A streamlined and readily accessible sample preparation protocol has been developed to enable TMT-based proteomic profiling of relatively low numbers of cells directly from a flow cytometer. These methods were applied to 12 freshly isolated immune cell types from mice to a depth of over 7000 quantified proteins. These data recapitulate many aspects of known immunology, nominate new cell type specific protein markers, and provide evidence for post-transcriptional regulation of gene expression across the immune system.
Keywords: Immunology*, Animal models*, Cell sorting, Gene Expression*, Systems biology*, FACS, Low Input, Post-transcriptional regulation, TMT
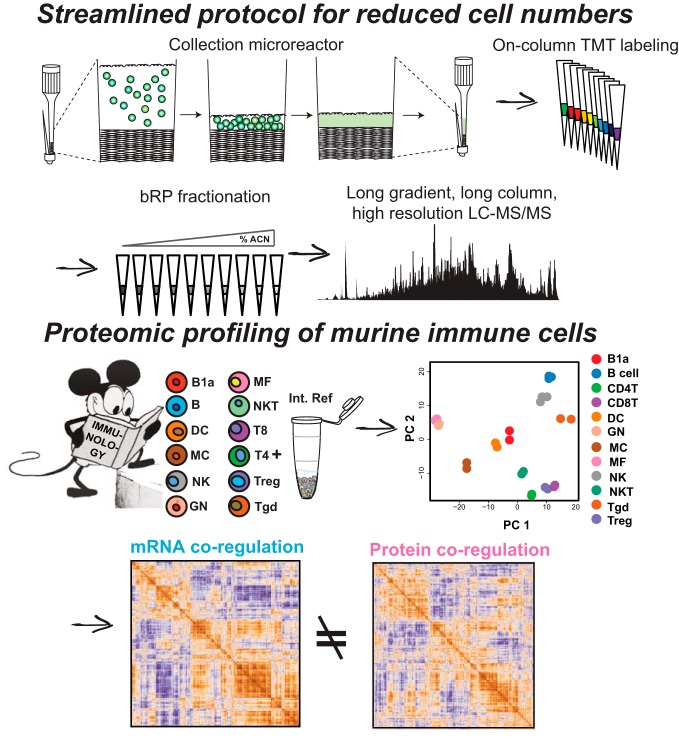
Graphical Abstract
Highlights
A low input proteomic profiling sample preparation workflow has been developed.
The protocol components are accessible and are widely applicable.
Over 7000 proteins across 12 immune cell types were quantified from 3e5 cells.
The data provide evidence for global post-transcriptional regulation.
Abstract
Proteomic profiling describes the molecular landscape of proteins in cells immediately available to sense, transduce, and enact the appropriate responses to extracellular queues. Transcriptional profiling has proven invaluable to our understanding of cellular responses; however, insights may be lost as mounting evidence suggests transcript levels only moderately correlate with protein levels in steady state cells. Mass spectrometry-based quantitative proteomics is a well-suited and widely used analytical tool for studying global protein abundances. Typical proteomic workflows are often limited by the amount of sample input that is required for deep and quantitative proteome profiling. This is especially true if the cells of interest need to be purified by fluorescence-activated cell sorting (FACS) and one wants to avoid ex vivo culturing. To address this need, we developed an easy to implement, streamlined workflow that enables quantitative proteome profiling from roughly 2 μg of protein input per experimental condition. Utilizing a combination of facile cell collection from cell sorting, solid-state isobaric labeling and multiplexing of peptides, and small-scale fractionation, we profiled the proteomes of 12 freshly isolated, primary murine immune cell types. Analyzing half of the 3e5 cells collected per cell type, we quantified over 7000 proteins across 12 key immune cell populations directly from their resident tissues. We show that low input proteomics is precise, and the data generated accurately reflects many aspects of known immunology, while expanding the list of cell-type specific proteins across the cell types profiled. The low input proteomics methods we developed are readily adaptable and broadly applicable to any cell or sample types and should enable proteome profiling in systems previously unattainable.
Proteome-wide measurements provide a more functionally relevant snapshot of cell states than transcriptional profiling alone. There is increasing evidence that steady-state measurements of mRNA levels only partially reflect the functional potential of a cell (1–5), whereas proteins are immediately available to sense and transduce extracellular cues and activate transcriptional responses to ultimately remodel the transcriptome/proteome. When used in combination, proteome profiling can reveal insights into regulatory steps such as the post-transcriptional, translational, and the post-translational levels (referred to hereafter as post-transcriptional) that can be missed with exome sequencing alone (6–9).
A major drawback for proteomic analyses is the high amount of protein input required, which can be too demanding for many biological systems. Typical sample preparation for mass spectrometry-based proteomics requires relatively large amounts of protein per sample (≥ 50 μg) per experimental condition. Samples such as cells purified by fluorescence-activated cell sorting (FACS)1, needle-core biopsies, and laser capture micro-dissected (LCM) tissue samples often yield low micrograms of protein per condition, preventing deep and quantitative global protein measurements using conventional proteomic sample preparation and analysis methods.
Immune cells comprise a wide variety of functionally distinct cell types and are most often characterized and classified by their transcriptional profiles, or a small set of protein surface markers (10–14). Previous studies profiling immune cell proteomes with liquid chromatography-mass spectrometry (LC-MS) have either not been input limited (human peripheral blood immune cells) or have expanded and differentiated purified murine immune cells in culture (15–19). Having relative protein abundances across the mouse immune system would provide a useful resource for future immunological studies in a genetically tractable organism.
Although powerful alternative approaches have been demonstrated for low input proteomics they require highly specialized equipment or expertise, or fall short of reaching an appreciable depth of coverage (20–22). de Graaf and colleagues developed a quantitative microproteomics approach for analysis of LCM samples and applied it to the analysis of murine kidney cells (23). A depth of around 4500 distinct proteins was achieved from low micrograms of total protein when samples were prepared using an automated liquid handling system (23). To date, no approaches suitable for deep, quantitative profiling of FAC-sorted cells have been reported. Here, we describe a simple to implement sample preparation protocol for TMT-based proteomic analysis of FAC-sorted cells that minimizes sample handling steps and processing time. The method combines efficient cell collection from a cell sorter, improved peptide labeling for sample multiplexing, and small-scale fractionation. Specialized equipment such as liquid handling robotics are not needed for these sample processing steps, making the method adaptable by most proteomics laboratories. Using the approach, we profiled 12 primary, murine immune cell types straight from FACS purification to a depth of over 7000 proteins, collecting 3e5 cells (∼2 μg/sample) and injecting half for analysis. Low input proteomics of freshly isolated cells provides high quality, quantitative proteomic data that strongly reflects known aspects of the immune system. Comparing our proteomic data to publicly available Immunological Genome Consortium (Immgen) transcriptional profiles, we find evidence for post-transcriptional regulation at the global scale.
EXPERIMENTAL PROCEDURES
Cell Isolation
Five-week-old male C57BL/6J mice were purchased from the Jackson Laboratory (Bar Harbor, ME) and were analyzed at 6 weeks of age. Immunocytes from pools of 3 mice were double sorted for high purity by flow cytometry (double-sorting) on a BD Aria instrument according to ImmGen protocols (33 and immgen.org). B cells, T4 cells, T8 cells, regulatory T cells, γδT cells, NKT cells, NK cells, and GNs were all obtained from spleens homogenized through a 100 μm cell strainer and treated with ACK lysing buffer, 1 ml per 108 cells. Macrophages and B1a cells were obtained by lavage of the peritoneal cavity with 7 ml PBS. Peritoneal mast cells were obtained by lavage of the peritoneal cavity with 7 ml HBSS containing 1 mm EDcell TA (11). All cells were stained and washed in DMEM using the indicated antibodies described below. For mass spectrometry, 300,000 cells were sorted into collection microreactors (described below), which were prepared by running 90 μl of phosphate buffered saline (PBS) through the tips by centrifuge. Quartz filter tips with sorted cells were briefly spun on a Galaxy Mini centrifuge (VWR #C1213, Radnor, PA) for 30 s allow the FACS sheath fluid (PBS) to run through the tip until only approximately one to five microliters of PBS from the cell suspension remained and leaving the cells on the filter. Finally, tips were frozen on dry ice before storage and cell lysis step. If 300,000 cells could not be collected from one sort, multiple tip digests were spun onto the same Stage tip to achieve this total.
Antibodies and Markers
Antibodies used included: anti-CD19 (ebio1D3, eFluor450 for FACS), anti-CD43 (eBioR2/60, PE), anti-CD5 (53–7.3, APC), anti-ICAM2 (3C4, AlexaFluor488), anti-F4/80 (BM8, PE-Cy7), anti-TCRb (H57–597, eFluor450), anti-CD4 (RM4–5, APC-eFluor780), anti-CD8 (53–6.7, APC), anti-IgM (eB121–15F9, FITC), anti-CD19 (ebio1D3, PE-Cy7), anti-B220 (RA3–6B2, eFluor450), anti-CD11c (N418, AlexaFluor700), anti-MHCII (M5/114.15.2, FITC), anti-Flt3 (A2F10, PE), anti-Ter199 (TER-119, APC-eFluor780), anti-Ly6G (R86–8C5, APC), anti-Ly6G (R86–8C5, APC-eFluor780), anti-NK1.1 (PK136, PE), anti-CD25 (PC61.5, APC), anti-TCRgd (ebioGL3, PE-Cy7), anti-CD19 (ebio1D3, APC-eFluor780), anti-CD4 (RM4–5, FITC), anti-CD8 (53–6.7, APC-eFluor780) from eBioscience, San Diego, CA, and anti-CD45 (30-F11, APC-Cy7), anti-CD11c (N418, APC), anti-CD11b (M1/70, APC), anti-CD8 (53–6.7, PacBlue), anti-CD19 (6D5, PacBlue), anti-CD4 (GK1.5, PacBlue), anti-FcεR1α (MAR-1, PE), anti-CD117 (2B8, PE-Cy7) from BioLegend, San Diego, CA.
Collection Microreactor Tips
Gel loading tips (Ranin LTS 20 uL) were packed with three to five punches of Whatman QM-A grade SiO2 mesh (GE Life Sciences, Marlborough, MA) with a 16-gauge blunt end needle to act as a small filter onto which cells accumulate (49). For FACS collection, microreactor tips were inserted into the cap of 1.5 ml centrifuge tubes that had a cross cut into them. This allows the tip and tube to be spun in various types of benchtop centrifuges. The quartz mesh was pre-wet with PBS prior to cell collection by spinning in a Galaxy minicentrifuge for a quick pulse. Cells were directly sorted into the microreactor tips and spun as above every 200 μl or so to remove the sheath fluid. This can be repeated two to three times until the tip builds up backpressure from the high number of cells. The cells are then washed with ice cold PBS and spun until near dry. The tip is then kinked right below the quartz membrane and is held shut by a 3–6 mm slice of a 200 μl pipette tips. For storage, the kinked tip's top is covered with parafilm and the entire device is stored in a 1.5 ml microfuge tube.
For cell lysis, the tip remained kinked for the entire digestion protocol. Ten microliters of 8 m urea, 10 mm TCEP and 10 mm iodoacetamide in 50 mm ammonium bicarbonate (ABC) is added using a gel loading tip to the surface of the quartz membrane. Pipetting up and down provided the shear forces to lyse the cells. For many cell types, especially T cells, the lysate becomes viscous because of genomic DNA, indicating cell lysis. Lysis, reduction, and alkylation is allowed to happen at room temperature for 30 min, shaking in the dark. 50 mm ABC is used to dilute the urea to less than 2 m and the appropriate amount of trypsin for a 1:100 enzyme to substrate ratio is added and allowed to incubate at 37 °C overnight. Once digestion is completed the pipette tip ring is removed from the kinked microreactor tip. The tip will remain partially kinked. The kinked end of the tip is inserted into a premade, equilibrated Stage tip. The microreactor-tip-in-a-Stage-tip contraption is spun at 3500 × g until the entire digest passes through the C18 resin. 75 μl 0.1% formic acid (FA) is then added to the microreactor tip washing the quartz membrane onto the C18 resin. This is repeated a second time. At this stage, the digest has now been loaded directly from the microreactor tip to the Stage tip C18 resin and is ready to be washed and eluted or can go directly to on-column TMT labeling, starting at the HEPES/TMT step (see below).
On-column TMT Labeling
Stage tips were packed with one punch of C18 mesh (Empore) with a 16-gauge blunt end needle. Resin was conditioned with 50 μl methanol (MeOH), followed by 50 μl 50% acetonitrile (ACN)/0.1% FA, and equilibrated with 75 μl 0.1% FA twice. The digest was loaded by spinning at 3500 × g until the entire digest passed through. If needed, the bound peptides were washed twice with 75 μl 0.1% FA. One μl of TMT reagent (∼19 μg when resuspended according to manufacturer's instructions) in 100% ACN was added to 100 μl freshly made HEPES, pH 8, and passed over the C18 resin at 350 × g until the entire solution has passed through. The HEPES and residual TMT was washed away with two applications of 75 μl 0.1% FA and peptides were eluted with 50 μl 50% ACN/0.1% FA followed by a second elution with 50% ACN/20 mm ammonium formate (NH4HCO2), pH 10. Peptide concentrations were estimated using an absorbance reading at 280 nm and checking of label efficiency was performed on 1/20th of the elution. We strongly recommend using freshly prepared HEPES and TMT reagents when possible. Older reagents can cause singly charged contaminants which become a predominant signal when low amounts of peptides (≤ 1 μg) are loaded onto the LC-MS. These contaminants can be partially avoided via bSDB fractionation, where the contaminants mainly elute above an ACN percentage of ∼33%. After using 1/20th of the elution to test for labeling efficiency, the samples are mixed before fractionation and analysis.
Stage tip bSDB Fractionation
200 μl pipette tips were packed with two punches of sulfonated divinylbenzene (SDB-RPS, Empore) with a 16 gauge needle. The Stage tip was conditioned and equilibrated as described above. After loading ∼20 μg peptides, a pH switch was performed using 25 μl 20 mm NH4HCO2, pH 10, and was considered part of fraction one. Then, step fractionation was performed according to the amount of peptide material and assuming equal mass distribution. For example, 20 μg of peptides were fractionated into nine fractions of 20 mm NH4HCO2, pH 10, with ACN concentrations of 5, 10, 15, 20, 25, 30, 35, 40, and 90%. Each fraction was transferred to autosampler vials and dried via vacuum centrifugation and stored at −80 °C until needed. As a rule of thumb, we determine the number of Stage tip fractions by the amount of total TMT labeled peptides going into the fractionation, (total peptide input/2) - 1. For example, if one has 2 μg/sample and ten TMT labeled samples, assuming equal mass distribution, we would have nine final fractions. Injecting half of each fractions leads to ∼1 μg on-column for LC-MS data acquisition.
For comparison, bC18 and SCX (both Empore) tips were made and fractionated in the same way as bSDB. SCX resin was conditioned with MeOH, followed by 50% ACN, 0.1% FA, 0.1% FA, and NH4OH, pH 11, 25% ACN, and was equilibrated with 0.1% FA. Prior to SCX fractionation, digests were desalted via Stage tip and were loaded onto the SCX resin in the C18 elution buffer, 50% ACN, 0.1% FA. SCX step fractionation was performed using 50 mm NH4C2H3O2, 25% ACN, pH 4.5, followed by 50 mm NH4C2H3O2, 25% ACN, pH 5.5, 50 mm NH4C2H3O2, 25% ACN, pH 6.5, NH4OH, 25% ACN, pH 8.0, 50 mm NH4HCO2, 50 mm 25% ACN, pH 9.0, and NH4OH, 25% ACN, pH 11.0. Each SCX fraction was then desalted via C18 Stage tip, dried via vacuum centrifugation and stored at −80 °C. For gradient extension experiments, only the separation gradient was extended by the stated amounts. A 1× gradient represents a 110-min method with an 84-min separation gradient. For these experiments, we kept the final fractions to six, rather than the nine as suggested above. This was to keep the gradient length extension consistent and, thus, comparisons more interpretable. All individual fractions were analyzed using the 1x gradient described above.
Protein Input Determination
We decided that sacrificing enough sample for each cell type analyzed to determine protein levels was undesirable. Instead, we prepared separate sorts for representative cell types for the sole purpose of protein input determination. GN, B, MF and CD4+ T cells were sorted into collection microreactors, lysed and digested as described above. The desalted peptides were measured using absorbance at 280 nm on a Nanodrop spectrophotometer. The 1 μg/μl Jurkat digest we use for instrument QC was serially diluted in half for a five-point standard curve. These protein yields were then assumed for all remaining cell collections.
Experimental Design and Statistical Rationale-Common Reference
A “common reference” sample used as an internal standard for each LC-MS run was prepared by mixing 200,000 cells each from the B.1a peritoneal cavity (PC), MF.PC, B.Sp, T4.spleen (Sp), T8.Sp, DC.Sp, GN.Sp, Treg.Sp, γδT.Sp, NK.Sp, and NKT.Sp populations,100,000 total CD45- mesenteric lymph node cells, 100,000 total CD45+ and total CD45− splenocytes three hours after subcutaneous injection of 10,000 U IFNα, 200,000 total CD45+ bone marrow cells, 200,000 total CD45+ gut cells, 200,000 total CD45+ peritoneal cavity cells, and 500,000 total CD45+ splenocytes. Sample layout for all other TMT channels can be found in supplemental Table S7.
Data Acquisition
Chromatography was performed using a Proxeon UHPLC at a flow rate of 200 nl/min. Peptides were separated at 50 °C using a 75 μm i.d. PicoFrit (New Objective, Woburn, MA) column packed with 1.9 μm AQ-C18 material (Dr. Maisch, Germany) to 50 cm in length over a 235 min run. The on-line LC gradient went from 6% B at 1 min to 30% B in 204 mins, followed by an increase to 60% B by minute 214, then to 90% by min 215, and finally to 50% B until the end of the run. Mass spectrometry was performed on a Thermo Scientific Q Exactive Plus mass spectrometer. After a precursor scan from 300 to 2000 m/z at 70,000 resolution, the top 12 most intense multiply charged precursors were selected for higher energy collisional dissociation (HCD) at a resolution of 35,000. Precursor isolation width was set to 1.7 m/z and the maximum MS2 injection time was 100 msecs for an automatic gain control of 5e4. Dynamic exclusion was set to 20 s and only charge states two to six were selected for MS2. Half of each fraction was injected for each data acquisition run.
Data Processing
Data were searched all together with Spectrum Mill (Agilent) using the Uniprot Mouse database (17 Oct. 2014, 41,309 entries), containing common laboratory contaminants. A fixed modification of carbamidomethylation of cysteine and variable modifications of N-terminal protein acetylation, oxidation of methionine, and TMT-10plex labels were searched. The enzyme specificity was set to trypsin and a maximum of three missed cleavages was used for searching. The maximum precursor-ion charge state was set to six. The MS1 and MS2 mass tolerance were set to 20 ppm. All TMT reporter ions were ratioed to the common reference channel of 131. Peptide and protein FDRs were calculated to be less than 1% using a reverse, decoy database.
Proteins were only reported if they were identified with at least two distinct peptides and a Spectrum Mill score protein level score ≥ 20. In all supplemental tables, the identifier column includes the Uniprot accession number_Gene symbol_Protein name_#unique peptides. Protein inference was performed in two different ways. For reporting number of proteins detected, protein subgroups were expanded if a subgroup specific peptide was identified. This reflects the number of various proteoforms that can be created from a single gene. For downstream analyses protein subgroups were collapsed to the proteoform with the most or best evidence so that all proteins were only represented by their gene name a single time. Peptides common between subgroups were used for quantitation.
TMT10 reporter ion intensities in each MS/MS spectrum were corrected for isotopic impurities by the Spectrum Mill protein/peptide summary module using the afRICA correction method which implements determinant calculations according to Cramer's Rule (50) and general correction factors obtained from the reagent manufacturer's certificate of analysis (https://www.thermofisher.com/order/catalogue/product/90406)
Fractional intensities (frINT) were calculated as the sum of precursor ion chromatographic peak areas in MS1 spectra for all PSMs contributing to the protein. A similar approach has been previously reported (36). frINT for each protein was calculated by splitting the combined precursor ion abundance in proportion to its individual normalized protein-level reporter ion ratio. For example, the amount of “fractional intensity” of protein A from one TMT channel (e.g. 126) can be calculated as the summed MS1 intensity for all peptides from protein A multiplied by the fraction that the 126 channel contributed to the summed MS1 intensity. Formally, frINT of 126 is written as:
where frINTprotA is the fractional intensity of protein A, IprotA_MS1, is the summed MS1 intensity of all peptides attributed to protein A, i126 is the intensity of the 126 reporter ion for all protein A peptides, and Σiall is the summed intensity of all reporter ions used in the experiment.
The peak area for the extracted ion chromatogram (XIC) of each precursor ion subjected to MS/MS was calculated automatically by the Spectrum Mill software in the intervening high-resolution MS1 scans of the LC-MS/MS runs using narrow windows around each individual member of the isotope cluster. Peak widths in both the time and m/z domains were dynamically determined based on MS scan resolution, precursor charge and m/z, subject to quality metrics on the relative distribution of the peaks in the isotope cluster versus theoretical. Although the determined protein abundances are generally reliable, several experimental factors contribute to variability in the determined abundance for a protein. These factors include incomplete digestion of the protein; widely varying response of individual peptides because of inherent variability in ionization efficiency as well as interference/suppression by other components eluting at the same time as the peptide of interest, and sampling of the chromatographic peak between MS/MS scans. The number of observable tryptic peptides/protein can be used to correct summed peptide abundances for protein length. The frINT values used for all analyses were calculated by dividing each protein's frINT value by the median histone intensity (HIST2H2BB, H2AFX, HIST1H4A, HIST1H3A, and HIST1H1C) for that sample/TMT channel, the sum of the TIC for all fractions for a 10-plex, as well as the number of observable tryptic peptides for a given protein. This was then multiplied by 1e6 to give ppm Histone. These values provide rough estimates for abundance differences between proteins. The ratiometric data should be used for comparing a protein's abundance level across cell types.
To generate samples for testing frINT, we cultured E14 mouse embryonic stem cells in serum containing medium. For differentiation, LIF was removed for 48 h, followed by 500 μm retinoic acid (Sigma) for 48 h more. Cells were lysed and digested as described above. After desalting, the sample was split and 5 μg was desalted for LFQ analysis, or 5 μg was on-column TMT labeled, mixed and analyzed as described above. Roughly 1 μg was analyzed for both sample formats. Label free data was acquired as described above with the exception that the MS2 resolution was set to 17,500 and the normalized collision energy was 27. Data were searched on MaxQuant version 1.3.0.5. against the Uniprot Mouse Canonical database, 2014Apr03 (43,427 entries) with a precursor and product ion tolerance of 20 ppm. Trypsin/P was set for digestion conditions allowing up to two missed cleavages. Variable peptide modifications allowed for Met oxidation and protein N-term acetylation, Cys carbamidomethylation was considered a constant modification. Match between runs within a 2 min RT window was enabled and both the peptide and protein FDR was set to < 0.01.
Data Analysis
Data Imputation
After replicate recall correlation analysis, log2 FC values were normalized by two component Gaussian mixture model-based normalization (32)). Missing values between 10-plex cassettes were imputed if the protein was detected in at least half of all samples using k-nearest neighbor (k-NN) imputation (51). Imputed data sets were only used for the moderated F-test and marker selection analysis (described below). This is noteworthy as exclusively expressed proteins may only be detected in a single cell type and would have been ignored for downstream analysis.
Top 20 Differentially Expressed Genes from Microarray
The top 20 differentially expressed genes were calculated by taking the 20 largest fold change values from one cell type compared with all the others (supplemental Table S2). For protein fold change mapping, duplicate gene names and genes not detected in this study were removed.
Differential Abundance Analysis
The normalized and imputed data set was subjected to a moderated F-test (Smyth, 2004), followed by Benjamini-Hochberg Procedure correcting for multiple hypothesis testing. We drew an arbitrary cutoff at adj. P val < 0.01. The heatmap shows the log2 fold change to the common reference and was clustered using one minus Pearson correlation metric (https://software.broadinstitute.org/morpheus/).
Hierarchical Clustering and Marker Selection
Hierarchical clustering and marker selection analysis was performed in Morpheus (https://software.broadinstitute.org/morpheus/). Clustering was performed on rows and/or columns indicated by the dendrogram for individual analyses, using one minus Pearson correlation as the metric. For Marker Selection analysis, the signal to noise metric using a one cell type versus all approach was performed. The number of permutations was set to 10,000 and only proteins with an FDR of less than 0.15 were kept, except for mast cells (MC), where there FDR cutoff to be included in the heatmap was 0.05 for size considerations.
Principal Component Analysis
Principal component analysis (PCA) was performed on an arbitrary cutoff of the 666 proteins (top 13%) with the highest F-statistics (highest FC, high precision). 15% is the cutoff typically used in Immgen related studies (10–13, 33, 44). To analyze gene sets contributing to the resultant projections, the loadings from PC1 and PC2 were analyzed using Gene Set Enrichment Analysis (GSEA v2.2.3, broadinstitute.org/gsea) with the c2.cp.v5.2.symbols.gmt[Curated] gene matrix. Enriched gene sets (adj. P val < 0.05) are displayed with their respective normalized enrichment score (NES).
Analysis of RNA/Protein Relationships
5125 gene products (collapsed to gene name) were identified in both the transcriptomics and proteomics and were used for all subsequent RNA/protein analyses. To calculate the cell type specific RNA/protein correlations, we performed Spearman's rank correlation on the ppm histone and microarray values (12, 33).
For analysis of co-regulation, the Euclidean distance between all pairwise mRNA or protein (log2 FC values) measurements was calculated and hierarchically clustered. To visualize the discrepancies seen between mRNA co-regulation at the protein level, the protein ρ's were mapped in order of the mRNA clustering.
Expression ranks were calculated by ranking expression level of each gene product for each cell type using either the microarray data or the frINT values. The most abundant gene product was given a value of 1, the lowest, 5,125. Delta-rank scores were calculated by subtracting the protein rank from the RNA rank. Thus, large negative values suggest relatively high protein levels and low RNA levels. Large positive numbers, high RNA, low protein. We calculated the delta-rank score for every gene product for every cell.
RESULTS
Development of Low Input Proteomic Sample Preparation Methods
To adapt proteomics sample preparation protocols for low cell numbers directly from flow cytometry we focused on four main steps: efficient capturing of FAC-sorted cells, minimizing sample handling, improving sample multiplexing, and optimizing fractionation for low amounts of TMT-labeled peptides (< 10 μg) (Fig. 1). We concentrated on primary immune cells from mice as they are well-characterized at the transcriptome and single protein level with antibodies but are poorly studied at the proteome level because of their relatively poor availability from a small animal.
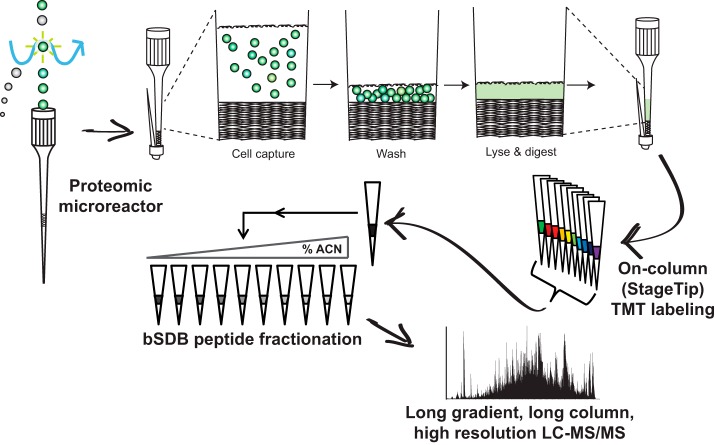
Fig. 1.
Low input proteomics workflow. Cells are FAC-sorted into collection microreactors, where they can be easily washed and stored until lysis. Lysis, reduction, alkylation, and digestion all happen in the microreactor. Digestions are then loaded onto a C18 Stagetip for desalting and on-column TMT labeling. After labeling, samples are mixed and subjected to small-scale sulfonated polystyrenedivinylbenzene at pH 10 (bSDB) fractionation. The resultant fractions are then analyzed by LC-MS/MS using 50 cm columns and long on-line gradients into a high mass accuracy/resolution mass spectrometer.
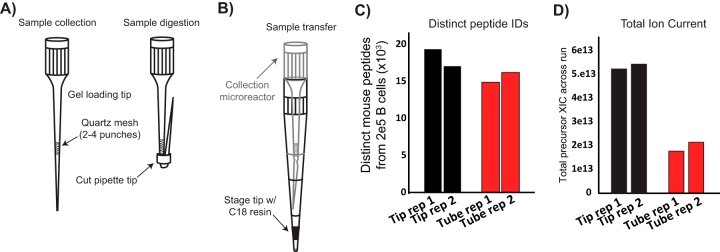
Because FAC-sorting and washing cells in microfuge tubes can lead to significant cell loss (24), we developed a simple-to-make collection microreactor that enables efficient capture and washing of FAC-sorted cells, as well as lysis and digestion of low cell numbers (Fig. 2A). We fabricated collection microreactors for facile collection and washing of FAC-sorted cells. After the final column-format washing of cells, i.e. not in batch format, cells are lysed, and proteins digested. The digest can then be directly transferred to a C18 Stage tip for sample desalting (25) (Fig. 2B). To compare the collection microreactors to standard centrifuge tubes we sorted 2e5 primary B cells directly into tubes or tips and compared the number peptides identified with either method. Collection microreactor tips outperformed tubes in distinct peptides identified in single shot, label free LC-MS/MS mode, as well as having greater total ion current (TIC), indicating greater overall peptide intensity (Fig. 2C–2D). Given the improvements in peptide IDs, together with the ease of washing and avoiding disruption of a non-visible cell pellet, we incorporated the collection microreactors into the low input proteomics workflow.
Fig. 2.
Collection microreactors A, Collection microreactor design and usage. B, Transfer of digest to Stage tip is accomplished by inserting the microreactor into a pre-equilibrated Stage tip. C, Comparison of number of mouse peptides identified using either standard microfuge tubes or the collection microreactors from 2e5 B cells. Two collection replicates are shown. D, Total ion current (TIC) is a proxy for the amount of material loaded on-column, from either standard microfuge tubes or the collection microreactors from 2e5 B cells.
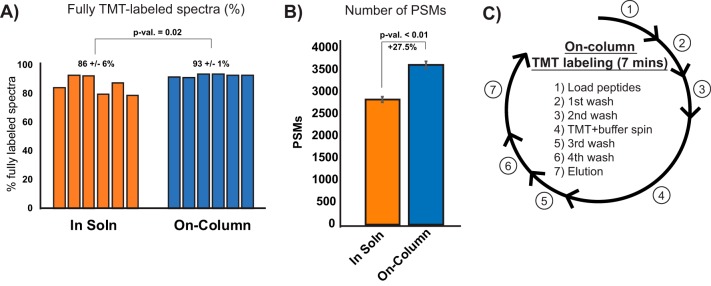
Isobaric labeling of peptides enables multiplexed peptide/protein identification and MS2 level quantitation for up to eleven samples using tandem mass tags (TMT) (26, 27). Multiplexing low input samples provides a boost in MS1 signal intensity by combining signal from multiple experiments, improving overall sensitivity of data acquisition (28). In-solution TMT labeling requires an additional desalting step post-quenching, which often results in sample loss. To circumvent these sample handling steps we tested whether we could directly couple desalting and TMT labeling of small peptide amounts when adsorbed to the C18 resin (23, 29, 30). Two micrograms of whole cell lysate digest was labeled either in-solution in a microfuge tube or pre-adsorbed to C18 resin. On-column TMT labeling yielded a higher percentage of fully-labeled peptides (all possible primary amines coupled to TMT) and was more reproducible than the standard protocol (Fig. 3A). The on-column labeling method also yielded a 27% increase in peptide spectral matches (PSMs) compared with in-solution labeling (Fig. 3B). Finally, on-column TMT labeling avoided an extra desalting step and reduced the entire labeling protocol from about three hours to less than 10 mins (Fig. 3C). These results demonstrate that the on-column TMT labeling strategy is faster and more efficient for labeling low microgram levels of peptides.
Fig. 3.
On-column TMT labeling A, Labeling efficiency of on-column TMT labeling compared with in-solution labeling for low peptide amounts. Mean and standard deviation is shown, Student's t test p value. B, Mean and standard deviation of number of PSMs for in-solution or on-column TMT labeling, n = 6 Student's t test p value. C, Steps and timing for desalting coupled with on-column TMT labeling.
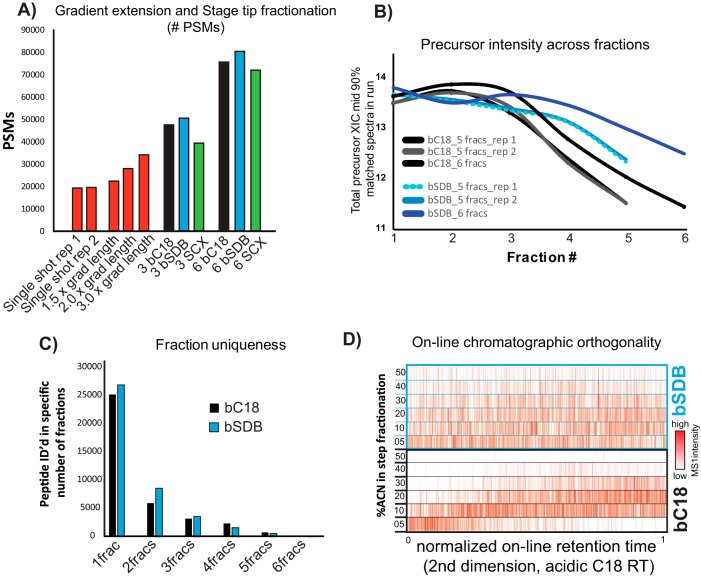
To reduce sample complexity of the full proteome digest for deep proteome profiling, we explored Stage tip fractionation strategies for TMT-labeled peptide mixtures (25, 31). Comparing strong cation exchange (SCX), C18 at pH 10 (bC18), or sulfonated polystyrenedivinylbenzene at pH 10 (bSDB) we found that fractionation improved the number of identified TMT-labeled peptides compared with gradient extension alone (Fig. 4A). Both basic reversed phase formats consistently identified more peptides than SCX fractionation (Fig. 4A). bSDB fractionation outperformed bC18 fractionation in number of PSMs, had higher precursor intensities for analytes identified in later fractions, and greater fraction uniqueness (Fig. 4A–4C). Comparison of retention times (RT) of peptides identified in their respective bC18 or bSDB step fractions compared with their on-line, acidic RT in LC-MS/MS of an unfractionated sample showed a more even distribution of peptides across fractions with bSDB (Fig. 4D). These data show bSDB is an orthogonal, well-suited method for fractionating of small amounts of TMT-labeled peptides.
Fig. 4.
Characterization of small-scale peptide fractionation. A, Comparison of the number of PSMs from extended on-line LC-MS/MS gradients, and different small-scale fractionation modalities to a single shot analysis. Assuming equal mass distribution, ∼1 μg was loaded on-column. B, Total ion current (TIC) for identified peptides across fractions for two different hydrophobicity-based fractionation modalities. C, Fraction uniqueness between basic reversed phase on C18 packing material (bC18) and sulfonated polystyrenedivinylbenzene at pH 10 (bSDB) fractionation. Bar graph shows the number of peptides identified in a single fraction (1frac), in two fractions (2frac) and so on. D, Orthogonality between fractionation modalities to online acidic C18 chromatography. For the six fractions generated and analyzed via LC-MS/MS, the intensity of each peptide (red color scale) is plotted on the x axis according to their on-line, normalized RT (nRT) and are compared with the individual fractions in increasing acetonitrile (ACN) percentage along the y axis.
Deep, Quantitative Proteomic Profiling of 3e5 FAC-sorted Immune Cells
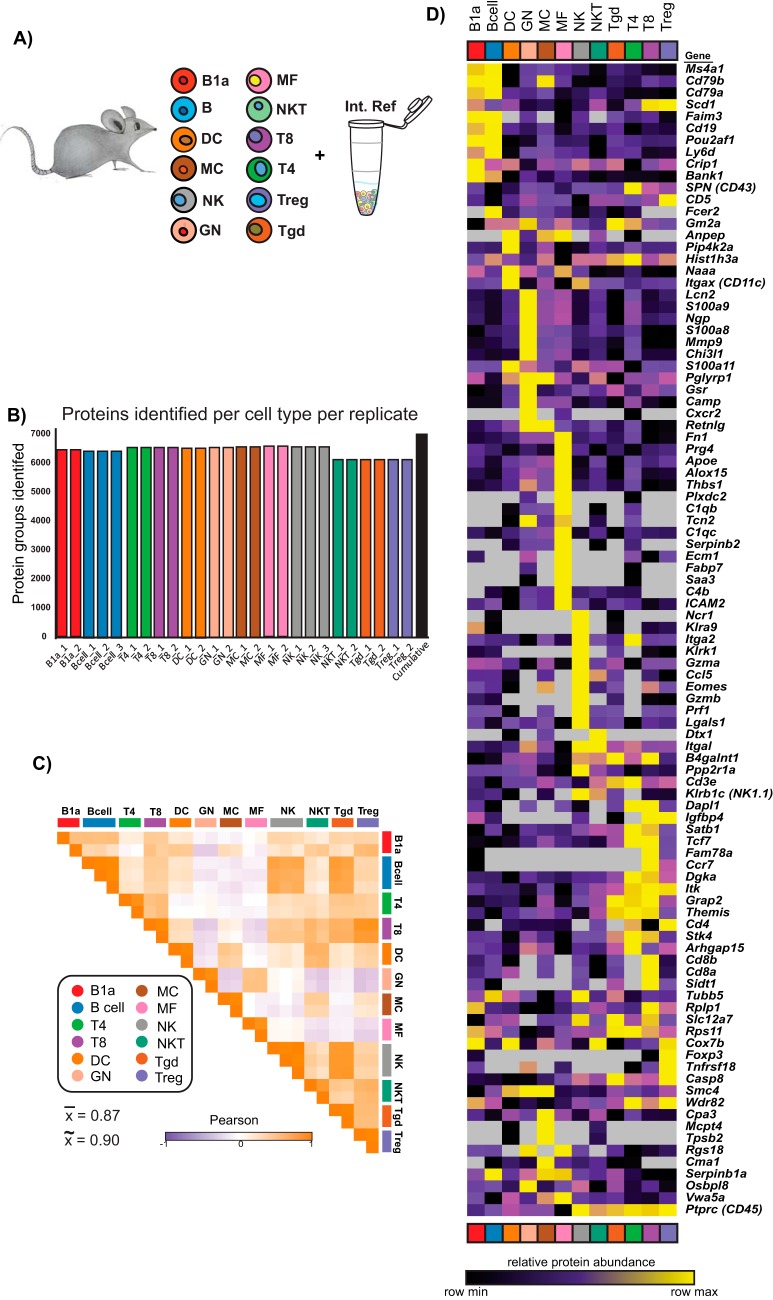
Having developed a streamlined sample preparation protocol, we performed quantitative proteome profiling on FAC-sorted primary murine immune cells. 12 cell types were chosen spanning a range of abundance levels in the mouse (Fig. 5A). To mitigate large numbers of mice and long FACS times, we chose to collect 300,000 cells from each cell type in duplicate or triplicate. 3e5 cells yielded a range of protein amounts after urea lysis and digestion from about 4 μg for neutrophils (GN) and B cells to approximately 2 μg for T cell subtypes. To have a common reference connecting the separate TMT 10-plexes to enable cross-plex analyses, we created a sample comprised of a mixture of immune cell types (supplemental Table S1) prepared at the beginning of the study (32). This common reference sample was labeled with the TMT-131 reagent and was included in all TMT 10-plex experiments (Fig. 5A).
Fig. 5.
Low input proteomics of 12 primary murine immune cell types. A, Twelve immune cell types were doubly FAC-sorted according to Immgen protocols. B1a, a.k.a. B-1; B, a.k.a. B-2; DC, dendritic cell; MC, mast cell; NK, natural killer cell; GN, neutrophils; MF, macrophage; NKT, natural killer cell; T8, CD8+ T cells; T4, CD4+ T cells; Treg, regulatory T cells; γδT, gamma delta T cells. The common reference was created as a mixture of immune cell types listed in supplemental Table S1. All replicates are shown in supplemental Fig. S1. B, Number of protein groups identified for each cell type used in the final analyses. The cumulative number of protein groups identified across all TMT 10-plexes is shown in black. C, Pairwise Pearson correlation analysis of all samples included in downstream analyses in log2 protein FC space. The mean and median Pearson r for biological replicates, plotted along the diagonal, are 0.87 and 0.90, respectively. All replicates are shown in supplemental Fig. S1. D, Heatmap of global log2 protein FC values for the 20 most differentially expressed mRNAs (see methods) for the cell types profiled in this study. Duplicate genes were collapsed and gene products not detected at the protein level were excluded. Gene names are displayed on the right for reference.
We performed the low input proteomics protocol described above on four TMT 10-plex experiments of ∼2 μg peptides per sample. On average 6427 protein groups (including all proteoforms) were identified in each 10-plex by two or more peptides per protein (peptide and protein FDR < 1%); 7023 protein groups were identified across all samples (Fig. 5B and supplemental Fig. S1A). After performing correlation analysis across biological replicates (defined here as starting from different mice) of the log2 transformed protein ratios of the individual samples to the common reference, we excluded any cell type replicates with a Pearson r < 0.5, except for B1a cells as only duplicates were provided (supplemental Fig. S1B). The median and mean Pearson r across the included biological replicates were 0.90 and 0.87, respectively (Fig. 5C). A recent study from our laboratory showed the median and mean Pearson's r between preparation replicates for high inputs were both 0.9, (32) indicating our low input data is of high quality, comparable to standard sample preparation methods.
To test if protein measurements recapitulated known expression patterns of immune cell types, we mapped the protein fold change values for the top 20 differentially expressed mRNAs for each cell type ((33) and Immgen.org) (supplemental Table S2). We found that most canonical markers of specific cell types had expected and exclusive abundances (e.g. CD8a/b in CD8+ T cells) or near exclusive abundances (e.g. CD19 in B and B1a cells, and CD4 in CD4+ T cells and Treg cells) (Fig. 5D).
Identification of Differentially Abundant and Immune Cell Type-Specific Proteins
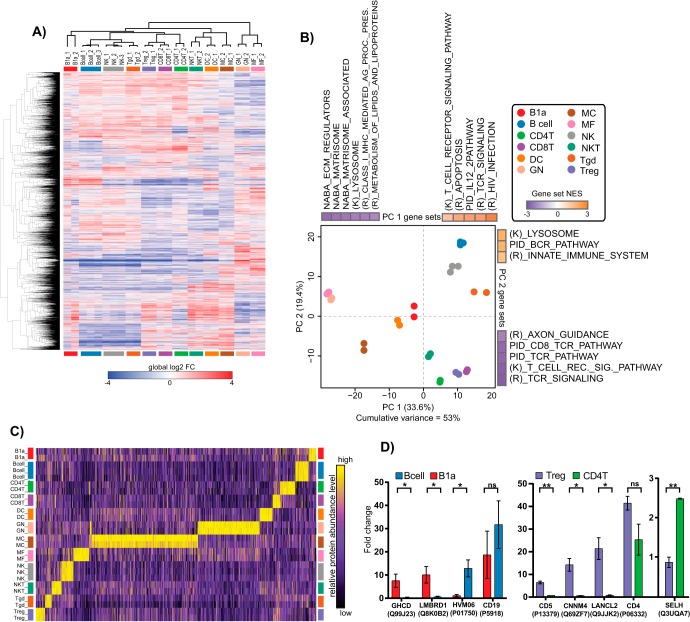
To identify differentially abundant (DA) proteins between different cell types, we performed a moderated F-test across all biological replicates. 4241 protein groups were found to be DA in at least one cell type with a false discovery rate (FDR) < 0.01 (Fig. 6A and supplemental Table S3). Principal component analysis (PCA) followed by gene set enrichment analysis (GSEA) was performed to visualize how the cell types clustered and which pathways drove their separation (Fig. 6B). When the top 13% (666 proteins, arbitrary cutoff) DA proteins were projected into PCA space there was high overlap between biological replicates, and cell types known to be similar (e.g. lymphocytes versus myeloid, T cells versus B cells, etc.) clustered together. GSEA of the loadings for PC1 and PC2 revealed eleven and eight pathways, respectively (adjusted p value <0.05), driving the separation. The gene sets separating the cell types along the PC1 axis showed enrichment for T cell receptor (TCR) and B cell-related pathways in the positive direction, whereas pathways associated with extracellular matrix remodeling were enriched in the negative direction. For PC2, gene sets involved in B cell receptor (BCR) signaling and the innate immune system were enriched in the positive direction, whereas TCR signaling was enriched in the negative direction. These data show that immunologically relevant pathways drive the differences between cell types at the protein level, demonstrating the fidelity of low input proteomics.
Fig. 6.
Differential protein abundances and pathway analysis of murine immune cell types. A, Heatmap of log2 FC of differentially abundant proteins as determined by a moderated F-test, FDR < 0.01. Row and columns were clustered with the one minus Pearson metric. B, Principle component analysis (PCA) of the top 13% (666 proteins, arbitrary cut-off) most differentially abundant proteins determined by the moderated F-test across cell type replicates. Enriched gene sets of PC1 and PC2 loadings (P val < 0.05) are displayed according to the directionality of the replicate measurement clusters. Normalized enrichment score (NES) is displayed for each enriched gene set. (K), Kegg; (R), Reactome. C, Column-relative heatmap for cell type specific proteins determined by Marker Selection Analysis and their expression levels across all immune cell types profiled. D, Select examples of plasma membrane annotated DA proteins between molecularly similar cell types. Mean and standard deviation is shown. p values calculated by the Student's t test are denoted as * < 0.05, ** < 0.01, and “ns” for not significant.
To identify proteins that may better distinguish closely related cell types, we employed data imputation (supplemental Table S4) followed by marker selection analysis, comparing one cell type to all others in an iterative fashion (34). Marker selection analysis displayed high cell-type specificity for DA proteins, with γδT cells showing the fewest markers, MCs the highest (Fig. 6C and supplemental Table S5). This analysis identified distinct plasma membrane-annotated proteins (Uniprot.org) between molecularly similar cell types that could provide alternative or additional markers (supplemental Fig. S2 and supplemental Table S6). For example, GHDC, LMBRD1, and region 102 of Ig heavy chain V distinguish B cells from B1a cells (both CD19+); or CD5, CNNM4, LANCL2, and SELH for CD4+ T cells from Tregs (both CD4+) (Fig. 6D). These data demonstrate that low input proteomics recapitulates many known aspects of immunology and may provide valuable new insights for future immunological studies.
Protein/RNA Relationships Reveal Evidence for Post-transcriptional Regulation Across the Immune System
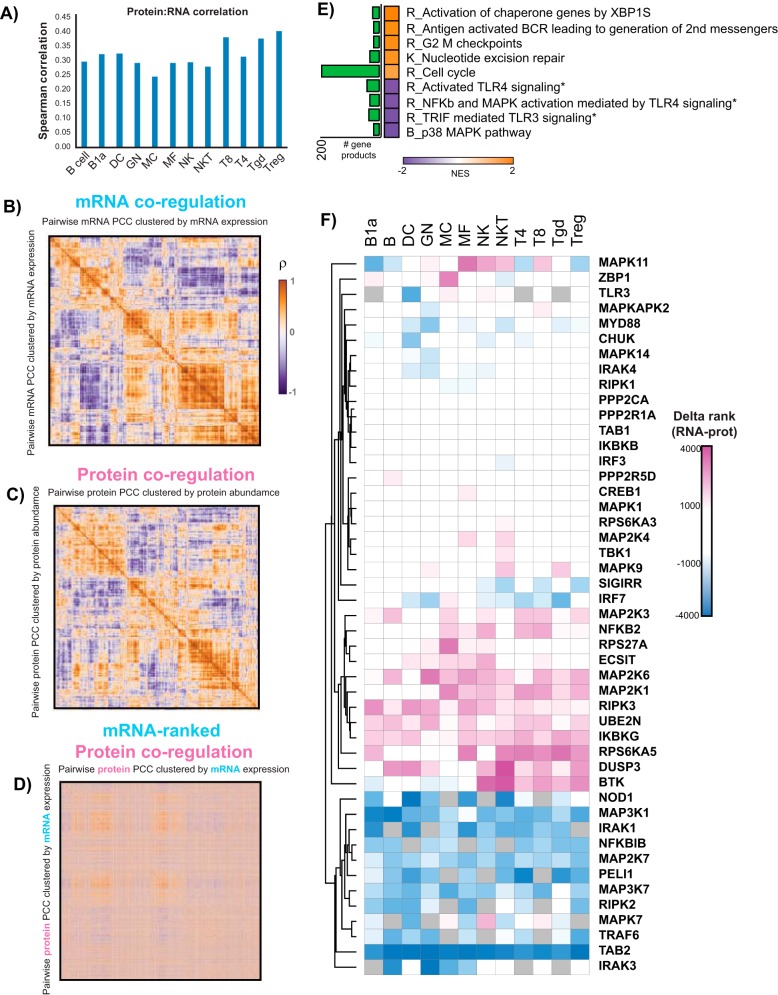
We performed FACS for this study based on Immgen's protocols and gating strategies (33) (www.immgen.org) to allow comparison of our proteomic data to previously acquired transcriptomic data (10–13, 33). Using collapsed protein groups (proteoforms from genes with the strongest evidence at the peptide level), we had overlapping measurements for 5,125 gene products. To test the appropriateness of investigating protein/RNA relationships, we calculated the Spearman's rank correlation coefficient (ρ) for all gene products for each individual cell type. We found all cell types had a correlation coefficient in agreement with previous reports for unperturbed cells, 0.25 < ρ < 0.50 (Fig. 7A) (1, 3–5).
Fig. 7.
Protein/RNA relationships in primary murine immune cells. A, Spearman's rank correlation coefficient (ρ, y axis) for protein/RNA levels using 5,125 overlapping protein/RNA measurements for each immune cell type (x axis). B, Heatmap of ρ for each pairwise mRNA comparison across all immune cell types profiled in this study. C, Heatmap of ρ for each pairwise protein log2 FC comparison across all immune cell types profiled in this study. D, Heatmap of ρ calculated from protein fold change values orders according to mRNA ρ clustered order. E, GSEA of gene product correlation coefficients across immune cells for all overlapping protein/RNA measurements (p value < 0.01). Normalized enrichment scores (NES) are plotted, along with the number of gene products identified in each gene set (green bar graph). * denotes that the gene products from these gene sets were combined and used for analysis in Fig. 7F. F, Clustering analysis of delta-rank gene product levels in the TLR gene sets from Fig. 7E for individual cell types. Gene products within a cell type with relatively low RNA levels and high protein levels will have a large, positive number (pink). Blue suggests relatively low protein levels and high RNA levels. Clustering was performed using the Euclidian distance metric.
To find evidence of post-transcriptional regulation across the immune system, we asked whether we could see global trends in altered protein/RNA levels by looking for co-regulation changes between mRNA and protein (35). We calculated ρ for each pairwise mRNA measurement across all the cell types profiled (13). Clustering of the ρ values for RNA expression showed blocks of positive correlations, strongly suggesting these transcripts are co-expressed across immune cell types (Fig. 7B). Performing the same analysis in log2 FC protein space, we again could see blocks of positive correlations, though the overall data structure between the RNA and protein was markedly different (Fig. 7C). To visualize and better contrast the differences between mRNA and protein co-regulation, we reorganized the matrix of protein ρ's using the ordering derived from hierarchical clustering of the mRNA ρ matrix. This mRNA-ordered, protein co-regulation matrix showed a gross loss of off-diagonal blocks of positive correlations (Fig. 7D), implying protein co-expression is governed by distinct processes from those underlying mRNA co-expression in primary immune cells.
To further investigate protein/RNA relationships, we next asked which pathways show evidence for post-transcriptional regulation across the immune cells analyzed. We calculated the Pearson correlation coefficient (r) for each gene product individually across all cell types using mean mRNA levels and mean relative protein abundances. GSEA of the genes' r values identified nine gene sets with a p value of < 0.01 (Fig. 7E). Five of the gene sets were enriched for positive protein/RNA correlation, whereas four were enriched for negative protein/RNA correlation. Among the positively-correlated gene sets, we found gene products within the cell cycle gene set among the positively correlated protein/RNA levels, consistent with previous studies (3, 32). We also found a BCR signaling pathway gene set to have positively correlated protein/RNA patterns across the 12 immune cell subtypes (Fig. 7E). On the other hand, the negatively-correlated gene sets included Toll-like receptor (TLR) and MAPK signaling pathways, suggesting that post transcriptional regulation may play a role in the innate immunity response.
To gain insight into the directionality of the negatively correlated TLR pathways, i.e. high protein levels/low RNA levels or vice versa, we rank-ordered the abundance level of each gene product within each cell type and calculated the difference in rank (delta-rank) between RNA and protein. First, to estimate relative inter-protein levels we took the summed precursor intensities for peptides belonging to each individual protein and scaled the individual TMT channel contributions to find how each sample contributed to the overall protein level (36) (supplemental Fig. S3A). These “fractional intensity” (frINT) measurements showed high agreement with MaxQuant's MaxLFQ, a widely-used label free MS1-based quantitation software package for proteomic data analysis (37) (supplemental Fig. S3B). After applying frINT to our immune cell dataset we rank ordered the protein and RNA levels for each individual cell type, where 1 was the most abundant gene product, 5125 the least abundant, for the delta-rank analysis. Clustering analysis of the delta-rank values for all gene products detected in the three TLR-related gene sets with negative RNA:protein correlations about one-third showed little to no change in the delta-rank changes (delta > 2000, e.g. the change in rank was not at least 2000 in either direction) (Fig. 7F). We were also able to identify subclusters of gene products that suggest low RNA/high protein levels (large, positive values, pink) or low protein/high RNA levels (large, negative values, blue). Some gene products also showed cell type specific delta-ranks, suggesting different immune cell types regulate specific gene products to establish their respective proteomes.
DISCUSSION
Proteome profiling offers new levels of information about cellular identity and regulation that can be missed with genomics or transcriptomics alone. The generation of genome-wide protein measurements, however, is often limited by the amount of input needed for deep and high-quality proteomic profiling. Here, we show improved sample preparation strategies for MS-based proteomic analysis of low cellular inputs from FAC-sorted cells. Using these low input proteomics methods, we were able to quantify over 7000 protein groups analyzing half of the 300,000 freshly isolated mouse immune cell-type collected directly from FACS.
The sensitivity of our low input methodology derives from a combination of three main improvements: fabrication of collection microreactors for easy cell collection and digestion; on-column TMT labeling coupled with desalting for faster and more efficient peptide labeling; and optimized small-scale fractionation of TMT-labeled peptides. The microreactors provided a versatile cell capture system with several benefits over microfuge tube or in-StageTip (25, 31) collection including the ability to wash cells in a column-like format without the need to see a hardly visible pellet, smaller lysis and digestion volumes, and a filter which can prevent StageTip clogging. On-column labeling of peptides with TMT reduces the time and number of sample handling steps needed for conventional in-solution labeling because the desalting and labeling occur on the same resin. On-column labeling also was more efficient specifically for fully TMT-labeled spectra for low microgram inputs compared with in solution, even without a desalting step prior to on-column labeling (23). Although solid-phase peptide labeling has been previously reported in acidic conditions (30), the on-column protocol reported here takes less time and has been optimized for 1000-fold less material. Finally, optimizing small-scale fractionation for TMT-labeled peptides allowed improved depth of coverage for low input proteomics. Previous work has shown that for medium levels inputs (between 5 and 50 μg peptide per state) of unlabeled peptides, SCX outperformed basic reversed-phase (bRP) chemistries in number of peptide identifications (25, 31). Our work here shows that both bRP modalities tested outperformed SCX, likely because of fact that TMT labeling converts primary amines into amides, dampening the positively charged character of tryptic peptides. Together, the reduced sample handling steps, combined with efficient TMT labeling and off-line, tip-based fractionation, enabled the deep, quantitative proteomic profiling of 300,000 murine primary immune cells isolated directly from FAC-sorting.
We applied our low input proteomic-sample preparation to 12 freshly isolated murine immune cell types sorted according to Immgen protocols. To our knowledge, this study provides the first proteome-wide characterization of the major immune cell types from their resident tissues rather than peripheral blood. Over 7000 proteins were quantified with high reproducibility as seen by the high replicate recall between mouse replicates. The median Pearson correlation between mouse replicates acquired using low input proteomics protocols matched that of the high input, preparation replicates used routinely by our laboratory for longitudinal quality assessments (Fig. 5C) (32).
Differential analysis of protein abundances found nearly two-thirds of proteins were DA in at least one cell type. This high proportion was expected as even pairwise comparisons of vastly different cell types at the RNA level reveal large differences genome-wide (13). When projected into PCA space, we found that the relevant cell types clustered together, and that the clustering was driven by expected pathways, such as BCR and IL12 pathways for B cells, and TCR signaling for T cells (Fig. 6B). This analysis also found that MF, GN, and to a lesser extent MCs, had low relatedness through PC1 to the other cell types, and that proteins associated with extracellular matrix remodeling were the main drivers of this separation (38, 39). As MF, GN, and MC are often associated with solid tissues, we hypothesize these cell types have mechanisms to move through the extracellular matrix.
Using marker selection analysis, we found that MCs were the most distinct cell type proteome-wise from the other immune cell types profiled, a finding consistent with mRNA expression profiling (11). Differentiating cell types based on DA proteins showed better discrimination between closely related cell types than protein levels chosen by their differential mRNA expression. For example, most protein products determined by their differential mRNA levels in B and B1a cells showed little difference between the two cell types (Fig. 5D). A similar situation was seen between T4 and T8 cells, and especially for PTPRC (CD45), which was present in high amounts across NK cells and all T cell subtypes. From the marker selection analysis, we were able to nominate several plasma membrane annotated proteins that could provide additional or alternative markers to distinguish B cells from B1a cells, or T4 from Tregs (Fig. 6D). Consistent with these results, CD5 and LANCL2 have established roles in Treg activity (40–43).
We adhered strictly to Immgen sorting protocols to enable the comparison of transcriptomic data to our proteomic data to gain insight into potential regulatory points of gene product expression (10–13, 44). Cell type specific protein/RNA correlations showed a relationship expected for steady state cells (1, 3–5). To establish evidence for post-transcriptional regulation genome-wide, we looked for changes in co-regulation at the protein or RNA levels. Recent work by Kustatscher and colleagues has shown that transcriptional co-regulation is largely driven by proximity to other actively transcribed genes on their respective chromosome (35). The phenomenon of proximal genes being co-regulated is commonly observed in topologically associated domains and gene expression neighborhoods molecularly characterized by 3D-interaction mapping of genomic DNA (45, 46). This co-regulation at the transcription level, however, was dampened at the proteome level (35), likely because of translational control, and to a lesser extent protein stability, to maintain levels of functionally related or physically interacting proteins (4, 47, 48). We looked for mRNA and protein co-regulation by calculating and clustering the p's for the 5125 gene products with overlapping measurements. Both mRNA and protein data showed respective and distinct co-clustering blocks, suggestive of co-regulation across the immune cell types profiled in this study (Fig. 7B–7C). Rearranging the protein p's in the order determined by hierarchical clustering of the mRNA data revealed a distinct loss in data structure and off-diagonal correlations (Fig. 7D) suggesting primary mouse immune cells show a degree of buffering transcriptional co-regulation to the desired protein landscape.
The low input proteomics described here extend the applicability of previous methodologies to FAC-sorted immune cells and highlight the value of reduced-handling sample preparation, especially those requiring analysis of limiting amounts of sample (22, 23, 36). Other applications of the methods we describe include profiling of other cell types, co-immunoprecipitation studies, and post-enrichment labeling. We envision these methods will enable researchers to perform proteomic experiments of scarce samples previously out of reach.
DATA AVAILABILITY
The original mass spectra may be downloaded from MassIVE (http://massive.ucsd.edu) under the identifier: MassIVE MSV000083180. The data are directly accessible via ftp://MSV000083180@massive.ucsd.edu.
Supplementary Material
Acknowledgments
We thank Christophe Beniost (Harvard Medical School) and the Immgen Consortium for providing the samples and experimental input. We also thank Shankha Satpathy, Inbal Benhar, Bea Hamilton, Filip Mundt-Suger, Namrata Udeshi, Philipp Mertins, and Egle Kvedaraite for useful discussions; and to Susan Klaeger, John Ray, and Jacob Jaffe for critical assessment of the manuscript.
Footnotes
* This work was supported by grants 5U24CA210986 and U01CA214125 (S.A.C.) and 5U24CA210979 (D.R.M.) from the US National Institutes of Health as part of the National Cancer Institute Clinical Proteomics Tumor Analysis Consortium Initiative.
 This article contains supplemental Figures and Tables.
This article contains supplemental Figures and Tables.
1 The abbreviations used are:
- FACS
- fluorescence-activated cell sorting
- LCM
- laser capture micro-dissected
- frINT
- fractional intensity
- XIC
- extracted ion chromatogram
- TIC
- total ion current.
REFERENCES
- 1. de Sousa Abreu R., Penalva L. O., Marcotte E. M., and Vogel C. (2009) Global signatures of protein and mRNA expression levels. Mol. Biosyst. 5, 1512–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rabani M., Levin J. Z., Fan L., Adiconis X., Raychowdhury R., Garber M., Gnirke A., Nusbaum C., Hacohen N., Friedman N., Amit I., and Regev A. (2011) Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat. Biotechnol. 29, 436–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jovanovic M., Rooney M. S., Mertins P., Przybylski D., Chevrier N., Satija R., Rodriguez E. H., Fields A. P., Schwartz S., Raychowdhury R., Mumbach M. R., Eisenhaure T., Rabani M., Gennert D., Lu D., Delorey T., Weissman J. S., Carr S. A., Hacohen N., and Regev A. (2015) Dynamic profiling of the protein life cycle in response to pathogens. Science 347, 1259038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schwanhausser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., and Selbach M. (2011) Global quantification of mammalian gene expression control. Nature 473, 337–342 [DOI] [PubMed] [Google Scholar]
- 5. Vogel C., Abreu Rde S., Ko D., Le S. Y., Shapiro B. A., Burns S. C., Sandhu D., Boutz D. R., Marcotte E. M., and Penalva L. O. (2010) Sequence signatures and mRNA concentration can explain two-thirds of protein abundance variation in a human cell line. Mol. Syst. Biol. 6, 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kronke J., Udeshi N. D., Narla A., Grauman P., Hurst S. N., McConkey M., Svinkina T., Heckl D., Comer E., Li X., Ciarlo C., Hartman E., Munshi N., Schenone M., Schreiber S. L., Carr S. A., and Ebert B. L. (2014) Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science 343, 301–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mahrus S., Trinidad J. C., Barkan D. T., Sali A., Burlingame A. L., and Wells J. A. (2008) Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell 134, 866–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mancias J. D., Wang X., Gygi S. P., Harper J. W., and Kimmelman A. C. (2014) Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature 509, 105–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Terenzio M., Koley S., Samra N., Rishal I., Zhao Q., Sahoo P. K., Urisman A., Marvaldi L., Oses-Prieto J. A., Forester C., Gomes C., Kalinski A. L., Di Pizio A., Doron-Mandel E., Perry R. B., Koppel I., Twiss J. L., Burlingame A. L., and Fainzilber M. (2018) Locally translated mTOR controls axonal local translation in nerve injury. Science 359, 1416–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bezman N. A., Kim C. C., Sun J. C., Min-Oo G., Hendricks D. W., Kamimura Y., Best J. A., Goldrath A. W., Lanier L. L., and Immunological Genome Project, C. (2012) Molecular definition of the identity and activation of natural killer cells. Nat. Immunol. 13, 1000–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dwyer D. F., Barrett N. A., Austen K. F., and Immunological Genome Project, C. (2016) Expression profiling of constitutive mast cells reveals a unique identity within the immune system. Nat. Immunol. 17, 878–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jojic V., Shay T., Sylvia K., Zuk O., Sun X., Kang J., Regev A., Koller D., Immunological Genome Project, C., Best A. J., Knell J., Goldrath A., Joic V., Koller D., Shay T., Regev A., Cohen N., Brennan P., Brenner M., Kim F., Rao T. N., Wagers A., Heng T., Ericson J., Rothamel K., Ortiz-Lopez A., Mathis D., Benoist C., Bezman N. A., Sun J. C., Min-Oo G., Kim C. C., Lanier L. L., Miller J., Brown B., Merad M., Gautier E. L., Jakubzick C., Randolph G. J., Monach P., Blair D. A., Dustin M. L., Shinton S. A., Hardy R. R., Laidlaw D., Collins J., Gazit R., Rossi D. J., Malhotra N., Sylvia K., Kang J., Kreslavsky T., Fletcher A., Elpek K., Bellemarte-Pelletier A., Malhotra D., and Turley S. (2013) Identification of transcriptional regulators in the mouse immune system. Nat. Immunol. 14, 633–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Painter M. W., Davis S., Hardy R. R., Mathis D., Benoist C., and Immunological Genome Project, C. (2011) Transcriptomes of the B and T lineages compared by multiplatform microarray profiling. J. Immunol. 186, 3047–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Villani A. C., Satija R., Reynolds G., Sarkizova S., Shekhar K., Fletcher J., Griesbeck M., Butler A., Zheng S., Lazo S., Jardine L., Dixon D., Stephenson E., Nilsson E., Grundberg I., McDonald D., Filby A., Li W., De Jager P. L., Rozenblatt-Rosen O., Lane A. A., Haniffa M., Regev A., and Hacohen N. (2017) Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hukelmann J. L., Anderson K. E., Sinclair L. V., Grzes K. M., Murillo A. B., Hawkins P. T., Stephens L. R., Lamond A. I., and Cantrell D. A. (2016) The cytotoxic T cell proteome and its shaping by the kinase mTOR. Nat. Immunol. 17, 104–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mertins P., Przybylski D., Yosef N., Qiao J., Clauser K., Raychowdhury R., Eisenhaure T. M., Maritzen T., Haucke V., Satoh T., Akira S., Carr S. A., Regev A., Hacohen N., and Chevrier N. (2017) An integrative framework reveals signaling-to-transcription events in toll-like receptor signaling. Cell Rep. 19, 2853–2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rieckmann J. C., Geiger R., Hornburg D., Wolf T., Kveler K., Jarrossay D., Sallusto F., Shen-Orr S. S., Lanzavecchia A., Mann M., and Meissner F. (2017) Social network architecture of human immune cells unveiled by quantitative proteomics. Nat. Immunol. 18, 583–593 [DOI] [PubMed] [Google Scholar]
- 18. Ron-Harel N., Santos D., Ghergurovich J. M., Sage P. T., Reddy A., Lovitch S. B., Dephoure N., Satterstrom F. K., Sheffer M., Spinelli J. B., Gygi S., Rabinowitz J. D., Sharpe A. H., and Haigis M. C. (2016) Mitochondrial biogenesis and proteome remodeling promote one-carbon metabolism for T cell activation. Cell Metab. 24, 104–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tan H., Yang K., Li Y., Shaw T. I., Wang Y., Blanco D. B., Wang X., Cho J. H., Wang H., Rankin S., Guy C., Peng J., and Chi H. (2017) Integrative proteomics and phosphoproteomics profiling reveals dynamic signaling networks and bioenergetics pathways underlying T cell activation. Immunity 46, 488–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhu Y., Piehowski P. D., Zhao R., Chen J., Shen Y., Moore R. J., Shukla A. K., Petyuk V. A., Campbell-Thompson M., Mathews C. E., Smith R. D., Qian W. J., and Kelly R. T. (2018) Nanodroplet processing platform for deep and quantitative proteome profiling of 10–100 mammalian cells. Nat. Commun. 9, 882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Di Palma S., Stange D., van de Wetering M., Clevers H., Heck A. J., and Mohammed S. (2011) Highly sensitive proteome analysis of FACS-sorted adult colon stem cells. J. Proteome Res. 10, 3814–3819 [DOI] [PubMed] [Google Scholar]
- 22. Hughes C. S., Foehr S., Garfield D. A., Furlong E. E., Steinmetz L. M., and Krijgsveld J. (2014) Ultrasensitive proteome analysis using paramagnetic bead technology. Mol. Syst. Biol. 10, 757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. de Graaf E. L., Pellegrini D., and McDonnell L. A. (2016) Set of novel automated quantitative microproteomics protocols for small sample amounts and its application to kidney tissue substructures. J. Proteome Res. 15, 4722–4730 [DOI] [PubMed] [Google Scholar]
- 24. Li S., Plouffe B. D., Belov A. M., Ray S., Wang X., Murthy S. K., Karger B. L., and Ivanov A. R. (2015) An integrated platform for isolation, processing, and mass spectrometry-based proteomic profiling of rare cells in whole blood. Mol. Cell. Proteomics 14, 1672–1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rappsilber J., Mann M., and Ishihama Y. (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nature Protocols 2, 1896–1906 [DOI] [PubMed] [Google Scholar]
- 26. McAlister G. C., Huttlin E. L., Haas W., Ting L., Jedrychowski M. P., Rogers J. C., Kuhn K., Pike I., Grothe R. A., Blethrow J. D., and Gygi S. P. (2012) Increasing the multiplexing capacity of TMTs using reporter ion isotopologues with isobaric masses. Anal. Chem. 84, 7469–7478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thompson A., Schafer J., Kuhn K., Kienle S., Schwarz J., Schmidt G., Neumann T., and Hamon C. (2003) Tandem mass tags: A novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 75, 1895–1904 [DOI] [PubMed] [Google Scholar]
- 28. Unwin R. D. (2010) Quantification of proteins by iTRAQ. Methods Mol. Biol. 658, 205–215 [DOI] [PubMed] [Google Scholar]
- 29. Boersema P. J., Raijmakers R., Lemeer S., Mohammed S., and Heck A. J. (2009) Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc. 4, 484–494 [DOI] [PubMed] [Google Scholar]
- 30. Bohm G., Prefot P., Jung S., Selzer S., Mitra V., Britton D., Kuhn K., Pike I., and Thompson A. H. (2015) Low-pH solid-phase amino labeling of complex peptide digests with TMTs improves peptide identification rates for multiplexed global phosphopeptide analysis. J. Proteome Res. 14, 2500–2510 [DOI] [PubMed] [Google Scholar]
- 31. Kulak N. A., Pichler G., Paron I., Nagaraj N., and Mann M. (2014) Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods 11, 319–324 [DOI] [PubMed] [Google Scholar]
- 32. Mertins P., Mani D. R., Ruggles K. V., Gillette M. A., Clauser K. R., Wang P., Wang X., Qiao J. W., Cao S., Petralia F., Kawaler E., Mundt F., Krug K., Tu Z., Lei J. T., Gatza M. L., Wilkerson M., Perou C. M., Yellapantula V., Huang K. L., Lin C., McLellan M. D., Yan P., Davies S. R., Townsend R. R., Skates S. J., Wang J., Zhang B., Kinsinger C. R., Mesri M., Rodriguez H., Ding L., Paulovich A. G., Fenyo D., Ellis M. J., Carr S. A., and Nci C. (2016) Proteogenomics connects somatic mutations to signalling in breast cancer. Nature 534, 55–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Heng T. S., Painter M. W., and Immunological Genome Project, C. (2008) The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 9, 1091–1094 [DOI] [PubMed] [Google Scholar]
- 34. Gould J., Getz G., Monti S., Reich M., and Mesirov J. P. (2006) Comparative gene marker selection suite. Bioinformatics 22, 1924–1925 [DOI] [PubMed] [Google Scholar]
- 35. Kustatscher G., Grabowski P., and Rappsilber J. (2017) Pervasive coexpression of spatially proximal genes is buffered at the protein level. Mol. Syst. Biol. 13, 937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Saltzman A. B., Leng M., Bhatt B., Singh P., Chan D. W., Dobrolecki L., Chandrasekaran H., Choi J. M., Jain A., Jung S. Y., Lewis M. T., Ellis M. J., and Malovannaya A. (2018) gpGrouper: A peptide grouping algorithm for gene-centric inference and quantitation of bottom-up proteomics data. Mol. Cell. Proteomics 17, 2270–2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 38. Naba A., Clauser K. R., Ding H., Whittaker C. A., Carr S. A., and Hynes R. O. (2016) The extracellular matrix: Tools and insights for the “omics” era. Matrix Biol. 49, 10–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Naba A., Clauser K. R., Hoersch S., Liu H., Carr S. A., and Hynes R. O. (2012) The matrisome: in silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices. Mol. Cell. Proteomics 11, M111.014647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bassaganya-Riera J., Guri A. J., Lu P., Climent M., Carbo A., Sobral B. W., Horne W. T., Lewis S. N., Bevan D. R., and Hontecillas R. (2011) Abscisic acid regulates inflammation via ligand-binding domain-independent activation of peroxisome proliferator-activated receptor gamma. J. Biol. Chem. 286, 2504–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Delpoux A., Yakonowsky P., Durand A., Charvet C., Valente M., Pommier A., Bonilla N., Martin B., Auffray C., and Lucas B. (2014) TCR signaling events are required for maintaining CD4 regulatory T cell numbers and suppressive capacities in the periphery. J. Immunol. 193, 5914–5923 [DOI] [PubMed] [Google Scholar]
- 42. Hawiger D., Masilamani R. F., Bettelli E., Kuchroo V. K., and Nussenzweig M. C. (2004) Immunological unresponsiveness characterized by increased expression of CD5 on peripheral T cells induced by dendritic cells in vivo. Immunity 20, 695–705 [DOI] [PubMed] [Google Scholar]
- 43. Henderson J. G., Opejin A., Jones A., Gross C., and Hawiger D. (2015) CD5 instructs extrathymic regulatory T cell development in response to self and tolerizing antigens. Immunity 42, 471–483 [DOI] [PubMed] [Google Scholar]
- 44. Narayan K., Sylvia K. E., Malhotra N., Yin C. C., Martens G., Vallerskog T., Kornfeld H., Xiong N., Cohen N. R., Brenner M. B., Berg L. J., Kang J., and Immunological Genome Project, C. (2012) Intrathymic programming of effector fates in three molecularly distinct gammadelta T cell subtypes. Nat. Immunol. 13, 511–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dixon J. R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J. S., and Ren B. (2012) Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ji X., Dadon D. B., Powell B. E., Fan Z. P., Borges-Rivera D., Shachar S., Weintraub A. S., Hnisz D., Pegoraro G., Lee T. I., Misteli T., Jaenisch R., and Young R. A. (2016) 3D chromosome regulatory landscape of human pluripotent cells. Cell Stem Cell 18, 262–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mathieson T., Franken H., Kosinski J., Kurzawa N., Zinn N., Sweetman G., Poeckel D., Ratnu V. S., Schramm M., Becher I., Steidel M., Noh K. M., Bergamini G., Beck M., Bantscheff M., and Savitski M. M. (2018) Systematic analysis of protein turnover in primary cells. Nat. Commun. 9, 689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zecha J., Meng C., Zolg D. P., Samaras P., Wilhelm M., and Kuster B. (2018) Peptide level turnover measurements enable the study of proteoform dynamics. Mol. Cell. Proteomics [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zougman A., Selby P. J., and Banks R. E. (2014) Suspension trapping (STrap) sample preparation method for bottom-up proteomics analysis. Proteomics 14, 1006–1000 [DOI] [PubMed] [Google Scholar]
- 50. Shadforth I. P., Dunkley T. P., Lilley K. S., and Bessant C. (2005) i-Tracker: for quantitative proteomics using iTRAQ. BMC Genomics 6, 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Troyanskaya O., Cantor M., Sherlock G., Brown P., Hastie T., Tibshirani R., Botstein D., and Altman R. B. (2001) Missing value estimation methods for DNA microarrays. Bioinformatics 17, 520–525 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original mass spectra may be downloaded from MassIVE (http://massive.ucsd.edu) under the identifier: MassIVE MSV000083180. The data are directly accessible via ftp://MSV000083180@massive.ucsd.edu.