in mice. Nat Neurosci 8:1586–1594. 10.1038/nn1562 [DOI] [PubMed] [Google Scholar]
- Petrovsky N, Ettinger U, Hill A, Frenzel L, Meyhöfer I, Wagner M, Backhaus J, Kumari V (2014) Sleep deprivation disrupts prepulse inhibition and induces psychosis-like symptoms in healthy humans. J Neurosci 34:9134–9140. 10.1523/JNEUROSCI.0904-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping L, Duong DM, Yin L, Gearing M, Lah JJ, Levey AI, Seyfried NT (2018) Global quantitative analysis of the human brain proteome in Alzheimer's and Parkinson's disease. Sci Data 5:180036. 10.1038/sdata.2018.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince JA, Yassin MS, Oreland L (1997) Neuroleptic-induced mitochondrial enzyme alterations in the rat brain. J Pharmacol Exp Ther 280:261–267. [PubMed] [Google Scholar]
- Regehr WG. (2012) Short-term presynaptic plasticity. Cold Spring Harb Perspect Biol 4:a005702. 10.1101/cshperspect.a005702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikhy R, Ramaswami M, Krishnan KS (2003) A temperature-sensitive allele of Drosophila sesB reveals acute functions for the mitochondrial adenine nucleotide translocase in synaptic transmission and dynamin regulation. Genetics 165:1243–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld M, Brenner-Lavie H, Ari SG, Kavushansky A, Ben-Shachar D (2011) Perturbation in mitochondrial network dynamics and in complex I dependent cellular respiration in schizophrenia. Biol Psychiatry 69:980–988. 10.1016/j.biopsych.2011.01.010 [DOI] [PubMed] [Google Scholar]
- Rutkowski TP, Schroeder JP, Gafford GM, Warren ST, Weinshenker D, Caspary T, Mulle JG (2017) Unraveling the genetic architecture of copy number variants associated with schizophrenia and other neuropsychiatric disorders. J Neurosci Res 95:1144–1160. 10.1002/jnr.23970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro MR, Bray SM, Warren ST (2012) Molecular mechanisms of fragile X syndrome: a twenty-year perspective. Annu Rev Pathol 7:219–245. 10.1146/annurev-pathol-011811-132457 [DOI] [PubMed] [Google Scholar]
- Schneider M, Debbané M, Bassett AS, Chow EW, Fung WL, van den Bree M, Owen M, Murphy KC, Niarchou M, Kates WR, Antshel KM, Fremont W, McDonald-McGinn DM, Gur RE, Zackai EH, Vorstman J, Duijff SN, Klaassen PW, Swillen A, Gothelf D, et al. (2014) Psychiatric disorders from childhood to adulthood in 22q11.2 deletion syndrome: results from the International Consortium on Brain and Behavior in 22q11.2 Deletion Syndrome. Am J Psychiatry 171:627–639. 10.1176/appi.ajp.2013.13070864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw PJ, Cirelli C, Greenspan RJ, Tononi G (2000) Correlates of sleep and waking in Drosophila melanogaster. Science 287:1834–1837. [DOI] [PubMed] [Google Scholar]
- Sheng ZH, Cai Q (2012) Mitochondrial transport in neurons: impact on synaptic homeostasis and neurodegeneration. Nat Rev Neurosci 13:77–93. 10.1038/nrn3156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark KL, Xu B, Bagchi A, Lai WS, Liu H, Hsu R, Wan X, Pavlidis P, Mills AA, Karayiorgou M, Gogos JA (2008) Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat Genetics 40:751–760. 10.1038/ng.138 [DOI] [PubMed] [Google Scholar]
- Sullivan PF, Daly MJ, O'Donovan M (2012) Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat Rev Genet 13:537–551. 10.1038/nrg3240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztainberg Y, Zoghbi HY (2016) Lessons learned from studying syndromic autism spectrum disorders. Nat Neurosci 19:1408–1417. 10.1038/nn.4420 [DOI] [PubMed] [Google Scholar]
- Taylor EB. (2017) Functional properties of the mitochondrial carrier system. Trends Cell Biol 27:633–644. 10.1016/j.tcb.2017.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstreken P, Ly CV, Venken KJ, Koh TW, Zhou Y, Bellen HJ (2005) Synaptic mitochondria are critical for mobilization of reserve pool vesicles at Drosophila neuromuscular junctions. Neuron 47:365–378. 10.1016/j.neuron.2005.06.018 [DOI] [PubMed] [Google Scholar]
- Vos M, Lauwers E, Verstreken P (2010) Synaptic mitochondria in synaptic transmission and organization of vesicle pools in health and disease. Front Synaptic Neurosci 2:139. 10.3389/fnsyn.2010.00139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan C, Borgeson B, Phanse S, Tu F, Drew K, Clark G, Xiong X, Kagan O, Kwan J, Bezginov A, Chessman K, Pal S, Cromar G, Papoulas O, Ni Z, Boutz DR, Stoilova S, Havugimana PC, Guo X, Malty RH, et al. (2015) Panorama of ancient metazoan macromolecular complexes. Nature 525:339–344. 10.1038/nature14877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeber EJ, Levy M, Sampson MJ, Anflous K, Armstrong DL, Brown SE, Sweatt JD, Craigen WJ (2002) The role of mitochondrial porins and the permeability transition pore in learning and synaptic plasticity. J Biol chem 277:18891–18897. 10.1074/jbc.M201649200 [DOI] [PubMed] [Google Scholar]
- Wen Z, Nguyen HN, Guo Z, Lalli MA, Wang X, Su Y, Kim NS, Yoon KJ, Shin J, Zhang C, Makri G, Nauen D, Yu H, Guzman E, Chiang CH, Yoritomo N, Kaibuchi K, Zou J, Christian KM, Cheng L, et al. (2014) Synaptic dysregulation in a human iPS cell model of mental disorders. Nature 515:414–418. 10.1038/nature13716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesseling H, Xu B, Want EJ, Holmes E, Guest PC, Karayiorgou M, Gogos JA, Bahn S (2017) System-based proteomic and metabonomic analysis of the Df(16)A(+/−) mouse identifies potential miR-185 targets and molecular pathway alterations. Mol Psychiatry 22:384–395. 10.1038/mp.2016.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Candille SI, Choi Y, Xie D, Jiang L, Li-Pook-Than J, Tang H, Snyder M (2013) Variation and genetic control of protein abundance in humans. Nature 499:79–82. 10.1038/nature12223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaleski C, Bassett AS, Tam K, Shugar AL, Chow EW, McPherson E (2009) The co-occurrence of early onset parkinson disease and 22q11.2 deletion syndrome. Am J Med Genet A 149A:525–528. 10.1002/ajmg.a.32650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zuo X, Yang B, Li Z, Xue Y, Zhou Y, Huang J, Zhao X, Zhou J, Yan Y, Zhang H, Guo P, Sun H, Guo L, Zhang Y, Fu XD (2014) MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell 158:607–619. 10.1016/j.cell.2014.05.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YQ, Roote J, Brogna S, Davis AW, Barbash DA, Nash D, Ashburner M (1999) stress sensitive B encodes an adenine nucleotide translocase in Drosophila melanogaster. Genetics 153:891–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatic SA, Vrailas-Mortimer A, Gokhale A, Carey LJ, Scott E, Burch R, McCall MM, Rudin-Rush S, Davis JB, Hartwig C, Werner E, Li L, Petris M, Faundez V (2018) Rare disease mechanisms identified by genealogical proteomics of copper homeostasis mutant pedigrees. Cell Syst 6:368–380.e6. 10.1016/j.cels.2018.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
22q11.2 Microdeletion Genealogical Proteome Comparisons Among Pedigrees and Mass Spectrometry Quantitation Strategies. A-D) Cellular Component Gene ontologies (GO:CC) obtained using the ENRICHR engine. Data are depicted as canvases where every tile is occupied by an individual GO category whose p value significance is depicted by color intensity. A) First canvas to the left depicts a comparison for genealogical proteomes obtained in one pedigree by TMT (green) and to Label Free Quantification (LFQ, red). Other canvases in A show experiments comparing TMT with two independent SILAC experiments and combinations of TMT, LFQ and SILAC. Overlap of GO:CC terms is presented as yellow. B) Comparison of GO:CC terms obtained by genealogical proteomics from three pedigrees. C) Represents GO:CC term tiles overlapping among pedigrees in B). D) Shows gene ontology terms obtained by pooling into one dataset the proteomes from all pedigrees in B). E. Presents relevant GO terms in D. Individual pedigree and collective bioinformatics data can be found in Fig. 1-3. Download Figure 1-1, TIF file (1.2MB, tif)
Quantitative Mass Spectrometry Data for 22q11.2 Genealogical Proteomic Studies. Download Figure 1-2, XLSB file (10.7MB, xlsb)
Comparative Bioinformatic Analysis of 22q11.2 Genealogical Proteomes. Download Figure 1-3, XLSX file (68.7KB, xlsx)
The Drosophila Transcriptome Encoding Mitochondrial Proteins is Cell Type Specific. A-B) mRNA from single neuron types isolated from Drosophila mushroom bodies were analyzed by RNAseq. The transcriptome encoding mitochondrial proteins, as defined by Chen et al (Chen et al., 2015), was analyzed by principal component analysis (A) and hierarchical clustering using 1-Pearson correlation clustering (B) of columns (cells) and rows (transcripts). Cell types were identified as in Crocker et al (Crocker et al., 2016). Note the robust segregation of Kenyon cells from other cell types by the expression of the transcriptome encoding mitochondrial proteins. Download Figure 2-1, TIF file (673.8KB, tif)
Comparative Bioinformatics of the 22q11.2 Proteome and Two Independent Df(16)A-/+ Brain Proteomes. A) Venn diagrams depict from top to bottom: a comparison of common hits between our Df(16)A-/+ brain proteome and the Df(16)A-/+ brain proteome reported by Wesseling et al. PMID: 27001617. The Wesseling Df(16)A-/+ brain proteome and our 22q11.2 proteome. The Wesseling Df(16)A-/+ brain proteome and the mouse Mitocarta 2.0 dataset. B) Cellular Component gene ontology analysis of GO CC generated with the ENRICHR engine using the Wesseling Df(16)A-/+ brain proteome dataset and a similarly sized random mouse gene dataset. Random gene list was generated with the engine RandomGeneSetGenerator. C) Cellular Component gene ontology analysis (GO CC) was performed with the ENRICHR engine using the Wesseling Df(16)A-/+ brain proteome dataset either by itself, or in combination with our 22q11.2 proteome, or with 1500 (1x) or 3000 (2x) randomly generated genes. See discussion. Download Figure 2-2, TIF file (883.6KB, tif)
Bioinformatic Analysis of 22q11.2 Genealogical Proteomes and Df(16)A-/+ brain proteomes. Download Figure 2-3, XLSX file (146KB, xlsx)
Quantitative Mass Spectrometry Data for Df(16)A-/+ brains. Download Figure 2-4, XLSX file (3.7MB, xlsx)
Comprehensive in silico Interactome of the SLC25A1 and SLC25A4 Transporters. A) Comprehensive in silico interactome of the SLC25A1 and SLC25A4 mitochondrial transporters. Complexes I to V of the respiratory chain as well as SLC25A transporter family members are color coded. All nodes colored gray represent hits in the 22q11.2 proteome. B) The comprehensive interactome was analyzed with graph theory to determine high connectivity nodes predictive of essential genes using the closeness and betweeness centrality coefficients. Note the high connectivity of SLC25A4 in the comprehensive interactome. Download Figure 4-1, TIF file (4.4MB, tif)
Reduced Expression of Drosophila dSLC25A1-dSLC25A4 does not Affect Cellular ATP/ADP ratios. ATP/ADP ratios were measured in third instar larvae, adult heads, and human Hap1 cells of the indicated genotypes. n=3 for Drosophila Tissues and n=6 for Hap1 cells, One Way ANOVA followed by Fisher’s Least Significant Difference Comparison. Download Figure 5-1, TIF file (227.2KB, tif)
Drosophila SLC25A1 Orthologue Sea is Required in Catecholaminergic Neurons for Sleep. A) Individual hypnograms of Canton S control, sea RNAi controls, and catecholaminergic-specific sea RNAi (Ddc>RNAi) flies (n=2 each) illustrates sleep-wake activity patterns across the 12:12 hour light (zeitgeber times ZT1 to 12) and dark (zeitgeber times ZT12 to 24) periods. B) Heat maps of sleep-wake activity (gray and teal, respectively) in Ddc driver control (Ddc>CS, n=56), sea RNAi control (n=40), and catecholaminergic-specific sea RNAi animals (Ddc>RNAi, n=40) depict activity for each animal averaged across one hour bins. Each column is one zeitgeber hour and each row is one animal. C-H) Probability plots of sleep parameters per 24 hours (C, D and G) or 12 hours light/dark periods (E, F and H) from animals depicted in B. TST is total sleeping time. G-H) The number of sleep bouts is decreased in catecholaminergic-specific sea RNAi animals. I-J) No effect of glutamatergic-specific sea RNAi (VGlut>CS=38, sea RNAi=40, VGlut>RNAi= 44 animals) p values were estimated with the Kolmogorov–Smirnov test. Download Figure 9-1, TIF file (2.6MB, tif)