Abstract
Lipid metabolism requires CoA, an essential cofactor found in multiple subcellular compartments, including the peroxisomes. In the liver, CoA levels are dynamically adjusted between the fed and fasted states. Elevated CoA levels in the fasted state are driven by increased synthesis; however, this also correlates with decreased expression of Nudix hydrolase (Nudt)7, the major CoA-degrading enzyme in the liver. Nudt7 resides in the peroxisomes, and we overexpressed this enzyme in mouse livers to determine its effect on the size and composition of the hepatic CoA pool in the fed and fasted states. Nudt7 overexpression did not change total CoA levels, but decreased the concentration of short-chain acyl-CoAs and choloyl-CoA in fasted livers, when endogenous Nudt7 activity was lowest. The effect on these acyl-CoAs correlated with a significant decrease in the hepatic bile acid content and in the rate of peroxisomal fatty acid oxidation, as estimated by targeted and untargeted metabolomics, combined with the measurement of fatty acid oxidation in intact hepatocytes. Identification of the CoA species and metabolic pathways affected by the overexpression on Nudt7 in vivo supports the conclusion that the nutritionally driven modulation of Nudt7 activity could contribute to the regulation of the peroxisomal CoA pool and peroxisomal lipid metabolism.
Keywords: beta-oxidation; bile acid metabolism; peroxisomes, metabolomics; Nudix hydrolase 7; coenzyme A
The liver is a metabolically flexible organ that plays a key role in the maintenance of whole-body lipid and glucose homeostasis (1–3). Several pathways, including gluconeogenesis, ketogenesis, and bile acid synthesis, occur primarily in this organ and rely on the availability of the essential cofactor, CoA. CoA is the major acyl group carrier in mammalian cells. Formation of acyl-CoAs allows for the delivery of activated substrates for a multitude of reactions and the modulation of metabolic pathways through allosteric regulation or posttranslational modification of histones and key metabolic enzymes (4–9). Cells contain limited and regulated amounts of CoA that are sequestered in multiple subcellular compartments, including the cytosol, peroxisomes, mitochondria, and endoplasmic reticulum (10–13). Within each subcellular compartment, free CoA and acyl-CoAs support specific pathways and reactions. These include ketogenesis, the TCA cycle, fatty acid β-oxidation in the mitochondria, fatty acid synthesis in the cytosol, autophagy in the endoplasmic reticulum, and the last steps in the bile acid biosynthetic pathway and fatty acid α- and β-oxidation in the peroxisomes (3, 12, 14).
In the liver, CoA levels change in response to the nutritional state to support the switch between glucose and fatty acid oxidation (11, 15, 16). This dynamic modulation occurs through a balance between CoA synthesis and degradation, which leads to a net increase in the concentration of hepatic CoA in the fasted state and to a net decrease in the fed state (16–18). CoA synthesis requires five enzymatic steps and is regulated through feedback inhibition of the first and rate-limiting enzyme of the pathway, pantothenate kinase, by free CoA and acyl-CoAs (19–21). The only currently known CoA-degrading enzymes are Nudix hydroxylase (Nudt)7 and Nudt19 (22, 23). Both enzymes are members of the Nudt superfamily, specifically hydrolyze CoA species to produce 3′,5′-ADP and (acyl-)phosphopantetheine, and reside in the peroxisomes, organelles that play a key role in lipid metabolism (18, 22–26). Nudt19 is the major isoform in the kidneys, where it contributes to the regulation of total CoA levels in these organs (18). Nudt7 is the major CoA-degrading enzyme in the liver. Recombinant Nudt7 hydrolyzes a broad range of CoA substrates, including free CoA, short- and medium-chain acyl-CoAs, and the CoA thioesters of bile acids (17, 18). Unlike Nudt19, Nudt7 is not inhibited by bile acids and its expression varies with fasting and feeding. In particular, Nudt7 levels decrease with fasting, correlating with the increase in CoA observed in fasted livers. While these findings have established that Nudt7 can degrade CoA in vitro, it has yet to be determined whether Nudt7 is actively involved in regulating hepatic CoA levels in vivo. Indeed, the peroxisomal CoA pool is estimated to represent only 4–10% of total liver CoA (10, 11), and it is currently unclear to what extent the activity of Nudt7 in liver peroxisomes could affect total CoA levels in the fed and fasted states. Furthermore, the localization, substrate specificity, and nutritional regulation of Nudt7 expression suggest that this enzyme may be involved in the regulation of peroxisomal pathways (27), but direct evidence is lacking.
To gain insight into the physiological role of Nudt7 and its ability to regulate liver CoA levels in vivo, we overexpressed the enzyme in the liver of C57BL/6J mice. Overexpression of Nudt7 in the fasted state, when endogenous Nudt7 activity was lowest, did not prevent the increase in total hepatic CoA levels that occurs with fasting, but it changed the acyl-CoA composition of the liver by significantly decreasing the concentration of short-chain acyl-CoAs and choloyl-CoA, a peroxisomal intermediate in bile acid synthesis. Untargeted and targeted metabolomics analysis revealed that fasted livers overexpressing Nudt7 contained significantly reduced levels of primary and secondary bile acids, in addition to an accumulation of medium- and long-chain acylcarnitines, which correlated with a lower rate of peroxisomal fatty acid oxidation. Our data show that overexpression of Nudt7 specifically interfered with the output of peroxisomal fatty acid oxidation and the bile acid biosynthetic pathway in the fasted state. Thus, changes in Nudt7 activity between the fed and fasted states could act to regulate the peroxisomal CoA pool and, in turn, peroxisomal lipid metabolism.
MATERIALS AND METHODS
Reagents were purchased from the following suppliers: HRP-conjugated GFP antibody, Alexa Fluor 647- and HRP-conjugated goat anti-rabbit IgG from Thermo Fisher Scientific; the antibodies against GAPDH and the FLAG epitope (DYKDDDK) from Cell Signaling; Waymouth MB 752/1 medium, medium 199 (M199), and the antibody against ATP-binding cassette subfamily D member 3 (PMP70) from Sigma-Aldrich. The Nudt7 antibody was generated as previously described (15). The fluorescent CoA derivative of monobromobimane (mBB), mBB-CoA, was synthesized as previously described (18). All other reagents were of analytical grade or better and were purchased from Sigma-Aldrich or Fisher Scientific, unless otherwise stated.
Adeno-associated virus production
Plasmid pscAAV-LP1-EGFP, carrying the enhanced green fluorescent protein coding sequence (henceforth referred to as GFP), was developed by John T. Gray and provided by St. Jude Children’s Research Hospital, Memphis, TN. Plasmid pscAAV-LP1-mNudt7, obtained by replacing the coding sequence of GFP with the sequence of mouse Nudt7 containing a FLAG tag at the N terminus, was a kind gift of Suzanne Jackowski, St. Jude Children’s Research Hospital, Memphis, TN. To produce adeno-associated virus (AAV), HEK 293T cells were transiently transfected with a 1:1:2.5 (mass ratio) mixture of plasmids pHGTI-adeno1 (developed by John T. Gray and provided by Harvard College, Cambridge, MA), pLTAAVhelp2-8 (developed by John T. Gray and provided by St. Jude Children’s Research Hospital, Memphis, TN), and either pscAAV-LP1-mNudt7 or pscAAV-LP1-EGFP using polyethylenimine. Transfected cells were harvested 72 h post transfection and lysed in 50 mM Tris-HCl, 50 mM NaCl, 1 mM MgCl2, 2% Triton X-100, 50 U/ml Pierce™ Universal Nuclease (pH 8.0), incubating for 20 min at 37°C. The lysate was then supplemented with 0.05 vol of sodium deoxycholate (10% solution in water) and further incubated at 37°C for 10 min. Following the addition of 0.1 vol of 5 M NaCl, the lysate was clarified by centrifugation at 4,000 g for 20 min at 20°C. The cleared lysate was layered on top of a discontinuous sucrose density gradient generated by layers of 60, 40, 25, and 15% sucrose in 20 mM diethanolamine hydrochloride (pH 9.0). Following centrifugation at 104,000 g (Beckman Optima LE-80) for 3 h at 20°C, the virus was recovered from the 40% sucrose layer and the 40–60% sucrose interface region of the gradient. The AAV particles were further purified by ion exchange chromatography on a POROS 50 HQ column (Thermo Fisher Scientific). AAV elution was monitored by measuring the absorbance at both 260 and 280 nm. Fractions containing the AAV particles were buffer exchanged into phosphate-buffered saline containing 0.01% Pluronic F-68, filter-sterilized, and quantified using the agarose gel method previously described (28).
Animal studies
Mice were fed a standard chow diet (Tekland 2018S) and maintained at a room temperature of 22.2 ± 0.2°C, room humidity of 40 ± 2%, and a 12 h light/12 h dark cycle, with the dark cycle starting at 6:00 PM. Six-week-old C57BL/6J male mice were purchased from Jackson Laboratory. Following a 2 week acclimation period, these mice were injected with 2.5 × 1011 genome copies of GFP- or Nudt7-AAV in 150 μl of sterile phosphate-buffered saline and, unless otherwise indicated, were euthanized 3–4 weeks after injection. Unless otherwise stated, fasting experiments were started at 7:00 AM, and mice were placed in cages with grids without food for the indicated amount of time. To measure food consumption, mice were individually housed in cages with grids and provided with a preweighed amount of food. Seventy-two hours later, the leftover food was weighed and used to calculate the average food consumption per mouse per 24 h. Serum levels of aspartate aminotransferase (AST) and alanine aminotransferase (ALT) were measured using Stanbio kits (EKF Diagnostic USA) and 3-hydroxybutyrate was determined using a 3-hydroxybutyrate enzyme solution and colorimetric detector from Cayman Chemicals, as per the manufacturer’s instructions. All studies were approved by the Institutional Animal Care and Use Committees of West Virginia University.
CoA analysis, immunoblotting, and RT-PCR
The concentration of total CoA (free CoA plus CoA thioesters) in liver homogenates was determined after conversion of the cofactor to the mBB derivative, as previously described (24). For Western blot analysis, flash-frozen tissues were homogenized in ice-cold radioimmunoprecipitation assay buffer supplemented with protease inhibitors (Biotool), and centrifuged at 10,000 g for 10 min at 4°C. Proteins (5 or 20 μg) were fractionated on 4–12% bis-Tris polyacrylamide gels and transferred onto nitrocellulose membranes using an iBlot dry transfer system (Thermo Fisher Scientific). The GAPDH and Nudt7 antibodies were used at a 1:5,000 and 1:3,000 dilution, respectively. Bound primary antibodies were detected by chemiluminescence with HRP-conjugated goat anti-rabbit IgG at a 1:45,000 dilution. The HRP-conjugated GFP antibody was used at a 1:7,000 dilution. RNA was isolated from flash-frozen tissue as previously described (29). Following the removal of genomic DNA with Turbo DNA-free kit (Thermo Fisher Scientific), mRNA levels were quantified in triplicate by RT-PCR using the Quantitect SYBR Green RT-PCR kit (Qiagen) and the primers previously reported (29). The relative abundance of each gene was calculated using the CT method, and the amount (2-ΔCT) reported relative to the average of ribosomal protein L22 (Rpl22) and β2-microglobulin (B2m). Primers 5′-GTGCCTTTCTCCAAAAGGTATTT-3′ and 5′-CTCTCTTTGCTGTTGGCGAC-3′ were used as forward and reverse primers, respectively, to amplify Rpl22. Primers 5′-ACTGACCGGCCTGTATGCTA-3′ and 5′-ATGTTCGGCTTCCCATTCTCC-3′ were used as forward and reverse primers, respectively for B2m.
RNA-Seq
Total RNA from mouse liver tissue was isolated as described above and delivered to the West Virginia University Genomics Core Facility for quality control and library preparation. The concentration of extracted total RNA was determined using a Qubit fluorometer (Thermo Fisher Scientific), while RNA quality was ascertained on an Agilent 2100 bioanalyzer using an RNA Nano 6000 chip. All of the samples used for the library preparation had RNA integrity number (RIN) values >8.0. Each library was built using 500 ng of RNA as input material for the stranded mRNA library preparation kit from KAPA Biosystems, following the manufacturer’s recommended protocol with nine cycles of PCR. The cDNA libraries were quantified via Qubit, pooled in equimolar ratios, and sent to the Genomics Core Facility at Marshall University where the libraries were sequenced on four lanes generating PE50bp reads with the Illumina HiSeq1500 system (16–36M reads per sample, ∼175M total reads). Read quality was assessed using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc) and deemed to require no trimming. The reads were quantified using Salmon (30) with transcripts obtained from Ensembl, version GRCm38.84. Using tximport (31), the data was imported into R and differential expression was computed using DESeq2 (32). We removed all genes that did not have at least a count of 10 in at least three samples. Genes were considered to be significantly changed if the Benjamini-Hockburg adjusted P-value was less than 0.01. WebGestalt (http://www.webgestalt.org) was used to conduct gene ontology and pathway enrichment analyses. The RNA-Seq raw data were deposited and are accessible through NCBI Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) with the GEO Series accession number GSE125237.
Enzymatic assays and lipid analyses
Total liver CoA diphosphohydrolase activity was measured in liver extracts obtained by homogenizing flash-frozen tissue (∼20 mg) in 750 μl of 20 mM Tris-HCl (pH 8.0). The homogenates were incubated on ice for 10 min before centrifuging at 20,000 g for 10 min. The supernatants were removed and increasing amounts of protein (6.25–50 μg) were incubated with 100 μM of mBB-CoA for 10 min at 37°C. The reactions were then stopped and analyzed as described previously (18).
Serum cholesterol and triglycerides were measured using Stanbio kits (EKF Diagnostics USA), as per the manufacturer’s instructions. Liver lipids were extracted from ∼50 mg of liver using a modification of the Bligh and Dyer procedure (33) optimized for lipid quantitation by the LipidMaps group (34). Following extraction, a portion of the lipids was dried and resuspended in 100 μl of 5% NP-40 for cholesterol analysis using the same Stan-Bio kit utilized for the determination of cholesterol in the serum. A second portion of the total lipid extract was fractionated for triglyceride analysis on HPTLC silica plates developed in hexane:ether:acetic acid (80:20:1, v:v:v). Resolved lipid bands and serial dilutions of a tripalmitin standard were visualized and quantitated by spraying with a 0.002% primulin solution followed by fluorescence analysis using G:BOX Chemi XX9 imaging system (Syngene).
Targeted and untargeted metabolomics
Global metabolomics profiling, combined with statistical and pathway enrichment analyses, were conducted by Metabolon, Inc. Raw data and metabolite fold-changes between groups are reported in supplemental Table S1. Principal component analysis, normalization, and scaling of selected metabolites for the generation of heat plots were conducted using MetaboAnalyst 4.0 (https://www.metaboanalyst.ca/) (35). Liver, small intestine, and bile samples were analyzed for their bile acid composition using a targeted assay by Metabolon, Inc. For the analysis of the acyl-CoA pool composition, CoA species were extracted as described by Minkler et al. (36). Briefly, flash-frozen livers (∼50 mg) were homogenized in 1.5 ml of a 3:1 (v:v) mixture of acetonitrile:isopropanol containing 0.625 μM of heptanoyl- and heptadecanoyl-CoA standards. Following the addition of 0.5 ml of 0.1 M KH2PO4 (pH 6.7), the homogenates were centrifuged at 16,000 g for 5 min. The supernatants were acidified by the addition of 0.25 vol of glacial acetic acid and applied onto 2-(2-pyridyl)ethyl-functionalized silica columns (100 mg; Sigma-Aldrich) previously conditioned with 1 ml of a 9:3:4:4 (v:v) mixture of acetonitrile:isopropanol:water:ace-tic acid. The columns were washed with 1 ml of the same solvent mixture and 1 ml of water before eluting the bound compounds with 4 ml of 90% methanol containing 15 mM ammonium hydroxide. The samples were dried at room temperature under nitrogen flow and stored at −20°C until analyzed. The LC/MS/MS analysis was conducted in the Metabolome Analysis Facility at West Virginia University on an AB Sciex QTrap 5500 mass spectrometer connected to an AB Sciex Exion UPLC and controlled by Analyst® version 1.6.3 software (Sciex). Frozen extracts were resuspended in 300 μl of 50% methanol and fractionated at 0.4 ml/min onto an Acquity BEH amide column (2.1 × 50 mm, 1.7 μm particle size; Waters) kept at 25°C and equilibrated with 10% solvent A (50 mM ammonium formate, pH 9.5) and 90% solvent B (acetonitrile). Following the injection of 1.5 μl of sample, the concentration of A was maintained at 10% for an additional 1.5 min, followed by a linear gradient that increased the concentration of A to 50% in 6.5 min and then to 65% in 30 s. The concentration of A was kept at 65% for 1.5 min before returning it to 10% in 3.5 min, followed by 4.5 min of column reequilibration. Positive ESI-MS/MS was performed using the following parameters: ion spray voltage of 5.5 kV, temperature of 500°C, ion source gas 1 set at 25, ion source gas 2 and curtain gas set at 20, collision gas set at medium. The entrance potential (EP), collision energy (CE), and collision cell exit potential (CXP) resulting in the neutral loss of 507.0 Da from the (M+H)+ precursor (37, 38) were individually optimized for free CoA (EP = 10 V, CE = 37 V, CXP = 15 V), acetyl-CoA (EP = 10 V, CE = 40 V, CXP = 15 V), malonyl-CoA (EP = 12 V, CE = 40 V, CXP = 15 V), succinyl-CoA (EP = 10 V, CE = 45 V, CXP = 17 V), butyryl-CoA (EP = 12 V, CE = 41 V, CXP = 20 V), heptanoyl-CoA (EP = 10 V, CE = 45 V, CXP = 20 V), octanoyl-CoA (EP = 7 V, CE = 46 V, CXP = 23 V), lauroyl-CoA (EP = 9 V, CE = 47 V, CXP = 20 V), heptadecanoyl-CoA (EP = 10 V, CE = 47 V, CXP = 20 V), and stearoyl-CoA (EP = 9 V, CE = 50 V, CXP = 23 V). The declustering potential was consistently set at 170 V. For all other CoA species, the parameters, EP, CE, and CXP, were set at 10, 45, and 17 V, respectively. Retention times for the detected CoA species are reported in supplemental Table S2. Peak analysis was conducted using MultiQuant 3.0.2 (Sciex). Peak areas corresponding to free CoA and acyl-CoAs with an acyl chain up to 10 carbons were normalized to the heptanoyl-CoA internal standard. Peak areas corresponding to longer acyl-CoAs and choloyl-CoA were normalized to the heptadecanoyl-CoA internal standard. Corrected peak areas were further normalized to the tissue weight.
Hepatocyte isolation
For immunofluorescence studies, primary hepatocytes were isolated from fed mice using the two-step liver perfusion method described by Klaunig et al. (39) with minor modifications. All perfusion steps were conducted at a flow rate of 7 ml/min with buffers kept at 37°C. The livers were first perfused with 50 ml of a modified Krebs buffer (120 mM NaCl, 24 mM NaHCO3, 20 mM glucose, 5 mM HEPES, 4.8 mM KCl, 1.2 mM MgSO4, 1.2 mM KH2PO4, 100 U/ml penicillin, 100 U/ml streptomycin, 50 μg/ml gentamycin) containing 0.1 mM EGTA (pH 7.4), followed by 50 ml of modified Krebs buffer supplemented with 1.4 mM CaCl2 and 10 μg/ml of Liberase TM (Sigma-Aldrich). Intermittent clamping of the inferior vena cava for about 10 s each time was conducted during this latter step to increase total cell yield (40). After the perfusion, the livers were excised, separated from the gall bladder, and transferred to a sterile dish containing 20 ml of the second perfusion buffer, ice-cold, without Liberase. The cells were released by tearing the liver with forceps. The cell suspension was filtered through a 100 μm strainer (Fisher Scientific) and washed three times with Waymouth medium [Waymouth MB 752/1 medium supplemented with 0.5 mM serine, 0.5 mM alanine, 20 mM HEPES (pH 7.4), 100 U/ml penicillin, 100 U/ml streptomycin, 50 μg/ml gentamycin] containing 1.9 mg/ml of fatty acid-free BSA and pelleted by spinning at 50 g for 2 min at 4°C. After the washes, the cells were resuspended in Waymouth medium containing 5% newborn calf serum, and 7.5 × 105 cells were plated on each well of collagen-coated 6-well plates. Viability (consistently >85%) and yield were assessed by counting the cells that excluded trypan blue. After a 5 h incubation, the medium was replaced with fresh Waymouth medium lacking the newborn calf serum and an overlay of Matrigel (0.2 mg/ml), and insulin (60 nM) was added as described by Cyphert et al. (41). The hepatocytes were incubated under these conditions overnight before being processed for immunofluorescence studies.
For the measurement of the rate of β-oxidation, the procedure for the isolation of hepatocytes was modified as follows. First, hepatocytes were isolated from mice fasted for 15 h (food removed at 4:00 PM) using perfusion buffers containing a reduced amount of glucose at 5.5 mM. Second, after releasing the cells from the liver capsule, M199 containing 25 mM of HEPES (pH 7.4) (M199/HEPES) was used for all the wash steps and to resuspend the cells at a final concentration of 1.0 × 106 cells/ml. This cell suspension was immediately used for β-oxidation studies.
Immunofluorescence and β-oxidation
Confocal microscopy was used as previously described (18) to determine the subcellular localization of the exogenous FLAG-tagged Nudt7 expressed in primary hepatocytes isolated from mice injected with Nudt7-AAV. The DYKDDDK antibody, the PMP70 antibody used to visualize the peroxisomes, and an Alexa Fluor 647 donkey-anti rabbit secondary antibody were used at a 1:1,000 dilution. β-Oxidation studies were conducted in triplicate on freshly isolated hepatocytes in suspension by measuring the production of 14C-labeled acid-soluble metabolites from [1-14C]palmitic acid (American Radiolabeled Chemicals) in the presence or absence of etomoxir, an inhibitor of mitochondrial fatty acid oxidation. Pilot experiments were conducted to determine the optimal combination of cell number and incubation time that would ensure linear product formation. Furthermore, pilot studies comparing the delivery of [1-14C]palmitic acid as a solution in ethanol or as a dry powder thoroughly resuspended in M199/HEPES containing 0.5% fatty acid-free BSA ruled out any adverse effect of 17 mM final ethanol concentration on the rate of fatty acid oxidation. Specifically, 7.5 × 105 cells were preincubated for 15 min at 37°C in 1.5 ml of M199/HEPES ± 45 μM etomoxir before starting the reaction with the addition of 0.5 ml of substrate mixture containing 0.5% fatty acid-free BSA and 0.4 mM of [1-14C]palmitic acid (0.4 μCi, specific activity 2 mCi/mmol, from a 111 mM stock solution in ethanol) in M199/HEPES. The hepatocytes were incubated with gentle shaking for 15 min at 37°C before stopping the reaction by transferring 400 μl of cell suspension to a tube containing 133 μl of 1 M perchloric acid. This acidified suspension was centrifuged at 20,000 g for 10 min and the radioactivity in 300 μl of supernatant was measured by liquid scintillation counting. Typical raw counts in samples incubated without etomoxir were between 4,000 and 9,000 cpm, while typical raw counts in samples incubated with etomoxir were between 1,500 and 3,000 cpm. Background radioactivity (350–700 cpm), estimated in reactions stopped immediately after the addition of the substrate mixture, was subtracted from each sample. For each mouse, peroxisomal β-oxidation was calculated from the samples incubated with etomoxir. Mitochondrial fatty acid oxidation was calculated as the difference between total fatty acid oxidation (no etomoxir) and peroxisomal fatty acid oxidation.
Statistical analysis
All data are reported as the mean ± the standard error. Unless otherwise stated, statistical significance (P < 0.05) was calculated using the Welch’s two-sample t-test (Metabolon Inc.) or the unpaired two-tailed Student’s t-test and two-way ANOVA using GraphPad Prism 7 (GraphPad Software).
RESULTS
Overexpression of Nudt7 does not prevent the increase in total CoA levels that occurs in fasted livers
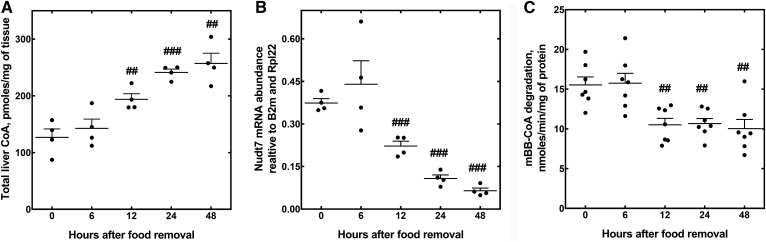
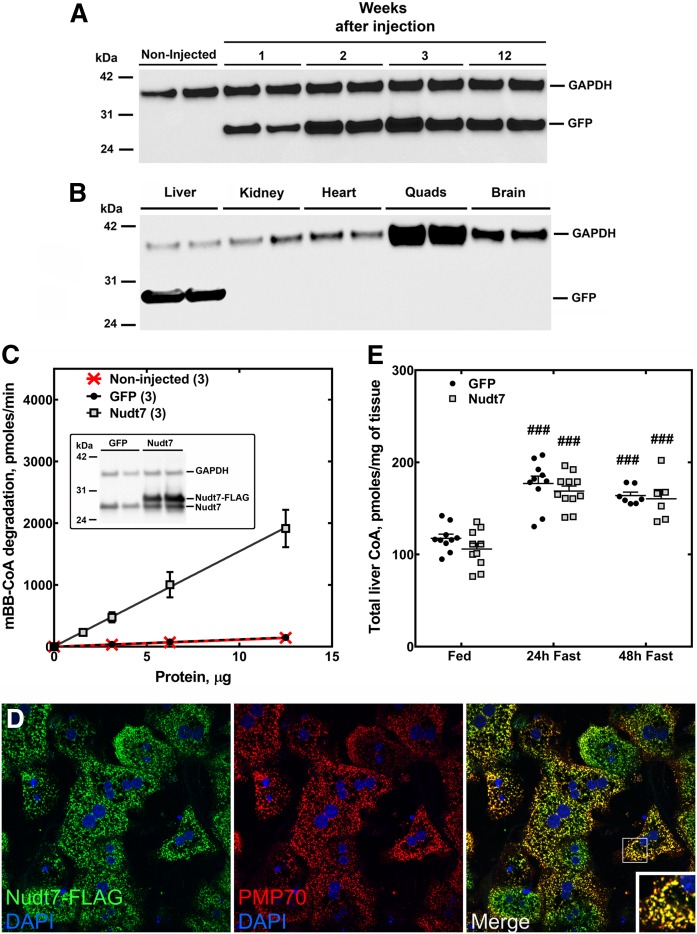
Compared with fed mice, the livers of mice fasted for 24 or 48 h exhibit a higher concentration of total CoA (free CoA plus acyl-CoAs) and lower expression of the CoA-degrading enzyme, Nudt7, the major CoA diphosphohydrolase isoform in this organ (17, 18, 22). To examine the inverse correlation between the cofactor concentration and Nudt7 expression in more detail, we measured liver CoA levels and Nudt7 mRNA and activity during a fasting time course, starting as early as 6 h after the removal of food (Fig. 1). We found that CoA levels increased significantly after a 12 h fast (Fig. 1A) and plateaued after a 24 h fast, closely matching the time course of the decrease in Nudt7 mRNA (Fig. 1B) and CoA-degrading activity (Fig. 1C) in the liver. To determine whether suppression of Nudt7 activity contributed to this accumulation of CoA, we overexpressed Nudt7 with an N-terminal FLAG tag in the livers of C57BL/6J mice and analyzed the effect of this manipulation on the concentration of hepatic CoA in fasted mice (Fig. 2). Transgene expression was obtained using AAV particles carrying the Nudt7 coding sequence under the control of the liver-specific LP1 promoter (42). Mice expressing GFP from the same AAV construct were used as controls and to characterize the transgene expression from this system, which was stable for up to 12 weeks (Fig. 2A) and confirmed to be liver-specific (Fig. 2B). All analyses were conducted 3–4 weeks after the AAV injection. Under these conditions, values for serum AST and ALT were in the normal range (43) and similar between GFP and Nudt7 mice, indicating the absence of liver damage (Table 1). Furthermore, injection of Nudt7-AAV did not affect food intake or body weight, while the weight of the liver normalized to body weight was modestly elevated in the Nudt7 mice under fed but not fasting conditions (Table 1). Exogenous Nudt7 was robustly overexpressed compared with the endogenous levels (Fig. 2C, inset) and measurement of the CoA-degrading activity in Nudt7 mice fasted for 24 h revealed a 10-fold increase in activity compared with either GFP mice or mice that were not injected with AAV (Fig. 2C). Nudt7 is a peroxisomal enzyme (23, 24). To determine whether the exogenously expressed Nudt7 was correctly localized to the peroxisomes, we isolated primary hepatocytes from mice injected with Nudt7-AAV and used immunofluorescence and confocal microscopy to visualize the subcellular localization of the FLAG epitope fused to the transgene. Nuclei were stained with DAPI and peroxisomes were visualized with an antibody against the endogenous protein PMP70. FLAG-tagged Nudt7 was found to colocalize with PMP70 with no detectable fluorescence in the cytoplasm (Fig. 2D), confirming efficient import of this protein into the peroxisomes. Nudt7 and GFP mice were then analyzed for total hepatic CoA levels in the fed state and following a 24 and 48 h fast. The hepatic concentration of CoA tended to be lower in fed and 24 h fasted Nudt7 mice compared with the GFP controls, but the difference did not reach statistical significance (Fig. 2E). Furthermore, total CoA levels increased with fasting in both Nudt7 and GFP livers, indicating that overexpression of Nudt7 was not able to blunt the accumulation of CoA driven by activation of the biosynthetic pathway. Overall, these results supported the conclusion that the fasting-induced decrease in Nudt7 activity was unlikely to significantly contribute to the increase in the total CoA content normally observed, at the whole tissue level, in the liver (Fig. 1).
Fig. 1.
Changes in hepatic CoA content, Nudt7 mRNA, and activity with fasting. Mice were fasted for 0–48 h and livers were harvested for the measurement of total hepatic CoA levels (A), Nudt7 mRNA (B), and CoA-degrading activity (C) using the Nudt7 substrate, mBB-CoA, as described in the Materials and Methods. Data are reported as the mean (horizontal lines) of measurements on individual mice (circles) ± SEM. ##P < 0.01, ###P < 0.001 relative to the fed state at t = 0.
Fig. 2.
Liver-specific overexpression of Nudt7 and GFP and effect on hepatic CoA levels. C57BL/6J mice were injected with AAV particles to achieve liver-specific overexpression of FLAG-tagged Nudt7 or the control protein GFP. A: Western blot analysis of liver homogenates (5 μg of protein) for GFP expression at different time points, following the intravenous injection of AAV particles. B: Three weeks after GFP-AAV injection, Western blot analysis of different tissue homogenates (5 μg of protein) confirmed that the transgene expression was liver-specific. C: Liver homogenates obtained from noninjected mice or from mice injected with Nudt7- and GFP-AAV were assayed for total CoA-degrading activity. C (inset): Western blot of liver homogenates (20 μg of protein) obtained from GFP and Nudt7 mice using the Nudt7 antibody. GAPDH was used as loading control in (A), (B), and (C, inset). Numbers in parenthesis represent the number of animals analyzed; data are reported as the mean ± SEM. D: Primary hepatocytes were isolated from Nudt7 mice, fixed, and used to visualize the localization of the enzyme using the FLAG antibody and confocal microscopy. Nudt7 is shown in green and the PMP70 protein, which was used as a marker for the peroxisomes, is shown in red. Cell nuclei were stained with DAPI, shown in blue. The inset in the merged image shows details at a higher magnification. The results are representative of two independent experiments. E: Total CoA measured in the liver of Nudt7 and GFP mice fed ad libitum or fasted for 24 and 48 h. Data are reported as the mean (horizontal lines) of measurements on individual mice (circles and squares) ± SEM. ###P < 0.001 relative to the fed state.
TABLE 1.
Selected features of fed and 24 h fasted GFP and Nudt7 mice
| Fed | 24 h Fast | |||
| GFP | Nudt7 | GFP | Nudt7 | |
| Serum ALT (mg/dl) | 21 ± 2 | 18 ± 1 | 35 ± 4a | 24 ± 3 |
| Serum AST (mg/dl) | 48 ± 6 | 36 ± 7 | 67 ± 9 | 62 ± 9a |
| Blood glucose (mg/dl) | 98 ± 6 | 105 ± 6 | 61 ± 3b | 55 ± 3b |
| Serum 3-hydroxybutyrate (mM) | 0.31 ± 0.04 | 0.38 ± 0.08 | 1.63 ± 0.15b | 1.57 ± 0.08b |
| Serum triglycerides (mg/dl) | 119 ± 18 | 144 ± 10 | 86 ± 6 | 83 ± 4b |
| Serum cholesterol (mg/dl) | 106 ± 8 | 90 ± 6 | 67 ± 6c | 68 ± 3c |
| Food intake (g/mouse/day) | 4.4 ± 0.5 | 4.9 ± 0.2 | N/A | N/A |
| Body weight (g) | 30.0 ± 0.5 | 30.8 ± 0.2 | 24.2 ± 0.3b | 24.3 ± 0.6b |
| Liver weight/body weight (mg/g) | 44.6 ± 1.9 | 50.3 ± 1.7d | 35.5 ± 1.4c | 39.4 ± 1.4b |
| Liver triglycerides (mg/g) | 0.9 ± 0.2 | 0.9 ± 0.1 | 19.4 ± 1.9b | 24.1 ± 6.0c |
| Liver cholesterol (mg/g) | 0.6 ± 0.1 | 0.5 ± 0.1 | 1.1 ± 0.1b | 0.9 ± 0.10c |
Values are expressed as the mean of seven to eight mice per condition ± SEM. N/A, not applicable.
P ≤ 0.05 relative to the fed state within GFP or Nudt7 groups.
P ≤ 0.001 relative to the fed state within GFP or Nudt7 groups.
P ≤ 0.01 relative to the fed state within GFP or Nudt7 groups.
P ≤ 0.05 relative to GFP under the same nutritional conditions.
Increased Nudt7 activity in the fasted state changes the acyl-CoA composition and affects lipid metabolism in the liver
Nudt7 is a promiscuous CoA diphosphohydrolase that hydrolyzes free CoA and a large range of short- and medium-chain acyl-CoAs in vitro (17, 18). To determine whether overexpression of Nudt7 changed the concentration of individual acyl-CoA species, we analyzed the relative composition of the acyl-CoA pool in Nudt7 and GFP livers by LC/MS/MS.
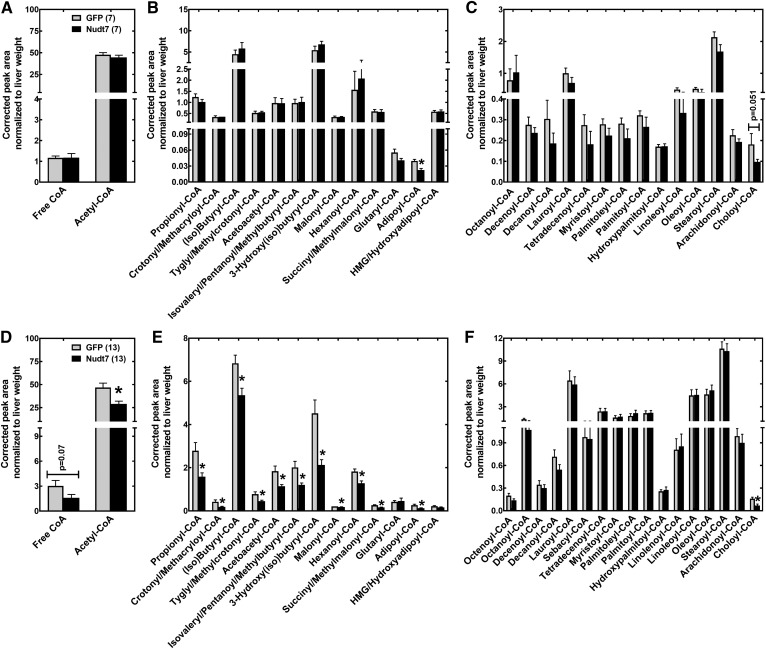
We found that, upon fasting, the vast majority of the short-chain acyl-CoAs monitored were significantly decreased in Nudt7 mice compared with the GFP controls (Fig. 3D–F). These included CoA thioesters known to be excellent in vitro substrates for Nudt7, such as acetyl-, propionyl-, (iso)butyryl-, hexanoyl-, malonyl-, and succinyl-CoA. Acetoacetyl-, 3-hydroxyl(iso)butyryl-, crotonyl/methacryloyl-, and tyglyl/methylcrotonyl-CoA, which are products of fatty acid and amino acid metabolism, were also decreased in the Nudt7 livers (Fig. 3E). The drop in the concentration of free CoA observed in fasted Nudt7 livers narrowly missed statistical significance (Fig. 3D). Choloyl-CoA, an intermediate in bile acid synthesis and known substrate of recombinant Nudt7 (17), was significantly decreased by Nudt7 overexpression in the fasted state and tended to be lower even in the fed state (Fig. 3C, F). Overexpression of Nudt7 had minimal effects on the acyl-CoA composition in the fed state, with adipoyl-CoA, a six-carbon dicarboxylic fatty acyl-CoA, being the only CoA thioester significantly decreased in fed Nudt7 mice compared with the GFP controls (Fig. 3A–C). Adipoyl-CoA levels remained significantly lower in the Nudt7 livers following a 24 h fast.
Fig. 3.
Liver acyl-CoA analysis. Livers isolated from fed (A–C) and 24 h-fasted (D–F) Nudt7 and GFP mice were analyzed by LC/MS/MS to determine the relative abundance of free and acetyl-CoA (A, D), short-chain acyl-CoAs (B, E), and medium- and long-chain acyl-CoAs (C, F). Numbers in parenthesis represent the number of animals analyzed; data are reported as the mean ± SEM. *P < 0.05.
Combined, these results showed that while the overexpression of Nudt7 did not significantly affect total CoA levels, it changed the acyl-CoA composition of the liver by decreasing the concentration of short-chain acyl-CoAs and choloyl-CoA. This effect was more pronounced in the fasted state, when the expression and activity of the endogenous Nudt7 in the liver were lowest (Fig. 1B, C).
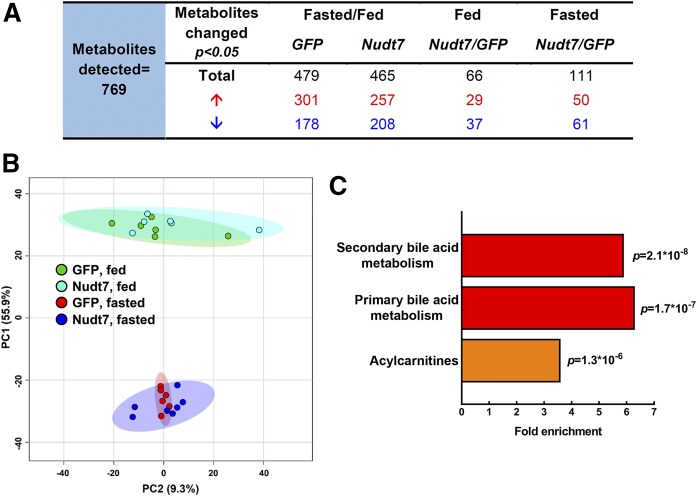
Mono- and dicarboxylic short-chain acyl-CoAs are products and intermediates of a variety of metabolic processes, including lipid, carbohydrate, and amino acid metabolism. To identify pathways that could be potentially altered by Nudt7 overexpression, we conducted global metabolic profiling of Nudt7 and GFP livers harvested from fed and fasted mice (Fig. 4, supplemental Table S1). As expected, the vast majority of the 769 metabolites detected was changed by the nutritional state, regardless of the specific transgene expressed (Fig. 4A). The separation between the fed and fasted mice, together with the significant overlap between GFP and Nudt7 groups, was also highlighted by principal-component analysis (Fig. 4B). In the fed state, 66 metabolites were significantly different between Nudt7 and GFP livers, while this number almost doubled to 111 in the fasted state (Fig. 4A). Only a few CoA species, CoA precursors, and metabolites were detected by the untargeted metabolic profiling. Among the latter were the products of the Nudt7 reaction, phosphopantetheine and 3′,5′-ADP, which were not significantly different between Nudt7 and GFP livers, although the higher levels of phosphopantetheine trended toward significance in the fasted Nudt7 livers (supplemental Table S1). The larger effect of Nudt7 expression on liver metabolism in the fasted state was consistent with the results from the acyl-CoA analysis (Fig. 3E) and led us to focus all subsequent analyses on this nutritional state. Under fasting conditions, pathway enrichment analysis revealed that the three most significantly and highly (>3-fold) enriched pathways were primary and secondary bile acid metabolism, henceforth considered as a single pathway, and acylcarnitines (Fig. 4C). These pathways, which are both part of lipid metabolism and either dependent on or tightly linked to CoA, were further analyzed (see below).
Fig. 4.
Global metabolic profiling of 24 h-fasted Nudt7 and GFP livers. A: Number of metabolites significantly changed in the Nudt7 and GFP livers by fasting and, within a given nutritional state, by Nudt7 overexpression. Under each condition, the numbers in red represent metabolites whose concentrations increased and the numbers in blue represent metabolites whose concentrations decreased. B: Score plot of the principal-component analysis (PCA) obtained using MetaboAnalyst 4.0. Colored ellipses represent 95% confidence intervals for each group. Colored dots represent individual mice. C: Top three most significantly enriched pathways that emerged from the global metabolic profiling.
We also conducted RNA-Seq analysis of the livers of fasted Nudt7 and GFP mice. This analysis revealed a modest effect of Nudt7 overexpression on the global transcriptome, identifying a total of 62 genes (33 upregulated, 29 downregulated) whose expression was significantly different in the Nudt7 livers compared with the GFP controls (fold-change >1.50 or ≤0.66, adjusted P value <0.01) (supplemental Table S3). Gene ontology analysis on this list of differentially expressed genes identified carboxylic ester hydrolase activity and fibronectin binding as significantly enriched molecular functions (Table 2), but no significantly enriched biological process. Pathway analysis using the Wikipathway database (https://www.wikipathways.org) identified the PPAR signaling pathway as significantly enriched (Table 2). The same result was obtained using the KEGG database (https://www.genome.jp/kegg/).
TABLE 2.
Over-representation enrichment analysis of the genes differentially expressed between Nudt7 and GFP mice in the fasted state
| Gene Symbol | Gene Name | Fold Change, Nudt7/GFP |
| Molecular function: carboxylic ester hydrolase activity (term: GO: 000052689); fold enrichment: 10.7, P = 0.0000171, FDR = 0.004 | ||
| Car2 | Carbonic anhydrase 2 | 2.11 |
| Ces1e | Carboxylesterase 1E | 1.61 |
| Ces1f | Carboxylesterase 1F | 1.98 |
| Ces1g | Carboxylesterase 1G | 2.11 |
| Ces2c | Carboxylesterase 2C | 3.17 |
| Abhd6 | Abhydrolase domain containing 6 | 1.81 |
| Molecular function: fibronectin binding (term: GO: 0001968); fold enrichment: 29.4, P = 0.00013, FDR = 0.015 | ||
| Itgb3 | Integrin β3 | 0.48 |
| Sdc4 | Syndecan 4 | 0.66 |
| Lrrc15 | Leucine rich repeat containing 15 | 0.48 |
| Pathway: PPAR signaling pathway (ID: WP2316); fold enrichment: 9.4, P = 0.00015, FDR = 0.025 | ||
| Cd36 | CD36 antigen | 2.32 |
| Cyp4a12b | Cytochrome P450 family 4 subfamily a polypeptide 12B | 1.96 |
| Pltp | Phospholipid transfer protein | 1.55 |
| Scd1 | Stearoyl-CoA desaturase 1 | 2.20 |
| Acaa1b | Acetyl-CoA acyltransferase 1B | 2.02 |
The analysis was conducted using WebGestalt, as described in the Materials and Methods section. FDR, false discovery rate; GO, gene ontology; ID, identifier.
Effect of increased Nudt7 activity on peroxisomal bile acid metabolism
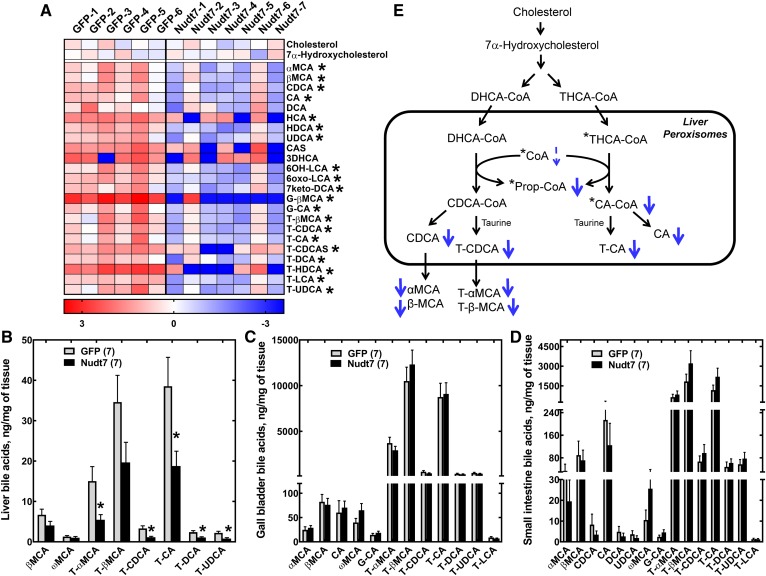
A heat plot of all the primary and secondary bile acids detected by the untargeted metabolomics analysis is shown in Fig. 5A. Out of the 23 bile acids quantified, 20 were significantly decreased in the livers of Nudt7 mice, with no difference in the levels of the bile acid precursors, cholesterol and 7α-hydroxycholesterol (44). The concentration of liver and serum cholesterol was also confirmed to be similar between GFP and Nudt7 mice by an independent assay (Table 1). To corroborate the results obtained from the untargeted metabolomics analysis, we injected another cohort of mice with GFP- and Nudt7-AAV and examined the bile acid composition of their livers with a targeted bile acid assay. This analysis was also extended to the gall bladder and small intestine of the mice, as these organs contain a large portion of the body bile acid pool. In the Nudt7 livers, the targeted analysis confirmed a significant ∼50% decrease in selected primary and secondary bile acids, including tauro-α-muricholic acid, taurocholic acid, taurodeoxycholic acid, and tauroursodeoxycholic acid, compared with the GFP controls (Fig. 5B). Hepatic levels of β-muricholic acid and tauro-β-muricholic acid also tended to be lower in the Nudt7 mice but did not reach statistical significance in the targeted analysis. No difference in the bile acid composition of the gall bladder and small intestine was detected (Fig. 5C, D), indicating that the effect of the increased activity of Nudt7 was limited to the liver, the site of bile acid synthesis and Nudt7 overexpression.
Fig. 5.
Nudt7 overexpression in the fasted state decreases the hepatic bile acid content. A: Heat plot of all primary and secondary bile acids, plus the bile acid precursors, cholesterol and 7α-hydroxycholesterol, detected by global metabolic profiling. Targeted bile acid analysis of livers (B), gall bladder extracts (C), and small intestine (D). In B–D, data are reported as the mean ± SEM and numbers in parenthesis represent the number of animals analyzed. *P < 0.05. E: Scheme of the conversion of DHCA-CoA and THCA-CoA to chenodeoxycholic acid (CDCA) and cholic acid (CA) derivatives in the peroxisomes. Known substrates of Nudt7 are marked by an asterisk. Overexpression of Nudt7 was associated with a decrease (thick blue arrows) in the concentration of choloyl-CoA (CA-CoA), propionyl-CoA (Prop-CoA), and of several primary and secondary bile acids, as detected by either untargeted or targeted bile acid analyses. The decrease in free CoA, which narrowly missed statistical significance (P = 0.07), is indicated by the thin dashed arrow. CA, cholic acid; CAS, cholic acid sulfate; CDCA, chenodeoxycholic acid; DCA, deoxycholic acid; 7keto-DCA, 7-keto-deoxycholic acid; HDCA, hyodeoxycholic acid; 3DHCA, 3-dehydrocholic acid; 6OH-LCA, 6-hydroxy-lithocholic acid; 6oxo-LCA, 6-oxo-lithocholic acid; G-CA, glycocholic acid; G-βMCA, glyco-β-muricholic acid; HCA, hyocholic acid; HDCA, hyodeoxycholic acid; LCA, lithocholic acid; αMCA, α-muricholic acid; βMCA, β-muricholic acid; ωMCA, ω-muricholic acid; T-CA, taurocholic acid; T-CDCA, taurochenodeoxycholic acid; T-CDCAS, taurochenodeoxycholic acid sulfate; T-DCA, taurodeoxycholic acid; T-αMCA, tauro-α-muricholic acid; T-βMCA, tauro-β-muricholic acid; T-LCA, taurolithocholic acid; T-HDCA, taurohyodeoxycholic acid; T-UDCA, tauroursodeoxycholic acid; UDCA, ursodeoxycholic acid.
Conversion of the primary bile acid precursors, trihydroxycholestanoic acid (THCA) and dihydroxycholestanoic acid (DHCA), to cholic and chenodeoxycholic acid, respectively, requires their activation as CoA thioesters and import into the peroxisomes, where their branched side chain undergoes a cycle of β-oxidation (14) (Fig. 5E). This process requires free CoA and releases propionyl-CoA and either chenodeoxycholoyl-CoA (CDCA-CoA) (from DHCA-CoA) or choloyl-CoA (from THCA-CoA), which are then primarily conjugated to taurine in mice. In the Nudt7 livers, the observed drop in the concentration of cholic acid and its derivatives was consistent with a robust decrease in the levels of choloyl-CoA (Fig. 3F). Similar to choloyl-CoA, THCA-CoA is a known substrate of Nudt7 (17); however, the concentration of THCA-CoA was below the detection limit of our assay and we could not determine whether the Nudt7 livers contained less of this bile acid precursor as well. At the gene expression level, Cyp39a1 was the only gene involved in bile acid metabolism that was differentially expressed and, specifically, downregulated, in the Nudt7 livers (supplemental Table S3). Cyp39a1 encodes a microsomal cytochrome P450 enzyme that possesses 7α-hydroxylase activity against 24(S)-hydroxycholesterol (45), but the contribution of this enzyme to bile acid synthesis in the liver is currently unknown.
Effect of Nudt7 overexpression on fatty acid β-oxidation
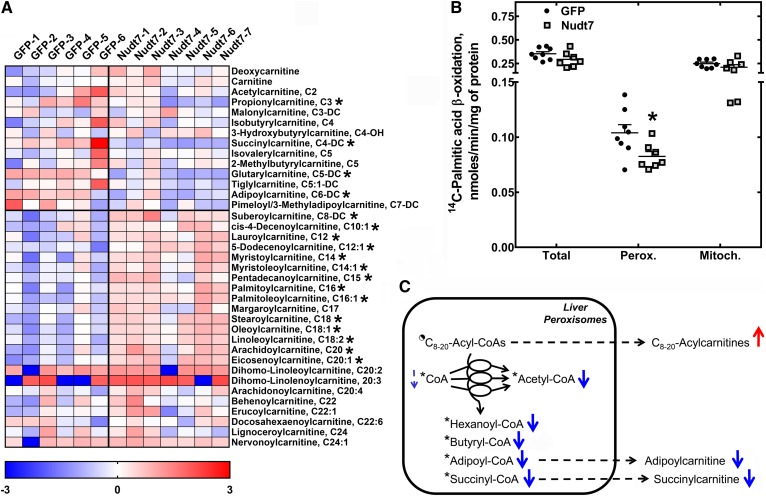
In addition to bile acids, the metabolic profiling of the livers identified 18 acylcarnitines that were significantly changed between Nudt7 and GFP mice in the fasted state (Fig. 6A). Acylcarnitines are in close equilibrium with acyl-CoAs due to the activity of carnitine acyltransferases that transfer the fatty acid from an acyl-CoA to free carnitine to prevent a buildup of acyl-CoAs at the expense of free CoA (46). Among the acylcarnitines significantly changed by the overexpression of Nudt7, short-chain acylcarnitines, such as propionylcarnitine, succinylcarnitine, glutarylcarnitine, and adipoylcarnitine, were significantly decreased, correlating with the global drop in the concentration of short-chain acyl-CoAs observed in the Nudt7 livers (Fig. 3E). Conversely, acylcarnitines with acyl chains between 8 and 20 carbons in length (C8–20-acylcarnitines) were found to accumulate, suggesting increased transfer of medium- and long-chain acyl groups from the correspondent acyl-CoAs to free carnitine. The concentration of free carnitine, which is the most abundant of the carnitine species in the liver, was not affected by these changes in the composition of the acylcarnitine pool (16, 47, 48).
Fig. 6.
Nudt7 overexpression in the fasted state decreases the rate of peroxisomal fatty acid oxidation. A: Heat plot of all the short-, medium-, and long-chain acylcarnitines, plus free carnitine and deoxycarnitine, detected by global metabolic profiling. B: Primary hepatocytes were isolated from fasted Nudt7 and GFP mice and incubated with [1-14C]palmitic acid in the presence or absence of etomoxir, as described in the Materials and Methods. Data are reported as the mean (horizontal lines) of measurements on individual mice (circles and squares) ± SEM. *P < 0.05. C: Schematic representation of the oxidation of medium- and long-chain acyl-CoAs (C8–20-acyl-CoAs) to short-chain acyl-CoAs in the peroxisomes. Known substrates of Nudt7 are marked by an asterisk. The half-filled circle indicates that some C8–20-acyl-CoAs (i.e., octanoyl- and lauroyl-CoA) are good substrates of Nudt7, but some others (i.e., stearoyl-CoA) are not. A decrease in the rate of peroxisomal fatty acid oxidation in the Nudt7 livers correlated with a decrease in the concentration of short-chain acyl-CoAs and correspondent acylcarnitines (thick blue arrows), together with the accumulation of C8–20-acylcarnitines. The decrease in free CoA missed statistical significance (P = 0.07), and this is indicated by a thin dashed arrow. Acylcarnitines derived from peroxisomal acyl-CoAs can be formed directly in the peroxisomes or outside the organelles following the release of the acyl groups as free fatty acids. Unlike acyl-CoAs, which are too bulky, the acylcarnitines formed inside the peroxisomes can leave the organelles (71).
Because this metabolic signature was consistent with a potential decrease in fatty acid oxidation (16, 49, 50), we measured the rate of 14C-labeled palmitic acid oxidation in primary hepatocytes isolated from fasted Nudt7 and GFP mice. To discriminate between the mitochondrial and peroxisomal pathways, we incubated the hepatocytes with and without etomoxir, an inhibitor of mitochondrial carnitine palmitoyltransferase 1 and, thus, mitochondrial fatty acid oxidation. Additionally, we measured the serum concentration of 3-hydroxybutyrate and glucose, as the circulating levels of these metabolites are linked to the oxidation of fatty acids in liver mitochondria through the production of NADH and acetyl-CoA in this subcellular compartment. We did not detect any significant difference in the rate of mitochondrial fatty acid oxidation between groups (Fig. 6B), and this result correlated with similar levels of 3-hydroxybutyrate and glucose in the serum of Nudt7 and GFP mice (Table 1). Conversely, we found that increased Nudt7 activity reduced the rate of fatty acid oxidation in the peroxisomes (Fig. 6B), the subcellular compartment where Nudt7 localized. Peroxisomes are known to preferentially shorten fatty acids instead of completely oxidizing them to acetyl-CoA (51). Indeed, octanoate undergoes only one to two cycles of peroxisomal fatty acid oxidation to produce hexanoate and butyrate (52). Consistent with this notion, the Nudt7 livers accumulated C8–20-acylcarnitines while exhibiting a lower concentration of acyl-CoAs and acylcarnitines with six carbons or less, which are the final products of the oxidation of monocarboxylic fatty acids (hexanoyl- and butyryl-CoA) and dicarboxylic fatty acids (adipoyl- and succinyl-CoA) in the peroxisomes (Fig. 6C). The decrease in the levels of these short-chain acyl-CoAs could have also been further exacerbated by direct degradation of these products by exogenous Nudt7.
Overexpression of Nudt7 led to a 2-fold increase in the expression of Acaa1b, the gene encoding the peroxisomal enzyme, acetyl-CoA acyltransferase 1B, also known as 3-ketoacyl-CoA thiolase B (Table 2). Acca1b catalyzes the last free CoA-dependent step in the oxidation of straight-chain fatty acids (25, 53). No other gene involved in either peroxisomal or mitochondrial fatty acid oxidation was differentially expressed in the Nudt7 livers. Although peroxisomal fatty acid oxidation was blunted in the Nudt7 livers, hepatic triglyceride levels were similar between Nudt7 and GFP mice (Table 1). Interestingly, this was associated with the upregulation of genes involved in the hydrolysis of triglycerides (Ces1g, Ces1f, and Ces2c) (54–56) and monoacylglycerols (Abhd6) (57–59) (Table 2).
Taken together with the effect on bile acid metabolism, the above data indicated that overexpression of Nudt7 predominantly affected peroxisomal lipid metabolism.
DISCUSSION
In this study, we determined the metabolic consequences of increasing the CoA-degrading activity in the liver by overexpressing mouse Nudt7, the most abundant of the currently known CoA diphosphohydrolases in this organ (17, 18, 22). One of our major findings was that the overexpression of the enzyme did not interfere with the increase in total liver CoA levels that occurs in the transition from the fed to the fasted state (Fig. 2E). This supports the conclusion that, upon fasting, the rise in CoA observed at the whole tissue level is driven by increased synthesis (16), with negligible contribution from a decrease in Nudt7 expression and activity (Fig. 1B, C).
The exogenously expressed Nudt7 was correctly localized to the peroxisomes (Fig. 2D). Based on subcellular fractionation studies on rat livers, these organelles are estimated to contain only about 4–10% of the total hepatic CoA, depending on the nutritional state (11, 60). This subcellular localization combined with the fact that overexpression of the enzyme had no significant effect on total liver CoA levels but decreased the concentration of choloyl-CoA, a CoA thioester specifically generated in the peroxisomes, suggests that the activity of this enzyme may have been restricted to the peroxisomal CoA pool. This conclusion is further supported by the second major finding of this study: increased expression of Nudt7 in the fasted state specifically blunted peroxisomal processes, including peroxisomal fatty acid β-oxidation (Fig. 6).
Overexpression of Nudt7 in the fed state, when the endogenous Nudt7 levels are highest, had a very modest effect on the composition of the hepatic acyl-CoA pool. Conversely, overexpression of Nudt7 in the fasted state led to a 15–50% decrease in the concentration of most of the short-chain acyl-CoAs detected (Fig. 3E). Short-chain acyl-CoAs, which comprise a small portion of total liver CoA (15, 60, 61), are excellent substrates for the enzyme (17, 18); thus, increased degradation of these CoA species likely contributed to lowering their concentration in the Nudt7 livers. However, this mechanism alone would not explain the distinctive accumulation of C8–20-acylcarnitines observed in the same samples (Fig. 6A). This metabolic signature was associated with a reduced rate of peroxisomal fatty acid oxidation, which, in turn, likely blunted product formation. Among the products whose concentrations were curtailed were hexanoyl- and butyryl-CoA, which derive from the shortening of straight-chain fatty acids, acetyl-CoA, which is produced at each cycle of fatty acid oxidation, and both adipoyl- and succinyl-CoA, which are the final products of the oxidation of long-chain dicarboxylic fatty acids, a process that preferentially occurs in the peroxisomes (52, 62–64). Propionyl-CoA, whose concentration was also significantly decreased in the fasted Nudt7 livers, can be produced from the oxidation of 2-methyl-branched-chain fatty acids, another process that is restricted to the peroxisomes (14). The accumulation of acylcarnitines with a wide range of acyl chain lengths also suggests that the flux through the whole peroxisomal β-oxidation pathway, and not just at a specific step, was decreased in the Nudt7 livers. This, in turn, correlated with a strong trend toward a lower concentration of free CoA, which is required to complete each cycle of fatty acid oxidation and might have been limiting in the peroxisomes of the Nudt7 livers due to increased hydrolysis (Fig. 3D). Analysis of the acyl-CoA pool composition in the peroxisomes would be required to confirm this hypothesis; however, the measurement of individual CoA species, and even total CoA, in mitochondria-free peroxisomal fractions is still technically challenging in mice due to the small sample size. As observed in other mouse models, inhibition of peroxisomal fatty acid oxidation in the Nudt7 livers was also associated with the activation of PPARα and with increased transcription of genes involved in lipid transport and metabolism (Table 2) (65, 66).
Efficient off-loading of acyl chains from CoA to carnitine likely explains why, unlike medium- and long-chain acylcarnitines, no change in the concentration of the correspondent acyl-CoAs was detected in the Nudt7 livers at the whole tissue level. For octanoyl- and, possibly, decanoyl-CoA, this process could occur inside the peroxisomes by the action of carnitine octanoyltransferase, which is abundant in mouse liver (67–69). For acyl-CoAs with a C10 acyl group or longer, the formation of acylcarnitines would likely be catalyzed by carnitine palmitoyltransferase 1, following hydrolysis of long-chain acyl-CoAs by peroxisomal thioesterases, such as Acot3 or Acot8, and reactivation of the released free fatty acids as acyl-CoAs outside the peroxisomes (27, 67, 69, 70). Because CoA species are too bulky to diffuse through the peroxisomal membrane (71), the presence of multiple peroxisomal thioesterases and carnitine octanoyltransferase allows the acyl group attached to CoA to leave the peroxisomes as free fatty acids or acylcarnitines and enter metabolic pathways in other subcellular compartments. For example, acetate produced from acetyl-CoA inside the peroxisomes contributes to the synthesis of malonyl-CoA (52, 72), whose levels were correspondingly decreased in the Nudt7 livers. Similarly, the succinyl group derived from the oxidation of dicarboxylic fatty acids in the peroxisomes can feed into the TCA cycle (62). In the livers of the fasted Nudt7 mice, the levels of succinyl-CoA and succinylcarnitine were acutely decreased by 40% and 80%, respectively. In spite of this, the concentration of succinate, fumarate, malate, and oxaloacetate was not significantly different between Nudt7 and GFP livers (supplemental Table S1), indicating that anaplerosis from amino acid metabolism was sufficient to support the portion of the TCA cycle that, together with pyruvate carboxylation, produces mitochondrial oxaloacetate for gluconeogenesis. Accordingly, fasting blood glucose levels were similar between Nudt7 and GFP mice (Table 1). Interestingly, Taniguchi et al. (73) identified an association between Nudt7 expression and the redness of pork meat, and subsequently showed that overexpression of porcine Nudt7, which is about 70% identical to the murine isoform, leads to a significant decrease in heme content during rat L6 myoblast differentiation (74). Heme biosynthesis starts from the condensation of succinyl-CoA and glycine in the mitochondria (75). The authors suggested that the effect on heme levels could be due to reduced availability of succinyl-CoA, a conclusion supported by our data. Overall, our results strongly suggest that Nudt7 activity contributes to the regulation of the levels of acetyl-CoA, succinyl-CoA, and other short-chain acyl-CoAs produced by fatty acid oxidation in the peroxisomes.
In addition to peroxisomal fatty acid oxidation, Nudt7 overexpression in the fasted state also altered bile acid metabolism. In particular, the fasted Nudt7 livers contained lower amounts of both primary and secondary bile acids (Fig. 5). This phenotype was associated with a robust decrease in the concentration of choloyl-CoA (Fig. 3F), a bile acid precursor produced via β-oxidation of the 2-methylacyl side chain of THCA-CoA in the peroxisomes (14, 44). Both THCA-CoA and choloyl-CoA are known substrates for Nudt7 (17), and the decrease in the concentration of choloyl-CoA could be due to a combination of mechanisms. These include an increase in direct degradation of choloyl-CoA by Nudt7, increased degradation of THCA-CoA, and/or decreased β-oxidation of THCA-CoA, which could not be detected in our samples. No information is available on the ability of Nudt7 to hydrolyze DHCA-CoA and CDCA-CoA in vitro, and neither compound was detected in the GFP or Nudt7 livers. In spite of this, the robust decrease in chenodeoxycholic acid derivatives, including conjugated and unconjugated α- and β-muricholic acids, suggests the possibility that these CoA thioesters could be Nudt7 substrates as well. Combined, the drop in the availability of choloyl-CoA and the decrease in the hepatic concentration of both cholic acid and chenodeoxycholic acid derivatives suggest that increased Nudt7 activity may have blunted bile acid synthesis by interfering with the final peroxisomal steps of the pathway.
Once synthesized in the liver, bile acids are immediately secreted and stored in the gall bladder until food ingestion leads to the contraction of this small organ to empty the bile into the intestinal tract. Most of the bile acids are then reabsorbed in the terminal ileum, secreted into the portal blood circulation, and transported back to the liver, where the small amount of bile acids lost in the feces is replenished by de novo synthesis (76). Only about 5% of the total bile acid pool needs to be synthesized by the liver at each cycle; thus, a defect in bile acid synthesis would take several fasting-to-refeeding cycles to affect the size of the total bile acid pool. This might explain why, in the fasted Nudt7 mice, the decrease in the bile acid content was only detected at the site of synthesis, but it did not affect the significantly larger pools contained in the gall bladder and small intestine (Fig. 5B–D).
Interestingly, Nudt7 overexpression in the liver also decreased the concentration of secondary bile acids, independent of any compositional change in the intestinal bile acid pool. Secondary bile acids are produced in the intestine by deconjugation and dehydroxylation of primary bile acids by the gut microbiome and arrive at the liver through the enterohepatic circulation (44, 76). Once in the liver, the secondary bile acids are activated as acyl-CoAs and reconjugated to taurine or glycine. Due to the dual localization of the enzyme that catalyzes this amidation reaction, bile acid-CoA:amino acid N-acyltransferase (Baat), the subcellular site of the secondary bile acid conjugation has been debated to be the cytosol or, as for the primary bile acids, the peroxisomes (77–79). Because there was no detectable amount of Nudt7 in the cytosol, the decrease in the concentration of secondary bile acids suggests that at least a portion of the correspondent acyl-CoAs was imported in the peroxisomes for conjugation and was hydrolyzed by Nudt7. Combined with the ability of Nudt7 to hydrolyze choloyl-CoA, this would support the conclusion that Nudt7 and Baat compete for substrates. Bile acids are signaling molecules that bind to multiple G protein-coupled receptors and nuclear receptors, such as the farnesoid X receptor and pregnane X receptor. In spite of this, the decrease in the bile acid content of the liver did not cause any significant changes in the expression of those receptors’ known targets, including small heterodimer partner (Nr0b2, SHP) and cholesterol 7α hydroxylase (Cyp7a1) (44, 76) (supplemental Table S3). Overall, gene expression was only modestly affected in the Nudt7 livers compared with the GFP controls, supporting the conclusion that the observed changes in peroxisomal metabolism were a direct consequence of the increased CoA-degrading activity of Nudt7.
Peroxisomal fatty acid oxidation and bile acid synthesis are normally activated in the fasted state (80, 81), when Nudt7 expression is at its lowest. Altogether, our data support the conclusion that modulation of Nudt7 activity by the nutritional state could contribute to the regulation of the output of both pathways. Analysis of these processes in the livers of Nudt7−/− mice will be required to further corroborate the role of Nudt7 in the regulation of peroxisomal lipid metabolism in this organ. Generation of the Nudt7−/− mice has recently been reported, but the phenotyping of these mice was limited to cartilage and immature murine articular chondrocytes, which, in the presence of phytol, showed decreased levels of catalase, suggesting a connection to peroxisomal function (82). Our data also strongly suggest that Nudt7-dependent CoA degradation may be limited to the quantitatively small pool of peroxisomal CoA. Analysis of the total CoA concentration in the livers of the fed and fasted Nudt7−/− mice will also be important to determine whether Nudt7, and its increased activity in the fed state, is indeed responsible for the decrease in CoA levels that occurs, at the whole tissue level, in the transition from the fasted to the fed state.
Supplementary Material
Acknowledgments
The authors thank Dominique Saporito, Deborah Corbin, and Terence McManus from the West Virginia University Metabolome Analysis Facility, Ryan Percifield from the West Virginia University Genomics Core Facility, and Donald Primerano from the Genomics Core Facility at Marshall University for their expert technical assistance.
Footnotes
Abbreviations:
- AAV
- adeno-associated virus
- ALT
- alanine aminotransferase
- AST
- aspartate aminotransferase
- CDCA-CoA
- chenodeoxycholoyl-CoA
- CE
- collision energy
- CXP
- collision cell exit potential
- DHCA
- dihydroxycholestanoic acid
- HRP
- horseradish peroxidase EP, entrance potential
- M199
- medium 199
- mBB
- monobromobimane
- Nudt
- Nudix hydrolase
- PMP70
- ATP-binding cassette subfamily D member 3
- THCA
- trihydroxycholestanoic acid
This work was supported by West Virginia University School of Medicine Startup Foundation Funding (R.L.) and National Institutes of Health Grants R35GM119528 (R.L.) and F31GM126838 (S.A.S.). The Metabolome and Genomics Core Facilities at West Virginia University and the Genomics Core Facility at Marshall University were supported by the WV Clinical and Translational Science Institute (CTSI) program, which is funded by National Institutes of Health Grant U54GM104942. The West Virginia University Genomics Core Facility was also supported by the West Virginia University Research Corporation and Eberly College of Arts and Sciences. The Bioinformatics Core was funded by WV IDea Network for Biomedical Research Excellence (INBRE), National Institutes of Health Grants P20GM103434 and U54GM104942. The West Virginia University Imaging Facilities were supported by the West Virginia University Cancer Institute and National Institutes of Health Grants P20RR016440, P30RR032138/P30GM103488, and U54GM104942. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Weickert M. O., and Pfeiffer A. F.. 2006. Signalling mechanisms linking hepatic glucose and lipid metabolism. Diabetologia. 49: 1732–1741. [DOI] [PubMed] [Google Scholar]
- 2.Bechmann L. P., Hannivoort R. A., Gerken G., Hotamisligil G. S., Trauner M., and Canbay A.. 2012. The interaction of hepatic lipid and glucose metabolism in liver diseases. J. Hepatol. 56: 952–964. [DOI] [PubMed] [Google Scholar]
- 3.Rui L. 2014. Energy metabolism in the liver. Compr. Physiol. 4: 177–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leonardi R., Zhang Y. M., Rock C. O., and Jackowski S.. 2005. Coenzyme A: back in action. Prog. Lipid Res. 44: 125–153. [DOI] [PubMed] [Google Scholar]
- 5.Choudhary C., Weinert B. T., Nishida Y., Verdin E., and Mann M.. 2014. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat. Rev. Mol. Cell Biol. 15: 536–550. [DOI] [PubMed] [Google Scholar]
- 6.Hirschey M. D., and Zhao Y.. 2015. Metabolic regulation by lysine malonylation, succinylation, and glutarylation. Mol. Cell. Proteomics. 14: 2308–2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Resh M. D. 2016. Fatty acylation of proteins: the long and the short of it. Prog. Lipid Res. 63: 120–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daniotti J. L., Pedro M. P., and Valdez Taubas J.. 2017. The role of S-acylation in protein trafficking. Traffic. 18: 699–710. [DOI] [PubMed] [Google Scholar]
- 9.Sabari B. R., Zhang D., Allis C. D., and Zhao Y.. 2017. Metabolic regulation of gene expression through histone acylations. Nat. Rev. Mol. Cell Biol. 18: 90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horie S., Ishii H., and Suga T.. 1981. Changes in peroxisomal fatty acid oxidation in the diabetic rat liver. J. Biochem. 90: 1691–1696. [DOI] [PubMed] [Google Scholar]
- 11.Van Broekhoven A., Peeters M. C., Debeer L. J., and Mannaerts G. P.. 1981. Subcellular distribution of coenzyme A: evidence for a separate coenzyme A pool in peroxisomes. Biochem. Biophys. Res. Commun. 100: 305–312. [DOI] [PubMed] [Google Scholar]
- 12.Peng Y., and Puglielli L.. 2016. N-lysine acetylation in the lumen of the endoplasmic reticulum: a way to regulate autophagy and maintain protein homeostasis in the secretory pathway. Autophagy. 12: 1051–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cooper D. E., Young P. A., Klett E. L., and Coleman R. A.. 2015. Physiological consequences of compartmentalized acyl-CoA metabolism. J. Biol. Chem. 290: 20023–20031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferdinandusse S., Denis S., Faust P. L., and Wanders R. J.. 2009. Bile acids: the role of peroxisomes. J. Lipid Res. 50: 2139–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leonardi R., Rock C. O., and Jackowski S.. 2014. Pank1 deletion in leptin-deficient mice reduces hyperglycaemia and hyperinsulinaemia and modifies global metabolism without affecting insulin resistance. Diabetologia. 57: 1466–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leonardi R., Rehg J. E., Rock C. O., and Jackowski S.. 2010. Pantothenate kinase 1 is required to support the metabolic transition from the fed to the fasted state. PLoS One. 5: e11107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reilly S. J., Tillander V., Ofman R., Alexson S. E., and Hunt M. C.. 2008. The nudix hydrolase 7 is an acyl-CoA diphosphatase involved in regulating peroxisomal coenzyme A homeostasis. J. Biochem. 144: 655–663. [DOI] [PubMed] [Google Scholar]
- 18.Shumar S. A., Kerr E. W., Geldenhuys W. J., Montgomery G. E., Fagone P., Thirawatananond P., Saavedra H., Gabelli S. B., and Leonardi R.. 2018. Nudt19 is a renal CoA diphosphohydrolase with biochemical and regulatory properties that are distinct from the hepatic Nudt7 isoform. J. Biol. Chem. 293: 4134–4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y. M., Rock C. O., and Jackowski S.. 2006. Biochemical properties of human pantothenate kinase 2 isoforms and mutations linked to pantothenate kinase-associated neurodegeneration. J. Biol. Chem. 281: 107–114. [DOI] [PubMed] [Google Scholar]
- 20.Vallari D. S., Jackowski S., and Rock C. O.. 1987. Regulation of pantothenate kinase by coenzyme A and its thioesters. J. Biol. Chem. 262: 2468–2471. [PubMed] [Google Scholar]
- 21.Rock C. O., Calder R. B., Karim M. A., and Jackowski S.. 2000. Pantothenate kinase regulation of the intracellular concentration of coenzyme A. J. Biol. Chem. 275: 1377–1383. [DOI] [PubMed] [Google Scholar]
- 22.Ofman R., Speijer D., Leen R., and Wanders R. J.. 2006. Proteomic analysis of mouse kidney peroxisomes: identification of RP2p as a peroxisomal nudix hydrolase with acyl-CoA diphosphatase activity. Biochem. J. 393: 537–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gasmi L., and McLennan A. G.. 2001. The mouse Nudt7 gene encodes a peroxisomal nudix hydrolase specific for coenzyme A and its derivatives. Biochem. J. 357: 33–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shumar S. A., Fagone P., Alfonso-Pecchio A., Gray J. T., Rehg J. E., Jackowski S., and Leonardi R.. 2015. Induction of neuron-specific degradation of coenzyme a models pantothenate kinase-associated neurodegeneration by reducing motor coordination in mice. PLoS One. 10: e0130013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Veldhoven P. P. 2010. Biochemistry and genetics of inherited disorders of peroxisomal fatty acid metabolism. J. Lipid Res. 51: 2863–2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wanders R. J., Vreken P., Ferdinandusse S., Jansen G. A., Waterham H. R., van Roermund C. W., and Van Grunsven E. G.. 2001. Peroxisomal fatty acid alpha- and beta-oxidation in humans: enzymology, peroxisomal metabolite transporters and peroxisomal diseases. Biochem. Soc. Trans. 29: 250–267. [DOI] [PubMed] [Google Scholar]
- 27.Hunt M. C., Tillander V., and Alexson S. E.. 2014. Regulation of peroxisomal lipid metabolism: the role of acyl-CoA and coenzyme A metabolizing enzymes. Biochimie. 98: 45–55. [DOI] [PubMed] [Google Scholar]
- 28.Fagone P., Wright J. F., Nathwani A. C., Nienhuis A. W., Davidoff A. M., and Gray J. T.. 2012. Systemic errors in quantitative polymerase chain reaction titration of self-complementary adeno-associated viral vectors and improved alternative methods. Hum. Gene Ther. Methods. 23: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia M., Leonardi R., Zhang Y. M., Rehg J. E., and Jackowski S.. 2012. Germline deletion of pantothenate kinases 1 and 2 reveals the key roles for CoA in postnatal metabolism. PLoS One. 7: e40871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patro R., Duggal G., Love M. I., Irizarry R. A., and Kingsford C.. 2017. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 14: 417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soneson C., Love M. I., and Robinson M. D.. 2015. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000 Res. 4: 1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Love M. I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bligh E. G., and Dyer W. J.. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917. [DOI] [PubMed] [Google Scholar]
- 34.Ivanova P. T., Milne S. B., Byrne M. O., Xiang Y., and Brown H. A.. 2007. Glycerophospholipid identification and quantitation by electrospray ionization mass spectrometry. Methods Enzymol. 432: 21–57. [DOI] [PubMed] [Google Scholar]
- 35.Chong J., Soufan O., Li C., Caraus I., Li S., Bourque G., Wishart D. S., and Xia J.. 2018. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 46: W486–W494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Minkler P. E., Kerner J., Ingalls S. T., and Hoppel C. L.. 2008. Novel isolation procedure for short-, medium-, and long-chain acyl-coenzyme A esters from tissue. Anal. Biochem. 376: 275–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haynes C. A. 2011. Analysis of mammalian fatty acyl-coenzyme A species by mass spectrometry and tandem mass spectrometry. Biochim. Biophys. Acta. 1811: 663–668. [DOI] [PubMed] [Google Scholar]
- 38.Magnes C., Sinner F. M., Regittnig W., and Pieber T. R.. 2005. LC/MS/MS method for quantitative determination of long-chain fatty acyl-CoAs. Anal. Chem. 77: 2889–2894. [DOI] [PubMed] [Google Scholar]
- 39.Klaunig J. E., Goldblatt P. J., Hinton D. E., Lipsky M. M., Chacko J., and Trump B. F.. 1981. Mouse liver cell culture. I. Hepatocyte isolation. In Vitro. 17: 913–925. [DOI] [PubMed] [Google Scholar]
- 40.Zhang W., Sargis R. M., Volden P. A., Carmean C. M., Sun X. J., and Brady M. J.. 2012. PCB 126 and other dioxin-like PCBs specifically suppress hepatic PEPCK expression via the aryl hydrocarbon receptor. PLoS One. 7: e37103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cyphert H. A., Alonge K. M., Ippagunta S. M., and Hillgartner F. B.. 2014. Glucagon stimulates hepatic FGF21 secretion through a PKA- and EPAC-dependent posttranscriptional mechanism. PLoS One. 9: e94996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nathwani A. C., Gray J. T., Ng C. Y., Zhou J., Spence Y., Waddington S. N., Tuddenham E. G., Kemball-Cook G., McIntosh J., Boon-Spijker M., et al. 2006. Self-complementary adeno-associated virus vectors containing a novel liver-specific human factor IX expression cassette enable highly efficient transduction of murine and nonhuman primate liver. Blood. 107: 2653–2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Otto G. P., Rathkolb B., Oestereicher M. A., Lengger C. J., Moerth C., Micklich K., Fuchs H., Gailus-Durner V., Wolf E., and Hrabe de Angelis M.. 2016. Clinical chemistry reference intervals for C57BL/6J, C57BL/6N, and C3HeB/FeJ mice (Mus musculus). J. Am. Assoc. Lab. Anim. Sci. 55: 375–386. [PMC free article] [PubMed] [Google Scholar]
- 44.Russell D. W. 2003. The enzymes, regulation, and genetics of bile acid synthesis. Annu. Rev. Biochem. 72: 137–174. [DOI] [PubMed] [Google Scholar]
- 45.Li-Hawkins J., Lund E. G., Bronson A. D., and Russell D. W.. 2000. Expression cloning of an oxysterol 7alpha-hydroxylase selective for 24-hydroxycholesterol. J. Biol. Chem. 275: 16543–16549. [DOI] [PubMed] [Google Scholar]
- 46.Ramsay R. R., and Zammit V. A.. 2004. Carnitine acyltransferases and their influence on CoA pools in health and disease. Mol. Aspects Med. 25: 475–493. [DOI] [PubMed] [Google Scholar]
- 47.Kivilompolo M., Ohrnberg L., Oresic M., and Hyotylainen T.. 2013. Rapid quantitative analysis of carnitine and acylcarnitines by ultra-high performance-hydrophilic interaction liquid chromatography-tandem mass spectrometry. J. Chromatogr. A. 1292: 189–194. [DOI] [PubMed] [Google Scholar]
- 48.Giesbertz P., Ecker J., Haag A., Spanier B., and Daniel H.. 2015. An LC-MS/MS method to quantify acylcarnitine species including isomeric and odd-numbered forms in plasma and tissues. J. Lipid Res. 56: 2029–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Vlies N., Tian L., Overmars H., Bootsma A. H., Kulik W., Wanders R. J., Wood P. A., and Vaz F. M.. 2005. Characterization of carnitine and fatty acid metabolism in the long-chain acyl-CoA dehydrogenase-deficient mouse. Biochem. J. 387: 185–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spiekerkoetter U., Tokunaga C., Wendel U., Mayatepek E., Ijlst L., Vaz F. M., van Vlies N., Overmars H., Duran M., Wijburg F. A., et al. 2005. Tissue carnitine homeostasis in very-long-chain acyl-CoA dehydrogenase-deficient mice. Pediatr. Res. 57: 760–764. [DOI] [PubMed] [Google Scholar]
- 51.Mannaerts G. P., and van Veldhoven P. P.. 1996. Functions and organization of peroxisomal beta-oxidation. Ann. N. Y. Acad. Sci. 804: 99–115. [DOI] [PubMed] [Google Scholar]
- 52.Kasumov T., Adams J. E., Bian F., David F., Thomas K. R., Jobbins K. A., Minkler P. E., Hoppel C. L., and Brunengraber H.. 2005. Probing peroxisomal beta-oxidation and the labelling of acetyl-CoA proxies with [1-(13C)]octanoate and [3-(13C)]octanoate in the perfused rat liver. Biochem. J. 389: 397–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arnauld S., Fidaleo M., Clemencet M. C., Chevillard G., Athias A., Gresti J., Wanders R. J., Latruffe N., Nicolas-Frances V., and Mandard S.. 2009. Modulation of the hepatic fatty acid pool in peroxisomal 3-ketoacyl-CoA thiolase B-null mice exposed to the selective PPARalpha agonist Wy14,643. Biochimie. 91: 1376–1386. [DOI] [PubMed] [Google Scholar]
- 54.Li Y., Zalzala M., Jadhav K., Xu Y., Kasumov T., Yin L., and Zhang Y.. 2016. Carboxylesterase 2 prevents liver steatosis by modulating lipolysis, endoplasmic reticulum stress, and lipogenesis and is regulated by hepatocyte nuclear factor 4 alpha in mice. Hepatology. 63: 1860–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Okazaki H., Igarashi M., Nishi M., Tajima M., Sekiya M., Okazaki S., Yahagi N., Ohashi K., Tsukamoto K., Amemiya-Kudo M., et al. 2006. Identification of a novel member of the carboxylesterase family that hydrolyzes triacylglycerol: a potential role in adipocyte lipolysis. Diabetes. 55: 2091–2097. [DOI] [PubMed] [Google Scholar]
- 56.Quiroga A. D., Li L., Trotzmuller M., Nelson R., Proctor S. D., Kofeler H., and Lehner R.. 2012. Deficiency of carboxylesterase 1/esterase-x results in obesity, hepatic steatosis, and hyperlipidemia. Hepatology. 56: 2188–2198. [DOI] [PubMed] [Google Scholar]
- 57.Blankman J. L., Simon G. M., and Cravatt B. F.. 2007. A comprehensive profile of brain enzymes that hydrolyze the endocannabinoid 2-arachidonoylglycerol. Chem. Biol. 14: 1347–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao S., Mugabo Y., Iglesias J., Xie L., Delghingaro-Augusto V., Lussier R., Peyot M. L., Joly E., Taib B., Davis M. A., et al. 2014. α/β-Hydrolase domain-6-accessible monoacylglycerol controls glucose-stimulated insulin secretion. Cell Metab. 19: 993–1007. [DOI] [PubMed] [Google Scholar]
- 59.Thomas G., Betters J. L., Lord C. C., Brown A. L., Marshall S., Ferguson D., Sawyer J., Davis M. A., Melchior J. T., Blume L. C., et al. 2013. The serine hydrolase ABHD6 is a critical regulator of the metabolic syndrome. Cell Reports. 5: 508–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Horie S., Isobe M., and Suga T.. 1986. Changes in CoA pools in hepatic peroxisomes of the rat under various conditions. J. Biochem. 99: 1345–1352. [DOI] [PubMed] [Google Scholar]
- 61.Tokutake Y., Iio W., Onizawa N., Ogata Y., Kohari D., Toyoda A., and Chohnan S.. 2012. Effect of diet composition on coenzyme A and its thioester pools in various rat tissues. Biochem. Biophys. Res. Commun. 423: 781–784. [DOI] [PubMed] [Google Scholar]
- 62.Jin Z., Bian F., Tomcik K., Kelleher J. K., Zhang G. F., and Brunengraber H.. 2015. Compartmentation of metabolism of the C12-, C9-, and C5-n-dicarboxylates in rat liver, investigated by mass isotopomer analysis: anaplerosis from dodecanedioate. J. Biol. Chem. 290: 18671–18677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bian F., Kasumov T., Thomas K. R., Jobbins K. A., David F., Minkler P. E., Hoppel C. L., and Brunengraber H.. 2005. Peroxisomal and mitochondrial oxidation of fatty acids in the heart, assessed from the 13C labeling of malonyl-CoA and the acetyl moiety of citrate. J. Biol. Chem. 280: 9265–9271. [DOI] [PubMed] [Google Scholar]
- 64.Suzuki H., Yamada J., Watanabe T., and Suga T.. 1989. Compartmentation of dicarboxylic acid beta-oxidation in rat liver: importance of peroxisomes in the metabolism of dicarboxylic acids. Biochim. Biophys. Acta. 990: 25–30. [DOI] [PubMed] [Google Scholar]
- 65.Seedorf U., Raabe M., Ellinghaus P., Kannenberg F., Fobker M., Engel T., Denis S., Wouters F., Wirtz K. W., Wanders R. J., et al. 1998. Defective peroxisomal catabolism of branched fatty acyl coenzyme A in mice lacking the sterol carrier protein-2/sterol carrier protein-x gene function. Genes Dev. 12: 1189–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fan C. Y., Pan J., Usuda N., Yeldandi A. V., Rao M. S., and Reddy J. K.. 1998. Steatohepatitis, spontaneous peroxisome proliferation and liver tumors in mice lacking peroxisomal fatty acyl-CoA oxidase. Implications for peroxisome proliferator-activated receptor alpha natural ligand metabolism. J. Biol. Chem. 273: 15639–15645. [DOI] [PubMed] [Google Scholar]
- 67.Westin M. A., Hunt M. C., and Alexson S. E.. 2008. Short- and medium-chain carnitine acyltransferases and acyl-CoA thioesterases in mouse provide complementary systems for transport of beta-oxidation products out of peroxisomes. Cell. Mol. Life Sci. 65: 982–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Farrell S. O., Fiol C. J., Reddy J. K., and Bieber L. L.. 1984. Properties of purified carnitine acyltransferases of mouse liver peroxisomes. J. Biol. Chem. 259: 13089–13095. [PubMed] [Google Scholar]
- 69.Miyazawa S., Ozasa H., Osumi T., and Hashimoto T.. 1983. Purification and properties of carnitine octanoyltransferase and carnitine palmitoyltransferase from rat liver. J. Biochem. 94: 529–542. [DOI] [PubMed] [Google Scholar]
- 70.Westin M. A., Alexson S. E., and Hunt M. C.. 2004. Molecular cloning and characterization of two mouse peroxisome proliferator-activated receptor alpha (PPARalpha)-regulated peroxisomal acyl-CoA thioesterases. J. Biol. Chem. 279: 21841–21848. [DOI] [PubMed] [Google Scholar]
- 71.Antonenkov V. D., and Hiltunen J. K.. 2006. Peroxisomal membrane permeability and solute transfer. Biochim. Biophys. Acta. 1763: 1697–1706. [DOI] [PubMed] [Google Scholar]
- 72.Reszko A. E., Kasumov T., David F., Jobbins K. A., Thomas K. R., Hoppel C. L., Brunengraber H., and Des Rosiers C.. 2004. Peroxisomal fatty acid oxidation is a substantial source of the acetyl moiety of malonyl-CoA in rat heart. J. Biol. Chem. 279: 19574–19579. [DOI] [PubMed] [Google Scholar]
- 73.Taniguchi M., Hayashi T., Nii M., Yamaguchi T., Fujishima-Kanaya N., Awata T., and Mikawa S.. 2010. Fine mapping of quantitative trait loci for meat color on Sus scrofa chromosome 6: analysis of the swine NUDT7 gene. J. Anim. Sci. 88: 23–31. [DOI] [PubMed] [Google Scholar]
- 74.Taniguchi M., Hayashi T., Nii M., Yamaguchi T., Fujishima-Kanaya N., Awata T., and Mikawa S.. 2010. Overexpression of NUDT7, a candidate quantitative trait locus for pork color, downregulates heme biosynthesis in L6 myoblasts. Meat Sci. 86: 728–732. [DOI] [PubMed] [Google Scholar]
- 75.Labbe R. F., Kurumada T., and Onisawa J.. 1965. The role of succinyl-CoA synthetase in the control of heme biosynthesis. Biochim. Biophys. Acta. 111: 403–415. [DOI] [PubMed] [Google Scholar]
- 76.Chiang J. Y. 2013. Bile acid metabolism and signaling. Compr. Physiol. 3: 1191–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.O’Byrne J., Hunt M. C., Rai D. K., Saeki M., and Alexson S. E.. 2003. The human bile acid-CoA:amino acid N-acyltransferase functions in the conjugation of fatty acids to glycine. J. Biol. Chem. 278: 34237–34244. [DOI] [PubMed] [Google Scholar]
- 78.Pellicoro A., van den Heuvel F. A., Geuken M., Moshage H., Jansen P. L., and Faber K. N.. 2007. Human and rat bile acid-CoA:amino acid N-acyltransferase are liver-specific peroxisomal enzymes: implications for intracellular bile salt transport. Hepatology. 45: 340–348. [DOI] [PubMed] [Google Scholar]
- 79.Styles N. A., Falany J. L., Barnes S., and Falany C. N.. 2007. Quantification and regulation of the subcellular distribution of bile acid coenzyme A:amino acid N-acyltransferase activity in rat liver. J. Lipid Res. 48: 1305–1315. [DOI] [PubMed] [Google Scholar]
- 80.Hunt M. C., Yang Y. Z., Eggertsen G., Carneheim C. M., Gafvels M., Einarsson C., and Alexson S. E.. 2000. The peroxisome proliferator-activated receptor alpha (PPARalpha) regulates bile acid biosynthesis. J. Biol. Chem. 275: 28947–28953. [DOI] [PubMed] [Google Scholar]
- 81.Shin D. J., Campos J. A., Gil G., and Osborne T. F.. 2003. PGC-1alpha activates CYP7A1 and bile acid biosynthesis. J. Biol. Chem. 278: 50047–50052. [DOI] [PubMed] [Google Scholar]
- 82.Song J., Baek I. J., Chun C. H., and Jin E. J.. 2018. Dysregulation of the NUDT7-PGAM1 axis is responsible for chondrocyte death during osteoarthritis pathogenesis. Nat. Commun. 9: 3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.