Abstract
Endogenous synthesis of PUFAs is mediated by genes controlling fatty acid elongases 2 and 5 (ELOVL2 and ELOVL5) and by exogenous DHA intake. Associations between elongases and PUFA levels probably involve genetic variants of ELOVL and changes in DHA intake, but data about their combined effect on PUFA levels are sparse. We hypothesized that each factor would directly affect PUFAs and that interactions between haplotypes and DHA intake would influence PUFAs. We explored four levels of DHA intake in pregnant Chinese Han women and 10 SNPs in the ELOVL genes to determine associations with PUFAs in breast milk. The SNP rs3798713 and 3-SNP haplotype (rs2281591, rs12332786, and rs3798713) in ELOVL2 were associated with linoleic acid (LA) concentrations. However, carriers of the 3-SNP haplotype with higher DHA intake (second quartile: 14.58–43.15 mg/day) had higher concentrations of LA, arachidonic acid, EPA, and DHA compared with the interaction baseline. In ELOVL5, five SNPs (rs2294867, rs9357760, rs2397142, rs209512, and rs12207094) correlated with PUFA changes. Compared with those who had the 5-SNP haplotype C-A-C-G-A and low DHA intake (<14.58 mg/day), carriers with other haplotypes (A-A-C-A-A or C-A-C-A-A) and high DHA intake (≥118.82 mg/day) had increased EPA levels after adjustments for age and BMI. This study showed that maternal genetic variants in ELOVL2 and ELOVL5 were associated with PUFA levels in breast milk and that the combination of SNP haplotypes and higher DHA intake increased PUFA concentrations.
Keywords: docosahexaenoic acid, gene polymorphism, diet, fatty acid synthesis, nutrition, lactation, clinical studies, haplotype
PUFAs, especially LC-PUFAs, are essential nutrients that are associated with the development of the human brain and neurotransmitter function (1–3). In particular, the n-3 fatty acid DHA (22:6n-3), which is present in breast milk, has been identified in animal and human studies as being crucial for the development of the central nervous system during fetal life and early infancy (2, 4). Findings from epidemiological and observational studies substantiate the association between maternal seafood consumption in pregnancy and breastfeeding and improved infant neurodevelopment (5–9). This may be explained partly by the resultant increase in the early supply of LC-PUFAs, especially DHA, which accumulate in the brain during early growth spurts in children (2, 10). Pregnant women who consumed high amounts of n-3 LC-PUFAs in their diet showed positive effects on pregnancy outcomes, such as longer gestational duration and greater birth weight (11). Additionally, LC-PUFAs may increase infant growth and enhance the short- and long-term development of the offspring (11–13). A randomized controlled trial showed that supplementation with n-3 LC-PUFAs in the third trimester of pregnancy reduced the absolute risk of persistent wheeze or asthma and infections of the lower respiratory tract in offspring (14). The fetus is mainly supplied with LC-PUFAs by transfer from the maternal circulation via the placenta (15). Therefore, an adequate supply of LC-PUFAs during pregnancy is critical for fetal life onward.
Elongation of LC-PUFAs in the n-3 family is made possible by enzymes called elongases, which are encoded by the elongation of a very long-chain fatty acid (ELOVL) gene family on chromosome 6. Elongases catalyze the elongation of aliphatic carbon chains leading to the formation of LC-PUFAs (16, 17). Fatty acid elongases 2 and 5, encoded by the ELOVL2 (6p24.2) and ELOVL5 (6p12.1) genes, respectively, are involved in LC-PUFA synthesis. Plasma percentages of n-3 LC-PUFAs have been shown to be associated with SNPs in ELOVL2 (18) and ELOVL5 (19, 20), and the enzyme activities are influenced by SNPs within the ELOVL gene family after n-3 LC-PUFA supplementation (21, 22).
The mechanisms underlying the observed associations between elongases and PUFA levels are inconsistent but probably involve genetic variants within ELOVL and changes in levels of DHA intake. Consequently, we hypothesized that ELOVL polymorphisms and DHA intake would each be directly associated with PUFA concentrations. We also hypothesized that the interactions of haplotypes within ELOVL2 and ELOVL5 with DHA consumption during pregnancy may influence PUFA concentration in the breast milk of healthy lactating Chinese Han women.
MATERIALS AND METHODS
Sociodemographic and study design
We recruited 422 healthy Chinese Han pregnant women, 22–40 years of age, who registered for postpartum care at Shirentang House in Changchun from March 2012 to December 2014. Only participants with no maternal pregnancy complications were included. Other exclusion criteria included metabolic diseases (including diabetes) and communicable diseases. The sample size varied from a total of 422 participating women to 420 in analyses of ELOVL SNP versus breast-milk PUFAs as one outcome and from 422 to 370 in analyses of DHA intake versus breast-milk PUFAs as another outcome. All participants gave informed consent according to the procedures approved by the ethics committee of Jilin University. The study abided by the Declaration of Helsinki principles.
Questionnaire survey and breast-milk collection
We gave each participant a face-to-face interview and a semistructured food-frequency questionnaire. The questionnaire was used to assess the dietary intake of enrolled subjects during their pregnancy. It included specific questions about the consumption of sources containing DHA, such as freshwater fish, seafood, and canned tuna. To better understand the data of DHA intake, in the interview, we also asked participants the DHA supplement’s brand, the supplementing time, and daily doses. The investigators checked the content of DHA in supplements and calculated the daily doses of DHA of the enrolled subjects. Five kinds of PUFA dietary intakes were calculated based on Yang (23). In addition, DHA dietary intakes were calculated as milligrams per day. Twenty milliliters of breast milk were collected between 9:00 AM and 11:00 AM between the 22nd and 25th day after delivery. The first few drops of milk were discarded, and then the mature breast milk was collected. The samples were stored at −80°C (24) for approximately 1 month.
Fatty acid analysis
The levels of eight kinds of fatty acids in breast milk were determined by direct methylation (25) with subsequent analysis by gas chromatography-flame ionization detection. A GC-14B gas chromatograph (Shimadzu Corp., Kyoto, Japan) was equipped with an sp-2560 capillary column (100 m × 0.25 mm × 0.20 μm; Supelco, Bellefonte, PA). The internal standard method was used to calculate the levels of fatty acid methyl esters. Fatty acid methyl esters were prepared from milk by combining 0.2 ml fat with 2 ml methanol and benzene (4:1; v/v), 33 µl internal standard (daturic acid, C17:0), and 200 µl acetyl chloride in a 10 ml glass tube. Specific experimental steps have been described previously (26).
SNP selection and genotyping
SNPs in ELOVL2 and ELOVL5 were identified using the International HapMap Project SNP database and the NCBI database (http://www.ncbi.nlm.nih.gov/snp). Selected SNPs of the ELOVL gene cluster (rs2281591, rs12332786, rs3798713, rs3778166, rs9468304, rs2294867, rs9357760, rs2397142, rs209512, rs12207094) have been genotyped using the Sequenom MassARRAY system (BO MIAO Biological Technological Co., Beijing, China) with validated primers, and the genotyping success rate was above 96%. ELOVL2 and ELOVL5 are located on chromosome 6 (10.99–11.05 Mb and 53.28–53.34 Mb, respectively), and the selected SNPs are all intron variants. Each SNP has a minor allele frequency above 10% in the Asian population according to the SNP database of the NCBI. The milk samples were thawed at 4°C, and genomic DNA was extracted from 300 µl of the cellular layer using a DNA kit (TIANGEN, Beijing, China) according to the manufacturer’s instructions.
Statistical analysis
Normal distribution of the fatty acids was tested by the Kolmogorov-Smirnov test and distribution plots. Data were expressed as means ± SDs for normal distributed variables and as medians (25th to 75th percentiles) for skewed distribution data. The skewed measurements of γ-linolenic acid (GLA; 18:3n-6), EPA (20:5n-3), and DHA concentrations were expressed as square roots to obtain a normal distribution. Hardy-Weinberg equilibrium was tested by a Chi-square goodness-of-fit test for each SNP locus. The genotype association with PUFA concentrations was tested using SNPstats software (https://www.snpstats.net/start.htm). Linear-regression analysis was used to investigate the associations of ELOVL gene polymorphisms with levels of PUFAs. Statistical analysis was performed using SPSS version 16.0 (SPSS Inc., Chicago, IL). Haplotype analyses play an important role in genetic studies (27, 28). It is impossible to define the combination of haplotypes carried by any one individual, but all possible combinations can be computed, and techniques such as the EM algorithm incorporated in the haplo.stats package of R software (https://cran.r-project.org/src/contrib/Archive/haplo.stats) can be used to assign a probability to each haplotype pair. To explore potential effects of multi-SNP and DHA intake, the interactions between the ELOVL2/5 haplotype and DHA intake on LC-PUFA levels were performed by the general linear model using R software version 3.5.0 adjusted for confounding factors. P < 0.05 (two-tailed) was considered statistically significant.
RESULTS
Characteristics of the study subjects
The 422 healthy lactating mothers included in this study had an average age of 30.29 ± 3.40 years and mainly came from middle-income households (54.09%). Gestational age was 39.27 ± 1.00 weeks. The preconception BMI was 20.95 ± 3.33 kg/m2, which was within the normal range. A total of 31.10% of subjects had a vaginal delivery, while the rest had cesareans. A total of 54.39% of the subject breastfed exclusively, while the remaining 45.61% opted for mixed feeding. A total of 50.75% of the mothers had a university education (Table 1).
TABLE 1.
Background characteristics of the lactating mothers
| Variable | Value |
| Age (years) | 30.29 ± 3.40 |
| Gestational age (weeks) | 39.27 ± 1.00 |
| Preconception BMI (kg/m2) | 20.95 ± 3.33 |
| Gestational weight gain (kg) | 18.88 ± 6.30 |
| Feeding patterns [n (%)] | |
| Exclusive breastfeeding | 223 (54.39) |
| Mixed feeding | 187 (45.61) |
| Delivery patterns [n (%)] | |
| Vaginal | 130 (31.10) |
| Cesarean | 288 (68.90) |
| Education level [n (%)] | |
| Graduate school | 37 (9.25) |
| University | 203 (50.75) |
| High school | 140 (35.00) |
| Middle school | 17 (4.25) |
| Primary school | 3 (0.75) |
| Household income [n (%)] | |
| Low | 57 (16.67) |
| Middle | 185 (54.09) |
| High | 100 (29.24) |
Effect of DHA intake during pregnancy on the concentration of PUFAs in breast milk
We investigated the effect of DHA intake (including dietary and supplemental DHA) during pregnancy on the concentration of eight PUFAs in breast milk and sought to determine whether there was a dose-response relationship. The lactating mothers were classified into four subgroups depending on the quartiles of DHA intake (Q1: < 14.58; Q2: 14.58–43.15; Q3: 43.16–118.82; and Q4: ≥118.82 mg/day) (Table 2). However, the results showed that there was no significant difference for PUFA composition of breast milk among the four groups.
TABLE 2.
Association between DHA intake and breast-milk PUFAs by quartile of DHA intake
| PUFAs (g/100 g) | DHA intake (mg/day) | P | ||||
| Q1 (<14.58) | Q2 (14.58–43.15) | Q3 (43.16–118.82) | Q4 (≥118.82) | |||
| n-6 | LA | 0.359 ± 0.198 | 0.386 ± 0.190 | 0.397 ± 0.168 | 0.369 ± 0.207 | 0.52 |
| GLA | 0.039 (0.021–0.056) | 0.039 (0.026–0.061) | 0.042 (0.025–0.053) | 0.036 (0.022–0.053) | 0.67 | |
| DGLA | 0.052 (0.030–0.071) | 0.054 (0.037–0.071) | 0.052 (0.036–0.078) | 0.049 (0.027–0.075) | 0.64 | |
| ARA | 0.076 ± 0.039 | 0.085 ± 0.041 | 0.086 ± 0.039 | 0.076 ± 0.044 | 0.20 | |
| DTA | 0.019 (0.011–0.024) | 0.018 (0.012–0.025) | 0.019 (0.012–0.027) | 0.016 (0.010–0.024) | 0.26 | |
| n-3 | ALA | 0.143 (0.084–0.197) | 0.137 (0.096–0.185) | 0.154 (0.103–0.189) | 0.149 (0.086–0.208) | 0.87 |
| EPA | 0.007 (0.004–0.011) | 0.008 (0.005–0.012) | 0.007 (0.005–0.011) | 0.007 (0.005–0.011) | 0.56 | |
| DHA | 0.044 (0.028–0.061) | 0.049 (0.032–0.072) | 0.053 (0.033–0.068) | 0.048 (0.028–0.063) | 0.15 | |
Values are means ± SDs for normal distribution or medians (25th to 75th percentiles) for skewed distributed variables.
Frequency of SNPs
The distributions of genotype frequencies in the 422 subjects were in accordance with Hardy-Weinberg equilibrium (P > 0.05). The selected SNPs were all in introns, and the genotyping success rate was >96%. The characteristics of the detected SNPs, including their positions on chromosome 6 and their genotypes, are summarized in Table 3. Minor allele frequencies ranged from ∼10.7% to 46.8% of the population.
TABLE 3.
Characteristics of 10 polymorphisms in the ELOVL gene cluster
| Gene | SNP | Position (bp) | Alleles | Genotypea | Alleleb | Genotyping Success Rate (%) | |||
| M/m | MM | Mm | mm | M | m | ||||
| ELOVL2 | rs2281591 | 10990260 | A/G | 264 | 135 | 21 | 663 | 177 | 99.53 |
| rs12332786 | 10998735 | C/G | 236 | 160 | 24 | 632 | 208 | 99.53 | |
| rs3798713 | 11008389 | C/G | 180 | 186 | 45 | 546 | 276 | 97.39 | |
| rs3778166 | 11032931 | G/A | 131 | 206 | 79 | 468 | 364 | 98.58 | |
| rs9468304 | 11041932 | A/G | 207 | 171 | 41 | 585 | 253 | 99.29 | |
| ELOVL5 | rs2294867 | 53289156 | C/A | 156 | 186 | 66 | 498 | 318 | 96.68 |
| rs9357760 | 53325336 | A/G | 183 | 179 | 50 | 545 | 279 | 97.63 | |
| rs2397142 | 53335501 | C/G | 186 | 177 | 44 | 549 | 267 | 96.45 | |
| rs209512 | 53338779 | A/G | 114 | 217 | 87 | 445 | 391 | 99.05 | |
| rs12207094 | 53339377 | A/T | 335 | 82 | 4 | 750 | 90 | 99.76 | |
M, major allele; m, minor allele; MM, major allele homozygote; mm, minor allele homozygote; Mm, heterozygote.
Number of women carrying MM, Mm, or mm.
Number of women carrying M or m.
Association between ELOVL2 and ELOVL5 genotypes and PUFA concentrations
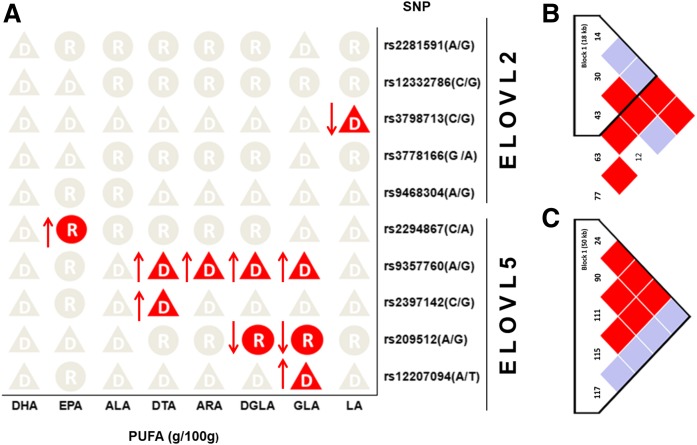
The association of three genetic models (codominant, dominant, and recessive) for each of the 10 SNPs with breast-milk PUFAs was analyzed using SNPstats software and adjusted for age, preconception BMI, and DHA intake of subjects. The best-fit genetic model of the 10 SNPs was chosen based on the Akaike information criterion and Bayesian information criterion (Fig. 1). Carriers of the minor allele of rs3798713 (P = 0.019) in the ELOVL2 gene had lower linoleic acid (LA; 18:2n-6) concentrations of breast milk than homozygous subjects for the major allele. The subjects carrying the minor allele homozygote of rs2294867 (P = 0.036) within ELOVL5 had higher EPA concentrations than those who carried the major allele. Subjects who carried the minor allele of rs9357760 in the ELOVL5 gene had higher GLA (P = 0.043), dihomo-γ-linolenic acid (DGLA; 20:3n-6) (P = 0.013), arachidonic acid (ARA; 20:4n-6) (P = 0.014), and docosatetraenoic acid (DTA; 22:4n-6) (P = 0.009) concentrations than homozygotes for the major allele. Carriers of the minor allele of rs2397142 had higher DTA levels compared with homozygotes for the major allele (P = 0.027). The subjects homozygous for the minor allele of rs209512 had lower GLA (P = 0.042) and DGLA (P = 0.039) concentrations than those carrying the major allele. A significant association was also observed between rs12207094 (P = 0.029) in ELOVL5 and the level of GLA; carriers of the minor allele had higher levels compared with homozygotes for the major allele. However, no statistically significant differences were found between the other SNPs and fatty acid levels. The full details of the analytical results are provided in supplemental Table S1.
Fig. 1.
Breast-milk PUFA levels in optimal genotype model (A) and linkage-disequilibrium block plots of SNPs in ELOVL2 (B) and ELOVL5 (C). The letter “D” in the triangle represents the dominant model (PUFA changes of Mm + mm compared with MM); the letter “R” in the circle represents the recessive model (PUFA changes of mm compared with MM + Mm). MM, major allele homozygote; mm, minor allele homozygote; Mm, heterozygote.
Interaction effect between ELOVL2 and ELOVL5 haplotypes and DHA intake on breast-milk PUFA levels
The results of the ELOVL2 and ELOVL5 haplotype frequencies can be found in supplemental Tables S2 and S3. Table 4 shows that a 3-SNP haplotype (H2: A-G-G) in ELOVL2 (rs2281591 A/G, rs12332786 C/G, and rs3798713 C/G) was associated with a decline in LA (P = 0.046), EPA (P = 0.022), and DHA (P = 0.008) concentrations in breast milk compared with carriers of the baseline haplotype (H1: A-C-C) after adjusting for age and BMI. However, we found that the interaction of haplotype H2 with the second quartile of DHA intake (Q2: 14.58–43.15 mg/day) increased the concentration of LA (P = 0.038), ARA (P = 0.025), EPA (P = 0.034), and DHA (P = 0.004) compared with carriers of haplotype H1 with low DHA intake (Q1: <14.58 mg/day) after adjusting for age and BMI.
TABLE 4.
Interaction between ELOVL2/5 haplotype and DHA intake on breast-milk PUFA level
| LA | GLA | DGLA | ARA | DTA | ALA | EPA | DHA | |||||||||
| coef | P | coef | P | coef | P | coef | P | coef | P | coef | P | coef | P | coef | P | |
| ELOVL2 | ||||||||||||||||
| Q1a | Reference | |||||||||||||||
| Q2a | −0.0243 | 0.644 | −0.0161 | 0.344 | −0.0047 | 0.567 | −0.0053 | 0.635 | −0.0012 | 0.654 | −0.0099 | 0.643 | −0.0084 | 0.333 | −0.0031 | 0.876 |
| H1b | Reference | |||||||||||||||
| H2b | −0.0723 | 0.046 | −0.0205 | 0.093 | −0.0065 | 0.253 | −0.0125 | 0.107 | −0.0022 | 0.251 | −0.0253 | 0.083 | −0.0143 | 0.022 | −0.0364 | 0.008 |
| Q1-H1c | Reference | |||||||||||||||
| Q2-H2c | 0.1107 | 0.038 | 0.0329 | 0.060 | 0.0108 | 0.192 | 0.0255 | 0.025 | 0.0040 | 0.143 | 0.0385 | 0.072 | 0.0190 | 0.034 | 0.0586 | 0.004 |
| ELOVL5 | ||||||||||||||||
| Q1a | Reference | |||||||||||||||
| Q2a | −0.0144 | 0.790 | −0.0144 | 0.406 | −0.0023 | 0.780 | 0.0005 | 0.967 | −0.0004 | 0.891 | −0.0327 | 0.128 | −0.0102 | 0.246 | −0.0193 | 0.342 |
| Q4a | −0.0450 | 0.374 | −0.0126 | 0.437 | −0.0072 | 0.345 | −0.0136 | 0.204 | −0.0024 | 0.351 | −0.0223 | 0.267 | −0.0020 | 0.812 | 0.0036 | 0.851 |
| H1b | Reference | |||||||||||||||
| H4b | −0.0546 | 0.376 | −0.0280 | 0.157 | −0.0098 | 0.295 | −0.0145 | 0.264 | −0.0040 | 0.205 | −0.0131 | 0.595 | −0.0168 | 0.095 | −0.0329 | 0.157 |
| H5b | −0.0086 | 0.886 | 0.0081 | 0.672 | 0.0031 | 0.736 | −0.0043 | 0.735 | −0.0011 | 0.721 | −0.0034 | 0.886 | −0.0106 | 0.273 | −0.0172 | 0.473 |
| Q1-H1c | Reference | |||||||||||||||
| Q2-H4c | 0.0341 | 0.699 | 0.0213 | 0.455 | 0.0030 | 0.829 | 0.0093 | 0.615 | 0.0003 | 0.953 | 0.0679 | 0.053 | 0.0072 | 0.611 | 0.0434 | 0.185 |
| Q4-H4c | 0.1994 | 0.068 | 0.0603 | 0.084 | 0.0385 | 0.020 | 0.0326 | 0.159 | 0.0046 | 0.409 | 0.0998 | 0.022 | 0.0359 | 0.042 | 0.0621 | 0.129 |
| Q2-H5c | 0.1158 | 0.238 | 0.0250 | 0.452 | 0.0318 | 0.036 | 0.0382 | 0.069 | 0.0083 | 0.099 | 0.0527 | 0.182 | 0.0424 | 0.011 | 0.0550 | 0.141 |
| Q4-H5c | −0.0068 | 0.944 | 0.0446 | 0.162 | 0.0059 | 0.694 | 0.0029 | 0.887 | 0.0020 | 0.695 | −0.0055 | 0.885 | 0.0393 | 0.015 | 0.0600 | 0.124 |
Quartile level of DHA intake. Q1: < 14.58 mg/day (reference); Q2: 14.58–43.15 mg/day; and Q4: ≥118.82 mg/day.
Type of haplotype. ELOVL2, H1: A-C-C (reference); ELOVL2, H2: A-G-G; ELOVL5, H1: C-A-C-G-A (reference); ELOVL5, H4: A-A-C-A-A; and ELOVL5, H5: C-A-C-A-A. Haplotypes are named according to their frequency.
Effect of interaction between DHA intake and haplotype on PUFA levels compared with the reference.
Compared with those who had a 5-SNP haplotype (H1: C-A-C-G-A) (rs2294867 C/A, rs9357760 A/G, rs2397142 C/G, rs209512 A/G, and rs12207094A/T) in ELOVL5 with low DHA intake (Q1), carriers with the haplotype H4: A-A-C-A-A who consumed high levels of DHA (Q4: ≥118.82 mg/day) had higher levels of DGLA, α-linolenic acid (ALA; 18:3n-3), and EPA (P = 0.020, P = 0.022, and P = 0.042, respectively) in breast milk after adjusting for age and BMI. Another interaction between the haplotype H5: C-A-C-A-A and DHA intake (Q2) increased DGLA and EPA concentrations (P = 0.036 and P = 0.011, respectively). Likewise, haplotype H5 interacting with DHA intake in Q4 increased EPA concentrations (P = 0.015) in breast milk (Table 4). Table 4 lists the significant results of the haplotype analysis for ELOVL2 and ELOVL5 (P < 0.05). Full details of related results can be found in supplemental Tables S4 and S5.
DISCUSSION
In this study, we analyzed polymorphisms in genes encoding the elongases involved in LC-PUFA synthesis in lactating mothers to disentangle their role in modifying potential nutritional advantages of LC-PUFAs in breastfeeding. Maternal genetic variants in the ELOVL gene family were associated with breast-milk levels of LC-PUFAs. Furthermore, DHA intake during pregnancy, including dietary intake and supplementation, were not associated with the concentrations of eight LC-PUFAs that were regulated by ELOVL2 and ELOVL5 gene variations. However, we showed an interaction effect between ELOVL2 and ELOVL5 haplotypes and DHA intake on breast-milk LC-PUFA levels.
We observed that rs3798713 of the ELOVL2 gene influenced the LC-PUFA concentration; carriers of the minor allele had lower LA concentrations than homozygotes for the major allele. Our previous study, however, found no significant association between ELOVL2 variants and LC-PUFAs (19), which may be due to a small sample size. Higher EPA levels have been shown in individuals containing minor alleles of rs953413 (29), rs2236212 (18), and rs3798719 (20), with another study showing no effect of the rs3734398 (21) within the ELOVL2 gene, while lower DHA levels have been observed in individuals carrying rs953413 (29), rs2236212 (18), and rs3734398 (21), with another study showing no effect of the rs3798719 (20). Inconsistencies between our study and other studies may be because the subjects in our study were all Chinese Han lactating women, and population heterogeneity among studies such as variations in SNP frequencies across the major population groups may also lead to differences in study outcomes.
Regarding the ELOVL5 gene, we found that subjects carrying the minor allele homozygote of rs2294867 had higher EPA concentrations than those who carried the major allele. GLA, DGLA, ARA, and DTA concentrations were significantly affected by rs9357760 in the ELOVL5 gene, and the subjects who carried the minor allele had higher concentrations than those who were homozygous for the major allele. Carriers of the minor allele of rs2397142 had higher DTA levels compared with major allele homozygotes. The subjects homozygous for the minor allele of rs209512 had lower GLA and DGLA concentrations than those carrying the major allele. A significant association was also observed between the minor allele of rs12207094 and a higher level of GLA. In Spanish population-based birth cohorts, a trend was observed for the minor allele of rs12207094 and higher EPA levels. Additionally, the minor alleles of rs17544159, rs9395855, and rs12207094 were associated with a high EPA-ARA ratio. This study also found that children of mothers carrying the rs17544159-C or rs12207094-T allele had higher cognition scores compared with children of mothers homozygous for the major allele (20). Our results mainly suggest that minor alleles of specific SNPs within ELOVL5 could previously be related to n-6 PUFA levels, probably due to increased transcription or to more enzyme activity of ELOVL5.
In the second half of pregnancy, DHA accumulates rapidly in neural cortex tissues (30) and retinal membrane synapses (31). DHA comes from the diet or adipose stores and is synthesized from precursor fatty acids circulating in the maternal bloodstream for uptake by the placenta and fetus. It is not clear how much DHA is released from adipose stores or is synthesized, but a remarkable association of maternal DHA intake during pregnancy with maternal circulating DHA shows that the diet may be a major source of DHA for the developing fetus (32–34). Thus, we examined the effect of maternal DHA intake during pregnancy on breast-milk LC-PUFAs. However, we did not find any difference. It may be the lower intake of DHA (<200 mg/d) (35) among most subjects (86.49%) in our study that affected the generalizability of the findings to a wider population.
Haplotypes effectively capture joint marker correlations and evolutionary history; a progressive knowledge of haplotype structures holds great promise for the use of haplotype information to understand genetic factors (36). Our results suggested an association of a haplotype (A-G-G) of ELOVL2 with lower LA, EPA, and DHA levels but not ARA levels. An analysis of the impact of DHA intake on these FA levels, however, showed no differences. Interestingly, there was a significant interaction effect between this haplotype and the second quartile of DHA intake on higher LA, EPA, DHA, and ARA levels.
Zhang et al. reported that minor allele carriers of a 3-SNP haplotype in ELOVL2 were associated with decreased DHA levels (37). However, the association between this haplotype and the plasma percentages of EPA, docosapentaenoic acid [22:5(n-3)], and DHA were not significant in the Modulation of Atherosclerosis Risk by Increasing Doses of N-3 Fatty Acids trial (21).
In the present study, carriers with the haplotype A-A-C-A-A of 5-SNPs in ELOVL5 who consumed high levels of DHA (Q4) had a higher level of DGLA, ALA, and EPA concentrations compared with those who had the haplotype C-A-C-G-A with low DHA intakes (Q1) adjusted for age and BMI. The interaction between the haplotype C-A-C-A-A and DHA intake (Q2 and Q4, but not Q3) increased EPA concentrations. The reason may be due to the lower frequency of the haplotype in Q3 compared with Q2 and Q4, resulting in lower power. It is known that DHA supplementation results in an increase in EPA concentrations (38, 39). Metabolically, increases in EPA concentration may be due to increased EPA biosynthesis or decreased EPA degradation. Increased EPA in our study is assumed to result from higher DHA intake via the slowed elongation of EPA. However, there is no significant effect of high DHA intake or a haplotype of ELOVL5 on EPA level in our study until high DHA intake interacts with a haplotype. This suggests a role for specific ELOVL haplotypes that may affect EPA elongation when DHA intake is high.
We speculate that DHA intake for mothers during pregnancy may modulate ELOVL2 and ELOVL5 gene expression through several mechanisms, such as epigenetic modifications, leading to a change in phenotypes. Consistent with our findings, observational studies have shown gene-diet interaction effects with SNPs within the FADS gene cluster and the ELOVL gene family. These interaction effects could potentially modulate the enzyme activities of desaturases and elongases following n-3 fatty acid supplementation (22).
Limitations of our study included the reliance on estimates of n-3 LC-PUFA consumption from food-frequency questionnaires and recorded DHA intake rather than using controlled doses of DHA. In addition, DHA intake in our study was relatively low across all quartiles, which may be due to the fact that participants came from inland areas and consumed less DHA-rich seafood in their daily diet. It also explained why there were no differences in breast-milk PUFA levels among the lactating mothers with different quartiles of DHA intake. Therefore, low DHA intake in the study population limits the generalizability of our results, and further studies that include a DHA-supplemented group are needed in the future. This would allow us to explore the interaction between DHA intake and ELOVL genes. Most genes contributing to quantitative phenotypes confer modest effects, requiring large sample sizes for detection with high power. Our finding of significant gene and diet interactions as a determinant of the breast-milk PUFA concentration in 422 subjects must therefore be regarded with caution. Furthermore, we anticipate that the increase in dietary variability would tend to blur the potential effects of ELOVL genes. Studies carry the risk of a false-positive finding, which can arise by chance, for genotyping errors or failure to correct for multiple testing across the number of SNPs or phenotypes tested. However, our genotyping success rate was high, and the genotypes met Hardy-Weinberg equilibrium.
In conclusion, we showed that ELOVL2 and ELOVL5 genetic variants were associated with alterations in PUFA levels in Han Chinese lactating mothers. Additionally, we observed interactions between DHA intake during pregnancy and specific haplotypes within ELOVL2 and ELOVL5 SNPs and an increased level of PUFA levels, especially EPA.
Supplementary Material
Acknowledgments
The authors thank all of the pregnant women who participated in this study. The authors also thank the data-collection team.
Footnotes
Abbreviations:
- ALA
- α-linolenic acid
- ARA
- arachidonic acid
- DGLA
- dihomo-γ-linolenic acid
- DTA
- docosatetraenoic acid
- ELOVL
- elongation of the very long-chain fatty acid
- GLA
- γ-linolenic acid
- LA
- linoleic acid
- Q
- quartile
This work was supported by the Education Department of Jilin Province Grant JJKH20170875KJ and the Department of Science and Technology of Jilin Province Grant 20180101122JC.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Lauritzen L., Sorensen L. B., Harslof L. B., Ritz C., Stark K. D., and Astrup A.. 2017. Mendelian randomization shows sex-specific associations between long-chain PUFA-related genotypes and cognitive performance in Danish schoolchildren. Am. J. Clin. Nutr. 106: 88–95. [DOI] [PubMed] [Google Scholar]
- 2.Lauritzen L., Brambilla P., Mazzocchi A., Harslof L. B., Ciappolino V., and Agostoni C.. 2016. DHA effects in brain development and function. Nutrients. 8: E6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campoy C., Escolano-Margarit M. V., Anjos T., Szajewska H., and Uauy R.. 2012. Omega 3 fatty acids on child growth, visual acuity and neurodevelopment. Br. J. Nutr. 107: S85–S106. [DOI] [PubMed] [Google Scholar]
- 4.Anderson G. J., Neuringer M., Lin D. S., and Connor W. E.. 2005. Can prenatal N-3 fatty acid deficiency be completely reversed after birth? Effects on retinal and brain biochemistry and visual function in rhesus monkeys. Pediatr. Res. 58: 865–872. [DOI] [PubMed] [Google Scholar]
- 5.Hibbeln J. R., Davis J. M., Steer C., Emmett P., Rogers I., Williams C., and Golding J.. 2007. Maternal seafood consumption in pregnancy and neurodevelopmental outcomes in childhood (ALSPAC study): an observational cohort study. Lancet. 369: 578–585. [DOI] [PubMed] [Google Scholar]
- 6.Oken E., Osterdal M. L., Gillman M. W., Knudsen V. K., Halldorsson T. I., Strom M., Bellinger D. C., Hadders-Algra M., Michaelsen K. F., and Olsen S. F.. 2008. Associations of maternal fish intake during pregnancy and breastfeeding duration with attainment of developmental milestones in early childhood: a study from the Danish National Birth Cohort. Am. J. Clin. Nutr. 88: 789–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Victora C. G., Bahl R., Barros A. J., Franca G. V., Horton S., Krasevec J., Murch S., Sankar M. J., Walker N., and Rollins N. C.. 2016. Breastfeeding in the 21st century: epidemiology, mechanisms, and lifelong effect. Lancet. 387: 475–490. [DOI] [PubMed] [Google Scholar]
- 8.Cope M. B., and Allison D. B.. 2008. Critical review of the World Health Organization’s (WHO) 2007 report on ‘evidence of the long-term effects of breastfeeding: systematic reviews and meta-analysis’ with respect to obesity. Obes. Rev. 9: 594–605. [DOI] [PubMed] [Google Scholar]
- 9.Innis S. M. 2014. Impact of maternal diet on human milk composition and neurological development of infants. Am. J. Clin. Nutr. 99: 734S–741S. [DOI] [PubMed] [Google Scholar]
- 10.Gould J. F., Smithers L. G., and Makrides M.. 2013. The effect of maternal omega-3 (n-3) LCPUFA supplementation during pregnancy on early childhood cognitive and visual development: a systematic review and meta-analysis of randomized controlled trials. Am. J. Clin. Nutr. 97: 531–544. [DOI] [PubMed] [Google Scholar]
- 11.Carlson S. E., Colombo J., Gajewski B. J., Gustafson K. M., Mundy D., Yeast J., Georgieff M. K., Markley L. A., Kerling E. H., and Shaddy D. J.. 2013. DHA supplementation and pregnancy outcomes. Am. J. Clin. Nutr. 97: 808–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olsen S. F., and Joensen H. D.. 1985. High liveborn birth weights in the Faroes: a comparison between birth weights in the Faroes and in Denmark. J. Epidemiol. Community Health. 39: 27–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leventakou V., Roumeliotaki T., Martinez D., Barros H., Brantsaeter A. L., Casas M., Charles M. A., Cordier S., Eggesbo M., van Eijsden M., et al. 2014. Fish intake during pregnancy, fetal growth, and gestational length in 19 European birth cohort studies. Am. J. Clin. Nutr. 99: 506–516. [DOI] [PubMed] [Google Scholar]
- 14.Bisgaard H., Stokholm J., Chawes B. L., Vissing N. H., Bjarnadóttir E., Schoos A. M., Wolsk H. M., Pedersen T. M., Vinding R. K., and Thorsteinsdóttir S.. 2016. Fish oil-derived fatty acids in pregnancy and wheeze and asthma in offspring. N. Engl. J. Med. 375: 2530–2539. [DOI] [PubMed] [Google Scholar]
- 15.Duttaroy A. K. 2009. Transport of fatty acids across the human placenta: a review. Prog. Lipid Res. 48: 52–61. [DOI] [PubMed] [Google Scholar]
- 16.Jakobsson A., Westerberg R., and Jacobsson A.. 2006. Fatty acid elongases in mammals: their regulation and roles in metabolism. Prog. Lipid Res. 45: 237–249. [DOI] [PubMed] [Google Scholar]
- 17.Barman M., Nilsson S., Torinsson Naluai A., Sandin A., Wold A. E., and Sandberg A. S.. 2015. Single nucleotide polymorphisms in the FADS gene cluster but not the ELOVL2 gene are associated with serum polyunsaturated fatty acid composition and development of allergy (in a Swedish birth cohort). Nutrients. 7: 10100–10115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lemaitre R. N., Tanaka T., Tang W., Manichaikul A., Foy M., Kabagambe E. K., Nettleton J. A., King I. B., Weng L. C., Bhattacharya S., et al. 2011. Genetic loci associated with plasma phospholipid n-3 fatty acids: a meta-analysis of genome-wide association studies from the CHARGE Consortium. PLoS Genet. 7: e1002193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li X., Gan Z. W., Ding Z., Wu Y. X., Chen X. Y., Tian H. M., Liu G. L., Yang Y. T., and Xie L.. 2017. Genetic variants in the ELOVL5 but not ELOVL2 gene associated with polyunsaturated fatty acids in Han Chinese breast milk. Biomed. Environ. Sci. 30: 64–67. [DOI] [PubMed] [Google Scholar]
- 20.Morales E., Bustamante M., Gonzalez J. R., Guxens M., Torrent M., Mendez M., Garcia-Esteban R., Julvez J., Forns J., Vrijheid M., et al. 2011. Genetic variants of the FADS gene cluster and ELOVL gene family, colostrums LC-PUFA levels, breastfeeding, and child cognition. PLoS One. 6: e17181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alsaleh A., Maniou Z., Lewis F. J., Hall W. L., Sanders T. A., and O’Dell S. D.. 2014. ELOVL2 gene polymorphisms are associated with increases in plasma eicosapentaenoic and docosahexaenoic acid proportions after fish oil supplement. Genes Nutr. 9: 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cormier H., Rudkowska I., Lemieux S., Couture P., Julien P., and Vohl M. C.. 2014. Effects of FADS and ELOVL polymorphisms on indexes of desaturase and elongase activities: results from a pre-post fish oil supplementation. Genes Nutr. 9: 437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang Y. X. 2009. China Food Composition. 2nd edition Beijing University Medical Press, Beijing, China. [Google Scholar]
- 24.Innis S. M., Gilley J., and Werker J.. 2001. Are human milk long-chain polyunsaturated fatty acids related to visual and neural development in breast-fed term infants? J. Pediatr. 139: 532–538. [DOI] [PubMed] [Google Scholar]
- 25.Lepage G., and Roy C. C.. 1986. Direct transesterification of all classes of lipids in a one-step reaction. J. Lipid Res. 27: 114–120. [PubMed] [Google Scholar]
- 26.Ding Z., Liu G. L., Li X., Chen X. Y., Wu Y. X., Cui C. C., Zhang X., Yang G., and Xie L.. 2016. Association of polyunsaturated fatty acids in breast milk with fatty acid desaturase gene polymorphisms among Chinese lactating mothers. Prostaglandins Leukot. Essent. Fatty Acids. 109: 66–71. [DOI] [PubMed] [Google Scholar]
- 27.Schaid D. J. 2004. Evaluating associations of haplotypes with traits. Genet. Epidemiol. 27: 348–364. [DOI] [PubMed] [Google Scholar]
- 28.Tzeng J. Y., Wang C. H., Kao J. T., and Hsiao C. K.. 2006. Regression-based association analysis with clustered haplotypes through use of genotypes. Am. J. Hum. Genet. 78: 231–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanaka T., Shen J., Abecasis G. R., Kisialiou A., Ordovas J. M., Guralnik J. M., Singleton A., Bandinelli S., Cherubini A., Arnett D., et al. 2009. Genome-wide association study of plasma polyunsaturated fatty acids in the InCHIANTI Study. PLoS Genet. 5: e1000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinez M. 1992. Tissue levels of polyunsaturated fatty acids during early human development. J. Pediatr. 120: S129–S138. [DOI] [PubMed] [Google Scholar]
- 31.Fleith M., and Clandinin M. T.. 2005. Dietary PUFA for preterm and term infants: review of clinical studies. Crit. Rev. Food Sci. Nutr. 45: 205–229. [DOI] [PubMed] [Google Scholar]
- 32.Innis S. M. 2005. Essential fatty acid transfer and fetal development. Placenta. 26 (Suppl A): S70–S75. [DOI] [PubMed] [Google Scholar]
- 33.Helland I. B., Saugstad O. D., Saarem K., Van Houwelingen A. C., Nylander G., and Drevon C. A.. 2006. Supplementation of n-3 fatty acids during pregnancy and lactation reduces maternal plasma lipid levels and provides DHA to the infants. J. Matern. Fetal Neonatal Med. 19: 397–406. [DOI] [PubMed] [Google Scholar]
- 34.Decsi T., Campoy C., and Koletzko B.. 2005. Effect of N-3 polyunsaturated fatty acid supplementation in pregnancy: the Nuheal trial. Adv. Exp. Med. Biol. 569: 109–113. [DOI] [PubMed] [Google Scholar]
- 35.Chinese Nutrition Society. 2014. Chinese Dietary Reference Intakes (DRIs) Handbook (2013). Standards Press of China, Beijing, China. [Google Scholar]
- 36.Tanaka T. 2005. International HapMap project. Nihon Rinsho 63 (Suppl. 12): 29–34. [PubMed] [Google Scholar]
- 37.Zhang J. Y., Kothapalli K. S., and Brenna J. T.. 2016. Desaturase and elongase-limiting endogenous long-chain polyunsaturated fatty acid biosynthesis. Curr. Opin. Clin. Nutr. Metab. Care. 19: 103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arterburn L. M., Eileen Bailey H., and Harry O.. 2006. Distribution, interconversion, and dose response of n-3 fatty acids in humans. Am. J. Clin. Nutr. 83: 1467S–1476S. [DOI] [PubMed] [Google Scholar]
- 39.Allaire J., Harris W. S., Vors C., Charest A., Marin J., Jackson K. H., Tchernof A., Couture P., and Lamarche B.. 2017. Supplementation with high-dose docosahexaenoic acid increases the omega-3 index more than high-dose eicosapentaenoic acid. Prostaglandins Leukot. Essent. Fatty Acids. 120: 8–14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.