Abstract
Objectives
Recent studies have demonstrated that primordial germ cells (PGC) can be differentiated from human umbilical cord mesenchymal stem cells (hUC‐MSCs), and embryonic stem cells (ESCs) in vitro. Nevertheless, efficiencies were low and unstable. Here, whether hUC‐MSCs can be induced to differentiate into germ‐like cells with the aid of bone morphogenetic protein (BMP4) was investigated.
Materials and methods
Human umbilical cord mesenchymal stem cells were freshly isolated and cultured with BMP4. SSEA‐1+/− cells were purified using magnetic‐activated cell sorting (MACS) from the hUC‐MSCs, and further induced with BMP4. Quantitative real‐time PCR (qRT‐PCR) and immunofluorescence analysis were used to determine PGC and germ‐like cell‐specific markers.
Results
Human umbilical cord mesenchymal stem cells differentiated into SSEA‐1+ spherical PGC‐like cells efficiently with 12.5 ng/ml BMP4. qRT‐PCR and immunofluorescence analysis demonstrated that SSEA‐1+ cells expressed higher levels of PGC‐specific markers than SSEA‐1− cells. Furthermore, SSEA‐1+ cells were induced with BMP4 to differentiate into STRA8, SCP3, DMRT1 and PLZF‐positive male germ‐like cells, and some sperm‐like cells were obtained by 7–14 days after induction.
Conclusion
These results suggest that SSEA‐1+ hUC‐MSCs can differentiate into male germ‐like cells in the presence of BMP4. This study provides an efficient protocol to study germ‐cell development using hUC‐MSCs.
Introduction
Mesenchymal stem cells (MSCs) derived from bone marrow are a well‐characterized population of adult stem cells, which have the ability to differentiate into various cell types, including adipose cells, cartilage, bone, tendon and ligament, muscle, skin and even neural cells 1, 2, 3. Recently, it has been reported that mouse and human bone marrow‐derived MSCs can differentiate into gametes (sperm or oocyte) in vivo and in vitro 4. These induced cells share characteristics of typical germ cells and the authors proposed that bone marrow stem cells could migrate 5 and colonize ovaries to maintain a plentiful stock for reproduction, and may differentiate into sperm or oocytes, in mice 4. However, aspirating bone marrow from a donor is invasive; in addition, differentiation potential and number of bone marrow‐derived MSCs decreases gradually with age of the donor 4. Thus, many scientists search for alternative sources of MSCs. Recently, many have found that hUC‐MSCs are capable of differentiating into cells of three different connective tissue lineages, bone, cartilage and adipose tissues. hUC‐MSCs are one of the best candidates for tissue engineering of musculoskeletal phenotypes, even capable of differentiating into cardiomyocytes, neuron‐like cells, hepatocytes, and even germ‐like cells 4, 6, 7.
During mouse embryonic differentiation, the ectoderm begins to secrete BMP4 and BMP8b at embryo 5.5 (E5.5) stage, and after that, some ectoderm cells express PRDM1. By E6.0‐6.25, secretion of BMP4 peaks and PRDM1‐positive cells become precursors of PGCs. Thereafter follows formation of PGCs, and PGCs migrate into the genital ridge to differentiate into germ cells 8. It has been demonstrated that BMP4 plays a regulatory role in the process of PGC migration and specification 9. Under induction of BMP4, mouse and human embryonic stem cells can successfully differentiate into PGCs 9. Concerning the function of germ cell induction, BMP4 is as efficient as retinoic acid (RA). And it has also been reported that BMP4 can stimulate bone marrow mesenchymal stem cells (BMSCs) to express some early germ cell‐specific markers 10. BMP4 is also beneficial to formation of PGC derived from ES cells and BMSCs 6, 10, 11, 12. Our previous work has demonstrated that hUC‐MSCs have the potential to differentiate into germ‐like cells under treatment with retinoic acid or follicle fluid. However, the mechanisms were not clear and efficiencies tended to be low and unstable 7.
As BMP4 is of great importance in germ cell development, we investigated whether hUC‐MSCs could differentiate into germ cells by induction of BMP4. Thus, we found that a number of MSCs expressed SSEA‐1, an important marker of PGC. We wondered whether MACS‐purified SSEA‐1+ cells could upregulate formation of male germ‐like cells from hUC‐MSCs. The aim of this study has been to discover whether BMP4 would efficiently promote SSEA‐1+ hUC‐MSC differentiation into male germ‐like cells in vitro.
Materials and methods
Isolation and culture of hUC‐MSC
Four umbilical cords were used in this study informed consent having been obtained from each donor; all procedures were approved by our institutional ethics committee. Fresh umbilical cord tissues were soaked in phosphate‐buffered saline (PBS) containing penicillin and streptomycin (100 U/ml; Sigma, St. Louis, MO, USA) for 30 s, during which they were pressed hard to remove blood cells. Tissues were then changed to fresh PBS and cut to expose the wrapped Wharton jelly. The two arteries and one vein were then isolated from the Wharton jelly and it was peeled away from the cortex. Wharton jelly was then washed twice in fresh PBS and cut into 5 mm pieces. After being cultured inversely for 2 h at 37 °C, culture medium was added to the plates. In the order of 5 days later, appropriate cells would migrate out. When these cells reached 80% confluence at 8–14 days, they were dissociated using trypsin, and primary hUC‐MSCs were obtained.
The hUC‐MSCs were cultured as previously described 13, in DMEM/F12 (Invitrogen, Carlsbad, CA, USA) culture medium, containing 15% foetal bovine serum (FBS, Hyclone, Logan, UT, USA), supplemented with 0.1 mm 2‐mercaptoethanol (Invitrogen), 2 mm l‐glutamine (Invitrogen), 1% non‐essential amino acids (Invitrogen), under humidified conditions at 37 °C with 5% CO2. Culture plates had been treated with gelatin overnight, and cells were passaged every 2 days.
Isolation and culture of SSEA‐1+/− cell‐derived hUC‐MSCs
SSEA‐1+ hUC‐MSCs were obtained by MACS using a MiniMACS separation unit (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany) with SSEA‐1‐specific MicroBeads (Miltenyi Biotec GmbH). Briefly, hUC‐MSCs (~1.0 × 107 cells) were resuspended in 80 μl MACS buffer (PBS containing 2 mm EDTA) and 0.5% bovine serum albumin, then mixed with 20 μl magnetically labelled anti‐SSEA‐1 MicroBeads for 15 min at 4 °C. Labelled cells were then resuspended in 500 μl MACS buffer and loaded on a MACS column previously washed several times in MACS buffer. Thus, SSEA‐1+ hUC‐MSCs were retained within the columns while the SSEA‐1− hUC‐MSCs were run through. After removing columns from the magnetic field, magnetically retained SSEA‐1+ hUC‐MSCs were obtained by being eluted into centrifuge tubes.
Immunofluorescence staining
Cells were fixed in 4% paraformaldehyde for 15 min, then rinsed twice in PBS for 3 min × 2. After blocking with 1% BSA for 30 min, cells were incubated in primary antibody overnight. Information concerning primary antibodies is as follows: C‐KIT (mouse monoclonal antibody, 1:200; Biolegend, San Diego, CA, USA), PRDM1 (mouse monoclonal antibody, 1:400; Biolegend), OCT‐4 (mouse monoclonal antibody, 1:500; Chemicon, Billerica, MA, USA), SSEA‐1 (mouse monoclonal antibody, 1:100; Chemicon), VASA (Rabbit polyclonal antibody, 1:200; Abcam, Cambridge, UK) and ACROSIN (mouse monoclonal antibody, 1:400; Santa Cruz Biotechnology, Dallas, TX, USA). Thereafter, cells were rinsed 3 times in PBS each ×3 min, and fluorochrome‐conjugated secondary antibody (1:500; Chemicon) was added and incubated at 37 °C for 1 h. Cells were then rinsed 3 times in PBS 3 min ×3. Hoechst 33342 (Sigma) was used to stain cell nuclei, 2 min at room temperature (RT), after being rinsed twice for 3 min each in PBS; cells were examined using a fluorescence microscope.
hUC‐MSC induction
Induction medium was made with a range of concentrations of human BMP4 (12.5 ng/ml, 25 ng/ml; PeproTech, Rocky Hill, NJ, USA) in normal culture medium 10.
For EB formation, 2 × 105 cells were seeded into 35‐mm plates with 1.5 ml culture medium. Cells were resuspended for 10 h, and EBs formed after a further 3 days. Appropriate EBs were added to 96‐well plates and 12‐well plates; after adherence overnight, culture medium was replaced with induction medium, which was changed every 2 days 14.
Purified SSEA‐1+/− hUC‐MSCs were plated into 0.1% gelatin‐coated 96‐well plates (1000 cells/well) and 12‐well (2 × 104 cells/well) plates with induction medium. In addition, SSEA‐1+/− cells were seeded on mitotically inactive mouse embryonic fibroblasts (MEFs) (1 × 104/well) in 96‐well tissue culture plates with induction medium.
Quantitative real‐time PCR analysis
Total RNA was extracted from 1.0 × 106 cells using Trizol reagent (Qiagen, Beijing, China) according to the manufacturer's instructions. Total RNA was reverse transcribed into cDNA using M‐MLV Reverse Transcriptase Reagent kit (Thermo Scientific, Waltham, MA, USA) according to the manufacturer's instructions. qRT‐PCR analysis was carried out in triplicate on a CFX96 Real‐Time PCR system (Bio‐Rad Ltd, Berkeley, CA, USA) at final volume 15 μl, containing 0.5 μl cDNA (1:10 diluted), 7.5 μl SYBR (Bioer Co.Ltd., Hangzhou, China), 6.3 μl ddH2O, 0.3 μl forward primer, 0.3 μl reverse primer, and 0.1 μl Taq DNA polymerase. Expression of β‐actin was used as house‐keeping control. qRT‐PCR procedures were set as follows: 5 min at 94 °C, followed by 40 cycles of amplification consisting denaturation for 20 s at 94 °C, 30 s at 58 °C for annealing and 10 s at 70 °C for elongation. The comparative CT method was used to measure relative gene expression. Primers used are listed in Table 1.
Table 1.
Primers for qRT‐PCR
| Gene name | Sense primer | Antisense primer | Tm (°C) | GenBank accession number |
|---|---|---|---|---|
| GAPDH | AGGTCGGTGTGAACGGATTTG | TGTAGACCATGTAGTTGAGGTCA | 58 | NM_008084.2 |
| SSEA‐1 | ACGGATAAGGCGCTGGTACTA | GGAAGCCATAGGGCACGAA | 59 | NM_010242.3| |
| PRDM1 | TGGAGGACGCTGATATGACT | CTTACCACGCCAATAACCTC | 59 | NM_007548.3 |
| STELLA | GACCCAATGAAGGACCCTGAA | GCTTGACACCGGGGTTTAG | 59 | NM_139218.1 |
| STRA8 | AGCAGCTTAGAGGAGGTCAAGA | TACTCGGAACCTCACTTTTGTC | 57 | XM_006505830.1 |
| SCP3 | CTAGAATTGTTCAGAGCCAGAGA | GTTCAAGTTCTTTCTTCAAAG | 57 | NM_011517.2 |
| C‐KIT | TGACTTACGACAGGCTCGTG | AAGGAGTGAACAGGGTGTGG | 57 | NM_001122733 |
| PRDM14 | ACAGCCAAGCAATTTGCACTAC | TTACCTGGCATTTTCATTGCTC | 57 | NM_001081209.2 |
| SOX2 | GCCCAGGAGAACCCCAAGAT | GGGTGCCCTGCTGCGAGTA | 58 | NM_011443.3 |
| DMRT1 | TGGGTTCTGGAAGCAAGAAG | CTGTCTTCTCAGGGCCACCT | 58 | NM_015826.5 |
| GFRa1 | GACCGTCTGGACTGTGTGAAAG | TTAGTGTGCGGTACTTGGTGC | 58 | XM_006526685.1 |
| VASA | TATGTGCCTCCCAGCTTCAGTA | CTGGATTGGGAGCTTGTGAAGA | 59 | XM_006517526.1 |
| AP2r | TGAAGATGAAGCTGGGCTTT | TCCATTCTCTTCCGGTTCAG | 58.5 | NM_009335.2 |
| NANOG | ATGAAGTGCAAGCGGTGGCAGAAA | CCTGGTGGAGTCACAGAGTAGTTC | 59 | NM_028016.2 |
BrdU incorporation assay
Cell proliferation was determined by BrdU incorporation assay. Briefly, BrdU (Sigma) was added to the cell culture medium at 40 ng/ml for 2 h. Then, cells were fixed in carbinol and acetone (1:1) for 15 min at RT and washed twice in PBS (pH7.4). They were then treated with 2 m HCl for 45 min at RT and washed 2 twice in PBS. Cells were treated with boric acid at RT for 15 min and were incubated with anti‐BrdU antibody (mouse monoclonal antibody, 1:100; Santa Cruz) at 4 °C overnight. Cells were then incubated in FITC‐conjugated secondary antibody (1:500; Millipore, Schwalbach, Germany) for 1 h at RT. After being washed twice in PBS, cells were observed using an immunofluorescence microscope. To determine level of cell proliferation, each group was performed in triplicate, and 5 fields (100×) were randomly chosen in which to count the percentage of BrdU‐positive cells.
Statistical analysis
One‐way analysis of variance (one‐way ANOVA) was used and post‐tests were conducted using Newman–Keuls multiple range test, if P‐values were significant. Students' t‐test was used when only two pairs of data were compared. All data were represented as mean ± SD, and statistical significance was expressed as follows: *P < 0.05; **P < 0.01; ***P < 0.001. All data were representative of at least three different experiments and were analysed using Graphpad Prism software (La Jolla, CA, USA).
Results
BMP4 induced hUC‐MSCs to differentiate into PGC‐like cells
Presence of typical MSC markers was determined by flow cytometry as described previously, and performed along with adipogenic and osteogenic differentiation potential in our laboratory 13, 15. hUC‐MSCs exhibited spindle fibroblast‐like outline or were irregular in shape. Embryoid bodies (EBs) were formed in suspension culture, and were inoculated into 96‐well plates coated with gelatin. Typical EBs grew adherently and some peripheral cells migrated out. After 1 week BMP4 treatment, some migrating cells became spherical (Fig. 1a), similar to primordial germ cells. Nucleus‐cytoplasmic ratio of these cells was high. 12.5 ng/ml BMP4 treatment resulted in more spherical PGC‐like cells than 25 ng/ml BMP4 (Fig. 1b). These results indicated that 12.5 ng/ml BMP4 could induce hUC‐MSCs to transdifferentiate into PGC‐like cells.
Figure 1.
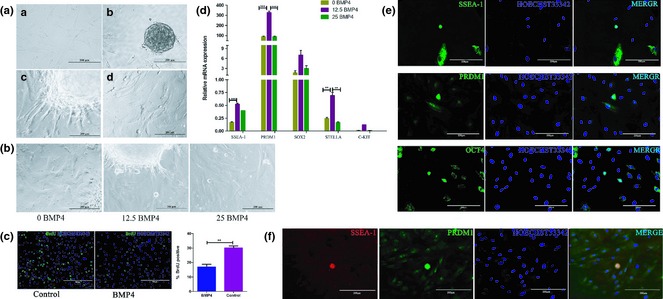
BMP4 effect on differentiation of hUC ‐MSCs into PGC‐like cells. (a) Morphology of hUC‐MSCs under induction of BMP4. a. Untreated hUC‐MSCs cultured in normal culture medium. b. EBs were formed from hUC‐MSCs. c. Adherent EB before induction. d. Cell morphology after 7 days induction. (b) After 7 days induction with BMP4, round‐shaped cells migrated out from EBs. (c) Proliferation profile of BMP4 induced cells or untreated cells was determined by BrdU incorporation assay. (d) qRT‐PCR analysis examined relative expression levels of SSEA‐1, C‐KIT, PRDM1, SOX2 and STELLA. These PGC‐specific markers were expressed at higher levels in the 12.5 ng/ml BMP4‐induced group. (e) Immunofluorescence staining for SSEA‐1, PRDM1 and OCT4. Induced round‐shaped cells highly expressed these PGC‐specific markers and had higher nucleus‐cytoplasmic ratio. (f) Co‐staining for SSEA‐1 and PRDM1. Round‐shaped cells were double‐positive for these two markers.
Cells from induction and control groups were subjected to BrdU incorporation assay, and we found proliferation level of induced cells reduced significantly (Fig. 1c). Our observations are in accordance with one former report, which showed that proliferation rate of germ cells was much lower than that of MSCs 4.
To clarify whether the induced round‐shaped cells were really PGCs, we conducted qRT‐PCR and immunofluorescence staining to test for expression of PGC‐specific markers at mRNA and protein levels. The results showed that levels of SSEA‐1, PRDM1, STELLA, SOX2 and C‐KIT were all significantly higher in 12.5 ng/ml BMP4 compared to 25 ng/ml, even though levels of some markers were also to some extent higher in 25 ng/ml BMP4 induction medium compared to control (Fig. 1d). Concerning these results, we conducted immunofluorescence staining of these induced cells. As expected, round‐shaped cells were triple‐positive for SSEA‐1, PRDM1 and OCT4 (Fig. 1e). To further prove that these cells had PGC‐specific characteristics, SSEA‐1 and PRDM1 were co‐stained in these cells. As expected, these cells expresed SSEA‐1 and PRDM1 simultaneously. These results provide evidence that induced spherical cells were PGC‐like.
hUC‐MSCs differentiated into male germ‐like cells under BMP4 induction
As PGCs can develop into germ cells in vivo 16 and moreover hUC‐MSCs can be induced as PGC‐like cells according to our work, we then investigated whether they could also differentiate into male germ cells. A range of specific markers of meiosis and male germ cells was tested to evaluate differentiation status of induced cells derived from hUC‐MSCs. We found that mRNA expression levels of STRA8 and PLZF increased significantly when the cells were treated with 12.5 ng/ml BMP4 (Fig. 2a). Immunofluorescence staining results also demonstrated that some spindle‐like cells were highly positive for VASA and SSEA‐1 simultaneously (Fig. 2b). This indicates that 12.5 ng/ml BMP4 not only induced hUC‐MSC differentiation to PGC‐like cells but also further induced them to differentiate into male germ‐like cells.
Figure 2.
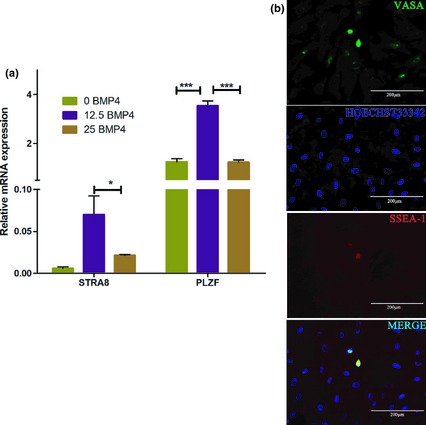
Expression levels of male germ cell‐specific markers of hUC ‐MSCs induced by different concentrations of BMP4. (a) Under induction of 12.5 ng/ml BMP4, expression levels of STRA8 and PLZF increased significantly, as determined by qRT‐PCR analysis. (b) Shrinking SSEA‐1+ cells were highly positive for VASA, as demonstrated by immunofluorescence staining.
Purification of SSEA‐1+ and SSEA‐1− cells from hUC‐MSCs with MACS
It has been reported that SSEA‐1 is a marker of early PGCs 16 and that epiblasts can differentiate into PGC‐like cells in vitro, under treatment of BMP4 17. Similarly, we found that a fraction of hUC‐MSCs expressed SSEA‐1 when subjected to BMP4 induction. Then, we supposed it was likely that BMP4 sensitive cells could be those which were SSEA1‐positive. Thus, we sorted SSEA‐1+ cells by MACS. Results were first 20 thousand SSEA‐1+ cells from 3.6 million cells, second time, 52.8 thousand from 9.6 million, and the third time, 43 thousand from 7.2 million cells. Percentage of SSEA‐1+ cells was in the order of 5.6% hUC‐MSCs. By qRT‐PCR analysis, as well as immunofluorescence staining, SSEA‐1 mRNA and protein were both highly expressed in putative SSEA‐1+ cells. Moreover, after counting numbers of SSEA‐1 positive cells, we found that more than (95 ± 4)% cells in MASC SSEA‐1+ cells were highly positive for SSEA‐1 (Fig. 3a), whereas less than (15 ± 5)% SSEA‐1− cells were present (Fig. 3a).
Figure 3.
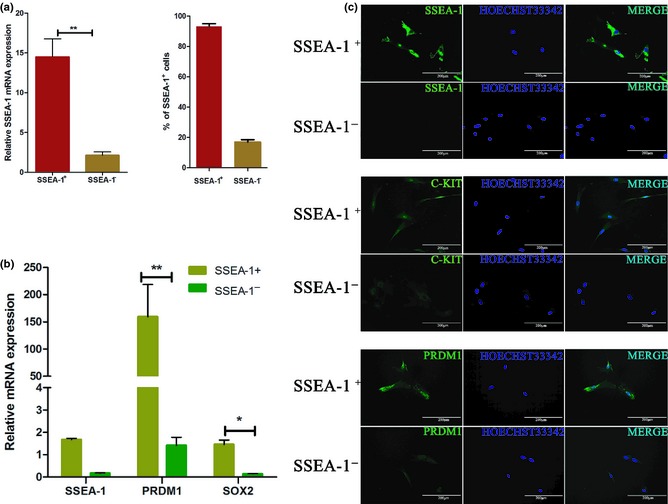
Examination of SSEA‐1 + and SSEA‐1 − cells after MACS. (a) Analysis of sorting efficiency by immunofluorescence staining and qRT‐PCR of SSEA‐1+ cell percentage and SSEA‐1 expression level respectively. (b) qRT‐PCR analysis of sorted SSEA‐1+ and SSEA‐1− cells. Expression levels of PRDM1, SSEA‐1 and SOX2 in SSEA‐1+ cells were significantly higher than those of SSEA‐1− cells. (c) Immunofluorescence staining of expression levels of SSEA‐1, PRDM1 and C‐KIT in sorted SSEA‐1+ and SSEA‐1− cells.
As SSEA‐1 is an important marker of PGC, we wondered whether other PGC‐specific markers were also highly expressed in MACS sorted SSEA‐1+ cells. Using immunofluorescence staining (Fig. 3c) and qRT‐PCR analysis (Fig. 3b), we found that some PGC core markers, such as PRDM1, SSEA‐1 and SOX2, were expressed significantly higher in SSEA‐1+ than in SSEA‐1− cells. These results demonstrate that sorted SSEA‐1+ cells expressed higher levels of PGC‐specific markers compared to SSEA‐1− cells. Thus, SSEA‐1+ cells were closer to PGCs than SSEA‐1− cells. Based on this, we assumed that in the induction process of hUC‐MSCs, SSEA‐1+ cells were the major phenotype that differentiated to PGCs and finally to male germ‐like cells.
SSEA‐1+ and SSEA‐1− hUC‐MSCs responded differently to BMP4, and SSEA‐1+ cells differentiated into PGC‐like cells
To test the response of SSEA‐1+ and SSEA‐1− cells to BMP4, we treated the two groups with BMP4 (12.5 ng/ml). One week later, SSEA‐1+ cells were observed to have become smaller in size compared to SSEA‐1− cells. Moreover, edges of SSEA‐1+ cells were smooth, and intercellular spaces could be seen more clearly. In contrast, SSEA‐1− cells did not seem to grow were relatively larger in volume, and had obscure intercellular spaces whose features were very close to those of hUC‐MSCs (Fig. 4a). We also analysed cell proliferation rates with BrdU incorporation assay and the results indicated that SSEA‐1+ cells had lower proliferation levels compared to SSEA‐1− cells (Fig. 4b). These results demonstrated that SSEA‐1+ cells were the main cell type responsive to BMP4, which further supported our hypothesis.
Figure 4.
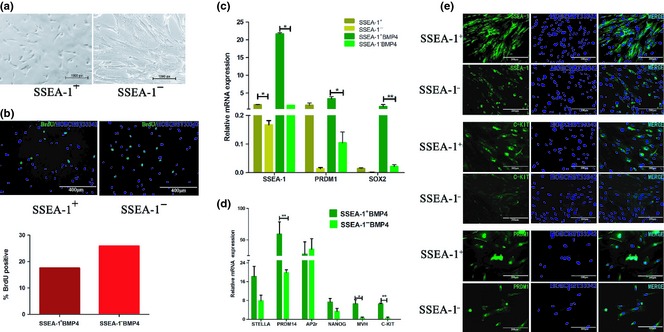
SSEA‐1 + hUC ‐MSCs differentiated into PGC‐like cells under induction of BMP4. (a) Morphological changes of sorted SSEA‐1+ and SSEA‐1− hUC‐MSCs. Under the induction medium, SSEA‐1+ cells visibly shrank, but SSEA‐1− cells had invariant sizes. (b) Proliferation levels of BMP4 induced cells were determined by BrdU immunofluorescence staining. SSEA‐1+ cells had higher proliferation level compared to SSEA‐1− cells. (c) qRT‐PCR analysis of PRDM1, SSEA‐1 and SOX2 expression levels in SSEA‐1+ and SSEA‐1− hUC‐MSCs induced by BMP4. Expression levels of these were increased in both groups, but this trend was more evident in the SSEA‐1+ group. (d) qRT‐ PCR analysis demonstrated that STELLA, PRDM14, NANOG, VASA and C‐KIT expression levels had a similar trend with SSEA‐1 expression, but AP2γ maintained a relatively stable level in both groups. (e) Immunofluorescence staining of SSEA‐1, C‐KIT and PRDM1 in BMP4‐induced SSEA‐1+ and SSEA‐1− hUC‐MSCs.
Expression levels of SSEA‐1, PRDM1 and SOX2 mRNA in SSEA‐1+ as well as SSEA‐1− cells had an increasing tendency to respond to BMP4, as shown by qRT‐PCR analysis (Fig. 4c). Moreover, SSEA‐1 level was much higher in SSEA‐1+ cells compared to that in SSEA‐1− cells, which indicated that SSEA‐1+ cells were more sensitive to BMP4. We then tested mRNA levels of PGC markers, STELLA, PRDM14, NANOG, VASA and C‐KIT 17, and found that they were also highly expressed in the SSEA‐1+ group. However, a further PGC marker, AP2γ, was not significantly changed between these two groups (Fig. 4d). Additionally, immunofluorescence showed that protein levels of SSEA‐1, PRDM1 and C‐KIT were also higher in the SSEA‐1+ group than in the SSEA‐1− group (Fig. 4e). Based on reports that PRDM1 and PRDM14 could promote formation of PGCs 18, we draw the conclusion that SSEA‐1+ cells were the main cell type reprogrammed into PGC‐like cells under BMP4 induction.
SSEA‐1+ hUC‐MSCs differentiated into male germ‐like cells
Sorted SSEA‐1+ cells were seeded on MEF feeder cell layers, then were induced by BMP4. Consequently, morphology of these cells changed in addition to appearance of round‐shaped cells and some germ‐like cells, as had appeared obviously (Fig. 5a). As sperm‐like in morphology, polarized cells could be seen occasionally amongst SSEA‐1+ cells, and expression levels of PLZF and STRA8 were both increased when hUC‐MSCs were induced by BMP4, we speculated that SSEA‐1+ cells transdifferentiated into PGC‐like cells under BMP4 induction, then differentiated towards male germ cells. To test this hypothesis, we analysed mRNA levels of GFRa1, DMRT1 and SCP3. Our results showed that these male germ‐cell and meiosis markers mRNA levels were all significantly increased in the SSEA‐1+ group compared to SSEA‐1− cells (Fig. 5b). Sperm‐like cells were also subjected to Acrosin immunofluorescence staining (Fig. 5c) and results showed that they were stained positively. Thus, we concluded that SSEA‐1+ UC‐MSCs went through the process towards PGCs under BMP4 induction and thereafter, differentiated into male germ‐like cells.
Figure 5.
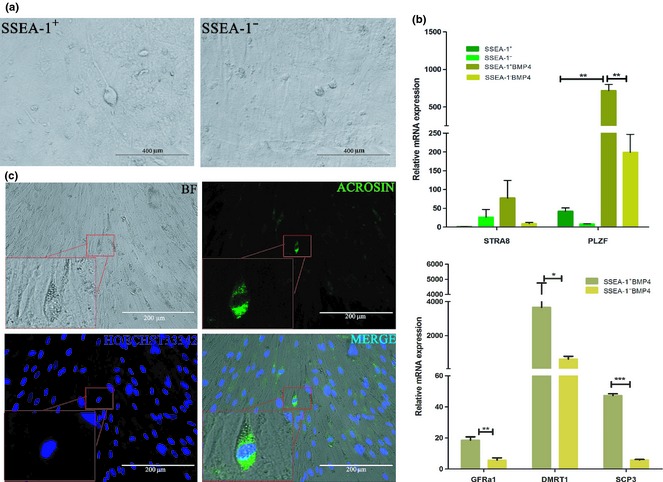
Analysis of male germ cell‐specific markers. (a) SSEA‐1+ and SSEA‐1− hUC‐MSCs were cultured on MEF feeder cells, and were induced by BMP4. Sperm‐like cells were observed in SSEA‐1+ cell groups. (b) qRT‐PCR analysis of expression levels of STRA8, PLZF, GFRa1, DMRT1 and SCP3 between the BMP4‐induced and untreated SSEA‐1+ as well as SSEA‐1− cells. Expression levels of these were increased significantly in SSEA‐1+ cells. (c) Immunofluorescence staining of sperm‐like cells on feeder cells, positive for Acrosin.
Discussion
Multipotential MSCs have the ability to differentiate into a variety of lineages, including bone, adipocyte, osteoblast and hepatocyte‐like cells and even gametes in vivo or in vitro 1, 16, 19, 20, 21, 22. UC‐derived MSCs may be an excellent alternative source of bone marrow stem cells as they may be considered to be ‘younger’ than other adult stem cells 4, 13; moreover, the human umbilical cords is easy to access, and would be free of immuno‐rejection if applied autologously, clinically 23.
The BMP signal is of great importance for inhibition of ES cell differentiation towards neurons. First, both BMP and LIF up‐regulate Id family proteins to maintain the self‐renewal ability of ES cells 21; secondly, BMP4 can bind to the second function domain of ovol2 through smad1/5/8 to upregulate expression of ovol2 and up‐regulation of ovol2 inhibits ES cell differentiation towards neurons to some extent 24. Considering these findings, presence of BMP4 may also inhibit hUC‐MSCs to differentiate towards neurons. Also, BMP4 can promote ES cell differentiation to PGC 25, 26, and it can also stimulate BMSCs to express some early markers of germ cells 10. Based on this knowledge, we took advantage of BMP4 to induce differentiation of hUC‐MSC. As expected, BMP4‐induced cells expressed germ cell‐specific markers; these cells were mainly SSEA‐1+. Our results demonstrated an efficient method for generating germ‐like cells derived from hUC‐MSC.
For maintenance of human embryonic stem (hES) cells, signalling molecules, such as FGF, Wnt and TGF‐β, all play important regulatory roles to retain high expression levels of Oct4, Sox2 and other transcription factors, hence maintaining pluripotency of ES cells. As an important member of the TGF‐β superfamily, BMP has dual functions for human ES cells, as it can not only suppress self‐renewal ability of hES cells, but also induce hES cell differentiation, the effects of which are all dependent on concentration of BMP. Some reports have shown that during the process of embryonic differentiation, BMP4 can regulate hESC differentiation to trophoblast cells 11, 27. It has also been demonstrated that by molecular switching such as Nanog, BMP4 can activate the FGF signalling pathway, and through regulating downstream MEK‐ERK to control hESC differentiation to mesendoderm 28.
Recently, it has been reported that during the differentiation process of murine ES cells, BMP4 can inhibit the ERK signalling pathway by upregulating ERK‐specific dual‐specificity phosphatase 9 (an important downstream molecule of MAPK) 29; this inhibition affects the MAPK signalling pathway of ES cells to some extent. As MAPK signalling is present in various types of cells, to regulate cell differentiation 30, BMP4 may be of great importance for controlling differentiation of ES cells. ES cells and BMSCs can differentiate to PGCs effectively with induction by BMP4 10, 25. Human PGCs have similar origins as murine PGCs, both are differentiated from ES cells, and SSEA‐1 expression level would be upregulated during the process of this ES differentiation 31. In our study, hUC‐MSCs shrank in volume under BMP4 treatment, and some spherical cells appeared. Further analysis revealed that these round‐shaped cells expressed high levels of PGC‐specific markers, SSEA‐1 31, PRDM1, STELLA, PRDM14, NANOG, VASA 17, 26, 32, 33, 34 and C‐KIT 34. Based on this, they were regarded to be PGC‐like cells. MACS is a technique beneficial for maintaining high vitality of cells during the sorting process and does not affect following culture of the isolated cells 4. By MACS, we sorted SSEA‐1+ and SSEA‐1− cells and found that SSEA‐1+ cells were efficiently purified from hUC‐MSCs, and were the main cell type sensitive to BMP4 induction.
As SSEA‐1 is an important marker of PGC, and considering formation of PGCs 8, 16, 35 when the BMP4 signal is activated, PRDM1 begins to be expressed; when BMP4 secretion level peaks, PRDM1‐positive cells are viewed as being precursor PGCs 26. Subsequently, other markers, including C‐KIT 36, begin to be expressed. Thus, we postulated that in our model, under induction by BMP4, SSEA‐1+ UC‐MSCs expressed PRDM1 and later, expression levels of C‐KIT, STELLA, PRDM14, NANOG and VASA were elevated accordingly, then forming PGC‐like cells. As SSEA‐1 is a PGC‐specific marker, we postulated that SSEA‐1+ UC‐MSCs were very close to PGCs, and SSEA‐1+ UC‐MSCs easily transdifferentiated into PGC‐like cells, which then differentiated to the direction of male germ‐like cells. Plzf is a critical transcriptional factor for self‐renewal of spermatogonial stem cells (SSCs) 37, and we observed its expression in BMP4‐treated male germ‐like cells. During the differentiation process of PGCs to germ cells, STRA8 was considered to be the first putative marker and switch to meiosis in mammals 38. SCP‐3 is a sister chromatid arm cohesin of mammalian meiosis I, and first appears in leptotene‐stage spermatocytes and disappears in late meiotic cells 33. Sex‐determining gene‐DMRT1 39 and Acrosin a major protease, are present in the acrosome of mature mammalian spermatozoa. Our results indicate that MACS sorted SSEA‐1+ cells differentiated into PLZF, STRA8, SCP‐3, GFRa1, DMRT1, SCP3 and even Acrosin‐positive male germ‐like cells when treated with BMP4. We concluded that under induction of BMP4, PGC‐like cells derived from SSEA‐1+ hUC‐MSCs differentiated into male germ‐like cells by upregulating expression of STRA8, and thence its downstream genes DMRT1, PLZF and SCP3 were upregulated. Thus, we proposed the possible pathway of hUC‐MSC differentiation towards male germ‐like cells under induction of BMP‐4 (Fig. 6).
Figure 6.
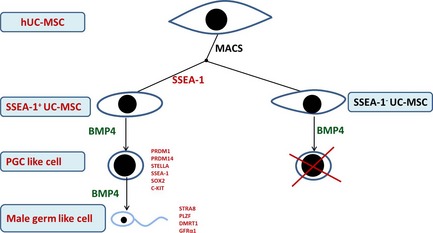
Possible pathway of hUC ‐MSC differentiation towards male germ‐like cells under induction of BMP4. In the presence of BMP4, SSEA‐1+ cells in hUC‐MSCs upregulated PGC‐associated transcriptional factors, PRDM1, PRDM14, STELLA, SSEA‐1, C‐KIT and SOX2. Meanwhile, these cells shrank in size, became round‐shaped in morphology, increased nucleus‐cytoplasmic ratio. Thereafter, male germ‐cell markers, STRA8, SCP3, DMRT1 and PLZF, greatly increase. Finally, these cells were induced to differentiate towards male germ cells.
In conclusion, we found that BMP4 efficiently induced hUC‐MSCs to differentiate into SSEA‐1+, round‐shaped PGC‐like cells, and that SSEA1+ cells further differentiated into sperm‐like cells. These results indicate that SSEA‐1+ UC‐MSCs differentiated into male germ cells under induction of BMP4.
Acknowledgements
This work was supported by the grants from the Program of National Natural Science Foundation of China (31272518), the Key Project of Chinese Ministry of Science and Technology (2013CB947900).
References
- 1. Jiang YH, Jahagirdar BN, Reinhardt RL, Schwartz RE, Keene CD, Ortiz‐Gonzalez XR et al (2002) Pluripotency of mesenchymal stem cells derived from adult marrow. Nature 418, 41–49. [DOI] [PubMed] [Google Scholar]
- 2. Sanchez‐Ramos J, Song S, Cardozo‐Pelaez F, Hazzi C, Stedeford T, Willing A et al (2000) Adult bone marrow stromal cells differentiate into neural cells in vitro . Exp. Neurol. 164, 247–256. [DOI] [PubMed] [Google Scholar]
- 3. Gopalakrishnan V, Vignesh R, Arunakaran J, Aruldhas M, Srinivasan N (2006) Effects of glucose and its modulation by insulin and estradiol on BMSC differentiation into osteoblastic lineages. Biochem. Cell Biol. 84, 93–101. [DOI] [PubMed] [Google Scholar]
- 4. Nayernia K, Lee JH, Drusenheimer N, Nolte J, Wulf G, Dressel R et al (2006) Derivation of male germ cells from bone marrow stem cells. Lab. Invest. 86, 654–663. [DOI] [PubMed] [Google Scholar]
- 5. Nakaki F, Hayashi K, Ohta H, Kurimoto K, Yabuta Y, Saitou M (2013) Induction of mouse germ‐cell fate by transcription factors in vitro . Nature 501, 222–226. [DOI] [PubMed] [Google Scholar]
- 6. Kee K, Gonsalves JM, Clark AT, Pera RAR (2006) Bone morphogenetic proteins induce germ cell differentiation from human embryonic stem cells. Stem Cells Dev. 15, 831–837. [DOI] [PubMed] [Google Scholar]
- 7. Qiu P, Bai Y, Liu C, He X, Cao H, Li M et al (2012) A dose‐dependent function of follicular fluid on the proliferation and differentiation of umbilical cord mesenchymal stem cells (MSCs) of goat. Histochem. Cell Biol. 138, 593–603. [DOI] [PubMed] [Google Scholar]
- 8. Ohinata Y, Ohta H, Shigeta M, Yamanaka K, Wakayama T, Saitou M (2009) A signaling principle for the specification of the germ cell lineage in mice. Cell 137, 571–584. [DOI] [PubMed] [Google Scholar]
- 9. Dudley BM, Runyan C, Takeuchi Y, Schaible K, Molyneaux K (2007) BMP signaling regulates PGC numbers and motility in organ culture. Mech. Dev. 124, 68–77. [DOI] [PubMed] [Google Scholar]
- 10. Mazaheri Z, Movahedin M, Rahbarizadeh F, Amanpour S (2011) Different doses of bone morphogenetic protein 4 promote the expression of early germ cell‐specific gene in bone marrow mesenchymal stem cells. In Vitro Cell. Dev. Biol. Anim. 47, 521–525. [DOI] [PubMed] [Google Scholar]
- 11. Erb TM, Schneider C, Mucko SE, Sanfilippo JS, Lowry NC, Desai MN et al (2011) Paracrine and epigenetic control of trophectoderm differentiation from human embryonic stem cells: the role of bone morphogenic protein 4 and histone deacetylases. Stem Cells Dev. 20, 1601–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hiller M, Liu C, Blumenthal PD, Gearhart JD, Kerr CL (2011) Bone morphogenetic protein 4 mediates human embryonic germ cell derivation. Stem Cells Dev. 20, 351–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hua J, Pan S, Yang C, Dong W, Dou Z, Sidhu KS (2009) Derivation of male germ cell‐like lineage from human fetal bone marrow stem cells. Reprod. Biomed. Online 19, 99–105. [DOI] [PubMed] [Google Scholar]
- 14. Desbaillets I, Ziegler U, Groscurth P, Gassmann M (2000) Embryoid bodies: an in vitro model of mouse embryogenesis. Exp. Physiol. 85, 645–651. [PubMed] [Google Scholar]
- 15. Hua J, Qiu P, Zhu H, Cao H, Wang F, Li W (2011) Multipotent mesenchymal stem cells (MSCs) from human umbilical cord: potential differentiation of germ cells. Afr. J. Biochem. Res. 5, 113–123. [Google Scholar]
- 16. Saitou M, Yamaji M (2012) Primordial germ cells in mice. Cold Spring Harb. Perspect. Biol. 4, a008375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hayashi K, Ohta H, Kurimoto K, Aramaki S, Saitou M (2011) Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell 146, 519–532. [DOI] [PubMed] [Google Scholar]
- 18. Magnusdottir E, Dietmann S, Murakami K, Gunesdogan U, Tang F, Bao S et al (2013) A tripartite transcription factor network regulates primordial germ cell specification in mice. Nat. Cell Biol. 15, 905–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rocha B, Calamia V, Mateos J, Fernandez‐Puente P, Blanco FJ, Ruiz‐Romero C (2012) Metabolic labeling of human bone marrow mesenchymal stem cells for the quantitative analysis of their chondrogenic differentiation. J. Proteome Res. 11, 5350–5361. [DOI] [PubMed] [Google Scholar]
- 20. Liu Y, Berendsen AD, Jia S, Lotinun S, Baron R, Ferrara N et al (2012) Intracellular VEGF regulates the balance between osteoblast and adipocyte differentiation. J. Clin. Invest. 122, 3101–3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang X, Liao S, Nelson ER, Schmalzigaug R, Spurney RF, Guilak F et al (2012) The cytoskeletal regulatory scaffold protein GIT2 modulates mesenchymal stem cell differentiation and osteoblastogenesis. Biochem. Biophys. Res. Commun. 425, 407–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yu X, Wang N, Qiang R, Wan Q, Qin M, Chen S et al (2014) Human amniotic fluid stem cells possess the potential to differentiate into primordial follicle oocytes in vitro. Biol. Reprod. 113, 112920. [DOI] [PubMed] [Google Scholar]
- 23. Romanov YA, Svintsitskaya VA, Smirnov VN (2003) Searching for alternative sources of postnatal human mesenchymal stem cells: candidate MSC‐like cells from umbilical cord. Stem Cells 21, 105–110. [DOI] [PubMed] [Google Scholar]
- 24. Zhang T, Zhu Q, Xie Z, Chen Y, Qiao Y, Li L et al (2013) The zinc finger transcription factor Ovol2 acts downstream of the bone morphogenetic protein pathway to regulate the cell fate decision between neuroectoderm and mesendoderm. J. Biol. Chem. 288, 6166–6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Makoolati Z, Movahedin M, Forouzandeh‐Moghadam M (2011) Bone morphogenetic protein 4 is an efficient inducer for mouse embryonic stem cell differentiation into primordial germ cell. In Vitro Cell. Dev. Biol. Anim. 47, 391–398. [DOI] [PubMed] [Google Scholar]
- 26. Saitou M, Barton SC, Surani MA (2002) A molecular programme for the specification of germ cell fate in mice. Nature 418, 293‐300. [DOI] [PubMed] [Google Scholar]
- 27. Xu R‐H, Chen X, Li DS, Li R, Addicks GC, Glennon C et al (2002) BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat. Biotechnol. 20, 1261–1264. [DOI] [PubMed] [Google Scholar]
- 28. Yu P, Pan G, Yu J, Thomson JA (2011) FGF2 sustains NANOG and switches the outcome of BMP4‐induced human embryonic stem cell differentiation. Cell Stem Cell 8, 326–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li Z, Fei T, Zhang J, Zhu G, Wang L, Lu D et al (2012) BMP4 Signaling Acts via dual‐specificity phosphatase 9 to control ERK activity in mouse embryonic stem cells. Cell Stem Cell 10, 171–182. [DOI] [PubMed] [Google Scholar]
- 30. Ying QL, Wray J, Nichols J, Batlle‐Morera L, Doble B, Woodgett J et al (2008) The ground state of embryonic stem cell self‐renewal. Nature 453, 519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thomson JA, Odorico JS (2000) Human embryonic stem cell and embryonic germ cell lines. Trends Biotechnol. 18, 53–57. [DOI] [PubMed] [Google Scholar]
- 32. Park TS, Galic Z, Conway AE, Lindgren A, van Handel BJ, Magnusson M et al (2009) Derivation of primordial germ cells from human embryonic and induced pluripotent stem cells is significantly improved by coculture with human fetal gonadal cells. Stem Cells 27, 783–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tunyaplin C, Shapiro MA, Calame KL (2000) Characterization of the B lymphocyte‐induced maturation protein‐1 (Blimp‐1) gene, mRNA isoforms end basal promoter. Nucleic Acids Res. 28, 4846–4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vincent JJ, Li ZW, Lee SA, Liu X, Etter MO, Diaz‐Perez SV et al (2011) Single cell analysis facilitates staging of Blimp1‐dependent primordial germ cells derived from mouse embryonic stem cells. PLoS ONE 6, e28960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bikoff EK, Morgan MA, Robertson EJ (2009) An expanding job description for Blimp‐1/PRDM1. Curr. Opin. Genet. Dev. 19, 379–385. [DOI] [PubMed] [Google Scholar]
- 36. Mauduit C, Hamamah S, Benahmed M (1999) Stem cell factor/c‐kit system in spermatogenesis. Hum. Reprod. Update 5, 535–545. [DOI] [PubMed] [Google Scholar]
- 37. Song W, Zhu H, Li M, Li N, Wu J, Mu H et al (2013) Promyelocytic leukaemia zinc finger maintains self‐renewal of male germline stem cells (mGSCs) and its expression pattern in dairy goat testis. Cell Prolif. 46, 457–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhou Q, Li Y, Nie R, Friel P, Mitchell D, Evanoff RM et al (2008) Expression of stimulated by retinoic acid gene 8 (Stra8) and maturation of murine gonocytes and spermatogonia induced by retinoic acid in vitro. Biol. Reprod. 78, 537–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Llera‐Herrera R, Garcia‐Gasca A, Abreu‐Goodger C, Huvet A, Ibarra AM (2013) Identification of male gametogenesis expressed genes from the scallop Nodipecten subnodosus by suppressive subtraction hybridization and pyrosequencing. PLoS ONE 8, e73176. [DOI] [PMC free article] [PubMed] [Google Scholar]


