Abstract
Pathogenic organisms may be sensitive to inhibitors of sterol biosynthesis, which carry antimetabolite properties, through manipulation of the key enzyme, sterol methyltransferase (SMT). Here, we isolated natural suicide substrates of the ergosterol biosynthesis pathway, cholesta-5,7,22,24-tetraenol (CHT) and ergosta-5,7,22,24(28)-tetraenol (ERGT), and demonstrated their interference in Acanthamoeba castellanii steroidogenesis: CHT and ERGT inhibit trophozoite growth (EC50 of 51 nM) without affecting cultured human cell growth. Washout experiments confirmed that the target for vulnerability was SMT. Chemical, kinetic, and protein-binding studies of inhibitors assayed with 24-AcSMT [catalyzing C28-sterol via Δ24(28)-olefin production] and 28-AcSMT [catalyzing C29-sterol via Δ25(27)-olefin production] revealed interrupted partitioning and irreversible complex formation from the conjugated double bond system in the side chain of either analog, particularly with 28-AcSMT. Replacement of active site Tyr62 with Phe or Leu residues involved in cation-π interactions that model product specificity prevented protein inactivation. The alkylating properties and high selective index of 103 for CHT and ERGT against 28-AcSMT are indicative of a new class of steroidal antibiotic that, as an antimetabolite, can limit sterol expansion across phylogeny and provide a novel scaffold in the design of amoebicidal drugs. Animal studies of these suicide substrates can further explore the potential of their antibiotic properties.
Keywords: anti-amoeba drugs, suicide substrate, sterol biosynthesis
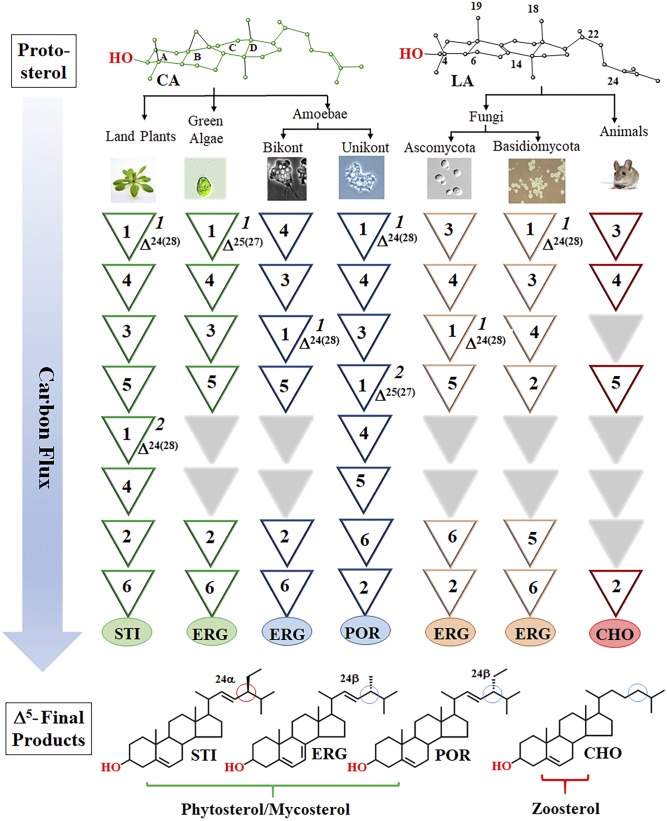
As a result of evolution, sterol synthesis proceeds across eukaryotic kingdoms as a series of organized chemical steps affording flat-amphipathic Δ5-sterols that typically function as an architectural component of membranes (1–3). However, while there are relatively few sterolic genes involved in the conversion of 2,3-oxidosqualene to Δ5-sterol (4–6), there are several lineage-specific enzymes of distinct substrate preference and product specificity that, through their catalytic efficiency, establish the particular metabolite order preserved intact throughout nature (7–10). In some cases, organisms use linear pathways between metabolites that converge to produce the same final product, as in C28-ergosterol (11), whereas others diverge to yield chemical diversity, as in the epimeric pair of C29 α/β-sterols, stigmasterol and poriferasterol (Fig. 1; supplemental Fig. S1A, B) (2). Notably, the pattern of sterol complexity is determined by sterol methyltransferase (SMT), namely, 24-SMT (SMT1), which catalyzes the first C1-transfer reaction, and C28-SMT (SMT2), which catalyzes the second C1-transfer reaction. These enzymes distinguished mechanistically through a coupled methylation-deprotonation reaction that spawns Δ24(28)-olefins or Δ25(27)-olefins (supplemental Fig. S1C) capable of generating branched routes to products different in stereochemistry and supernumerary carbon (2, 12). While the sterol methylation reaction to form the C24-methyl or C28-methyl group in ergostane, stigmastane, and poriferastane skeletons requires common cationic intermediates, the possibility for natural substrate analogs potentiating disruptive SMT activity through misplaced intermediate reactivity remains enigmatic.
Fig. 1.
Variations in metabolite order in sterol biosynthesis. The numbers in inverted triangles indicate one or more enzymatic steps: step 1, sterol 24- or 28-methylation (1, 24-SMT; 2, 28-SMT); step 2, sterol 25(27)- or 24(25)-, or 24(28)-reduction; step 3, sterol C14-demethylation; step 4, - sterol C4-demethylation; step 5, several biosynthetic steps that cluster after C14-demethylation involve B/C-ring metabolism of sterol C8 to C7 isomerization, sterol C5-desaturation, or C7-reduction to give the typical sequence of Δ8 to Δ7 to Δ5,7 to Δ5; step 6, sterol C22-desaturation (compare supplemental Fig. S1A). The Δ5-sterol products: STI, stigmasterol; ERG, ergosterol; CHO, cholesterol. The six representative sterol biosynthesis pathways are based on chemical identifications, labeling studies, and kinetic properties of pure enzymes isolated from land plants, green algae, bikont and unikont amoebae, ascomycete and basidiomycete fungi, and vertebrate animals (2, 8–11).
This revised understanding for plasticity in pathway topology raises new questions about the predicted sensitivities of pathogenic organisms to inhibitors of sterol biosynthesis and the rationale for divergence and disappearance of ancestral SMT in various lineages. In practice, one could identify antimetabolites, otherwise used as anticancer drugs (13), which target chokepoint enzymes in sterol biosynthesis through knowledge of endogenous metabolite structures that control growth. To date, all naturally occurring sterol intermediates are considered safe potential substrates for sterolic enzymes, so long as they possess the appropriate functional group(s) for turnover. Despite that, substrate analogs prepared with a toxic functional group to inactivate enzymes catalyzing the sterol C14-demethylation (CYP51) and sterol C24-methylation (SMT) reactions have proven harmful to protozoa viability by their ability to covalently bind CYP51 or SMT (14–17). These analogs may be considered a synthetic chemotype of antimetabolite for blocking ergosterol biosynthesis via protein alkylation.
During the course of our investigations of the Acanthamoeba castellanii responsible for blinding keratitis and granulomatous amebic encephalitis (16, 17), we observed natural C27- and C28-yeast sterols (supplemental Fig. S2) (10, 18) harboring the uncommon side chain diene group of Δ22,24, which can interfere with trophozoite growth by depleting cells of essential C28- and C29-phytosterols. In contrast, cholesterol supplementation to the medium does just the opposite; it can stimulate amoeba growth without effect on steroidogenesis. Intriguingly, feeding the steroidal Δ22,24-dienes to human epithelial kidney (HEK) cells has no effect on growth or cholesterol biosynthesis. The importance of these heretofore unrecognized observations is twofold. Foremost, Δ22,24-sterols, recognized now as a new class of antibiotic, could affect metabolic challenges as variables in sterol genealogy/biosynthesis driven by SMT gene gain. Likewise, these antimetabolites could replace intermediates or serve as product to compromise an evolving cholesterol biosynthesis pathway in animals capable of Δ22-introduction but constrained by reduced SMT gene expression or loss (19–21). Here, we report a comprehensive picture for SMT as a key mechanistic node to parasite termination and establish that substrate mimics synthesized in yeast as steroidal antimetabolites in Acanthamoeba potentially exist in the biosynthetic toolkit of other species to interfere with the normal metabolic processes within pathogenic organisms. Quite unexpectedly, we found the newly identified fungal antibiotics capable of protein alkylation in amoeba sterol biosynthesis provide a mechanism to limit the C28-/C29-sterol assemblage across phylogeny.
MATERIALS AND METHODS
Materials
The source of reagents and sterol substrates/standards cycloartenol (soybean seed), 24(28)-methylene lophenol (corn pollen), cyclolaudenol (Chlamydomonas reinhardtii cells), 24(28)-methylene cycloartanol (product of cloned soybean 24-SMT), cholesta-5,7,24-trienol (CTO) (ERG6 yeast mutant), cholesta-5,7,22,24-tetraenol (CHT) (ERG6 yeast mutant), ergosta-5,7,22,24(28)-tetraenol (ERGT) (product of cloned Saccharomyces cerevisiae SMT), cholesta-5,7,22-trienol (from incubation of GL7 yeast mutant with cholesta-5,7-dienol), ergosterol (A. castellanii cells), ergosta-5,7,24(28)-trienol (Trypanosoma brucei cells), 7-dehydroporiferasterol (A. castellanii cells), protothecasterol (Prototheca wickerhamii cells), and other ergostane, stigmastane and poriferastane monols from our sterol collection (19, 22, 23) shown in supplemental Table S1 (A. castellanii or C. reinhardtii). The [3H3-methyl]SAM (10 μCi/μM), [2H3-methyl]SAM (99.3% 2H3 enrichment-S/D/N isotopes; Ponte-Claire), and SAM chloride salt together with chromatographic materials, the Bradford protein assay kit, and the QuikChange site-directed mutagenesis kit (Strategene) were as described (19, 22). The [28-2H2]ERGT was prepared from deuterated SAM paired with CHT incubated against 24-AcSMT.
Metabolite analysis and structure identification
Amoeba cultures or HEK cells were harvested at one or more points during growth in the presence and absence of steroidal inhibitor. Cell pellets were split with an internal standard of 5α-cholestane added to one of the cell pellets for determination of sterol amounts in cells. Cells were saponified with 10% methanolic KOH and extracted with hexanes to yield a nonsaponifiable lipid fraction (NLF). The neutral lipids were analyzed by GC-MS and, for some samples, by TLC or HPLC-UV against cholesterol as a chromatographic reference. Ac strain ATCC 30010 was cultivated in tissue culture T-25 ml flasks, prepared with ATTC medium 712 in amounts of 10 ml, and then inoculated with pure trophozoites and incubated at 25°C statically with daily hand shaking. After a 4 day incubation, cells were pelleted and washed with phosphate buffer (10 ml ×3). HEK cells from ATCC were cultured in RPMI medium supplemented with 20% heat-inactivated fetal bovine serum in humidified atmosphere containing 5% CO2 as described (15). Growth was monitored by determining fresh weights of pellet and cell count with a hemocytometer. Pelleted cells were saponified in aqueous methanolic KOH (10% KOH, 5% water, 85% methanol, w/v) at reflux temperature for 1 h to produce a NLF of total sterol and a mixture of steroidal monols and steroidal diols.
Instrumental methods have been reported previously (11, 16, 17, 19). Briefly, proton and carbon NMR spectra were recorded in CDCl3 at ambient temperature using a 500 MHz spectrophotometer with the chemical shifts referenced to chloroform resonating at 7.265 ppm and reported as δ in parts per million. Mass spectra were obtained on a Hewlett-Packard 6890 GC-HP 5973 MSD instrument (electron impact, 70 eV; scan range, m/z 50–550 amu). HPLC was carried out at room temperature using a Phenomenex Luna C18-column (250 mm × 4.6. mm × 5 μM) connected to a diode array multiple wavelength diode array detector with 5% aqueous methanol as eluant. Capillary GC (0.25 mm internal diameter, by 30 m fused silica column coated with Zebron ZB-5 from Phenomenex) was operated at a flow rate of He set at 1.2 ml/min, injector port set at 250°C, and a temperature program of initial 170°C, held for 1 min and increased at 20°C/min to 280°C. Retention times of sterols were normalized to their retention time relative to that of cholesterol in GC (RRTc) of 13.8 min (or a bit longer to 14.5 min subject to column clipping) or in HPLC (αc) of 20 min. and were compared with those of authentic standards in our sterol collection.
In sterol analysis, product distributions were determined by approximate integration of chromatographic peaks. The sterol was routinely examined as the 3-OH compound; but in some cases to show the number of hydroxyl groups in the structure, total sterol in the NLF was prepared as the TMS derivative as follows: The extracted sterol was converted to the TMS ester using 15 μl of N,O-Bis(trimethylsilyl)trifluoroacetamide with trimethylchlorosilane (99:1) and 15 μl of pyridine as catalyst at room temperature for 5 min. The resulting TMS derivative (1 μl) was injected directly into the gas chromatograph mass spectrometer for analysis.
Time-kill and washout assays
Minimum amoebicidal concentration (MAC) (defined as the lowest concentration of inhibitor with no visible live trophozoites), IC50 (50% effective dose value of inhibitor), and time-kill assay were determined at 25°C in 24-well plates containing a total volume of 1 ml as previously described (17). Experiments were performed in triplicate (or in duplicate with excellent agreement between biological replicates of less than 10% difference in cell count) using a light microscope to count cells and trypan blue staining to establish cell death. Target vulnerability can be assessed through cell washout experiments where free drug (or in our case, steroidal inhibitor) is “removed” from the system by washing or dilution, while continued occupancy of the drug after washing can be due to protein alkylation. Here, 25 ml T-flasks supplemented with 10 ml of medium and treated with 10 μM of exogenous sterol were inoculated with 5 × 104 cells/ml and cultured for 96 h to early growth arrest, which for control yields 1–5 × 106 cells/ml. Arrested cultures were pelleted. A 10-fold serially diluted suspension was used as inoculum for further growth experiments in 24-well plates prepared as in the MAC experiments. An untreated sample was used as a negative control. At 96 h growth, following inoculation of 106 cells/ml, 105 cells/ml, 104 cells/ml, and 103 cells/ml, cell number was determined using a hemocytometer. Experiments were performed in triplicate (N = 3 ± 10%). In a separate experiment, [28-3H2]ERGT, prepared from CHT and [3H3-methyl]SAM against cloned 24-AcSMT then purified by HPLC, was added to cell cultures at 20 μM. The cultures were inoculated with 5 × 104 to 1 × 105 cells/ml and grown for 4 days, and then the cells were pelleted for further processing.
Kinetic and product determination of AcSMT catalysis
General methods for heterologous expression, lysate preparation, steady-state experiments for substrate conversion to product, and inhibitor (IC50) determination were as reported (16, 17). Assays were performed at 35°C for 45 min (predetermined initial velocity conditions) in triplicate with less than 10% deviation. Conversion of the IC50 value (based on dose-response plots of increasing inhibitor concentration ranging from 4 to 150 μM against saturating amounts of 100 μM native substrate cycloartenol for 24-AcSMT; Km = 48 μM or 24(28)-methylene lophenol for 28-AcSMT; Km = 25 μM) and 150 μM SAM to Ki value was accomplished using the Cheng-Prussoff equation (12). To promote maximum conversion of substrate by SMT catalysis, preparative incubations were carried out overnight with saturating sterol (100 μM) and excess SAM (300 μM) against 2.5 or 5.0 mg/ml total lysate protein.
Site-directed mutagenesis and AcSMT purification
The A. castellanii sterol C24-methyltransferase genes (24-SMT, XP_004336540 and 28-SMT, XP_004335307) were synthesized by Eurofin MWG Operon (Huntsville, AL) incorporating an Nde1 restriction site at the 5′ end and a BAMHI restriction site at the 3′ end of the open reading frames. Genes were cloned in pET11a expression vector (Novagen, Madison, WI). Gene integrity was verified by PCR using gene-specific primers and by DNA sequencing. To ease protein purification, two new constructs of 24-AcSMT and 28-AcSMT were prepared using pET30b vector with 6xHis3 tags on both termini and an S-tag on the N terminus to enhance protein solubility. The vectors were transferred to E. coli BL21 (DE3) cells for protein expression. Cells were grown in Luria-Bertani broth (pH 7.5) supplemented with kanamycin (50 μg/ml) and grown for 3.5 h at 30°C at 200 rpm shaking. Expression was induced by the addition of IPTG (400 uM) followed by incubation for another 18 h. Protein isolation and lysate preparation were performed according to our previous methods (16–18). The activities of the new recombinant SMTs were similar to those of their nontagged counterparts, indicating that the native conformation of wild-type enzyme was retained following protein expression of His12-tagged SMT.
SMT was purified using Ni-NTA chromatography as follows: The total broken cell soluble preparation (25,000 g supernatant) of 5–20 mg total protein was loaded onto HisPur NTA resin (2 ml) packed into a HisPur Ni-NTA spin column (Thermo Scientific) and washed with high salt buffer to remove unspecific binding, then low salt washing to remove non-His-tagged protein (total 200 ml). The SMT was eluted with a step-wise gradient by elution buffer containing imidazole concentrations of 50, 75, 100, and 150 mM (2 ml each), respectively. The levels of expression, purity, and size of the recombinant protein were monitored chromatographically on a 10% SDS-PAGE gel electrophoresis followed by Coomassie Blue R250 staining. Total protein concentrations were determined by the Bradford method with commercial reagents (Bio-Rad) using bovine γ-globulin as standard. Preliminary evaluation of the enzyme activity from lysate preparation by GC-MS analysis confirmed that the cloned enzymes harboring the two His-tags were equally active to those of wild-type protein. The molecular mass of the new proteins differed: wild-type 24-AcSMT, 39.0 kDa and 28-AcSMT, 39.3 kDa versus His6-constructs of 24-AcSMT, 45.7 kDa and 28-AcSMT, 46.2 kDa. Point mutations were generated using the QuickChange II site-directed mutagenesis kit (Agilent Technologies) according to the manufacturer’s instructions from 24-AcSMT and 28-AcSMT wild-type plasmids that contained two His6-tags, one at the N terminus and the other at the C terminus. The oligonucleotide sequences of mutagenic primers are listed in supplemental Table S2. Mutations were introduced at position 60 of 24-AcSMT and position 64 of 28-AcSMT of Phe or Leu to replace the conserved amino acid tyrosine (supplemental Fig. S10, supplemental Table S2). In contrast to wild-type SMTs, which expressed to similar levels, the His12-tagged 28-AcSMT expressed to about 50% level of 24-AcSMT (supplemental Fig. S11).
Inactivation studies
Partitioning experiments to measure the C24-methyl monol sterol [ergosta-5,7,22,24(28)-tetraen-3-ol and ergosta-5,7,22,25(27)-tetraen-3-ol] to C24-methyl diol sterol [ergosta-5,7,23(24),3,22-diol] ratio were carried out in standard assay conditions at 10 min, 20 min, 45 min, 1 h, 2 h, 3 h, 6 h, and 24 h using 100 μM substrate and 150 μM SAM. The reaction was terminated by addition of methanolic KOH and the total sterol recovered in the usual manner for GC-MS analysis. Quantification of the monol and diol products was determined by measuring the abundance of each ion (expressed as area under the curve) for the relevant molecular ion in the mass spectrum generated by the selected ion monitoring (SIM) technique at the high mass end for M+ 380 (substrate), 394 (product), and 412 (product) amu, respectively. The enzyme-generated sterol composition of the incubation mixture was analyzed by GS-MS.
To measure time- and concentration-dependent generation of C24-methyl monol C28-sterol [ergosta-5,7,22,24(28)-tetraen-3-ol and ergosta-5,7,22,25(27)-tetraen-3-ol] to C24-methyl diol sterol [ergosta-5,7,23(24),3,22-diol] from CHT incubation with 28-AcSMT, assays were carried out under initial velocity conditions using the standard protocol at 10 min, 20 min, 45 min, 1 h, 2 h, 3 h, 6 h, and 24 h of lysate: SMT (approximately 100 μg SMT per assay) was varied from 5 to 50 μM against fixed CHT at 100 μM and SAM at 150 μM. These conditions limit SAM and therefore limit the enzyme from generating C29-sterol, thereby simplifying the analysis. Time-course studies were carried out from 0 to 300 min. The reaction was terminated by addition of methanolic KOH and the total sterol recovered in the usual manner for GC-MS analysis. Quantification of the monol and diol products was determined by measuring the abundance of each ion (expressed as area under the curve) for the relevant molecular ion in the mass spectrum generated by the SIM technique at the high mass end for M+ 380 (substrate), 394 (product), and 412 (product) amu, respectively.
For separate tests of enzyme inactivation, small scale (600 ml) incubations of SMT performed under standard assay conditions against 100 μM CTO as preferred substrate because it can bind effectively to both 24-AcSMT and 28-AcSMT and is close in structure to the analog inhibitor. Substrate protection against SMT2 was determined by preincubation of CTO or 24(28)-methylene lophenol at 50 μM and 100 μM concentrations for 10 min, and then 20 μM of analog CHT was added to the reaction vial and incubated for another 1 h at 35°C. The reaction was terminated by the addition of methanolic KOH. The resulting organic extract was analyzed for product formation by GC-MS.
Covalent binding experiments
A soluble preparation containing 100 μM ERGT or DHL, 300 μM SAM amended with catalytic amounts of [3H3-methyl]SAM (3 × 106 dpm), and 5 mg total protein (approximately 250 μg 28-SMT or 500 μg 24-SMT) was incubated overnight under standard assay conditions, affording a 3H-analog-SMT complex. The sterol-protein complex was purified using Ni-NTA chromatography eluted with imidazole in 5 ml fractions as described above. An aliquot of each fraction (200 μl) was added to scintillation fluid and counted on a scintillation counter. Most radioactivity (>80–95%, depending on the source of SMT) was recovered in the wash.
Proteomic analysis
His12-tagged recombinant wild-type 28-AcSMT and 28-AcSMT complexed with analog were purified using the standard protocol. The imidazole was removed from the eluted protein solution and the buffer was changed to 20 mM Tris-HCl (pH 8.0) using ultrafiltration. After buffer change, the protein was diluted to 1 mg/ml (based on the calculation of absorption at optical density at UV 280) and incubated with ERGT and SAM using the standardized assay condition as described (22). A parallel assay with sterol and no SAM was used as control. After overnight incubation at 35°C, the assay mixture was subjected to Ni-NTA to remove loosely bound sterol and SAM. The pure alkylated and control proteins were concentrated followed by a buffer change to 50 mM ammonium bicarbonate buffer (pH 8.0). The protein concentrations were determined at OD280 and purity confirmed by SDS-PAGE. The molecular mass of both native and alkylated proteins was determined using an MDS SCIEX 4800 MALDI TOF/TOF™ analyzer (Applied Biosystems, Foster City, CA) with sinapinic acid as matrix for assisting ionization.
RESULTS
Unmasking amoeba steroidogenesis vulnerability to substrate mimicry
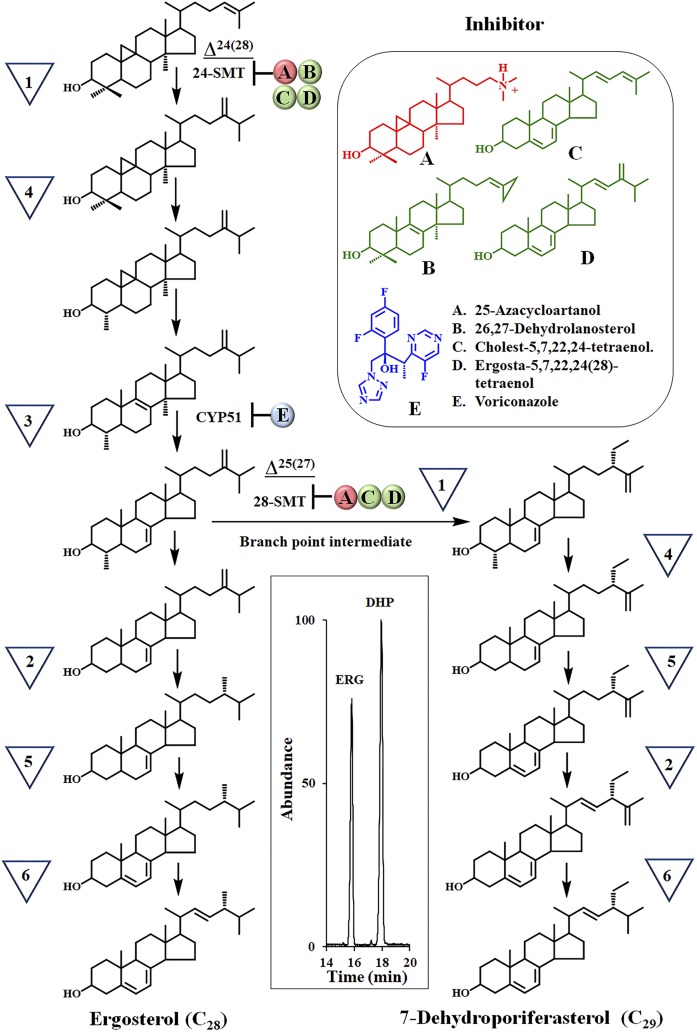
Our previous studies of A. castellanii cells revealed that: i) two functional classes of SMT can operate in tandem, such that the Δ24(28)-olefin pathway catalyzed by SMT1 precedes the Δ25(27)-olefin pathway catalyzed by SMT2; ii) C29-sterol biosynthesis proceeds as a branch pathway of the canonical C28-sterol biosynthesis pathway to ergosterol (Fig. 2); and iii) the C28- and C29-sterol composition can undergo marked changes as trophozoites (that synthesize ergosterol and 7-dehydroporiferasterol) differentiate into resting cysts (that synthesize brassicasterol and poriferasterol) or lyse/die (from synthesis of 6-methyl aromatic sterols) (16, 17). We further discovered that the proportion of C28- and C29-sterols at growth arrest can change by supplementing the medium with either a tight binding inhibitor, such as 25-azacycloartanol, or a suicide substrate inhibitor, such as 26,27-dehydrolanosterol, that selectively target the AcSMTs that, thereby, kill trophozoites (Fig. 2, supplemental Fig. S1C).
Fig. 2.
A. castellanii sterol biosynthesis pathway. The inverted triangles represent biosynthetic steps as defined in Fig. 1. Inhibitors were tested against Ac cell growth and isolated enzymes, SMT and CYP51, as reported in (16, 17) and results of studies herein. The inset is a GC profile of total sterols isolated from 4 day growth-arrested trophozoites. ERG, ergosterol; DHP, 7-dehydroporiferasterol.
Initially, 10 μM of cholesterol in the form of fetal bovine serum or 10 μM solubilized in DMSO were tested against Ac trophozoites cultured in T-flasks and observed to stimulate growth (supplemental Fig. S3). GC-MS analysis of 4 day growth-arrested Ac cultures showed that cholesterol from the serum-treated cells represents about 15% total sterol, while the endogenous amoeba sterol composition remain unchanged. CTO supplemented to the medium at 10 μM had no effect on growth or amoeba steroidogenesis. In contrast, Ac cultured on 10 μM yeast sterol, CHT or ERGT, for 4 days inhibited cell proliferation, affording approximately 15% (±5%) of the cell number or fresh weight of the pellet relative to control. GC-MS analysis of total sterols showed the C28-ergosterol to C29-7-dehydroporiferasterol ratio in the control was 44:55 [Fig. 2, inset (GC profile)], while the C28-ergosterol to C29-7-dehydroporiferasterol ratio in CHT- and ERGT-treated cells was slightly modified, 36:64 and 38:62, respectively; the amount of endogenous ergosterol and 7-dehydroporiferasterol in treated cells decreased by approximately 15%, while approximately 10% of the dietary sterol was accumulated into the cell. Although these growth experiments of treated cells were repeated several times and the sterol composition of 4 day cells analyzed by GC-MS, in no case was there detectable substrate accumulation of either cycloartenol or 24(28)-methylene lophenol or of steroidal diol formation, even when using SIM of the relevant ions to check for trace sterol in the chromatogram.
The [28-3H2]ERGT was prepared enzymatically from CHT and 3H-SAM (specific activity 1 × 105 dpm/70 μg) and incubated with trophozoites cultured as above for 4 days. The radioactivity of the resulting cell pellet was initially 1 × 104 dpm. After five washes against phosphate buffer, the cell pellet radioactivity dropped to 0 dpm, showing excess 3H completely removed by the washout. The pellet was then extracted with methanol/chloroform to remove 3H-sterol accumulated by cells yielding 2.6 × 103 dpm. These results indicate approximately 9% uptake of 3H-sterol into the cell, which is in good agreement with the quantification of ERGT into the cells by GC-MS analysis above. The pellet was subsequently saponified in aqueous methanol KOH and then extracted with hexane yielding an organic extract of 286 dpm, consistent with protein alkylation. In a separate experiment, we investigated to determine whether an extended duration of action can be observed through the growth response even when the drug has been cleared. Here, control and treated cells were supplemented with 10 μM ERGT and incubated for 4 days as in the radioactivity experiment. The resulting cell pellet was washed to remove unbound analog, and then divided into three samples of inoculum of 1 × 103 cells/ml, 1 × 104 cells/ml, and 1 × 105 cells/ml added to 1 ml of medium in a 24-well plate system. The cells were cultured for 96 h to permit adequate time for 100% growth recovery to 1 × 106 cells/ml. At the conclusion of the growth experiment, the percentage of cells in the treated cultures compared with control was 0, 20, and 40%, respectively. Thus, selectivity for target engagement was confirmed.
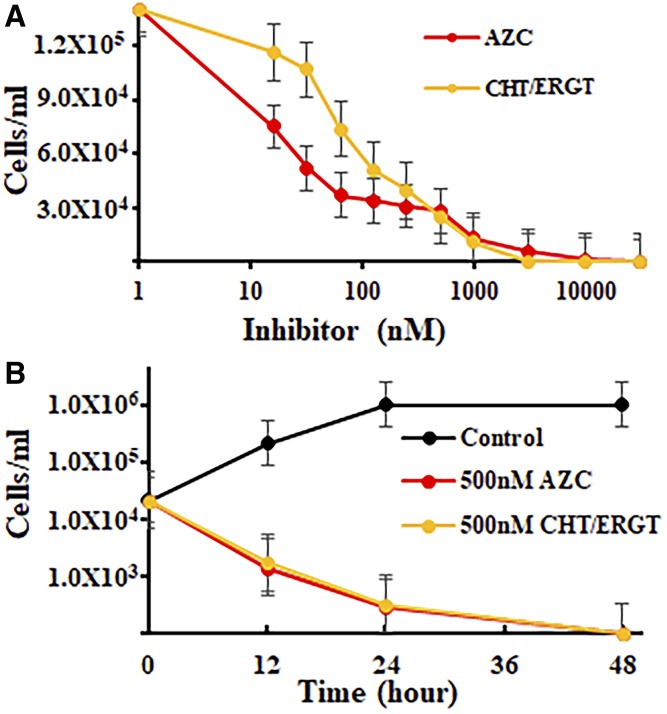
Given these results, we determined the Ac growth response to CHT and ERGT using the standard protocol of amoebae seeded into 24/96-well plate systems. We used this system successfully in our evaluation of medical azoles and steroidal transition state analogs that bind CYP51 and SMT (16, 17, 24). Both CHT and ERGT generated IC50s of approximately 51 nM and a MAC of approximately 5 μM, while 25-azacycloartanol, used as reference (17), inhibits growth with similar potency (Fig. 3A). Alternatively, time-kill studies of CHT or ERGT led to cell death by 48 h (Fig. 3).
Fig. 3.
A. castellanii growth response to the suicide substrates, CHT and ERGT, in comparison to our published report of inhibition by 25-azacycloartanol (AZC) (17). The SEMs are less than 5% of three independent experiments. Ac trophozoites (90–95% pure) were inoculated into 24-well plates as described in the Materials and Methods at varied concentrations of inhibitor (A) or at fixed concentration of inhibitor (B).
CHT and ERGT: neutral effect on cultured HEK cell growth and cholesterol biosynthesis
CHT and ERGT were incubated in HEK cells and showed no effect on cell proliferation to 40 μM. GC-MS analysis of 40 μM treated HEK cells showed that both dietary supplements were accumulated by cells (supplemental Fig. S3). However, while neither compound had an effect on cholesterol biosynthesis, both of them were metabolized to a steroidal triene, as noted by a gain in two hydrogen atoms (M+ 380 to M+ 382) (supplemental Fig. S5) (17–25). MS showing loss of the m/z 143/157 fragment for the nucleus Δ5,7-system (2, 25) and ions at 109/269/271 amu showing retention of the conjugated diene side chain system support that metabolism occurred in the nucleus. Moreover, the chromatographic mobility of cholesta-5,22,24-trienol is different from authentic samples of 5,7,22-trienol and CTO, while the chromatographic mobility of ergosta-5,22,24(28)-trienol is different from authentic samples of ergosta-5,7,22-trienol and ergosta-5,7,24(28)-trienol (supplemental Table S1). These results suggest that the cholesterol biosynthesis pathway in HEK cells utilizes a Δ7-sterol reductase enzyme in metabolism of CHT and ERGT, while the Δ24-reductase fails to bind the two yeast sterols productively. Comparing the IC50 growth values for CHT or ERGT supplied to Ac with the growth response of HEK cells to CHT or ERGT at 40 μM, the highest concentration tested affording no effect on growth, yields a selective index of approximately 103, which shows that the steroidal inhibitors are of equal potency to azolic CYP51 inhibitors as anti-amoeba agents (24).
Leveraging SMT catalysis for steroidal antibiotic characterization
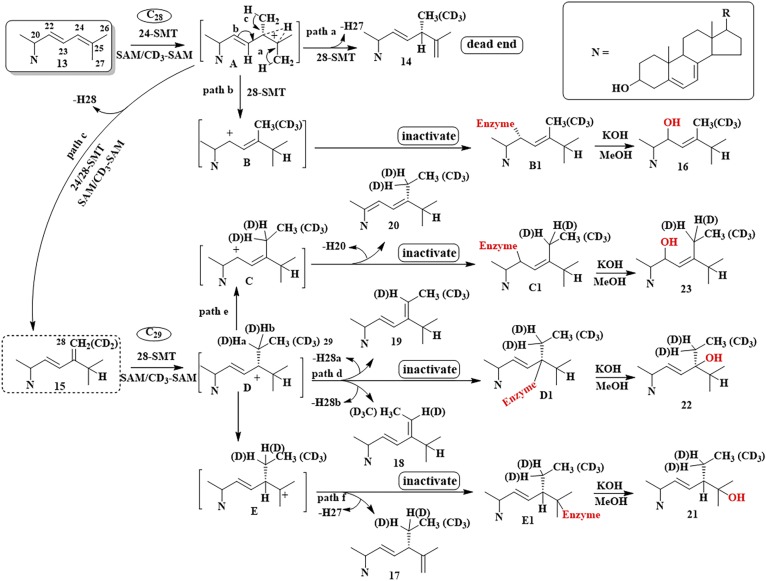
With the availability of our previously cloned 24-AcSMT and 28-AcSMT (9), we could determine the structural specificity of the enzymes to substrate analogs and the sterol methylation reaction pathway that determine turnover versus protein alkylation. As a control, we started with 24-AcSMT exposed to CTO, which does not possess a Δ22-bond. GC-MS analysis of the sterol region from 13 to 20 min revealed that the substrate converts to a single product, ergosta-5,7,24(28)-trienol 9 (100%) (supplemental Fig. S6). However, 28-AcSMT incubated with CTO generates C28- and C29-sterol products representing the first and second C1-transfer reactions, consistent with the established SMT2 substrate acceptance of CTO (9). The product of the first C1-transfer reaction is the same as in 24-AcSMT catalysis. The C28-product then undergoes a second C28-methylation yielding a triplet of C29-steroidal monol 10, 11, and 12 products with the Δ25(27)-olefin 10 representing the major product (supplemental Figs. S6, S7). To determine whether the individual C1- and C2-transfer reactions are dependent on a common cationic intermediate, we conducted an isotopically sensitive branching experiment of 24-AcSMT and 28-AcSMT incubated with CTO paired with [2H3-methyl]SAM. Against 24-AcSMT, CTO converts to [28-2H2]ergosta-5,7,24(28)-trienol in 100% yield. However, against 28-AcSMT, the amount of enzyme-generated C29-sterol is reduced markedly in the product mixture to 50% and the [28-2H]C29-olefinic product ratio altered such that Δ24(28)-olefin formation 11/12 is suppressed, while Δ25(27)-olefin formation 10 is increased (supplemental Figs. S1C, S6). The results show a strong kinetic isotope effect on C28 deprotonation mated to the intermediate C24-cation that determines product turnover in the step-wise C2-transfer reaction. The inability to detect a KIE in the first C1-transfer reaction is consistent with a nonstop methylation-deprotonation reaction pathway at this level of sterol methylation. Intriguingly, the kinetic disruption did not alter branching in the direction of enzyme inactivation from which steroidal diol should be evident by GC-MS analysis of the saponified sample.
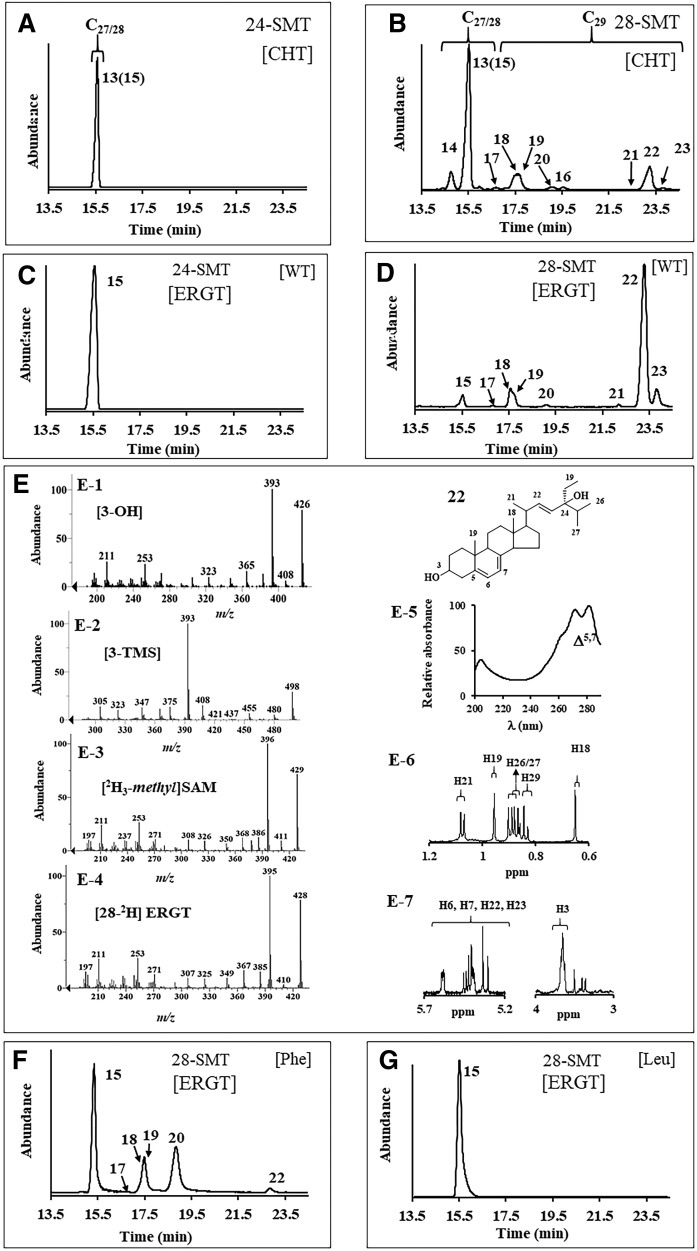
Next, 24-AcSMT and 28-AcSMT were assayed with CHT and ERGT. GC-MS analysis of enzyme-generated sterol profiles show that 24-AcSMT converts CHT to ERGT in 50% yield (based on the ratio of molecular ions m/z 380 and 394, Fig. 4A) without any accompanying byproduct, in agreement with our incubation of CHT with yeast S. cerevisiae SMT, which catalyzes the Δ24(28)-olefin pathway (data not shown) yielding ERGT in 100% yield; ERGT is not productively bound to either 24-AcSMT (Fig. 4C) or ScSMT (data not shown). Alternatively, 28-AcSMT converts CHT to a set of expected C28- and C29-sterols in the “sterol region” of the chromatogram between 13 and 20 min and a second set of unexpected peaks that elute late in the GC chromatogram between 19.5 and 24 min (Fig. 4B). Mass ions for these new compounds corresponded to hydroxylation derivatives suggesting C28- and C29-steroidal Δ22-diols (25) (supplemental Fig. S5). These diols appear only in GC analysis following saponification of cell extract, while chloroform-methanol extractions yield a mixture of steroidal monols. Consequently, we have adopted the SMT-generated diol product as an indicator of protein alkylation.
Fig. 4.
Chromatographic and spectral analysis of SMT-generated C28- and C29-sterol products. A–D: GC profile of total sterol from 24-AcSMT or 28-AcSMT incubated with CHT or ERGT. E: Mass spectrum of compound 22 prepared as indicated for panels E-1 to E-4. Panel E-5 is the UV spectrum of 22, while panels E-6 and E-7 are the NMR with assignment of 22. F, G: GC profiles for total sterol generated from mutant 28-AcSMT assayed with SAM and ERGT.
Incubation of 28-AcSMT with CHT led to six steroidal monols eluting in GC between 14 and 19.5 min, each of which possess a similar parent M+ peak, m/z 394 (from C24-methylation) or 408 (from C28-methylation). In GC, the isomeric C28- and C29-sterol products showed relevant ions at M+, M+-CH3, M+-H2O, and M+-CH3-H2O, and a base peak diagnostic of their side chain structures as reported (2, 23, 25) (supplemental Fig. S5). MS and GC RRTc values of compounds 14 and 15 matched those of authentic standards (16, 17, 26, 27), while the identification of the other compounds required more analysis. In HPLC, 17, 18, 19, and 20 elute as a wide-fraction αc 1.65–1.70. The sterols possessed a similar fingerprint λmax at 282 nm typical of a Δ5,7-system in the nucleus. The UV spectrum for 17 also possessed end absorption showing the side chain double bonds were not in conjugation, while the UV spectra for 18, 19, and 20 possessed an additional λmax at approximately 230 nm, indicative of a conjugated double bond in the side chain, which, for biosynthetic reasons, placed the double bonds toward C20 (supplemental Table S1). Product 20 possessed an electron impact mass spectrum showing an ion at m/z 137 for fragment C10H17 (not present in MS of 17, 18, or 19) representing cleavage of the side chain through the C17(20) bond due to a double bond at C20(22) (supplemental Fig. S5). This product 20 could be unambiguously distinguished from its isomers, 18 and 19, through incubation of 28-AcSMT with [28-2H2]ERGT and SAM. Enzyme-generated 28-2H2 20 retains two deuterium atoms in the side chain (M+ 410), proving that deprotonation does not occur at C28, while 28-2H 18 and 28-2H 19 each have lost one deuterium atom in the side chain, proving deprotonation at C28 (M+ 409) (supplemental Fig. S8). For mechanistic reasons, sterol methylation to form 18 and 19 follows the Δ24(28)-olefin pathway while 20 formation involves a new route (Fig. 5).
Fig. 5.
Proposed sterol methylation pathways of Δ22,24-sterol substrates incubated with 24-SMT and 28-SMT enzymes. As discussed in the text, SMTs that favor the Δ24(28)-olefin route are more likely to forward these substrates to productive products, while SMTs that favor the Δ25(27)-olefin route are more likely to forward these substrates into protein alkylation.
In the case of the putative steroidal diols eluting late in the GC trace, MS analysis of 21, 22, and 23 following incubation of 28-AcSMT with ERGT paired with [2H3-methyl]SAM demonstrated that these products originated catalytically in methylation by SAM (supplemental Figs. S8, S9). In the incubation of 28-AcSMT with CHT, a peak appeared in the GC chromatogram for 16, previously identified as a product of CHT incubation with T. brucei SMT that catalyzes the Δ25(27)-olefin pathway (23, 27). In the incubation of 28-AcSMT with CHT or ERGT, identification of the late eluting peaks in GC corresponding to 21, 22, and 23 was a bit more challenging because no reference standards are available for these compounds. Their retention time and mass of m/z 426 are consistent with formation of a C29-steroidal diol of molecular formula C29H46O2 (25–27). In HPLC, 21 elutes at αc 0.52, 22 elutes at αc 0.54, and 23 elutes at αc 0.50 before ERGT, which elutes late at αc 1.35. In UV, 21, 22, and 23 possess similar spectra showing a typical finger print for a Δ5,7-system in the sterol nucleus of λmax of 282 nm (Fig. 4, panel E-5).
The identity of the isomers could be firmly established by a combination of approaches in which the products were TMS-derivatized, isotopically labeled at C28 with one or two deuterium atoms, or analyzed by NMR: Thus, the TMS-derivative of 21 and 22 possessed m/z M+ 498, while 23 possessed M+ 570 (Fig. 4), consistent with 21 and 22 having one secondary and one tertiary alcohol in the structure, while 23 has two secondary alcohols in the structure. MS of products of 28-AcSMT incubated with SAM paired with [28-2H2]ERGT showed m/z M+ 428 for compounds 21, 22, and 23, indicating that no C28-deprotonation occurred (Fig. 4; supplemental Figs. S8, S9). The 1H-NMR spectrum of 22 indirectly located the tertiary OH group at C24 rather than at C25 through relevant doublets for C26/C27 resonating at 0.87 (d) and 0.89 (d). Of the four olefinic protons, two in the side chain were coupled to one another (j =15.0 1H, dd; 15.4 1H, dd) indicating the presence of an E-disubstituted alkene originating in the substrate. A signal for the C3β-OH group (Fig. 4, panels E-5–E-7) and signals for side chain methyl/methylene groups at H18 (singlet), H19 (singlet), H21 (doublet), and H29 (triplet) (Fig. 4) confirm the identification of 22 distinct from 21. Structure 23 was initially distinguished from 21 and 22 by its TMS-derivative. MS of 23 showed a fragment for side chain cleavage at m/z 127 consistent with the location of C22 in the side chain (25, 27). NMR analysis provided additional proof of structure showing relevant signals for H18 0.636 (s), H19 0.954 (s), H21/H26/27 0.96 to 1.019 (m), H22 (bearing OH) 5.320 (d), and H23 5.30 (m). On the basis of these results, several new inactivation metabolites have been characterized and a new sterol methylation pathway has been proposed that recognize alternate trajectories for the methylated Δ22,24-intermediate to proceed, either to turnover or covalent bind to enzyme (Fig. 5).
Active-site mutagenesis reveals a residue switch that models reaction channeling
Recent site-directed mutagenesis experiments of SMT enzymes have demonstrated the importance of tyrosine residues in the substrate binding segment Region 1 lined with aromatic residues [Region 1, YXYWGWGXXFHF (supplemental Fig. S10)], and together with homology modeling of the SMT active site suggested the tyrosine60/64 in the amoeba SMTs can play a role in binding and catalysis (27, 28). Specifically, this conserved residue may stabilize carbocation intermediates generated during sterol methylation through cation-π interactions that channel deprotonation routes into Δ25(27)- or Δ24(28)-olefin products and mediate the C1- to C2-transfer abilities of the enzyme, affording turnover rather than protein alkylation (28, 29). With this in mind, Tyr60 in 24-AcSMT and corresponding Tyr64 in 28-AcSMT were mutated to Phe or Leu and the resulting recombinant proteins expressed in E. coli in similar fashion to wild-type AcSMT (supplemental Fig. S11). GC-MS analysis of the mutants, Tyr60Phe or Tyr60Leu, incubated with CTO, CHT, or ERGT showed products of molecular ion peaks of m/z 396 and 394, consistent with the M+ ions of ergosta-5,7,24(28)-trienol and ERGT, respectively, in yields ranging from 100% against CTO to 3–7% against CHT, while ERGT was recovered in unchanged form. Tyr64Phe and Tyr64Leu mutants incubated with CTO generated the same set of C28- and C29-steroidal products generated by wild-type, but in different proportions. In contrast to control 28-AcSMT that converts CTO to C28- and C29-sterols in a ratio of 0:100 (supplemental Fig. S6), the Phe mutant produces a C28- to C29-sterol ratio of 16:84, while the Leu mutant produces a C28- to C29-sterol ratio of 82:18, showing the importance of Tyr64 in mediating the first and second C1-transfer reaction. A different picture emerges when CHT and ERGT are incubated with the Tyr64 mutants. As shown in Fig. 4F, G, Phe mutant produces an altered C28- to C29-sterol product ratio that includes blockage of C28-steroidal monol conversion and a significant reduction in inactivation products of the C29-steroidal diol pathway compensated by an increase in turnover products, including 18, 19, and 20. Leu mutation led to the abolishment of activity such that ERGT was recovered in unchanged form, consistent with the importance of the aromatic residue in stabilizing cation-π interactions. The anomalous methylation increase in 20 by the Try64Phe mutant is presumably made possible by the relaxed control over substrate and intermediate conformations in the active site to favor the Δ24(28)-olefin pathway. The deprotonation leading to the formation of 20 may involve protons chemically and geometrically distinct from those lost in generation of the native substrate-24(28)-methylene lophenol, indicating that multiple amino acid residues (or peptide bonds) could be acting as adventitious active site bases in the mutant methyltransferase.
CHT and ERGT are selective irreversible inhibitors of 28-AcSMT
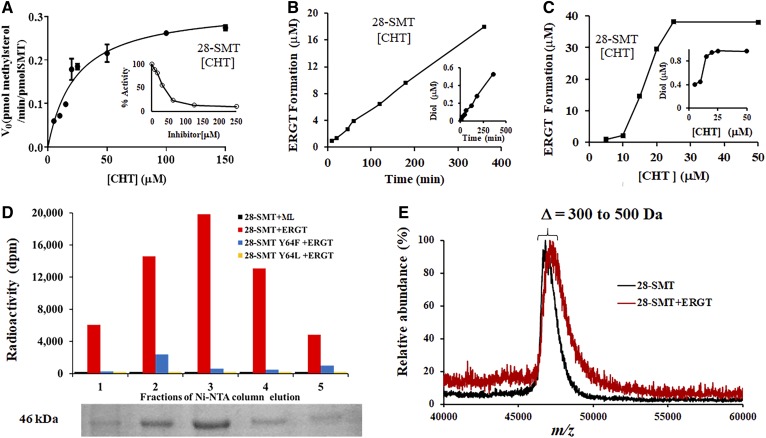
To investigate the requirement for covalence in a complementary fashion to that previously observed for 26,27-dehydrolanosterol against 24-AcSMT where irreversible binding was established (supplemental Fig. S1C) (16), we studied the kinetics and mechanism-based covalent binding of 28-AcSMT incubated with CHT and ERGT. In preliminary GC-MS analyses, we observed that CHT or ERGT incubated with 28-AcSMT generated C28- and C29-steroidal diol products of molecular mass m/z 412 and 426, as expected for aborted methylation products of SMT turnover. Here, we evaluated CHT as substrate of 28-AcSMT. CHT afforded a Km 25 μM and kcat 0.23 min−1 (Fig. 6A). These values compare with a Km 44 μM and kcat 1.5 min−1 for 28-AcSMT assayed with its favored substrate, 24(28)-methylene lophenol. In a separate experiment, the concentration of CHT varied against fixed concentration (Km) 24(28)-methylene lophenol generated an IC50 38 μM (Ki 8 μM). For comparison purposes, the IC50 of CHT against cycloartenol incubated with 24-AcSMT was determined to be 55 μM (Ki 18 μM). The observed IC50s are comparable to those obtained for inhibition of protozoan and fungal SMTs by product analogs of the reaction.
Fig. 6.
Kinetic, radiochemical, and proteomic analyses of yeast sterol analogs complexed with 28-AcSMT. A: Plot of the initial velocity of a SMT-catalyzed reaction versus the substrate concentration (inset is the IC50 of CHT against the Km of 24(28)-methylene lophenol substrate). B: Plot of time of incubation against CHT conversion to C28-steroidal monol and diol. C: Plot of increasing concentration of CHT against product formation of C28-steroidal monol and diol. To limit C29-sterol formation, the SAM concentration is fixed at 100 μM (supplemental Fig. S12). D: SDS-PAGE gel of different Ni-NTA column fractions containing 28-AcSMT complexed with ERGT versus radioactivity detected in each fraction (see the Materials and Methods for details of the binding experiment). F: Mass spectral analysis of ligand-free and ligand-bound (ERGT) 28-AcSMT.
We next incubated 28-AcSMT with CHT paired with SAM at 100 μM in order to limit the second C1-transfer reaction (supplemental Fig. S12). SMT methylation of CHT yields multiple cations that when eliminated by deprotonation or quenched through protein alkylation led to a linear formation of C28-steroidal monol and diol (Fig. 6B, C). The 24(28)-methylene lophenol assayed at 50 and 100 μM concentrations of [3H3-methyl]SAM against CHT at 100 μM for 1 h was shown to protect the enzyme from inactivation, generating 10% and 25% C-methylation activity, respectively, relative to the C-methylation activity of a control incubation. These results indicate that inactivation is active site directed. Alternatively, no C28-steroidal diol is formed when 24-AcSMT is incubated with CHT and no substrate protection observed when the experiment is carried out against cycloartenol as favored substrate.
Rate changes in sterol C24-methylation leading to distinct intermediate cations that convert to C28-steroidal monol and diol were used to establish the partition ratio. Thus, time course experiments of 28-AcSMT assayed with initial concentrations of CHT at 0, 20, and 45 μM revealed the catalytic turnover of C24-methyl sterol monol formation (r = 0.99, Y = 0.0495x + 0.4001) to C24-methyl sterol diol formation (r = 0.88, Y = 0.0013x + 0.014) yield a partition ratio of 34 (Fig. 6B). This measure for formation of C24-methyl monol product per inactivation event is somewhat higher but no less deadly to the amoeba than the partition ratio of 3 established previously for 26-flourocholesta-5,7,24-trienol against the protozoan parasite, TbSMT (15).
To verify covalent inhibition of 28-AcSMT, we followed retention of the enzyme-generated 3H-intermediate bound to SMT through enzyme purification. Figure 6 shows that after Ni-NTA chromatography, neither wild-type SMT incubated with 24(28)-methylene lophenol nor mutant SMTs incubated with ERGT retain significant radioactivity in the homogenous protein, while wild-type SMT incubated with ERGT yields a complex of covalent binding (Fig. 6D). The irreversible nature of the ERGT-28-AcSMT complex was further confirmed by MALDI-TOF analysis showing an increase in molecular mass of Δ300–500 Da (Fig. 6E), where the mass increase in the protein-sterol complex agrees with a single intermediate 22 forming the covalent enzyme-substrate adduct. These results support the GC-MS analysis of 28-AcSMT mutants incubated with ERGT showing that either mutation shifted the partitioning away from the Δ25(27)-product necessary for proper C28- to C29-sterol balance and away from C29 steroidal diol formation indicative of protein alkylation.
Steroidal antibiotic potency across kingdoms
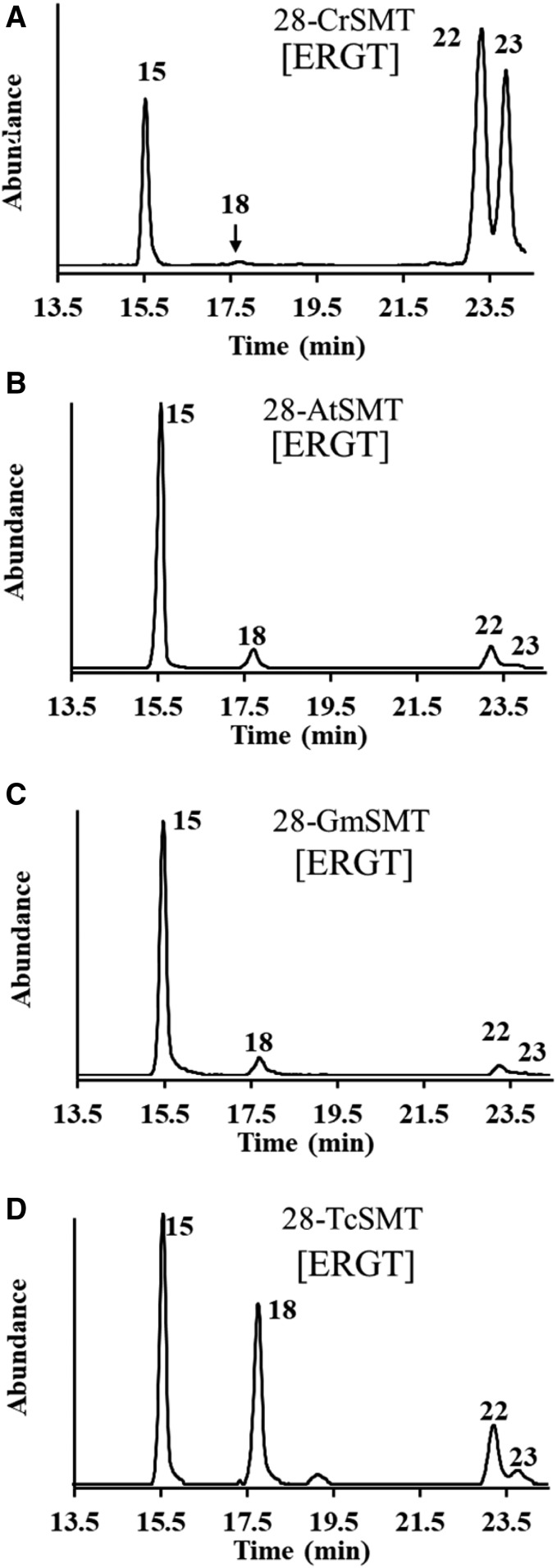
To determine whether the effect of the newly discovered steroidal antibiotics could be effective in regulating steroidogenesis beyond the amoeba, we examined ERGT as a substrate of multiproduct SMT2-type enzymes synthesized in green algae, land plants, and protozoa. The natural substrates of these enzymes are obtusifoliol (green alga SMT2), 24(28)-methylene lophenol (land plant SMT2), and ergosta-5,7,24(28)-trienol (trypanosome SMT2), which convert to 24-ethyl(idene)-Δ24(28)- or -Δ25(27)-olefin products (19, 30–33). Incubation of the four SMT2 enzymes with ERGT produced similar sterol profiles, as observed in the incubation of 28-AcSMT with ERGT (Fig. 7). We observed SMT2 enzymes that favor the Δ25(27)-olefin pathway, as in the green alga and Acanthamoeba SMTs, produce significant C29-steroidal diol, suggesting extensive protein alkylation, while SMT2 enzymes that favor the Δ24(28)-olefin pathway, as in the trypanosome and land plant SMT2s, produce minor amounts of C29-steroidal diol, thereby minimizing the antimetabolite toxicity on C29-steroidogenesis. The different behavior of the SMT2 enzymes toward suicide substrate, as well as significant differences in sterol methylation reaction pathways, suggest species differences in the structures of the active site that employ distinct partitioning in response to an active-site-directed affinity label.
Fig. 7.
Sterol profiles of Chlamaydomonas reinhardtii (A), Arabidopsis thaliana (B), Glycine max (C), and Trypanosoma cruzi (D) SMT2-type enzymes incubated with ERGT. The compounds identified in the chromatograms are: 15, ERGT; 18, stigmast-5,7,22,24(28)-tetraenol; 22, poriferasta-5,7,22(23)-trien-3,34-diol; and 23, poriferasta-5,7,22(23)-trien-3,22-diol.
DISCUSSION
SMT1 and SMT2 are the core components of phytosterol and mycosterol biosynthetic pathways and together are responsible for the generation of the vast array of 24-alkyl sterol structures. The fine-tuning of these methyltransferase enzymes under the pressure of natural selection can be correlated to sterol genealogy on the basis of substrate acceptance and the reactive carbocationic intermediate generated during methylation of the substrate Δ24-double bond (21, 31). As a result, initial substrate-enzyme complementarity to achieve a methylation-competent side chain conformation likely governs the specific reaction pathway to product variability and serves as driver of new 24-alkyl sterol traits. These findings together with the structural similarity in the primary sequences and tetrameric subunit organization of a wide-range of SMTs reflect common active-site topography and support the suggestion that these enzymes, each of strict substrate preference and catalyzing distinct canonical or branched reaction pathways in the construction of C28-ergostane and C29-stigmastane skeletons, respectively, evolved divergently from a common ancestral protein, presumed substrate promiscuous and catalytically diverse in product profile (6, 19).
With five examples of independently evolved SMT2 enzymes that show differential sensitivities to Δ24-monoene and Δ22,24-diene sterols represented by obtusifoliol, 24(28)-methylene lophenol, and ERGT (supplemental Fig. S2), it can be appreciated that lineage-specific steroidogenesis involving specific metabolites follows a common order of central catalysis where SMT2 enzymes seem to have shaped Δ22-C29-sterol production. When SMT2 enzyme cannot exercise sufficient control over productive side chain conformations for sterol methylation, catalysis is impaired and the normal course and equilibrium of metabolic sequences in steroidogenesis are inhibited, a consequence of which is to become an evolutionary dead end that prevents further expansion of C29-sterols. The process is seemingly hindered under physiological conditions, because the cell has adapted the biosynthetic sequence to avoid introduction of the Δ22-bond until after methylation of the Δ24-bond.
The substrate-specific SMT2 inactivation observed for CHT and ERGT of this work indicates a somewhat restricted binding region around the bound intermediate, and it is likely that the unique electronics and side chain orientation of Δ22,24-sterol analogs coordinated to sterol methylation of Δ25(27)-olefin production might have contributed to the SMT sensitivities that shaped sterol diversity early in evolution. Though land plant SMT1 enzymes can bind the substrate analog, 24-methyl cycloartanol (= cyclobranol) (34, 35), the analog tested against soybean SMT1 led to enzyme inactivation (35). Typically in land plants, cycloartenol is converted to 24-methyl desmosterol, which is converted by the sterol 24-reductase to campesterol (24α-methyl cholesterol). Intriguingly, 24-methyl desmosterol binds nonproductively to SMT1 showing the importance of substrate acceptability against steroidal enzymes competing for the side chain Δ24-bond (2, 34). Thus, as for CHT and ERGT capable to destroy Acanthamoeba SMT2 cyclobranol presents side chain features to land plant SMT1 that can terminate the enzyme in a time-dependent manner (35). It is worth mentioning that the sterol methylation reaction for the amoeba and land plant SMT differ; one operates the ∆25(27) -olefin pathway while the other operates the Δ24(28) -olefin pathway, suggesting mechanistic specificity toward protein alkylation is linked to the enzyme recognition of substrate shape and structure of the functional group. Consequently, when these particular side chains are coupled to an ineffective substrate, then metabolites are produced out of order, thereby compromising steroidogenesis. While still fragmented, these new observations of SMT acceptance of potential steroidal antimetabolites depict individual patterns of metabolite order in sterol biosynthesis (Fig. 1) may have evolved in response to specific enzyme sensitivities for suicide substrates. We surmise that, if the SMT reactions operating far from equilibrium dictate the direction and regulatory capacity of the sterol metabolic pathway, then successful catalysis is not random but requires specific partnering of substrate with SMT enzyme.
By following a lead from synthetic suicide substrates that can inactivate SMT and knowledge of function of aromatic amino acids in the substrate binding segment, a specific tyrosine64 to phenyalanine change in 28-AcSMT (SMT2) was found to be sufficient to essentially switch the reaction channeling from one that inhibits catalysis in the presence of ERGT to one that enables protein to synthesize utilizable products used in remodeling the C28- to C29-sterol profile during the course of evolution. The promiscuity of such single mutants at position-64 originating from the altered methylation template could have potentiated evolution of the ancestral protein, leading to the production of Δ22,24-substrates. Many of the SMT1 enzymes, such as in fungi, soybean, Arabidopsis, and amoeba, are high-fidelity SMTs that generate one product rapidly through a nonstop methylation-deprotonation reaction coupled to the Δ24(28)-olefin pathway. However, SMT2 enzymes, such as in green algae, land plants, protozoa, and amoeba, generate one major methyl product and (usually) minor quantities of one or more isomeric methyl products through a step-wise mechanism that proceeds through the ∆25(27) - or ∆24(28) - olefin pathways. Thus, when SMT2s accommodate a more permissive substrate, the enzyme-generated intermediate can undergo premature quenching and, according to off-pathway conformations, fail to convert productively. For these reasons, Δ22,24-substrates shown to be catalytically productive against different SMT2-type enzymes cannot prevail in C29-sterol biosynthesis pathways. It is worth mentioning that the work described here provides long-sought direct evidence for the first 24-ethyl intermediate in SMT2 catalysis represented by compound 22 that converts to multiple 24-ethyl(idene) products (36, 37).
A surprising discovery was the observation of turnover-dependent inactivation of 28-AcSMT in vivo, suggesting that the sterol methylation catalyst generating C29-sterols could be a select target in antibiotherapies, while the natural products could be a new class of antibiotic considered to be biosynthetic antimetabolites. These newly identified antimetabolites that can masquerade as essential metabolites and promote catalytic destruction may now be considered natural poisons of select parasitic organisms. The sterol analysis of treated Ac cells cultured in T-flasks indicated that the antimetabolites inhibit overall steroidogenesis. However, the slight increase in C29-sterol in 96 h-treated cells suggests a metabolic response of overexpression of SMT2 enzyme, consistent with the long exposure of the suicide substrate on target enzyme.
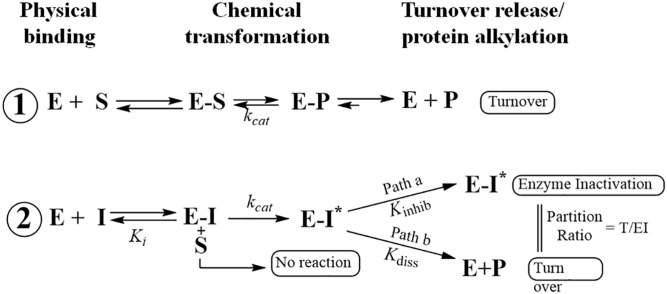
Although CHT can outcompete the natural substrate and inhibit 24-AcSMT and 28-AcSMT with similar Ki values, the loss of inactivation power against 24-AcSMT can be attributed to the catalytic Δ24(28)-olefin pathway operated by most SMT1-type enzymes (33, 37). The in vitro potency of the CHT established via the ratio of the kinetic values Km of natural substrate/Ki of inhibitor against 28-AcSMT of 25 μM/8 μM = 3, compare favorably with the in vivo amoebicidal potency of 5 μM. Interestingly, the thrust of CHT-induced inactivation appears to come from inhibition of the C29 reaction intermediate trapped by an active site base. Mechanistically, extended conjugation of Δ22-bond in Δ24-substrates exerts a dramatic increase in the reactivity of substrate by increasing the transition state for formation of the 24-methyl/ethyl C24-cation intermediate. This structure becomes a transition-state/reactive intermediate analog affording the suicide substrate represented by E-I* species that proceeds down path a in Scheme 1. In contrast, the transition state/high energy intermediate analogs represented by 25-azacycloartanol undergo reversible (albeit tight) binding as indicated in Scheme 1, where E-I ultimately returns to E + I.
In summary, we have succeeded in identifying a new class of antibiotic, elucidating its mode of action and uncovering the origin of its remarkable specificity. This work highlights the importance of using a combination of chemical, enzymatic, and microbiological approaches (38) in antimetabolite discovery and reveals a conceptually elegant mechanism of action. Encouraged by the steroidal antimetabolite depriving the parasite of essential sterol products without effect on the host and then to show the antimetabolite acts as a catalytically incompetent dead-end analog, we anticipate that CHT and ERGT will serve as prototype antibiotic inhibitors that will enable further preclinical validation and will inspire a search for a wider range of steroidal antimetabolites targeting alternate sterolic enzymes that could be developed into leading pharmaceuticals. In order to gain insight into the translational aspects of the current research, the portfolio of protozoan diseases needs to be further expanded, and comparison of in vitro assays, animal models of infection, and clinical data must be performed with the new steroidal antibiotics, all of which are currently underway.
Supplementary Material
Footnotes
Abbreviations:
- CHT
- cholesta-5,7,22,24-tetraenol
- CTO
- cholesta-5,7,24-trienol
- ERGT
- ergosta-5,7,22,24(28)-tetraenol
- HEK
- human epithelial kidney
- MAC
- minimum amoebicidal concentration
- NLF
- nonsaponifiable lipid fraction
- SIM
- selected ion monitoring
- SMT
- sterol methyltransferase
This work was supported by National Institutes of Health Grant R21/R33 AI119782 (to W.D.N.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Bloch K. 1991. Cholesterol: evolution of structure and function. In Biochemistry of Lipids, Lipoproteins and Membranes. D. E. Vance and J. E. Vance, editors. Elsevier, Amsterdam. 363–381. [Google Scholar]
- 2.Nes W. R., and McKean M. L.. 1977. Biochemistry of Steroids and Other Isopentenoids. University Park Press, Baltimore, MD. [Google Scholar]
- 3.Krause M. R., and Regen S. L.. 2014. The structural role of cholesterol in cell membranes: from condensed bilayers to lipid rafts. Acc. Chem. Res. 47: 3512–3521. [DOI] [PubMed] [Google Scholar]
- 4.Fügi M. A., Gunasekera K., Ochsenreiter T., Guan X., Wenk M. R., and Mäser P.. 2014. Genome profiling of sterol synthesis shows convergent evolution in parasites and guides chemotherapeutic attack. J. Lipid Res. 55: 929–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desmond E., and Gribaldo S.. 2009. Phylogenomics of sterol synthesis: insights into the origin, evolution, and diversity of a key eukaryotic feature. Genome Biol. Evol. 1: 364–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gold D. A., Grabenstatter J., de Mendoza A., Riesgo A., Ruiz-Trillo I., and Summons R. E.. 2016. Sterol and genomic analyses validate the sponge biomarker hypothesis. Proc. Natl. Acad. Sci. USA. 113: 2684–2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hargrove T. Y., Wawrzak Z., Liu J., Nes W. D., Waterman M. R., and Lepesheva G. I.. 2011. Substrate preferences and catalytic parameters determined by structural characteristics of sterol 14α-demethylase (cyp51) from Leishmania infantum. J. Biol. Chem. 286: 26838–26848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nes W. D. 2011. Biosynthesis of cholesterol and other sterols. Chem. Rev. 111: 6423–6451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouvier F., Rahier A., and Camara B.. 2005. Biogenesis, molecular regulation and function of plant isoprenoids. Prog. Lipid Res. 44: 357–429. [DOI] [PubMed] [Google Scholar]
- 10.Lees N., Bard M., and Kirsch D. R.. 1997. Biochemistry and molecular biology of sterol synthesis in Saccharomyces cerevisiae In Biochemistry and Function of Sterols. E. J. Parish and W. D. Nes, editors. CRC Press, Boca Raton, FL. 85–99. [PubMed] [Google Scholar]
- 11.Miller M. B., Haubrich B. A., Wang Q., Snell W. J., and Nes W. D.. 2012. Evolutionarily conserved Δ25(27)-olefin ergosterol biosynthesis pathway in the alga Chlamydomonas reinhardtii. J. Lipid Res. 53: 1636–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nes W. D. 2000. Sterol methyl transferase: enzymology and inhibition. Biochim. Biophys. Acta. 1529: 63–88. [DOI] [PubMed] [Google Scholar]
- 13.Peters G. J. 2014. Novel developments in the use of antimetabolites. Nucleosides Nucleotides Nucleic Acids. 33: 358–374. [DOI] [PubMed] [Google Scholar]
- 14.Hargrove T. Y., Wawrzak Z., Liu J., Waterman M. R., Nes W. D., and Lepesheva G. I.. 2012. Structural complex of sterol 14α-demethylase (CYP51) with 14α-methylenecyclopropyl-Δ7-24,25-dihydrolanosterol. J. Lipid Res. 53: 311–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leaver D. J., Patkar P., Singha U. K., Miller M. B., Haubrich B. A., Chaudhuri M., and Nes W. D.. 2015. Fluorinated sterols are suicide inhibitors of ergosterol biosynthesis and growth in Trypanosoma brucei. Chem. Biol. 22: 1374–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kidane M. E., Vanderloop B. H., Zhou W., Thomas C. D., Ramos E., Singha U., Chaudhuri M., and Nes W. D.. 2017. Sterol methyltransferase a target for anti-amoeba therapy: towards transition state analog and suicide substrate drug design. J. Lipid Res. 58: 2310–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou W., Warrilow A. G., Thomas C. D., Ramos E., Parker J. E., Price C. L., Vanderloop B. H., Fisher P. M., Loftis M. D., and Kelly D. E.. 2018. Functional importance for developmental regulation of sterol biosynthesis in Acanthamoeba castellanii. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 1863: 1164–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu S. H., and Nes W. D.. 1988. Biosynthesis of cholesterol in the yeast mutant erg6. Biochem. Biophys. Res. Commun. 155: 509–517. [DOI] [PubMed] [Google Scholar]
- 19.Haubrich B. A., Collins E. K., Howard A. L., Wang Q., Snell W. J., Miller M. B., Thomas C. D., Pleasant S. K., and Nes W. D.. 2015. Characterization, mutagenesis and mechanistic analysis of an ancient algal sterol C24-methyltransferase: implications for understanding sterol evolution in the green lineage. Phytochemistry. 113: 64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kodner R. B., Summons R. E., Pearson A., King N., and Knoll A. H.. 2008. Sterols in a unicellular relative of the metazoans. Proc. Natl. Acad. Sci. USA. 105: 9897–9902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Najle S. R., Molina M. C., Ruiz-Trillo I., and Uttaro A. D.. 2016. Sterol metabolism in the filasterean Capsaspora owczarzaki has features that resemble both fungi and animals. Open Biol. 6: 160029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nes W. D., McCourt B. S., Zhou W. X., Ma J., Marshall J. A., Peek L. A., and Brennan M.. 1998. Overexpression, purification, and stereochemical studies of the recombinant (S)-adenosyl-L-methionine: delta 24(25)- to delta 24(28)-sterol methyl transferase enzyme from Saccharomyces cerevisiae. Arch. Biochem. Biophys. 353, 297–311. [DOI] [PubMed] [Google Scholar]
- 23.Zhou W., Lepesheva G. I., Waterman M. R., and Nes W. D.. 2006. Mechanistic analysis of a multiple product sterol methyltransferase from Trypanosoma brucei implicated in ergosterol biosynthesis. J. Biol. Chem. 281: 6290–6296. [DOI] [PubMed] [Google Scholar]
- 24.Lamb D. C., Warrilow A. G., Rolley N. J., Parker J. E., Nes W. D., Smith S. N., Kelly D. E., and Kelly S. L.. 2015. Azole antifungal agents to treat the human pathogens Acanthamoeba castellanii and Acanthamoeba polyphaga through inhibition of sterol 14α-demethylase (CYP51). Antimicrob. Agents Chemother. 59: 4707–4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goad L. J., and Akihisa T.. 1997. Analysis of Sterols. Blackie Academic, London. [Google Scholar]
- 26.Xu S. H., Norton R. A., Crumley F. G., and Nes W. D.. 1988. Comparison of the chromatographic properties of sterols, select additional steroids and triterpenoids: gravity-flow column liquid chromatography, thin-layer chromatography, gas-liquid chromatography and high-performance liquid chromatography. J. Chromatogr. 452: 377–398. [DOI] [PubMed] [Google Scholar]
- 27.Liu J., Ganapathy K., Wywial E., Bujnicki J. M., Nwogwugwu C. A., and Nes W. D.. 2011. Effect of substrate features and mutagenesis of active site tyrosine residues on the reaction course catalysed by Trypanosoma brucei sterol C-24-methyltransferase. Biochem. J. 439: 413–422. [DOI] [PubMed] [Google Scholar]
- 28.Ganapathy K., Jones C. W., Stephens C. M., Vatsyayan R., Marshall J. A., and Nes W. D.. 2008. Molecular probing of the Saccharomyces cerevisiae sterol 24-C methyltransferase reveals multiple amino acid residues involved with C 2-transfer activity. Biochim. Biophys. Acta. 1781: 344–351. [DOI] [PubMed] [Google Scholar]
- 29.Nes W. D., Song Z., Dennis A. L., Zhou W., Nam J., and Miller M. B.. 2003. Biosynthesis of phytosterols. Kinetic mechanism for the enzymatic C-methylation of sterols. J. Biol. Chem. 278: 34505–34516. [DOI] [PubMed] [Google Scholar]
- 30.Zhou W., and Nes W. D.. 2003. Sterol methyltransferase2: purification, properties, and inhibition. Arch. Biochem. Biophys. 420: 18–34. [DOI] [PubMed] [Google Scholar]
- 31.Neelakandan A. K., Song Z., Wang J., Richards M. H., Wu X., Valliyodan B., Nguyen H. T., and Nes W. D.. 2009. Cloning, functional expression and phylogenetic analysis of plant sterol 24C-methyltransferases involved in sitosterol biosynthesis. Phytochemistry. 70: 1982–1998. [DOI] [PubMed] [Google Scholar]
- 32.Sowa M. 2016. Purification, Characterization, and Inhibition of C-24 Sterol Methyltransferase from Trypanosoma cruzi. MS Dissertation. Texas Tech University, Lubbock, TX. [Google Scholar]
- 33.Bouvier-Navé P., Husselstein T., Desprez T., and Benveniste P.. 1997. Identification of cDNAs encoding sterol methyl-transferases involved in the second methylation step of plant sterol biosynthesis. Eur. J. Biochem. 246: 518–529. [DOI] [PubMed] [Google Scholar]
- 34.Nes W. D., Janssen G. G., and Bergenstrahle A.. 1991. Structural requirements for transformation of substrates by the (S)-adenosyl-L-methionine:delta 24(25)-sterol methyl transferase. J. Biol. Chem. 266: 15202–15212. [PubMed] [Google Scholar]
- 35.Wang J., and Nes W. D.. 2008. Cyclobranol: a substrate for C25-methyl sterol side chains and potent mechanism-based inactivator of plant sterol methyltransferase. Bioorg. Med. Chem. Lett. 18: 3878–3881. [DOI] [PubMed] [Google Scholar]
- 36.Fujimoto Y., Sato N., Iwai K., Hamada H., Yamada J., and Morisaki M.. 1997. Stereochemistry of the reduction of 24-methyldesmosterol to campesterol and dihydrobrassicasterol in higher plants. Chem. Commun. (Camb.). 7: 681–682. [Google Scholar]
- 37.Kerr R. G., and Baker B. J.. 1991. Marine sterols. Nat. Prod. Rep. 8: 465–497. [Google Scholar]
- 38.Tonge P. J. 2018. Drug-target kinetics in drug discovery. ACS Chem. Neurosci. 9: 29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.