Abstract
Objectives
Many natural products have pharmacological or biological activities that can be of therapeutic benefit in treating diseases, and are also an important source of inspiration for development of potential novel drugs. The past few decades have witnessed extensive study of natural products for their promising prospects in application of medicinal chemistry, molecular biology and pharmaceutical sciences.
Materials and methods
Natural product databases have provided systematic collection of information concerning natural products and their derivatives, including structure, source and mechanisms of action, which significantly support modern drug discovery.
Results
Currently, a considerable number of natural product databases, such as TCM Database@Taiwan, TCMID, CEMTDD, SuperToxic and SuperNatural, have been developed, providing data such as integrated medicinal herbs, ingredients, 2D/3D structures of the compounds, related target proteins, relevant diseases, and metabolic toxicity and more.
Conclusions
We focus on an analytical overview of current natural product databases, and further discuss the good, bad or imperfection of current ones, in the hope of better integrating existing relevant outcomes, thus providing new routes for future drug discovery.
Introduction
Natural products, often defined as compounds derived from natural sources that possess biological activities 1, have historically been known to include active components of many traditional medicines and have continuously received attention for their extensive pharmacological and/or biological activities, that can be of great therapeutic value and market potential 2. Realizing inherent therapeutic potential of natural products, much effort has been made in their study, from which considerable numbers of bioactive compounds have been isolated and investigated every year, compiling a vast resource for future investigation 3. Meanwhile, it is recognised that natural products and their derivatives represent more than half FDA‐approved drugs, and continue to be rich sources for drug discovery 4. However, data show that even as technology develops and investment increases substantially, approvals for new drugs from natural products have not increased accordingly, laying the mainstream pharmaceutical industry unexpectedly uninformed 5.
Rapid development of natural product drugs makes systematic collection of information in the form of natural product databases crucial for existing outcome integration. Here, we will mainly introduce five representative natural product (NP) databases to summarize how their development has progressed over the last few years. We will include TCM Database@Taiwan 6, traditional Chinese medicine integrative database (TCMID) 7, Chinese ethnic minority traditional drug database (CEMTDD) 8, SuperToxic 9 and SuperNatural 10. These databases collect and provide data from integrated medicinal herbs, ingredients, 2D/3D structures of compounds, related target proteins, relevant diseases and metabolic toxicity, which are essential in natural product studies for scientists, clinical physicians and pharmacists. We balance their advantages and disadvantages while specifically mentioning their unique functions and software package integrations. For example, TCM Database@Taiwan has the largest traditional Chinese medicine data source that exhibits relationships between herbs, ingredients and compounds, but excludes relevant diseases, target proteins and interaction networks 6.
Here, we present an analytical overview of current natural product databases (Table.1), focusing on their strengths and weaknesses, then discuss their limitations as well as trends in building future ones, in the hope of better integration of existing outcomes, thus leading to stronger support for future drug development from natural products.
Table 1.
Comparisons of current natural product databases
| Database | Data source | Advantage | Disadvantage | Web service | Web link |
|---|---|---|---|---|---|
| NPACT | Chinese herb | Experimentally determined in vitro and in vivo biological activity | Only 1574 entries |
Similarity search Java‐based Molecular Editor tool draw structure to be searched in the database |
http://crdd.osdd.net/raghava/npact/ |
| Database specified for cancer therapy | Online submission | ||||
| HIT | Chinese herbs | Protein targets related to herbs are included | All information is covered by other DB except the key feature | Text mining and curation(scan abstracts) | http://lifecenter.sgst.cn/hit/ |
| Compound similarity search (Tanimoto coefficient) | |||||
| AfroDb | African medicinal plants | Covers the entire continent of Africa | Data from traditional healers have no words based on | MOE software generated 3D structure | N/A |
| Classified with ‘Drug‐like’, ‘lead‐like’ and ‘fragment‐like’ subsets | |||||
| CVDHD | Herbs(FDA approved drugs derived from natural products) | Contains integrated medicinal herbs, natural products, CVD‐related target proteins, docking results, diseases and clinical biomarkers | \ | Available for downloading docking scores of all molecules and single target proteins | http://pkuxxj.pku.edu.cn/ |
| API for Cytoscape is integrated | |||||
| CamMedNP | Plants | Plant sources ware collected from geographical collection sites | \ | Generate 3D structures using MOE and energyz MMFF94 | N/A |
| NuBBE | Brazilian plants | ‘Rule of five’ drug‐likeness evaluate compounds were grouped by acquisition source | No functions or diseases related to these compounds | Molecular drawing interface WebME | http://nubbe.iq.unesp.br/nubbeDB.html |
| Substructure search engine:CDK | |||||
| 3DMET | Natural product | 3D structure display | Short for drug features of compounds | JME molecular editor applet for a graphical input | http://www.3dmet.dna.affrc.go.jp/ |
| TTD | drug data | data sources of interaction with targets comparison with succeed drugs | Absence of interactions of targets | N/A | http://bidd.nus.edu.sg/group/ttd/ttd.asp |
| SuperToxic | Toxic compounds | Be used to evaluate the risk of use for compounds | Mechanisms of toxin is absent | MyChem/OpenBabel chemical function | http://bioinformatics.charite.de/supertoxic |
| MarvinSketch drawing and uploading function | |||||
| JMol inspection compound | |||||
| SuperNatural | Various sources | Large and comprehensive | \ | ChemDoodle integrated for structure drawing | http://bioinformatics.charite.de/supernatural |
| Chemistry development kit calculate fingerprint | |||||
| Tanimoto coefficient similarity measure | |||||
| TCMID | Chinese herb | Having the largest data set for related field | Unable to display network on the web page | Self‐developed network‐display tools which can show Herb–disease network, Herbal ingredients—targets interaction network, Herbal ingredient–target–disease–drug network | http://www.megabionet.org/tcmid/ |
| Compounds and disease‐specific functions interaction networks bridge the gap between TCM and modern western medicine | |||||
| Chem‐TCM | Chinese herb | \ | \ | http://www.chemtcm.com/ | |
| CEMTDD | Chinese ethnic minorities’ herb | The most complete database structure among database above include herb‐compound, compound‐target, herb‐disease | Disease cannot be used as the keyword for query | Cytoscape Web integrated for network displaying | http://www.cemtdd.com/ |
| TCMDB@Taiwan | Chinese herbs | Possess the largest traditional Chinese medicine data source | Only shows the relation between herb, ingredients and compound | ChemAxon: user can draw compound structure and query | http://tcm.cmu.edu.tw/ |
| Users can download the structure of the molecule in cdx (2D) or mol2 (3D) |
Databases
TCM Database@Taiwan
TCM Database@Taiwan is a database for Chinese traditional medicinal compounds, which aim to facilitate virtual screening for researchers conducting computer‐aided drug design (CADD) by providing freely downloadable 3D compound structures of ingredients used in traditional Chinese medicine. Currently, update of the database is comprised of 61 000 compounds with 20 000 ingredients and 453 herbs, all of which can be easily downloaded in cdx (2D) and Tripos mol2 (3D) formats. The basic search is architected in two parts, in chemical composition and Traditional Chinese Medicine (herbs), where traditional Chinese theories are carefully involved in classification. Data queries can be conducted within the TCM database via either basic or advanced options that specify search clauses, such as molecular properties, substructures, TCM ingredients and TCM classification. Additionally, ChemAxon plugin has been integrated for chemical drawing, favourable to structure searching. TCM Database@Taiwan has been conceived as an important project dedicated to building a comprehensive TCM library and largely improves TCM query system available up to now. It can serve as both resource for virtual screening and for reference for biochemical assays. We highly expect that the upcoming version of the database will integrate more information regarding mechanisms of compound activation, which may further facilitate study by biologists or pharmacologists 6. This database is accessible at: http://tcm.cmu.edu.tw/
TCMID: traditional Chinese medicine integrative database
TCMID is a database of Chinese traditional medicine, built up of six independent modules, namely prescription, herb, ingredients, target, disease and drugs. Aiming for establishing connections between herbal ingredients and diseases they are meant to treat, through disease related genes/proteins. The TCMID database was developed accordingly to bridge the gap between TCM and modern western medicine, previously a daunting task to undertake. To virtually display interacting networks between herbs, herbal ingredients, targets and diseases related to them, the TCMID self‐developed tool has been integrated within the website for network browsing. To better illustrate the network displaying function, an official example has been provided in the literature referring to building the herbal ingredient–target–disease–drug network. The basic assumption stands that an interaction is built between herbal ingredient and disease‐related protein target; a potential mechanism of disease treatment with the ingredient is highly suggested, and vice versa. The most attractive aspect of TCMID comes as it enables integration of TCM theories such as ‘Pattern’ or ‘Zang‐Fu’ (that profoundly affect the prescriptions), with modern medicine, where molecular mechanisms are extensively included. Up to now, 46 914 prescriptions, 8159 herbs, 25 210 compounds, 6826 drugs, 3791 diseases and 17 521 related targets have been collected in TCMID, making it the largest data set in the field 7. This database is accessible at: http://www.megabionet.org/tcmid/
CEMTDD: Chinese ethnic minority traditional drug database
CEMTDD, known as the database for Chinese minority herbs, has been built on data retrieved from various resources (mainly from Kazakh and Uygur traditional drugs), and is composed of modules including plants, metabolites, indications, active compounds, targeted proteins, mechanism and diseases. Users log into the website and easily access the compound–target–disease network by searching the database through a range of simple steps, as shown in Fig. 1. The network display function is inherently integrated on the web, and employs software package Cytoscape that enables graphic display of interactions between herbs, compounds, targets and other proteins, in a simple and convenient way. Online graphic network integration through Cytoscape Web greatly facilitates its users in understanding mechanisms in a modern scientific way. However, some amelioration is still required, for example, ‘disease’ was not included in keywords for information retrieval 8. Completion of the database fills a vacancy in databases in CEMTDs and establishes a sound foundation for future exploitation and utilization of CEMTDs. CEMTDD also provides a new perspective in exploring more candidate compounds from less known resources as potential drugs in the coming future. Generally, though lack of completeness in some information within it, this can be seen as a first effort towards Chinese ethnic minority medicine web resources, and provides good examples for database developers from this point of view. Hopefully more workers will keep an eye on fields of natural products with less popularity and exploit their potential in future utilization. This database is accessible at: http://www.cemtdd.com/
Figure 1.
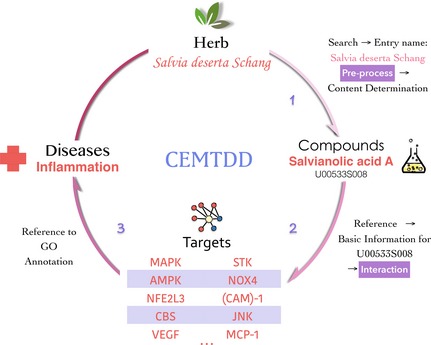
Exemplary information retrieval process through CEMTDD . CEMTDD allows a simple and fast query for Chinese ethnic minority herbs as described in this figure. Users' access to this website should first log in the searching interface located on the menu bar, where they are able to see the entry for herbs, compounds and targets. In this case, Salvia deserta Schang, a traditional herb from Xinjiang, was typed and ready for information retrieval, where herb‐related targets, chemicals and disease are exhibited and provide clues for scientific research.
SuperToxic
SuperToxic is a database comprised of comprehensive substances that exhibit proved toxicity coming from different sources including animals, plants and artificial synthetics. It is assumed that the natural products within these toxins, whose functions are originally being applied in protecting their producers, are also of scientific significance for disease treatment. Users can easily access approximately 60 000 structures with corresponding properties including toxicity information by options through structure search, name search or property search, while browsing mode is also provided which allows users to browse toxins starting with any alphabet characters and numbers, or through other naming systems such as CAS and NSC. Toxicity information within the retrieved data comprises of dose, testing type (measurement for toxicity, such as LD50 and nlog (LC50)) and cell line or organism that was assayed for toxicity. The database also integrated software packages that are extensively employed in modern compound database building, such as MarvinSketch (compound sketching and uploading, here applied for structure retrieval), JMol (compound inspection) and MyChem/OpenBabel (computing properties for the structures that user submit) 9. In summary, SuperToxic composes a comprehensive source for toxicology data, which strongly combines structural, functional and chemical information, while clarification of how the toxin affects life is still absent in the current version of this database. Its proper employment could greatly facilitate experimental optimizations and contract time spent on animal testing as used for risk evaluation. This database is accessible at: http://bioinformatics.charite.de/supertoxic/
SuperNatural
Built upon various sources, SuperNatural is the first public resource that contains 3D structures and conformers of natural compounds 10. Recently, the updated version of SuperNatural (here refers to Super Natural II) has been released. Super Natural II has now significantly expanded its scale, as it is now comprised of 325 508 natural compounds, a noticeable 7‐fold expansion from its original version in 2006. SuperNatural II offers a comprehensive review of natural product information that outperforms many other. It can provide the most important structural and physicochemical properties, predicted toxicity class can be retrieved for around 170 000 compounds in a single platform, while vendor information is also retrievable for the vast majority of compounds. Super Natural II database basically allows five options for data retrieval. Besides the commonly seen search via name and structure (supported by ChemDoodle), it also provides search in substructure, which allows user to retrieve data through specific data; MoA (mechanism of action), which enables users to screen compounds on their putative targets through specifying a target protein as term for searching; and pathways, which allow users to browse the overall KEGG pathways that were previously mapped with natural products stored in the database. Besides the searching service provides above, compound clustering is also supported in SuperNatural II database calculated using MyChem based on Tanimoto coefficient, through which results are very clearly visualized in a heat map where red colour corresponds to high similarity 11. To sum up, the SuperNatural II database is a free resource with embedded screening functions for bioactive natural compounds, whose comprehensive information as well as the newly integrated functions makes the database a very handy tool for scientists studying this field. This database is accessible at: http://bioinf-applied.charite.de/supernatural_new/index.php
Other natural product databases
As information technology rises, data explosions have been met in all fields within cultures, technologies and science. Under such circumstances, potential numbers of databases serving natural product queries is far beyond our imagination. Above, we have introduced some representative databases regarding natural products. Here, we collected some other notable natural product databases, which are greatly diversified in their specificity either regionally or disciplinarily. TDD: Therapeutic target database, a resource for facilitating target‐oriented drug discovery 12, 3DMET: Three‐Dimensional Structure Database of Natural Metabolites, a novel database of curated 3D structures 13, NuBBE: Development of a Natural Products Database from the Biodiversity of Brazil, is a web source for natural products and derivatives from the Brazilian biodiversity containing the compounds obtained by the academic group NuBBE 14, CVDHD: a cardiovascular disease herbal database for drug discovery and network pharmacology 15, AfroDb: A Select Highly Potent and Diverse Natural Product Library from African Medicinal Plants 16, CamMedNP: Building the Cameroonian 3D structural natural products database for virtual screening 17, HIT: Herb Ingredients' targets, linking herbs to their targets 18, and NPACT: Naturally Occurring Plant‐based Anti‐cancer Compound‐Activity‐Target database 19 (Table 1).
Conclusions
For thousands of years, natural products have been demonstrated to be a rich source of therapeutic agents and thus play a key role in treating diverse types of human disease, such as cancer 20, 21. Chemical structure and bio‐activities of these compounds varies greatly, for which reason they incessantly offer inspiration to innovations in medicine, nutrition, agrochemical research and life sciences. Rapid advancing development of database technologies has strongly propelled systematic collection of natural products regarding their detailed information integrated from existing outcomes, which significantly assist new drug discoveries. As mentioned above, numerous natural product databases, focusing on different aspects of natural products, have already been developed. These databases have served as important tools for natural product studies and continuously provide specialized information to the worldwide scientific community, hence, to a certain degree, a key prompter for new drug discovery and development (Fig. 2).
Figure 2.
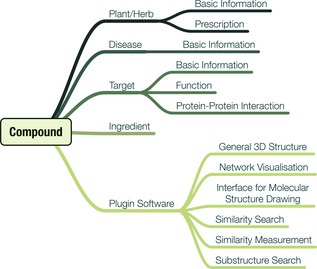
Natural product database structure.
Despite positive aspects of current natural product databases, we still have to realize their limitations and thus seek for improvement. Above all, limitations should be realized upon currently built natural product databases, which have laid more focus on integrated medicinal herbs, ingredients, 2D/3D structures of compounds, rather than related diseases and mechanisms (for example, target proteins), which as well occupy an important role in drug discovery and development. Currently, the main strategy in drug discovery is based on a ‘one gene–one drug–one disease’ paradigm. However, a drug's efficacy is impaired by robustness of any protein interaction network in the treated objectives. To overcome limitations, the concept of effective combinatorial drugs and drugs with multiple targets has led to increased interest in systems‐oriented approaches for drug discovery. Thus, systems‐oriented approaches such as interaction networks need to be more highly valued in design of natural product databases. Secondly, databases produced upon western and eastern natural products have disparate emphases. Eastern databases tend to place more emphasis on herbal ingredients and therapeutic effect, as discussions concerning mechanisms are always absent from display, while most western databases are compound‐based and focus more on mechanisms than origin or folk use. As the description of diseases between eastern and western medicine has diversified greatly, this is also embodied in databases built upon them.
Scalability and comprehensiveness must also be concerned in database construction of natural products. To some degree, comprehensiveness stands as the future for natural product web sources, which suggests that databases like TCM databases@Taiwan or SuperNatural are naturally ahead of many others. However, though enchanting the comprehensive dream of human in building databases, much similar to humanities obsessiveness for skyscrapers, values are still appreciated for small databases focusing on specificities, for example, CEMTDD, which is the most valued web resource in the related field to now. Comprehensiveness may cause problems such as difficulties in selecting desired modules, inability in quickly locating information within a database and relatively steep curve for familiarity on utilization. We are presently seeing skyscrapers rising from the ground, while appreciating living in houses deep in the forest with lodgeable rooms and delicate designs as well. Encouragement should be awarded for databases built from minority resources, believing these efforts will finally be paid off and integrate into the comprehensive databases for a profounder use. Natural product consist a special resource that have been extensively studied yet have not been comprehensively collected, which means, we are climbing, we are still on our way.
Taken together, imperfections and divergences still exist in current natural product databases. To develop better natural product databases and thus better support drug discovery and development, effort should be made to view natural product drugs in networks and put more emphasis on systems‐oriented approaches. Involvement of the interaction networks between herbs, ingredients, compounds, related target proteins and diseases in natural product databases might be key factors to bridge the East–West gap.
Conflict of interest
We declare that none of the authors have a financial interest related to this work.
Acknowledgements
We are grateful to Prof. Jin‐hui Wang for his critical review of this manuscript. This work was supported in part by grants from the Key Projects of the National Science and Technology Pillar Program (No. 2012BAI30B02), National Natural Science Foundation of China (No. 81260628, 81303270 and 81473091), and Shenyang Science and Technology Project (F12‐157‐9‐00).
References
- 1. Baker DD, Chu M, Oza U, Rajgarhia V (2007) The value of natural products to future pharmaceutical discovery. Nat. Prod. Rep. 24, 1225–1244. [DOI] [PubMed] [Google Scholar]
- 2. Koehn FE, Carter GT (2005) The evolving role of natural products in drug discovery. Nat. Rev. Drug Discov. 4, 206–220. [DOI] [PubMed] [Google Scholar]
- 3. De Luca V, Salim V, Atsumi SM, Yu F (2012) Mining the biodiversity of plants: a revolution in the making. Science 336, 1658–1661. [DOI] [PubMed] [Google Scholar]
- 4. Paterson I, Anderson EA (2005) Chemistry. The renaissance of natural products as drug candidates. Science 310, 451–453. [DOI] [PubMed] [Google Scholar]
- 5. Li JW, Vederas JC (2009) Drug discovery and natural products: end of an era or an endless frontier? Science 325, 161–165. [DOI] [PubMed] [Google Scholar]
- 6. Chen CY (2011) TCM Database@Taiwan: the world's largest traditional Chinese medicine database for drug screening in silico. PLoS ONE 6, e15939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xue R, Fang Z, Zhang M, Yi Z, Wen C, Shi T (2013) TCMID: Traditional Chinese Medicine integrative database for herb molecular mechanism analysis. Nucleic Acids Res. 41, D1089–D1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huang J, Wang JH (2014) CEMTDD: Chinese Ethnic Minority Traditional Drug Database. Apoptosis 19, 1419–1420. [DOI] [PubMed] [Google Scholar]
- 9. Schmidt U, Struck S, Gruening B, Hossbach J, Jaeger IS, Parol R et al (2009) SuperToxic: a comprehensive database of toxic compounds. Nucleic Acids Res. 37, D295–D299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dunkel M, Fullbeck M, Neumann S, Preissner R (2006) SuperNatural: a searchable database of available natural compounds. Nucleic Acids Res. 34, D678–D683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Banerjee P, Erehman J, Gohlke BO, Wilhelm T, Preissner R, Dynkel M. (2015) Super Natural II – a database of natural products. Nucleic Acids Res. 43, D935–D939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mangal M, Sagar P, Singh H, Raghava G, Agarwal S (2012) Therapeutic target database update 2012: a resource for facilitating target‐oriented drug discovery. Nucleic Acids Res. 40, D1128–D1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Maeda MH, Kondo K (2013) Three‐dimensional structure database of natural metabolites (3DMET): a novel database of curated 3D structures. J. Chem. Inf. Model. 53, 527–533. [DOI] [PubMed] [Google Scholar]
- 14. Valli M, Santos RN, Figueira LD, Nakajima CH, Castro‐Gamboa L, Andricopulo AD (2013) Development of a natural products database from the biodiversity of Brazil. J. Nat. Prod. 76, 439–444. [DOI] [PubMed] [Google Scholar]
- 15. Gu JY, Gui YS, Chen LR, Yuan G, Xu X (2013) CVDHD: a cardiovascular disease herbal database for drug discovery and network pharmacology. J. Cheminform. 5, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ntie‐Kang F, Zofou D, Babiaka SB, Meudom R, Scharfe M, Lifongo LL et al (2013) AfroDb: a select highly potent and diverse natural product library from African medicinal plants. PLoS ONE 8, e78085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ntie‐Kang F, Mbah JA, Mbaze LM, Lifongo LL, Scharfe M, , Ngo HJ et al (2013) CamMedNP: building the Cameroonian 3D structural natural products database for virtual screening. BMC Complement. Altern. Med. 13, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ye H, Ye L, Kang H, Zhang DF, Lin T, Tang KL et al (2011) HIT: linking herbal active ingredients to targets. Nucleic Acids Res. 39, D1055–D1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mangal M, Sagar P, Singh H, Raghava GPS, Agarwal SM (2013) NPACT: Naturally Occurring Plant‐based Anti‐cancer Compound‐Activity‐Target database. Nucleic Acids Res. 41, D1124–D1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang X, Chen LX, Ouyang L, Cheng Y, Liu B (2012) Plant natural compounds: targeting pathways of autophagy as anti‐cancer therapeutic agents. Cell Prolif. 45, 466–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ouyang L, Luo Y, Tian M, Zhang SY, Lu R, Wang JH et al (2014) Plant natural products: from traditional compounds to new emerging drugs in cancer therapy. Cell Prolif. 47, 506–515. [DOI] [PMC free article] [PubMed] [Google Scholar]


