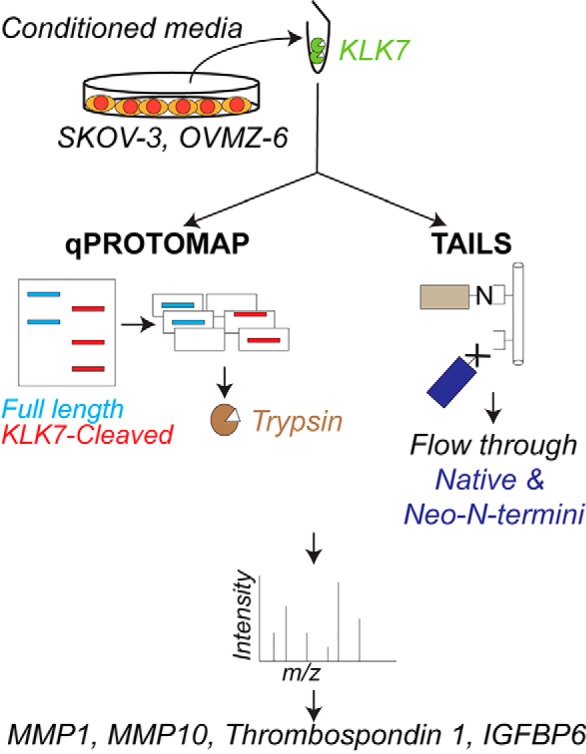
KLK7 secreted by ovarian cancer cells into the peritoneal fluid may aid in cancer progression. To evaluate the functional role of KLK7 in this context, knowledge on its substrate repertoire is crucial. Herein, using two proteomics approaches, qPROTOMAP and TAILS, we identified pro-MMP10, thrombospondin 1 and IGFBP6 as putative KLK7 substrates in an in vitro setting.
Keywords: Degradomics*, Extracellular matrix*, Proteases*, Ovarian cancer, Secretome, Substrate identification, Kallikrein-related Peptidases
Graphical Abstract
Highlights
Kallikrein-related peptidase 7 is over expressed in ovarian cancer.
Quantitative PROTOMAP and TAILS approaches identified putative substrates of KLK7.
Pro-MMP10 is activated by KLK7.
KLK7 cleaves thrombospondin 1 and IGFBP6 in vitro.
Abstract
Kallikrein-related peptidase 7 (KLK7) is a serine peptidase that is over expressed in ovarian cancer. In vitro functional analyses have suggested KLK7 to play a cancer progressive role, although monitoring of KLK7 expression has suggested a contradictory protective role for KLK7 in ovarian cancer patients. In order to help delineate its mechanism of action and thereby the functional roles, information on its substrate repertoire is crucial. Therefore, in this study a quantitative proteomics approach—PROtein TOpography and Migration Analysis Platform (PROTOMAP)—coupled with SILAC was used for in-depth analysis of putative KLK7 substrates from a representative ovarian cancer cell line, SKOV-3, secreted proteins. The Terminal Amine Isotopic Labeling of Substrates (TAILS) approach was used to determine the exact cleavage sites and to validate qPROTOMAP-identified putative substrates. By employing these two technically divergent approaches, exact cleavage sites on 16 novel putative substrates and two established substrates, matrix metalloprotease (MMP) 2 and insulin growth factor binding protein 3 (IGFBP3), were identified in the SKOV-3 secretome. Eight of these substrates were also identified on TAILS analysis of another ovarian cancer cell (OVMZ-6) secretome, with a further seven OVMZ-6 substrates common to the SKOV-3 qPROTOMAP profile. Identified substrates were significantly associated with the common processes of cell adhesion, extracellular matrix remodeling and cell migration according to the gene ontology (GO) biological process analysis. Biochemical validation supports a role for KLK7 in directly activating pro-MMP10, hydrolysis of IGFBP6 and cleavage of thrombospondin 1 with generation of a potentially bioactive N-terminal fragment. Overall, this study constitutes the most comprehensive analysis of the putative KLK7 degradome in any cancer to date, thereby opening new avenues for KLK7 research.
Kallikrein-related peptidase (KLK1) 7 is a secreted serine peptidase that is over expressed in pathological conditions, including ovarian cancer. Ovarian cancer is the leading cause of death from gynecological malignancies despite its relatively low level of occurrence (1). Ascites plays a major role in ovarian cancer progression by accumulating shed malignant cells as aggregates, promoting cell survival and leading to peritoneal dissemination (2–5). These cells floating within the ascites harbor a metastatic preference and secrete factors, such as peptidases, that either facilitate disease progression or inhibition by modulating the surrounding microenvironment. KLK7 is secreted by the cells that are present in the ascites (6).
Functional studies performed to date have suggested KLK7 has cancer progressive properties (4), although, in clinical studies the increased expression of KLK7 has been shown to be associated with a protective impact on overall survival of ovarian cancer patients (7, 8). Knowledge of the putative KLK7 substrate repertoire secreted by cancer cells in the ascites microenvironment is thus fundamental to determine the functional role of KLK7 in cancer.
To date, KLK7 substrates have been identified, a priori, in a biochemical context by incubating recombinant proteins with the KLK7 peptidase in vitro, with minimal representation of cellular context and physiological conditions. No studies to date have assessed KLK7-mediated proteolysis in ovarian cancer cells using in-depth proteomics approaches (9). To this end, we sought to use two proteomic profiling approaches namely PROtein TOpography and Migration Analysis Platform (PROTOMAP) (10) and Terminal Amine Isotopic Labeling of Substrates (TAILS) (11) to identify putative KLK7 substrates and their exact cleavage sites in ovarian cancer using the secretome of representative ovarian cancer cells. This study thereby identified 23 putative novel KLK7 substrates increasing the number of currently identified putative KLK7 substrates, thus providing an excellent base for future research into the functional role of KLK7 in ovarian cancer.
EXPERIMENTAL PROCEDURES
Cell Lines and Cell Culture
The SKOV-3 (the American Type Culture Collection, ATCC1) cell line, derived from a patient with moderately well differentiated adenocarcinoma (12, 13), was maintained (37 °C, 5% (v/v) CO2) in phenol red-free RPMI 1640 media with 2 mm l-glutamine (Life Technologies, VIC, Australia) supplemented with 10% (v/v) fetal bovine serum (FBS; Sigma-Aldrich, Carlsbad, CA). The OVMZ-6 cell line derived from a FIGO Stage IV poorly differentiated serous adenocarcinoma (14) was maintained in DMEM with high glucose, Glutamax-I and pyridoxine (Invitrogen, Life Technologies), with 10% FBS, 10 mm HEPES, 0.55 mm l-arginine, 0.272 mm l-asparagine and 20 μg/ml gentamycin. All cell lines were regularly screened for mycoplasma contamination.
Conditioned Media Collection
SKOV-3 and OVMZ-6 cells were grown to 70% confluence, depleted of serum proteins and after 24 h conditioned media (CM) was collected. Cleared CM in the presence of 50 μm EDTA was concentrated and buffer-exchanged into assay buffer (1× PBS pH 8.0, Amicon Ultra-15 centrifugal filter units). The protein concentration was measured using the BCA method (Sigma Aldrich) and adjusted to 1 mg/ml using assay buffer.
Production of Recombinant KLK7 and dmKLK7
Recombinant active KLK7 and a catalytically inactive double mutant KLK7 (dmKLK7) were produced and purified in the Pichia pastoris expression system and the percent active material in the produced KLK7 sample was measured using active site titration against human (rh) Serpin A3/α1-Antichymotrypsin as described elsewhere (15). Throughout the study KLK7 concentration was calculated considering the percent active material as determined by active site titration.
Experimental Design and Statistical Rationale
Three independent biological replicates were used for each proteomics analysis. Criteria for protein identification: for qPROTOMAP analysis MaxQuant (version 1.5.0.30) was used. For TAILS analysis, X!Tandem (release 2009.10.01.1 LabKey, Insilicos, Institute for Systems Biology) contained within the TPP (v4.6) was used. Quantitation was based on unique peptides only and a minimum of two ratio counts was required. In both approaches peptide identifications were accepted if they could be established at a FDR ≤1%. Data from each of three biological replicates from two proteomics approaches was analyzed independent of other replicates and proteomics approaches. MS data have been deposited to the ProteomeXchange consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (16). Zipped MaxQuant “.txt” files including “.wiff” files have been deposited with the dataset identifier PXD003812 for SKOV-3 qPROTOMAP analysis. The TAILS MS files have been deposited with the dataset identifier PXD003752 or DOI: 10.6019/PXD003752 for SKOV-3 and PXD004591 or DOI: 10.6019/PXD004591 for OVMZ-6.
Screening for KLK7 Substrates in SKOV-3 Cell Secretome Using qPROTOMAP
Stable Isotope Labeling by Amino acids in Cell culture (SILAC) Media Preparation and Cell Culture
For SKOV-3 labeling, RPMI 1640 media (deficient in l-Lysine, and l-Arginine; Sigma-Aldrich) was prepared by adding either l-Lysine (40.00 mg/L, Sigma-Aldrich) and l-arginine (50.0825 mg/L, Sigma-Aldrich) or l-D4-Lysine (K4, 40.8632 mg/L, Sigma-Aldrich) and l-Arginine.HCl [U-13C6, 99%] (R6, 62.2466 mg/L, Cambridge Isotope Laboratories, Tewksbury, MA) or l-Lysine.2HCl [U-13C6, 99%; U-15N2, 99%] (K8, 41.7063 mg/L, Cambridge Isotope Laboratories) and l-Arginine.HCl [U-13C6, 99%; U-15N4, 99%] (R10, 63.365 mg/L, Cambridge Isotope Laboratories) to make Light, Medium or Heavy labeling media, respectively. Cells were passaged as in section Cell Lines and Cell Culture with 10% dialyzed FBS (Invitrogen, US Origin) and at passage 6 cell lysates were MS analyzed to determine >95% of SILAC incorporation. Note: a quarter (0.2875 mm) of the arginine present in commercial RPMI 1640 media (1.1494 mm) was used for SILAC labeling in order to prevent arginine to proline conversion under extra arginine concentrations (17).
Treating SILAC-SKOV-3 Cell Conditioned Media with KLK7 and dmKLK7
Light, medium and heavy CM (20 μg each) was treated with KLK7 (1:50; active site-titrated KLK7:CM; w/w) or equivalent amounts of dmKLK7 or assay buffer as controls (37 °C, 18 h) in three biological replicates as below.
| Experiment | Light Labelled | Medium Labelled | Heavy Labelled |
|---|---|---|---|
| Biological Replicate 1 | assay buffer | KLK7 treated | dmKLK7 treated |
| Biological Replicate 2 | dmKLK7 treated | assay buffer | KLK7 treated |
| Biological Replicate 3 | KLK7 treated | dmKLK7 treated | assay buffer |
Reactions were stopped by heating with the sample buffer and alkylated with iodoacetamide (IAA, 12.5 mm, RT, 1 h, light-protected, Sigma-Aldrich).
SDS-PAGE, Gel Slicing and In-gel Trypsin Digestion
Half of each treatment (10 μg) in each biological replicate was pooled and resolved on a 12% SDS-PAGE gel (supplemental Fig. S1A). To identify KLK7 substrates, the PROTOMAP approach was followed (10), where gel lanes were partitioned into 20 slices, then further sectioned into ∼1 mm3 pieces and pooled before drying with neat acetonitrile. In-gel trypsin digestion was performed as per Shevchenko et al. (18), omitting reduction and alkylation steps as these were performed before SDS-PAGE. Concentrated peptide extracts were re-suspended in 20 μl of 0.1% (v/v) formic acid containing 2% acetonitrile.
qPROTOMAP LC-MS/MS
Tryptic peptides were analyzed on a nano ACQUITY UPLC system (Waters, Milford, MA) coupled to a TripleTOF 5600plus mass spectrometer (AB Sciex, Framingham, MA) via a Nanospray III Ion Source (AB Sciex). A sample aliquot of 5 μl was loaded onto a trap column (Symmetry C18 (5 μm) 0.18 mm x 20 mm; Waters), washed for 3 min at a rate of 15 μl/min and then separated on a BEH130 C18 column (1.7 μm, 75 μm x 200 mm; Waters) at a rate of 300 nL/min. Eluent A was 0.1% formic acid in milliQ water and eluent B was 0.1% formic acid in acetonitrile. The sample was trapped at 2% B and separated using the following gradient: 0 min (2% B) - 1 min (5% B) - 61 min (30% B) - 71 min (45% B) - 78 min (95% B) - 88 min (95% B) - 90 min (2% B) - 110 min (2% B). MS data was acquired in a data dependent manner.
The TripleTOF 5600plus mass spectrometer was operated in positive ion and high-resolution mode with an approximate resolution of 35,000. The mass window for precursor selection in the quadrupole was set to unit resolution (m/z 0.7 window). Parameter settings for the ion source were as follows: gas 1 = 25, gas 2 = 0, curtain gas = 25, ion spray voltage = 2300 V. Ion optics parameters were set as follows: Declustering potential = 80 V, collision energy (survey scan) = 10 V, rolling collision energy voltage was used for CID (collision-induced dissociation) fragmentation with a collision energy spread of 3. Nitrogen was used as the collision gas. Each cycle (1.3 s) consisted of a TOF-MS survey scan (mass range: 300–1800 Da, dwell time: 250 ms) followed by sequential fragmentation (mass range: 100–2000 Da, dwell time: 50 ms) of the 20 most intense precursors selected according to IDA (Information Dependent Acquisition) criteria. IDA criteria were specified as follows: Ions with m/z 300–1250 and charge 2–5 above a threshold of 20 cps were selected for fragmentation and excluded thereafter for 8 s. Mass spec recalibration was performed after eight hours of data acquisition using 50 fmol of a bovine serum albumin digest standard.
MS data was analyzed using MaxQuant version 1.5.0.30 (19). Each gel band analyzed was defined as a separate experiment in the experimental design section. MS/MS spectra were searched with Andromeda (20) against a SwissProt database (taxonomy: Homo sapiens; 20,267 entries, downloaded August 2013) and the MaxQuant contaminant database (247 entries) containing frequently observed contaminants such as keratins and proteases. Database searches were performed with the following search parameters: SILAC triplex labels were specified as light (Arg0Lys0), medium (Arg6Lys4) and heavy (Arg10Lys8). Strict enzyme specificity was required for trypsin with two missed cleavages allowed. Variable modifications included M oxidation (+ 15.994919), N and Q deamidation (+ 0.984016) and N-terminal acetylation of proteins (+ 42.010565). Carbamidomethylation of C (+ 57.021464) was set to fixed modification. AB Sciex TOF was set as instrument type using default settings. Precursor tolerance was set to 0.07 Da and 0.006 Da for the first and main search, respectively. Fragment tolerance was set at 40 ppm. False discovery rates (FDRs) at the peptide and protein levels were fixed at 1%. Quantitation was based on unique peptides only and a minimum of two ratio counts was required. The requantification feature was enabled.
Peptograph Generation and Analysis
The MaxQuant “txt” results files were further processed and quantitative peptographs were created with in-house Perl scripts generated based on qPROTOMAP from Dix et al. (21). In brief, cleaved proteins were identified based on the distribution of peptide ratios in each band. The distribution of peptide ratios in each band were organized into quartiles. The band was flagged as control specific, indicative of a parental degradation event, if the ratios in the upper three quartiles were more than 3-fold elevated in the control sample. The band was flagged as KLK7 specific, indicative of a cleaved fragment, if the ratios in the lower three quartiles were more than 3-fold elevated in the KLK7-treated samples. At this stage, peptides with MaxQuant score <40 or Posterior Error Probability (PEP) >0.05 and MaxQuant score <60 and PEP >0.01 were not included in the peptographs. Peptographs of those proteins with cell surface or extracellular localization, as denoted by Ingenuity Pathway Analysis software (IPA, Qiagen), were interrogated for evidence of KLK7-mediated proteolysis. A description of the peptograph interpretation can be found in supplemental Fig. S2. Peptographs were assessed to identify putative KLK7 substrates in KLK7-treated versus buffer control as no cleavages were observed in the dmKLK7-treated compared with the buffer control. Briefly, a protein to be considered a putative KLK7 substrate, KLK7-generated protein fragments were required to be abundantly identified in the KLK7-treated group; i.e. have a log2 SILAC label ratio ≥ 2 compared with the buffer-treated group, isolated from a single gel slice. In addition, only one KLK7-generated product was required for the protein to be considered a putative KLK7 substrate, although many KLK7 substrates were identified with multiple KLK7-generated protein fragments.
Screening for cleavage sites of qPROTOMAP-identified putative KLK7 substrates in ovarian cancer representative cell secretions using TAILS
Treating Cell Conditioned Media with KLK7 and dmKLK7
CM (200 μg) was treated with either KLK7, dmKLK7 (1:200; active site-titrated KLK7: CM; w/w) or assay buffer as controls (37 °C, 18 h) in three biological replicates (supplemental Fig. S1B). The reaction was quenched (1 m guanidine hydrochloride, 5 mm DTT; boiling 5–10 min), followed by alkylation (chloroacetamide 12 mm, RT, 30 min, light-protected).
TAILS Sample Preparation
Samples were prepared as described (22) with a few modifications as given below. The labeling reaction was quenched by adding 100 mm glycine and vortexing (RT, 30 min). Pooled samples were trypsin digested (supplemental Fig. S1C) and desalted using the Oasis HLB cartridges (Waters, United Kingdom) as per manufacturer's instructions. The peptide samples were depleted of the HPG-ALD polymer (available through www.flintbox.ca) by using the C18 StageTips, which were preconditioned by forcing 100 μl of 100% methanol through the disks with a syringe fitted to the end of the pipette tip. Any remaining organic solvent was removed from the column by forcing through 100 μl of buffer A (0.1% formic acid in milliQ water or acidified water) twice. The acidified peptide sample (pH <3.5) was forced through the column three times followed by two washes with buffer A. Peptides were eluted with 40 μl of buffer B (0.1% formic acid, 80% acetonitrile in water), vacuum dried and re-dissolved in 2% ACN, 0.1% formic acid by sonicating for 10 min and spin-vortexing for another 10 min.
TAILS LC-MS/MS
In brief, the resulting peptides were analyzed by LC-MS/MS using the Q Exactive mass spectrometer (Thermo Scientific, Bremen, Germany) coupled online with a RSLC nano UPLC (Ultimate 3000, Thermo Scientific). Samples were loaded on a nanoViper PepMap100 C18 trap column (100 μm × 2 cm) in 2% (v/v) acetonitrile/0.1% (v/v) formic acid, at a flow rate of 15 μl/min. Peptides were eluted and separated at a flow rate of 250 μl/min on a PepMap C18 nanocolumn (3 μm 100 Å pore size, 75 μm × 50 cm, Thermo), where acetonitrile was elevated from 2% (v/v) to 8% (v/v) over 1 min, followed with a linear acetonitrile gradient from 8% (v/v) to 24% (v/v) in 0.1% (v/v) formic acid for 24 min, followed by a linear increase to 30% (v/v) acetonitrile in 0.1% (v/v) formic acid over 3 min, an additional increase up to 80% (v/v) acetonitrile in 0.1% (v/v) formic acid over 3 min, followed by reduction of acetonitrile back to 2% (v/v) and re-equilibration. The eluent was nebulized and ionized using a Thermo nano electrospray source with a distal coated fused silica emitter (New Objective, Woburn, MA). Typical mass spectrometric conditions were as follows: spray voltage, 1.7 kV; no sheath and auxiliary gas flow; heated capillary temperature, 275 °C.
The Q Exactive instrument was operated in the data dependent mode to automatically switch between full scan MS and MS/MS acquisition. Survey full scan MS spectra (m/z 375–1800) were acquired in the Orbitrap with 70,000 resolution (m/z 200) after accumulation of ions to a 3 × 106 target value with maximum injection time of 30 ms. Dynamic exclusion was set to 15 s. The 10 most intense multiply charged ions (z ≥ 2) were sequentially isolated and fragmented in the octopole collision cell by higher energy collision dissociation (HCD, normalized collision energy 27%) with a maximum injection time of 60 ms. MS/MS spectra were acquired in the Orbitrap with 17,500 resolution, automatic gain control target of 5 × 104 counts and 2.0 Da isolation width. Underfill ratio was at 1%.
TAILS Data Analysis
Spectra were converted to mzXML files using MSconvert (Profile setting, facilitated by ProteoWizard release 3.0.9134 (23), and were searched using X!Tandem [release 2009.10.01.1 LabKey, Insilicos, Institute for Systems Biology, (24)] contained within the TPP [v4.6; (25)] with high-resolution k-score against the UniProt Knowledgebase (UniProtKB) human protein database [Released February 2015; SwissProt 42077 entries (26)] supplemented with known contaminants [cRAP database released Feb 2012; 115 entries (27)]. Reverse sequences (Decoys) informed false positive identification frequency. Search parameters were as follows: precursor ion mass (monoisotopic) tolerance ± 20 ppm; MS/MS tolerance ± 0.02 Da; and semi-Arg-C cleavage allowing for up to one missed cleavage. Fixed modifications included C carbamidomethylation (+ 57.021464) and light dimethylation (+ 28.031300) of N termini and K residues. Variable modifications included M oxidation (+ 15.994919) and the mass difference of H and L dimethylation (+ 6.031817) for N termini and K residues. Peptide Prophet (v4.6) was used to curate peptide-spectrum matches of FDR ≤ 1% and assign representative proteins/protein isoforms. XPRESS software (28) within the TPP was used for relative quantification of H : L peptide ratios.
Peptide analysis was performed according to Kleifeld et al. (22). Briefly, for TAILS-identified peptides in each replicate, mean for the log2 (H:L) values in “XPRESS” column was calculated and that value was subtracted from log2(H:L) ratios calculated for each peptide in the data set. Histogram of the log2-ratios was plotted to check for normal distribution and deviation from the expected center at 0 using Wessa.net73 (http://www.wessa.net/rwasp_fitdistrnorm.wasp) (supplemental Fig. S3). The standard deviation of the corrected log2 ratios for natural N termini was calculated and peptides with the corrected log2(H:L) ratios that are higher than three times the standard deviation (i.e. Log2 (H:L) ≥ 2) are considered to be high-probability substrates of the protease of interest in the substrate with a cleavage site as identified by the neo-N-terminal peptide. KLK7-generated neo-N termini were selected if (a) Log2 (H:L) ≥ 2; (b) identified by at least two spectra or in at least two replicates (with different variable modifications included) (29); and (c) annotated to be extracellular or of cell surface origin in the Ingenuity Pathway Analysis (IPA). Data from each of three biological replicates was analyzed independent of other replicates.
The KLK7 cleavage site specificity profile, based on TAILS-identified cleavage events within all putative KLK7 substrates, was generated using iceLogo (https://iomics.ugent.be/icelogoserver/). Annotation of domains and binding/interaction regions spanning the identified KLK7 cleavage sites, as annotated in the UniProtKB, was performed using TopFINDer [TopFIND 3.0; (30)]. Biological process annotations in the Gene Ontology Database that were significantly associated with putative KLK7 substrates identified by both qPROTOMAP and TAILS were calculated using a modified Fisher Exact test embedded in the functional annotation tool provided by The Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7 (31).
Validation of KLK7-mediated Proteolysis of Selected Substrates Identified in Both Proteomics Platforms
Recombinant Protein Digestion and SDS-PAGE/Silver Staining or Western Blotting Analyses
Human recombinant proteins of THBS1 (Merck, NSW, Australia), pro-MMP10, pro-MMP1 (R&D Systems, Minneapolis, MN) and IGFBP6 (Thermo Fisher Scientific, VIC, Australia) were digested with KLK7 in assay buffer (37 °C/18 h) using 1:10–1000 molar ratios (similar to ∼1:50–5000 w/w ratios) of active site-titrated KLK7: substrate. As controls, recombinant proteins were treated with buffer or dmKLK7 to the highest concentration of KLK7, and KLK7 or dmKLK7 also were incubated alone (37 °C/18 h) to distinguish any bands generated by auto-degradation of the enzymes. Reactions were terminated and reduced by boiling with Laemmli sample buffer containing β-mercaptoethanol. Samples were resolved by SDS-PAGE and visualized by silver staining. Western blot analysis was also performed on respective KLK7-treated recombinant proteins or CM with antibodies targeting THBS1 (full length: ab1823, Abcam, Cambridge, MA; N-20: sc-12312, Santa Cruz, California), MMP1 (full length: ab89767, Abcam), KLK7 (32), MMP2 (amino acids 475–490: ab37150, Abcam) and IGFBP6 (amino acids 128–141 of human IGFBP6, ab109765, Abcam).
MMP10 Activity Assay
Pro-MMP10 was treated with KLK7 (1:100 molar ratio; active site-titrated KLK7:pro-MMP10) or assay buffer or equivalent concentrations of dmKLK7 as controls (37 °C/18 h). Relative fluorescence units (RFU) was measured after adding Mca-R-P-K-P-V-E-Nval-W-R-K(Dnp)-NH2, the MMP10-specific peptide substrate (10 μm; R&D Systems) in MMP10 assay buffer using a X-Mark Micro plate Spectrophotometer (Bio-Rad, Philadelphia, PA; Ex. 320 nm, Em. 405 nm, 37 °C). A blank sample, consisting only of MMP10 assay buffer and peptide substrate, served as a background control. RFU of all samples were corrected for RFU of the blank at 0 s. Results presented correspond to mean of corrected RFU obtained in nine technical replicates.
Confirmation of IGFBP6 Cleavage Site
Recombinant IGFBP6 was incubated with active KLK7 (active site-titrated KLK7:IGFBP6 = 1:50 molar ratio) for 18 h at 37 °C in KLK7 activity buffer. Protein mixture was loaded on C18 column (A5700310 OMIX, 96 C18 10 μl from Agilent Technologies Australia, VIC, Australia) and N termini demethylated with Formaldehyde. After washes and elution, proteins were trypsin digested overnight at 37 °C (ratio trypsin/protein of 1:10, Promega Sequencing grade Trypsin (V5111)). Peptides were cleaned using C18 column and analyzed on an Easy nLC - Q Exactive Plus (configurations are as above). MS data have been analyzed using Proteome Discoverer 2.2.0.388 (Thermo Scientific). MS/MS spectra were searched with Sequest HT engine against a SwissProt database (Homo Sapiens, 20,153 sequences). Database search was performed with the following search parameters: semi-trypsin peptides with two missed cleavages allowed, variable modifications included M oxidation (+ 15.994 Da), N-terminal acetylation (+ 42.010 Da) and N-terminal Dimethylation (+28.031 Da). Carbamidomethylation of C (+ 57.021 Da) and Dimethylation of K (+28.031 Da) were set to fixed modification. Precursor tolerance was set to 10 ppm and fragment tolerance was set at 0.02 Da. False discovery rates (FDRs) at the peptide and protein levels were fixed at 1%.
RESULTS
Novel Putative KLK7 Substrates Were Identified in SKOV-3 Cell CM by qPROTOMAP
Eighty-seven proteins were determined to be putative extracellular KLK7 substrates among a total of 261 putative substrates identified across all three samples (Table I, supplemental Table S1). Notably, established KLK7 substrates, fibronectin 1 (FN1), desmoglein 2 (DSG2, (33)) and matrix metalloprotease 2 (MMP2, (34)) were also cleaved in SKOV-3 cell CM herein, validating the qPROTOMAP approach. The remaining other 84 KLK7 substrates have not been determined as ovarian cancer cell-derived KLK7 substrates by other published work, and thus are deemed novel putative KLK7 substrates. Some are established substrates of other KLKs, thus their determination as KLK7 substrates was not unexpected. These include basement or extracellular matrix proteins, such as collagen I and IV, and other proteins, such as amyloid beta (A4) precursor protein (APP), IGFBP2, thrombospondin 1 (THBS1), and vinculin (VCL). Thus, apart from the established KLK7 substrates, FN1, DSG2, and MMP2 and the six above known to be cleaved by other KLKs, KLK7 cleaved an additional 78 novel substrates.
Table I. Eighty-seven putative extracellular KLK7 substrates identified in the qPROTOMAP analysis of SKOV-3 cell conditioned media.
| Accession | Description | Protein Length(AA) | Protein Size (kDa) | Protein Coverage (%) | Unique Peptides |
|---|---|---|---|---|---|
| O00468 | Agrin | 2045 | 214.8 | 40.8 | 61 |
| P61160 | Actin-related protein 2 | 394 | 44.8 | 18 | 7 |
| P61158 | Actin-related protein 3 | 418 | 47.4 | 29.2 | 12 |
| P04083 | Annexin A1 | 346 | 38.7 | 68.5 | 28 |
| P08133 | Annexin A6 | 673 | 75.9 | 21.1 | 13 |
| P05067 | Amyloid beta A4 protein | 770 | 86.9 | 35.5 | 24 |
| P30530 | Tyrosine-protein kinase receptor UFO | 894 | 98.3 | 7.3 | 5 |
| Q6YHK3 | CD109 antigen | 1445 | 161.7 | 11.7 | 16 |
| P55285 | Cadherin-6 | 790 | 88.3 | 9 | 7 |
| P00751 | Complement factor B | 764 | 85.5 | 23.3 | 17 |
| Q9Y696 | Chloride intracellular channel protein 4 | 253 | 28.8 | 67.6 | 16 |
| O94985 | Calsyntenin-1 | 981 | 109.8 | 39.8 | 37 |
| Q00610 | Clathrin heavy chain 1 | 1675 | 191.6 | 38 | 55 |
| Q99715 | Collagen alpha-1(XII) chain | 3063 | 333.1 | 52.9 | 153 |
| P39060 | Collagen alpha-1(XVIII) chain | 1754 | 178.2 | 17 | 24 |
| P02452 | Collagen alpha-1(I) chain | 1464 | 138.9 | 13.9 | 17 |
| P02462 | Collagen alpha-1(IV) chain | 1669 | 160.6 | 9.6 | 10 |
| P08572 | Collagen alpha-2(IV) chain | 1712 | 167.6 | 10.5 | 13 |
| P20908 | Collagen alpha-1(V) chain | 1838 | 183.6 | 13.6 | 19 |
| P12109 | Collagen alpha-1(VI) chain | 1028 | 108.5 | 17.7 | 15 |
| P12110 | Collagen alpha-2(VI) chain | 1019 | 108.6 | 9 | 10 |
| Q02388 | Collagen alpha-1(VII) chain | 2944 | 295.2 | 16.8 | 37 |
| P00450 | Ceruloplasmin | 1065 | 122.2 | 10.3 | 9 |
| O00622 | Protein CYR61 | 381 | 42 | 17.1 | 6 |
| Q14118 | Dystroglycan | 895 | 97.4 | 22 | 16 |
| Q9UBP4 | Dickkopf-related protein 3 | 350 | 38.4 | 28 | 7 |
| *Q14126 | Desmoglein-2 | 1118 | 122.3 | 3.8 | 3 |
| Q16610 | Extracellular matrix protein 1 | 540 | 60.7 | 26.3 | 13 |
| O43854 | EGF-like repeat and discoidin I-like domain | 480 | 53.8 | 36 | 18 |
| P20827 | Ephrin-A1 | 205 | 23.8 | 7.3 | 2 |
| P15311 | Ezrin | 586 | 69.4 | 41.1 | 25 |
| Q92520 | Protein FAM3C | 227 | 24.7 | 50.2 | 10 |
| P23142 | Fibulin-1 | 703 | 77.2 | 19.9 | 10 |
| P98095 | Fibulin-2 | 1184 | 126.6 | 24.5 | 22 |
| P35555 | Fibrillin-1 | 2871 | 312.2 | 20.4 | 51 |
| Q14512 | Fibroblast growth factor-binding protein 1 | 234 | 26.3 | 23.9 | 6 |
| *P02751 | Fibronectin | 2386 | 262.6 | 36.1 | 66 |
| Q12841 | Follistatin-related protein 1 | 308 | 35 | 69.2 | 27 |
| O95633 | Follistatin-related protein 3 | 263 | 27.7 | 17.1 | 4 |
| Q14393 | Growth arrest-specific protein 6 | 721 | 79.7 | 9.2 | 5 |
| P56159 | GDNF family receptor alpha-1 | 465 | 51.5 | 11 | 5 |
| P35052 | Glypican-1 | 558 | 61.7 | 41.9 | 18 |
| P28799 | Granulins | 593 | 63.5 | 30.5 | 16 |
| P06396 | Gelsolin | 782 | 85.7 | 16.2 | 9 |
| P98160 | Basement membrane-specific heparan sulfate protein | 4391 | 468.8 | 34.2 | 110 |
| Q92743 | Serine protease HTRA1 | 480 | 51.3 | 22.1 | 12 |
| P18065 | Insulin-like growth factor-binding protein 2 | 325 | 34.8 | 30.8 | 9 |
| Q16270 | Insulin-like growth factor-binding protein 7 | 282 | 29.1 | 57.8 | 20 |
| P26006 | Integrin alpha-3 | 1051 | 116.6 | 8.8 | 10 |
| P55268 | Laminin subunit beta-2 | 1798 | 196 | 30 | 42 |
| P11047 | Laminin subunit gamma-1 | 1609 | 177.6 | 48.9 | 73 |
| P09382 | Galectin-1 | 135 | 14.7 | 64.4 | 10 |
| Q08380 | Galectin-3-binding protein | 585 | 65.3 | 40.9 | 19 |
| Q9Y4K0 | Lysyl oxidase homolog 2 | 774 | 86.7 | 28.3 | 19 |
| Q8N2S1 | Latent-transforming growth factor beta-binding protein 4 | 1624 | 173.4 | 6.3 | 9 |
| O00339 | Matrilin-2 | 956 | 106.8 | 12.9 | 10 |
| P08581 | Hepatocyte growth factor receptor | 1390 | 155.5 | 12.1 | 14 |
| P03956 | Interstitial collagenase | 469 | 54 | 50.1 | 30 |
| P09238 | Stromelysin-2 | 476 | 54.2 | 26.5 | 14 |
| *P08253 | 72 kDa type IV collagenase | 660 | 73.9 | 24.4 | 14 |
| P26038 | Moesin | 577 | 67.8 | 48 | 31 |
| Q14112 | Nidogen-2 | 1375 | 151.3 | 40 | 40 |
| P48745 | Protein NOV homolog | 357 | 39.2 | 37.3 | 11 |
| P19021 | Peptidyl-glycine alpha-amidating monooxygenase | 973 | 108.3 | 11 | 11 |
| Q8NBP7 | Proprotein convertase subtilisin/kexin type | 692 | 74.3 | 22.8 | 13 |
| Q9NRA1 | Platelet-derived growth factor C | 345 | 39 | 18 | 7 |
| P00749 | Urokinase-type plasminogen activator | 431 | 48.5 | 52.9 | 21 |
| P07225 | Vitamin K-dependent protein S | 676 | 75.1 | 13.3 | 10 |
| P07602 | Prosaposin | 524 | 58.1 | 39.9 | 17 |
| P10586 | Receptor-type tyrosine-protein phosphatase F | 1907 | 212.9 | 23.7 | 35 |
| Q13332 | Receptor-type tyrosine-protein phosphatase S | 1948 | 217 | 17.3 | 23 |
| Q92626 | Peroxidasin homolog | 1479 | 165.3 | 38.6 | 49 |
| P63000 | Ras-related C3 botulinum toxin substrate 1 | 192 | 21.5 | 25 | 5 |
| O00560 | Syntenin-1 | 298 | 32.4 | 44.6 | 10 |
| P05121 | Plasminogen activator inhibitor 1 | 402 | 45.1 | 34.6 | 14 |
| P50454 | Serpin H1 | 418 | 46.4 | 41.4 | 10 |
| P09486 | SPARC | 303 | 34.6 | 31 | 11 |
| Q13813 | Spectrin alpha chain, non-erythrocytic 1 | 2472 | 284.5 | 36.7 | 79 |
| Q01082 | Spectrin beta chain, non-erythrocytic 1 | 2364 | 274.6 | 32.1 | 63 |
| P52823 | Stanniocalcin-1 | 247 | 27.6 | 34 | 9 |
| Q15582 | Transforming growth factor-beta-induced protein | 683 | 74.7 | 43.5 | 26 |
| P07996 | Thrombospondin-1 | 1170 | 129.4 | 15.9 | 14 |
| P16035 | Metalloproteinase inhibitor 2 | 220 | 24.4 | 62.3 | 18 |
| Q9Y490 | Talin-1 | 2541 | 269.8 | 23.6 | 42 |
| Q99536 | Synaptic vesicle membrane protein VAT-1 homolog | 393 | 41.9 | 8.7 | 3 |
| P13611 | Versican core protein | 3396 | 372.8 | 6.8 | 23 |
| P18206 | Vinculin | 1134 | 123.8 | 49.1 | 52 |
Accession, reviewed UniProtKB accession; Description, UniProtKB protein description;
*Established KLK7 substrates.
TAILS Approach Identifies Exact Cleavage Sites OfqPROTOMAP-identified Putative KLK7 Substrates
To determine the exact cleavage sites of the qPROTOMAP-identified substrates TAILS analysis was employed with a lower KLK7:CM ratio (1:200), thereby allowing for identification of precise KLK7 cleavage sites. However, pre-TAILS analysis was not performed; putative cleavage site identification was limited to proteins that have been already identified in the qPROTOMAP approach. This analysis identified cleavage sites for 18 putative KLK7 substrates (Table II, supplemental Table S2). Notably, two established KLK7 substrates, MMP2 (34) and IGFBP3 (35) were cleaved in SKOV-3 cell CM, in the TAILS approach. Moreover, the two established substrates of KLK7 that were identified in the qPROTOMAP approach, DSG2 and FN1, were not detected to be cleaved by KLK7 in the TAILS approach where a 4-fold less concentration (1:200 of active site-titrated KLK7: CM w/w) of KLK7 was used compared with the qPROTOMAP approach (1:50 of active site-titrated KLK7: CM w/w).
Table II. Eighteen putative extracellular KLK7 substrates identified in the TAILS analysis of SKOV-3 cell conditioned media.
| Accession | Symbol | Protein Description | P1 | Peptides | m/z | Precursor charge | Probability |
|---|---|---|---|---|---|---|---|
| Peptidases and peptidase regulators | |||||||
| #P03956 | MMP1 | Matrix metallopeptidase 1 | 70 | F.FGLKVTGKPDAETLKVMKQPR.C | 838.5505 | 3 | 1 |
| 71 | F.GLKVTGKPDAETLKVMKQPR.C | 789.5278 | 3 | 1 | |||
| 116 | Y.RIENYTPDLPR.A | 469.9315 | 3 | 0.9889 | |||
| 149 | F.SGDVQLAQDDIDGIQAIYGR.S | 723.3728 | 3 | 1 | |||
| *#P08253 | MMP2 | Matrix metallopeptidase 2 | 80 | F.FGLPQTGDLDQNTIETMR.K | 1035.5226 | 2 | 1 |
| #P09238 | MMP10 | Matrix metallopeptidase 10 | 99 | F.SSFPGMPKWR.K | 420.9112 | 3 | 1 |
| Pathway inhibitors | |||||||
| #Q9UBP4 | DKK3 | Dickkopf WNT signaling pathway inhibitor 3 | 128 | F.SETVITSVGDEEGRR.S | 834.9341 | 2 | 0.9987 |
| ECM/basement membrane constituents and ECM/growth factor regulators | |||||||
| #Q02388 | COL7A1 | Collagen, type VII, alpha 1 | 2819 | Y.AADTAGSQLHAVPVLR.V | 547.3162 | 3 | 0.9991 |
| #Q99715 | COL12A1 | Collagen, type XII, alpha 1 | 1139 | D.NTVVLEELR.A | 553.8351 | 2 | 0.9452 |
| 1138 | Y.DNTVVLEELR.A | 611.3486 | 2 | 0.9986 | |||
| 2926 | M.LNQIPNDYQSSR.N | 734.8836 | 2 | 0.9924 | |||
| #P35555 | FBN1 | Fibrillin 1 | 1760 | Y.TGLPVDIDECR.E | 654.8376 | 2 | 0.9998 |
| #O95633 | FSTL3 | Follistatin-like 3 | 84 | L.GFLGLVHCLPCKDSCDGVECGPGKACR.M | 1051.1868 | 3 | 1 |
| *P17936 | IGFBP3 | Insulin-like growth factor binding protein 3 | 186 | Y.KVDYESQSTDTQNFSSESKR.E | 813.4225 | 3 | 1 |
| P24592 | IGFBP6 | Insulin-like growth factor binding protein 6 | 165 | R.HLDSVLQQLQTEVYR.G | 932.0126 | 2 | 1 |
| 179 | Y.RGAQTLYVPNCDHR.G | 860.9416 | 2 | 0.8584 | |||
| #Q14112 | NID2 | Nidogen 2 (osteonidogen) | 80 | N.GIISTQDFPR.E | 584.3326 | 2 | 0.8835 |
| 124 | Y.REDTSPAVLGLAAR.Y | 745.4311 | 2 | 0.9988 | |||
| #P09486 | SPARC | Secreted protein, acidic, cysteine-rich (osteonectin) | 179 | Y.ERDEDNNLLTEKQKLR.V | 1052.1161 | 2 | 0.918 |
| #P07996 | THBS1 | Thrombospondin 1 | 258 | Y.IGHKTKDLQAICGISCDELSSMVLELR.G | 1059.2505 | 3 | 1 |
| 665 | Y.LGHYSDPMYR.C | 636.8161 | 2 | 0.9809 | |||
| Cell adhesion molecules/regulators and signal transducers | |||||||
| #P55285 | CDH6 | Cadherin 6, type 2, K-cadherin | 343 | Y.TLKVEASNPYVEPR.F | 835.9911 | 2 | 0.9999 |
| Cytoskeletal proteins/regulators | |||||||
| #P15311 | EZR | Ezrin | 424 | Y.TAKIALLEEAR.R | 641.9221 | 2 | 0.9972 |
| Other (Ion channel, galactoside/carbohydrate binding and immunomodulatory proteins) | |||||||
| #O94985 | CLSTN1 | Calsyntenin 1 | 169 | Y.KATVIEGKQYDSILR.V | 912.0781 | 2 | 0.9999 |
| #Q08380 | LGALS3BP | Lectin, galactoside-binding, soluble, 3 binding protein | 442 | Y.SSDYFQAPSDYR.Y | 735.3416 | 2 | 0.9975 |
| #P07602 | PSAP | Prosaposin | 171 | F.MANIPLLLYPQDGPR.S | 866.4871 | 2 | 0.9998 |
Accession, reviewed UniProt KB accession; Symbol, UniProtKB/Swiss-Prot entry name prefix (all_HUMAN) with HUGO gene name for UniProtKB entries; Protein Description, UniProtKB protein description; P1, KLK7 cleaves C-terminal to; Peptides, TAILS-identified peptide cleavage sites with the “.” representing where the cleavage is identified; *Established KLK7 substrates.
KLK7 Hydrolyzes Substrates Into Bio-active Products
In the current study, three (calsyntenin-1, MMP10 and thrombospondin 1) of the 16 putative KLK7 substrates were identified to be hydrolyzed by KLK7 to produce potential bioactive by products (Table III) as in UniProtKB. Herein, KLK7-mediated hydrolysis of MMP10 and thrombospondin 1 was further assessed, as KLK7-generated bioactive fragments of these proteins have a high likelihood of exerting cancer promoting functions according to the literature. Interestingly, MMP1 which was also identified as a substrate of KLK7 could only be validated on Western blot analysis of SKOV-3 CM and not with recombinant proteins (supplemental Fig. S4).
Table III. Putative KLK7 substrates that may have hydrolyzed into bioactive by-products by KLK7 proteolysis in SKOV-3 cell conditioned media, as in UniProtKB.
| Description | Putative KLK7 cleavage product/chain | Annotated cleavage product bioactivity in UniProtKB |
|---|---|---|
| Calsyntenin-1 | A fragment of soluble calsyntenin alpha | None annotated |
| Matrix metallopeptidase 10 (stromelysin 2) | Stromelysin 2 | Peptidase |
| Thrombospondin 1 | N-terminal heparin binding domain | Regulation of angiogenesis |
Description, UniProtKB protein description.
KLK7 Directly Activates pro-MMP10
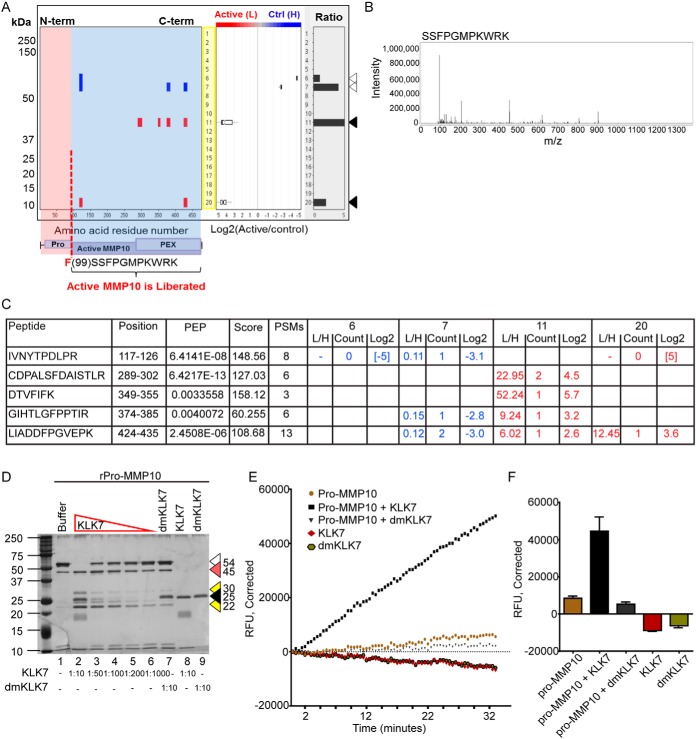
Pro-MMP10 was identified as a substrate of KLK7 in the qPROTOMAP and TAILS analysis of SKOV-3 cell CM. Peptides derived from pro-MMP10 were identified in both the control and KLK7-treated conditions (Fig. 1A and 1C, See supplementary Fig. S2 for peptograph description). In gel slices 6 and 7 at ∼55 kDa three protein fragments representing the full length pro-MMP10 were identified (blue). Protein fragments representative of the active MMP10 were retrieved in the control sample to a lesser extent, indicating that MMP10 remains in its inactive form in the control sample. In contrast, in the KLK7-treated condition (red), a larger number of protein fragments, representative of active MMP10, at ∼45 kDa (gel slice 11), were identified with high confidence showing much higher abundance in the KLK7-treated sample (Log2 KLK7/control = 2–5). Further in the KLK7-treated condition, two peptides in gel slice 20 were identified, which may have been the result of auto-cleavage of MMP10 post activation, or potential cleavage by another protease in the cell CM. Intriguingly, pro-MMP10 was also recognized as a substrate of KLK7 in the TAILS analysis. Only one cleavage site was identified cleaving C-terminal to phenylalanine99 (F99↓SSFPGMPKWR; Log2 KLK7/control = 4.32, 20-fold higher in the KLK7-treated condition) suggesting KLK7-mediated cleavage following the activation peptide, that spans from the residue 18 to 98 (Fig. 1A and 1B) of the prepro-MMP10. Further, the identified cleavage site (F99↓SSFPGMPKWR), as shown by the TAILS analysis, suggests direct cleavage of pro-MMP10 by KLK7, noting the chymotryptic-like activity of KLK7 (15, 36).
Fig. 1.
KLK7 directly activates pro-MMP10. A, Peptograph (replicate 2) representing KLK7-mediated hydrolysis of pro-MMP10 in SKOV-3 cell CM. See supplemental Fig. S2 for peptograph description. The higher molecular weight (MW) peptides (blue - Heavy labeled) in gel slice 7 (Log2 KLK7/control = −3 to −5) represent the full length pro-MMP10 identified in the buffer-treated sample. The lower MW fragments in gel slices 11 and 20 (Log2 KLK7/control = 3 to 5) represent the KLK7 cleavage fragments identified in the KLK7-treated sample (red - Light labeled). A schematic of selected protein domains, based on annotation in the UniProtKB, is shown beneath the X-axis (purple boxes), aligned with the appropriate residues. Pro, activation peptide; PEX, hemopexin domain; the molecular weight of the protein standard (kDa) is indicated to the left. Arrow heads in colors depict, open: pro-MMP10 full length; black, KLK7-generated MMP10 fragments. The box plot in the middle panel represents the distribution of ratios found in each gel slice. B, Spectrum for the TAILS identified KLK7 cleavage site C-terminal to F99 and is also shown beneath the domain structure in a red dotted vertical line in A. C, Peptides identified in the qPROTOMAP analysis in gel slices 6, 7, 11 and 20 (PEP, posterior error probability; Score, the sum of the ion scores of all peptides identified; PSMs, peptide spectrum matches) with their respective Light/Heavy ratio, count and Log2 value. D, Silver-stained 12% SDS-PAGE showing hydrolysis of recombinant (r) pro-MMP10 (400 ng) by recombinant active KLK7 (1/10 −1/1000 molar ratio to rpro-MMP10); buffer and dmKLK7 treatments were used as controls. Arrow heads in colors depict, open: pro-MMP10 full length protein; red: 45 kDa cleavage fragment; yellow: cleavage fragment ∼22 and 30 kDa; black: KLK7 or dmKLK7. E, Pro-MMP10 treated with KLK7 (black squares) showed increasing fluorescence emission over time compared with the controls. Pro-MMP10 (yellowish brown circles) showed a slight increase in fluorescence emission with the fluorogenic peptide substrate over time, indicating partial activity, because of residual active MMP10 in the recombinant pro-MMP10 protein purchased. KLK7 (red diamonds) or dmKLK7 (green circles) did not show activity with the MMP10 fluorogenic peptide substrate, confirming that fluorescence intensity increases in the KLK-treated pro-MMP10 is because of putative KLK7-activated-pro-MMP10, but not because of residual KLK7 activity. F, Bar graph represents end point (at 35 min) of the reactions performed with triplicate biological replicates (n = 3) with the error bar representing the standard error of the mean. In all instances blank corrected fluorescence values were plotted.
On biochemical validation (Fig. 1D), KLK7 completely degraded the pro-MMP10 band at 54 kDa (1:10 molar ratio, lane 2), generating 45 kDa active MMP10 protein, as noted by an increase in intensity compared with lane 1. To further assess the direct activation of pro-MMP10 by KLK7, a peptide substrate assay was performed. KLK7-treated pro-MMP10 showed a clear increase in RFU over time compared with that of the dmKLK7-treated pro-MMP10 or pro-MMP10 alone, indicating KLK7-mediated activation of pro-MMP10 (Fig. 1E, 1F).
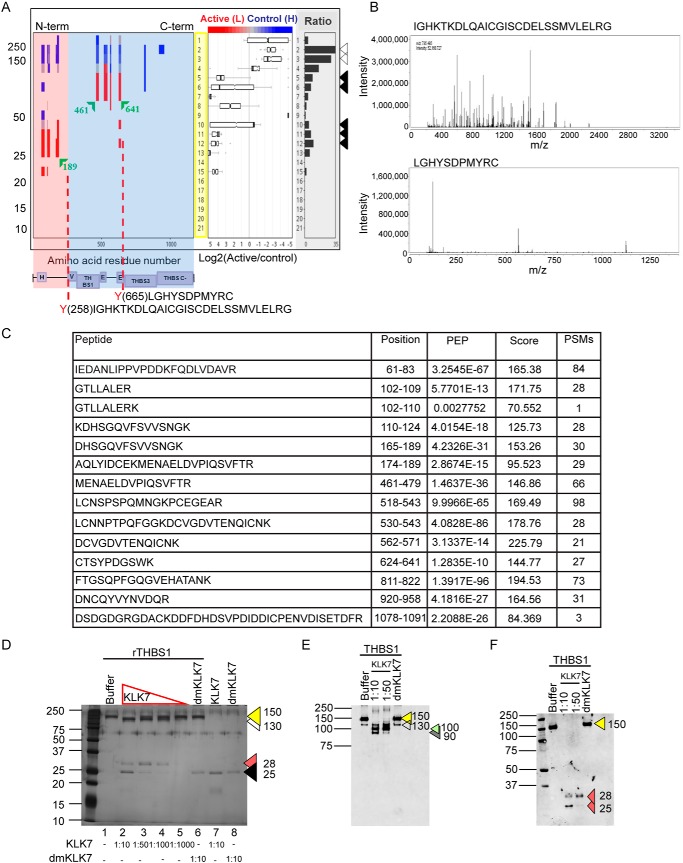
KLK7 liberates the N terminus of THBS1
THBS1 was identified as a substrate of KLK7 in the qPROTOMAP and TAILS analysis of SKOV-3 cell CM. Peptides representing THBS1 were identified in both the control and KLK7-treated conditions, with a high number of high molecular weight peptides identified in the former (Fig. 2A and 2C, See supplemental Fig. S2 for peptograph description). In the control group (blue), THBS1 protein fragments were identified in the gel slices 2 and 3, representing full length THBS1 (150 kDa). In the KLK7-treated condition (red), N-terminal THBS1 fragments spanning the heparin binding domain (H) were identified in the gel slices 11, 12, and 13 (26–28 kDa) and also peptides spanning the von Willebrand factor-type C (VWFC) domain, THBS1-like domain 1 (THBS-1) and the EGF-like domains (E) were identified in gel slices 5–6 representing a 75–100 kDa fragment.
Fig. 2.
KLK7-mediated cleavage of thrombospondin 1. A, Peptograph (replicate 1) representing KLK7-mediated hydrolysis of THBS1 in SKOV-3 cell CM See supplemental Fig. S2 for peptograph description. Arrowheads to the right indicate the migration of THBS1 retrieved from the control (open) and KLK7-generated fragments of THBS1 (filled). The higher molecular weight (MW) peptides (blue and gray) are abundant in gel slices 1–4 (Log2 KLK7/control = 0 to −5), representing the fragments derived from the full-length protein identified in both KLK7- and buffer-treated samples. The lower MW fragments are abundant in gel slices 5–15 (Log2 KLK7/control = 0–5), representing the KLK7 cleavage fragments (red) found in the KLK7-treated sample. A schematic of selected protein domains, based on annotation in the UniProtKB, is shown beneath the X-axis (purple boxes), aligned with the appropriate residues. H, heparin-binding; V, von Willebrand factor, type-C; THBS1/3, THBS type-1/3 repeat; E, epidermal growth factor-like; THBS C-, THBS C-terminal. The TAILS identified KLK7 cleavage sites C-terminal to Y258 and Y665 are shown by red dotted vertical lines and B, represents the respective spectrums. C, Peptides identified in the qPROTOMAP analysis (PEP, posterior error probability; Score, The sum of the ion scores of all peptides identified; PSMs, peptide spectrum matches). D, Silver-stained 12% SDS-PAGE showing hydrolysis of recombinant (r) THBS1 (500 ng) by recombinant active KLK7 (1/10 −1/1000 molar ratio to rTHBS1); buffer and dmKLK7 treatments were used as controls. Arrow heads in colors depict, yellow: THBS1 full length protein; white: cleavage fragment ∼130 kDa; red: 28 kDa cleavage fragment; black: KLK7 or dmKLK7. Western blot analysis of KLK7-treated rTHBS1 using antibodies targeting E, full length and F, N-terminal THBS1 confirmed the KLK7-mediated generation of N-terminal heparin binding domain containing fragment. Arrowheads indicate different protein products: yellow: full length THBS1, 150 kDa; white: 130 kDa; green: 100 kDa; gray: 90 kDa; red: 28 and 25 kDa fragments. The MW of the protein standard (kDa) is indicated to the left.
The TAILS analysis confirmed qPROTOMAP observations by identifying two peptides representing KLK7-mediated cleavage of THBS1 at two distinct sites. The peptide depicting the cleavage of THBS1 after tyrosine (Y258) Y258↓IGHKTKDLQAICGISCDELSSMVLER (Fig. 2A and 2B; Log2 KLK7/control = 9.96) possibly generates an N-terminal 28 kDa fragment containing the heparin binding domain. The second cleavage site was identified C-terminal to Y665 (Y665↓LGHYSDPMYR), suggesting a cleavage in the EGF-like 2 domain generating a C-terminal THBS-like domain 3 (THBS-3) and THBS C-terminal (THBS-C) containing fragment and a fragment containing the Von Willebrand Factor-type C (VWFC) domain, THBS-1 and the EGF-like domains (E), in accordance with the qPROTOMAP observation.
In the biochemical validation (Fig. 2D), KLK7 hydrolyzed rTHBS1 at molar ratios of 1/10 1/50, and 1/100 and to a lesser extent at the 1/1000 molar ratio generating 130 kDa and 28 kDa fragments. Importantly, the 28 kDa and 130 kDa fragments were of high intensity, reflecting KLK7-mediated liberation of an N-terminal smaller fragment and another higher molecular weight mid-terminal fragment. The above observations were further validated by performing Western blot analysis using antibodies targeting the full length (Fig. 2E) and N-terminal sequence (Fig. 2F) of THBS1.
Heterogeneity of Cell Secreted Proteins Identified in Ovarian Cancer Cell Lines
TAILS analysis of the OVMZ-6 cell secretome identified 37 putative extracellular KLK7 substrates (Table IV, supplemental Table S3). Interestingly, on comparison of the TAILS analyses of the SKOV-3 and OVMZ-6 cell secretomes to determine any heterogeneity between the cell lines, only eight putative extracellular KLK7 substrates were identified in both SKOV-3 and OVMZ-6 cell CM. These substrates included calsyntenin-1 (CLSTN1), cystatin-C (CST3), dickkopf-related protein 3 (DKK3), ezrin (EZR), insulin-like growth factor-binding protein 6 (IGFBP6), lectin, galactoside-binding, soluble, 3 binding protein (LGALS3BP), prosaposin (PSAP) and osteonectin (SPARC) (Table IV, proteins denoted with a #). Moreover, comparison of the qPROTOMAP and TAILS-identified substrates in SKOV-3 and OVMZ-6 cells, respectively, revealed another seven substrates exclusive to the nine common substrates listed above (Table IV, see proteins denoted with a ¥), increasing the list of common substrates to 16. Of these sixteen commonly identified substrates we sought to further biochemically validate IGFBP6, an established substrate of KLK4 and KLK5 (37), cleavage of which is known to activate IGF signaling and enhance cancer cell proliferation (38). Notably, the established KLK7 substrate, midkine (MDK) was cleaved in OVMZ-6 cell CM.
Table IV. Thirty-seven putative extracellular KLK7 substrates identified in the TAILS analysis of OVMZ-6 cell conditioned media.
| Accession | Symbol | Protein Description | P1 | Peptides | m/z | Precursor charge | Probability |
|---|---|---|---|---|---|---|---|
| Peptidases and peptidase regulators | |||||||
| #P01034 | CST3 | cystatin C | 125 | F.QIYAVPWQGTMTLSKSTCQD.A | 1227.1276 | 2 | 0.9996 |
| 128 | Y.AVPWQGTMTLSKSTCQD.A | 1025.0246 | 2 | 1 | |||
| ¥Q92743 | HTRA1 | HtrA serine peptidase 1 | 286 | F.SLQNTVTTGIVSTTQR.G | 870.4891 | 2 | 1 |
| P06744 | GPI | glucose-6-phosphate isomerase | 426 | L.ANFLAQTEALMR.G | 699.8846 | 2 | 0.9971 |
| Q9Y287 | ITM2B | integral membrane protein 2B | 254 | F.ENKFAVETLIC.S | 739.9136 | 2 | 0.9999 |
| P10646 | TFPI | tissue factor pathway inhibitor | 247 | Y.SGCGGNENNFTSKQECLR.A | 1063.5051 | 2 | 1 |
| 209 | F.EFHGPSWCLTPADR.G | 853.9121 | 2 | 1 | |||
| Pathway inhibitors | |||||||
| #Q9UBP4 | DKK3 | dickkopf WNT signaling pathway inhibitor 3 | 128 | F.SETVITSVGDEEGRR.S | 834.9341 | 2 | 0.9998 |
| ECM/basement membrane constituents and ECM/growth factor regulators | |||||||
| ¥Q14512 | FGFBP1 | fibroblast growth factor binding protein 1 | 97 | A.GNPTSCLKLKDER.V | 810.4851 | 2 | 0.9996 |
| P51858 | HDGF | hepatoma-derived growth factor | 79 | R.KGFSEGLWEIENNPTVKASGY.Q | 810.1128 | 3 | 0.9991 |
| #P24592 | IGFBP6 | insulin like growth factor binding protein 6 | 219 | G.KSLPGSPDGNGSSSCPTGSS.G | 1002.4841 | 2 | 0.9999 |
| 218 | M.GKSLPGSPDGNGSSSCPTGSS.G | 1030.9951 | 2 | 1 | |||
| 179 | Y.RGAQTLYVPNCDHR.G | 860.9416 | 2 | 0.9365 | |||
| *P21741 | MDK | midkine (neurite growth-promoting factor 2) | 88 | F.ENWGACDGGTGTKVR.Q | 838.4281 | 2 | 1 |
| 87 | K.FENWGACDGGTGTKVR.Q | 911.9626 | 2 | 1 | |||
| 29 | K.KGGPGSECAEWAWGPCTPSSKDCGVGFR.E | 1048.1725 | 3 | 0.9963 | |||
| 86 | Y.KFENWGACDGGTGTKVR.Q | 993.0416 | 2 | 1 | |||
| #P09486 | SPARC | secreted protein, acidic, cysteine-rich (osteonectin) | 179 | Y.ERDEDNNLLTEKQKLR.V | 1052.1161 | 2 | 0.9983 |
| Cell adhesion molecules/regulators and signal transducers/cell surface receptors | |||||||
| ¥P05067 | APP | amyloid beta precursor protein | 433 | Q.HFQEKVESLEQEAANER.Q | 1056.5531 | 2 | 1 |
| P63092 | GNAS | GNAS complex locus | 318 | Y.TTPEDATPEPGEDPR.V | 823.3916 | 2 | 1 |
| Q13421 | MSLN | mesothelin | 67 | L.GFPCAEVSGLSTER.V | 772.3851 | 2 | 0.9957 |
| O95631 | NTN1 | netrin 1 | 251 | F.SRLHTFGDENEDDSELAR.D | 1063.0036 | 2 | 0.9983 |
| 254 | L.HTFGDENEDDSELAR.D | 884.8951 | 2 | 1 | |||
| Q99985 | SEMA3C | semaphorin 3C | 499 | Y.VSSNEGVSQVSLHR.C | 766.9156 | 2 | 1 |
| Q08629 | SPOCK1 | sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1 | 22 | R.HLDALAGGAGPNHGN.F | 717.8686 | 2 | 1 |
| Q9UDY2 | TJP2 | tight junction protein 2 | 459 | F.KSTGDIAGTVVPETNKEPR.Y | 1051.1211 | 2 | 0.9998 |
| Cytoskeletal proteins/regulators | |||||||
| ¥P61160 | ACTR2 | ARP2 actin-related protein 2 homolog (yeast) | 356 | L.GGAVLADIMKDKDNFWMTR.Q | 1135.6296 | 2 | 1 |
| #P15311 | EZR | ezrin | 547 | R.THNDIIHNENMR.Q | 764.3811 | 2 | 0.9998 |
| 579 | R.IDEFEA.L | 764.3811 | 2 | 0.9998 | |||
| 295 | R.KPDTIEVQQM.K | 628.8631 | 2 | 0.9973 | |||
| O75781 | PALM | paralemmin | 56 | L.EGTPSSASEGDEDLRR.Q | 870.4161 | 2 | 1 |
| ¥Q9Y490 | TLN1 | talin 1 | 1575 | F.ASNPEFSSIPAQISPEGR.A | 960.9971 | 2 | 1 |
| Other (Ion channel, galactoside/carbohydrate/lipid binding and transporters | |||||||
| #P61769 | B2M | beta-2-microglobulin | 85 | L.YYTEFTPTEKDEYACR.V | 1071.0121 | 2 | 0.9999 |
| 87 | Y.TEFTPTEKDEYACR.V | 907.9486 | 2 | 1 | |||
| 86 | Y.YTEFTPTEKDEYACR.V | 989.4806 | 2 | 1 | |||
| 108 | S.QPKIVKWDRD.M | 759.4716 | 2 | 0.9996 | |||
| P54105 | CLNS1A | chloride channel, nucleotide-sensitive, 1A | 168 | Y.TYEEGLSHLTAEGQATLER.L | 1070.0426 | 2 | 0.9998 |
| #O94985 | CLSTN1 | calsyntenin 1 | 169 | Y.KATVIEGKQYDSILR.V | 912.0781 | 2 | 1 |
| Q6UVK1 | CSPG4 | chondroitin sulfate proteoglycan 4 | 1856 | L.SVVDPDSAPGEIEYEVQR.A | 1012.5051 | 2 | 1 |
| P17931 | LGALS3 | lectin, galactoside-binding, soluble, 3 | 107 | Y.GAPAGPLIVPYNLPLPGGVVPR.M | 1094.6566 | 2 | 1 |
| #Q08380 | LGALS3BP | lectin, galactoside-binding, soluble, 3 binding protein | 442 | Y.SSDYFQAPSDYR.Y | 735.3416 | 2 | 0.9997 |
| P29122 | PCSK6 | proprotein convertase subtilisin/kexin type 6 | 302 | Y.SASWGPDDDGKTVDGPGR.L | 942.9716 | 2 | 1 |
| ¥Q8NBP7 | PCSK9 | proprotein convertase subtilisin/kexin type 9 | 418 | F.SAKDVINEAWFPEDQR.V | 987.0236 | 2 | 1 |
| 400 | L.SAEPELTLAELR.Q | 681.8881 | 2 | 0.9976 | |||
| 183 | Y.LLDTSIQSDHREIEGR.V | 952.0081 | 2 | 0.9919 | |||
| #P07602 | PSAP | prosaposin | 171 | F.MANIPLLLYPQDGPR.S | 866.4871 | 2 | 0.9995 |
| 172 | M.ANIPLLLYPQDGPR.S | 800.9671 | 2 | 1 | |||
| O95810 | SDPR | serum deprivation response | 395 | Y.EGSYALTSEEAER.S | 738.3571 | 2 | 0.9998 |
| Q9ULF5 | SLC39A10 | solute carrier family 39 (zinc transporter), member 10 | 80 | F.GLEKLLTNLGLGER.K | 791.0041 | 2 | 1 |
| O14745 | SLC9A3R1 | solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 1 | 180 | R.SVDPDSPAEASGLR.A | 717.8676 | 2 | 0.9942 |
| Oxidative stress-related | |||||||
| O43827 | ANGPTL7 | angiopoietin like 7 | 271 | F.STKDKDNDNCLDKCAQLR.K | 773.0898 | 3 | 0.9987 |
| Inflammatory response | |||||||
| ¥P04083 | ANXA1 | annexin A1 | 56 | M.VKGVDEATIIDILTKR.N | 625.0787 | 3 | 0.9983 |
| O95407 | TNFRSF6B | tumor necrosis factor receptor superfamily member 6b | 233 | L.QALEAPEGWGPTPR.A | 771.9096 | 2 | 0.9891 |
| 261 | L.TELLGAQDGALLVR.L | 745.4436 | 2 | 0.9998 | |||
| 90 | Y.CNVLCGEREEEAR.A | 828.3881 | 2 | 0.944 |
Accession, reviewed UniProt KB accession; Symbol, UniProtKB/Swiss-Prot entry name prefix (all _HUMAN) with HUGO gene name for UniProtKB entries; Protein Description, UniProtKB protein description; P1, KLK7 cleaves C-terminal to; Peptides, TAILS-identified peptides with “.” indicating respective semi-tryptic cleavage sites; *Established KLK7 substrate; #Substrates identified in both SKOV-3 and OVMZ-6 cell CM by TAILS analysis; ¥Substrates identified in qPROTOMAP and TAILS analysis of SKOV-3 and OVMZ-6 cell CM respectively.
KLK7-mediated Cleavage of IGFBP6
Cleavage C-terminal to Y179 was detected in both SKOV-3 and OVMZ-6 cell lines (Y179↓RGAQTLYVPNCDHRG [Log2 KLK7/control = 9.96 in OVMZ-6]). In SKOV-3 TAILS analysis two cleavages cleaving C-terminal to arginine (R)165 and Y179 (R165↓HLDSVLQQLQTEVYRG [Log2 KLK7/control = 2.94] and Y179↓RGAQTLYVPNCD HRG [Log2 KLK7/control = 4.64]) were identified (Fig. 3A and 3B). However, in OVMZ-6 cell CM, two cleavages additional to the above were detected cleaving C-terminal to G219 and M218 (G219↓KSLPGSPDGNGSSSCPTGSSG [Log2 KLK7/control = 9.96], M218↓GKSLPGSPDGN GSSSCPTGSSG [-log2 = 4.64]). This observation suggests that other individually identified cleavage sites, such as G219 and M218 are potentially differentially influenced by other factors in these two cell lines or missed cleavages by trypsin. Further, the identified cleavage sites display either KLK7-mediated direct cleavages of IGFBP6 or indirect cleavages, as noted by the chymotryptic- (Y179) or tryptic-like (R165) cleavages and other cleavage specificities, such as G219 respectively.
Fig. 3.
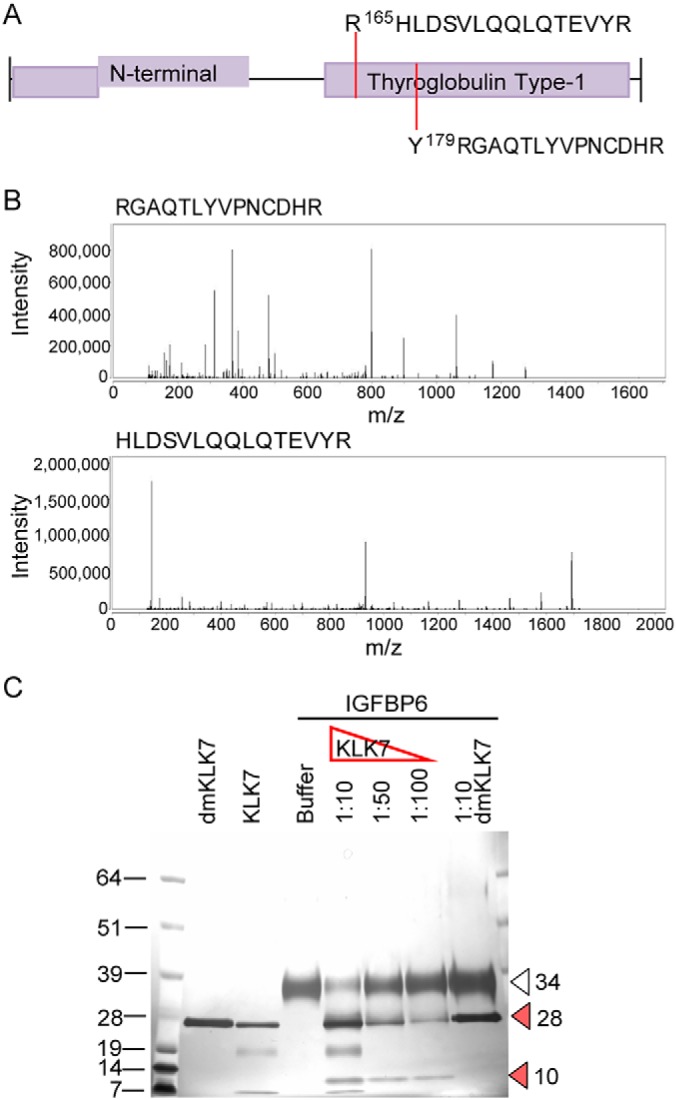
KLK7-mediated hydrolysis of IGFBP6 in SKOV-3 and OVMZ-6 cell CM. A, TAILS-identified cleavage sites are indicated by dotted lines on the schematic of selected protein domains, based on annotation in the UniProtKB. B, Respective spectra of the TAILS-identified peptides. C, Silver-stained 12% SDS-PAGE showing hydrolysis of recombinant IGFBP6. Arrowheads indicate: open: full length IGFBP6, 34 kDa; red: cleaved IGFBP6 fragments, 10 and 28 kDa. The molecular weight (MW) of the protein standard (kDa) is indicated to the left.
In the biochemical validation (Fig. 3C), KLK7 hydrolyzed rIGFBP6 at molar ratios of 1/10 1/50, and 1/100 molar ratio generating a 10 kDa fragment. For further verification, KLK7-generated major cleavage product of 10 kDa was sequenced to identify the primary KLK7 cleavage site on IGFBP6. This analysis revealed that the Y179 is the most abundant N-terminal apart from the natural N terminus of IGFBP6 (RCPGCGQGVQAGCPGGCVEEEDGGSPAEGCAEAEGCLRRpeptide) (Table V), further verifying the TAILS-identified cleavage site. Apart from the Y179 cleavage site, R165 cleavage was also identified with low abundance, although a fragment representing this cleavage had not been observed in the biochemical validation.
Table V. Peptides identified in MS analysis of KLK7-generated IGFBP6 fragment.
| Sequence | Modifications | # PSMs | # Missed Cleavages | Theo. MH+ [Da] | Abundances (Grouped): Sample | XCorr | Top Apex RT [min] | |
|---|---|---|---|---|---|---|---|---|
| A182 | QTLYVPNCDHR | 1×Carbamidomethyl [C8]; 1×Dimethyl [N-Term] | 1 | 0 | 1430.685 | 2.88 | ||
| G81 | LQCHPPKDDEAPLR | 1×Carbamidomethyl [C3]; 1×Dimethyl [K7]; 1×Dimethyl [N-Term] | 1 | 1 | 1731.885 | 3.78 | ||
| Q156 | DTEMGPCR | 1×Carbamidomethyl [C7]; 1×Dimethyl [N-Term] | 1 | 0 | 993.4128 | 2.34 | ||
| N151 | SAGVQDTEMGPCR | 1×Carbamidomethyl [C12]; 1×Oxidation [M9]; 1×Dimethyl [N-Term] | 1 | 0 | 1451.625 | 4.2 | ||
| A27 | RCPGCGQGVQAGCPGGCVEEEDGGSPAEGCAEAEGCLRR | 6×Carbamidomethyl [C2; C5; C13; C17; C30; C36]; 1×Dimethyl [N-Term] | 15 | 2 | 4164.71 | 1.00E+09 | 9.79 | 15.16 |
| *Y179 | RGAQTLYVPNCDHR | 1×Carbamidomethyl [C11]; 1×Dimethyl [N-Term] | 3 | 1 | 1714.844 | 3.00E+08 | 4.32 | 14.05 |
| Y74 | TPNCAPGLQCHPPKDDEAPLR | 2×Carbamidomethyl [C4; C10]; 1×Dimethyl [K14]; 1×Dimethyl [N-Term] | 3 | 1 | 2429.17 | 2.00E+08 | 6.1 | 14.8 |
| A27 | RCPGCGQGVQAGCPGGCVEEEDGGSPAEGCAEAEGCLR | 6×Carbamidomethyl [C2; C5; C13; C17; C30; C36]; 1×Dimethyl [N-Term] | 10 | 1 | 4008.609 | 2.00E+08 | 9.75 | 15.54 |
| L185 | YVPNCDHR | 1×Carbamidomethyl [C5]; 1×Dimethyl [N-Term] | 3 | 0 | 1088.494 | 9.00E+07 | 2.65 | 11.1 |
| N51 | SAGVQDTEMGPCRR | 1×Carbamidomethyl [C12]; 1×Dimethyl [N-Term] | 2 | 1 | 1591.732 | 7.00E+07 | 4.38 | 13.95 |
| N151 | SAGVQDTEMGPCRR | 1×Carbamidomethyl [C12]; 1×Oxidation [M9]; 1×Dimethyl [N-Term] | 2 | 1 | 1607.726 | 5.00E+07 | 3.42 | 13.62 |
| Q37 | AGCPGGCVEEEDGGSPAEGCAEAEGCLRR | 4×Carbamidomethyl [C3; C7; C20; C26]; 1×Dimethyl [N-Term] | 2 | 1 | 3065.245 | 5.00E+07 | 5.58 | 15.26 |
| N77 | CAPGLQCHPPKDDEAPLR | 2×Carbamidomethyl [C1; C7]; 1×Dimethyl [K11]; 1×Dimethyl [N-Term] | 3 | 1 | 2117.027 | 2.00E+07 | 6.33 | 14.69 |
| N151 | SAGVQDTEMGPCR | 1×Carbamidomethyl [C12]; 1×Dimethyl [N-Term] | 1 | 0 | 1435.63 | 2.00E+07 | 3.9 | 14.62 |
| A38 | GCPGGCVEEEDGGSPAEGCAEAEGCLRR | 4×Carbamidomethyl [C2; C6; C19; C25]; 1×Dimethyl [N-Term] | 3 | 1 | 2994.208 | 9.00E+06 | 5.14 | 15.24 |
| Q37 | AGCPGGCVEEEDGGSPAEGCAEAEGCLR | 4×Carbamidomethyl [C3; C7; C20; C26]; 1×Dimethyl [N-Term] | 1 | 0 | 2909.144 | 7.00E+06 | 7.56 | 15.84 |
| *R165 | HLDSVLQQL | 1×Dimethyl [N-Term] | 1 | 0 | 1080.605 | 5.00E+06 | 2.09 | 18.92 |
| A111 | VAEENPKESKPQAGTARPQDVNRR | 2×Dimethyl [K7; K10]; 1×Dimethyl [N-Term] | 1 | 2 | 2761.47 | 5.00E+06 | 4.75 | 13.6 |
| R209 | GPCWCVDR | 2×Carbamidomethyl [C3; C5]; 1×Dimethyl [N-Term] | 1 | 0 | 1077.46 | 4.00E+06 | 2.16 | 15.55 |
| G51 | SPAEGCAEAEGCLRR | 2×Carbamidomethyl [C6; C12]; 1×Dimethyl [N-Term] | 1 | 1 | 1690.764 | 2.00E+06 | 2.81 | 14.13 |
| R208 | RGPCWCVDR | 2×Carbamidomethyl [C4; C6]; 1×Dimethyl [N-Term] | 1 | 1 | 1233.562 | 2.00E+06 | 2.94 | 14.43 |
| Q34 | GVQDTEMGPCRR | 1×Carbamidomethyl [C10]; 1×Dimethyl [N-Term] | 1 | 1 | 1433.662 | 2.00E+06 | 3.2 | 13.83 |
| G142 | TSTTPSQPNSAGVQDTEMGPCRR | 1×Carbamidomethyl [C21]; 1×Dimethyl [N-Term] | 1 | 1 | 2505.146 | 2.00E+06 | 4.3 | 14.57 |
| A38 | GCPGGCVEEEDGGSPAEGCAEAEGCLR | 4×Carbamidomethyl [C2; C6; C19; C25]; 1×Dimethyl [N-Term] | 1 | 0 | 2838.107 | 1.00E+06 | 5.81 | 15.81 |
| R180 | GAQTLYVPNCDHR | 1×Carbamidomethyl [C10]; 1×Dimethyl [N-Term] | 1 | 0 | 1558.743 | 9.00E+05 | 3.6 | 14.8 |
| G51 | SPAEGCAEAEGCLR | 2×Carbamidomethyl [C6; C12]; 1×Dimethyl [N-Term] | 1 | 0 | 1534.662 | 5.00E+05 | 2.02 | 14.61 |
| Q183 | TLYVPNCDHR | 1×Carbamidomethyl [C7]; 1×Dimethyl [N-Term] | 1 | 0 | 1302.626 | 4.00E+05 | 2.57 | 14.84 |
PSMs, peptide spectrum matches; Theo MH+, protonated monoisotopic mass of the peptides in Daltons; XCorr, A search-dependent score - it scores the number of fragment ions that are common to two different peptides with the same precursor mass and calculates the cross-correlation score for all candidate peptides queried from the database by SEQUEST searches; RT (min), the peptide's retention time during chromatographic separation; and *peptides identified in TAILS analysis. Peptides are sorted according to the “Abundances (grouped samples)” column.
Annotated Cellular Processes of the Identified Putative Extracellular KLK7 Substrates in SKOV-3 and OVMZ-6 Cell CM
Putative extracellular KLK7 substrates identified in both cell lines are predicted to be primarily involved in cell-cell adhesion, extracellular matrix organization, and cell surface receptor-linked signal transduction gene ontology (GO)-annotated biological processes (Table VI, supplemental Table S4–S5), equivalent to the established functions of KLK7. The GO analysis revealed that, albeit the differences in the secreted proteome of these two cell lines, KLK7 exerts similar functional roles by cleaving extracellular and membrane proteins involved in these processes.
Table VI. Gene Ontology-annotated biological processes for TAILS-identified putative extracellular KLK7 substrates in SKOV-3 and OVMZ-6 cell conditioned media.
| GOTERM_BP_FAT | P Value | Symbol |
|---|---|---|
| SKOV-3 | ||
| GO:0007155∼cell adhesion | 4.31E-06 | COL7A1, EZR, NID2, CDH6, COL12A1, LGALS3BP, THBS1, CLSTN1 |
| GO:0022610∼biological adhesion | 4.35E-06 | COL7A1, EZR, NID2, CDH6, COL12A1, LGALS3BP, THBS1, CLSTN1 |
| GO:0030574∼collagen catabolic process | 2.16E-04 | MMP2, MMP10, MMP1 |
| GO:0044243∼multicellular organismal catabolic process | 3.67E-04 | MMP2, MMP10, MMP1 |
| GO:0032963∼collagen metabolic process | 4.27E-04 | MMP2, MMP10, MMP1 |
| GO:0044259∼multicellular organismal macromolecule metabolic process | 5.24E-04 | MMP2, MMP10, MMP1 |
| GO:0044236∼multicellular organismal metabolic process | 7.47E-04 | MMP2, MMP10, MMP1 |
| GO:0001501∼skeletal system development | 0.004787827 | MMP2, SPARC, COL12A1, FBN1 |
| OVMZ-6 | ||
| GO:0006928∼cell motion | 1.80E-04 | ACTR2, ANXA1, TLN1, SEMA3C, NTN1, SPOCK1, APP |
GOTERM_BP_FAT, summarized version of biological processes in the Gene Ontology (GO) database; P Value, calculated using a modified Fisher Exact test; Symbol, UniProtKB/Swiss-Prot entry name prefix (all _HUMAN) with HUGO gene name for UniProtKB entries
TAILS Analysis Determined A Chymotryptic-like Cleavage Specificity Profile for KLK7
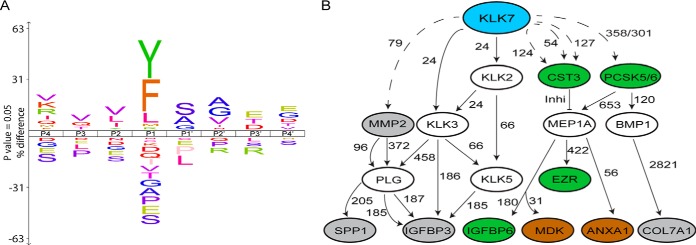
The KLK7 cleavage site specificity profile (Fig. 4A), based on TAILS-identified cleavage events within all putative KLK7 substrates (before sorting for extracellular proteins), predominantly showed chymotryptic-like cleavage specificity of KLK7 by cleaving after tyrosine (Y), phenylalanine (F), leucine (L) and methionine (M) at the P1 position, where the scissile peptide bond is located. Chymotryptic-like cleavages of KLK7 and tryptic-like cleavage specificities cleaving c-terminal to arginine/R were observed in accordance with the KLK7 cleavage site specificity profile obtained in other studies (15, 36, 39, 40). Additionally, nonchymotryptic-like cleavage site specificities, such as serine (S), proline (P), glutamine (E), alanine (A), glycine (G), threonine (T), isoleucine (I), valine (V), glutamine (Q) and aspartic acid (D) were observed, which could be proteolytic effects of other proteases activated by KLK7-mediated hydrolysis in cell CM.
Fig. 4.
KLK7 subsite preferences and potential role in the greater proteolytic web. A, To determine KLK7 subsite preferences, TAILS-identified cleavage sites are displayed as IceLogos representation by comparing both the positive and negative frequency percentage of an amino acid (vertical axis) at a certain location in the multiple sequence alignment (horizontal axis) and the Swiss-Prot Homo sapiens protein database. KLK7 showed chymotryptic-like specificities by cleaving C-terminal to tyrosine (Y), phenylalanine (F) and leucine (L). The height of the single amino acid residue reflects its occurrence rate for each position in P4-P4′. Amino acid residues are color coded according to their physico-chemical properties. Sequence logo x axis indicates percent difference at a p value of 0.05%. B, Potential association of KLK7 with TAILS-identified (11) substrates, determined using the TopFINDer (v3.0) software. Blue oval, protease of interest, KLK7; gray ovals, TAILS-identified KLK7 substrates in SKOV-3 cell line; brown ovals, TAILS-identified KLK7 substrates in OVMZ-6 cell line; green ovals, TAILS-identified substrates in both cell lines; open ovals, KLK7 substrates known to date and putative intermediate proteins; dotted line, cleavages identified in TAILS; solid lines, recorded cleavage events in MEROPS; and numbers, amino acid residues at cleavage sites.
Potential KLK7 Role in the Greater Proteolytic Web
The TAILS analysis identified non-KLK7 cleavage preferences at the P1 position, which has not been detected in the literature (36). Thus, these non-KLK7 cleavage preferences were thought to have originated from indirect cleavages that occurred upon KLK7-mediated proteolytic activation of other zymogens or cleavage of inhibitors of other proteases. In view of this, we utilized the TopFINDer web tool to determine potential protease cascades activated by KLK7 (supplemental Table S13, supplemental Table S6). TopFINDer predicted potential KLK7-activated protease cascades involving eleven of the novel substrates, including insulin-like growth factor binding protein 3 (IGFBP3), IGFBP6, ezrin (EZRI), matrix-metalloprotease 2 (MMP2), osteopontin (SPP1), cystatin C (CST3), proprotein convertase subtilisin/kexin type 5 (PCSK5), collagen, type VII, alpha 1 (COL7A1), midkine (MDK) and annexin-1 (ANXA1) (Fig. 4B). Interestingly, substrates that are directly cleaved by KLK7, such as IGFBP3, IGFBP6, MMP2 and MDK, as shown in this as well as other studies are also suggested to be cleaved by indirect pathways. Perhaps in a complex in vivo microenvironment both direct and indirect pathways are plausible, and study of these cascades provides the opportunity to understand potential relationships between identified or established KLK7 substrates and putative protein/protease intermediates in the in vivo microenvironment.
DISCUSSION
The present study combined two in-depth quantitative proteomics approaches for profiling of the putative substrate repertoire of the KLK7 peptidase in vitro as an initial screening for potential physiological targets. Considering that KLK7 is a secreted protease that is activated in the extracellular microenvironment, and the role of the ascites in metastatic ovarian cancer, we used conditioned media (CM) from two ascites-derived cell lines, SKOV-3 and OVMZ-6, to identify the KLK7 degradome and exact cleavage sites using qPROTOMAP and/or TAILS analysis, respectively. We identified 16 novel substrates and two known substrates with high confidence in the secretome of the SKOV-3 cell line using both approaches. On TAILS analysis of the OVMZ-6 secretome, eight substrates were among the 16 SKOV-3 substrates noted above with a further seven OVMZ-6 substrates also identified by the qPROTOMAP approach in SKOV-3 cells. A further 83 putative substrates were identified overall although these still require more comprehensive verification. This is the first study to conduct a comprehensive analysis of the KLK7 degradome in ovarian cancer cell secretomes to define the putative substrate repertoire of KLK7 in this microenvironment.
qPROTOMAP and TAILS approaches, although allows for global substrate identification, are linked with technical bias. For instance, the most abundant proteins are favored, and some proteins were not detected because of peptide loss causing false negative identifications (41, 42). In TAILS it is necessary to perform a LC-MS/MS analysis with pre-TAILS samples (prior to negative selection), to ascertain protein identifications (22). In this study, TAILS protein identifications were limited to those identified in the qPROTOMAP analysis, where protein identifications were made with the protein fragments detected in each gel slice across all replicates spanning the length of the protein. Moreover, in the qPROTOMAP analysis we have used a 1:50 KLK7 to CM (w/w), whereas a 1:200 (w/w) ratio was used in the TAILS analysis, potentially leading to identification of a lesser number of cleavage events in the TAILS experiment than in the qPROTOMAP approach.
A large percentage of the proteins identified were of intracellular origin, which may have been released upon cell lysis. In fact, the cancer cell secretome has been shown to consist of classical intracellular proteins released through unconventional pathways (43). Further, intracellular proteins may get secreted into the extracellular microenvironment through vesicles, such as exosomes (43). Although such intracellular proteins secreted into the extracellular microenvironment may still function as signaling molecules that regulate cell-cell communication, only the substrates with an extracellular origin were considered in this study.
In addition to the known KLK7 substrate, MMP2, we have now shown that KLK7 activates MMP10, which in turn, and like that of KLK7, can cleave ECM and basement membrane components (44). Thus, proteolytic processing of these proteins by MMP10 suggests both KLK7-mediated direct and indirect functional effects in cell shedding and cell adhesion to the peritoneal wall initiating metastasis (45). Interestingly, in the KLK7 TAILS analysis COL12A1, a MMP10 substrate, was also cleaved after D1139NTVVLEELRA. This may suggest a potential MMP10-mediated cleavage of COL12A1, because MMP10 cleaves C-terminal to aspartic acid (D) (46).
MMP1, a known cancer promoting peptidase (56) was also identified to be cleaved by KLK7 in the complex secretome microenvironment but not in the biochemical assay (supplemental Fig. S4), suggesting a KLK7-mediated proteolytic event induced by other factors available in cell CM but not in the biochemical assay. Perhaps pro-MMP1 is cleaved by another protease activated by KLK7-mediated proteolytic activity or another factor may have stimulated the KLK7-mediated cleavage of pro-MMP1 in the complex pool of proteins secreted by SKOV-3 cells. The cleavage sites identified for MMP1 in the TAILS assay have chymotryptic-like cleavage site preferences, suggesting a KLK7-mediated activation of another chymotryptic-like protease in the SKOV-3 cell CM. This observation implies the potential of KLK7-activated protease cascades and the bias associated with the use of biochemical assays to determine peptidase substrates emphasizing the importance of proteomics analysis of complex protein pools, as in the present study, to determine putative substrates of peptidases that are aberrantly expressed in cancer. However, any functional effect of this KLK7-mediated cleavage of MMP1 is not clear and requires further functional validation. Nonetheless, these combined findings on KLK7 cleavage of several MMPs point to a potentially critical proteolytic interaction in the ovarian tumor microenvironment.
The current study reported a KLK7-mediated cleavage of THBS1, producing a 28 kDa N-terminal fragment, using both the qPROTOMAP and TAILS approaches. Cleavage of THBS1 N terminus has been already reported in the literature, where A disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS)-1 (47), thrombin (48), and cathepsin G (49) were shown to cleave THBS1, generating similar N-terminal THBS1 fragments, of 36 kDa, 25 kDa, and 28 kDa, respectively. Interestingly, the N-terminal 28 kDa fragment was previously shown to promote angiogenesis through a syndecan-4proteoglycan-dependent pathway (50), although full length THBS1 actively inhibits angiogenesis by inducing receptor-mediated apoptosis in activated endothelial cells (51). Further, high levels of THBS1 in high grade ovarian cancer patients are associated with improved survival because of inhibition of angiogenesis (52).
Insulin-like growth factor binding protein (IGFBP) 6 was proteolytically processed in both SKOV-3 and OVMZ-6 cell CM. IGFBP6 has been shown to promote cell migration (53), suppress cellular proliferation (54) and inhibit angiogenesis (55) in other cancers, suggesting a cancer promoting as well as a suppressive role, but has not been widely studied in the ovarian cancer context. However, IGFBP6 have been shown to be lowly expressed in ovarian cancer patients compared with the healthy controls (56). TAILS-determined cleavage of IGFBP6 suggests a cleavage in the thyroglobulin type-1 domain, the consequences of which is yet to be interrogated.
We observed a significant difference in the pool of cell secreted proteins by the two cell lines employed. Importantly, the OVMZ-6 line is from a FIGO Stage IV poorly differentiated serous adenocarcinoma (14) whereas the SKOV-3 cell line is derived from a patient with moderately well differentiated adenocarcinoma (12, 13) so heterogeneity of cell secreted proteins would be expected (57). Despite the heterogeneity of cell secreted proteins, the GO-annotated biological functions suggested that KLK7-mediated proteolytic processing of these different substrates ultimately led to classical KLK7 functions that had been identified to date, such as cell shedding and ECM remodeling. Notably, we still identified 15 common KLK7 substrates in the secretome of these 2 cell lines.
In accordance with the KLK7 cleavage site specificity profile obtained in other studies, KLK7-derived peptides showed primarily chymotryptic- (Y, F, L and M) and tryptic-like (R) cleavage site specificities (36, 39, 40). This is the first study to determine KLK7 cleavage site specificities using human-derived full-length proteins as the library and thus determining the physiological cleavage specificities of KLK7. In this analysis, specificities other than chymotryptic- and tryptic-like cleavages were also observed, which could have originated from indirect cleavage events from proteases activated by KLK7-mediated proteolytic processing, such as MMP10.
The present study has contributed greatly to the knowledge of the KLK7 degradome in the ovarian cancer microenvironment by increasing the existing list of KLK7 substrates. The novel putative extracellular KLK7 substrates identified in the secretome of two distinct ovarian cancer ascites-derived cell lines using two proteomics approaches will help determine the physiological substrates of KLK7 and mechanisms underlying the functional roles of KLK7. Further, it provides a large data source of 83 putative substrates, which can be interrogated for future studies analyzing the mechanistic and functional role of KLK7 in this cancer and may be extrapolated to other cancers as well. Similar studies should also be conducted on other KLKs and other proteases, which may help in delineating the role of these peptidases in the greater proteolytic web.
DATA AVAILABILITY
MS data have been deposited to the ProteomeXchange consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository. Zipped MaxQuant “.txt” files including “.wiff” files have been deposited with the dataset identifier PXD003812 for SKOV-3 qPROTOMAP analysis. The TAILS MS files have been deposited with the dataset identifier PXD003752 or DOI: 10.6019/PXD003752 for SKOV-3 and PXD004591 or DOI: 10.6019/PXD004591 for OVMZ-6.
Supplementary Material
Footnotes
* This work was funded by grants-in-aid from the National Health and Medical Research Council of Australia (553045), Australia-India Strategic Research Fund (BF060023), the Cancer Council Queensland (1051318, 1105922), Wesley Research Institute Foundation (2010-07) and the DAAD Deutsch-Australisches Netzwerk der Personalisierten Krebsmedizin. Access to proteomic infrastructure in the QIMR Berghofer Protein Discovery Centre was made possible by funding from Bioplatforms Australia and the Queensland State Government provided through the Australian Government National Collaborative Infrastructure Strategy (NCRIS) and EIF Fund. JAC is a Principal Research Fellow of the National Health and Medical Research Council of Australia (1005717).
 This article contains supplemental Figures and Tables.
This article contains supplemental Figures and Tables.
1 The abbreviations used are:
- ATCC
- American Type Culture Collection
- dmKLK7
- double mutant Kallikreinrelated peptidase 7
- BCA
- bicinchoninic acid assay
- BSA
- bovine serum albumin
- CM
- conditioned media
- COL
- collagen
- DPBS
- Dulbecco's phosphate-buffered saline
- ECM
- extracellular matrix
- FBS
- fetal bovine serum
- FDR
- false discovery rate
- HPG-ALD
- hyper branched polyglycerol-aldehyde
- IAA
- iodoacetamide
- IDA
- Information Dependent Acquisition
- IGFBP
- insulin-like growth factor binding protein
- IPA
- ingenuity pathway analysis
- MMP
- matrix metalloprotease
- qPROTOMAP
- quantitative PROtein TOpography and Migration Analysis Platform
- SILAC
- stable isotopic labelling by amino acids in cell culture
- TAILS
- Terminal Amine Isotopic Labelling of Substrates
- TPP
- trans proteomics pipeline
- THBS1
- thrombospondin 1
- UniProtKB
- UniProt Knowledgebase.
REFERENCES
- 1. Siegel R. L., Miller K. D., and Jemal A. (2016) Cancer statistics, 2016. CA Cancer J. Clin. 66, 7–30 [DOI] [PubMed] [Google Scholar]
- 2. Bhoola S., and Hoskins W. J. (2006) Diagnosis and Management of Epithelial Ovarian Cancer. Obstet. Gynecol., 107, 1399–1410 [DOI] [PubMed] [Google Scholar]
- 3. Puls L. E., Duniho T., Hunter J. E., Kryscio R., Blackhurst D., and Gallion H. (1996) The prognostic implication of ascites in advanced-stage ovarian cancer. Gynecol. Oncol. 61, 109–112 [DOI] [PubMed] [Google Scholar]
- 4. Dong Y., Tan O. L., Loessner D., Stephens C., Walpole C., Boyle G. M., Parsons P. G., and Clements J. A. (2010) Kallikrein-related peptidase 7 promotes multicellular aggregation via the α5β1 integrin pathway and paclitaxel chemoresistance in serous epithelial ovarian carcinoma. Cancer Res. 70, 2624. [DOI] [PubMed] [Google Scholar]
- 5. Ahmed N., and Stenvers K. L. (2013) Getting to know ovarian cancer ascites: opportunities for targeted therapy-based translational research. Front. Oncol. 3, 256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Prezas P., Arlt M. J., Viktorov P., Soosaipillai A., Holzscheiter L., Schmitt M., Talieri M., Diamandis E. P., Krüger A., and Magdolen V. (2006) Overexpression of the human tissue kallikrein genes KLK4, 5, 6, and 7 increases the malignant phenotype of ovarian cancer cells. Biol. Chem. 387, 807–811 [DOI] [PubMed] [Google Scholar]
- 7. Dorn J., Bronger H., Kates R., Slotta-Huspenina J., Schmalfeldt B., Kiechle M., Diamandis E. P., Soosaipillai A., Schmitt M., and Harbeck N. (2015) OVSCORE - a validated score to identify ovarian cancer patients not suitable for primary surgery. Oncol. Lett. 9, 418–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dorn J., Beaufort N., Schmitt M., Diamandis E.P., Goettig P., and Magdolen V. (2014) Function and clinical relevance of kallikrein-related peptidases and other serine proteases in gynecological cancers. Crit. Rev. Clin. Lab. Sci. 51, 63–84 [DOI] [PubMed] [Google Scholar]
- 9. Fuhrman-Luck R. A., Silva M. L., Dong Y., Irving-Rodgers H., Stoll T., Hastie M. L., Loessner D., Gorman J. J., and Clements J. A. (2014) Proteomic and other analyses to determine the functional consequences of deregulated kallikrein-related peptidase (KLK) expression in prostate and ovarian cancer. Proteomics Clin. Appl. 8, 403–415 [DOI] [PubMed] [Google Scholar]
- 10. Dix M. M., Simon G. M., and Cravatt B. F. (2008) Global mapping of the topography and magnitude of proteolytic events in apoptosis. Cell 134, 679–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kleifeld O., et al. (2010) Isotopic labeling of terminal amines in complex samples identifies protein N-termini and protease cleavage products. Nat. Biotechnol. 28, 281–288 [DOI] [PubMed] [Google Scholar]
- 12. Fogh J., Fogh J. M., and Orfeo T. (1977) One hundred and twenty-seven cultured human tumor cell lines producing tumors in nude mice. J. Natl. Cancer Inst. 59, 221–226 [DOI] [PubMed] [Google Scholar]
- 13. Fogh J., Wright W. C., and Loveless J. D. (1977) Absence of HeLa cell contamination in 169 cell lines derived from human tumors. J. Natl. Cancer Inst. 58, 209–214 [DOI] [PubMed] [Google Scholar]
- 14. Mobus V., Gerharz C. D., Press U., Moll R., Beck T., Mellin W., Pollow K., Knapstein P. G., and Kreienberg R. (1992) Morphological, immunohistochemical and biochemical characterization of 6 newly established human ovarian carcinoma cell lines. Int. J. Cancer 52, 76–84 [DOI] [PubMed] [Google Scholar]
- 15. Silva L. M., Stoll T., Kryza T., Stephens C. R., Hastie M. L., Irving-Rodgers H. F., Dong Y., Gorman J. J., and Clements J. A. (2017) Mass spectrometry-based determination of Kallikrein-related peptidase 7 (KLK7) cleavage preferences and subsite dependency. Sci. Rep. 7, 6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vizcaino J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Ríos D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P. A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R. J., Kraus H. J., Albar J. P., Martinez-Bartolomé S., Apweiler R., Omenn G. S., Martens L., Jones A. R., and Hermjakob H. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ong S. E., Kratchmarova I., and Mann M. (2003) Properties of 13C-substituted arginine in stable isotope labeling by amino acids in cell culture (SILAC). J. Proteome Res. 2, 173–181 [DOI] [PubMed] [Google Scholar]
- 18. Shevchenko A., Tomas H., Havlis J., Olsen J. V., and Mann M. (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 [DOI] [PubMed] [Google Scholar]
- 19. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 20. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J.V., and Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 21. Dix M. M., Simon G. M., Wang C., Okerberg E., Patricelli M. P., and Cravatt B. F. (2012) Functional interplay between caspase cleavage and phosphorylation sculpts the apoptotic proteome. Cell 150, 426–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kleifeld O., Doucet A., Prudova A., auf dem Keller U., Gioia M., Kizhakkedathu J. N., and Overall C. M. (2011) Identifying and quantifying proteolytic events and the natural N terminome by terminal amine isotopic labeling of substrates. Nat. Protocols 6, 1578–1611 [DOI] [PubMed] [Google Scholar]
- 23. Kessner D., Chambers M., Burkem R., Agus D., and Mallick P. (2008) ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics 24, 2534–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Craig R., and Beavis R. C. (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 [DOI] [PubMed] [Google Scholar]
- 25. Pedrioli P. G. (2010) Trans-proteomic pipeline: a pipeline for proteomic analysis. Methods Mol. Biol. 604, 213–238 [DOI] [PubMed] [Google Scholar]
- 26. Magrane M., and U. Consortium, (2011) UniProt Knowledgebase: a hub of integrated protein data. Database (Oxford), 2011, p. bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mellacheruvu D., Wright Z., Couzens A. L., Lambert J. P., St-Denis N. A., Li T., Miteva Y. V., Hauri S., Sardiu M. E., Low T. Y., Halim V. A., Bagshaw R. D., Hubner N. C., Al-Hakim A., Bouchard A., Faubert D., Fermin D., Dunham W. H., Goudreault M., Lin Z. Y., Badillo B. G., Pawson T., Durocher D., Coulombe B., Aebersold R., Superti-Furga G., Colinge J., Heck A. J., Choi H., Gstaiger M., Mohammed S., Cristea I. M., Bennett K. L., Washburn M. P., Raught B., Ewing R. M., Gingras A. C., and Nesvizhskii A. I. (2013) The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat. Methods 10, 730–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Keller A., Eng J., Zhang N., Li X. J., and Aebersold R. (2005) A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol. Syst. Biol. 1, 0017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. auf dem Keller U., Prudova A., Gioia M., Butler G. S., and Overall C. M. (2010) A statistics-based platform for quantitative N-terminome analysis and identification of protease cleavage products. Mol. Cell. Proteomics 9, 912–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fortelny N., Yang S., Pavlidis P., Lange P. F., and Overall C. M.. Proteome TopFIND 3.0 with TopFINDer and PathFINDer: database and analysis tools for the association of protein termini to pre- and post-translational events. Nucleic Acids Res. 43, D290–D297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huang da W, Sherman B. T., and Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 32. Sondell B., Dyberg P., Anneroth G. K., Ostman P. O., and Egelrud T. (1996) Association between expression of stratum corneum chymotryptic enzyme and pathological keratinization in human oral mucosa. Acta Dermato-venereologica 76, 177–181 [DOI] [PubMed] [Google Scholar]
- 33. Ramani V. C., and Haun R. S. (2008a) The extracellular matrix protein fibronectin is a substrate for kallikrein 7. Biochem. Biophys. Res. Commun. 369, 1169–1173 [DOI] [PubMed] [Google Scholar]
- 34. Ramani V. C., Kaushal G. P., and Haun R. S. (2011) Proteolytic action of kallikrein-related peptidase 7 produces unique active matrix metalloproteinase-9 lacking the C-terminal hemopexin domains. Biochim. Biophys. Acta 1813, 1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yu Y., Prassas I., Dimitromanolakis A., and Diamandis E.P. (2015) Novel biological substrates of human kallikrein 7 identified through degradomics. J. Biol. Chem., 290, 17762–17775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Skytt A., Stromqvist M., and Egelrud T. (1995) Primary substrate specificity of recombinant human stratum corneum chymotryptic enzyme. Biochem. Biophys. Res. Commun. 211, 586–589 [DOI] [PubMed] [Google Scholar]
- 37. Lawrence M. G., Lai J., and Clements J. A. (2010) Kallikreins on steroids: structure, function, and hormonal regulation of prostate-specific antigen and the extended kallikrein locus. Endocrine reviews, 31, 407–446 [DOI] [PubMed] [Google Scholar]
- 38. Borgono C. A., and Diamandis E. P. (2004) the emerging roles of human tissue kallikreins in cancer. Nature Rev. 4, 876–890 [DOI] [PubMed] [Google Scholar]
- 39. Debela M., Magdolen V., Schechter N., Valachova M., Lottspeich F., Craik C. S., Choe Y., Bode W., and Goettig P. (2006) Specificity profiling of seven human tissue kallikreins reveals individual subsite preferences. J. Biol. Chem. 281, 25678–25688 [DOI] [PubMed] [Google Scholar]
- 40. Oliveira J. R., Bertolin T. C., Andrade D., Oliveira L. C., Kondo M. Y., Santos J. A., Blaber M., Juliano L., Severino B., Caliendo G., Santagada V., and Juliano M. A. (2015) Specificity studies on Kallikrein-related peptidase 7 (KLK7) and effects of osmolytes and glycosaminoglycans on its peptidase activity. Biochim. Biophys. Acta 1854, 73–83 [DOI] [PubMed] [Google Scholar]
- 41. Speicher K. D., Kolbas O., Harper S., and Speicher D. W. (2000) Systematic analysis of peptide recoveries from in-gel digestions for protein identifications in proteome studies. J. Biomol. Tech. 11, 74–86 [PMC free article] [PubMed] [Google Scholar]
- 42. Rappsilber J., Mann M., and Ishihama Y. (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 [DOI] [PubMed] [Google Scholar]
- 43. Sinha A., Ignatchenko V., Ignatchenko A., Mejia-Guerrero S., and Kislinger T. (2014) In-depth proteomic analyses of ovarian cancer cell line exosomes reveals differential enrichment of functional categories compared to the NCI 60 proteome. Biochem. Biophys. Res. Commun. 445, 694–701 [DOI] [PubMed] [Google Scholar]
- 44. Schlage P., Kockmann T., Kizhakkedathu J. N., and auf dem Keller U. (2015) Monitoring matrix metalloproteinase activity at the epidermal-dermal interface by SILAC-iTRAQ-TAILS. Proteomics 15, 2491–2502 [DOI] [PubMed] [Google Scholar]
- 45. Justilien V., Regala R. P., Tseng I. C., Walsh M. P., Batra J., and Radisky E. S., Murray N. R., Fields A. P. (2012) Matrix metalloproteinase-10 is required for lung cancer stem cell maintenance, tumor initiation and metastatic potential. PLoS ONE 7, e35040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schlage P., Kockmann T., Kizhakkedathu J. N., and auf dem Keller U. (2015) Monitoring matrix metalloproteinase activity at the epidermal-dermal interface by SILAC-iTRAQ-TAILS. Proteomics 15, 2491–2502 [DOI] [PubMed] [Google Scholar]
- 47. Lee N. V., Sato M., Annis D. S., Loo J. A., Wu L., Mosher D. F., and Iruela-Arispe M. L. (2006) ADAMTS1 mediates the release of antiangiogenic polypeptides from TSP1 and 2. EMBO J. 25, 5270–5283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lawler J., Connolly J. E., Ferro P., and Derick L. H. (1986) Thrombin and chymotrypsin interactions with thrombospondin. Ann. N.Y. Acad. Sci. 485, 273–287 [DOI] [PubMed] [Google Scholar]
- 49. Rabhi-Sabile S., Pidard D., Lawler J., Renesto P., Chignard M., and Legrand C. (1996) Proteolysis of thrombospondin during cathepsin-G-induced platelet aggregation: functional role of the 165-kDa carboxy-terminal fragment. FEBS Lett. 386, 82–86 [DOI] [PubMed] [Google Scholar]
- 50. Ferrari do Outeiro-Bernstein M. A., Nunes S. S., Andrade A. C., Alves T. R., Legrand C., and Morandi V. (2002) A recombinant NH(2)-terminal heparin-binding domain of the adhesive glycoprotein, thrombospondin-1, promotes endothelial tube formation and cell survival: a possible role for syndecan-4 proteoglycan. Matrix Biol. 21, 311–324 [DOI] [PubMed] [Google Scholar]
- 51. Jimenez B., Volpert O. V., Crawford S. E., Febbraio M., Silverstein R. L., and Bouck N. (2000) Signals leading to apoptosis-dependent inhibition of neovascularization by thrombospondin-1. Nat. Med. 6, 41–48 [DOI] [PubMed] [Google Scholar]
- 52. Alvarez A. A., Axelrod J. R., Whitaker R. S., Isner P. D., Bentley R. C., Dodge R. K., and Rodriguez G. C. (2001) Thrombospondin-1 expression in epithelial ovarian carcinoma: association with p53 status, tumor angiogenesis, and survival in platinum-treated patients. Gynecol. Oncol. 82, 273–278 [DOI] [PubMed] [Google Scholar]
- 53. Fu P., Thompson J. A., and Bach L. A. (2007) Promotion of cancer cell migration: an insulin-like growth factor (IGF)-independent action of IGF-binding protein-6. J. Biol. Chem. 282, 22298–22306 [DOI] [PubMed] [Google Scholar]
- 54. Koike H., Ito K., Takezawa Y., Oyama T., Yamanaka H., and Suzuki K. (2005) Insulin-like growth factor binding protein-6 inhibits prostate cancer cell proliferation: implication for anticancer effect of diethylstilbestrol in hormone refractory prostate cancer. Br. J. Cancer 92, 1538–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang C., Lu L., Li Y., Wang X., Zhou J., Liu Y., Fu P., Gallicchio M. A., Bach L. A., and Duan C. (2012) IGF binding protein-6 expression in vascular endothelial cells is induced by hypoxia and plays a negative role in tumor angiogenesis. Int. J. Cancer 130, 2003–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gunawardana C., Kuk C., Smith C. R., Batruch I., Soosaipillai A., and Diamandis E. P. (2009) Comprehensive analysis of conditioned media from ovarian cancer cell lines identifies novel candidate markers of epithelial ovarian cancer. J. Proteome Res. 8, 4705–4713 [DOI] [PubMed] [Google Scholar]
- 57. Coscia F., Watters K. M., Curtis M., Eckert M. A., Chiang C. Y., Tyanova S., Montag A., Lastra R. R., Lengyel E., and Mann M. (2016) Integrative proteomic profiling of ovarian cancer cell lines reveals precursor cell associated proteins and functional status. Nat. Commun. 7, 12645. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
MS data have been deposited to the ProteomeXchange consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository. Zipped MaxQuant “.txt” files including “.wiff” files have been deposited with the dataset identifier PXD003812 for SKOV-3 qPROTOMAP analysis. The TAILS MS files have been deposited with the dataset identifier PXD003752 or DOI: 10.6019/PXD003752 for SKOV-3 and PXD004591 or DOI: 10.6019/PXD004591 for OVMZ-6.