Abstract
Isotope labels are frequently used tools to track metabolites through complex biochemical pathways and to discern the mechanisms of enzyme-catalyzed reactions. Isotopically labeled l-serine is often used to monitor the activity of the first enzyme in sphingolipid biosynthesis, serine palmitoyltransferase (SPT), as well as labeling downstream cellular metabolites. Intrigued by the effect that isotope labels may be having on SPT catalysis, we characterized the impact of different l-serine isotopologues on the catalytic activity of recombinant SPT isozymes from humans and the bacterium Sphingomonas paucimobilis. Our data show that S. paucimobilis SPT activity displays a clear isotope effect with [2,3,3-D]l-serine, whereas the human SPT isoform does not. This suggests that although both human and S. paucimobilis SPT catalyze the same chemical reaction, there may well be underlying subtle differences in their catalytic mechanisms. Our results suggest that it is the activating small subunits of human SPT that play a key role in these mechanistic variations. This study also highlights that it is important to consider the type and location of isotope labels on a substrate when they are to be used in in vitro and in vivo studies.
Keywords: sphingolipid, biosynthesis, mechanism, regulation, membrane protein
Sphingolipids (SLs) are a family of l-serine-derived molecules that have been shown to play a range of functional roles in various species (1, 2). Their importance is reflected in the expanding list of diseases that are now linked with SLs and their metabolites (3). Serine palmitoyltransferase (SPT) is a pyridoxal 5′-phosphate (PLP)-dependent enzyme that catalyzes the first step in SL biosynthesis, the Claisen-like condensation of l-serine with palmitoyl-CoA to form 3-ketodihydrosphingosine (3-KDS) (Fig. 1A) (4). Although SPT is conserved across all SL-producing organisms studied to date, there are structural differences between SPTs from different taxa. In bacteria such as Sphingomonas paucimobilis, SPT is a soluble homodimer, whose structure was determined in 2007, revealing a three-domain architecture (5). Although all three domains are required for dimerization of the enzyme, the central domain is required for catalysis, and the active site is formed at the dimer interface (5, 6). In contrast to bacterial isoforms, the SPT homologs from higher eukaryotes are heterodimers of two core subunits, long-chain base 1 (LCB1) and LCB2, also referred to as SPT1 and SPT2. The LCB1 and LCB2 proteins share significant sequence homology, suggesting shared ancestry, but display key differences in the residues proposed to be involved in PLP binding and enzyme catalysis (7–11).
Fig. 1.
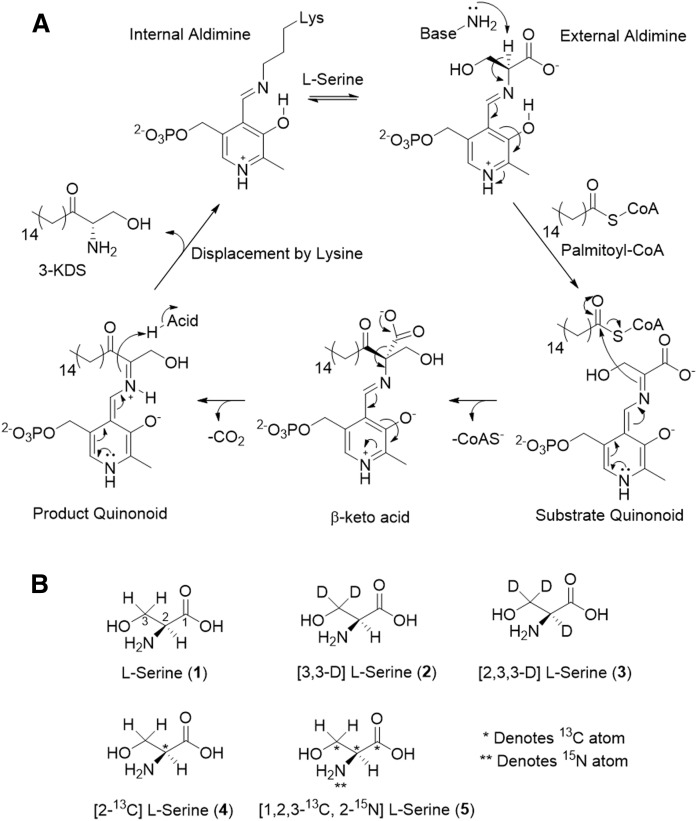
A: The proposed mechanism of SPT. Briefly, l-serine binds to the PLP-bound, internal aldimine, displacing the active-site lysine to form the SPT PLP: l-serine external aldimine complex. Binding of palmitoyl-CoA induces a conformational change that causes removal of the α-proton from l-serine to form the PLP: l-serine quinonoid. The quinonoid attacks the palmitoyl-CoA thioester and leads to C-C bond formation, which, after electron rebound, results in CoAS release and formation of the β-keto acid intermediate. Decarboxylation leads to the PLP-bound product quinonoid, which is reprotonated by an active-site acid. The active-site lysine then displaces the 3-KDS product and reforms the internal aldimine with PLP bound. B: Isotope labeling patterns of l-serine substrates used in this study. Each isotopologue is labeled with appropriate heavy atom (deuterium D), 13C, or 15N denoted by an asterisk. l-serine 1 and 5 are referred to as light and heavy, respectively.
Moreover, the eukaryotic SPTs are integral membrane proteins that reside in the ER membrane. In humans, additional complexity is added by the presence of two isoforms of LCB2 (named LCB2a and LCB2b), as well as of two isoforms of a third, small subunit of SPT (ssSPTa and ssSPTb), whose functionally equivalent small protein in Saccharomyces cerevisiae is Tsc3p (12, 13). These ssSPTs have a single spanning transmembrane domain and have been shown to increase the in vitro activity of the SPT complex by up to ∼100-fold (13, 14). To compound this, in eukaryotes, SPT forms a complex with regulatory partners known as the orosomucoid-like protein (ORMs), which are negative regulators of SPT activity (15, 16). In yeast, this complex is called “SPOTS,” consisting of Lcb1, Lcb2, Tsc3p, Orm1, Orm2, and Sac1 (15). The molecular mechanisms by which this complex controls SPT activity are unclear, but phosphorylation of Orm1 and Orm2 by a kinase cascade is in part responsible for the regulation of SPT (15, 16). In humans, the equivalent ORMDL proteins lack the phosphorylation sites found in the yeast ORMs, and so it is unknown how these proteins regulate the activity of SPT (17, 18). A recent study has suggested that the ORMs and ORMDLs respond directly to ceramide concentrations to control SPT enzymatic turnover, but the mechanistic details have yet to be revealed (19). Mutations of both human SPTs are also linked with a rare genetic disease, hereditary sensory neuropathy type 1 (HSAN1; reviewed in ref. 20). These mutations result in a gain of function and increased promiscuity with respect to the amino-acid specificity of the enzyme; mutant SPTs generate so-called “deoxy SLs” from glycine and l-alanine (21). Despite their acknowledged link to HSAN1, the exact mechanism of how these atypical SLs exert their damage on neurons remains unclear. Also, there is growing evidence that ORMDLs are linked to various disease pathologies, but their exact roles are under current investigation (22).
SPT belongs to the α-oxoamine synthase (AOS) family of PLP-dependent enzymes, whose enzymology has been extensively studied, and a mechanism has been proposed for SPT (Fig. 1A) (5, 23–27). Catalysis begins with displacement of the active-site lysine from the PLP-bound internal aldimine (also known as the holoform) form of the enzyme by the l-serine substrate to generate the SPT PLP:l-serine external aldimine complex. Binding of palmitoyl-CoA is thought to induce a conformational change, which allows an active-site base to abstract the α-proton of l-serine, forming a substrate quinonoid. Electron rebound from the PLP-bound quinonoid results in C-C bond formation with liberation of free CoA (CoASH). Decarboxylation of the PLP:β-keto acid follows, and the product quinonoid is reprotonated by an active-site acid. Displacement by the active-site lysine releases the product 3-KDS from the PLP:product external aldimine and returns the SPT to the PLP-bound, holoform.
Isotope labels are frequently used in metabolic and natural product biosynthesis studies, where they can be used to track the incorporation of small-molecule precursors into complex metabolites in a cellular environment (28–31). The inherent differences in zero-point energies between different isotopes can be used to elucidate the mechanistic details of enzyme-catalyzed reactions. This is due to the fact that these energy differences result in increased bond strength, which can be observed as an effect on the rate of an enzyme-catalyzed reaction—a kinetic isotope effect (KIE). As such, this effect has been used to great effect in mechanistic studies (32–35).
Given the increase in the use of isotopically labeled l-serine in SL analysis, especially the use of the [2,3,3-D]l-serine isotopologue (compound 3 in Fig. 1B), we wanted to investigate what effect, if any, these labels have on the SPT-catalyzed reaction. We were concerned that, without an understanding of the underlying biochemistry, a misplaced isotope label could lead to unintended consequences for two reasons—1) incorrect rate measurements and 2) using a mass label that is lost in the reaction. For metabolic tracking, it is advantageous to have as big a mass difference as possible in the products of interest to allow one to clearly differentiate those derived from the labeled substrate. However, these labels are of no value if they are lost during the enzyme-catalyzed steps of a pathway. Herein, we report the use of a series of isotopically labeled l-serine substrates and their effects on the kinetics of the SPT-catalyzed reaction. Our data show that with soluble, homodimeric S. paucimobilis SPT, the presence of a deuterium label on the α-carbon results in a significant KIE, implicating abstraction of this proton as being a rate-determining step. In contrast, surprisingly, in the multisubunit, membrane-bound human homolog of the enzyme, no KIE is observed for the same substrate. Our results are in agreement with a recent report by Hannun and colleagues on the yeast SPT complex (34) and suggest that there are subtle mechanistic differences between the cytoplasmic and membrane-bound forms of the enzymes that are dependent on the activating small subunit.
MATERIALS AND METHODS
Source of l-serine isotopologues
The structures and labels of the l-serine substrates used are shown in Fig. 1B. Nonisotopically labeled l-serine (1) was purchased from Sigma (catalog no. S4500). The [3,3-D] l-serine (2) (catalog no. DLM-161), [2,3,3-D]l-serine (3) (catalog no. DLM-582), and [1,2,3,-13C, 2-15N]l-serine (5) (catalog no. CNLM-474) were purchased from Cambridge Isotope Laboratories. The [2-13Cl-serine (4) (catalog no. 9370) was purchased from Icon Isotopes.
Expression and purification of S. paucimobilis SPT
The N-terminal His6-tagged S. paucimobilis SPT (SpSPT) was expressed in Escherichia coli and purified from a pET-28a expression plasmid essentially as previously described (36). Briefly, a single colony was picked into 200 ml of LB medium and grown overnight at 37°C with 35 µg·ml−1 kanamycin. The overnight culture was diluted back to OD600 = 0.05. Cells were grown to OD600 = 0.6 at 37°C with 35 µg·ml−1 kanamycin. At OD600 = 0.6, cells were induced with 0.1 mM isopropyl β-d-thiogalactopyranoside and incubated at 30°C for a further 5 h before harvesting at 4,122 g. Cells were washed in PBS buffer before resuspension in 1:1 (wt/vol) wash buffer (50 mM HEPES, pH 7.8, 150 mM NaCl, 10 mM imidazole, and 25 µM PLP). Cells were lysed by sonication (10 min, 30 s on, 30 s off, 10 μm), and cell-free extract was prepared by centrifugation (24,446 g, 30 min). The cell-free extract was then applied to a 1 ml His-Trap column (GE Life Sciences) and washed with wash buffer until the baseline was flat. SpSPT was then eluted by an imidazole gradient (10–300 mM) over 1 h. Fractions containing protein were pooled and concentrated in a 30 kDa spin filter to a final volume of 1 ml. Concentrated protein was finally purified by gel filtration chromatography (Superdex S200 column, GE Life Sciences), eluting with 50 mM HEPES (pH 7.8), 150 mM NaCl, and 25 µM PLP. Fractions containing protein were pooled and concentrated in a 30 kDa spin filter, before storage at −80°C. Protein concentrations were determined by the method of Bradford (1976) (37).
Expression and purification of single-chain human SPT
The construction and expression of an active single-chain human SPT (scSPT) has been described previously (14, 17, 38), and a full report on its biochemical and structural characterization is to follow (Somashekarappa et al, unpublished observations). This construct contains the three human SPT subunits (LCB2a, ssSPTa, and LCB1) linked together in a head-to-tail fashion into a single-chain protein that can be expressed in yeast, as well as CHO-LyB and HEK293 mammalian cells. Most recently, this construct has also been used in HeLa cells to study SPT regulation (19). The plasmid encoding single-chain SPT was used to transform the S. cerevisiae Δlcb1 strain. A single yeast colony was then used to inoculate 200 ml of YPD medium, which was grown overnight at 26°C. The overnight culture was diluted back to OD600 = 0.05, and 0.5 mM CuSO4 was added. Cells were grown overnight at 26°C before harvesting at 5,000 g. Harvested cells were washed with PBS before resuspension in buffer (50 mM Tris, pH 8.0, 0.25 M sorbitol, 0.3 M NaCl, 2 mM EDTA, 2 mM EGTA, 1 mM PMSF, 0.5 mM benzamidine, and 25 µM PLP). Cells were lysed by bead-beating (0.5 mm diameter zirconia beads), and cellular debris was removed by centrifugation at 5,000 g. The supernatant was then collected, and membranes were harvested by ultracentrifugation at 100,000 g for 1 h. Harvested membranes were washed in membrane buffer (50 mM HEPES, pH 8, 150 mM NaCl, 10% glycerol, 1 mM PMSF, and 25 µM PLP) before reharvesting by ultracentrifugation at 164,244 g for 1 h. Membranes were solubilized in 1% n-dodecyl β-d-maltoside (DDM; Generon) for 1 h before unsolubilized material was removed by ultracentrifugation at 164,244 g for 1 h. Solubilized protein was added to 1 ml of equilibrated nickel resin (GE Life Sciences) and incubated for 1 h at 4°C. Resin was washed with 10 column volumes (CV) wash buffer (50 mM HEPES, pH 8, 150 mM NaCl, 30 mM imidazole, 10% glycerol, 1 mM PMSF, and 25 µM PLP) before elution into 5 CV elution buffer (0.024% DDM, 50 mM HEPES, pH 8, 150 mM NaCl, 300 mM imidazole, 10% glycerol, 1 mM PMSF, and 25 µM PLP). The eluted protein was concentrated and stored at −80°C until required.
Kinetic analysis of purified bacterial SpSPT and human scSPT
All assays were conducted in a total volume of 100 µl and were performed using a BioTek Synergy HT plate reader. Assays contained l-serine (100 µM to 40 mM), palmitoyl-CoA (250 µM), DTNB (ε412 = 14,150, 400 µM), SpSPT or scSPT (SpSPT, 400 nM; scSPT, 5 µM), assay buffer [50 mM HEPES, pH 7.8, 100 mM NaCl, (0.05% DDM for scSPT), to 100 µl]. Measurements were taken at 412 nm over the course of 1 h. Statistical analysis was performed by unpaired t-test using the GraphPad online calculator (39).
LCB formation by human SPTs in yeast microsomes
Microsomal membrane preparation.
For these experiments, we used two forms of the human SPT complex expressed in yeast microsomes. One was the fused gene construct scSPT described in the section “Expression and purification of single-chain human SPT” without purification. The other used human SPT from yeast microsomes where the human LCB1, LCB2a, and ssSPTa genes were independently coexpressed as described previously (13). Cells were harvested at exponential phase, pelleted at 5,000 g, washed with water, repelleted, and then washed with TEGM buffer (50 mM Tris, pH 7.5, 1 mM EGTA, 1 mM β-mercaptoethanol, 1 mM PMSF, 1 μg·ml−1 Leupeptin, 1 μg·ml−1 Pepstatin A, and 1 μg·ml−1 Aprotinin) buffer. The cell pellets were resuspended in TEGM buffer at 1 ml/50 OD600 nm cells, and glass beads (0.5 mm diameter) were added to ∼1/4 inch from the meniscus. The cells were disrupted by vortexing 4 × 1 min with 1 min on ice in between, transferred to a new tube with extensive washing of the beads, and pelleted at 8,000 g for 10 min. The supernatant was transferred to new tubes and spun at 100,000 g for 30 min. The resulting pellet was resuspended by a Dounce homogenizer in at least 10× volume and repelleted at 100,000 g. The final membrane pellet was resuspended in TEGM buffer containing 33% glycerol and stored at −80°C. Total protein concentration of the microsomal membranes was determined by Bio-Rad dye reagent using IgG as a standard.
Microsomal SPT assay
The reaction was started by adding 400 μg of microsomal membrane to a reaction cocktail (final volume 200 μl) containing 50 mM HEPES, pH 8.1, 50 μM PLP, 10 mM l-serine (1) or deuterated serine stable isotope (2 or 3; Fig. 1B), and 100 μM pentadecanoyl coenzyme-A (Avanti Polar Lipids). After a 10 min reaction at 37°C, 100 μl of 2 N NH4OH and 100 μl of 1 M NaBH4 were added, and the reaction was kept at 37°C for an additional 10 min. LCBs were extracted by the addition of 2.0 ml of CHCl3:methanol (1:2) containing 250 nM C17-sphingosine as an internal standard, followed by the addition of 1 ml of CHCl3 and 2 ml of 0.5 M NH4OH, vortexing, and centrifuging briefly. The upper aqueous layer was aspirated off, and the lower layer was washed with 2 ml of 60 mM KCl and centrifuged. The washing was carried out twice, and 1.3 ml of the sample was dried and subjected to derivatization for HPLC analysis.
Analysis of LCBs from microsomal SPT assays
The LCB extract was redissolved in 80 μl of methanol:190 mM triethylamine (20:3) and 20 μl of AccQ-Fluor Reagent (Waters). After incubation at room temperature for at least 60 min, 10 μl of 1.0 M KOH (methanol) was added. A total of 80 μl of the sample (corresponding to 0.24 mg of protein) was injected on a Genesis C18 4 μM HPLC column (Jones Chromatography) and resolved on an Agilent 1100 series HPLC equipped with a fluorescence detector. The LCBs were resolved using an isocratic mobile phase of acetonitrile:methanol:H2O:acetic acid:triethylamine (48:32:16.5:3.0:0.7) at 1.5 ml·min−1 and detected by the AccQ fluorescence (excitation 244 nm/emission 398 nm).
Mass-spectrometry analysis of 3-KDS products
An aliquot (50 µl) from each SPT kinetic assay end-point reaction was desalted on C18 resin and eluted in 100% acetonitrile. Samples were dried and resuspended in 50% acetonitrile/0.1% formic acid before direct infusion ESI mass-spectrometry analysis using a TriVersa NanoMate (Advion) coupled to a 12T FTICR (Bruker). Twenty 4-megaword transients were averaged over m/z 200–500, providing a KDS resolution of at least 300,000. Data were analyzed using Bruker Compass DataAnalysis 4.4 for detection of compounds and their accurate monoisotopic masses. The theoretical isotope envelope was predicted using empirical formula and the Bruker IsotopePattern tool. Raw data were exported using Bruker CompassXport for replotting in Origin.
RESULTS
Kinetic analysis of bacterial and human SPTs
We began by comparing the kinetics of the purified bacterial (SpSPT) and human (scSPT) forms of SPT using “light” l-serine (1) and the commercially available, isotopically labeled “heavy” l-serine derivatives (2–5) (Fig. 1B). We have carried out comprehensive structural, biochemical, and mechanistic studies of SpSPT (5, 36) since it was first isolated from bacterial cells (26). The catalytic activity of this recombinant, homodimeric, soluble SpSPT is relatively easy to monitor using a convenient, continuous spectrophotometric assay with the DTNB reagent reacting with released CoASH (6). Importantly, we also confirmed the production of the KDS product using high-resolution mass spectrometry to track the incorporation or loss of isotopic labels derived from the various l-serine substrates (see supplemental Fig. S1).
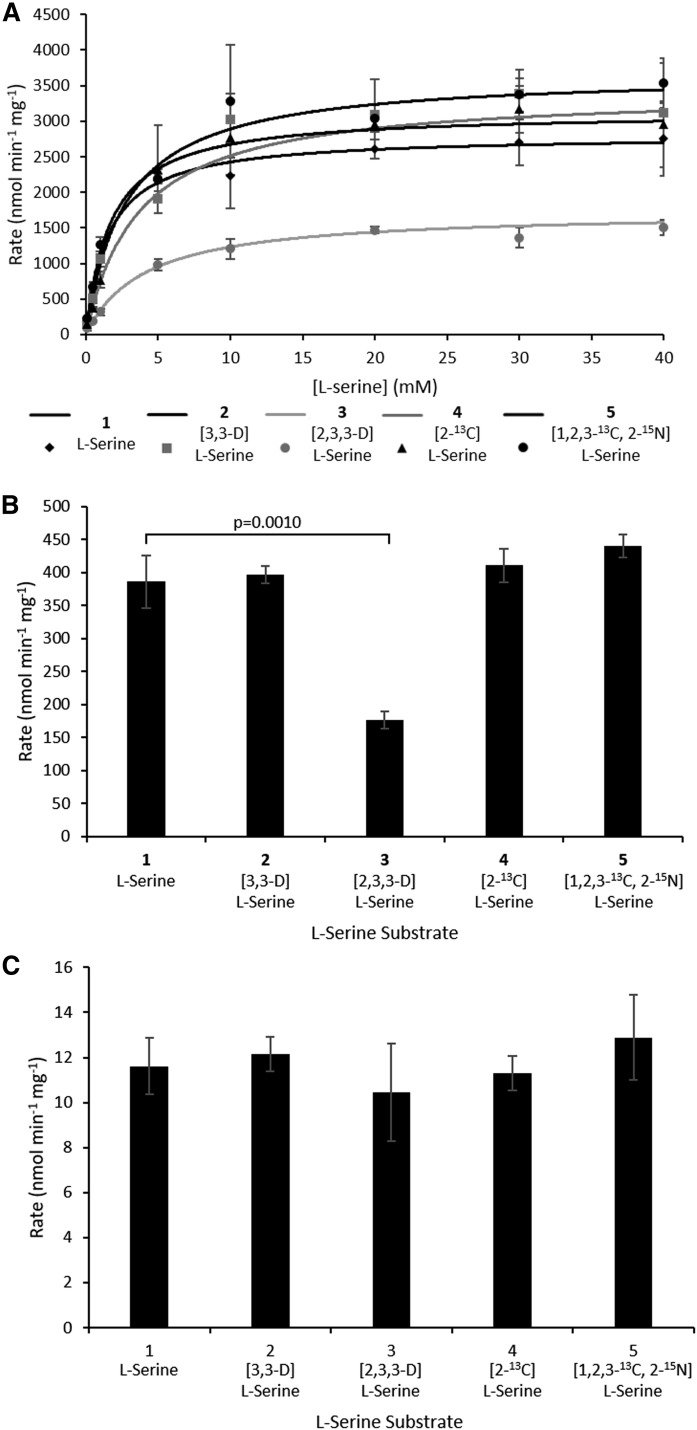
For the SpSPT, we have kinetic data using the “light, unlabeled” l-serine (1) from previous studies, and the data generated here are in good agreement (l-Ser KM = 1.56 ± 0.10 mM, specific activity = 386.3 ± 39.7 nmol·min−1·mg−1; Fig. 2A, B, Table 1). Carrying out the same analysis with D-, 13C-, and 15N-labeled l-serine substrates allowed us to determine the impact of the isotopic labels in the substrate on the catalytic activity of the enzyme. As expected, [3,3-D]l-serine (2) behaved essentially the same as the light l-serine version with a KM = 2.72 ± 0.29 mM and specific activity 396.7 ± 13.1 nmol·min−1·mg−1 (P = 0.6888; Fig. 2A, B, Table 1). Similarly, [2-13C]l-serine (4) also gave equivalent values (KM = 3.64 ± 0.61 mM, specific activity = 410.6 ± 25.5 nmol·min−1·mg−1, P = 0.4228; Fig. 2A, B, Table 1). The “heaviest” l-serine {[1,2,3-13C, 2-15N] (5))}, with all three carbons and the amine nitrogen isotopically labeled, also gave comparable data to the lightest version (KM = 1.79 ± 0.20 mM, specific activity = 440.5 ± 17.9 nmol·min−1·mg−1, P = 0.0974; Fig. 2A, B, Table 1). In contrast, [2,3,3-D]l-serine (3), with deuterium labels at C2 and C3, showed a substantial decrease in rate and the highest KM of all substrates tested (KM = 4.09 ± 0.32 mM and specific activity = 176.9 ± 13.0 nmol·min−1·mg−1, P = 0.0093; Fig. 2A, B, Table 1).
Fig. 2.
Rates of SpSPT- and scSPT-catalyzed reactions in the presence of l-serine isotopologues 1–5. A: Initial rate of SpSPT-catalyzed reaction of l-serine isotopologues at varying concentrations with 250 µM palmitoyl-CoA. 1, l-serine; 2, [3,3-D]l-serine; 3, [2,3,3-D]l-serine; 4, [2-13C]l-serine; 5, [1,2,3-13C, 2-15N]l-serine. Rate of purified SpSPT (B) and rate of purified scSPT (C). Both B and C were carried out with 10 mM l-serine substrates 1–5 and 250 µM palmitoyl-CoA.
TABLE 1.
Kinetic parameters (KM and rates for spSPT and scSPT) versus different serine isotopologues
| SpSPT | scSPT | ||
| KM (mM) | Rate (nmol·min−1·mg−1) | Rate (nmol·min−1·mg−1) | |
| 1: l-serine | 1.56 ± 0.10 | 386.3 ± 39.7 | 11.6 ± 1.3 |
| 2: [3,3-D]l-serine | 2.72 ± 0.29 | 396.7 ± 13.1 | 12.2 ± 0.8 |
| 3: [2,3,3-D]l-serine | 4.09 ± 0.32 | 176.9 ± 13.0 | 10.4 ± 2.2 |
| 4: [2-13C]l-serine | 3.64 ± 0.61 | 410.6 ± 25.5 | 11.3 ± 0.8 |
| 5: [1,2,3-13C, 2-15N]l-serine | 1.79 ± 0.20 | 440.5 ± 17.9 | 12.9 ± 1.9 |
We also measured the accurate masses of the KDS products derived from each l-serine substrate (Table 2, supplemental Fig. S1). Unlabeled l-serine gives KDS with m/z 300.2905, which is in very good agreement with the empirical formula for the [M+H]+ ion (C18H37NO2, expected m/z [M+H]+ = 300.2897). The heaviest l-serine substrate, with four isotope labels {[1,2,3-13C, 2-15N]l-serine, (5)}, gives a KDS m/z 303.2953 (13C2C16H3715NO2, expected m/z [M+H]+ = 303.2935), which has a nominal Δmass = +3 compared with the lightest substrate and is consistent with the loss CO2 as is expected during the SPT-catalyzed reaction (Fig. 1A). The [3,3-D]l-serine (2) gives a KDS product with m/z 302.3043 (expected m/z [M+H]+ for C18H35D2NO2 = 302.3023) that confirms retention of both labels. Similarly, the singularly labeled [2-13C]l-serine (4) gave a KDS with m/z 301.2944 (expected m/z [M+H]+ for 13C1C17H37NO2 = 301.2931), again consistent with retention of the label. Importantly, the [2,3,3-D]l-serine (3)-derived KDS product gave a m/z 302.3042 (expected m/z [M+H]+ for C18H35D2NO2 = 302.3023), which clearly indicates the loss of a deuterium during the SPT-catalyzed reaction (Fig. 1A).
TABLE 2.
Expected (assuming retention of all isotope labels) and observed masses of 3-KDS products
| Substrate | Expected mass of 3-KDS product [M+H]+ | Observed mass of 3-KDS product [M+H]+ | Molecular formula of product | |
| SpSPT | scSPT | |||
| 1: l-serine | 300.2897 | 300.2905 | 300.2928 | C18H37NO2 |
| 2: [3,3-D]l-serine | 302.3023 | 302.3044 | 302.3045 | C18H35D2NO2 |
| 3: [2,3,3-D]l-serine | 302.3023 | 302.3042 | 302.3048 | C18H34D3NO2 |
| 4: [2-13C]l-serine* | 301.2931 | 301.2944 | 301.2955 | 13C1C17H37NO2 |
| 5: [1,2,3-13C, 2-15N]l-serine# | 303.2935 | 303.2932 | 303.2955 | 13C2C16H3715N1O2 |
*Mass of KDS product derived from [2-13C]l-serine (4) accounts for loss of α-carbon deuterium.
Mass of KDS product derived from [1,2,3-13C, 2-15N]l-serine (5) accounts for loss of 13C isotope as a result of decarboxylation during KDS formation.
Unlike the bacterial SPT, it is technically difficult to isolate the multisubunit, ER-membrane-bound human SPT complex (15). However, we have been able to purify milligram quantities of recombinant “fused” human single-chain SPT (formed by linking the subunits LCB2-ssSPTa-LCB1) from a yeast expression system (Somashekarappa et al., unpublished observations) (14, 17, 38, 40). This purified scSPT is catalytically active and able to support growth of yeast cells where endogenous SPT activity has been knocked out (38). We used the same l-serine substrate panel (1–5) to measure the rate of the SPT-catalyzed reaction for each substrate, but, unfortunately, it was not possible to measure the KM for scSPT using the convenient DTNB assay because of the difficulty in detecting activity at low l-serine concentrations—purified scSPT is ∼33-fold less active than its purified bacterial SpSPT counterpart (Fig. 2B, C, Table 1). However, the rate for each l-serine substrate 1–5 could be determined by having the amino acid substrate at a final concentration of 10 mM in the reaction. The rate with the light l-serine substrate 1 was 1161.7 ± 125.2 pmol·min−1·mg−1 (Fig. 2C, Table 1). Using a 3H L-serine radiolabeled assay, we previously reported the activity of scSPT in yeast microsomes (∼250 pmol·min−1·mg−1) and CHO-LyB cells (∼1,500 pmol·min−1·mg−1) (14), as well as HEK293 microsomal preparations (∼1,500 pmol·min−1·mg−1) (17). This shows that the purified scSPT is highly active upon removal from the microsomal environment. Then, when we measured the rates (12.2 ± 0.8, 10.4 ± 2.2, 11.3 ± 0.8, and 12.9 ± 1.9 nmol·min−1·mg−1) using each of the “heavy” l-serine substrates 2–5, respectively, we observed no significant differences (P = 0.5524, 0.4621, 0.7185, and 0.3863, respectively) when compared with l-serine 1. The MS analysis of the scSPT KDS products (supplemental Fig. S1) verified that the human enzyme gave the same incorporation patterns as that observed for the bacterial SPT, including the loss of a deuterium with the [2,3,3-D]l-serine substrate 3 (Table 2).
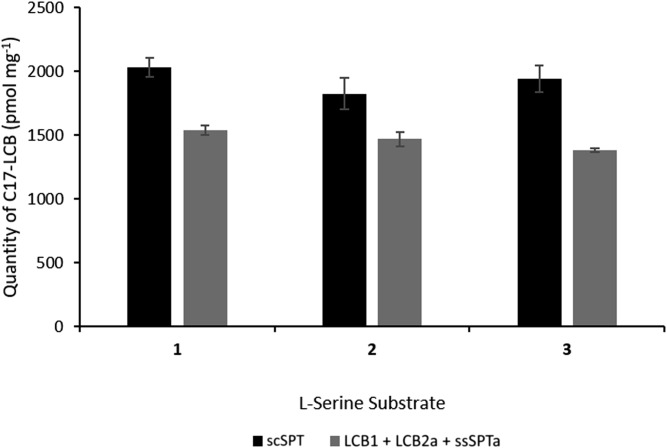
To confirm that the observed kinetic data for the single-chain SPT were not a result of the enzyme being removed from its membrane-bound environment or due to the constraints imposed by the fusion strategy, we also carried out LCB analysis of yeast microsomes containing the overexpressed three-subunit, single-chain scSPT form and the human SPT complex produced by independent coexpression of the three subunits LCB1, LCB2, and ssSPTa. This analysis was carried out over a 10-min period and measured the levels of C17 LCB production using a C15-CoA substrate and l-serine isotopologues 1, 2, and 3 (l-serine, [3,3-D]l-serine, and [2,3,3-D]l-serine, respectively). The use of this C15 substrate ensures that the LCB production is derived from a de novo, SPT-catalyzed in vitro reaction. The LCBs were detected using fluorescent LC derivatization (Fig. 3) (38). Over this time course, we found that the levels of C17 LCBs produced in microsomal preparations from the scSPT (2,031.6 ± 76.1, 1,825.9 ± 125.0, and 1,943.5 ± 104.0 pmol mg−1) or the coexpressed subunits (1,538.1 ± 34.9, 1,468.3 ± 58.9, and 1,381.2 ± 14.2 pmol mg−1) with l-serine (1), [3,3-D]l-serine (2) and [2,3,3-D]l-serine (3), respectively, were closely matched. The coexpressed SPT complex displayed ∼71–80% activity when compared with the fusion scSPT, but, importantly, the rates were not different when comparing each of l-serine (1), [3,3-D]l-serine (2), and [2,3,3-D]l-serine (3) (Fig. 3). These data confirm that neither the fusing together of the three SPT subunits nor the extraction from the microsomal membrane environment is detrimental to the activity of SPT complex.
Fig. 3.
Quantification of C17-LCBs produced by yeast membranes. The membranes were prepared from cells expressing scSPT (black bars) or coexpressing LCB1 + LCB2a + ssSPTa (gray bars). Yeast membranes were incubated with l-serine isotopologues 1, 2, and 3, along with 100 μM pentadecanoyl coenzyme-A (C15-CoA) to generate the C17-LCBs. The products were derivatized with AccQ-Fluor reagent and analyzed by HPLC and fluorescent detection.
KIE of isotopically labeled l-serine on human and bacterial SPT
From the primary rate data for both enzymes, we extracted KH/KIsotope values with each of the four isotope-labeled substrates according to Cleland (32, 33). These KIE values are summarized in Table 3. Interestingly, with human scSPT, we did not observe any significant KIE with KH/KD values ranging from 0.95 ± 0.12 for [3,3-D]l-serine (2), 1.11 ± 0.23 ([2,3,3-D]l-serine (3), 1.03 ± 0.14 ([2-13C]l-serine (4), and 0.90 ± 0.18 ([1,2,3-13C, 2-15N]l-serine (5), when compared with l-serine (1). In contrast, the bacterial SpSPT displayed different kinetics to the human enzyme. Although no significant effect on the reaction rate was observed for labeled l-serine substrates [3,3-D]l-serine (2), [2-13C]l-serine (4), and [1,2,3-13C, 2-15N]l-serine (5), with values of 0.97 ± 0.11, 0.94 ± 0.12, and 0.87 ± 0.11, respectively, we did observe a KH/KIsotope isotope value of 2.19 ± 0.13 for the [2,3,3-D]l-serine substrate (3), which has a deuterium label at C2 (Table 3).
TABLE 3.
KIE values (KH/KIsotope) determined for the reactions of purified SpSPT and ScSPT against isotopically labeled l-serine
| Substrate | KH/KIsotope | |
| SpSPT | scSPT | |
| 2: [3,3-D]l-serine | 0.97 ± 0.11 | 0.95 ± 0.12 |
| 3: [2,3,3-D]l-serine | 2.18 ± 0.13 | 1.11 ± 0.23 |
| 4: [2-13C]l-serine | 0.94 ± 0.12 | 1.03 ± 0.14 |
| 5: [1,2,3-13C, 2-15N]l-serine | 0.88 ± 0.11 | 0.90 ± 0.18 |
DISCUSSION
Isotopically labeled amino acids have proved useful in studies of SL biosynthesis in various species and cell lines (34, 41–44). However, it is important to recognize that the type and position of the label can significantly influence the underlying kinetics of the pathway being investigated. We used five different isotopically labeled versions of unlabeled light l-serine (1) and a range of heavy l-serine substrates with deuterium, 13C, and 15N labels in specific positions ([3,3-D]l-serine, [2,3,3-D]l-serine, [2-13C]l-serine, and [1,2,3-13C, 2-15N]l-serine (2–5) (Fig. 1B), in order to observe what effect, if any, the type and position of these labels have on the kinetics of the enzyme-catalyzed reaction. We used purified bacterial SpSPT, whose mechanism, kinetics, and X-ray structure have been well characterized (5, 6, 23, 24),d and compared this cytoplasmic homodimer with the more complex, membrane-bound SPT from humans. For this, we fused the subunits of human SPT together to form recombinant scSPT, allowing us to control the stoichiometry of the LCB1:ssSPTa:LCB2a subunits as 1:1:1, although the exact natural ratio of the subunits within any eukaryotic SPT complex is still not known.
With SpSPT, we observed no noticeable effect on the rate with labeled l-serine substrates [3,3-D]l-serine, [2-13C]l-serine, and [1,2,3-13C, 2-15N]l-serine (2, 4, and 5), which was expected because none of the isotopes were in a position where they would influence catalysis based on the proposed SPT mechanism (Fig. 1A). However, in the presence of [2,3,3-D]l-serine (3), the rate of the reaction was decreased by ∼50%. We derived a KIE (KH/KD) of 2.18 ± 0.13 for this reaction using this substrate. When compared with the reaction in the presence of [3,3-D]l-serine (2), for which we derived no discernible KIE, it can clearly be seen that the presence of a deuteron at the C-α position (C2) of the amino acid affects the rate of the reaction. This demonstrates that this deprotonation step is partially rate-determining in the SpSPT-catalyzed reaction. In contrast to the bacterial homolog, the rates of the human scSPT-catalyzed reaction were unaffected, regardless of the type or position of the isotope in the l-serine substrate.
We confirmed that the absence of a KIE with human scSPT was not an artifact of our fusion strategy nor was it due to having removed the enzyme from its native membrane environment. We did this by measuring in vitro SPT activity in microsomes prepared from yeast lacking endogenous SPT and expressing either the scSPT or the three independent subunits (LCB1, LCB2a, and ssSPTa) of human SPT. Using C15 acyl-CoA as substrate, de novo SPT activity was measured by quantitating the C17 LCBs because yeast does not produce C15-CoA and C17 LCBs. Our data show no difference in the levels of C17 LCBs, regardless of whether l-serine (1), [3,3-D]l-serine (2), or [2,3,3-D]l-serine (3) was used with either of the microsomal SPTs, validating the results obtained with purified human scSPT.
Examination of the proposed SPT reaction mechanism and comparison with those put forward for the other AOS family members provides a rationale for the observed KIEs (Fig. 1A). Formation of the substrate quinonoid requires deprotonation by an active-site base at C2, a process that is initiated by binding of the second substrate, palmitoyl-CoA (25). As such, it is not surprising that replacement of the α-proton with a deuterium {with [2,3,3-D]l-serine (3)} would alter the rate of the reaction. In SpSPT, from the KIE data, it is clear that this deprotonation is at least a partially, if not completely, rate-determining step. However, it is surprising that this is not the case in human SPT. As well as being membrane-bound, the other major difference between the SPTs from prokaryotes and eukaryotes is the influence of a set of ssSPT isoforms (ssSPTa/b in mammals and Tsc3p in yeast). These small subunits have been shown to not only increase the rate of the mammalian SPT-catalyzed reaction by ∼100-fold, but also control the acyl-CoA substrate specificity of the enzyme with the ssSPTa subunit conferring C16–CoA specificity and ssSPTb conferring both C16 and C18 acyl-CoA chain selectivity (13, 14, 45). We speculate whether the presence of the ssSPTa subunit in our fusion SPT construct is responsible for changing the rate-limiting step of the reaction. However, an atomic-level structure is needed to confirm that this is the case. With regards to substrates [3,3-D]l-serine (2), [2-13C]l-serine (3), and [1,2,3-13C, 2-15N]l-serine (5), none of these compounds have deuterium labels in bond-making or -breaking positions, so no KIE would be expected. The effect, if any, of the 15N and 13C labels is either low or not observed, because KIEs for 15N and 13C are not as pronounced, due to the smaller change in mass between 14N-15N and 12C-13C (44, 45). Although it might be argued that the fusion SPT construct does not behave like native SPT, several lines of evidence indicate that it does. For example, changing a single amino acid in the ssSPTa within the scSPT shifts the acyl-CoA substrate preference analogously to what is observed for the heterotrimeric SPT (14). Similarly, introducing the HSAN1 SPTLC1 C133W mutation into the scSPT confers promiscuity for the amino acid substrate (38). Finally, the fusion SPT is regulated by the ORMDLs similarly to the heterotrimeric SPT (17, 19).
Isotopically labeled substrates have been used to investigate the mechanism of other AOS family members, such as E. coli KBL, in which R/S [2-3H, 2-14C]-labeled glycine and acetyl-CoA were used to show that the enzyme selectively removes the pro-R proton at C-α (46). Ferreira and colleagues have used deuterated glycine (at C-α) to investigate the Rhodococcus capsulata ALAS-catalyzed reaction of glycine with succinyl-CoA (47, 48). In these reactions, a KIE of 1.2 (KH/KD) was determined, indicating that in ALAS abstraction of the C-α proton was not rate-limiting. Similarly with the AONS from Bacillus subtilis, Ploux and Marquet used deuterium-labeled C-α l-alanine to calculate a KIE (KH/KD) of 1.3 for the AONS-catalyzed reaction of l-alanine with pimeloyl-CoA (49). Both the KIEs for AONS and ALAS are noticeably lower than for SPT, indicating that the SPT mechanism is subtly different when compared with other members of the AOS family.
In a recent complementary study of the yeast SPT, Hannun and colleagues used tandem mass spectrometry to observe SPT-catalyzed KDS formation and reduction to dihydrosphingosine (presumably by the endogenous KDS-reductase) in yeast microsomes in which the SPT subunits (LCB1, LCB2, and small subunit Tsc3p) were not overexpressed but present at endogenous levels (34). Interestingly, they did not observe an effect on the rate of the S. cerevisiae SPT-catalyzed reaction for either [3,3-D]l-serine (2) or [2,3,3-D]l-serine (3), in agreement with the data we obtained for human scSPT. Because these assays were conducted in a native microsomal environment, we assume that Tsc3p, the yeast small subunit, would have been present. Taken together, this suggests that the respective small subunits, Tsc3p in yeast SPT and ssSPTa/b in human SPT, could well be responsible for the difference in kinetics we observe between the bacterial and eukaryotic SPTs. Another intriguing aspect of the SPT complex is the impact of HSAN1 disease-causing mutations in SPTLC1 and SPTLC2 that increase the basal activity of the SPTLC1/SPTLC2 heterodimer (14, 50, 51). Based on these observations, we have argued that the penalty for mutationally increasing SPT activity of the heterodimer is loss of amino-acid substrate specificity and that a major role of the ssSPTs is to increase enzymatic activity without compromising amino-acid selectivity. This is consistent with the possibility that deprotonation is rate-limiting for the heterodimer and that the HSAN1 mutations lead to a change in the rate-limiting step enhanced deprotonation of l-serine, as well as of alanine and glycine. Indeed, we have reported that the SPTLC1 C133W HSAN1 mutation does not affect the binding affinity of l-alanine, but rather the rate of condensation of l-alanine with palmitoyl-CoA (38).
In conclusion, the use of labeled serine derivatives is a powerful tool for studying SL biosynthesis in various organisms (31, 41, 42, 52). Our studies with the human enzyme [and by others with the yeast isoform (34)] validate [2,3,3-D]l-serine (3) as a useful tool. However, using purified bacterial SPT, we have observed that there is a kinetic penalty for the use of a deuterium at Cα. In addition, because the label is lost, regardless of the SPT isoform, we conclude that there is no practical use in SPT assays for an l-serine substrate with a deuterium label on C2. Although bacterial SpSPT and human scSPT catalyze the formation of the same KDS product, there are clearly different kinetics at play, and these appear to be dependent on the presence of the small subunit. This also demonstrates that, without first investigating the effects of a label on the reaction, one could possibly pay a heavy kinetic price, as was observed with SpSPT (45). Where a label is required, we recommend that substrates containing either 13C or 15N labels be explored, because if these atoms are involved in rate-determining steps, the kinetic penalty is less.
Acknowledgments
Mass spectrometry analysis was acquired on an instrument funded by Engineering and Physical Sciences Research Council Grant EP/K039717/1 (to the University of Edinburgh.
Footnotes
Abbreviations:
- 3-KDS
- 3-ketodihydrosphingosine
- AOS
- α-oxoamine synthase
- DDM
- n-dodecyl β-d-maltoside
- HSAN1
- hereditary sensory neuropathy type 1
- KIE
- kinetic isotope effect
- LCB
- long-chain base
- ORM
- orosomucoid-like protein
- PLP
- pyridoxal 5′-phosphate
- scSPT
- single-chain serine palmitoyltransferase
- SL
- sphingolipid
- SPT
- serine palmitoyltransferase
- spSPT
- Sphingomonas paucimobilis serine palmitoyltransferase
- ssSPT
- small subunit of serine palmitoyltransferase
This work was supported by Biotechnology and Biological Sciences Research Council Grant BB/M003493/1 (to D.J.C. and J.H.N.) and Uniformed Services University of the Health Sciences Collaborative Health Initiative Research Program Grant 70-3155 (to T.M.D.).
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Harrison P. J., Dunn T. M., and Campopiano D. J.. 2018. Sphingolipid biosynthesis in man and microbes. Nat. Prod. Rep. 35: 921–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Merrill A. H. 2011. Sphingolipid and glycosphingolipid metabolic pathways in the era of sphingolipidomics. Chem. Rev. 111: 6387–6422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hannun Y. A., and Obeid L. M.. 2018. Sphingolipids and their metabolism in physiology and disease. Nat. Rev. Mol. Cell Biol. 19: 175–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lowther J., Naismith J. H., Dunn T. M., and Campopiano D. J.. 2012. Structural, mechanistic and regulatory studies of serine palmitoyltransferase. Biochem. Soc. Trans. 40: 547–554. [DOI] [PubMed] [Google Scholar]
- 5.Yard B. A., Carter L. G., Johnson K. A., Overton I. M., Dorward M., Liu H., McMahon S. A., Oke M., Puech D., Barton G. J., et al. 2007. The structure of serine palmitoyltransferase; gateway to sphingolipid biosynthesis. J. Mol. Biol. 370: 870–886. [DOI] [PubMed] [Google Scholar]
- 6.Raman M. C. C., Johnson K. A., Yard B. A., Lowther J., Carter L. G., Naismith J. H., and Campopiano D. J.. 2009. The external aldimine form of serine palmitoyltransferase: structural, kinetic, and spectroscopic analysis of the wild-type enzyme and HSAN1 mutant mimics. J. Biol. Chem. 284: 17328–17339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pinto W. J., Wells G. W., and Lester R. L.. 1992. Characterization of enzymatic synthesis of sphingolipid long-chain bases in Saccharomyces cerevisiae: mutant strains exhibiting long-chain-base auxotrophy are deficient in serine palmitoyltransferase activity. J. Bacteriol. 174: 2575–2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao C., Beeler T., and Dunn T.. 1994. Suppressors of the Ca(2+)-sensitive yeast mutant (csg2) identify genes involved in sphingolipid biosynthesis. Cloning and characterization of SCS1, a gene required for serine palmitoyltransferase activity. J. Biol. Chem. 269: 21480–21488. [PubMed] [Google Scholar]
- 9.Nagiec M. M., Lester R. L., and Dickson R. C.. 1996. Sphingolipid synthesis: Identification and characterization of mammalian cDNAs encoding the Lcb2 subunit of serine palmitoyltransferase. Gene. 177: 237–241. [DOI] [PubMed] [Google Scholar]
- 10.Hanada K., Hara T., Nishijima M., Kuge O., Dickson R. C., and Nagiec M. M.. 1997. A mammalian homolog of the yeast LCB1 encodes a component of serine palmitoyltransferase, the enzyme catalyzing the first step in sphingolipid synthesis. J. Biol. Chem. 272: 32108–32114. [DOI] [PubMed] [Google Scholar]
- 11.Weiss B., and Stoffel W.. 1997. Human and murine serine-palmitoyl-CoA transferase. Eur. J. Biochem. 249: 239–247. [DOI] [PubMed] [Google Scholar]
- 12.Gable K., Slife H., Bacikova D., Monaghan E., and Dunn T. M.. 2000. Tsc3p is an 80-amino acid protein associated with serine palmitoyltransferase and required for optimal enzyme activity. J. Biol. Chem. 275: 7597–7603. [DOI] [PubMed] [Google Scholar]
- 13.Han G., Gupta S. D., Gable K., Niranjanakumari S., Moitra P., Eichler F., Brown R. H., Harmon J. M., and Dunn T. M.. 2009. Identification of small subunits of mammalian serine palmitoyltransferase that confer distinct acyl-CoA substrate specificities. Proc. Natl. Acad. Sci. USA. 106: 8186–8191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harmon J. M., Bacikova D., Gable K., Gupta S. D., Han G., Sengupta N., Somashekarappa N., and Dunn T. M.. 2013. Topological and functional characterization of the ssSPTs, small activating subunits of serine palmitoyltransferase. J. Biol. Chem. 288: 10144–10153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Breslow D. K., Collins S. R., Bodenmiller B., Aebersold R., Simons K., Shevchenko A., Ejsing C. S., and Weissman J. S.. 2010. Orm family proteins mediate sphingolipid homeostasis. Nature. 463: 1048–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han S., Lone M. A., Schneiter R., and Chang A.. 2010. Orm1 and Orm2 are conserved endoplasmic reticulum membrane proteins regulating lipid homeostasis and protein quality control. Proc. Natl. Acad. Sci. USA. 107: 5851–5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta S. D., Gable K., Alexaki A., Chandris P., Proia R. L., Dunn T. M., and Harmon J. M.. 2015. expression of the ORMDLS, modulators of serine palmitoyltransferase, Is regulated by sphingolipids in mammalian cells. J. Biol. Chem. 290: 90–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis D., Kannan M., and Wattenberg B.. 2018. Orm/ORMDL proteins: gate guardians and master regulators. Adv. Biol. Regul. 70: 3–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis D. L., Gable K., Suemitsu J., Dunn T. M., and Wattenberg B. W.. 2019. The ORMDL/Orm-serine palmitoyltransferase (SPT) complex is directly regulated by ceramide: reconstitution of SPT regulation in isolated membranes. J. Biol. Chem. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunn T. M., Tifft C. J., and Proia R. L.. 2019. A perilous path: the inborn errors of sphingolipid metabolism. J. Lipid Res. 60: 475–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Penno A., Reilly M. M., Houlden H., Laurá M., Rentsch K., Niederkofler V., Stoeckli E. T., Nicholson G., Eichler F., Brown R. H., et al. 2010. Hereditary sensory neuropathy type 1 is caused by the accumulation of two neurotoxic sphingolipids. J. Biol. Chem. 285: 11178–11187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paulenda T., and Draber P.. 2016. The role of ORMDL proteins, guardians of cellular sphingolipids, in asthma. Allergy. 71: 918–930. [DOI] [PubMed] [Google Scholar]
- 23.Beattie A. E., Clarke D. J., Wadsworth J. M., Lowther J., Sin H-L., and Campopiano D. J.. 2013. Reconstitution of the pyridoxal 5′-phosphate (PLP) dependent enzyme serine palmitoyltransferase (SPT) with pyridoxal reveals a crucial role for the phosphate during catalysis. Chem. Commun. (Camb.). 49: 7058–7060. [DOI] [PubMed] [Google Scholar]
- 24.Lowther J., Charmier G., Raman M. C., Ikushiro H., Hayashi H., and Campopiano D. J.. 2011. Role of a conserved arginine residue during catalysis in serine palmitoyltransferase. FEBS Lett. 585: 1729–1734. [DOI] [PubMed] [Google Scholar]
- 25.Ikushiro H., Fujii S., Shiraiwa Y., and Hayashi H.. 2008. Acceleration of the substrate Cα deprotonation by an analogue of the second substrate palmitoyl-CoA in serine palmitoyltransferase. J. Biol. Chem. 283: 7542–7553. [DOI] [PubMed] [Google Scholar]
- 26.Ikushiro H., Hayashi H., and Kagamiyama H.. 2001. A water-soluble homodimeric serine palmitoyltransferase from Sphingomonas paucimobilis EY2395T strain: purification, characterization, cloning, and overproduction. J. Biol. Chem. 276: 18249–18256. [DOI] [PubMed] [Google Scholar]
- 27.Ikushiro H., Hayashi H., and Kagamiyama H.. 2004. Reactions of serine palmitoyltransferase with serine and molecular mechanisms of the actions of serine derivatives as inhibitors. Biochemistry. 43: 1082–1092. [DOI] [PubMed] [Google Scholar]
- 28.Mahmud T. 2007. Isotope tracer investigations of natural products biosynthesis: the discovery of novel metabolic pathways. J. Labelled Compounds Radiopharmaceuticals. 50: 1039–1051. [Google Scholar]
- 29.Rinkel J., and Dickschat J. S.. 2015. Recent highlights in biosynthesis research using stable isotopes. Beilstein J. Org. Chem. 11: 2493–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clendinen C. S., Stupp G. S., Ajredini R., Lee-McMullen B., Beecher C., and Edison A. S.. 2015. An overview of methods using 13C for improved compound identification in metabolomics and natural products. Front. Plant Sci. 6: 611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ziv C., Malitsky S., Othman A., Ben-Dor S., Wei Y., Zheng S., Aharoni A., Hornemann T., and Vardi A.. 2016. Viral serine palmitoyltransferase induces metabolic switch in sphingolipid biosynthesis and is required for infection of a marine alga. Proc. Natl. Acad. Sci. USA. 113: E1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cleland W. W. 2003. The use of isotope effects to determine enzyme mechanisms. J. Biol. Chem. 278: 51975–51984. [DOI] [PubMed] [Google Scholar]
- 33.Cleland W. W. 1987. The use of isotope effects in the detailed analysis of catalytic mechanisms of enzymes. Bioorg. Chem. 15: 283–302. [Google Scholar]
- 34.Ren J., Snider J., Airola M. V., Zhong A., Rana N. A., Obeid L. M., and Hannun Y. A.. 2018. Quantification of 3-ketodihydrosphingosine using HPLC-ESI-MS/MS to study SPT activity in yeast Saccharomyces cerevisiae. J. Lipid Res. 59: 162–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kohen A., Roston D., Stojković V., and Wang Z.. 2011. Kinetic isotope effects in enzymes. In Encyclopedia of Analytical Chemistry: Applications, Theory, and Instrumentation. R. A. Meyers, editor. John Wiley & Sons, Ltd; Chichester, UK. 77–99. [Google Scholar]
- 36.Wadsworth J. M., Clarke D. J., McMahon S. A., Lowther J. P., Beattie A. E., Langridge-Smith P. R. R., Broughton H. B., Dunn T. M., Naismith J. H., and Campopiano D. J.. 2013. The chemical basis of serine palmitoyltransferase inhibition by myriocin. J. Am. Chem. Soc. 135: 14276–14285. [DOI] [PubMed] [Google Scholar]
- 37.Bradford M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254. [DOI] [PubMed] [Google Scholar]
- 38.Gable K., Gupta S. D., Han G., Niranjanakumari S., Harmon J. M., and Dunn T. M.. 2010. A disease-causing mutation in the active site of serine palmitoyltransferase causes catalytic promiscuity. J. Biol. Chem. 285: 22846–22852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.QuickCalcs. GraphPad, San Diego, CA. Accessed November 28, 2017, at https://www.graphpad.com/quickcalcs/.
- 40.Han G., Gable K., Yan L., Allen M. J., Wilson W. H., Moitra P., Harmon J. M., and Dunn T. M.. 2006. Expression of a novel marine viral single-chain serine palmitoyltransferase and construction of yeast and mammalian single-chain chimera. J. Biol. Chem. 281: 39935–39942. [DOI] [PubMed] [Google Scholar]
- 41.Ernst D., Murphy S. M., Sathiyanadan K., Wei Y., Othman A., Laurá M., Liu Y-T., Penno A., Blake J., Donaghy M., et al. 2015. Novel HSAN1 mutation in serine palmitoyltransferase resides at a putative phosphorylation site that is involved in regulating substrate specificity. NeuroMol. Med. 17: 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hannich J. T., Mellal D., Feng S., Zumbuehl A., and Riezman H.. 2017. Structure and conserved function of iso-branched sphingoid bases from the nematode Caenorhabditis elegans. Chem. Sci. 8: 3676–3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Merrill A. H. 1983. Characterization of serine palmitoyltransferase activity in Chinese hamster ovary cells. Biochim. Biophys. Acta. 754: 284–291. [DOI] [PubMed] [Google Scholar]
- 44.Sprinson D. B., and Coulon A.. 1954. The precursors of sphingosine in brain tissue. J. Biol. Chem. 207: 585–592. [PubMed] [Google Scholar]
- 45.Zhao L., Spassieva S., Gable K., Gupta S. D., Shi L-Y., Wang J., Bielawski J., Hicks W. L., Krebs M. P., Naggert J., et al. 2015. Elevation of 20-carbon long chain bases due to a mutation in serine palmitoyltransferase small subunit b results in neurodegeneration. Proc. Natl. Acad. Sci. USA. 112: 12962–12967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bashir Q., Rashid N., and Akhtar M.. 2006. Mechanism and substrate stereochemistry of 2-amino-3-oxobutyrate CoA ligase: implications for 5-aminolevulinate synthase and related enzymes. Chem. Commun. (Camb.). 5065–5067. [DOI] [PubMed] [Google Scholar]
- 47.Gong J., Hunter G. A., and Ferreira G. C.. 1998. Aspartate-279 in aminolevulinate synthase affects enzyme catalysis through enhancing the function of the pyridoxal 5′-phosphate cofactor. Biochemistry. 37: 3509–3517. [DOI] [PubMed] [Google Scholar]
- 48.Hunter G. A., Zhang J., and Ferreira G. C.. 2007. Transient kinetic studies support refinements to the chemical and kinetic mechanisms of aminolevulinate synthase. J. Biol. Chem. 282: 23025–23035. [DOI] [PubMed] [Google Scholar]
- 49.Ploux O., and Marquet A.. 1996. Mechanistic studies on the 8-amino-7-oxopelargonate synthase, a pyridoxal-5′-phosphate-dependent enzyme involved in biotin biosynthesis. Eur. J. Biochem. 236: 301–308. [DOI] [PubMed] [Google Scholar]
- 50.Beattie A. E., Gupta S. D., Frankova L., Kazlauskaite A., Harmon J. M., Dunn T. M., and Campopiano D. J.. 2013. The pyridoxal 5′-phosphate (PLP)-dependent enzyme serine palmitoyltransferase (SPT): effects of the small subunits and insights from bacterial mimics of Human hLCB2a HSAN1 mutations. BioMed Res. Int. 2013: 194371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Monaghan E., Gable K., and Dunn T.. 2002. Mutations in the Lcb2p subunit of serine palmitoyltransferase eliminate the requirement for the TSC3 gene in Saccharomyces cerevisiae. Yeast. 19: 659–670. [DOI] [PubMed] [Google Scholar]
- 52.Krisnangkura K., and Sweeley C. C.. 1976. Studies on the mechanism of 3-ketosphinganine synthetase. J. Biol. Chem. 251: 1597–1602. [PubMed] [Google Scholar]