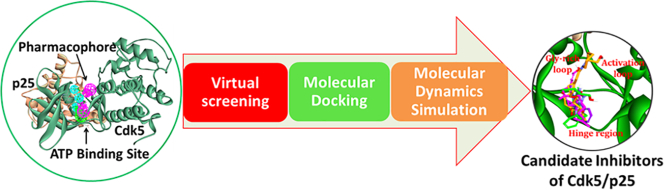
Abstract
Deregulation of Cdk5 is a hallmark in neurodegenerative diseases and its complex with p25 forms Cdk5/p25, thereby causes severe neuropathological insults. Cdk5/p25 abnormally phosphorylates tau protein, and induces tau-associated neurofibrillary tangles in neurological disorders. Therefore, the pharmacological inhibition of Cdk5/p25 alleviates tau-associated neurological disorders. Herein, computational simulations probed two candidate inhibitors of Cdk5/p25. Structure-based pharmacophore investigated the essential complementary chemical features of ATP-binding site of Cdk5 in complex with roscovitine. Resultant pharmacophore harbored polar interactions with Cys83 and Asp86 residues and non-polar interactions with Ile10, Phe80, and Lys133 residues of Cdk5. The chemical space of selected pharmacophore was comprised of two hydrogen bond donors, one hydrogen bond acceptor, and three hydrophobic features. Decoy test validation of pharmacophore obtained highest Guner-Henry score (0.88) and enrichment factor score (7.23). The screening of natural product drug-like databases by validated pharmacophore retrieved 1126 compounds as candidate inhibitors of Cdk5/p25. The docking of candidate inhibitors filtered 10 molecules with docking score >80.00 and established polar and non-polar interactions with the ATP-binding site residues of Cdk5/p25. Finally, molecular dynamics simulation and binding free energy analyses identified two candidate inhibitors of Cdk5/p25. During 30 ns simulation, the candidate inhibitors established <3.0 Å root mean square deviation and stable hydrogen bond interactions with the ATP-binding site residues of Cdk5/p25. The final candidate inhibitors obtained lowest binding free energies of −122.18 kJ/mol and − 117.26 kJ/mol with Cdk5/p25. Overall, we recommend two natural product candidate inhibitors to target the pharmacological inhibition of Cdk5/p25 in tau-associated neurological disorders.
Keywords: Cdk5/p25 inhibitors, Structure-based pharmacophore modeling, Tau-pathogenesis, Molecular docking, Molecular dynamics simulation
Abbreviations: Cdk5, Cyclin-dependent kinase 5; Cdks, Cyclin-dependent kinases; CGMC, Cyclin-dependent kinases, mitogen-activated protein kinases, glycogen synthase kinases, and Cdk-like kinases; AD, Alzheimer's disease; PD, Parkinson's disease; HD, Hungtington's disease; MPTP, 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine; P5, A 24-residues mimetic peptide of p35; TAT, Twin-arginine targeting; Aβ, Amyloid beta; ZNPD, Zinc Natural Product Database; IBS, InterBioScreen; DS, Discovery Studio; GH, Guner-Henry; EF, Enrichment factor; ROF, Rule of five; ADMET, Absorption, distribution, metabolism, excretion, and toxicity; BBB, Blood-brain barrier; GOLD, Genetic optimization of ligand docking; PLP, Piecewise linear potential; ASP, Astex statistical potential; MD, Molecular dynamics; PME, Particle mesh ewald; K, kelvin; TIP3P, Transferable intermolecular potential with 3 points; GROMACS, Groningen Machine for Chemical Simulation; NVT, Number of particles, volume, and temperature; NPT, Number particle, pressure, and temperature; PDB, Protein databank; H-bond, Hydrogen bond; HBA, Hydrogen bond acceptor; HBD, Hydrogen bond donor; HYP, Hydrophobic; RMSD, Root mean square deviation; 2D, Two-dimentional; 3D, Three-dimentional; GA, Genetic algorithm; GFA, Genetic Function Approximation
Graphical Abstract
Highlights
-
•
Structure-based drug designing identified two novel natural product inhibitors of Cdk5/p25.
-
•
The novel inhibitors could cross blood-brain barrier to abrogate Tau-associated neurological insults in brain.
-
•
The recommended inhibitors of Cdk5/p25 established strong polar interactions with Cys83, Glu81, Asn131 and Asn144 of Cdk5.
-
•
Candidate inhibitors showed higher binding affinity with Cdk5/p25.
1. Introduction
Cyclin-dependent kinases (Cdks) form protein kinase sub-family named CMGC (cyclin-dependent kinases, mitogen-activated protein kinases, glycogen synthase kinases, and Cdk-like kinases) [1]. Cdks are proline-directed serine/threonine protein kinases and need a regulatory subunit for activation. Human Cdks family is comprised of thirteen member Cdks (Cdk1-Cdk13) and preferentially express during mitosis [2]. Cdk enzymes regulate post-translational modification of proteins by transferring a phosphate molecule from ATP to substrate(s) [3].
Despite high sequence homology with other Cdks (~70%), Cdk5 is an atypical member of Cdks and is activated by its co-activator either p35 or p39 [4,5]. Cdk5 expression is indispensible in developmental biology of brain and neurons [6]. Cdk5 regulates several neuronal functions; for instance, synaptic plasticity, pain signaling, learning and memory formation, and drug addiction. Deregulation of Cdk5 induces neurodegenerative diseases such as Alzheimer's disease (AD), Parkinson's disease (PD), and Hungtington's disease (HD) [7]. Previously, Cdk5-mediated aberrant processing of amyloid beta (Aβ) has been investigated in AD samples [[8], [9], [10]]. In PD mouse models, the administration of 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) toxin accumulated high concentration of Cdk5/p25 complex in dopaminergic neurons and caused neuronal death. Further, Cdk5 inhibition attenuated MPTP-induced damages of nigrostriatal dopaminergic neurons and proposed as an essential regulator in Parkinson's disease [11]. Another study showed that abnormal phosphorylation of endophilin B1 by Cdk5 induces autophagy in PD model [12]. In HD, deregulation of Cdk5 is associated with striatal neuronal death and contributes to learning and memory deficits [13,14].
Cdk5 is an attractive therapeutic target to alleviate neurotoxicity and tau pathology in several neurodegenerative diseases [15]. For instance, Cdk5 inhibition has been targeted to protect neurons in stress condition. Chang et al. investigated that Cdk5 inhibition protects neuronal death in Aβ(1–42) toxicity [16]. In another study, Tan et al. (2015) demonstrated that P5 (a 24-residues peptide mimicking p35) can cross blood-brain barrier (BBB) and cell membrane which is facilitated by twin-arginine targeting (TAT) protein. This study reported that the administration of p5-TAT could not affect the cellular level of Cdk5 activators (p39, p35, and p25), but abrogated tau and glucocorticoid receptor (GR) phosphorylation [17]. Previously, a study has reported that (R)-roscovitine is a Cdk5 inhibitor and could efficiently attenuate microglial proliferation in ischemic rats [18]. The same study also described the interaction mechanisms of two other inhibitors including Aloisine A and Indirubin-3′-monoxime. Dinaciclib (SCH 727965) is another potent inhibitor of Cdks and obtained low IC50 value (1 nM) against Cdk5 with higher inhibition activity [19]. Dinaciclib could successfully exhibit promising therapeutic index in solid tumors, suppression of retinoblastoma phosphorylation and inhibition of bromodeoxyuridine incorporation in tumor cell lines. Sundaram et al. demonstrated selective inhibition of Cdk5 by overexpressing Cdk5 inhibitory peptide (CIP) and rescued neurodegenerative pathologies caused by Cdk5/p25 hyperphosphorylation. The same study showed that CIP-p25 tetra transgenic mice abrogated tau and amyloid pathologies and neuroinflammation [20]. TFP5 peptide is a 24-amino acid peptide (Lys254-Ala277), which is truncated from p35, and its intraperitoneal injection showed significant reduction of ischemia in adult ischemic stroke rats [21]. PHA-793887 is a selective inhibitor of Cdk5/p25 with IC50 value of 5 nM and currently entered the Phase 1 clinical trials [22]. PHA-793887 remained potent inhibitor in several cancer types such as human ovarian A2780, colon HCT-116 and pancreatic BX-PC3 carcinoma xenograft models.
Being a critical regulator of neurological disorders, pharmacotherpeutics of Cdk5 is very limited. The available inhibitors of Cdk5 are mainly tested in several cancer types. Nevertheless, involvement of Cdk5 in neurological disorders is essentially studied by mutation studies of either Cdk5 or its co-activators [[23], [24], [25]]. Therefore, the identification of novel drug-like molecules to target Cdk5/p25 is a fundamental approach to combat neurological disorders. Herein, we have exploited the conserved interaction pattern of Cdk5 and its state-of-the-art inhibitor (R)-roscovitine and identified promising natural product inhibitors of Cdk5/p25. In brief, a structure-based pharmacophore model was designed to reflect the essential chemical space of Cdk5/p25 inhibition. The pharmacophore model was validated by decoy test method. The natural product drug-like databases were built from InterBioScreen Database (IBSD) and Zinc Natural Product Database (ZNPD), and subsequently screened by the validated pharmacophore model. Molecular docking simulation was employed to identify the best docked drug-like molecules in the ATP-binding site of Cdk5/p25. Furthermore, molecular dynamics simulation was employed and the final candidate hit molecules of Cdk5/p25 were filtered. Finally, MM/PBSA approach was followed and the binding free energies of the final candidate inhibitors of Cdk5/p25 were identified.
2. Material and Methods
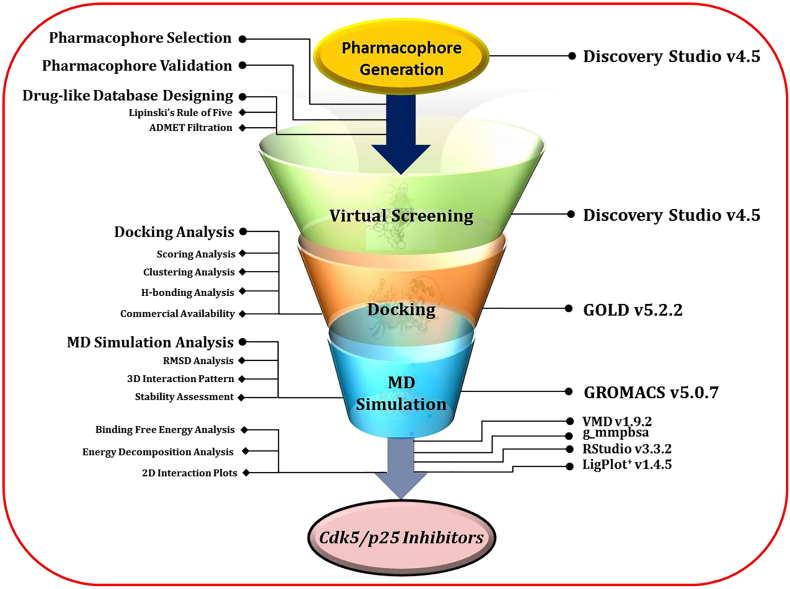
The schematic representation of pharmacophore modeling, virtual screening, molecular docking, molecular dynamics simulation, and binding free energy analysis is summarized in Fig. 1.
Fig. 1.
Schematic representation. Structure-based pharmacophore generation, drug-like database designing and virtual screening was performed in Discovery Studio v4.5. Molecular docking simulation was carried out through Genetic Optimization of Ligand Docking (GOLD v5.2.2) package. Molecular dynamics simulations were carried out in GROMACS v5.0.7. g_mmpbsa is a GROMACS plugin tool and estimated the binding free energy of Cdk5/p25 and the corresponding ligand(s). RStudio v3.3.2 and LigPlot+v1.4.5 were employed for energy decomposition and 2D interaction pattern, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.1. Generation of Pharmacophore Model
To get insight into Cdk5-inhibitor interaction, a pharmacophore model was generated. The crystal structure of human Cdk5 in complex with p25 and roscovitine was taken from protein data bank [26]. The structure of Cdk5/p25 was prepared and the unwanted molecules such as salts and water were removed. Residues of Cdk5 in closed vicinity of the (R)-roscovitine (8.0 Å) were identified by Binding and Edit module of Discovery Studio v4.5 (Accelrys Inc. San Diego, USA). The roscovitine-complemented pharmacophoric features of Cdk5 residues were identified by using Interaction Generation protocol of Discovery Studio v4.5 (hereafter DS). Afterwards, the Receptor-ligand Pharmacophore Generation tool of DS was employed and pharmacophore models were generated. The best pharmacophore model was selected on the basis of high selectivity score and compatible interactions with catalytic active residues of the ATP-binding site of Cdk5/p25. The selectivity score classifies pharmacophore models on the basis of sensitivity and specificity of novel ligands against the receptor, and the top-ranked models are returned [27]. During the selectivity score determination, the generated pharmacophore models are enumerated against a diverse 3D database of drug-like molecules available in DS and the selectivity is measured by introducing Genetic Function Approximation (GFA) approach [[27], [28], [29]]. The GFA model is built from a training set of 1544 pharmacophore models in DS. The number of pharmacophoric features varies from two to eight as per pharmacophore and each set is used to screen CapDiverse database in DS. The GFA model is trained on descriptors obtained from the total number of features in pharmacophore models and the inter-features distance bin values.
2.2. Validation of Pharmacophore Model
Decoy test method [30] was used to evaluate the suitability of pharmacophore to identify Cdk5/p25 inhibitors. In brief, the already known inhibitors of Cdk5 were collected by literature mining. The inhibitors whose experimental activities were determined by the same biological assays were included and assigned as test set. The test set was incorporated with inactive compounds of Cdk5, and assigned as the decoy test set and hence was comprised of active and inactive molecules of Cdk5/p25. The decoy test set was screened by the selected pharmacophore model to retrieve the best-fitted molecules. The screening was performed by Ligand Pharmacophore Mapping protocol, implanted in DS. The pharmacophore-retrieved molecules were used to calculate Guner-Henry (GH) score and enrichment factor (EF) score as:
| (1) |
| (2) |
where GH: Guner-Henry score, EF: enrichment factor, D: total number of molecules in the decoy test set, A: number of Cdk5/p25 inhibitors (active molecules) in the decoy test set, Ha: number of Cdk5/p25 inhibitors (active molecules) in the hit list, Ht: number of pharmacophore-retrieved hits.
Other parameters including percent yield (%Y), false positive, and false negative were also estimated during the validation of pharmacophore. The GH score ranges between 0 and 1 and indicates null and ideal models, respectively. Pharmacophore model with GH score higher than 0.70 is considered as valid model for virtual screening of large databases [31,32]. The enrichment factor (EF) determines the specificity and selectivity of the model to identify active compounds during the screening of decoy test set. The higher the EF value the higher will be the selectivity of the model [[33], [34], [35]].
2.3. Development of Drug-like Database and Virtual Screening
In this study, two natural product databases; InterBioScreen (https://www.ibscreen.com/) and Zinc Natural Product Database (http://zinc.docking.org/) were taken and a drug-like database was developed. Lipinski's Rule of Five (ROF) and ADMET Descriptors modules of DS were employed to evaluate the physiochemical and pharmacokinetics and pharmacodynamics properties of drug-like databases, respectively.
Subsequently, pharmacophore model was utilized as a 3D query and the drug-like database was screened by employing the Ligand Pharmacophore Mapping protocol of DS. During screening, the search option was assigned as Best/Flexible, while the Maximum Omitted Features option was set to “0” to ensure the mapping of all features of the pharmacophore.
2.4. Molecular Docking Simulation
For docking analysis, the previously well-established procedure was followed [[35], [36], [37]]. In brief, the crystal structure of Cdk5/p25 (PDB ID: 1UNL) was taken from Protein Data Bank (https://www.rcsb.org/) [26]. Prior to docking, the structure of Cdk5/p25 was cleaned and unwanted molecules were removed. The ligand binding coordinates of the ATP binding site of Cdk5/p25 were calculated from the inbound ligand (roscovitine) by Define and Edit Binding Site module, implanted in DS. The drug-like molecules from virtual screening were prepared and energy minimized in DS. Finally, the docking studies were carried out in Genetic Optimization of Ligand Docking (GOLD v5.2.2) package [38]. GOLD uses genetic algorithm for docking and allows full flexibility of small molecules and partial flexibility of protein during the docking operation. During docking, the hydrogen atoms of Cdk5/p25 were replaced by Add Hydrogen module of GOLD package. The scoring functions ChemPLP (piecewise linear potential) and ASP (astex statistical potential) were employed as default scoring and rescoring functions, respectively [39]. ChemPLP optimizes pose prediction and steric complementarity between protein and ligand. ASP score estimates inter-atomic potential and has comparable accuracy to Chemscore and Goldfitness scoring functions. Furthermore, the inherent Genetic Algorithm (GA) module of GOLD package was set to hundred (100) to produce one hundred poses for each pharmacophore-retrieved drug-like molecule. During docking, roscovitine and indirubin-3′-monoxime (another inhibitor of Cdk5) were used as reference compounds and referred as REF1 and REF2, respectively, throughout the analysis. ChemPLP and ASP scores, clustering analysis, and hydrogen bond interactions with catalytic active residues of Cdk5/p25 were used as selection parameters during the docking analysis.
2.5. Molecular Dynamics Simulation
Molecular dynamics (MD) simulation was employed to understand the mechanistic mechanism of Cdk5/p25 and candidate hit molecules in simulated physiological environment. Simulations were performed in Groningen Machine for Chemical Simulation (GROMACS v5.0.7) package [40]. The parameters and coordinate files of Cdk5/p25 and candidate hit molecules were generated by CHARMM36m forcefield [40] in GROMACS and SwissParam [41], respectively. Each simulation system was prepared in octahedral box, hydrated by TIP3P water model and neutralized by 0.1 M NaCl solution. Initially, each system was minimized by applying a maximum force of 10 kJ/mol to avoid steric hindrance. Prior to simulation, each system was equilibrated. An NVT ensemble (constant number of particles, volume, and temperature) was employed at 300 K for 100 ps by V-rescale thermostate and temperature was equilibrated. The pressure of each system was equilibrated at 1.0 bar by Parrinello-Rahman barostat [42]. Each equilibrated system was simulated for 30 ns. During simulation, particle mesh ewald (PME) was used to estimate long-range electrostatic interactions. V-rescale thermostat and Parrinello-Rahman barostat were employed to maintain the temperature and pressure at 300 K and 1.0 bar, respectively. Bond lengths were restrained by LINCS algorithm [43].
2.6. Binding Free Energy Analysis
Estimation of the binding affinity of inhibitor(s) towards target protein plays a crucial role in computational drug designing [44]. In aqueous solution, the protein-ligand binding free energy is calculated as:
| (3) |
Herein, GROMACS plugin tool “g_mmpbsa” [45] was employed to estimate binding free energy of the final hit molecules and Cdk5/p25. In brief, time-equidistance conformations of Cdk5/p25 in complex with hit molecule(s) were extracted. The representative data frames were used and several energetic terms including polar and non-polar factors were estimated by MM/PBSA approach [46].
Data preparation and analyses were done by Discovery Studio v4.5 (DS), GOLD v5.2.2 package, GROMACS v5.0.7, LigPlot+ v1.4.5, Visual Molecular Dynamics Viewer (VMD v1.9.2), and RStudiov v3.3.2 package.
3. Results
3.1. Pharmacophore Model Generation
The crystal structure of Cdk5/p25 in complex with roscovitine was taken from Protein Data Bank (PDB ID: 1UNL) [26]. During pharmacophore generation, Cdk5/p25 and roscovitine were treated as the protein and inhibitor structures, respectively. The Receptor-ligand Pharmacophore Generation module of DS was employed and ten pharmacophore models were generated. Chemical characterization of each pharmacophore is given in Table 1. The resultant pharmacophores suggested that polar and non-polar interactions are the pre-dominant features of Cdk5/p25 inhibition (Fig. 2). Since, our results observed that pharmacophore 1 (hereafter Pharm1) obtained highest selectivity score of 11.22 and established key polar and non-polar interactions with the essential residues of ATP-binding site of Cdk5/p25, therefore, we selected Pharm1 for further analyses (Table 1).
Table 1.
Characterization of pharmacophores.
| Pharmacophore summery | |||
|---|---|---|---|
| Pharmacophore | Number of features | Featuresa | Selectivity score |
| Pharmacophore 1 | 6 | HBA, HBD, HBD, HYP, HYP, HYP | 11.224 |
| Pharmacophore 2 | 5 | HBA, HBD, HBD, HYP, HYP | 9.7088 |
| Pharmacophore 3 | 5 | HBA, HBD, HBD, HYP, HYP | 9.7088 |
| Pharmacophore 4 | 5 | HBA, HBD, HBD, HYP, HYP | 9.7088 |
| Pharmacophore 5 | 5 | HBD, HBD, HYP, HYP, HYP | 9.7088 |
| Pharmacophore 6 | 5 | HBA, HBD, HYP, HYP, HYP | 8.7953 |
| Pharmacophore 7 | 5 | HBA, HBD, HYP, HYP, HYP | 8.7953 |
| Pharmacophore 8 | 4 | HBA, HBD, HBD, HYP | 8.1940 |
| Pharmacophore 9 | 4 | HBA, DHB, HBD, HYP | 8.1940 |
| Pharmacophore 10 | 4 | HBA, HBD, HBD, HYP | 8.1940 |
Features: HBA, hydrogen bond acceptor; HBD, hydrogen bond donor; HYP, hydrophobic.
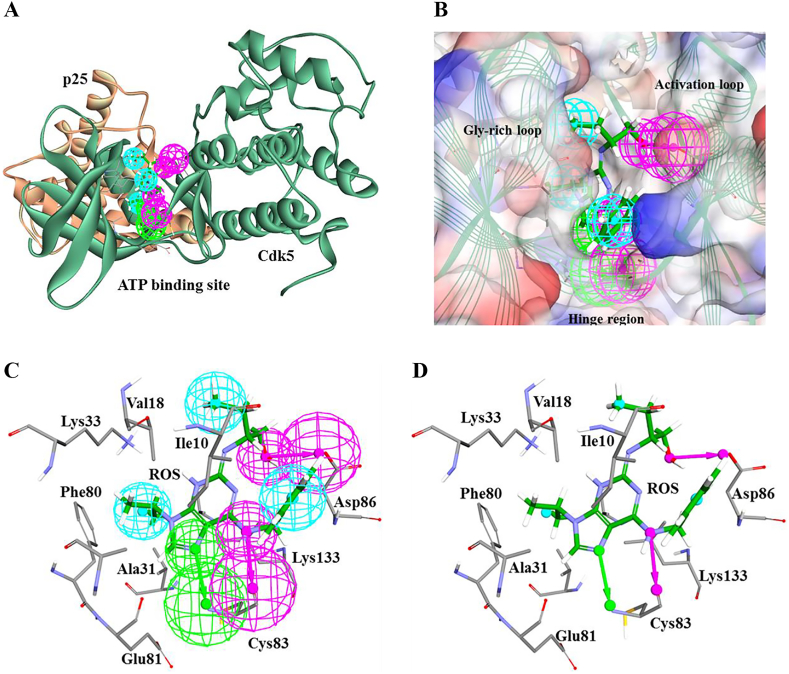
Fig. 2.
Generation and selection of suitable pharmacophore. A) Cdk5/p25 in complex with roscovitine was used and pharmacophores were generated. Cdk5 and p25 are shown as light green and light orange cartoon representations. Roscovitine is depicted as green stick representation and the correspondent pharmacophore is depicted as wire mesh representation in the ATP-binding site of Cdk5/p25. B) Represents the 3D orientation of selected pharmacophore in the ATP-binding site of Cdk5/p25. The ATP-binding site of Cdk5 is sub-divided into Gly-rich loop, activation loop, and the hinge region. C) and D) Illustrate the mapping of each pharmacohoric feature and the corresponding residue of Cdk5. Pharmacophore-associated residues of Cdk5 are depicted as stick representation and labeled. Roscovitine is shown as green colour stick representation and labeled as ROS. Green, magenta, and cyan colors depict hydrogen bond acceptor, hydrogen bond donor, and hydrophobic features of the pharmacophore, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Our results observed that Pharm1 occupied and oriented in the ATP-binding site of Cdk5/p25. Our modeling studies demonstrated that polar and non-polar interactions were pointed towards the hinge region (Cys83 residue of Cdk5) and glycine–rich (Gly-rich) loop of Cdk5, respectively (Fig. 2A, B). Our visual inspection observed that hydrogen bond donor and acceptor features of Pharm1 reflected polar interactions between the catalytic residue Cys83 of Cdk5 and roscovitine (Fig. 2C, D). An additional hydrogen bond donor was observed between the Asp86 residue of Cdk5 and roscovitine. Furthermore, a hydrophobic feature was sequestered by a residues triad (Lys33, Phe80, and Ala31 residues of Cdk5) and roscovitine. Another hydrophobic feature was associated with a residues dyad (Val18 and Ile10 residues of Cdk5) and hydrophobic tail of roscovitine (Fig. 2C, D).
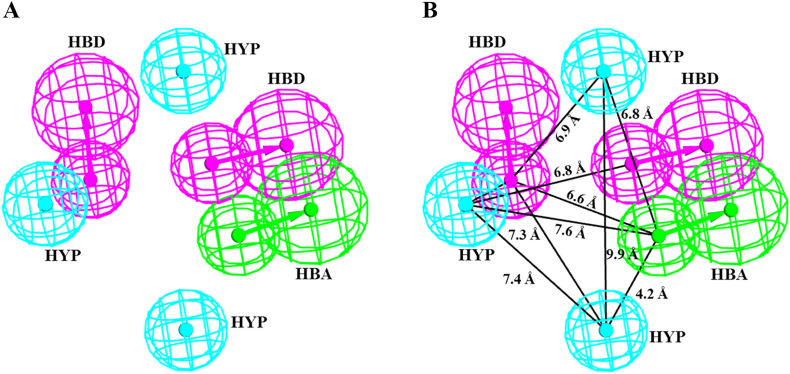
Our results observed that the chemical space of Pharm1 was comprised of six chemical features including two hydrogen bond donors, one hydrogen bond acceptor, and three hydrophobic features (Fig. 3A). The three dimensional (3D) spatial arrangement of the chemical features of Pharm1 reflected that one hydrogen bond acceptor and one hydrogen bond donor were oriented close to each other while the second hydrogen bond donor was distant oriented. Pharm1 suggested that the distribution of non-polar interactions (hydrophobic features) were quite random and covered the entire space (Fig. 3A). The inter-features distance of Pharm1 is depicted in Fig. 3B.
Fig. 3.
Chemical characterization of the selected pharmacophore. A) The selected pharmacophore has a total of six features including two hydrogen bond donors (HBD), one hydrogen bond acceptor (HBA), and three hydrophobic (HYP) features. HBA, HBD, and HYP are colored as green, magenta, and cyan, respectively. B) The inter-features distance of the selected pharmacophore is displayed in angstrom (Å). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. Quality Assessment Test (Validation) of Pharmacophore
Pharm1 was validated by employing as a 3D query to identify previously known inhibitors of Cdk5/p25 from a decoy test set. The decoy test set was comprised of randomly collected 453 molecules, where 57 molecules were incorporated as experimentally tested inhibitors of Cdk5 (Table 2).
Table 2.
Pharmacophore validation by decoy test method.
| Number | Parameter | Calculated values |
|---|---|---|
| 1 | Total number of molecules in the database (D) | 453 |
| 2 | Total number of active molecules of Cdk5 in the database (A) | 57 |
| 3 | Total number of active molecules of Cdk5 in the retrieved hits (Ha) | 51 |
| 4 | Number of retrieved hits by pharmacophore (Ht) | 56 |
| 5 | % Yield of actives [(Ha/Ht) × 100] | 0.91 |
| 6 | % Ratio of actives [(Ha/A) × 100] | 0.89 |
| 7 | False negative [A–Ha] | 6 |
| 8 | False positive [Ht – Ha] | 5 |
| 9 | Goodness of fit (GH) | 0.88 |
| 10 | Enrichment Factor (EF) | 7.23 |
During the decoy test set screening, Pharm1 mapped a total of 56 molecules, where 51 molecules were the active inhibitors of Cdk5 while 5 molecules were the inactive molecules of Cdk5. Our analysis suggested that Pharm1 achieved highest GH score and EF score of 0.88 and 7.23, respectively (Table 2). Furthermore, other parameters including percent yield, ratio of actives, and false positive and false negatives also suggested that Pharm1 preferentially mapped Cdk5 inhibitors only.
3.3. Designing and Virtual Screening of Natural Product Drug-like Databases by Pharm1
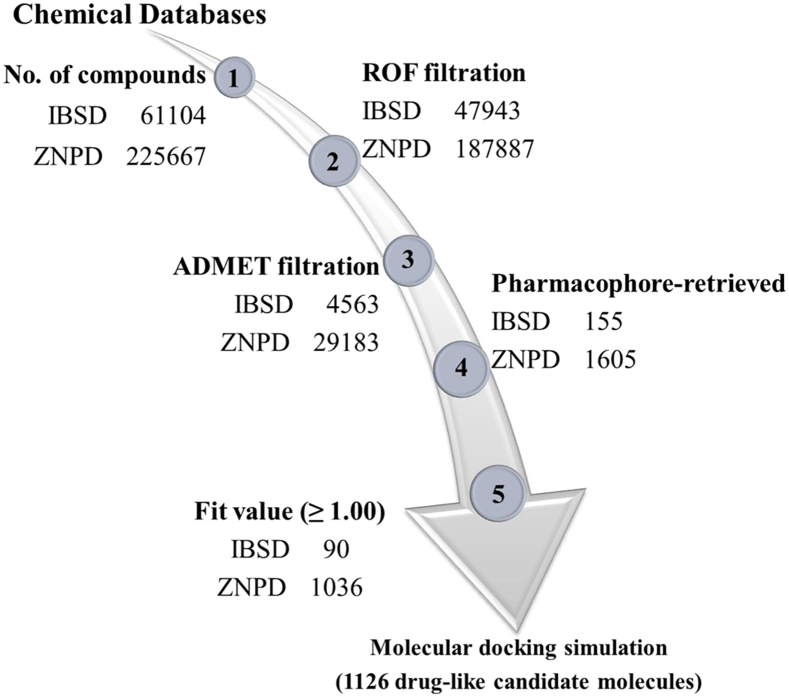
In computational drug designing, the physiochemical and pharmacokinetics and pharmacodynamics properties of chemical molecules are strictly evaluated. The Lipinski's Rule of Five and ADMET Descriptors modules of DS filtered 4563 and 29,183 compounds from natural product databases IBS and ZNPD, respectively (Fig. 4).
Fig. 4.
Designing and virtual screening of natural product drug-like databases. InterBioScreen (IBS) and Zinc natural product database (ZNPD) were used to design a drug-like database. Lipinski's rule of five (ROF) and absorption, distribution, metabolism, excretion, and toxicity (ADMET) filtration evaluated 4563 and 29,183 compounds as drug-like molecules from IBSD and ZNPD, respectively. The selected pharmacophore was used as a 3D query and a total of 155, and 1605 drug-like molecules were retrieved from IBSD and ZNPD, respectively. A fit value (≥ 1.00) was used and a total of 90 and 1036 natural product drug-like molecules were identified as the best-fitted molecules on pharmacophore.
The threshold values of parameters to evaluate the physiochemical and pharmacokinetics and pharmacodynamics properties of the natural product drug-like molecules are given in supplementary table S1. Since, our study has focused on the identification of natural product inhibitors to target Cdk5/p25 in brain. Therefore, we have strictly followed the hydrophobicity of candidate molecules to penetrate blood-brain barrier (BBB). The pharmacokinetics analysis of the final candidate hits obtained lowest BBB level of “1” as compared to the highest BBB level of “3” for both the reference compounds. Furthermore, the ADMET-BBB plot suggested that only the candidate hits occupied the BBB ellipses of high penetration (Fig. S4).
Pharm1 was employed as a 3D query to screen the natural product drug-like databases. The screening was carried out by Ligand Pharmacophore Mapping module of DS with Best/Flexible parameterizations. Pharm1 retrieved a total of 155 and 1605 compounds from IBSD and ZNPD, respectively (Fig. 4). Furthermore, a threshold fit value of ≥1.00 was used to filter the best-fitted molecules onto pharmacophore. Finally, a total of 90 and 1036 compounds were filtered from IBSD and ZNPD, respectively, as the best-fitted molecules for further analysis (Fig. 4).
3.4. Understanding the Mechanistic Interactions of Candidate Inhibitors and Cdk5/p25
Investigation of physical interaction of drug molecule and target protein is an essential step in computational drug designing [47]. The crystal structure of human Cdk5/p25 was taken as the receptor (Fig. S1) and the candidate hit molecules were treated as the ligands. The docking site of Cdk5/p25 was traced from inbound inhibitor (roscovitine) (Fig. S1). The docking results suggested that the reference compounds roscovitine (REF1) and indirubin-3′-monoxime (REF2) showed 84.03 and 32.49 and 62.23 and 29.18 as ChemPLP and ASP scores, respectively (Table 3).
Table 3.
Docking score analysis of REF1, REF2, Hit1 and Hit2 with Cdk5/p25.
| Compound | Docking score |
|
|---|---|---|
| ChemPLP | ASP | |
| REF1 | 84.03 | 32.49 |
| REF2 | 62.23 | 29.18 |
| Hit1 | 84.61 | 24.82 |
| Hit2 | 84.20 | 29.69 |
Since, the docking score of the reference compounds were used as an approximation, therefore, the threshold value of docking score was adjusted to ≥80.00 and the candidate hits were filtered (Table S2). Our results observed that only 52 candidate hit molecules from ZNPD could successfully follow the scoring criteria. Afterwards, the clustering analysis (conformational stability) observed 23 candidate inhibitors with cluster population >70% (Table S2). Finally, hydrogen bond (H-bond) analysis suggested that only 10 commercially available candidate hit molecules retained H-bonds with the catalytic active residue(s) of Cdk5/p25 (Table S2).
3.5. Molecular Dynamics Simulation
To understand the binding mode, stability, and molecular interaction pattern of the candidate hit molecules and Cdk5/p25 at simulated physiological condition, atomistic molecular dynamics simulation was carried out [48]. Herein, the 10 candidate molecules from docking simulation were simulated with Cdk5/p25 for 30 ns. Several parameters were evaluated to unveil the binding pattern, stability, and behavior of candidate hit molecules and Cdk5/p25. Among the ten candidate hits, two molecules (hereafter Hit1 and Hit2) were identified as the best candidate inhibitors of Cdk5/p25. The details of simulation systems of the reference compounds and final candidate hits are given in supplementary fig. S2 and table S3.
3.5.1. Root Mean Square Deviation (rmsd) and Potential Energy Analysis
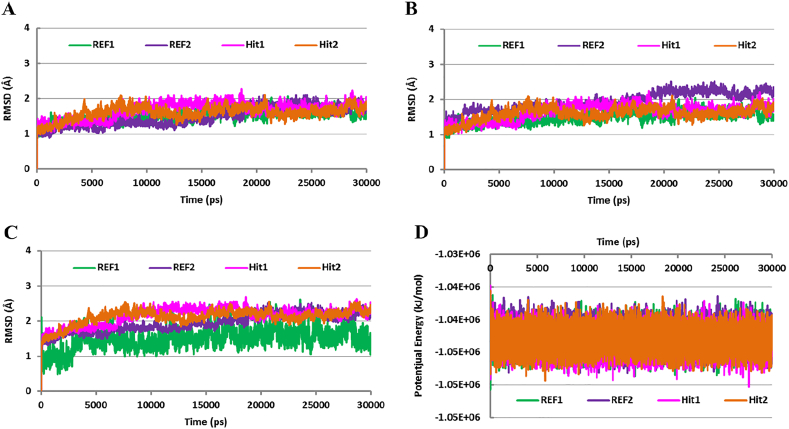
The convergence of each simulation system at approximately 5 ns advocates that Cdk5/p25 did not disintegrate during the simulation and behaved smoothly (Fig. 5). The stability of each system was evaluated by measuring the rmsd of alpha carbon atoms (hereafter Cα) and backbone atoms of Cdk5/p25 throughout the simulation period. Each system showed that the rmsd of Cα atoms obtained a steady-state after convergence and remained <3.0 Å during the production phase (Fig. 5A).
Fig. 5.
Root mean square deviation (rmsd) and potential energy analysis. The rmsd profile of the Cα atoms of Cdk5/p25 (A) and the backbone atoms of Cdk5/p25 (B). All the systems (REF1, REF2, Hit1, and Hit2) converged after 5 ns simulation and remained stable throughout the simulation period. C) Illustrates the rmsd profile of each candidate inhibitor (REF1, REF2, Hit1, and Hit2) in complex with Cdk5/p25. REF1, REF2, Hit1, and Hit2 showed stable rmsd (< 3.0 Å) after 5 ns simulation in the ATP-binding site of Cdk5/p25. D) Estimates the potential energy of each simulation system. Each system is represented by the presence of the corresponding inhibitor. Green, blue, magenta, and orange colors reflect the presence of REF1, REF2, Hit1, and Hit2, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The rmsd analysis of the backbone atoms was also carried out to infer the stability of the entire protein structure during the simulation period. Our findings suggested that the structure of Cdk5/p25 remained stable during the simulation time (Fig. 5B). The rmsd analysis of the Cα atoms and backbone atoms of Cdk5/p25 measured little amplitude (Fig. 5A, B). The details of minimum, maximum, and average rmsd values of the Cα atoms and backbone atoms of Cdk5/p25 after convergence are given in Supplementary Table S4.
The measurement of rmsd of Cdk5/p25 in complex with REF1, REF2, Hit1, and Hit2 mimics the stability of the correspondent complex. Our results obtained stable rmsd values for all the tested inhibitors in production phase (Fig. 5C). Each tested inhibitor showed <3.0 Å fluctuations during the production stage. Our results observed that the average rmsd value of REF1 is less than the average values of both the candidate hits (Table S4). But the rmsd of REF1 showed higher amplitude than the candidate inhibitors (Fig. 5C). Therefore, we perceived that the candidate inhibitors may slightly be more stable than REF1. The rmsd values of REF2 and both the candidate inhibitors (Hit1 and Hit2) were quite similar in all simulation systems. The average rmsd value of each simulated compound is given in supplementary table S4. Furthermore, the potential energy analysis reflected the stability of all the simulation systems (Fig. 5D). Our analysis confirmed that each simulation system obtained the lowest potential energy and remained stable throughout the simulation period. Furthermore, each inhibitor observed almost similar potential energy and suggested their stable binding with Cdk5/p25 (Fig. 5D).
3.5.2. Molecular Orientation and Molecular Interactions Analysis
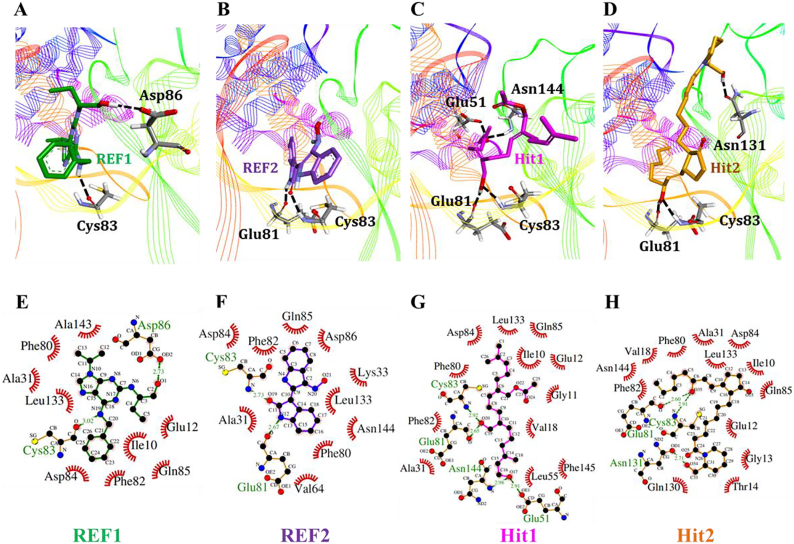
The primary substrate binding site of Cdk5 (ATP-binding site) is divided into three sub-sites including Gly-rich loop, activation loop and the hinge region (Fig. S3). The superimposition of all representative complexes observed that each inhibitor occupied the ATP-binding site of Cdk5/p25 and obtained similar orientation between the Gly-rich loop, activation loop, and hinge region (Fig. S3). A deeper insight suggests that Lys33, Phe80, Glu81, Cys83, and Asn144 are the essential residues to regulate the binding of ATP and facilitate the transfer of γ-phosphate of ATP to secondary substrate(s). Since, we observed that all the tested inhibitors occupied the ATP-binding site of Cdk5/p25. Therefore, our study asked the binding mechanism of candidate inhibitors in the ATP-binding site of Cdk5/p25. Our results observed that the final candidate inhibitors formed stable polar and non-polar interactions with the essential residues of ATP-binding site of Cdk5/p25 (Fig. 6, Table 4). The 3D and 2D interaction pattern of the reference compounds and candidate inhibitors with Cdk5/p25 is shown (Fig. 6).
Fig. 6.
3D and 2D molecular interaction pattern of the REF1, REF2, Hit1, and Hit2 with Cdk5/p25. Upper panel (A-D) displayed the 3D interaction representation of the REF1 (A), REF2 (B), Hit1 (C), and Hit2 (D) with Cdk5/p25. Cdk5/p25 is shown as thin wire representation. Hydrogen bond (H-bond) forming residues are shown as sticks and labeled. REF1, REF2, Hit1, and Hit2 are represented as green, blue, magenta, and orange stick models, respectively. Hydrogen bonds are represented as black colored dashed lines. Lower panel portrayed the 2D interaction pattern of the REF1 (E), REF2 (F), Hit1 (G), and Hit2 (H) with Cdk5/p25. Hydrogen bond forming residues of Cdk5 are labeled as green colour and H-bonds are depicted as green colored dashed lines with bond distance in angstrom (Å). Van der Waals and other hydrophobic interactions are represented as hemi-spheres and labeled. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 4.
Molecular interactions of the REF1, REF2, Hit1, and Hit2 with Cdk5/p25.
| Compound | Hydrogen bonds (≤ 3.5 Å) |
Non-polar interactions | |||
|---|---|---|---|---|---|
| Amino acid | Amino acid atom | Ligand atom | Distance (Å) | ||
| REF1 | Cys83 | O | N19 | 3.02 | Ile10, Glu12, Ala31, Phe80, Phe82, Asp84, Leu133, Ala143 |
| Asp86 | OD2 | O1 | 2.73 | ||
| REF2 | Glu81 | O | N12 | 2.67 | Ala31, Lys33, Val64, Phe80, Phe82, Asp84, Gln85, Asp86, Leu133, Asn144 |
| Cys83 | N | O19 | 2.73 | ||
| Hit1 | Glu51 | OE1 | O17 | 2.91 | Ile10, Gly11, Glu12, Val18, Ala31, Leu55, Phe80, Phe82, Asp84, Gln85, Leu133, Phe145 |
| Glu81 | O | O20 | 2.65 | ||
| Cys83 | N | O20 | 2.92 | ||
| Asn144 | N | O17 | 2.98 | ||
| Hit2 | Glu81 | O | O35 | 2.60 | Ile10, Glu12, Gly13, Thr14, Val18, Ala31, Phe80, Phe82, Asp84, Gln85, Leu133, Asn144 |
| Cys83 | N | O35 | 2.91 | ||
| Asn131 | OD1 | O34 | 2.71 | ||
The molecular interaction analyses suggested that REF1 and REF2 formed stable hydrogen bond interactions with Cys83 and Asp86 and Glu81 and Cys83 residues of Cdk5, respectively. (Fig. 6A, B). Our modeling results observed that Hit1 established H-bonds with Glu51, Glu81, Cys83 and Asn144 (Fig. 6C, G). Similarly, Hit2 also formed H-bonds with the catalytic residues Glu81 and Cys83 of the Cdk5. Additionally, Hit2 established another hydrogen bond with Asn131 residue of Cdk5 (Fig. 6D, H). The atomic details of polar and non-polar interactions of REF1, REF2, Hit1, and Hit2 are given in Table 4.
Furthermore, the comparative analyses of molecular interactions of Hit1 and Hit2 in complex with Cdk5/p25 were carried out before and after molecular dynamics simulation. Our results suggested that the mechanism making H-bonds remained stable (Fig. S4). In parallel, H-bond formation between the Hit1 and Asn144 also preserved before and after simulation. But the interaction pattern of additional H-bonds of Hit2 with Asp126 and Thr14 was changed and formed new H-bond with Asn131 of Cdk5 (Fig. S4). We proclaim that the spatial orientation of Asn131is close to Asp126 and Hit2 can form opportunistic H-bond with either of the side chains of these two residues. Apart from polar interactions, the hydrophobic interactions remained quite stable (Fig. S4). Since, docking procedure is very limited in terms of simulated physiological environment and produce small number of conformers. Therefore, we relied on molecular dynamics simulation which is more reliable and accurate approach to investigate molecular interaction pattern.
3.5.3. Hydrogen Bonds (H-bonds) Analysis
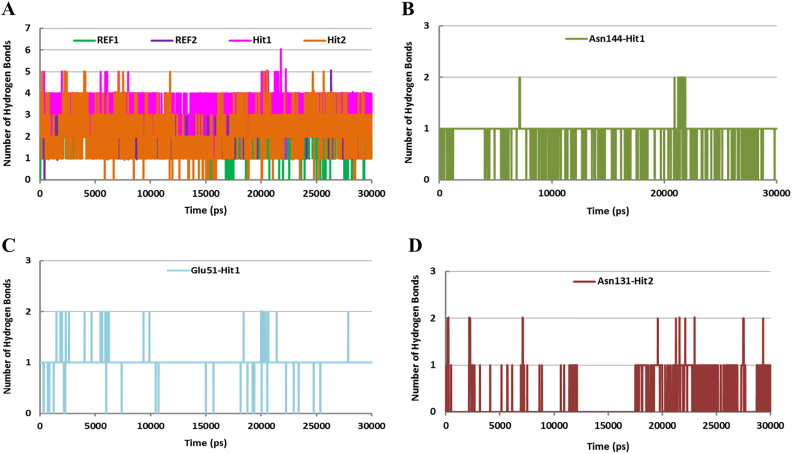
Furthermore, we asked the pattern of hydrogen bonds between the ligands and Cdk5/p25. Our results observed that the total number of H-bonds remained higher for Hit1 and Hit2 (Fig. 7).
Fig. 7.
Hydrogen bonds (H-bonds) analysis. A) Total number of H-bonds of REF1 (green), REF2 (blue), Hit1 (magenta), and Hit2 (Orange) with Cdk5/p25. Hit1 and Hit2 obtained higher number of H-bonds with Cdk5/p25. B) Reflects the consistency of H-bonds between the Asn144 residue of Cdk5 and Hit1. C) Represents that Glu51 of Cdk5 formed consistent H-bond with Hit1. D) Portrays the consistency of H-bond of Asn131 of Cdk5 and Hit2. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The comparative analysis of our results suggested that the average number of H-bonds of REF1 and REF2 with Cdk5/p25 remained two, while Hit1 and Hit2 established an average of four and three H-bonds with Cdk5/p25, respectively (Fig. 7). To address the question of slightly higher number of H-bonds by Hit1 and Hit2 proclaimed that non-catalytic residues of the ATP-binding site of Cdk5 are involved in inhibitors binding. Our results showed that Glu51 and Asn144 of Cdk5 formed additional H-bonds with Hit1. In parallel, the Asn131 residue of Cdk5 formed an additional H-bond with Hit2 (Fig. 7B-D). Intriguingly, our results identified that during the entire simulation period, Hit1 and Hit2 formed slightly stable H-bonds with Glu81 and Asn144, and Asn131, respectively (Fig. 7B-D). In prallel, Hit1 also formed consistent H-bond with Glu51 of the ATP-binding site of Cdk5/p25 (Fig. 7C).
3.6. Binding Free Energy Analysis
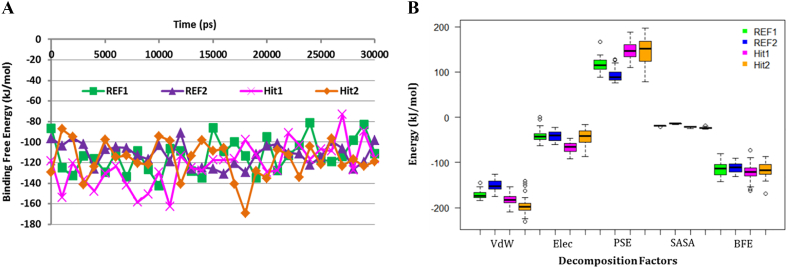
Binding free energies were calculated to understand the affinity of candidate inhibitors towards Cdk5/p25. The comparative analyses suggested that Hit1 and Hit2 obtained average binding free energy of −122.18 kJ/mol and − 117.26 kJ/mol, respectively (Fig. 8, Table 5). In contrast, the average values of binding free energy for REF1 and REF2 were estimated as −113.10 kJ/mol and − 111.77 kJ/mol, respectively (Fig. 8, Table 5).
Fig. 8.
Binding free energy analysis. A) g_mmpbsa calculated the binding free energies of REF1, REF2, Hit1, and Hit2 with Cdk5/p25 throughout the simulation time. Hit1 and Hit2 showed slightly low binding free energies than REF1 and REF2. The binding free energy of Cdk5/p25 with REF1, REF2, Hit1, and Hit2 has been depicted as green, blue, magenta, and orange, respectively. B) Energy decomposition analysis showed that Van der Waals (VdW) and polar solvation energies (PSE) played differential roles in the binding free energies of Hit1 and Hit2. Coloring scheme: REF1, Green; REF2, Blue, Hit1, Magenta; and Hit2, Orange. Energy decomposition factors: VdW, Van der Waals; Elec, Electrostatic energy; PSE, Polar solvation energy; SASA, Solvent accessible surface area energy; BFE, Binding free energy. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 5.
Analysis of binding free energy and decomposition factors of REF1, REF2, Hit1, and Hit2.
| Inhibitor | Energy decomposition factors (kJ/mol) |
Binding free energy (kJ/mol) |
|||
|---|---|---|---|---|---|
| VdWa | Elecb | PSEc | SASAd | ||
| REF1 | −171.37 | −40.60 | 117.77 | −18.89 | −113.10 |
| REF2 | −149.81 | −41.06 | 93.38 | −14.06 | −111.77 |
| Hit1 | −181.63 | −66.71 | 147.56 | −21.41 | −122.18 |
| Hit2 | −194.82 | −42.15 | 143.01 | −23.29 | −117.26 |
Van der Waals energy.
Electrostatic energy.
Polar solvation energy.
Solvent accessible surface area energy.
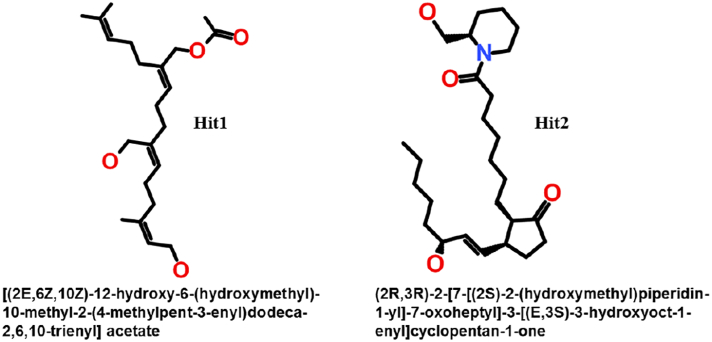
The decomposition of binding free energy showed that both polar and non-polar interactions participated in the inhibition of Cdk5/p25 (Fig. 8B). Among different energy components, van der Waals and polar solvation energies remained differential factors to contribute to binding affinity of the candidate inhibitors (Fig. 8B, Table 5). The average values of binding free energies of the reference compounds and candidate inhibitors are given in Table 5. Finally, the 2D structures of the final candidate inhibitors of the Cdk5/p25 are shown in Fig. 9.
Fig. 9.
The 2D structures of final candidate inhibitors of Cdk5/p25. The atomic compositions of both the structures are rich in oxygen contents and have extended conformation which is well-fitted with the ATP-binding site of Cdk5/p25.
Furthermore, an extensive literature mining was performed through the SciFinder [49] and Reaxys [50] servers and found that the newly identified compounds were not already evaluated against the Cdk5/p25. Therefore, we recommend the final hit compounds as novel natural product candidate inhibitors of Cdk5/p25 to attenuate tau-associated neurological disorders.
4. Discussion
Cyclin-dependent kinase 5 (Cdk5) is a vital kinase and essentially regulates neuronal development and physiology. In several neurological disorders, Cdk5 has been found to be abnormally activated upon its complex formation with p25– a proteolytic product of p35 [51]. Since, hyperactivated Cdk5/p25 abnormally phosphorylates tau and neurofilament proteins (NF-M/H), thereby inducing neurofibrillary tangles (NFTs) formation and eventually results in neuronal cell death [52,53]. Therefore, the pharmacological inhibition of Cdk5/p25 has been targeted to alleviate Cdk5/p25-mediated induction of neurofibrillary tangles and neurofilament formation in neurological disorders [20]. Our study has employed structure-based drug designing and probed novel natural product inhibitors of Cdk5/p25. Herein, the chemical hotspots of Cdk5-roscovitine were transformed into a 3D pharmacophore by Receptor-ligand Pharmacophore module of DS. Upon high selectivity score [54] and compatible interactions with catalytic active residues of Cdk5, the top-ranked pharmacophore was selected. The correspondent pharmacophore was comprised of six features including two hydrogen bond donors, one hydrogen bond acceptor and three hydrophobic features. The complementation analyses of the pharmacophoric features showed that closely oriented polar features comprising hydrogen bond donor and hydrogen bond acceptor are pointed towards the carboxyl oxygen and amino group of cysteine 83 (Cys83), respectively. It has already been reported that roscovitine, alosine A, and indirubin-3′-monoxime form polar interactions with the amide group and/or carboxyl group of the Cys83 residue of Cdk5 [26]. Our findings followed Ahn et al. that Cys83 of Cdk5 forms polar interactions with an ATP analogue inhibitor [55]. Our results observed that the third polar interaction feature (hydrogen bond donor) was complemented to carbonyl group of Asp86 of Cdk5. In parallel, Malmstrom et al. reported that 4-(1,3-benzothiazol-2-yl)-thiophene-2-sulfonamides forms polar interactions (H-bonds) with the carbonyl group of Asp86 of Cdk5 [56]. Our results observed that the three hydrophobic features of the selected pharmacophore established non-polar interactions with Lys33, Phe80, Ile10, and Val18. Previously, hydrophobic interactions have shown to play a critical role in the pharmacological inhibition of Cdk5 [26,55,56]. Therefore, we argued that the resultant pharmacophore is a suitable choice to use as a 3D query in virtual screening of the drug-like database for the identification of candidate inhibitors of Cdk5. The selected pharmacophore obtained highest GH and EF scores of 0.88 and 7.23, respectively, which validates its suitability in virtual screening practices [34,35]. Since, drug-like molecules are restricted to certain chemical properties such as physiochemical and pharmacokinetics and pharmacodynamics properties. Therefore, we set the blood-brain barrier (BBB) level to ≤2 with an intention to identify candidate molecules that can penetrate BBB and could target Cdk5/p25 in brain. ADMET-Blood Brain Barrier (ADMET-BBB) is a well-calibrated model in DS to predict blood-brain barrier penetration at 95% and 99% confidence ellipses and is derived over 800 compounds with known blood-brain barrier penetration values after oral administration [57]. This model evaluates a query molecule in one of the four prediction levels such as “level 0” (very high penetration), “level 1” (High penetration), “level 2” (medium penetration), “level 3” low penetration and “level 4” (undefined). Since, the evaluation of our candidate hits by ADMET-BBB model obtained level 1, therefore, we strongly proclaim that the candidate hits harbor enough hydrophobicity to penetrate the blood-brain barrier and could target Cdk5/p25 in brain.
The mechanistic studies of the pharmacophore-retrieved candidate molecules and Cdk5/p25 were carried out by docking. Several filters such as docking score ≥ 80.00, conformational stability, inhibitory molecular interactions with Cdk5/p25 and commercial availability of the candidate molecules were employed to identify the true positive candidate hit molecules of Cdk5/p25. Molecular docking studies obtained a total of 10 candidate molecules to inhibit Cdk5/p25. Since, molecular docking does not provide simulated physiological environment and real-time behavior of the drug-receptor interaction; therefore, molecular dynamics (MD) simulation was employed to identify the true positive inhibitors of Cdk5/p25 [35,36,58]. The root mean square deviation (rmsd) analysis is a key factor to evaluate the stability and successful execution of MD simulation [34,35,58]. Our results suggested that the final candidate inhibitors showed stable rmsd of the alpha carbon atoms (Cα) of Cdk5/p25, the backbone atoms of Cdk5/p25, and the Cdk5/p25 in complex with candidate inhibitors. Potential energy estimation is an essential component of molecular dynamics simulation analysis [34,35]. Our results estimated the lowest potential energy and confirmed that each simulation system behaved stable during the entire simulation period.
In computational drug designing, investigation of molecular interactions of drug-receptor complex is a pre-requisite to predict the binding mechanism of drug molecule. Our results suggested that the final candidate molecules formed polar interactions (H-bonds) with Cys83 residue of the ATP-binding site of Cdk5/p25. We argued that the similar binding pattern of Cdk5 inhibitors has already been reported [26,55,56]. Additionally, our results observed that the final candidate inhibitors formed hydrogen bonds with Glu81 of the Cdk5, which has already been reported that the ATP analogue inhibitor of Cdk5 forms hydrogen bond with Glu81 of the Cdk5 [55]. Likewise, the inspection of non-polar interactions between the candidate inhibitors and Cdk5/p25 suggested that Ile10, Val18, Phe80, Phe82, and Ile133 are the predominant residues to participate in inhibitors binding [26,55,56]. Finally, the binding free energy analyses affirmed that the candidate inhibitors showed higher affinity towards Cdk5/p25. The energy decomposition analysis suggested that both polar and non-polar interactions actively participated in the binding of candidate inhibitors and Cdk5/p25. Since, the candidate hit molecules are rich in oxygen contents and presents extended polymers of carbon atoms. Therefore, we proclaim that the differential factors of binding affinity of the candidate hit molecules with Cdk5/p25 may attribute to van der Waals and polar solvation energies. Overall, our study recommends a detailed account of structure-based drug designing and unveils the binding mechanism of novel natural product inhibitors of Cdk5/p25, thereby extending the interaction pattern of Cdk5/p25-inhibitor complex. Finally, we recommend two natural product candidate inhibitors of Cdk5/p25 as additional platforms to extend chemical space for the development of promising inhibitors of Cdk5/p25 in neurological disorders.
5. Conclusion
The abnormal hyperphosphorylation of tau protein by Cdk5/p25 results in neurological insults, thereby inducing neurofibrillary tangles (NFTs) formation, neuroinflammation, and neuronal death. To this end, the pharmacological inhibition of Cdk5/p25 abrogates tau-associated neurological disorders. Our study has employed computational simulations and identified two natural products as candidate inhibitors of Cdk5/p25 to attenuate tau-pathogenesis in neurological disorders. The molecular interactions information of Cdk5/p25 with Roscovitine was transformed into a 3D chemical space and a structure-based pharmacophore was designed and validated. Drug-like databases were designed from IBSD and ZNPD, screened by pharmacophore, and the candidate inhibitors of Cdk5/p25 were retrieved. Molecular docking followed by molecular dynamics simulation and binding free energy analysis identified two novel candidate inhibitors of Cdk5/p25. The candidate inhibitors occupied the ATP-binding site of Cdk5/p25, obtained stable root mean square deviation (rmsd) value and established hydrogen bonds with Cys83, Glu81, and either with Asn144 or Asn131. Furthermore, the newly identified candidate inhibitors showed higher affinity towards Cdk5/p25 in terms of binding free energy. Finally, we recommend these candidate inhibitors as fundamental platforms to attenuate Cdk5/p25-mediated tau pathogenesis in neurological disorders.
Conflict of Interest
The authors declare no competing interest.
Acknowledgments
Acknowledgments
N/A
Funding
This research was supported by Bio and Medical Technology Development Program of the National Research Foundation (NRF) and funded by the Korean government (MIST) (No. NRF-2018M3A9A7057263). Also supported by a grant of the Korean Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HI18C1728).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2019.04.010.
Contributor Information
Seok Ju Park, Email: psjgenesis@hanmail.net.
Keun Woo Lee, Email: kwlee@gnu.ac.kr.
Appendix A. Supplementary data
Supplementary material
References
- 1.Manning G., Whyte D.B., Martinez R., Hunter T., Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 2.Shah K., Lahiri D.K. Cdk5 activity in the brain - multiple paths of regulation. J Cell Sci. 2014;127:2391–2400. doi: 10.1242/jcs.147553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lim S., Kaldis P. Cdks, cyclins and CKIs: roles beyond cell cycle regulation. Development. 2013;140:3079–3093. doi: 10.1242/dev.091744. [DOI] [PubMed] [Google Scholar]
- 4.Dhavan R., Tsai L.-H. A decade of CDK5. Nat Rev Mol Cell Biol. 2001;2:749–759. doi: 10.1038/35096019. [DOI] [PubMed] [Google Scholar]
- 5.Sharma P., Sharma M., Amin N.D., Albers R.W., Pant H.C. Regulation of cyclin-dependent kinase 5 catalytic activity by phosphorylation. Proc Natl Acad Sci U S A. 1999;96:11156–11160. doi: 10.1073/pnas.96.20.11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohshima T., Ogura H., Tomizawa K., Hayashi K., Suzuki H., Saito T. Impairment of hippocampal long-term depression and defective spatial learning and memory in p35 mice. J Neurochem. 2005;94:917–925. doi: 10.1111/j.1471-4159.2005.03233.x. [DOI] [PubMed] [Google Scholar]
- 7.Pareek T.K., Keller J., Kesavapany S., Pant H.C., Iadarola M.J., Brady R.O. Cyclin-dependent kinase 5 activity regulates pain signaling. Proc Natl Acad Sci U S A. 2006;103:791–796. doi: 10.1073/pnas.0510405103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lopes J.P., Oliveira C.R., Agostinho P. Neurodegeneration in an Aβ-induced model of Alzheimer's disease: the role of Cdk5. Aging Cell. 2010;9:64–77. doi: 10.1111/j.1474-9726.2009.00536.x. [DOI] [PubMed] [Google Scholar]
- 9.Cruz J.C., Tseng H.-C., Goldman J.A., Shih H., Tsai L.-H. Aberrant Cdk5 activation by p25 triggers pathological events leading to neurodegeneration and neurofibrillary tangles. Neuron. 2003;40:471–483. doi: 10.1016/s0896-6273(03)00627-5. [DOI] [PubMed] [Google Scholar]
- 10.Wen Y., Yu W.H., Maloney B., Bailey J., Ma J., Marié I. Transcriptional regulation of beta-secretase by p25/cdk5 leads to enhanced amyloidogenic processing. Neuron. 2008;57:680–690. doi: 10.1016/j.neuron.2008.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith P.D., Crocker S.J., Jackson-Lewis V., Jordan-Sciutto K.L., Hayley S., Mount M.P. Cyclin-dependent kinase 5 is a mediator of dopaminergic neuron loss in a mouse model of Parkinson's disease. Proc Natl Acad Sci U S A. 2003;100:13650–13655. doi: 10.1073/pnas.2232515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong A.S.L., Lee R.H.K., Cheung A.Y., Yeung P.K., Chung S.K., Cheung Z.H. Cdk5-mediated phosphorylation of endophilin B1 is required for induced autophagy in models of Parkinson's disease. Nat Cell Biol. 2011;13:568–579. doi: 10.1038/ncb2217. [DOI] [PubMed] [Google Scholar]
- 13.Paoletti P., Vila I., Rifé M., Lizcano J.M., Alberch J., Ginés S. Dopaminergic and glutamatergic signaling crosstalk in Huntington's disease neurodegeneration: the role of p25/Cyclin-dependent kinase 5. J Neurosci. 2008;28:10090–10101. doi: 10.1523/JNEUROSCI.3237-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alvarez-Periel E., Puigdellívol M., Brito V., Plattner F., Bibb J.A., Alberch J. Cdk5 contributes to Huntington's disease learning and memory deficits via modulation of brain region-specific substrates. Mol Neurobiol. 2018;55:6250–6268. doi: 10.1007/s12035-017-0828-4. [DOI] [PubMed] [Google Scholar]
- 15.Piedrahita D., Hernández I., López-Tobón A., Fedorov D., Obara B., Manjunath B.S. Silencing of Cdk5 reduces neurofibrillary tangles in transgenic Alzheimer's mice. J Neurosci. 2010;30:13966–13976. doi: 10.1523/JNEUROSCI.3637-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang K.-H., Vincent F., Shah K. Deregulated Cdk5 triggers aberrant activation of cell cycle kinases and phosphatases inducing neuronal death. J Cell Sci. 2012;125:5124–5137. doi: 10.1242/jcs.108183. [DOI] [PubMed] [Google Scholar]
- 17.Tan X., Chen Y., Li J., Li X., Miao Z., Xin N. The inhibition of Cdk5 activity after hypoxia/ischemia injury reduces infarct size and promotes functional recovery in neonatal rats. Neuroscience. 2015;290:552–560. doi: 10.1016/j.neuroscience.2015.01.054. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Q., Chen C., Lü J., Xie M., Pan D., Luo X. Cell cycle inhibition attenuates microglial proliferation and production of IL-1β, MIP-1α, and NO after focal cerebral ischemia in the rat. Glia. 2009;57:908–920. doi: 10.1002/glia.20816. [DOI] [PubMed] [Google Scholar]
- 19.Parry D., Guzi T., Shanahan F., Davis N., Prabhavalkar D., Wiswell D. Dinaciclib (SCH 727965), a novel and potent cyclin-dependent kinase inhibitor. Mol Cancer Ther. 2010;9:2344–2353. doi: 10.1158/1535-7163.MCT-10-0324. [DOI] [PubMed] [Google Scholar]
- 20.Sundaram J.R., Poore C.P., Hazim N., Sulaimee B., Pareek T., Asad A.B.M.A. Specific inhibition of p25/Cdk5 activity by the Cdk5 inhibitory peptide reduces neurodegeneration in vivo. J Neurosci. 2013;33:334–343. doi: 10.1523/JNEUROSCI.3593-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ji Y.-B., Zhuang P.-P., Ji Z., Wu Y.-M., Gu Y., Gao X.-Y. TFP5 peptide, derived from Cdk5-activating cofactor p35, provides neuroprotection in early-stage of adult ischemic stroke. Sci Rep. 2017;7:40013. doi: 10.1038/srep40013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brasca M.G., Albanese C., Alzani R., Amici R., Avanzi N., Ballinari D. Optimization of 6,6-dimethyl pyrrolo[3,4-c]pyrazoles: identification of PHA-793887, a potent CDK inhibitor suitable for intravenous dosing. Bioorg Med Chem. 2010;18:1844–1853. doi: 10.1016/j.bmc.2010.01.042. [DOI] [PubMed] [Google Scholar]
- 23.Seo J., Kritskiy O., Watson L.A., Barker S.J., Dey D., Raja W.K. Inhibition of p25/Cdk5 attenuates tauopathy in mouse and iPSC models of frontotemporal dementia. J Neurosci. 2017;37:9917–9924. doi: 10.1523/JNEUROSCI.0621-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y., Wang L., Zhang X., Huang M., Li S., Wang X. Inhibition of Cdk5 rejuvenates inhibitory circuits and restores experience-dependent plasticity in adult visual cortex. Neuropharmacology. 2018;128:207–220. doi: 10.1016/j.neuropharm.2017.10.015. [DOI] [PubMed] [Google Scholar]
- 25.Kim C., Yun N., Lee J., Youdim M., Ju C., Kim W.-K. Phosphorylation of CHIP at Ser20 by Cdk5 promotes tAIF-mediated neuronal death. Cell Death Differ. 2015;23:333–346. doi: 10.1038/cdd.2015.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mapelli M., Massimiliano L., Crovace C., Seeliger M.A., Tsai L.-H., Meijer L. Mechanism of Cdk5/p25 binding by Cdk inhibitors. J Med Chem. 2005;48:671–679. doi: 10.1021/jm049323m. [DOI] [PubMed] [Google Scholar]
- 27.Arooj M., Sakkiah S., Kim S., Arulalapperumal V., Lee K.W. A combination of receptor-based pharmacophore modeling & QM techniques for identification of human chymase inhibitors. PLoS One. 2013;8 doi: 10.1371/journal.pone.0063030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rogers D., Hopfinger A.J. Application of genetic function approximation to quantitative structure-activity relationships and quantitative structure-property relationships. J Chem Inf Comput Sci. 1994;34:854–866. [Google Scholar]
- 29.Al-Balas Q., Hassan M., Al-Oudat B., Alzoubi H., Mhaidat N., Almaaytah A. Generation of the first structure-based pharmacophore model containing a selective “zinc binding group” feature to identify potential glyoxalase-1 inhibitors. Molecules. 2012;17:13740–13758. doi: 10.3390/molecules171213740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guner O. History and evolution of the pharmacophore concept in computer-aided drug design. Curr Top Med Chem. 2002;2:1321–1332. doi: 10.2174/1568026023392940. [DOI] [PubMed] [Google Scholar]
- 31.Jang C., Yadav D.K., Subedi L., Venkatesan R., Venkanna A., Afzal S. Identification of novel acetylcholinesterase inhibitors designed by pharmacophore-based virtual screening, molecular docking and bioassay. Sci Rep. 2018;8:14921. doi: 10.1038/s41598-018-33354-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gadhe C.G., Lee E., Kim M. Finding new scaffolds of JAK3 inhibitors in public database: 3D-QSAR models & shape-based screening. Arch Pharm Res. 2015;38:2008–2019. doi: 10.1007/s12272-015-0607-6. [DOI] [PubMed] [Google Scholar]
- 33.Sakkiah S., Thangapandian S., Lee K.W. Ligand-based Virtual Screening and Molecular Docking Studies to Identify the Critical Chemical Features of Potent Cathepsin D Inhibitors. Chem Biol Drug Dus. 2012;80:64–79. doi: 10.1111/j.1747-0285.2012.01339.x. n.d. [DOI] [PubMed] [Google Scholar]
- 34.Kumar R., Bavi R., Gi Jo M., Arulalapperumal V., Baek A., Rampogu S. New compounds identified through in silico approaches reduce the α-synuclein expression by inhibiting prolyl oligopeptidase in vitro. Sci Rep. 2017;7:10827. doi: 10.1038/s41598-017-11302-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeb A., Park C., Rampogu S., Son M., Lee G., Lee K.W. Structure-based drug designing recommends HDAC6 inhibitors to attenuate microtubule-associated Tau-pathogenesis. ACS Chem Neurosci. 2019;10:1326–1335. doi: 10.1021/acschemneuro.8b00405. n.d. [DOI] [PubMed] [Google Scholar]
- 36.Amin S.A., Bhattacharya P., Basak S., Gayen S., Nandy A., Saha A. Pharmacoinformatics study of Piperolactam A from Piper betle root as new lead for non steroidal anti fertility drug development. Comput Biol Chem. 2017;67:213–224. doi: 10.1016/j.compbiolchem.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 37.Amin S.A., Bhargava S., Adhikari N., Gayen S., Jha T. Exploring pyrazolo[3,4-d]pyrimidine phosphodiesterase 1 (PDE1) inhibitors: a predictive approach combining comparative validated multiple molecular modelling techniques. J Biomol Struct Dyn. 2018;36:590–608. doi: 10.1080/07391102.2017.1288659. [DOI] [PubMed] [Google Scholar]
- 38.Verdonk M.L., Cole J.C., Hartshorn M.J., Murray C.W., Taylor R.D. Improved protein-ligand docking using GOLD. Proteins Struct Funct Genet. 2003;52:609–623. doi: 10.1002/prot.10465. [DOI] [PubMed] [Google Scholar]
- 39.Li Y., Han L., Liu Z., Wang R. Comparative assessment of scoring functions on an updated benchmark: 2. Evaluation methods and general results. J Chem Inf Model. 2014;54:1717–1736. doi: 10.1021/ci500081m. [DOI] [PubMed] [Google Scholar]
- 40.Pronk S., Rd Pá ll S, Schulz R., Larsson P., Bjelkmar P., Apostolov R. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zoete V., Cuendet M.A., Grosdidier A., Michielin O. SwissParam: a fast force field generation tool for small organic molecules. J Comput Chem. 2011;32:2359–2368. doi: 10.1002/jcc.21816. [DOI] [PubMed] [Google Scholar]
- 42.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J Appl Phys. 1981;52:7182–7190. [Google Scholar]
- 43.Hess B., Bekker H., Berendsen H.J.C., Fraaije J.G.E.M. LINCS: a linear constraint solver for molecular simulations. J Comput Chem. 1997;18:1463–1472. [Google Scholar]
- 44.Yang C.-Y., Sun H., Chen J., Nikolovska-Coleska Z., Wang S. Importance of ligand reorganization free energy in protein-ligand binding-affinity prediction. J Am Chem Soc. 2009;131:13709–13721. doi: 10.1021/ja9039373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kumari R., Kumar R., Lynn A. g_mmpbsa—a GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model. 2014;54:1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- 46.Vorontsov I.I., Miyashita O. Crystal molecular dynamics simulations to speed up MM/PB(GB)SA evaluation of binding free energies of di-mannose deoxy analogs with P51G-m4-Cyanovirin-N. J Comput Chem. 2011;32:1043–1053. doi: 10.1002/jcc.21683. [DOI] [PubMed] [Google Scholar]
- 47.Guedes I.A., De Magalhães C.S., Dardenne L.E. Receptor-ligand molecular docking. Biophys Rev. 2014;6:75–87. doi: 10.1007/s12551-013-0130-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dodson G.G., Lane D.P., Verma C.S. Molecular simulations of protein dynamics: new windows on mechanisms in biology. EMBO Rep. 2008;9:144–150. doi: 10.1038/sj.embor.7401160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.SciFinder - Sign In, n.d.https://scifinder.cas.org/scifinder/login
- 50.Reaxys, n.d.https://www.reaxys.com/#/login
- 51.Patrick G.N., Zukerberg L., Nikolic M., de la Monte S., Dikkes P., Tsai L.H. Conversion of p35 to p25 deregulates Cdk5 activity and promotes neurodegeneration. Nature. 1999;402:615–622. doi: 10.1038/45159. [DOI] [PubMed] [Google Scholar]
- 52.Noble W., Olm V., Takata K., Casey E., Meyerson J., Gaynor K. Cdk5 is a key factor in tau aggregation and tangle formation in vivo. Neuron. 2003;38:555–565. doi: 10.1016/s0896-6273(03)00259-9. [DOI] [PubMed] [Google Scholar]
- 53.Ahlijanian M.K., Barrezueta N.X., Williams R.D., Jakowski A., Kowsz K.P., Mccarthy S. Hyperphosphorylated tau and neurofilament and cytoskeletal disruptions in mice overexpressing human p25, an activator of cdk5. Proc Natl Acad Sci U S A. 1999;97:2910–2915. doi: 10.1073/pnas.040577797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang J., Zhou H., Jiang Q., Sun L., Deng P. Novel transforming growth factor-beta receptor 1 antagonists through a pharmacophore-based virtual screening approach. Molecules. 2018;23 doi: 10.3390/molecules23112824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ahn J.S., Radhakrishnan M.L., Mapelli M., Choi S., Tidor B., Cuny G.D. Defining Cdk5 ligand chemical space with small molecule inhibitors of Tau phosphorylation. Cell Chem Biol. 2005;12:811–823. doi: 10.1016/j.chembiol.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 56.Malmström J., Viklund J., Slivo C., Costa A., Maudet M., Sandelin C. Synthesis and structure-activity relationship of 4-(1,3-benzothiazol-2-yl)-thiophene-2-sulfonamides as cyclin-dependent kinase 5 (cdk5)/p25 inhibitors. Bioorg Med Chem Lett. 2012;22:5919–5923. doi: 10.1016/j.bmcl.2012.07.068. [DOI] [PubMed] [Google Scholar]
- 57.Egan W.J., Lauri G. Prediction of intestinal permeability. Adv Drug Deliv Rev. 2002;54:273–289. doi: 10.1016/s0169-409x(02)00004-2. [DOI] [PubMed] [Google Scholar]
- 58.Rampogu S., Zeb A., Baek A., Park C., Son M., Lee K.W. Discovery of potential plant-derived peptide deformylase (PDF) inhibitors for multidrug-resistant bacteria using computational studies. J Clin Med. 2018;7:563. doi: 10.3390/jcm7120563. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material