Abstract
To better examine circuit mechanisms underlying perception and behavior, researchers need tools to enable temporally precise control of action-potential generation of individual cells from neuronal ensembles. Here we demonstrate that such precision can be achieved with two-photon (2P) temporally focused computer-generated holography to control neuronal excitability at the supragranular layers of anesthetized and awake visual cortex in both male and female mice. Using 2P-guided whole-cell or cell-attached recordings in positive neurons expressing any of the three opsins ReaChR, CoChR, or ChrimsonR, we investigated the dependence of spiking activity on the opsin's channel kinetics. We found that in all cases the use of brief illumination (≤10 ms) induces spikes of millisecond temporal resolution and submillisecond precision, which were preserved upon repetitive illuminations up to tens of hertz. To reach high temporal precision, we used a large illumination spot covering the entire cell body and an amplified laser at high peak power and low excitation intensity (on average ≤0.2 mW/μm2), thus minimizing the risk for nonlinear photodamage effects. Finally, by combining 2P holographic excitation with electrophysiological recordings and calcium imaging using GCaMP6s, we investigated the factors, including illumination shape and intensity, opsin distribution in the target cell, and cell morphology, which affect the spatial selectivity of single-cell and multicell holographic activation. Parallel optical control of neuronal activity with cellular resolution and millisecond temporal precision should make it easier to investigate neuronal connections and find further links between connectivity, microcircuit dynamics, and brain functions.
SIGNIFICANCE STATEMENT Recent developments in the field of optogenetics has enabled researchers to probe the neuronal microcircuit with light by optically actuating genetically encoded light-sensitive opsins expressed in the target cells. Here, we applied holographic light shaping and temporal focusing to simultaneously deliver axially confined holographic patterns to opsin-positive cells in the living mouse cortex. Parallel illumination efficiently induced action potentials with high temporal resolution and precision for three opsins of different kinetics. We extended the parallel optogenetic activation at low intensity to multiple neurons and concurrently monitored their calcium dynamics. These results demonstrate fast and temporally precise in vivo control of a neuronal subpopulation, opening new opportunities for revealing circuit mechanisms underlying brain functions.
Keywords: computer-generated holography, in vivo two-photon optogenetics, millisecond photoactivation, mouse visual cortex, temporal focusing
Introduction
The coordinated spike timing among neurons with precision in the range of milliseconds enables various synaptic mechanisms believed to play significant roles in regulating sensation, perception, and cognitive functions. Optogenetics, a field with a growing number of genetic tools (Mattis et al., 2011; Yizhar et al., 2011; Lin et al., 2013; Klapoetke et al., 2014), offers researchers new ways to investigate those mechanisms via fast and precise optical control of neuronal firing (Nagel et al., 2003; Boyden et al., 2005; Adamantidis et al., 2007; Zhang and Oertner, 2007; Li et al., 2013; Emiliani et al., 2015). However, until recently, researchers have been daunted by the challenge of obtaining optical control of excitability in one or several individually selected cells with millisecond resolution and precision into scattering tissue in vivo.
Wide-field illumination with visible light has been applied for driving neuronal activity reaching a high spiking rate, with millisecond peak latencies and submillisecond jitter, for a variety of opsins, including channelrhodopsin 2 (ChR2), ReaChR, Chronos, and ChrimsonR (Lin et al., 2013; Klapoetke et al., 2014). Even though single-photon (1P) illumination lacks optical sectioning and, in scattering samples, penetration depth, a significant number of studies have reported in vivo activity manipulation in depth through optical-fiber endoscopes (Adamantidis et al., 2007; Aravanis et al., 2007; Gradinaru et al., 2007). Single-photon patterned illumination, in contrast to wide-field illumination, provides improved spatial selectivity for activating neuronal ensembles, but is limited to shallow depths or transparent animals (Guo et al., 2009; Wyart et al., 2009; Szabo et al., 2014).
Two-photon (2P) optogenetic neuronal activation, which provides accurate targeting of neurons and reduced scattering in tissue (Helmchen and Denk, 2005), has been realized by employing one or a hybrid of two approaches: the scanning method (Packer et al., 2012, 2015; Carrillo-Reid et al., 2016; Yang et al., 2018) and the parallel method (Ronzitti et al., 2017a; Chen et al., 2018). Parallel methods for 2P optogenetic activation use phase-modulation techniques, such as computer-generated holography (CGH; Bègue et al., 2013; Dal Maschio et al., 2017), generalized phase contrast (Glückstad, 1996; Papagiakoumou et al., 2010, 2013), and extended Gaussian beams (Rickgauer et al., 2014; Straub et al., 2016; Pegard et al., 2017). Axial confinement of light patterns can be preserved for hundreds of micrometers by integrating these approaches with temporal focusing (TF; Andrasfalvy et al., 2010; Papagiakoumou et al., 2010, 2013; Bègue et al., 2013; Rickgauer et al., 2014; Baker et al., 2016; Straub et al., 2016; Pegard et al., 2017; Mardinly et al., 2018).
Until now, in vivo 2P optogenetic spike induction by using scanning-based excitation has been reported for the slow opsin C1V1 (Packer et al., 2015; Yang et al., 2018). Neuronal activation via 2P holographic spots or temporally focused Gaussian beams has been shown for C1V1 (Rickgauer et al., 2014), ChrimsonR, Chronos (Pegard et al., 2017) and its variant ChroME (Mardinly et al., 2018), and ChR2 as well as GtACR, an inhibitory opsin (Forli et al., 2018). Only one recent study reported the temporal properties of the fast opsin ChroME in vivo (Mardinly et al., 2018).
Here, we extended these investigations to three efficient ChR variants: ReaChR, CoChR, and ChrimsonR (Lin et al., 2013; Klapoetke et al., 2014). By using TF-CGH, we systematically characterized the achievable temporal resolution, precision, and spatial selectivity of these variants for in vivo parallel spike induction. The three opsins differ in their photocycle properties related to channel opening, closing, and inactivating (Nagel et al., 2003; Nikolic et al., 2009), which may play important roles in the temporal properties of 2P activation. ReaChR is a slow opsin, displaying the longest transition time spans between photocycle states (off-time constant: ∼90–150 ms; Lin et al., 2013; Chaigneau et al., 2016; Papagiakoumou et al., 2018). Meanwhile, ChrimsonR is the fastest of the three opsins (off-time constant: ∼15 ms). CoChR offers intermediate speed (off-time constant: ∼30 ms; Klapoetke et al., 2014; Shemesh et al., 2017). Using 2P-guided whole-cell or cell-attached recordings, we investigated the conditions of 2P parallel illumination of pyramidal cells and interneurons in the anesthetized mouse primary visual cortex (V1), which enabled reliable and temporally precise suprathreshold activation with <1 ms jitter, for the three opsins. By combining holographic photostimulation with electrophysiological recording or GCaMP6 calcium imaging, we investigated the spatial selectivity of holographic activation. Finally, using the same illumination conditions enabling fast and precise single-cell activation, we demonstrated multicell holographic activation in anesthetized and awake mouse V1.
Materials and Methods
Animals.
All animal experiments were performed in accordance with Directive 2010/63/EU of the European Parliament and of the Council of 22 September 2010. The protocols were approved by the Paris Descartes Ethics Committee for Animal Research with the registered number CEEA34.EV.118.12. Adult female or male C57BL/6J mice (Janvier Labs) were anesthetized with intraperitoneal injection of a ketamine–xylazine mixture (0.1 mg of ketamine and 0.01 mg of xylazine/g body weight) during stereotaxic injection and with isoflurane (2% for induction and 0.5–1% for experiment) during photostimulation experiments. Adult mice of both sexes from the transgenic line GP4.3 (The Jackson Laboratory), which express the calcium indicator GCaMP6s (Dana et al., 2014), were used in the all-optical experiment. Cortical neurons of 4-week-old mice were infected with viral vectors of opsins using stereotaxic injection. Holographic stimulation experiments were performed 5–12 weeks after injection.
Virus injection and surgical procedures.
The following viral constructs were used: AAV2/1-EF1α-ReaChR-tdTomato (for expressing ReaChR); AAV2/8-hSynapsin-CoChR-GFP and AAV2/8-hSynapsin-CoChR-mCardinal (for expressing CoChR); and AAV2/7m8-CAG-ChrimsonR-tdTomato (for expressing ChrimsonR). Through a craniotomy over the right V1 (3.5 mm caudal from bregma, 2.5 mm lateral from the midline), 1.5–2 μl of viral vectors were delivered via a cannula in L2/3 (250 μm deep) at a speed of 80–100 nL/min. For performing acute photostimulation experiments in vivo, a circular craniotomy of 2 mm diameter was made over V1 and the dura mater was removed. Agarose of 0.5–2% and a cover glass were applied on top of the craniotomy to dampen tissue movement. Partial all-optical experiments were conducted during successive sessions in lightly anesthetized or awake mice through a chronic cranial window (Holtmaat et al., 2009).
Optical system for 2P imaging and holographic stimulation in vivo.
The optical system was a custom-developed microscope combining galvo-based 2P scanning with temporally focused CGH (Chaigneau et al., 2016; Ronzitti et al., 2017b). A femtosecond pulsed beam delivered by a fiber amplifier system (Satsuma HP, Amplitude Systemes; pulse width, 250 fs; tunable repetition rate, 500–2000 kHz; maximum pulse energy, 20 μJ; maximum average power, 10 W; λ = 1030 nm) operated at 500 kHz, was widened through an expanding telescope (f1 = −25 mm; L2: f2 = 250 mm) and reflected on the sensitive area of a reconfigurable liquid-crystal-on-silicon spatial light modulator (SLM; X10468-07, Hamamatsu Photonics). The SLM was controlled by custom-designed software, Wavefront Designer IV (Lutz et al., 2008). The laser power was controlled with a λ/2 wave retarder (CVI Melles Griot) in combination with a polarizer cube (CVI Melles Griot). The reflected beam was projected at the back focal plane of the objective (40× W APO NIR, Nikon, 0.8 numerical aperture) with a telescope (f = 1000 mm; AC508-1000-B, Thorlabs; f = 500 mm, AC508-500-B, Thorlabs). The effect of the zero order in the sample was suppressed by introducing a cylindrical lens in front of the SLM (Hernandez et al., 2014). TF of the phase-modulated light pulses was performed through a reflective dispersion grating of 600 l/mm (Newport, Richardson Gratings 33009BK02-351R) and a lens (f = 500 mm, Thorlabs, AC508-500-B) collimating the dispersed spectral frequencies at the back aperture of the 40 × objective.
Two-photon imaging was performed with a Ti:sapphire laser (Coherent, Chameleon Vision II). A liquid crystal variable phase retarder (Meadowlark Optics, LRC-200-IR1) and a polarizer cube (Meadowlark Optics, BB-050-IR1) controlled the imaging power. The femtosecond pulsed beam was expanded (telescope lenses: f = 75 mm, f = 175 mm; Thorlabs) and raster-scanned on the sample via a pair of xy galvanometric mirrors (3 mm aperture; Cambridge Technology, 6215H series,) imaged at the back aperture of the microscope objective through a telescope (f = 100 mm; Thorlabs, AC508-300-B; tube lens, f = 300 mm; Thorlabs, AC508-100-B). Galvanometric mirrors were driven by two servo drivers (Cambridge Technology, MicroMax series 671) controlled by a digital–analog converter board (National Instrument, PCI-6110). Emitted fluorescence was reflected by a dichroic mirror (Semrock, FF705-Di02) toward two photomultiplier tubes (PMTs; Hamamatsu Photonics, R3896 and R9110 SEL) by a set of two matching aspheric lenses (f = 40 mm; Thorlabs, AL5040-A). Fluorescence light was filtered by an infrared light-blocking filter (Semrock, FF01-750sp), split into two channels by a dichroic mirror (Semrock, FF555-Di03) and detected through two emission filters (SemrockFF02-617/73 and FF01-510/84).
Two-photon imaging and photoactivation beams were combined through a large dichroic mirror (50 × 70 mm; Chroma Technology, T970dcspxr). Epifluorescence imaging was provided by an LED source (Thorlabs, M470L2) filtered by a bandwidth excitation filter (Semrock, FF01-473/10), coupled to a diffuser (Thorlabs, DG10-1500) and an achromatic lens (f = 30 mm; Thorlabs, #LA1805) to provide widefield illumination on the sample, after reflection on a dichroic mirror (Di01-R488). Single-photon excited fluorescence was collected through a tube lens (f = 180 mm) and detected by a CCD camera (Hamamatsu Photonics, Orca-05G) after passing through a visible bandwidth filter (Senrock, FF01-609/181). A switchable mirror in the detection pathway directed 2P-excited fluorescence to PMTs and 1P-excited fluorescence to a CCD.
Axial properties of holographic illumination.
The optical resolution obtained for a temporally focused holographic spot measuring 12 μm in diameter was fitted by a Lorentian curve with a full-width at half-maximum (FWHM) of 10.5 μm (see Fig. 5C). To estimate how this optical point spread function is affected by the size of the cell and the illumination conditions, we first fitted the experimental photocurrent-versus-illumination intensity for ReaChR previously obtained (Chaigneau et al., 2016) by an exponential function given by f(I) = 1 − , where I is the illumination intensity and Is = 0.03 mW/μm2, corresponding to the intensity at 60% of photocurrent saturation. Hence, the axial photocurrent distribution will be given by
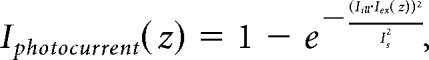 |
where Iill is the in-focus illumination intensity, and Iex(z) is the axial illumination profile (see Fig. 5C, magenta line). In our case, Iill has been estimated by calculating the average intensity used in the axial resolution experiments (see Fig. 5A) and assuming a scattering length at 1030 nm estimated as 175 μm (derived from simulations based on experimental data; Papagiakoumou et al., 2013; Picot et al., 2018), which results in a range of illumination intensity equal to 0.0630 ± 0.035 mW/μm2 (mean ± SD). The overall expected photoactivation axial distribution is then given by the convolution between the axial photocurrent distribution and the size of the cell, assumed as a 12 μm bandwidth rectangle function (see Fig. 5C).
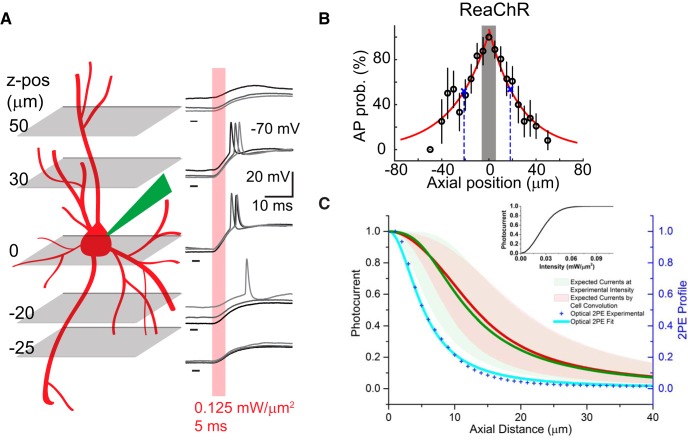
Figure 5.
Axial selectivity of holographic activation in vivo. A, Left, Schematic of the mapping of the axial resolution of holographic optogenetic activation in an opsin-positive cell by sequentially placing the excitation spot at different axial planes after manual defocusing. Right, Example whole-cell recordings of a ReaChR-expressing cell in response to 5-ms patterned illumination of 0.125 mW/μm2 at the corresponding axial positions. B, Axial resolution of AP generation in response to photostimulation through a circular holographic spot with a diameter of 12 μm at the threshold excitation intensity in neurons (n = 12) expressing ReaChR at L2/3 mouse visual cortex. Note the larger variability in AP generation when the excitation pattern is delivered deeper away from the focal plane. Shaded gray area indicates the assumed axial span of target soma. C, Estimated photocurrent distribution along the axial direction when illuminating at the intensity used for axial resolution experiments (I = 0.06 + 0.04 mW/μm2; mean ± SD) for ReaChR (green line). Expected photocurrent axial distribution after convolving the estimated distribution of photocurrents with a 12-μm-diameter step function, matching the assumed cell size (red line). Red and green shaded areas delimit the corresponding SD ranges, showing how the effective working excitation intensity can significantly affect the axial resolution. Inset, Photocurrent-versus-illumination intensity for ReaChR (adapted from Chaigneau et al., 2016). The 2P excitation (2PE) axial intensity profile of the illumination beam is also shown (blue line).
Two-photon-guided electrophysiology in vivo.
Neurons in L2/3 V1 of anesthetized mice were targeted with patch pipettes under the custom-built 2P microscope and the 40× water-immersion objective. The fluorophore labeling of GFP, dTomato, or td-Tomato in cells expressing any of the three opsins or GCaMP6s was visualized by excitation at 920 nm and the emitted fluorescence was collected in the corresponding channel, green or red, of the PMTs. Imaging data were acquired using ScanImage3 software (Pologruto et al., 2003; http://scanimage.org).
Whole-cell or cell-attached recordings were obtained by using microelectrodes fabricated from borosilicate glass (5–8 MΩ resistance) and filled with solution containing the following (in mM): 135 potassium gluconate, 10 HEPES, 10 sodium phosphocreatine, 4 KCl, 4 Mg-ATP, 0.3 Na3GTP, and 25–50 AlexaFlour-488 or AlexaFlour-594 for pipette visualization. The craniotomy was covered with the extracellular solution containing the following (in mM): 145 NaCl, 5.4 KCl, 10 HEPES, 1 MgCl2, and 1.8 CaCl2. Whole-cell membrane potentials recorded in the current-clamp mode were corrected for liquid junction potential (11.7 ± 0.3 mV, 16 measurements, mean ± SEM). Voltage recordings were acquired with a MultiClamp 700B amplifier and a Digidata 1550A digitizer, which were controlled by pCLAMP10 software (Molecular Devices). Electrophysiology data were filtered at 6 kHz and digitized at 20 kHz.
Holographic stimulation in vivo.
Two-photon photostimulation in anesthetized or awake mouse V1 was performed via a 12-μm-diameter holographic spot placed according to a high-resolution 2P reference image (512 × 512 pixels) including the target soma of an opsin-expressing cell. The spiking activity was recorded through a patch pipette (see above). For optical readout of calcium imaging upon photostimulation, single-target or multiple-target somata were mainly determined based on the ReaChR-dTomato expression. Through multiple 12-μm-diameter holographic spots, the targets were simultaneously illuminated while the population calcium activity in the 300 × 300 μm2 field of view (FOV) was monitored via the fluorescence changes of GCaMP6s, which were recorded using the imaging laser at 920 nm with 128 × 128 pixel resolution at a frame rate of 5.92 Hz.
Artifactual excitation of fluorophores was induced by the photostimulation laser during functional imaging. Photostimulation artifacts were displayed as consecutive lines of bright pixels. Specifically, when stimulating with 10 holographic light pulses of 10 ms at 11.84 Hz and scanning with 128 × 128 pixel resolution at a frame rate of 5.92 Hz (i.e., half of stimulation frequency), we expected two stimulation epochs to occur per frame, which would give rise to two bands of bright pixels 64 pixels apart. As we sampled at 128 × 128 pixel resolution, 10 ms illumination pulse corresponded to eight lines of artifact as follows: (10 * 0.001) s * (5.92 * 128) lines/s.
Two-photon scanning and stimulation onset were synchronized to leave a large central part of the FOV without any artifact. For example, we can align the two bright bands above and below the center of the FOV by triggering photostimulation 42.2 ms after a frame scan is initiated, so that the first bright band will appear after 32 line scans from the top and the second bright band will appear after 32 + 64 = 96 line scans, thus leaving the center FOV free from stimulation artifacts (see Figs. 6A, 8A). Artifactual excitation of fluorophores due to photostimulation laser was removed in postprocessing on the regions of interest (ROIs) affected by artifacts.
Figure 6.
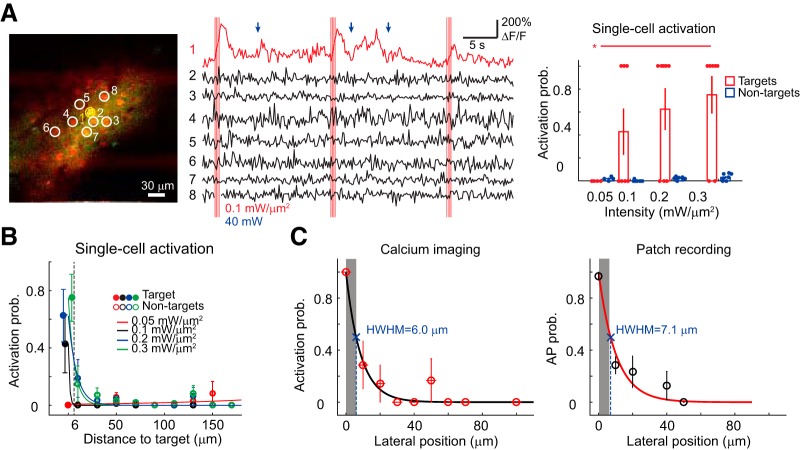
Optical readout of single-cell holographic activation. A, Left, An average intensity projection 2P image of neurons coexpressing the opsin ReaChR (red) and the calcium sensor GCaMP6s (green), with one target cell (yellow circle) and neighboring nontarget cells (white circles). Middle, The calcium signal from the corresponding target cell (red trace) and nontarget cells (black traces). Vertical red bars indicate photostimulation epochs. The excitation intensity is denoted as red. Blue arrows indicate spontaneous activity for the target cell. Blue denotes imaging power. Note two consecutive lines of bright pixels toward the upper and lower margins of the FOV are the artifacts induced by the photostimulation laser. Right, The activation probability (Activation prob.) of targets and nontargets as a function of excitation intensity in single-cell activation experiments (left; 8 FOVs). Each filled circle in red or blue indicates the mean activation probability of target or nontarget cells in one FOV. Asterisks denote significant difference in activation probability between excitation intensity (p = 0.013 between 0.05 and 0.3 mW/μm2). B, Activation probability based on calcium signal of target (solid circles at distance 0, with small x-jitter for visualization) and nontarget neurons (empty circles) as a function of distance from the target for single-cell activation experiments (8 FOVs). Dashed lines indicate the distance in which nontarget neurons displayed ∼50% activation probability compared to that of target neurons at an excitation intensity of 0.2 mW/μm2. The distance between nontarget cell and target cell is distributed in 20 μm bins. C, Lateral selectivity measured as target cells' activation probability based on calcium responses (left; n = 7 cells) and spiking probability (right; n = 8 cells) after moving the excitation spot laterally away from the target soma (n = 7 cells). Circles and error bars indicate mean ± SEM. Gray shaded area indicates the assumed lateral span of target soma.
Figure 8.
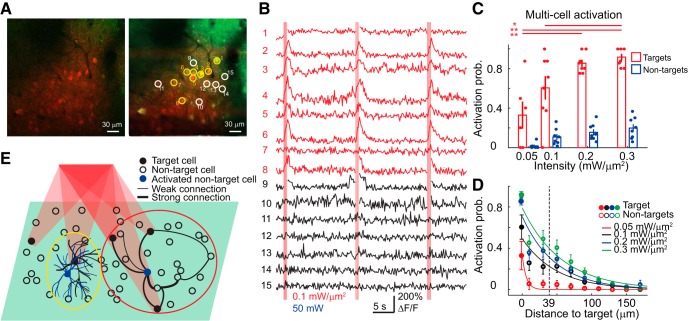
Holographic activation of multiple cells. A, An example multitarget experiment. Top left, A high-resolution 2P image of both red (ReaChR) and green (GCaMP6s) channels. Top right, Multiple target (yellow circles) and neighboring nontarget cells (white circles) on an average intensity profile. B, Plots of the calcium signal from the corresponding target (red traces) and nontarget (black traces) cells. Vertical red bars indicate photostimulation epochs. Red indicates excitation intensity. Blue indicates 2P scanning power. C, Activation probability of targets and nontargets as a function of excitation intensity in multicell activation experiments (8 FOVs). Each filled circle in red or blue indicates the mean activation probability of multiple targets or nontargets in one FOV. Asterisks denote significant difference in activation probability between excitation intensity (p < 0.0001 for target cells' activation probability between 0.05 and 0.2 mW/μm2, 0.05 and 0.3 mW/μm2, p = 0.12 and p = 0.021 between 0.1 and 0.2 mW/μm2, 0.1 and 0.3 mW/μm2; multiple comparisons after ANOVA). D, Activation probability based on calcium signal of target neurons (solid circles at distance 0) and nontarget (empty circles) as a function of distance from the target cell (8 FOVs). Dashed lines indicate the distance at which nontarget neurons displayed an activation probably that was ∼50% of that of target neurons at an excitation intensity of 0.3 mW/μm2. The distances between nontarget cells and target cells are distributed in 20 μm bins. E, Schematic illustration of out-of-target activation. Black filled circles indicate the target cells in a single cortical plane (green shade) activated by holographic stimulation. Empty circles indicate the nontarget cells located on the same plane. Black lines of varying thickness between neurons denote differences in connection strength. Two blue nontarget cells represent two cases that cause out-of-target activation. One nontarget cell (the blue cell in the yellow circle on the left) is activated because its neurites cross the excitation volume targeting the neighboring cell. The other nontarget cell (the blue cell in the red circle at the right) is activated because of its connections to other activated target cells.
Experimental design and statistical analysis.
Electrophysiology recordings of single-cell photoactivation were analyzed using custom-written scripts in Matlab (MathWorks). The latency of light-induced action potentials (APs) was defined as the time span between the illumination onset and the AP peak. The AP jitter was calculated as the SD of light-induced AP latencies in 3–6 repetitions. We measured the AP induction from eight male mice for ReaChR, six male mice for CoChR, 13 male mice for ChrimsonR, and three male transgenic ChrimsonR (ChrimsonR Tg) mice.
Image analysis was performed using ImageJ in combination with Matlab. For all-optical experiments, ROIs covering individual somata were manually selected based on the expression of both ReaChR-dTomato and GCaMP6s channels. We measured the calcium responses upon photostimulation from six mice (two females and four males). Image frames of the ROIs affected by the photostimulation artifacts were removed during analysis. Neuropil signal was estimated by measuring the intensity of pixels within a 42-μm-diameter (18 pixel) disk from the target ROI center and excluding ROI regions. The true calcium signal at the cell soma was calculated as follows:
where the contribution of neuropil contamination r was estimated here as 0.5 by comparing the fluorescence signal from the surrounding disk to that of a nearby small vessel (Kerlin et al., 2010). Neuropil contamination in our case is not substantial because the calcium transients were mainly the result of postsynaptic activation via holographic illumination. Relative percentage changes of fluorescence were computed for calcium signal as ΔF/F = (F − F0/F0), where F0 represents the average raw fluorescent signal of 1.5 s before photostimulation onset. A cell was considered activated when the mean ΔF/F 1.5 s after the first illumination onset was significantly different from that 1.5 s beforehand (right-tailed paired t-test with the significance level 0.01). Activation probability of target or nontarget cells for each FOV was determined as the average activation rate (1/1, 10/60, etc.). To determine the relationship between activation probability of a nontarget cell and its distance to a target, the activation probability of each nontarget cell was computed as the average activation rate relative to the distance to target, which is arranged in a 20 μm bin, for all target cells.
Statistical tests were conducted in Matlab. Data comparisons between neurons for opsin types or different photostimulation conditions (e.g., frequency of a train of light pulses, or excitation intensity) were performed using ANOVA and multiple comparisons of Tukey's method. Data between conditions were compared using a paired t-test (e.g., calcium signal before and after photostimulation onset in the same cell) or a two-sample t-test (e.g., fluorescence intensity of target and nontarget cells for all FOVs). Error ranges are indicated as SEM if not otherwise indicated.
Results
Submillisecond optical control of neuronal firing in vivo
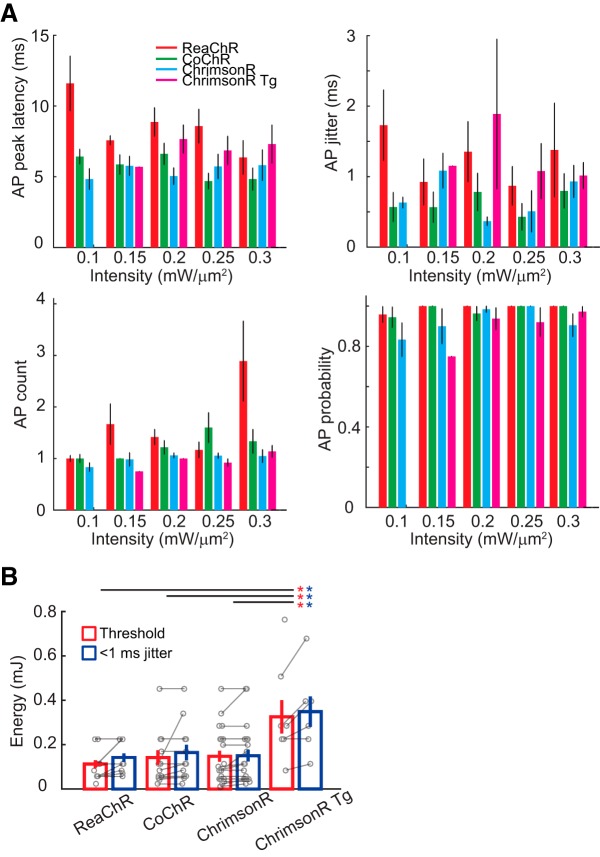
To investigate the role of channel kinetics on photostimulation temporal resolution and precision, we examined the 2P stimulation parameters (illumination intensity and duration) necessary for inducing an AP in V1 cells expressing ReaChR, CoChR, or ChrimsonR, via viral infections. Scanless optogenetic activation was achieved by combining 2P CGH and TF with a 2P scanning imaging microscope (see Materials and Methods; Papagiakoumou et al., 2008; Ronzitti et al., 2017a). Laser light pulses at a repetition rate of 500 kHz were delivered through a 12-μm-diameter holographic spot placed over the target cell soma. The intensity of the excitation laser was increased until the target cell elicited an AP, which was captured via 2P-guided whole-cell or cell-attached recordings (Fig. 1A). The temporal resolution and precision of AP induction were estimated respectively as the peak latencies' arithmetic mean and SD (i.e., the jitter). According to the spiking patterns of positive cells situated at L2/3 (117 ± 3 μm from the brain surface; mean ± SEM), we found that recordings for ReaChR (15 of 15 cells), CoChR (13 of 13 cells), and the majority of ChrimsonR (20 of 24 cells) were from putative excitatory pyramidal neurons, whereas few ChrimsonR recordings (4 of 24 cells) were from putative fast-spiking interneurons. Using brief pulse illumination of 2, 3, 5, and 10 ms, we found the threshold intensity in the range of 0.1–0.4 mW/μm2 (measured at the tip of objective, corresponding to 11–45 mW/cell), which is sufficient for triggering an AP with average latency of <10 ms and jitter faster than 2 ms for the three opsins (Fig. 1B; Table 1). By slightly increasing the excitation intensity above the threshold level, we obtained shortened AP latencies as well as submillisecond jitter (Figs. 1C,D, 2; Table 2). Such excitation intensity enabled AP generation in the target cell in response to 66 ± 11 light pulses (pulse duration, 0.5–15 ms; pulse intensity, 0.05–0.60 mW/μm2) for a recording time span of 32 ± 4 min (n = 54; Table 3).
Figure 1.
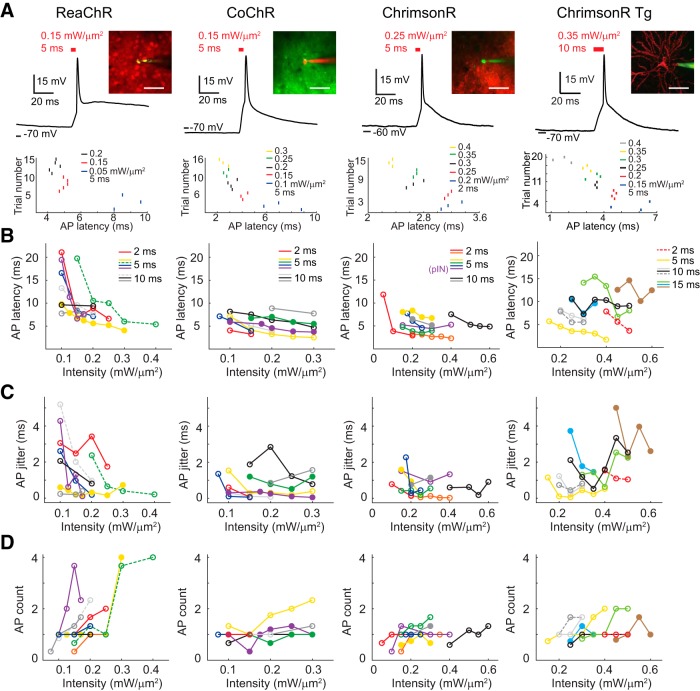
Precise 2P holographic activation of single cell in vivo. A, Top, Example traces of AP induction at the L2/3 of anesthetized mouse visual cortex upon brief pulse of holographic illumination in opsin-positive cells via viral expression of ReaChR, CoChR, and ChrimsonR and transgenic expression of ChrimsonR (ChrimsonR Tg) in mouse line Cux2-CreERT2;Ai167. Insets are 2P images of a target cell recorded via a glass pipette filled with AlexaFluor-488 (green) or AlexaFluor-592 (red). Bottom, Example raster plots of spike timing for each cell in response to different excitation intensities. Scale bar, 47 μm. B, AP peak latency in example individual cells (connected dots represent data from the same cell) in relation to the excitation intensity upon illumination of 2, 5, or 10 ms (n = 7, 8, 8, 7 for ReaChR, CoChR, ChrimsonR, and ChrimsonR Tg). For ReaChR and ChrimsonR Tg, data in dashed lines are from the same cell. Solid circles indicate whole-cell recordings. Open circles indicate cell-attached recordings. For ChrimsonR, three recordings were obtained from putative fast-spiking interneurons. C, Jitter of AP peak latencies as a function of illumination intensities from the same cells as in B. D, Spike count as a function of excitation intensity from the same cells as in B and C. For each opsin, AP properties from the same cell are indicated as the same color. Solid and dashed lines of the same color represent recordings under different illumination duration from the same cell.
Table 1.
Spiking properties upon 2P pulse stimulation with threshold power density
| Opsin | Light-pulse duration (ms) | AP latency (ms) | AP jitter (ms) | AP count | AP probability (%) | Laser-power densityb (mW/μm2) | Estimated laser-power densityc (mW/μm2) |
|---|---|---|---|---|---|---|---|
| ReaChR | 10 | 9.48 ± 1.41a (n = 8) | 1.84 ± 0.73a (n = 7) | 0.98 ± 0.065a (n = 8) | 90.58 ± 6.30a (n = 8) | 0.14 ± 0.018a (n = 8) | 0.072 ± 0.0083a (n = 8) |
| 5 | 11.16 ± 2.42a (n = 6) | 1.82 ± 0.60a (n = 6) | 1.11 ± 0.070a (n = 6) | 100.00 ± 0.00a (n = 6) | 0.13 ± 0.017a (n = 6) | 0.064 ± 0.0065a (n = 6) | |
| 2 | 21.10 (n = 1) | 3.05 (n = 1) | 1.00 (n = 1) | 100.00 (n = 1) | 0.10 (n = 1) | 0.054 (n = 1) | |
| CoChR | 10 | 8.18 ± 0.58a (n = 7) | 0.58 ± 0.13a (n = 6) | 1.00 ± 0.072a (n = 7) | 95.24 ± 4.76a (n = 7) | 0.19 ± 0.038a (n = 7) | 0.10 ± 0.018a (n = 7) |
| 5 | 6.74 ± 0.28a (n = 4) | 1.10 ± 0.27a (n = 4) | 1.08 ± 0.083a (n = 4) | 100.00 ± 0.00a (n = 4) | 0.11 ± 0.016a (n = 4) | 0.056 ± 0.0090a (n = 4) | |
| 3 | 6.93 (n = 1) | 0.36 (n = 1) | 1.00 (n = 1) | 100.00 (n = 1) | 0.15 (n = 1) | 0.058 (n = 1) | |
| 2 | 4.05 (n = 1) | 0.60 (n = 1) | 1.00 (n = 1) | 100.00 (n = 1) | 0.10 (n = 1) | 0.049 (n = 1) | |
| ChrimsonR | 10 | 8.50 ± 0.74a (n = 10) | 0.67 ± 0.23a (n = 8) | 0.89 ± 0.054a (n = 10) | 89.33 ± 5.46a (n = 10) | 0.24 ± 0.021a (n = 10) | 0.13 ± 0.015a (n = 10) |
| 5 | 6.21 ± 0.72a (n = 6) | 1.30 ± 0.26a (n = 6) | 0.99 ± 0.082a (n = 6) | 93.33 ± 5.77a (n = 6) | 0.15 ± 0.012a (n = 6) | 0.080 ± 0.0066a (n = 6) | |
| 2 | 5.24 ± 1.06a (n = 8) | 0.30 ± 0.13a (n = 5) | 0.96 ± 0.12a (n = 8) | 87.5 ± 6.10a (n = 8) | 0.15 ± 0.021a (n = 8) | 0.073 ± 0.011a (n = 8) | |
| ChrimsonR Tg | 15 | 13.32 ± 0.75a (n = 2) | 3.16 ± 1.85a (n = 2) | 0.82 ± 0.017a (n = 2) | 81.67 ± 1.67a (n = 2) | 0.38 ± 0.075a (n = 2) | 0.15 ± 0.026a (n = 2) |
| 10 | 9.55 ± 0.66a (n = 5) | 2.39 ± 1.08a (n = 4) | 0.82 ± 0.11a (n = 5) | 77.00 ± 10.20a (n = 5) | 0.22 ± 0.012a (n = 5) | 0.11 ± 0.013a (n = 5) | |
| 5 | 5.70 (n = 1) | 1.15 (n = 1) | 0.75 (n = 1) | 75.00 (n = 1) | 0.15 (n = 1) | 0.070 (n = 1) |
aMean ± SEM.
bLaser-power density measured at the tip of objective.
cEstimated power density by assuming the scattering length at 1030 nm as 175 μm, which is derived from simulations based on experimental data (Papagiakoumou et al., 2013).
Figure 2.
AP induction in response to different illumination intensities. A, Holography-induced spike properties of peak latency, jitter, AP count, and AP probability per stimulus in vivo as a function of excitation intensity (n = 8, 6, 9, 6, 3 for ReaChR upon photostimulation of 0.1, 0.15, 0.2, 0.25, 0.3 mW/μm2; n = 6, 8, 9, 5, 5 for CoChR, n = 4, 4, 16, 6, 7 for ChrimsonR, and n = 0, 1, 4, 5, 6 for ChrimsonR Tg of respective excitation intensity). B, Required energy using light pulses of 0.1–0.45 mW/μm2, 2–15 ms, for reaching spike threshold and submillisecond jitter (n = 15, 13, 24, 8 for ReaChR, CoChR, ChrimsonR, and ChrimsonR Tg respectively). Significant difference in energy-reaching threshold and <1 ms jitter is respectively denoted as red and blue asterisks for multiple comparisons between opsin types.
Table 2.
Spiking properties upon 2P pulse stimulation with submillisecond precision
| Opsin | Light-pulse duration (ms) | AP latency (ms) | AP jitter (ms) | AP count | AP probability (%) | Laser-power densityb (mW/μm2) | Estimated laser-power densityc (mW/μm2) |
|---|---|---|---|---|---|---|---|
| ReaChR | 10 (n = 7) | 8.80 ± 0.77a | 0.58 ± 0.15a | 1.24 ± 0.19a | 100.00 ± 0.00a | 0.16 ± 0.020a | 0.081 ± 0.0092a |
| 5 (n = 4) | 8.31 ± 1.68a | 0.47 ± 0.098a | 1.50 ± 0.29a | 100.00 ± 0.00a | 0.14 ± 0.021a | 0.073 ± 0.0087a | |
| CoChR | 10 (n = 7) | 7.69 ± 0.77a | 0.61 ± 0.12a | 1.05 ± 0.047a | 100.00 ± 0.00a | 0.22 ± 0.038a | 0.12 ± 0.018a |
| 5 (n = 4) | 5.84 ± 0.62a | 0.37 ± 0.15a | 0.92 ± 0.083a | 91.67 ± 8.33a | 0.14 ± 0.024a | 0.072 ± 0.014a | |
| 3 (n = 1) | 6.93 | 0.36 | 1.00 | 100.00 | 0.15 | 0.058 | |
| 2 (n = 1) | 4.05 | 0.60 | 1.00 | 100.00 | 0.10 | 0.049 | |
| ChrimsonR | 10 (n = 8) | 7.63 ± 0.55a | 0.48 ± 0.10a | 0.95 ± 0.05a | 95.00 ± 5.00a | 0.25 ± 0.033a | 0.13 ± 0.023a |
| 5 (n = 5) | 5.41 ± 0.87a | 0.60 ± 0.13a | 0.95 ± 0.05a | 95.00 ± 5.00a | 0.19 ± 0.033a | 0.098 ± 0.015a | |
| 2 (n = 8) | 3.82 ± 0.24a | 0.35 ± 0.10a | 1.11 ± 0.084a | 100.00 ± 0.00a | 0.18 ± 0.013a | 0.088 ± 0.0072a | |
| ChrimsonR Tg | 15 (n = 1) | 13.45 | 0.63 | 1.00 | 100.00 | 0.40 | 0.16 |
| 10 (n = 4) | 8.57 ± 0.77a | 0.66 ± 0.19a | 1.00 ± 0.00a | 100.00 ± 0.00a | 0.29 ± 0.038a | 0.15 ± 0.020a | |
| 5 (n = 1) | 4.58 | 0.10 | 1.00 | 100.00 | 0.20 | 0.094 |
aMean ± SEM.
bLaser-power density measured at the tip of objective.
cEstimated power density by assuming the scattering length at 1030 nm as 175 μm, which is derived from simulations based on experimental data (Papagiakoumou et al., 2013).
Table 3.
Sustainability of photostimulation experiments (mean ± SEM)
| ReaChR (n = 14) | CoChR (n = 11) | ChrimsonR (n = 21) | ChrimsonR Tg (n = 8) | |
|---|---|---|---|---|
| Number of light pulses | 80 ± 14 | 41 ± 12 | 65 ± 26 | 77 ± 15 |
| Recording time (min) | 43 ± 8 | 24 ± 7 | 27 ± 7 | 32 ± 4 |
| Energy (mJ) | 11.0 ± 2.6 | 5.2 ± 1.6 | 8.0 ± 2.2 | 27.9 ± 8.1 |
Further, using Cux2-CreERT2;Ai167 transgenic mice (Gil-Sanz et al., 2015; Madisen et al., 2015; Daigle et al., 2018), in which ChrimsonR-tdTomato was specifically expressed on the cell membrane of L2/3 neurons, APs were induced by shining short light pulses of 5, 10, or 15 ms with excitation intensity in the range of 0.15–0.45 mW/μm2, i.e., 17–51 mW/cell (138 ± 15 μm deep, n = 8; Fig. 1B; Table 1). As before, submillisecond AP jitter can be obtained by increasing the excitation intensity (Table 2).
The tendency of reduced AP latency and jitter with the increasing stimulation intensity was clearly observed in individual recordings (Fig. 1B,C). Variability in this trend likely arises from the up–down membrane potential fluctuation in vivo (Waters and Helmchen, 2006). Some recordings showed enlarged AP count with increasing stimulation intensity before saturation (Fig. 1D). Averaged data indicated that, while opsin type significantly affected spike latency, jitter, AP count, and AP probability, excitation intensity significantly modulated only AP count (for AP latency, jitter, count, and probability, the respective p values are p = 0.0001, p = 0.032, p < 0.0001, and p = 0.022 for opsin type; p = 0.66, p = 0.54, p = 0.0061, and p = 0.30 for excitation intensity; two-way ANOVA for opsin type and excitation intensity; Fig. 2A). Overall, we found that excitation intensity and opsin type play significant roles in holography-induced AP properties.
Compared with viral delivery of ChrimsonR, higher excitation energy (computed as the intensity over illumination time span) was required for reaching AP threshold and obtaining submillisecond jitter in ChrimsonR transgenic mice (ChrimsonR Tg) (Fig. 2B; p = 0.0023 and p = 0.010 for required energy reaching threshold and >1 ms jitter respectively, one-way ANOVA for opsin type and subsequent multiple comparisons). Because there is a higher proportion of recordings from ChrimsonR Tg compared with those from ChrimsonR showing >10 ms AP latency upon 10 ms illumination at the threshold intensity (3 of 10 recordings for ChrimsonR and three of five for ChrimsonR Tg), our viral infection of ChrimsonR may result in higher opsin expression compared with transgenic delivery.
These results suggest that parallel optogenetic excitation enables in vivo AP generation with <1 ms jitter for opsins of different channel kinetics. Moreover, our data demonstrate that the use of a laser with a low repetition rate enables low-intensity excitation.
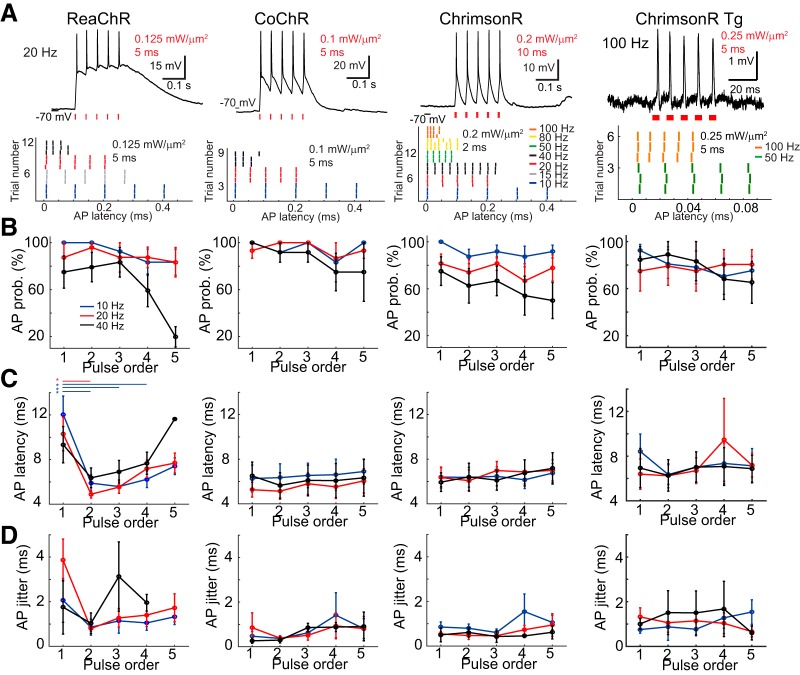
Temporally precise control of a train of APs
We investigated the stimulation conditions for precisely inducing a train of APs. Using illumination conditions according to the threshold intensity of each cell, we examined the spiking properties of a train of illuminations at different frequencies in vivo (Fig. 3A). Neurons expressing ReaChR, CoChR, or ChrimsonR, via both viral and transgenic delivery, fired APs following five consecutive pulse illuminations at 10, 20, and 40 Hz (Fig. 3B). We found that opsin type (p = 0.0067), stimulation frequency (p < 0.0001), and illumination pulse order (p = 0.0039) all significantly affect the probability of generating an AP (three-way ANOVA for opsin type, stimulation frequency, and pulse order on AP probability). Membrane potential remained the most depolarized between light pulses for ReaChR,intermediate for CoChR, and the least for ChrimsonR (membrane potential upon 20 Hz photostimulation after filtering out APs: 20.7 ± 7.9 mV for ReaChR, n = 3; 18.7 ± 3.3 mV for CoChR, n = 4; 4.1 ± 2.2 mV for ChrimsonR, n = 2). The different degrees of repolarization between light pulses are related to the different off-kinetics of the three opsins, with ChrimsonR the fastest, CoChR the middle, and ReaChR the slowest (Klapoetke et al., 2014; Chaigneau et al., 2016; Shemesh et al., 2017). For ChrimsonR, the fast repolarization between successive pulses enabled generation of an AP train at even higher frequencies (>40 Hz, n = 2 for ChrimsonR and ChrimsonR Tg; Fig. 3A).
Figure 3.
Precise generation of multiple APs with a train of patterned light pulses. A, Top, Representative whole-cell recordings of suprathreshold activation in vivo upon photostimulation of five illuminations at 20 Hz for ReaChR, CoChR, ChrimsonR, and cell-attached recordings of 100 Hz photostimulation for ChrimsonR Tg. Excitation intensity and light-pulse duration are indicated in red typeface. Bottom, Example raster plots of AP peak latencies relative to the onset of first light pulse from five or ten illuminations at different frequencies. Photostimulation condition indicated in black typeface. B–D, AP probability (AP prob.) AP latency, and jitter in relation to the pulse order of five illuminations at 10, 20, and 40 Hz for ReaChR, CoChR, ChrimsonR, and ChrimsonR Tg, respectively (mean ± SEM; n = 9, 8, 8 for ReaChR in response to 10, 20, 40 Hz photostimulation, n = 4, 5, 4 for CoChR, n = 8, 9, 8 for ChrimsonR, and n = 7, 6, 6 for ChrimsonR Tg). Note the significant decrease in AP latency and jitter upon the second stimulation for ReaChR. Asterisks denote significant difference for multiple comparison between pulse order.
The opsin photocycle also affected light-induced AP properties of peak latency (p = 0.0085 for opsin type, p = 0.74 for stimulation frequency, p = 0.014 for pulse order, three-way ANOVA) and jitter (p = 0.0001 for opsin type, p = 0.92 for stimulation frequency, p = 0.41 for pulse order, three-way ANOVA) upon repetitive illumination at 10, 20, and 40 Hz (Fig. 3C,D). Compared with the first illumination, the second light pulse induced significantly shortened AP latencies in ReaChR-positive cells (p < 0.0001 for 10 Hz; p = 0.0029 for 20 Hz in multiple comparisons). The slow off-kinetics of ReaChR keeps the cell membrane potential depolarized after the first light-induced AP for a longer period; in such an excited state, a shorter time span is required to reach the AP threshold; thus, the reduced AP latency when the second illumination is shone.
We further tested the capability of our system to precisely replicate the physiological activity patterns by stimulating ChrimsonR-expressing cells with an illumination sequence reproducing the same temporal pattern of spontaneous firing from the same (n = 1) or other neurons (n = 2). Photoevoked spikes were played back following the original time course with millisecond jitter (n = 3; Fig. 4).
Figure 4.
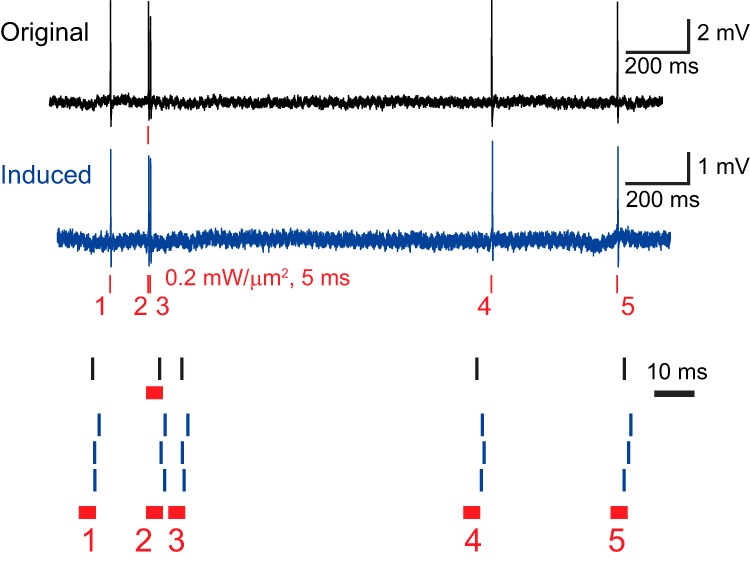
Mimicking spontaneous activity using holographic illumination. Top black trace shows cell-attached recordings of four spontaneous APs and one holography-induced AP in an example cell expressing ChrimsonR from a Cux2-CreERT2;Ai167 transgenic mouse. Middle blue trace shows the above spiking pattern precisely reproduced using five brief light pulses in the same cell. Bottom, Temporally zoomed-in raster plot of spike latencies in the original trial (black bars) and the three mimicking trials (blue bars) by aligning the light-pulse onset in the original trial with that of the second illumination in the mimicking trials. Note the submillisecond jitter of 0.78, 0.76, 0.78, 0.91, and 0.96 ms for five spikes in the four recordings.
In sum, we demonstrated that following multiple parallel holographic illuminations, positive cells elicited a train of spikes of ≤40 Hz, with <10 ms latency and <2 ms jitter. Higher firing rates induced by photostimulation are achievable by using a faster opsin, here ChrimsonR. This temporal precision enables playback of spontaneous neuronal activity.
Spatial selectivity of single-cell photoactivation
Because of the clear visualization of cytosol fluorophore in ReaChR-expressing cells (Fig. 1B), we chose to map the photoactivation spatial selectivity of holographic activation in vivo in mouse V1 infected with the ReaChR-dTomato construct. To characterize the photoactivation axial selectivity, we measured the spiking properties of a single target cell upon pulse stimulation at the threshold intensity after mechanically moving the objective so that the target cell would be out of the focal plane (Fig. 5A). The estimated AP probabilities were obtained by applying two exponential fits on the average AP probabilities above and below the focal plane (Fig. 5B).
Assuming the target soma is a 12-μm-diameter sphere (Fig. 5B, gray-shaded area), the target cell fired APs of ∼56% probability when the holographic spot was placed at a tangential position to the spot covering the target soma (axial positions of spot center ±12 μm; Fig. 5A,B). The evoked AP probability further decreased when the excitation pattern was moved outside the target cell (axial positions >12 μm or <−12 μm). The photoactivation axial selectivity, defined as the FWHM of the estimated AP probability curve along the z-axis, was 39 μm (n = 12). The spread of axial selectivity compared to the 2P axial intensity profile of holographic light patterns (FWHM, ∼10 μm; Fig. 5C) can result from the interaction of the optical resolution, the illumination intensity (which determines the level of opsin activation along the saturation curve; Fig. 5C, inset), and the distribution of opsin (which defines the spatial extent of the portion of the cell that may be activated). We calculated the range of illumination intensities experimentally used in Figure 5B (as mean ± SD) and simulated the corresponding axial distribution of ReaChR photocurrents (Fig. 5C, green area; see Materials and Methods). We then convolved this curve with a step function of 12 μm, assumed as the diameter of an opsin-expressing soma (Fig. 5C, red area). This results in a spread of the axial photocurrent profile (Fig. 5C, red line) whose range approaches experimental photoactivation axial selectivity. These results partially explain the extended axial selectivity obtained in experiments. However, out-of-focus activation of opsin-expressing neurites is likely to contribute to the widened axial selectivity. Moreover, out-of-focus illumination may modulate the temporal properties of AP generation in the target cell through back propagation of its dendritic activation or integration of dendritic signaling from neighboring neurons (Stuart and Spruston, 2015). Analogous measurements conducted for the more sparsely labeled opsin ChrimsonR gave rise to a similar axial selectivity of 35 μm (n = 5, data not shown).
We next asked whether spatially selective photoactivation could be obtained via optical read-out of calcium imaging. We sought to coexpress any of the three opsins in combination with the green calcium indicator GCaMP6s (Chen et al., 2013). As a result, we found a reliable combination of ReaChR–GCaMP6s by injecting the opsin viral vectors of ReaChR-dTomato at L2/3 of V1 in the transgenic mice GP4.3 (Dana et al., 2014). Because GCaMP6s was expressed in almost all cortical neurons in transgenic mice, we registered the spatial distribution of cells expressing ReaChR-dTomato in the red channel to identify double-positive cells of calcium sensor and opsin. The cytosol fluorophore expression initiated by the P2A sequence of the opsin construct largely enhanced the visualization of positive cells (Prakash et al., 2012; Baker et al., 2016; Mardinly et al., 2018), thus providing clear reference for placing the holographic spots over target soma.
In an FOV of 300 × 300 μm2, we selectively stimulated a single cell through chronic cranial windows in anesthetized or awake mice V1 (eight FOVs in two mice). We stimulated the target cell with 10 light pulses of 5 or 10 ms at 11.84 Hz (excitation intensity between 0.05 and 0.40 mW/μm2, corresponding to 7–45 mW/cell) and imaged the population neuronal activity at a frame rate of 5.92 Hz with a scanning power of 50 ± 3 mW (eight FOVs).
We observed that the single target cell displayed more prominent fluorescent changes of GCaMP6s signal with the increasing excitation intensity (Fig. 6A). We determined whether a neuron is activated based on its fluorescence change before and after photostimulation (see Materials and Methods). Activation probability differed between target and nontarget cells and was modulated by the excitation intensity (p < 0.0001 for target type and p = 0.064 for excitation intensity, two-way ANOVA). Upon illumination, ≤0.3 mW/μm2, nontarget cells located >6 μm away from the single target neuron were activated <50% (Fig. 6B). To further verify the spatial precision of holographic activation, we evaluated the lateral selectivity by measuring either the calcium signal or the spiking properties after placing the holographic spot at the AP threshold intensity laterally away from the target soma (Fig. 6C). We found comparable HWHM of 6–7 μm respectively for AP induction and calcium signal.
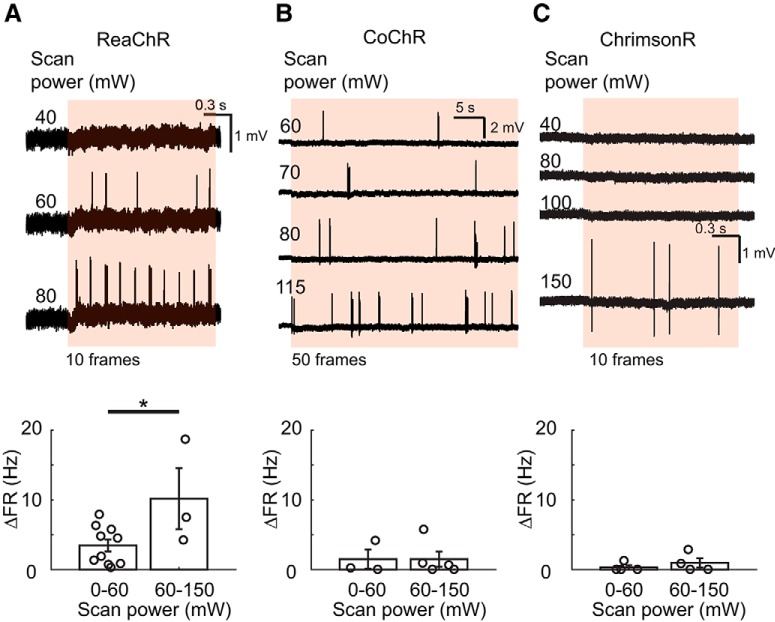
Notably, the slow channel closing of ReaChR renders it more prone to cross-talk activation while scanning at high imaging power of >60 mW (Fig. 7A,B; p = 0.064 and p = 0.0007, two-way ANOVA for scan power and opsin type, respectively; p = 0.035, p = 1.00, p = 1.00, multiple comparison of firing rate between scan power for ReaChR, CoChR, ChrimsonR in Fig. 7B). For optical readout of neuronal activation, we therefore used a scanning imaging power of 30–60 mW, which induced spurious AP firing 4.3 ± 0.9 Hz in ReaChR-positive cells (n = 9), for visualizing calcium events. A better signal-to-noise ratio of evoked calcium responses may be attained by using a fast opsin, whose cross-talk activation is less severe upon high scanning power (Fig. 7B,C). Of note, 2P scanning may still lead to subthreshold activation in opsin-positive cells, thus rendering increased network excitability (Ronzitti et al., 2017b).
Figure 7.
Cross-talk activation of opsins upon 2P scanning. Top, Representative cell-attached recordings of cross-talk activation upon 920 nm laser scanning at different scanning power for a 5.92 Hz frame rate involving cells positive for ReaChR (A), CoChR (B), or ChrimsonR (C) via viral infection. Bottom, Baseline-subtracted firing rate in relation to different scan power ranges (total n = 10, 6, 5 for ReaChR, CoChR, ChrimsonR) for scanning at 5.92 Hz frame rate. Elevated cross-talk firing is more pronounced for ReaChR when scanning at a power of >60 mW. Asterisk denotes significant difference for multiple comparison between scan power ranges.
Holographic activation of multiple cells in vivo
We tested simultaneous activation of multiple neurons, a major advantage of holographic stimulation, by using calcium imaging to monitor the population activity while placing multiple targets over double-positive somata of ReaChR and GCaMP6s cells. TF preserves the spatial selectivity of holographic spots (Papagiakoumou et al., 2018), even in scattering tissue (Bègue et al., 2013; Papagiakoumou et al., 2013). Using similar conditions (excitation intensity/cell, illumination duration, etc.) of single-cell activation experiments described above, we selectively photostimulated a group of 5–8 cells with bright cytosol dTomato expression (eight FOVs in five mice; Fig. 8A,B) while scanning at a power of 30–60 mW (on average, 44 ± 4 mW) for calcium imaging. Similarly, activation probability differed between target and nontarget cells and was modulated by the stimulation intensity (p < 0.0001 for target type and p < 0.0001 for excitation intensity; two-way ANOVA; Fig. 8C). We found the threshold excitation intensity of 0.125 ± 0.023 mW/μm2 (eight FOVs), defined as the intensity giving rise to ≥50% activation probability for multiple target cells, comparable with that obtained for spike induction for ReaChR (p = 0.83, two-sample t-test for threshold intensities of eight FOVs and those from 15 recordings in Table 1). Such threshold intensity for single-target activation was 0.19 ± 0.034 mW/μm2 (seven of eight FOVs in Fig. 6A–B). Hence, by using a moderate excitation intensity of 0.2 mW/μm2, we were able to effectively activate ∼85% of multiple target neurons (Fig. 8C).
We tested the effect of anesthesia on the threshold intensity of holographic stimulation. We obtained four awake recordings from two mice implanted with the chronic cranial window (three of eight FOVs for the multitarget experiment and one of eight FOVs for the single-target experiment). The threshold intensity of 0.15 ± 0.029 mW/μm2 for the four awake FOVs is very similar to that of 0.15 ± 0.030 mW/μm2 for the other 12 lightly anesthetized (∼0.5% isoflurane) FOVs (p = 1.0, two-sample t-test). Hence, the threshold intensity is independent of isoflurane anesthesia level.
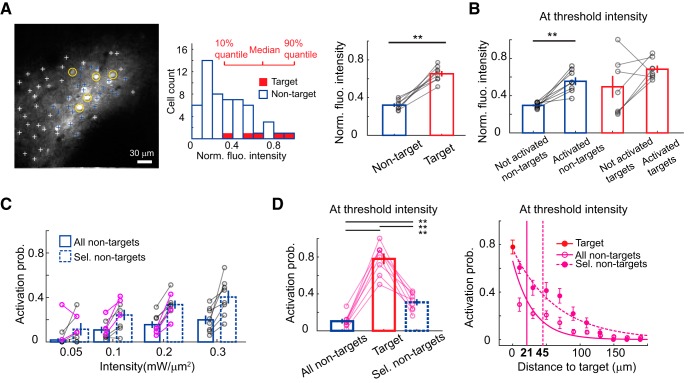
We estimated the lateral selectivity of multicell activation by calculating the activation probability of nontarget cells at various distances from target cells (Fig. 8; see Materials and Methods). Upon illumination at ≤0.3 mW/μm2, nontarget cells were activated <50% when they were >39 μm, a critical distance, from the target neuron (Fig. 8D). This distance is ∼5–10-fold farther than that for single-cell activation (Fig. 6B). Two effects contribute to the increased out-of-target responses: (1) activation of neurites from the nontarget cells near the target cells by defocused light; and (2) postsynaptic activation of nontarget cells by connections to multiple activated target cells (Fig. 8E).
During multicell activation experiments, we selected brighter cells from the red channel as target cells (Fig. 9A; p < 0.0001, two-sample t-test for medians of fluorescence intensity from target and nontarget somata) to ensure good enough opsin expression. We found that opsin-expression level, estimated as fluorescence intensity at the cell soma, significantly modulates the activation probability of both target and nontarget cells at the threshold intensity (Fig. 9B; p = 0.009 and p = 0.01, two-way ANOVA for activation type and target type). We selected nontarget cells that displayed fluorescence intensity within the 10–90% quantile of that from target cells (40 ± 7% of all nontarget cells, eight FOVs) and analyzed their activation probability with respect to the distance to targets. These nontarget subsets showed a trend of increased activation probability at different stimulation intensities (Fig. 9C; p < 0.0001, p < 0.0001, two-way ANOVA for excitation intensity and nontarget type). At the threshold intensity, we found that the out-of-target activation is more severe for the selected nontarget cells (Fig. 9D; p < 0.0001, one-way ANOVA for target type; p < 0.0001, p < 0.0001, p = 0.0062 for multiple comparisons between targets and all nontargets, targets and the selected nontargets, all and selected nontargets), probably due to increased neurite activation for nontarget cells in the vicinity of target cells. Whereas the critical distance upon threshold illumination increased from 21 to 45 μm for nontargets of matching opsin-expression level in multitarget experiments (Fig. 9D), it remained about the same for counterpart nontargets in single-target experiments (6 and 5 μm, for all and selected nontargets, respectively; seven of eight FOVs in Fig. 6A,B).
Figure 9.
Dependence of spatial selectivity on opsin expression level. A, Left, 2P image of ReaChR channel in an example FOV of five target cells (yellow circles). Middle, Histogram of normalized fluorescence intensity of target (red) and nontarget (blue edges) cells. Red lines indicate, respectively, the 10 and 90% quantiles as well as the median of target cells' fluorescence intensity. Among 54 nontarget cells in this example FOV, 24 nontargets had a fluorescence intensity that matched the intensity between the 10 and 90% quantiles of target cells (blue crosses on the left panel). The nontarget cells not selected are denoted as white crosses. Right, Averages of the medians of target and nontarget cells' fluorescence intensity (cell numbers of target vs nontarget cells are 5 and 50; 5 and 54; 5 and 74; 8 and 54; 8 and 58; 8 and 68; 7 and 57; 8 and 80 from 8 FOVs). Asterisks denote significant difference for two-sample t-test between target types. B, Averages of the medians of fluorescence intensity from target or nontarget cells based on their activation types at the threshold intensity (cell numbers of not-activated nontargets vs activated nontargets are 47 and 3; 49 and 5; 69 and 5; 53 and 1; 51 and 7; 50 and 18; 50 and 7; 73 and 7 from 8 FOVs; cell numbers of not-activated targets vs activated targets are 1 and 4; 0 and 5; 2 and 3; 4 and 4; 1 and 7; 1 and 7; 2 and 5; 1 and 7 from 8 FOVs). C, Activation probability of all and the selected nontargets at different excitation intensities in multicell activation experiments (p = 0.82, 0.99, 0.43, 0.93 at 0.05, 0.1, 0.2, 0.3 mW/μm2, cell numbers of selected vs all nontargets are 31 and 50; 24 and 54; 25 and 74; 3 and 54; 21 and 58; 43 and 68; 17 and 57; 39 and 80 cells from 8 FOVs; multiple comparisons after ANOVA). The average activation probability of nontargets in each FOV is represented as a circle, and those at the threshold intensity are indicated in magenta. D, Left, Activation probability (Activation prob.) of all and the selected nontarget cells (Sel. non-targets) compared with that of target cells (8 FOVs). Right, The relationship between activation probability and the distance to target cell for all and the selected nontarget cells (8 FOVs). The distances between target cells are distributed in 20 μm bins. The distance corresponding to 50% nontarget activation is indicated in bold font. B, D, Asterisks denote significant difference for multiple comparison between activation/target types. Bars and error bars indicate mean ± SEM.
In summary, the illumination conditions required to enable fast and precise light-evoked spike activation via TF-CGH enable the activation of several cells in parallel. These results indicate that the use of nonsoma-targeted opsins is limited to sparse multicell investigations. Homogeneous distribution of somatic opsins in a neuronal population is thus required for all-optical interrogation with unbiased single-cell resolution.
Discussion
We used TF-CGH to demonstrate optogenetic neuronal activation at L2/3 of anesthetized and awake mouse V1 using three opsins of different channel kinetics—ReaChR, CoChR, or ChrimsonR—reaching high temporal resolution and precision.
Specifically, we found the photostimulation conditions enabling, for the three opsins, AP generation with <9 ms temporal resolution and <1 ms temporal precision, even though spike induction in vivo depends on various factors of illumination intensity, opsin channel kinetics, opsin expression level, and intrinsic membrane potential fluctuation. AP generation was preserved at high stimulation frequencies until 40 Hz for faster opsins of CoChR and ChrimsonR. For the slow opsin ReaChR, the combined effect of delayed channel closing and inactivation led to decreases in AP probability and the temporal resolution and precision of spikes upon photostimulation of >20 Hz. Using the fast opsin ChrimsonR, we further demonstrated that the spontaneous activity can be precisely recalled in the target cell by designing a train of irregular light pulses. Hence, holographic illumination can be applied for precisely reproducing a burst of spikes, which is potentially very useful for triggering synchronous firing or mimicking spontaneous or evoked activity for closed-loop activity control in a neuronal ensemble of a specific spatial organization (Baranauskas, 2015; Grosenick et al., 2015). Such neuronal spatiotemporal coding has broad biological significance in areas ranging from synaptic mechanisms (Markram et al., 1997; Zhang et al., 1998; Stuart and Häusser, 2001), to sensory processing (Traub et al., 1996; Joris et al., 1998), to behavior (Schultz et al., 1997, 2017).
We demonstrated that the high peak power of laser pulses from a fiber amplifier with a low repetition rate enabled suprathreshold photoactivation in vivo using excitation intensity (on average ≤0.2 mW/μm2 or 23 mW/cell for the three opsins) considerably below typical nonlinear photodamage thresholds (Koester et al., 1999; Hopt and Neher, 2001; Olivié et al., 2008; Linz et al., 2016). By comparison, reaching comparable temporal resolution with a scanning approach would require excitation intensity 100–150 times higher (Picot et al., 2018; Yang et al., 2018). Yet the use of micrometer-sized spots, typical of holographic illumination, can induce thermal damage related to the linear absorption of light. However, we have recently demonstrated that by defining a fixed heat diffusion length with 3 ms illumination duration, holographic stimulation at an intensity of ≤0.2 mW/μm2 limits the induced temperature rise to <0.5 K for activating a single cell and <2 K for activating 100 cells with intersoma distances longer than the heat diffusion length (Picot et al., 2018). Hence, our optical system supplied with the high-energy laser should in principle enable precise playback of sensory-evoked responses encoded in multiple neurons without substantial heating.
We showed that holographic illumination effectively activated opsin-positive neurons not only by viral infection but also via a novel transgenic mouse line for layer-specific labeling, here ChrimsonR. The transgenic strategy is useful for expressing multiple constructs, e.g., combinations of dual opsins or opsin-activity reporters, in the same neuronal subpopulation, because it circumvents the issue of discrepant time windows for infecting neurons with two viral vectors. Up to now, in vivo all-optical studies have been mainly performed via viral expression of both opsin and calcium indicator (Rickgauer et al., 2014; Packer et al., 2015; Carrillo-Reid et al., 2016; Mardinly et al., 2018; Yang et al., 2018). Here, we carried out concurrent photostimulation and calcium imaging in double-positive cells by viral expression of ReaChR into V1 of transgenic mice expressing GCaMP6s. Alternative combinations are viral expression of calcium indicator in the transgenic opsin line or double transgenic line.
The channel off-kinetics is an important parameter that needs to be considered for concurrent holographic photostimulation and functional imaging. The longer the opsin off-time, the more important is the artifactual photostimulation induced by the imaging laser. The use of slow-kinetics opsin, such as ReaChR, thus requires a careful evaluation of imaging conditions (i.e., imaging power laser, frame rate, and FOV) to minimize these effects. Spurious activation of opsin upon scanning can be reduced by using a fast opsin in combination with GCaMP6 (Mardinly et al., 2018). Alternatively, cross-talk can be further reduced by using red-shifted calcium indicators (Inoue et al., 2015; Dana et al., 2016) with blue-shifted opsins, such as ChR2 (Forli et al., 2018). Nevertheless, there are no high-performance, high-energy/power laser sources emitting in the spectral range of ∼950 nm, where blue-shifted opsins peak for 2P excitation, thus limiting the number of target cells.
We have shown that parallel illumination combined with TF enables an optical axial resolution of ∼10 μm, although many parameters, such as the excitation intensity and opsin distribution, can cause the achievable spatial selectivity to significantly deteriorate. One strategy for achieving optical manipulation of single-cell resolution is to confine the opsin expression at the cell soma. Until now, such strategy has enabled single-cell resolution capability combined with ChR2 (Baker et al., 2016; Forli et al., 2018), CoChR (Shemesh et al., 2017), ChrimsonR, and a variant of Chronos (Pegard et al., 2017; Mardinly et al., 2018). Meanwhile, nonsomatic opsin-labeling neurites can be particularly useful for investigating neuronal projections between brain areas (Rajasethupathy et al., 2015; Leinweber et al., 2017), as well as the nonsomatic neuronal mechanisms, e.g., dendritic computation (Takahashi et al., 2016), of brain functions. Here, we have shown conditions (sparse staining or average distance between targets >∼40 μm) that for nonsomatic opsins enable near-cellular resolution.
Parallel optogenetic activation using TF-CGH could be extended in several directions. Our current optical system can integrate a second SLM for generating axially confined stimulation patterns in three dimensions (Hernandez et al., 2016; Accanto et al., 2018), which would be suitable for investigating the synaptic properties between cortical laminae or brain regions. By further incorporating the three-dimensional imaging methods (Grewe et al., 2011; Katona et al., 2012; Sofroniew et al., 2016; Lu et al., 2017), all-optical investigation of the functional wiring in a neuronal ensemble spanning a brain volume may be realized.
Footnotes
The authors declare no competing financial interests.
This work was supported by the European Union's Horizon 2020 research and innovation program under the Marie Skłodowska-Curie Grant Agreement no. 747598 (to I.-W.C.). E.P. and V.E. acknowledge support from the Agence Nationale de la Recherche ANR (Grants ANR-14-CE13-0016, Holohub and ANR-15-CE19-0001-01, 3DHoloPAc). V.E. acknowledges support from the Human Frontiers Science Program (Grant RGP0015/2016), the National Institutes of Health (Grant NIH U01NS090501-03), the Fondation Bettencourt Schueller (Prix Coups d'élan pour la recherche française), AXA Research Fund, and Getty Lab. E.R. received funding from the European Research Council Synergy Grant scheme (Helmholtz, ERC Grant Agreement 610110). The authors thank Edward S. Boyden for providing the CoChR viral constructs.
References
- Accanto N, Molinier C, Tanese D, Ronzitti E, Newman ZL, Wyart C, Isacoff E, Papagiakoumou E, Emiliani V (2018) Multiplexed temporally focused light shaping for high-resolution multi-cell targeting. Optica 5:1478–1491. 10.1364/OPTICA.5.001478 [DOI] [Google Scholar]
- Adamantidis AR, Zhang F, Aravanis AM, Deisseroth K, de Lecea L (2007) Neural substrates of awakening probed with optogenetic control of hypocretin neurons. Nature 450:420–424. 10.1038/nature06310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrasfalvy BK, Zemelman BV, Tang J, Vaziri A (2010) Two-photon single-cell optogenetic control of neuronal activity by sculpted light. Proc Natl Acad Sci U S A 107:11981–11986. 10.1073/pnas.1006620107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravanis AM, Wang LP, Zhang F, Meltzer LA, Mogri MZ, Schneider MB, Deisseroth K (2007) An optical neural interface: In vivo control of rodent motor cortex with integrated fiberoptic and optogenetic technology. J Neural Eng 4:S143–S156. 10.1088/1741-2560/4/3/S02 [DOI] [PubMed] [Google Scholar]
- Baker CA, Elyada YM, Parra A, Bolton MM (2016) Cellular resolution circuit mapping with temporal-focused excitation of soma-targeted channelrhodopsin. Elife 5:e14193. 10.7554/eLife.14193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranauskas G. (2015) Can optogenetic tools determine the importance of temporal codes to sensory information processing in the brain? Front Syst Neurosci 9:174. 10.3389/fnsys.2015.00174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bègue A, Papagiakoumou E, Leshem B, Conti R, Enke L, Oron D, Emiliani V (2013) Two-photon excitation in scattering media by spatiotemporally shaped beams and their application in optogenetic stimulation. Biomed Opt Express 4:2869–2879. 10.1364/BOE.4.002869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K (2005) Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci 8:1263–1268. 10.1038/nn1525 [DOI] [PubMed] [Google Scholar]
- Carrillo-Reid L, Yang W, Bando Y, Peterka DS, Yuste R (2016) Imprinting and recalling cortical ensembles. Science 353:691–694. 10.1126/science.aaf7560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaigneau E, Ronzitti E, Gajowa MA, Soler-Llavina GJ, Tanese D, Brureau AY, Papagiakoumou E, Zeng H, Emiliani V (2016) Two-photon holographic stimulation of ReaChR. Front Cell Neurosci 10:234. 10.3389/fncel.2016.00234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, Looger LL, Svoboda K, Kim DS (2013) Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499:295–300. 10.1038/nature12354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen IW, Papagiakoumou E, Emiliani V (2018) Towards circuit optogenetics. Curr Opin Neurobiol 50:179–189. 10.1016/j.conb.2018.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle TL, Madisen L, Hage TA, Valley MT, Knoblich U, Larsen RS, Takeno MM, Huang L, Gu H, Larsen R, Mills M, Bosma-Moody A, Siverts LA, Walker M, Graybuck LT, Yao Z, Fong O, Nguyen TN, Garren E, Lenz GH, et al. (2018) A suite of transgenic driver and reporter mouse lines with enhanced brain cell type targeting and functionality. Cell 174:465–480.e22. 10.1016/j.cell.2018.06.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Maschio M, Donovan JC, Helmbrecht TO, Baier H (2017) Linking neurons to network function and behavior by two-photon holographic optogenetics and volumetric imaging. Neuron 94:774–789.e5. 10.1016/j.neuron.2017.04.034 [DOI] [PubMed] [Google Scholar]
- Dana H, Chen TW, Hu A, Shields BC, Guo C, Looger LL, Kim DS, Svoboda K (2014) Thy1-GCaMP6 transgenic mice for neuronal population imaging in vivo. PLoS One 9:e108697. 10.1371/journal.pone.0108697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dana H, Mohar B, Sun Y, Narayan S, Gordus A, Hasseman JP, Tsegaye G, Holt GT, Hu A, Walpita D, Patel R, Macklin JJ, Bargmann CI, Ahrens MB, Schreiter ER, Jayaraman V, Looger LL, Svoboda K, Kim DS (2016) Sensitive red protein calcium indicators for imaging neural activity. Elife 2:1–24. 10.7554/eLife.12727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emiliani V, Cohen AE, Deisseroth K, Häusser M (2015) All-optical interrogation of neural circuits. J Neurosci 35:13917–13926. 10.1523/JNEUROSCI.2916-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forli A, Vecchia D, Binini N, Succol F, Bovetti S, Moretti C, Nespoli F, Mahn M, Baker CA, Bolton MM, Yizhar O, Fellin T (2018) Two-photon bidirectional control and imaging of neuronal excitability with high spatial resolution in vivo. Cell Rep 22:3087–3098. 10.1016/j.celrep.2018.02.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil-Sanz C, Espinosa A, Fregoso SP, Bluske KK, Cunningham CL, Martinez-Garay I, Zeng H, Franco SJ, Müller U (2015) Lineage tracing using Cux2-cre and Cux2-CreERT2 mice. Neuron 86:1091–1099. 10.1016/j.neuron.2015.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glückstad J. (1996) Phase contrast image synthesis. Opt Commun 130:225–230. [Google Scholar]
- Gradinaru V, Thompson KR, Zhang F, Mogri M, Kay K, Schneider MB, Deisseroth K (2007) Targeting and readout strategies for fast optical neural control in vitro and in vivo. J Neurosci 27:14231–14238. 10.1523/JNEUROSCI.3578-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewe BF, Voigt FF, van't Hoff M, Helmchen F (2011) Fast two-layer two-photon imaging of neuronal cell populations using an electrically tunable lens. Biomed Opt Express 2:2035–2046. 10.1364/BOE.2.002035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosenick L, Marshel JH, Deisseroth K (2015) Closed-loop and activity-guided optogenetic control. Neuron 86:106–139. 10.1016/j.neuron.2015.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo ZV, Hart AC, Ramanathan S (2009) Optical interrogation of neural circuits in Caenorhabditis elegans. Nat Methods 6:891–896. 10.1038/nmeth.1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmchen F, Denk W (2005) Deep tissue two-photon microscopy. Nat Methods 2:932–940. 10.1038/nmeth818 [DOI] [PubMed] [Google Scholar]
- Hernandez O, Guillon M, Papagiakoumou E, Emiliani V (2014) Zero-order suppression for two-photon holographic excitation. Opt Lett 39:5953–5956. 10.1364/OL.39.005953 [DOI] [PubMed] [Google Scholar]
- Hernandez O, Papagiakoumou E, Tanese D, Fidelin K, Wyart C, Emiliani V (2016) Three-dimensional spatiotemporal focusing of holographic patterns. Nat Commun 7:11928. 10.1038/ncomms11928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtmaat A, Bonhoeffer T, Chow DK, Chuckowree J, De Paola V, Hofer SB, Hübener M, Keck T, Knott G, Lee WC, Mostany R, Mrsic-Flogel TD, Nedivi E, Portera-Cailliau C, Svoboda K, Trachtenberg JT, Wilbrecht L (2009) Long-term, high-resolution imaging in the mouse neocortex through a chronic cranial window. Nat Protoc 4:1128–1144. 10.1038/nprot.2009.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopt A, Neher E (2001) Highly nonlinear photodamage in two-photon fluorescence microscopy. Biophys J 80:2029–2036. 10.1016/S0006-3495(01)76173-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue M, Takeuchi A, Horigane S, Ohkura M, Gengyo-Ando K, Fujii H, Kamijo S, Takemoto-Kimura S, Kano M, Nakai J, Kitamura K, Bito H (2015) Rational design of a high-affinity, fast, red calcium indicator R-CaMP2. Nat Methods 12:64–70. 10.1038/nmeth.3185 [DOI] [PubMed] [Google Scholar]
- Joris PX, Smith PH, Yin TC (1998) Coincidence detection in the auditory system: 50 years after Jeffress. Neuron 21:1235–1238. 10.1016/S0896-6273(00)80643-1 [DOI] [PubMed] [Google Scholar]
- Katona G, Szalay G, Maák P, Kaszás A, Veress M, Hillier D, Chiovini B, Vizi ES, Roska B, Rózsa B (2012) Fast two-photon in vivo imaging with three-dimensional random-access scanning in large tissue volumes. Nat Methods 9:201–208. 10.1038/nmeth.1851 [DOI] [PubMed] [Google Scholar]
- Kerlin AM, Andermann ML, Berezovskii VK, Reid RC (2010) Broadly tuned response properties of diverse inhibitory neuron subtypes in mouse visual cortex. Neuron 67:858–871. 10.1016/j.neuron.2010.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapoetke NC, Murata Y, Kim SS, Pulver SR, Birdsey-Benson A, Cho YK, Morimoto TK, Chuong AS, Carpenter EJ, Tian Z, Wang J, Xie Y, Yan Z, Zhang Y, Chow BY, Surek B, Melkonian M, Jayaraman V, Constantine-Paton M, Wong GK, et al. (2014) Independent optical excitation of distinct neural populations. Nat Methods 11:338–346. 10.1038/nmeth.2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koester HJ, Baur D, Uhl R, Hell SW (1999) Ca2+ fluorescence imaging with pico- and femtosecond two-photon excitation: signal and photodamage. Biophys J 77:2226–2236. 10.1016/S0006-3495(99)77063-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinweber M, Ward DR, Sobczak JM, Attinger A, Keller GB (2017) A sensorimotor circuit in mouse cortex for visual flow predictions. Neuron 95:1420–1432.e5. 10.1016/j.neuron.2017.08.036 [DOI] [PubMed] [Google Scholar]
- Li H, Penzo MA, Taniguchi H, Kopec CD, Huang ZJ, Li B (2013) Experience-dependent modification of a central amygdala fear circuit. Nat Neurosci 16:332–339. 10.1038/nn.3322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JY, Knutsen PM, Muller A, Kleinfeld D, Tsien RY (2013) ReaChR: a red-shifted variant of channelrhodopsin enables deep transcranial optogenetic excitation. Nat Neurosci 16:1499–1508. 10.1038/nn.3502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linz N, Freidank S, Liang XX, Vogel A (2016) Wavelength dependence of femtosecond laser-induced breakdown in water and implications for laser surgery. Phys Rev B 94:024113 10.1103/PhysRevB.94.024113 [DOI] [Google Scholar]
- Lu R, Sun W, Liang Y, Kerlin A, Bierfeld J, Seelig JD, Wilson DE, Scholl B, Mohar B, Tanimoto M, Koyama M, Fitzpatrick D, Orger MB, Ji N (2017) Video-rate volumetric functional imaging of the brain at synaptic resolution. Nat Neurosci 20:620–628. 10.1038/nn.4516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz C, Otis TS, DeSars V, Charpak S, DiGregorio DA, Emiliani V (2008) Holographic photolysis of caged neurotransmitters. Nat Methods 5:821–827. 10.1038/nmeth.1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Garner AR, Carandini M, Zeng H, Madisen L, Garner AR, Shimaoka D, Chuong AS, Klapoetke NC, Li L, van der Bourg A, Niino Y, Egolf L, Monetti C, Gu H, Mills M, Cheng A, Tasic B, Nguyen TN, Sunkin SM, Benucci A, Nagy A, Miyawaki A, Helmchen F, et al. (2015) Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron 85:942–958. 10.1016/j.neuron.2015.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardinly AR, Oldenburg IA, Pégard NC, Sridharan S, Lyall EH, Chesnov K, Brohawn SG, Waller L, Adesnik H (2018) Precise multimodal optical control of neural ensemble activity. Nat Neurosci 21:881–893. 10.1038/s41593-018-0139-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markram H, Lübke J, Frotscher M, Sakmann B (1997) Regulation of synaptic efficacy by coincidence of postsynaptic APs and EPSPs. Science 275:213–215. 10.1126/science.275.5297.213 [DOI] [PubMed] [Google Scholar]
- Mattis J, Tye KM, Ferenczi EA, Ramakrishnan C, O'Shea DJ, Prakash R, Gunaydin LA, Hyun M, Fenno LE, Gradinaru V, Yizhar O, Deisseroth K (2011) Principles for applying optogenetic tools derived from direct comparative analysis of microbial opsins. Nat Methods 9:159–172. 10.1038/nmeth.1808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel G, Szellas T, Huhn W, Kateriya S, Adeishvili N, Berthold P, Ollig D, Hegemann P, Bamberg E (2003) Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci U S A 100:13940–13945. 10.1073/pnas.1936192100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolic K, Grossman N, Grubb MS, Burrone J, Toumazou C, Degenaar P (2009) Photocycles of channelrhodopsin-2. Photochem Photobiol 85:400–411. 10.1111/j.1751-1097.2008.00460.x [DOI] [PubMed] [Google Scholar]
- Olivié G, Giguère D, Vidal F, Ozaki T, Kieffer JC, Nada O, Brunette I (2008) Wavelength dependence of femtosecond laser ablation threshold of corneal stroma. Opt Express 16:4121–4129. 10.1364/OE.16.004121 [DOI] [PubMed] [Google Scholar]
- Packer AM, Peterka DS, Hirtz JJ, Prakash R, Deisseroth K, Yuste R (2012) Two-photon optogenetics of dendritic spines and neural circuits. Nat Methods 9:1202–1205. 10.1038/nmeth.2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer AM, Russell LE, Dalgleish HW, Häusser M (2015) Simultaneous all-optical manipulation and recording of neural circuit activity with cellular resolution in vivo. Nat Methods 12:140–146. 10.1038/nmeth.3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papagiakoumou E, de Sars V, Oron D, Emiliani V (2008) Patterned two-photon illumination by spatiotemporal shaping of ultrashort pulses. Opt Express 16:22039–22047. 10.1364/OE.16.022039 [DOI] [PubMed] [Google Scholar]
- Papagiakoumou E, Anselmi F, Bègue A, de Sars V, Glückstad J, Isacoff EY, Emiliani V (2010) Scanless two-photon excitation of channelrhodopsin-2. Nat Methods 7:848–854. 10.1038/nmeth.1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papagiakoumou E, Bègue A, Leshem B, Schwartz O, Stell BM, Bradley J, Oron D, Emiliani V (2013) Functional patterned multiphoton excitation deep inside scattering tissue. Nat Photonics 7:274–278. 10.1038/nphoton.2013.9 [DOI] [Google Scholar]
- Papagiakoumou E, Ronzitti E, Chen I-W, Gajowa M, Picot A, Emiliani V (2018) Two-photon optogenetics by computer-generated holography. In: Optogenetics: A roadmap (Stroh A, ed). Heidelberg: Springer. [Google Scholar]
- Pégard NC, Mardinly AR, Oldenburg IA, Sridharan S, Waller L, Adesnik H (2017) Three-dimensional scanless holographic optogenetics with temporal focusing (3D-SHOT). Nat Commun 8:1228. 10.1038/s41467-017-01031-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picot A, Dominguez S, Liu C, Chen IW, Tanese D, Ronzitti E, Berto P, Papagiakoumou E, Oron D, Tessier G, Forget BC, Emiliani V (2018) Temperature rise under two-photon optogenetic brain stimulation. Cell Rep 24:1243–1253.e5. 10.1016/j.celrep.2018.06.119 [DOI] [PubMed] [Google Scholar]
- Pologruto TA, Sabatini BL, Svoboda K (2003) ScanImage: Flexible software for operating laser scanning microscopes. Biomed Eng Online 2:13. 10.1186/1475-925X-2-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash R, Yizhar O, Grewe B, Ramakrishnan C, Wang N, Goshen I, Packer AM, Peterka DS, Yuste R, Schnitzer MJ, Deisseroth K (2012) Two-photon optogenetic toolbox for fast inhibition, excitation and bistable modulation. Nat Methods 9:1171–1179. 10.1038/nmeth.2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasethupathy P, Sankaran S, Marshel JH, Kim CK, Ferenczi E, Lee SY, Berndt A, Ramakrishnan C, Jaffe A, Lo M, Liston C, Deisseroth K (2015) Projections from neocortex mediate top-down control of memory retrieval. Nature 526:653–659. 10.1038/nature15389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickgauer JP, Deisseroth K, Tank DW (2014) Simultaneous cellular-resolution optical perturbation and imaging of place cell firing fields. Nat Neurosci 17:1816–1824. 10.1038/nn.3866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronzitti E, Ventalon C, Canepari M, Forget BC, Papagiakoumou E, Emiliani V (2017a) Recent advances in patterned photostimulation for optogenetics. Journal of Optics 19:113001 10.1088/2040-8986/aa8299 [DOI] [Google Scholar]
- Ronzitti E, Conti R, Zampini V, Klapoetke NC, Tanese D, Foust AJ, Klapoetke N, Boyden ES, Papagiakoumou E, Emiliani V (2017b) Sub-millisecond optogenetic control of neuronal firing by two-photon holographic photoactivation of Chronos. J Neurosci 37:10679–10689. 10.1523/JNEUROSCI.1246-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz W, Dayan P, Montague PR (1997) A neural substrate of prediction and reward. Science 275:1593–1599. 10.1126/science.275.5306.1593 [DOI] [PubMed] [Google Scholar]
- Schultz W, Stauffer WR, Lak A (2017) The phasic dopamine signal maturing: from reward via behavioural activation to formal economic utility. Curr Opin Neurobiol 43:139–148. 10.1016/j.conb.2017.03.013 [DOI] [PubMed] [Google Scholar]
- Shemesh OA, Tanese D, Zampini V, Linghu C, Piatkevich K, Ronzitti E, Papagiakoumou E, Boyden ES, Emiliani V (2017) Temporally precise single-cell-resolution optogenetics. Nat Neurosci 20:1796–1806. 10.1038/s41593-017-0018-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofroniew NJ, Flickinger D, King J, Svoboda K (2016) A large field of view two-photon mesoscope with subcellular resolution for in vivo imaging. Elife:1–20. 10.7554/eLife.14472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub C, Saulnier JL, Bègue A, Feng DD, Feng DD, Huang KW, Sabatini BL (2016) Principles of synaptic organization of GABAergic interneurons in the striatum. Neuron 92:84–92. 10.1016/j.neuron.2016.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart GJ, Häusser M (2001) Dendritic coincidence detection of EPSPs and action potentials. Nat Neurosci 4:63–71. 10.1038/82910 [DOI] [PubMed] [Google Scholar]
- Stuart GJ, Spruston N (2015) Dendritic integration: 60 years of progress. Nat Neurosci 18:1713–1721. 10.1038/nn.4157 [DOI] [PubMed] [Google Scholar]
- Szabo V, Ventalon C, De Sars V, Bradley J, Emiliani V (2014) Spatially selective holographic photoactivation and functional fluorescence imaging in freely behaving mice with a fiberscope. Neuron 84:1157–1169. 10.1016/j.neuron.2014.11.005 [DOI] [PubMed] [Google Scholar]
- Takahashi N, Oertner TG, Hegemann P, Larkum ME (2016) Active cortical dendrites modulate perception. Science 354:1587–1590. 10.1126/science.aah6066 [DOI] [PubMed] [Google Scholar]
- Traub RD, Whittington MA, Stanford IM, Jefferys JG (1996) A mechanism for generation of long-range synchronous fast oscillations in the cortex. Nature 383:621–624. 10.1038/383621a0 [DOI] [PubMed] [Google Scholar]
- Waters J, Helmchen F (2006) Background synaptic activity is sparse in neocortex. J Neurosci 26:8267–8277. 10.1523/JNEUROSCI.2152-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyart C, Del Bene F, Warp E, Scott EK, Trauner D, Baier H, Isacoff EY (2009) Optogenetic dissection of a behavioural module in the vertebrate spinal cord. Nature 461:407–410. 10.1038/nature08323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Carrillo-Reid L, Bando Y, Peterka DS, Yuste R (2018) Simultaneous two-photon imaging and two-photon optogenetics of cortical circuits in three dimensions. Elife 7:e32671. 10.7554/eLife.32671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yizhar O, Fenno LE, Prigge M, Schneider F, Davidson TJ, O'Shea DJ, Sohal VS, Goshen I, Finkelstein J, Paz JT, Stehfest K, Fudim R, Ramakrishnan C, Huguenard JR, Hegemann P, Deisseroth K (2011) Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature 477:171–178. 10.1038/nature10360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LI, Tao HW, Holt CE, Harris WA, Poo M (1998) A critical window for cooperation and competition among developing retinotectal synapses. Nature 395:37–44. 10.1038/25665 [DOI] [PubMed] [Google Scholar]
- Zhang YP, Oertner TG (2007) Optical induction of synaptic plasticity using a light-sensitive channel. Nat Methods 4:139–141. 10.1038/nmeth988 [DOI] [PubMed] [Google Scholar]